ABSTRACT
The connections between the claustrum and the cortex in mouse are systematically investigated with adeno‐associated virus (AAV), an anterograde viral tracer. We first define the boundary and the three‐dimensional structure of the claustrum based on a variety of molecular and anatomical data. From AAV injections into 42 neocortical and allocortical areas, we conclude that most cortical areas send bilateral projections to the claustrum, the majority being denser on the ipsilateral side. This includes prelimbic, infralimbic, medial, ventrolateral and lateral orbital, ventral retrosplenial, dorsal and posterior agranular insular, visceral, temporal association, dorsal and ventral auditory, ectorhinal, perirhinal, lateral entorhinal, and anteromedial, posteromedial, lateroposterior, laterointermediate, and postrhinal visual areas. In contrast, the cingulate and the secondary motor areas send denser projections to the contralateral claustrum than to the ipsilateral one. The gustatory, primary auditory, primary visual, rostrolateral visual, and medial entorhinal cortices send projections only to the ipsilateral claustrum. Primary motor, primary somatosensory and subicular areas barely send projections to either ipsi‐ or contralateral claustrum. Corticoclaustral projections are organized in a rough topographic manner, with variable projection strengths. We find that the claustrum, in turn, sends widespread projections preferentially to ipsilateral cortical areas with different projection strengths and laminar distribution patterns and to certain contralateral cortical areas. Our quantitative results show that the claustrum has strong reciprocal and bilateral connections with prefrontal and cingulate areas as well as strong reciprocal connections with the ipsilateral temporal and retrohippocampal areas, suggesting that it may play a crucial role in a variety of cognitive processes. J. Comp. Neurol. 525:1317–1346, 2017. © 2016 Wiley Periodicals, Inc.
Keywords: connectivity, tract tracing, AAV, cortex, claustrum, RRID:SCR_008848, RRID:AB_10000344, RRID:AB_509998, RRID:AB_10048713, RRID:AB_306956
Abbreviations
- ACAd
anterior cingulate area, dorsal part
- ACAv
anterior cingulate area, ventral part
- AId
agranular insular area, dorsal part
- AIp
agranular insular area, posterior part
- AIv
agranular insular area, ventral part
- AUDd
dorsal auditory area
- AUDp
primary auditory area
- AUDpo
posterior auditory area
- AUDv
ventral auditory area
- CLA
claustrum
- ECT
ectorhinal area
- ENTl
entorhinal area, lateral part
- ENTm
entorhinal area, medial part, dorsal zone
- ENTmv
entorhinal area, medial part, ventral zone
- EP
endopiriform nucleus
- FRP
frontal pole, cerebral cortex
- GU
gustatory area
- ILA
infralimbic area
- LP
lateral posterior nucleus of the thalamus
- MD
mediodorsal nucleus of the thalamus
- MOp
primary motor area
- MOs
secondary motor area
- ORBl
orbital area, lateral part
- ORBm
orbital area, medial part
- ORBvl
orbital area, ventrolateral part
- PAR
parasubiculum
- PERI
perirhinal area
- PIR
piriform area
- PL
prelimbic area
- POST
postsubiculum
- PRE
presubiculum
- RSPagl
retrosplenial area, lateral agranular part
- RSPd
retrosplenial area, dorsal part
- RSPv
retrosplenial area, ventral part
- SSp
primary somatosensory area
- SSs
supplemental somatosensory area
- SUB
subiculum
- TEa
temporal association area
- VAL
ventral anterolateral complex of thalamus
- VISa
anterior visual area
- VISal
anterolateral visual area
- VISam
anteromedial visual area
- VISC
visceral area
- VISl
lateral visual area
- VISli
laterointermediate visual area
- VISp
primary visual area
- VISpl
posterolateral visual area
- VISpm
posteromedial visual area
- VISpor
postrhinal visual area
- VISrl
rostrolateral visual area
The claustrum is a subcortical gray matter structure located beneath the insular cortex and above the striatum in all eutherian mammalian brains examined to date (Edelstein and Denaro, 2004). Although this structure was first identified over 2 centuries ago, its function remains elusive. The uncertainty concerning claustral function is at least partially a result of its unique shape (thin, curved, and rostrocaudally elongated) and deep location (sandwiched between external and extreme capsules), making it difficult to image or inactivate by targeted lesioning.
Based on widespread reciprocal connections with cortical areas and physiological response properties, various hypotheses have been put forth regarding the function of claustrum, including multimodal integration of stimulus information into a single conscious percept (Crick and Koch, 2005), amplification of cortical oscillations (Smythies et al., 2012, 2014), salience detector (Remedios et al., 2010, 2014), and allocation of selective attention (Mathur, 2014; Goll et al., 2015).
Electrical recordings of claustral neurons have yielded inconsistent results. Some studies have indicated that most claustral neurons respond to multimodal stimuli (polymodal), whereas a minority responds only to single modality stimuli (unimodal; Segundo and Machne, 1956; Spector et al., 1970; Clarey and Irvine, 1986). Others have reported that the great majority of claustral neurons are unimodal and very few are polymodal (Olson and Graybiel, 1980; Sherk and LeVay, 1981; Remedios et al., 2010, 2014). Early studies focusing on claustral lesions and stimulations proved inconclusive (Ruiz, 1960; Frontera and Stiehl, 1963; Gabor and Peele, 1964; Chorazyna et al., 1965). One recent report described a patient implanted with electrodes for treatment of epilepsy. Electrical stimulation at one site of the white matter tract beneath the claustrum made the patient stare blankly ahead until the stimulation stopped, without subsequent recall (Koubeissi et al., 2014). This behavior has some resemblance to the inactivation syndrome seen in cats following claustrum electrical stimulation (Gabor and Peele, 1964). Two other clinical cases with transient, bilateral selective lesion of the claustrum include one patient with epileptic seizures and temporary loss of vision, speech, and hearing (Sperner et al., 1996) and a second patient with temporary visual and auditory hallucinations (Ishii et al., 2011).
Stimulation of the claustrum in animals resulted in either inhibition or excitation of cortical neurons (Ptito and Lassonde, 1981; Salerno et al., 1984; Cortimiglia et al., 1991). Deciphering detailed pre‐ and postsynaptic components of the interconnections between claustrum and other cortical and subcortical areas will facilitate our understanding of claustral function (Rahman and Baizer, 2007). Advanced technologies such as optogenetics, pharmacogenetics, and viral based tract tracing, in combination with electrophysiology and behavioral analyses in transgenic animals, can link specific cell types or neuronal circuits to behavioral output (Mathur et al., 2009; Huang and Zeng, 2013; Lim et al., 2013; Zhang et al., 2014). This is particularly true for the laboratory mouse. A mouse model may be quite informative in revealing claustral function, so it is imperative to study connections between the claustrum and the cortex in wild‐type and transgenic mice.
Extensive anatomical studies over the past few decades have used conventional tract‐tracing methods in different mammalian species to show that the claustrum has topographical and reciprocal connections with most cortical areas, including motor, premotor, orbitofrontal, prefrontal, parietal, cingulate, temporal, visual, perirhinal, and entorhinal cortices (Edelstein and Denaro, 2004; Druga, 2014; Zingg et al., 2014). Claustral neurons receive convergent inputs from various cortical areas and send divergent outputs back to the corresponding cortical areas (Minciacchi et al., 1985; Li et al., 1986; Smith and Alloway, 2010, 2014; Patzke et al., 2014).
However, most of these studies only qualitatively investigated a limited number of cortical areas in large animals, such as monkeys and cats, making a systematic and quantitative comparison of claustrocortical and corticoclaustral connections within and across species impossible. Recent studies using diffusion tensor imaging indicated that the claustrum is the most connected structure in the human brain per unit volume, with its strongest connection to frontal and cingulate cortices (Milardi et al., 2015; Torgerson et al., 2015). Because the connections between various cortical areas and the claustrum are unequally weighted, a quantitative assessment of connectivity weights is important for better predicting its function at the network level (Markov et al., 2014). The Allen Mouse Brain Connectivity Atlas, which allows whole‐brain comparison of connectivity weights of quantified injection experiments in an averaged three‐dimensional (3D) model of the adult laboratory mouse brain, provides a useful means to further our understanding of the claustrum in this respect (Oh et al., 2014).
In rodents, some studies divide the claustrum into dorsal and ventral parts (Druga et al., 2014; Smith and Alloway, 2014) or into central “core” and peripheral “shell” regions (Real et al., 2006), whereas others use a combination of molecular markers to define it. The best known example markers are Gng2 and Pvalb (Mathur et al., 2009). Note that endopiriform nucleus, a region just ventral to the claustrum, has sometimes been referred to as the “ventral claustrum.” However, because it has interconnections only with other olfactory areas, and therefore appears to be primarily related to olfaction, we do not regard it as part of the claustrum.
Our anatomical study in mice has three aims. First, we define the boundary of the claustrum using different data types, including in situ hybridization (ISH) of unique marker genes selected from the Allen brain atlas (http://mouse.brain-map.org), histological reference data sets from the Allen Mouse Brain Connectivity Atlas (http://connectivity.brain-map.org/static/referencedata), and newly generated whole‐brain fluorescent data sets from Cre driver‐reporter transgenic mice. Second, we reconstruct a 3D spatial model of claustrum within our high‐resolution 3D reference atlas space, the Common Coordinate Framework (CCF), to allow accurate quantification of the axonal projections within the claustrum. Finally, we systemically and quantitatively analyze the data of the Allen Mouse Brain Connectivity Atlas (http://connectivity.brain-map.org/, RRID:SCR_008848), which contains injections covering the entire cortex and the claustrum from both wild‐type and Cre driver mice with the GFP‐expressing adeno‐associated virus (AAV) tracer (Harris et al., 2012; Oh et al., 2014). Our findings of the reciprocity, topography and bilateral connectivity between claustrum and cortex will, we hope, shed light on the function of the claustrum and allow more precisely targeted rodent studies of this structure in the future.
MATERIALS AND METHODS
Mice
Experiments were performed on wild‐type (C57BL/6J) and Cre driver transgenic mice at postnatal day (P) 56 ± 3. Cre driver lines were generated at the Allen Institute or imported from external sources for characterization. Two new knockin Cre driver lines, Gnb4‐IRES2‐Cre and Ntng2‐IRES2‐Cre, were generated for this study, as described previously (Madisen et al., 2010, 2015). The former was generated by using a conventional targeting vector, whereas the latter used a CRISPR/Cas9‐based targeting vector. Transgenic mice were produced from B6/129 F1 ES clones and were back‐crossed to C57BL/6J for two or more generations, thus containing 87% or more C57BL/6J background. All experimental procedures were approved by the Allen Institute Animal Care and Use Committee and conform to NIH guidelines.
Histology and immunohistochemistry
A detailed description of the histology and double immunohistochemistry (IHC) procedures can be found at the Allen Mouse Brain Connectivity Atlas documentation page (http://help.brain-map.org//display/mouseconnectivity/Documentation). Briefly, adult mice were anesthetized with 5% isoflurane and intracardially perfused with 10 ml saline (0.9% NaCl) followed by 50 ml freshly prepared 4% paraformaldehyde (PFA). Brains were rapidly dissected, postfixed, and transferred to a 30% sucrose solution. After sinking, brains were embedded in OCT and sectioned at 25 µm on a Leica 3050 S cryostat equipped with an Instrumedics Tape Transfer System (plus UV light polymerization chamber and warming pad). Every other section was collected sequentially to give a final sampling density of 50 µm. For Nissl staining or dual immunostaining, entire coronal series were used. Nissl‐stained sections were delipidated with the xylene substitute Formula 83 (CBG Biotech, Columbus, OH; catalog No. CH0104) and ethanol rehydrated. After several washes in water, the sections were stained in 0.21% thionin for 3 minutes and dehydrated by sequential immersion in increasing concentrations of ethanol. Differentiation and monitoring were performed at 95% ethanol before completion with pure ethanol. Dehydrated sections were subsequently incubated in Formula 83 and coverslipped with the Curemount mounting medium (Instrumedics, Hackensack, NJ; catalog No. 475232). For imaging, sections were scanned with the ×10 objective on ScanScope, an automated image capture platform (Aperio Technologies, Vista, CA).
For double IHC, sections were washed after antigen retrieval with 10 mM sodium citrate and then incubated in blocking solution (4% normal goat serum + 0.3% Triton X‐100 in PBS) for 1 hour. After brief rinsing, each series of sections was incubated with one of the following primary antibody pairs overnight: parvalbumin (PV; SWant, Belinzona, Switzerland; catalog No. PV25, RRID:AB_10000344; final dilution 1:2,000 in blocking solution) and SMI‐32 (Covance, Berkeley, CA; catalog No. SMI‐32R‐500, RRID:AB_509998; 1:1,000), or NeuN (Millipore, Bedford, MA; catalog No. MAB377, RRID:AB_10048713; 1:1,000) and NF‐160 (Abcam, Cambridge, MA; catalog No. ab9034, RRID:AB_306956; 1:1,000). After rinsing primary antibodies thoroughly, each series of sections was incubated in a pair of the secondary antibodies: goat anti‐rabbit‐488 (final dilution 1:1,000 in blocking solution for PV, and 1:500 for NF‐160) and goat anti‐mouse‐594 (1:500) overnight. After rinsing, sections were counterstained with DAPI (Invitrogen, Carlsbad, CA; catalog No. D1306) and coverslipped with Fluoromount G medium (Southern Biotechnology, Birmingham, AL; catalog No. 0100‐01). For imaging, sections were scanned on a fully automated, high‐speed multichannel epifluorescence scanning system, VS110/120 (Olympus, Center Valley, PA) with × 10 objectives.
Antibody characterization
All antibodies used were purchased commercially. We relied on the published characterization for these antibodies, because all had been used in previously published work with comparable model systems and methods. In our hands, these antibodies stain the appropriate patterns of cellular morphology and distribution that are the same as demonstrated in previous publications, as detailed in Table 1.
Table 1.
Primary Antibodies Used
| Antigen | Description of immunogen | Source, host species, catalog No., clone or lot, RRID | Concentration used | Characterization |
|---|---|---|---|---|
| PV (parvalbumin) | Rat muscle parvalbumin | SWant, rabbit polyclonal, PV 25, AB_10000344 | 1:2,000 (IHC) | Bunce et al., 2013; Cai et al., 2013 |
| SMI‐32 | Homogenized rat hypothalamii; subsequently determined to recognize nonphosphorylated epitopes on the neurofilament heavy chain | Covance Research Products Inc., mouse monoclonal, SMI‐32R‐500, AB_509998 | 1 µg/ml (IHC) | Brown and Sawchenko, 2007; Stillman et al., 2009 |
| NeuN | Purified cell nuclei from mouse brain | Millipore, mouse monoclonal, MAB377, clone A60, AB_10048713 | 1 µg/ml (IHC) | Wang et al., 2006; Eskilsson et al., 2014 |
| NF‐160 | Recombinant C‐terminal fragment of the rat medium neurofilament subunit | Abcam, rabbit polyclonal, ab9034, AB_306956 | 1:1,000 (IHC) | (Deng et al., 2009; tegeman et al., 2013 |
Tracer injection and brain imaging
A detailed description for tracer injection and brain imaging can be found at the Allen Mouse Brain Connectivity Atlas documentation page (http://help.brain-map.org//display/mouseconnectivity/Documentation), as well as in other references (Harris et al., 2012; Oh et al., 2014; Wang et al., 2014). Surgery procedures for phase 1 and phase 2 were identical, with the exception of tracer and mouse types. All injections were made into the right hemisphere. For phase 1 experiments, a pan‐neuronal AAV vector expressing EGFP under the human synapsin I promoter (AAV2/1.pSynI.EGFP.WPRE.bGH) was used in wild‐type mice. For phase 2, a Cre‐dependent AAV (AAV2/1.pCAG.FLEX.EGFP.WPRE.bGH) was used in Cre driver mouse lines. All adult mice were anesthetized with 5% isoflurane briefly and secured to a stereotaxic frame (model 1900; Kopf, Tujunga, CA) prior to surgery. During surgery, anesthesia was maintained at 1.8–2% isoflurane. After skin incision, a small divot was made in the skull surface with a fine drill burr. To reveal the brain surface, a thin layer of bone was removed with miniature forceps. For cortex and claustrum injections, a glass pipette (inner tip diameter 10–20 µm) loaded with AAV was lowered to the desired depth, as measured from pial surface of the brain. Coordinates for cortical and claustral injections were based on the atlas of Paxinos and Franklin (2001). Cortical injection coordinates are listed on web documentations and coordinates for claustrum are –0.46–1.78 mm anteroposterior from Bregma, 2.40–4.00 mm mediolateral from the midline, 2.60–3.10 mm from pial surface. The AAV (serotype 1, produced by UPenn viral core; titer > 1012 GC/ml) was delivered by iontophoresis (current 3 µA and 7 seconds on/7 seconds off duty cycle) for 5 minutes. After 21 days of survival, mice were anesthetized with 5% isoflurane and intracardially perfused with 10 ml saline (0.9% NaCl) followed by 50 ml freshly prepared 4% PFA. Brains were rapidly dissected, postfixed in 4% PFA at room temperature for 3–6 hours and overnight at 4 °C, then rinsed briefly with PBS and stored in PBS with 0.1% sodium azide. After removal of residual surface moisture with Kimwipes, brains were subsequently placed in 4.5% oxidized agarose (made by stirring 10 mM NaIO4 in agarose), transferred to a phosphate buffer solution, and placed in a grid‐lined embedding mold for standardized orientation in an aligned coordinate space. Multiphoton image acquisition for the Allen Mouse Brain Connectivity Atlas was accomplished by using the TissueCyte 1000 system (TissueVision, Cambridge, MA) coupled with a Mai Tai HP DeepSee laser (Spectra Physics, Santa Clara, CA).
Imaging of Cre driver‐reporter mice
To take advantage of certain Cre lines' unique recombination patterns for more refined anatomical structure delineation for the CCF, these Cre lines were crossed with the Ai14 tdTomato‐expressing Cre reporter line (Madisen et al., 2010), and the resulting Cre driver‐reporter mice were used for TissueCyte imaging. The procedure for Cre mouse perfusion and brain imaging is the same as that described above, excluding the tracer injections.
3D reconstruction of the claustrum
Detailed information for the 3D reconstruction of brain structures can be found at the Allen Mouse Brain Connectivity Atlas documentation page (http://help.brain-map.org//display/mouseconnectivity/Documentation). To delineate the claustrum with the highest possible fidelity, Cre driver‐reporter mouse brains exhibiting differential tdTomato labeling in genetically defined cell types and processes, as well as some of the connectivity data that have strong cortical projections to the claustrum, were mapped to the CCF. This was done by aligning the image stack of each brain globally to a 3D average template brain model derived from red‐channel autofluorescence of 1,675 brains from the Allen Mouse Brain Connectivity Atlas. The global alignment process between the Cre mouse brains or AAV‐injected brains and the average template consists of three steps: 1) a coarse registration initialized by matching the image moments of the image stack and template, 2) a rigid registration (rotation plus translation), and 3) a 12‐parameter affine registration. Each step was based on maximizing the image similarity metric between the transgenic image stack and the template using a multiresolution algorithm. To increase alignment accuracy further, local registration was then performed. As with the global alignment, local registration was conducted sequentially from coarse to fine at four resolution levels with decreasing smoothness constraints. With the Cre and connectivity data registered and overlaid, the right claustrum was reconstructed on the average template using 10‐μm voxel sizes via the 3D annotation software ITK‐Snap.
Quantification of the projections
Projection quantification is a core component of the Informatics Data Processing Pipeline (IDP) for the Allen Mouse Brain Connectivity Atlas (Kuan et al., 2015). In brief, after image reprocessing, an alignment module of the IDP was used to align all injection experiments with the average 3D model brain. A signal algorithm, based on a combination of adaptive edge/line detection and morphological processing, was applied to each section image to differentiate positive fluorescent signal from background signal. Segmented signal pixels were counted as projection strength in the claustrum and cortical areas. It should be noted that the detection algorithm operates on a per‐image basis and that passing fibers and axon terminals were not distinguished. In addition, the detected signals in cortical areas or claustrum include occasional retrogradely labeled neurons because the viral tracer is not purely anterograde. Imperfect alignment of each injection image set with the Allen Reference Atlas may also affect the quantification of the projection (Oh et al., 2014). Improvement in anatomical regional delineation in the CCF compared with the original Allen Reference Atlas also improves the accuracy of quantification.
RESULTS
Defining the boundary of the claustrum
In primates and cats, the claustrum can be easily distinguished in Nissl‐ or myelin‐stained specimens as subcortical gray matter embedded between two fiber tracts, the external capsule medially and the extreme capsule laterally. However, in rats and mice, the boundaries of the claustrum are not well defined and are inconsistent across studies (Real et al., 2006; Mathur et al., 2009; Druga et al., 2014; Smith and Alloway, 2014; Zingg et al., 2014). We first studied the boundary of the claustrum using two brain‐wide double immunohistochemistry (IHC) data sets (Fig. 1) which were generated as histological reference data sets for the Allen Mouse Brain Connectivity Atlas; one set stained with antibodies against NeuN (a pan‐neuronal marker labeling nuclear protein Fox‐3) and NF‐160 (an axonal marker labeling neurofilament M chain), another with PV (marker for a subset of interneurons) and SMI‐32 (labeling neurofilament H chain that reveals cell bodies, dendrites, and some thick axons).
Figure 1.
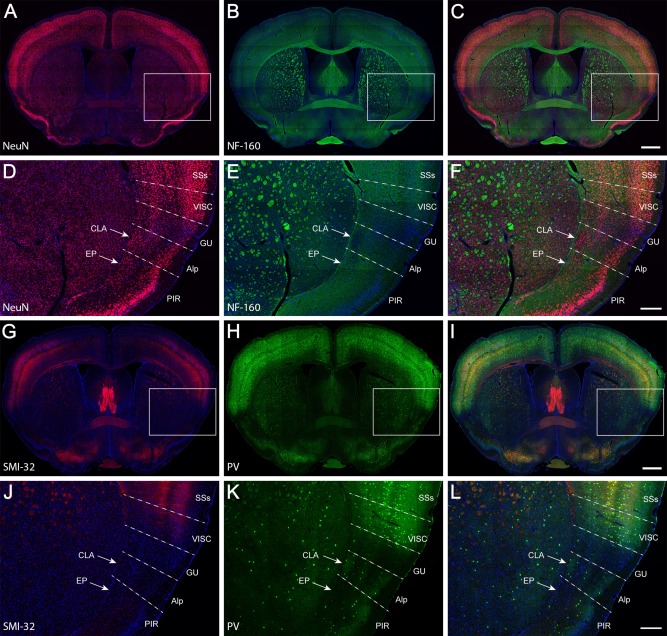
Double IHC for NeuN (red) and NF‐160 (green; A–F) and SMI‐32 (red) and parvalbumin (PV, green; G–L). Counterstaining with DAPI is in blue. Overlapping images from A and B, D and E, G and H, and J and K are, respectively, shown in C, F, I, and L. The boxes in A–C and G–I indicate, respectively, the enlargements in D–F and J–L. The dashed lines indicate approximate borders between cortical areas. For abbreviations see list. Scale bars = 737 μm in C (applies to A–C); 258 μm in F (applies to D–F); 737 μm in I (applies to G–I); 258 μm in L (applies to J–L).
In the NeuN/NF‐160 specimen, the claustrum appears as a densely packed group of neurons deep beneath the agranular insular cortex (Fig. 1A,C,D,F). It does not extend dorsally into the deep layers of the gustatory and visceral cortices, where neurons are loosely distributed. The densely packed group of neurons is coincidentally located in an oval region that is less well myelinated than its surrounding structures (Fig. 1B,C,E,F) such as the gustatory area, visceral area, and endopiriform nucleus (Fig. 1D–F). In the SMI‐32/PV specimen, SMI‐32 labeling (Fig. 1G,I,J,L) was not detected in the claustrum, the agranular insular area, or the gustatory area but was weakly present in the visceral area. PV labeling (Fig. 1H,I,K,L) was relatively stronger in the center of the claustrum than in its periphery but was barely seen in the gustatory area, visceral area, or endopiriform nucleus. In contrast to labeling in insular cortex, labeling of both PV and SMI‐32 was very strong in the rest of the neocortex.
Next, we further confirmed the size, shape and location of the claustrum with molecular markers. Genes specifically expressed in the claustrum were searched with AGEA Fine Structure Search and Differential Search tools of the Allen brain atlas (Ng et al., 2009) and then were manually selected. Mathur and colleagues (2009) identified guanine nucleotide binding protein gamma 2 (Gng2) as a marker gene for the rat claustrum. We confirmed this finding and extended it to a large set of genes with enriched expression in the mouse claustrum (Table 2). Netrin G2 (Ntng2), guanine nucleotide binding protein beta 4 (Gnb4), Gng2 and latexin (Lxn) are all expressed densely in the claustrum but relatively sparsely in nearby structures, such as the endopiriform nucleus and deep layers of the gustatory area and visceral area (Fig. 2). In the adjacent Nissl‐stained sections of the same specimen, the claustrum was revealed as a darkly stained group of neurons (Fig. 2C,F,I,L) corresponding to the densely ISH‐labeled neurons. These darkly stained neurons can also be seen in the two mouse atlases (Paxinos and Franklin, 2001; Dong, 2008).
Table 2.
Genes Enriched or Absent in the Claustruma
| Abbreviation | (+)/(−) | Full name |
|---|---|---|
| Adamtsl2 | (+) | ADAMTS‐like 2 |
| Bace1 | (+) | Beta‐site APP cleaving enzyme 1 |
| B3gat2 | (+) | Beta‐1,3‐glucuronyltransferase 2 |
| BC100451 | (+) | cDNA sequence BC100451 |
| Btg1 | (+) | B‐cell translocation gene 1, antiproliferative |
| Cadps2 | (+) | Ca2+‐dependent activator protein for secretion 2 |
| Car12 | (+) | Carbonic anyhydrase 12 |
| Cbln2 | (+) | Cerebellin 2 precursor protein |
| Chst11 | (+) | Carbohydrate sulfotransferase 11 |
| Cntnap3 | (+) | Contactin‐associated protein‐like 3 |
| Col11a1 | (+) | Collagen, type XI, alpha 1 |
| Cux2 | (+) | Cut‐like homeobox 2 |
| Gadd45g | (+) | Growth arrest and DNA damage‐inducible 45 gamma |
| Galnt14 | (+) | UDP‐N‐acetyl‐alpha‐D‐galactosamine:polypeptide N‐Acetylgalactosaminyltransferase 14 |
| Gfra1 | (+) | Glial cell line‐derived neurotrophic factor family receptor alpha 1 |
| Gnb4 | (+) | Guanine nucleotide binding protein (G protein), beta 4 |
| Gng2 | (+) | Guanine nucleotide binding protein (G protein), gamma 2 |
| Gnao1 | (+) | Guanine nucleotide binding protein, alpha O |
| Gpd2 | (+) | Glycerol phosphate dehydrogenase 2, mitochondrial |
| Gucy1a3 | (+) | Guanylate cyclase 1, soluble, alpha 3 |
| Id2 | (+) | Inhibitor of DNA binding 2 |
| Inpp5a | (+) | Inositol polyphosphate‐5‐phosphatase A |
| Itga7 | (+) | Integrin alpha 7 |
| Laptm4b | (+) | Lysosomal‐associated protein transmembrane 4B |
| LOC433093 | (+) | Similar to MAM domain‐containing glycosylphosphatidylinositol anchor 1; glycosyl‐phosphatidyl‐inositol‐MAM |
| Lypd6b | (+) | LY6/PLAUR domain containing 6B |
| Lxn | (+) | Latexin |
| Mt3 | (+) | Metallothionein 3 |
| Nfxl1 | (+) | Nuclear transcription factor, X‐box binding‐like 1 |
| Nmb | (+) | Neuromedin B |
| Nr4a2 | (+) | Nuclear receptor subfamily 4, group A, member 2 |
| Nsdhl | (+) | NAD(P)‐dependent steroid dehydrogenase‐like |
| Ntng2 | (+) | Netrin G2 |
| Oprk1 | (+) | Opioid receptor, kappa 1 |
| Pdia5 | (+) | Protein disulfide isomerase associated 5 |
| Plcl1 | (+) | Phospholipase C‐like 1 |
| Ppp1r1a | (+) | Protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| Rab3c | (+) | RAB3C, member RAS oncogene family |
| Rtn4rl2 | (+) | Reticulon 4 receptor‐like 2 |
| Sc4mol | (+) | Sterol‐C4‐methyl oxidase‐like |
| SStr2 | (+) | Somatostatin receptor 2 |
| Tmem163 | (+) | Transmembrane protein 163 |
| Tox | (+) | Thymocyte selection‐associated high mobility group box |
| Zfp804a | (+) | Zinc finger protein 804A |
| Col23a1 | (−) | Collagen, type XXIII, alpha 1 |
| Crym | (−) | Crystallin, mu |
| Ctgf | (−) | Connective tissue growth factor |
| Nxph3 | (−) | Neurexophilin 3 |
| Slit1 | (−) | Slit homolog 1 (Drosophila) |
| Rasal1 | (−) | RAS protein activator like 1 (GAP1 like) |
Gene enriched in the claustrum is indicated by plus (+), and gene expressed in its surrounding structures but not claustrum is indicted by minus (−). Genes shown in Figure 2 are in Italics.
Figure 2.
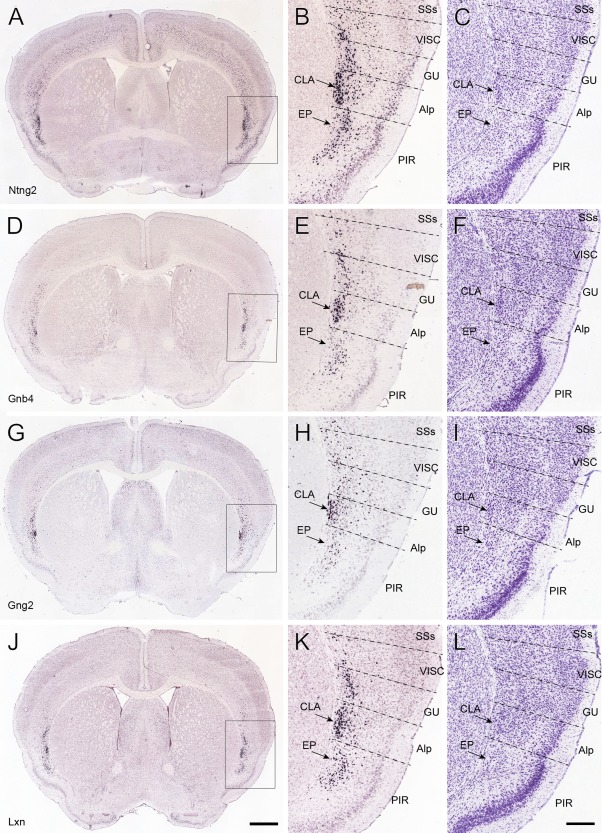
Molecular markers for mouse claustrum. A–C: Expression of Ntng2 by ISH, with the boxed area in A enlarged in B and a nearby Nissl‐stained section shown in C. D–F: Similarly for Gnb4. G–I: Similar for Gng2. J–L: Similar for Lxn. For abbreviations see list. Scale bars = 839 μm in J (applies to A,D,G,J); 300 μm in L (applies to B,C,E,F,H,I,K,L).
Finally, the boundary of the claustrum was delineated with six Cre driver‐reporter transgenic mouse lines that express tdTomato reporter positively or negatively in claustrum (Figs. 3, 4). In the Pvalb‐IRES‐Cre line, tdTomato expression pattern is very similar to that in the PV‐immunostained specimen (Fig. 3A,B). Pvalb‐positive neurons and fibers were labeled more strongly in the claustrum than in the endopiriform nucleus and were extensively labeled in the neocortex, except for the insular cortex. In the Cux2‐CreERT2 line, labeled neurons and their processes were observed in the claustrum and weakly in the agranular insular area and gustatory area but were absent in the endopiriform nucleus (Fig. 3C,D). The Cux2‐associated signal was very strong in layers 2/3 and 4 of the neocortex and weaker in layers 5 and 6. In the Rbp4‐Cre_KL100 line, tdTomato signal was found in processes (but not cell bodies) in the claustrum, and a few positive neurons were seen in the agranular insular area and gustatory area (Fig. 3E,F). The labeled neurons were dense in layer 5 of the neocortex, and their processes were labeled in layer 1 and weakly in layers 2/3 and 6. In the Ctgf‐2A‐dgCre line, tdTomato‐labeled cells were found in layer 6b of the neocortex, and their processes were seen strongly in layer 1; labeled cells were also found in the endopiriform nucleus but were absent in the claustrum (Fig. 4A,B). In the Rorb‐IRES2‐Cre line, strong signal was seen in layers 4 and 6 of the neocortex and structures surrounding the claustrum but not in the claustrum itself (Fig. 4C,D). In the Ntsr1‐Cre_GN220 line, signal was strong in the deep layers of the neocortex and weak in layer 1 of the agranular insular area and in the claustrum. No or very weak signal was detected in the endopiriform nucleus and piriform area (Fig. 4E,F). Taken together, these IHC, ISH, and Cre driver‐reporter data reveal very similar claustrum boundaries, which form the foundation for reconstructing the mouse claustrum in 3D below.
Figure 3.
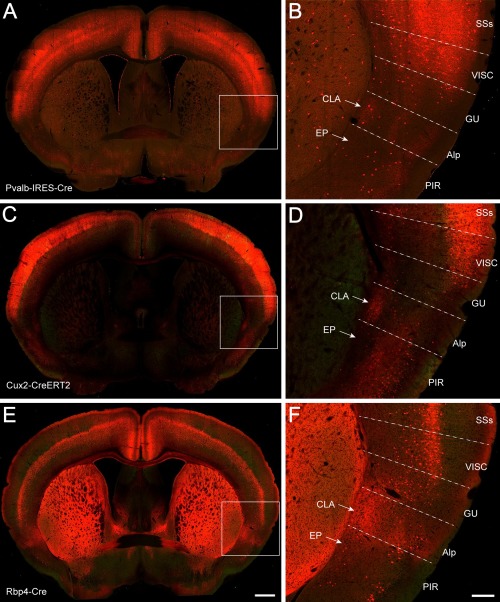
Transgenic Cre driver lines crossed to Ai14 reporter showing tdTomato signal in the claustrum. A,B: A TissueCyte section image from a Pvalb‐IRES‐Cre;Ai14 mouse, with the boxed area in A enlarged in B. Similarly for C,D (Cux2‐CreERT2;Ai14) and E,F (Rbp4‐Cre_KL100;Ai14). For abbreviations see list. Scale bars = 650 μm in E (applies to A,C,E); 200 μm in F (applies to B,D,F).
Figure 4.
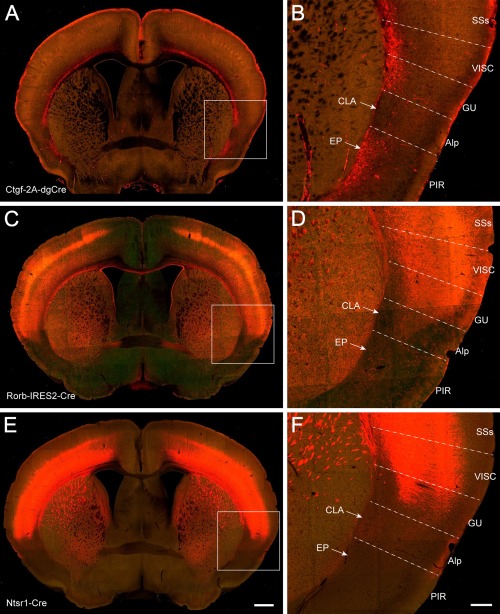
Transgenic Cre driver lines crossed to Ai14 reporter showing tdTomato signal absent or very weak in the claustrum. A,B: A TissueCyte section image from a Ctgf‐2A‐dgCre;Ai14 mouse, with the boxed area in A enlarged in B. Similarly for C,D (Rorb‐IRES2‐Cre;Ai14.) and E,F (Ntsr1‐Cre_GN220;Ai14). For abbreviations see list. Scale bars = 650 μm in E (applies to A,C,E); 200 μm in F (applies to B,D,F).
3D reconstruction of the mouse claustrum
To analyze quantitatively the connectivity strengths, the 3D shape of the claustrum was reconstructed within the CCF based on the Cre mice data described above, supplemented by some of the cortical injection data with strong projections into the claustrum (see below; experiments 286313491, 287769285, and 113226232). Figure 5 shows medial, dorsal, and lateral views of the reconstructed 3D claustrum. The claustrum (in one hemisphere) includes 275,448 voxels (10 × 10 × 10 μm3 per voxel in the CCF), equal to a volume of 0.275 mm3. It is 200 times smaller than one neocortical hemisphere, which contains 61,700,000 voxels. The volumetric ratio of the claustrum to neocortex is five times less than that in the cats, in which the volume of the claustrum is 1/42 of that of the neocortex (Sherk, 1986). In humans, the volumetric ratio of the claustrum to the cerebral cortex is 1/25 (Edelstein and Denaro, 2004; Crick and Koch, 2005). The claustrum appears at 1.74 mm from the frontal pole and is an elongated, thin sheet of gray matter stretching for ∼2.9 mm along the anteroposterior axis, larger in the anterior part and gradually tapering toward its posterior end. It curves forward medially around the striatum.
Figure 5.
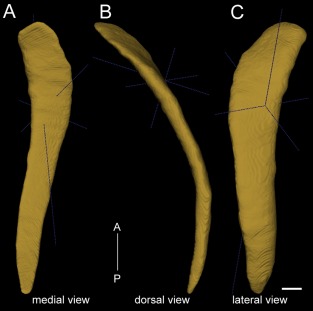
3D reconstruction of the mouse claustrum, shown in medial view (A), dorsal view (B), and lateral view (C). A‐P: Anterior to posterior direction. The dark blue lines depict anterior–posterior, medial–lateral, and dorsal–ventral axes in 3D. The mouse claustrum stretches for approximately 2.9 mm along the anterior–posterior axis, with a volume of 0.275 mm3. Scale bar = 250 μm.
Corticoclaustral projections
In total 113 neocortical and allocortical injections from the Allen Mouse Brain Connectivity Atlas (http://connectivity.brain-map.org/projection) were used to analyze corticoclaustral projections (Table 3). Among these, 85 were in wild‐type mice. Some injections in Rbp4‐Cre_KL100 and Cux2‐IRES‐Cre mice were included to obtain better coverage of certain cortical areas. Anatomical locations of the injection sites were identified and registered as in our previous study (Oh et al., 2014) and confirmed by examining their characteristic cortical and subcortical projection targets. Representative injection sites for 42 neocortical and allocortical areas are shown in coronal sections in Figure 6 and in top‐down surface views in Figure 7. Projections from these areas to the ipsilateral and contralateral claustrum are shown as projection density maps in Figure 8 and example images in Figure 9. Some of the large injections contaminated nearby structures, which are listed on the Web page as secondary injection sites. The nomenclature for cortical and subcortical structures was adapted from the Allen reference atlas (Dong, 2008), with modifications to higher visual areas (http://help.brain-map.org/download/attachments/2818171/MouseCCF.pdf; see also Abbreviations list).
Table 3.
Selected Cortical and Claustral Injections for Qualitative and Quantitative Analysesa
| Frontal cortex | ||||
| PL | 157711748b | 263106036 (Rbp4) | 278433737 (Cux2) | 294434161 (Cux2) |
| ILA | 157556400 | 286313491b (Rbp4) | 287494320 (Cux2) | |
| ORBm | 126860974 | 272781246b (Cux2) | 293471629 (Cux2) | |
| ORBvl | 112423392 | 167902586 (Rbp4) | 287769286b (Rbp4) | |
| ORBl | 112306316b | 156741826 (Rbp4) | 264708349 | |
| MOs | 100140756 | 112952510b | 157710335 | 166055636 (Rbp4) |
| 182793477 (Rbp4) | 180916954 | 168162771 (Rbp4) | ||
| MOp | 100141780 | 112670853 | 141602484 | 180709942b |
| 272697944 | 180719293 | |||
| Cingulate cortex | ||||
| ACAv | 112514202 | 126117554b (Rbp4) | 139520203 | 161458737 (Rbp4) |
| 286482701 (Rbp4) | ||||
| ACAd | 112458114 | 125833030 (Rbp4) | 139426984b | 146593590 |
| RSPd | 112424813 | 166054929b (Rbp4) | ||
| RSPv | 100140949 | 100148142b | 112595376 | 159832064 (Rbp4) |
| Insular cortex | ||||
| Ald | 112596790 | 166153483b (Rbp4) | 170721670 | 180709230 |
| 296048512 (Rbp4) | ||||
| Alp | 174361746b | 183330908 (Rbp4) | 272827141 (Cux2) | |
| GU | 272737914b | |||
| VISC | 180917660b | 180074890 | ||
| Olfactory area | ||||
| PIR | 112307046b | 127907465 | 131068390 | 146857301 |
| 157654069 | 146984209 | |||
| Retrohippocampal region | ||||
| PAR | 120280939b | |||
| PRE | 146984915b | |||
| POST | 100142354b | |||
| SUBd | 157063781b | |||
| ENTm | 113226232b | 127139568 | ||
| ENTl | 126116848b | |||
| Temporal cortex | ||||
| PERl | 142656218b | 180435652 | ||
| ECT | 293470216b (Cux2) | |||
| TEa | 127089669 | 166083557b (Rbp4) | 298178204 (Rbp4) | |
| AUDv | 112881858b | 116903230 | ||
| AUDp | 120491896b | |||
| AUDd | 120437703 | 146858006b | ||
| Parietal cortex | ||||
| SSs | 113036264 | 117298988b | 180717881 | |
| SSp‐tr | 100142655b | 112791318 | ||
| SSp‐ll | 112229814b | 180718587 | ||
| SSp‐ul | 126909424b | |||
| SSp‐bfd | 112951804 | 126908007 | 127866392b | |
| SSp‐m | 112936582 | 114290938b | 157654817 | |
| Occipital cortex | ||||
| VISpor | 157062358b | |||
| VISpl | 112373830b | |||
| VISli | 100141796b | |||
| VISp | 307558646 | 307137980 | 307297141 | 307321674 |
| 307593747 | 307557934b | 309003780 | 307296433 | |
| 307320960 | 307296433 | |||
| VISpm | 146077302b | 100141599 | ||
| VISam | 100148503b | 126861679 | 159753308 (Rbp4) | |
| VISal | 116903968b | |||
| VISrl | 272929308b (Rbp4) | |||
| CLA | ||||
| 180436360 | 296047806 (Cux2) | 485846989 b (Ntng2) | 485903475 (Gnb4) | |
| 513773998 (Ntng2) | 513775257 (Ntng2) |
Figure 6.
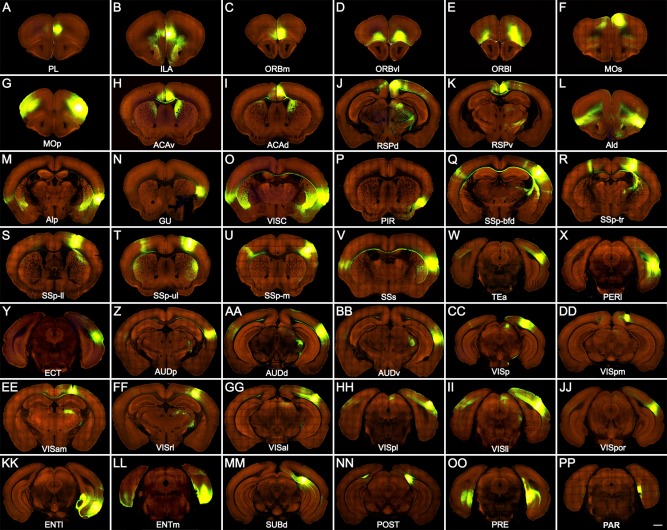
Representative images showing AAV‐GFP injection sites in 42 neocortical and allocortical areas of the right hemisphere. Injection sites are in prelimbic area (PL; A), infralimbic area (ILA; B), orbital area, medial part (ORBm; C), orbital area, ventrolateral part (ORBvl; D), orbital area, lateral part (ORBl; E), secondary motor area (MOs; F), primary motor area (MOp; G), anterior cingulate area, ventral part (ACAv; H), anterior cingulate area, dorsal part (AVAd; I), retrosplenial area, dorsal part (RSPd; J), retrosplenial area, ventral part (RSPv; K), agranular insular area, dorsal part (AId; L), agranular insular area, posterior part (AIp; M), gustatory area (GU; N), visceral area (VISC; O), piriform area (PIR; P), primary somatosensory area, barrel field (SSp‐bfd; Q), primary somatosensory area, trunk (SSp‐tr; R), primary somatosensory area, lower limb (SSp‐ll; S), primary somatosensory area, upper limb (SSp‐ul; T), primary somatosensory area, mouth (SSp‐m; U), supplemental somatosensory area (SSs; V), temporal association area (TEa; W), perirhinal area (PERI; X), ectorhinal area (ECT; Y), primary auditory area (AUDp; Z), dorsal auditory area (AUDd; AA), ventral auditory area (AUDv; BB), primary visual area (VISp; CC), posteromedial visual area (VISpm; DD), anteromedial visual area (VISam; EE), rostrolateral visual area (VISrl; FF), anterolateral visual area (VISal; GG), posterolateral visual area (VISpl; HH), laterointermediate visual area (VISli; II), postrhinal visual area (VISpor; JJ), entorhinal area, lateral part (ENTl; KK), entorhinal area, medial part (ENTm; LL), subiculum, dorsal part (SUBd; MM), postsubiculum (POST; NN), presubiculum (PRE; OO), and parasubiculum (PAR; PP). Fluorescent signal is green, and background is red. Scale bar = 800 μm.
Figure 7.
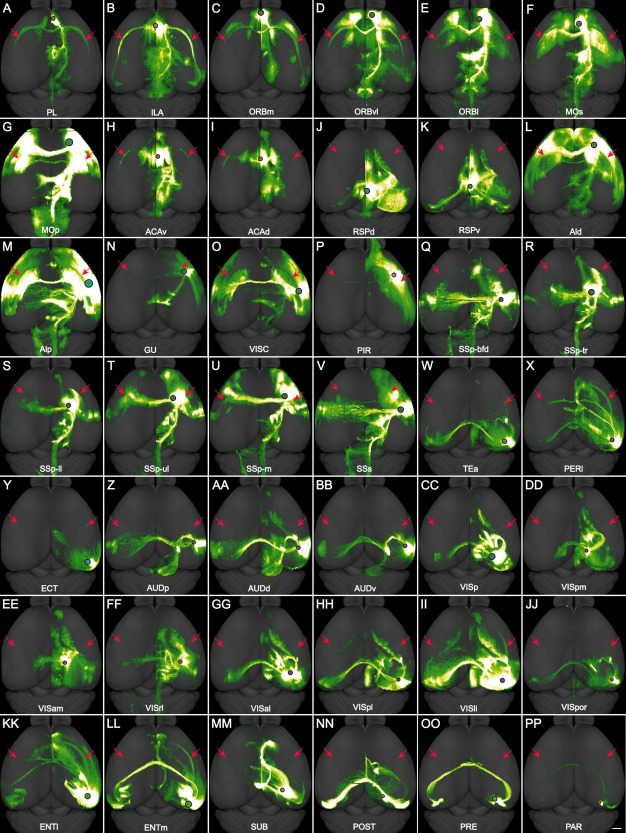
A–PP: Dorsal view of the AAV‐GFP injection sites in 42 neocortical and allocortical areas of the right hemisphere and their brain‐wide projections, corresponding to the panels in Figure 6. In each panel, the red cross within the black circle indicates the injection site, the size of the black circle is proportional to the size of the injection site, and the red arrows point to the claustrum in both hemispheres. For abbreviations see list. Scale bar = 1,000 μm.
Figure 8.
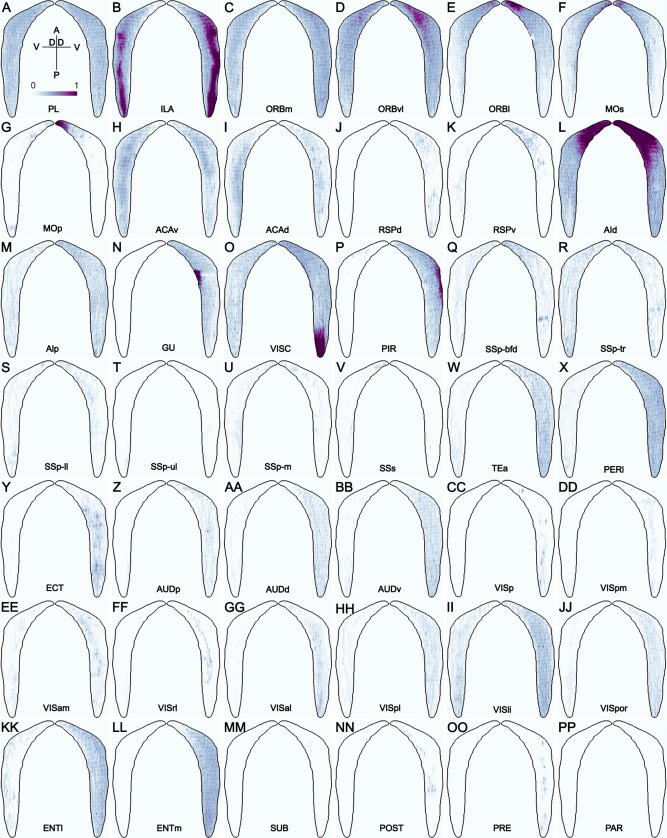
A–PP: Dorsal‐ventral view of the voxel‐level projection densities in ipsilateral claustrum (right) and contralateral claustrum (left) from the AAV‐GFP injections in 42 neocortical and allocortical areas of the right hemisphere, corresponding to the panels in Figures 6 and 7. D, dorsal; V, ventral; A, anterior; P, posterior. Color bar: white 0 to dark purple 1 in linear scale.
Figure 9.
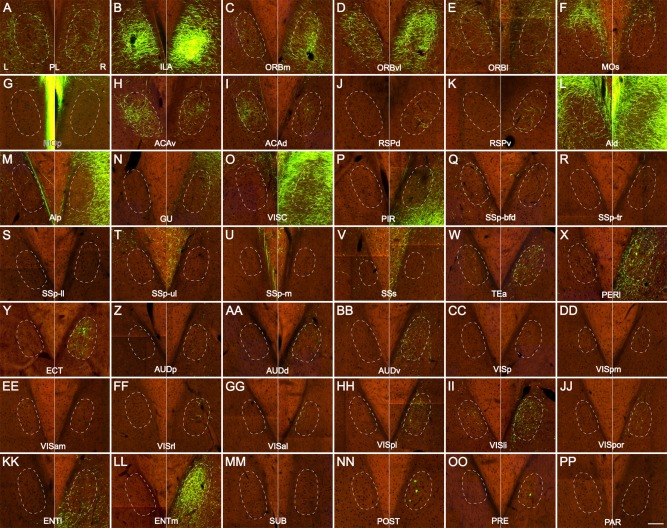
A–PP: Higher‐magnification coronal images of the ipsilateral (right) and contralateral (left) claustrum, showing incoming projections from the AAV‐GFP injections in 42 neocortical and allocortical areas of the right hemisphere, corresponding to the panels in Figures 6, 7, 8. Dashed ovals outline the proximate locations of the left and right claustrum. L, left claustrum; R, right claustrum. Scale bar = 100 μm.
Prelimbic and infralimbic areas
Four injections were made in the prelimbic (PL) and three in the infralimbic (ILA) areas (Table 3). Projections from both areas were seen across the entire anteroposterior extent of bilateral claustrum (Figs. 7, 8A,B). PL projected to the periphery of bilateral claustrum more than to its center part, with similar densities bilaterally (Fig. 9A). In contrast, ILA projected to the ventral part of bilateral claustrum with stronger input to the ipsilateral claustrum than to the contralateral one (Figs. 8, 9B).
Orbital areas
The orbital cortex consists of three subdivisions, medial (ORBm), lateral (ORBl), and ventrolateral (ORBvl). Three injections were made in each area (Table 3), and projections were observed across the entire anteroposterior extent of bilateral claustrum, with a higher density on the ipsilateral side (Figs. 7, 8, 9C–E). ORBm and ORBvl projections appeared stronger in the claustrum than those of the ORBl. Labeling density decreased gradually toward the posterior claustrum on both sides. Axon terminals mixed with passing fibers were denser in the periphery than in the center of the bilateral claustrum (Fig. 9C–E).
Motor areas
The motor cortex is composed of primary and secondary motor areas (MOp and MOs). Seven MOs and six MOp injections were selected (Table 3), spanning the elongated anteroposterior range. In the MOp injections, projections were not found in either ipsilateral or contralateral claustrum, although strong projections were seen in the motor thalamus, the VAL (ventral anterior–lateral complex of the thalamus; Figs. 7G, 9G). The dark purple part in Figure 8G is spillover from the injection site. Similarly, no projections were found in the bilateral claustrum of two anterior MOs injections (100140756 and 157710335, 0.5–0.8 mm posterior to the frontal pole), although projections from these injections were found in the thalamic nuclei VAL and MD (mediodorsal nucleus of the thalamus). This region was named as frontal association cortex in Paxinos and Franklin's (2001) atlas. In contrast to MOp and the anterior part of MOs, injections in the posterior part of MOs (112952510, 166055636,168162771, 180916954, and 182793477, 1.5–2.5 mm from the frontal pole) showed stronger projections in the contralateral than in the ipsilateral claustrum (Figs. 7, 8, 9F). The projections spanned nearly the entire anteroposterior extent of the bilateral claustrum (Fig. 8F), with the labeling density decreasing gradually toward the posterior. Axon terminals were distributed densely at the dorsal part of the bilateral claustrum (Figs. 8, 9F).
Cingulate areas
The cingulate cortex contains dorsal (ACAd) and ventral (ACAv) subdivisions, which received four and five injections, respectively (Table 3). ACAd and ACAv projections were similarly found in the entire extent of the bilateral claustrum, with a higher density in the contralateral side (Figs. 7, 8H,I). The input to claustrum appeared to be stronger from ACAv than from ACAd. Axon terminals were denser in the middle three‐fifths of the claustrum and decreased gradually toward most anterior and posterior parts (Fig. 8H,I). Axon terminals were distributed more densely in the dorsal part of the bilateral claustrum than in the ventral part (Fig. 9H,I).
Retrosplenial areas
The retrosplenial cortex consists of dorsal (RSPd), ventral (RSPv) and lateral agranular (RSPagl) parts. Four injections were made in RSPv and two in RSPd (Table 3). RSPv projections were extended moderately in the anterior one‐third of the ipsilateral claustrum and very sparsely in the contralateral (Figs. 7, 8, 9K). A few neurons were retrogradely labeled (Fig. 8K). RSPd projected to a location similar to that of RSPv but not to the contralateral side (Figs. 8, 9J).
Agranular insular areas
The agranular insular cortex is divided into dorsal (AId), ventral (AIv), and posterior (AIp) parts. Five injections were made into AId (with some possible contamination in AIv) and three in AIp (Table 3). Projections from both areas to claustrum were distributed throughout the anteroposterior extent of the bilateral claustrum at its periphery, with a higher density in the ipsilateral claustrum than in the contralateral (Figs. 7, 8, 9L,M). The darkest part was spillover from the injection site (Fig. 8L). Because both AId and AIp are strongly connected with each other on both hemispheres and are close to the claustrum, detected signal may contain both axonal terminals and passing fibers (Fig. 9L,M).
Gustatory and visceral areas
One injection was made in the gustatory (GU) area and two in the visceral (VISC) area (Table 3). Similar to the agranular insular areas, GU projected to the ipsilateral side (Figs. 7, 8, 9N), whereas VISC projected to the bilateral claustrum with relatively denser ipsilateral labeling (Figs. 7, 8, 9O). As with AId injections, the darkest parts were the result of spillover from the injection sites (Fig. 8N,O). Because the injection site of VISC was large, it is difficult to see projections in the immediate vicinity. Axon terminals mixed with passing fibers were distributed more in the dorsal periphery of the claustrum in the GU injection (Fig. 9N) and at the perimeter of the bilateral claustrum in the VISC injections (Fig. 9O).
Piriform area
The piriform area (PIR) is an olfactory area that received six injections (Table 3), each with similar projections in the claustrum. Labeling was very strong in the piriform area itself, as well as endopiriform nucleus, agranular insular cortex, and entorhinal cortex (Fig. 7P). Again, the darkest part was the result of spillover from the injection site (Fig. 8P). The projections of PIR were distributed more in the ventral part of the ipsilateral claustrum but not in the contralateral (Figs. 8, 9P).
Somatosensory areas
The somatosensory cortex consists of primary (SSp) and supplemental (SSs) somatosensory areas. Injections were made in defined regions of SSp, including barrelfields (SSp‐bfd), upper limb (SSp‐ul), lower limb (SSp‐ll), trunk (SSp‐tr), and mouth (SSp‐m), as well as in SSs (Table 3). In some of the SSp‐bfd injection cases in which injections were close to the posterior parietal area, projections were scattered in the ipsilateral claustrum and at the periphery of the contralateral claustrum (Figs. 7, 8, 9Q). In cases in which injections were made in the anterior part of SSp‐bfd and SSp‐ul, projection was barely found in either side of the claustrum (Fig. 8Q,T). Projections from SSp‐tr, SSp‐ll, and SSp‐m were sparse in the bilateral claustrum (Fig. 8R,S,U). In SSs injections, projection was barely found in either side of the claustrum (Figs. 7, 8, 9V).
Temporal association area
Three injections were made in the temporal association area (Tea; Table 3). In these cases, lateral posterior nucleus of the thalamus (LP) was densely labeled. Projections were found in the bilateral claustrum, with a higher density in the ipsilateral side than in the contralateral (Figs. 7, 8, 9W). Axon terminals were distributed in nearly the entire anteroposterior extent of the ipsilateral claustrum and the posterior one‐third of the contralateral (Fig. 8W).
Perirhinal and ectorhinal areas
Perirhinal (PERI) and ectorhinal (ECT) areas are located at the fundus and upper bank of rhinal sulcus, respectively, and are homologues to primate areas 35 and 36. As in primate, areas 35 and 36 together in mouse and rat are named as perirhinal cortex in some studies (Burwell, 2001; Beaudin et al., 2013). Two injections were made in PERI and one in ECT (Table 3). Like those from TEa, PERI projections were seen across the entire anteroposterior extent of the bilateral claustrum, denser in the ipsilateral side than in the contralateral (Figs. 7, 8, 9X). In contrast, ECT projected to the posterior three‐fourths of the ipsilateral claustrum (Figs. 7, 8, 9Y). Several retrogradely labeled neurons were found in the ipsilateral claustrum (Fig. 9Y).
Auditory areas
The auditory cortex consists of primary (AUDp), dorsal (AUDd), posterior (AUDpo), and ventral (AUDv) subdivisions. One injection was made in AUDp, two in AUDd, and two in AUDv (Table 3). No injection was made in AUDpo. Projections from AUDp were very sparse in nearly the entire anteroposterior extent of the ipsilateral claustrum but not in the contralateral (Figs. 7, 8, 9Z). Projections from AUDd and AUDv were found in the bilateral claustrum, stronger in the ipsilateral side than in the contralateral side, and were distributed across nearly the entire anteroposterior extent of ipsilateral claustrum (Figs. 7, 8, 9AA,BB). Although the injection sizes were similar between AUDd and AUDv, it seems likely that AUDv projected to the ipsilateral claustrum more than AUDd (Fig. 9AA,BB).
Visual areas
Recent studies have shown that the mouse visual cortex is composed of at least 10 areas (Wang and Burkhalter, 2007; Marshel et al., 2011; Garrett et al., 2014). From the topographic organization and virtual callosal connections, we have mapped these visual areas into the Common Coordinate Framework (see CCF document referenced above). The cortical area called posterior parietal cortex (PTLp), which is a mediolaterally running strip of cortex between the primary visual and the primary somatosensory areas, was replaced by three higher visual areas, namely, anterior (VISa), rostrolateral (VISrl), and anterolateral (VISal) visual areas, from medial to lateral. Lateral visual area (VISl) and posterolateral visual area (VISpl) are equivalent to areas LM and P, respectively, in other studies (Wang and Burkhalter, 2007; Marshel et al., 2011). Laterointermediate visual area (VISli) is a small strip running in the anteroposterior direction and located lateral to VISl. Postrhinal visual area (VISpor) is located posterolateral to VISli and above the rhinal sulcus and is posterior to TEa in the Allen reference atlas. We adopted existing nomenclature for the primary visual area (VISp), anteromedial visual area (VISam), and posteromedial visual area (VISpm), although their shapes and sizes were adjusted according to retinotopic maps.
Ten injections were made in VISp: two in VISpm, three in VISam, one in VISrl, one in VISal, one in VISpl, one in VISpor, and one in VISli (Table 3). No injections were made in VISa or VISl. The VISli and VISpor injections may have contaminations in VISl. All these visual areas projected to the bilateral claustrum (Figs. 8, 9CC–JJ). The projections were relatively denser in ipsilateral than in contralateral claustrum (Figs. 8, 9DD–JJ). Labeling was denser at posterodorsal portions of the ipsilateral claustrum from VISp, VISpm, and VISal (Figs. 8, 9CC,DD,GG), dorsally from VISam and VISrl (Figs. 8, 9EE,FF), and at its entire anteroposterior extent from VISpl, VISli, and VISpor (Figs. 8, 9HH–JJ). Remarkably, in all 10 injections in VISp, sparse projections were consistently seen in similar parts of the ipsilateral claustrum.
Retrohippocampal region
The retrohippocampal region includes entorhinal cortex, subiculum (SUB), postsubiculum (POST), presubiculum (PRE), and parasubiculum (PAR). The entorhinal cortex is divided into medial (ENTm), ventromedial (ENTmv), and lateral (ENTl) areas. One injection was made in ENTl and two in ENTm (Table 3). No injection was made in ENTmv. In both ENTl and ENTm injections, projections were seen in almost the entire anteroposterior extent of the ipsilateral claustrum (Figs. 7, 8, 9KK,LL). Sparse labeling was seen in the ventral part of the contralateral claustrum from the ENTl injection but not from the ENTm injections (Figs. 8, 9KK,LL).
One injection was made in each subicular area, including SUB, POST, PRE, and PAR (Table 3). Projections to the claustrum from these areas were not observed (Figs. 7, 8, 9MM–PP). Instead of axon terminals, a few retrogradely labeled neurons were found in the claustrum from POST and PRE injections (Figs. 8, 9NN,OO).
Claustrocortical projections
The projections from claustrum to cortex were analyzed in four cases with injection sites predominantly in the claustrum. Three were injected in Cre mice and one in a wild‐type mouse (Table 3). The three Cre mouse injections used two newly generated Cre lines to target claustrum‐selective marker genes (Fig. 2), Gnb4‐IRES2‐Cre and Ntng2‐IRES2‐Cre. We overlaid each image series onto the CCF, reconstructed the claustrum in 3D, and analyzed the anatomical locations of each injection site and its cortical and subcortical projection targets.
Injections in the Gnb4‐IRES2‐Cre and Ntng2‐IRES2‐Cre mice (Fig. 10A–D) are more specific and restrictive to the claustrum than in the wild‐type mouse. For these injections, projections were observed in all ipsilateral cortical areas as well as weakly in certain contralateral cortical areas that mirror the ipsilateral cortical areas that receive strong or moderate claustral inputs (Fig. 10E,F). The ipsilateral cortical areas receiving strong claustrum inputs include ORBm, ORBvl, ACAd, ACAv, PL, ILA, AId, AIv, AIp, ENT, and PERI. Those areas receiving moderate inputs are the frontal pole (FRP), MOs, ORBl, GU, VISC, RSPv, RSPd, RSPagl, VISam, VISpm, VISpl, VISpor, and TEa. Those receiving weak inputs are MOp, SSp, SSs, AUDp, AUDd, AUDv, AUDpo, VISp, VISl, VISli, VISal, VISrl, and VISa (Figs. 10E,F, 11).
Figure 10.
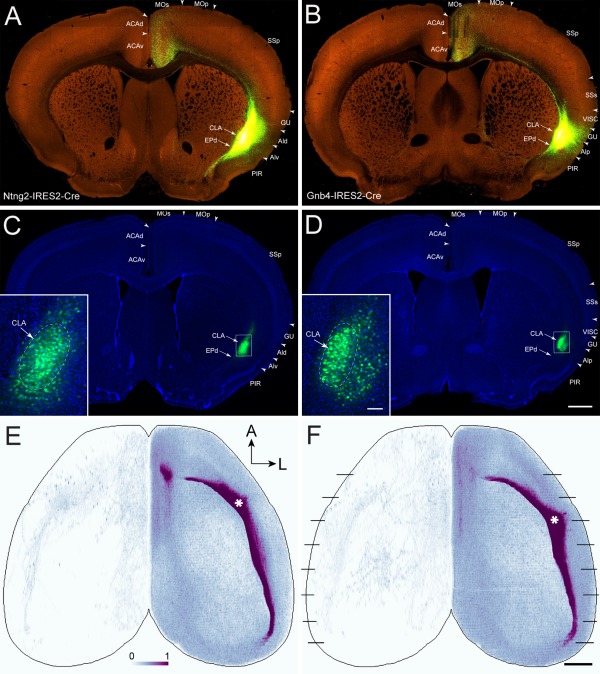
AAV‐GFP injections in the claustrum of the right hemisphere and their projections to the cortex. A,B: Low‐magnification TissueCyte images of injection sites in Ntng2‐IRES2‐Cre (485846989) and Gnb4‐IRES2‐Cre (485903475) mice, respectively. Note that laser excitation power during TissueCyte scanning was adjusted to maximize detection of single axon fibers anywhere in the brain, which led to oversaturation at the injection sites, preventing clear visualization of AAV‐infected cell bodies. C,D: Low‐magnification confocal images of the same sections shown in A and B, counterstained with DAPI. Arrows point to the claustrum and endopiriform nucleus in A–D. Insets: Higher‐magnification confocal images of the injection sites. Arrows point to the claustrum (outlined by the dashed lines). Note that laser excitation power during confocal imaging of the sections was adjusted to proper levels to distinguish AAV‐infected cell bodies from those uninfected. E,F: Dorsal view of the voxel‐level claustrocortical projection densities from injections shown in A and B, respectively. Asterisks indicate injection sites in the right hemisphere. Black lines in F indicate approximate levels of coronal sections shown in Figure 11. For abbreviations see list. A, anterior; L, lateral. Color bar: white 0 to dark purple 1 in linear scale. Scale bars = 700 μm in D (applies to A–D); 1,000 μm in E (applies to E,F); 80 μm in insets.
Figure 11.
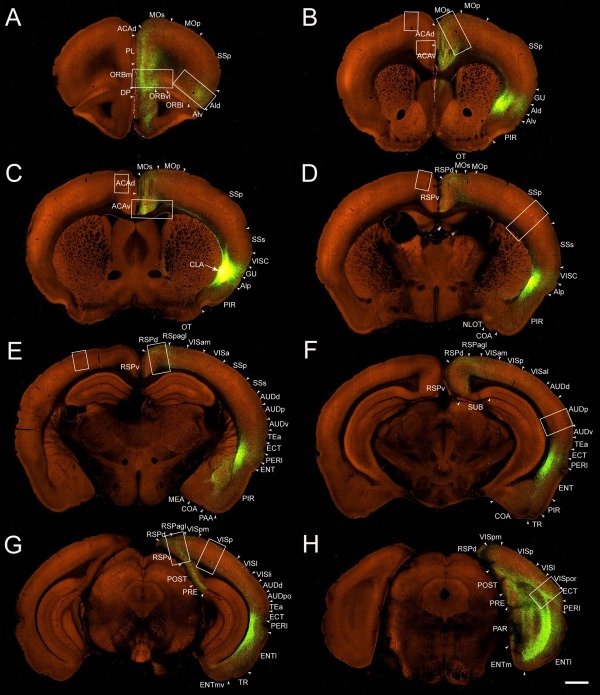
A–H: Low‐magnification coronal images of projections from the AAV‐GFP injection into the claustrum of the right hemisphere in a Gnb4‐IRES2‐Cre mouse (485903475). Sections in A–G are in 1‐mm intervals from anterior to posterior, and the interval between sections in G and H is 0.7 mm. Large and small boxes indicate regions where axon terminals are shown at higher magnification in Figure 12 for the right (ipsilateral) and left (contralateral) hemispheres, respectively. Arrow indicates injection site of the claustrum. Arrowheads indicate cortical area borders. For abbreviations see list. Scale bar = 800 μm.
The claustrum projected to different cortical areas with uneven densities and laminar distribution patterns. Areas such as ORBm, AId, MOs, ACAv, RSPd, RPSagl, and ECT that receive strong or moderate claustrum inputs shared similar laminar patterns. Here, labeled axons terminated most densely in layers 2/3, followed by 6 and 5, and sparsely in the inner portion of layer 1 (Figs. 11, 12). In cortical areas that receive weak claustral inputs such as SSp, AUDp, and VISp, projections terminated sparely from layer 1 to layer 6 (Figs. 11, 12).
Figure 12.
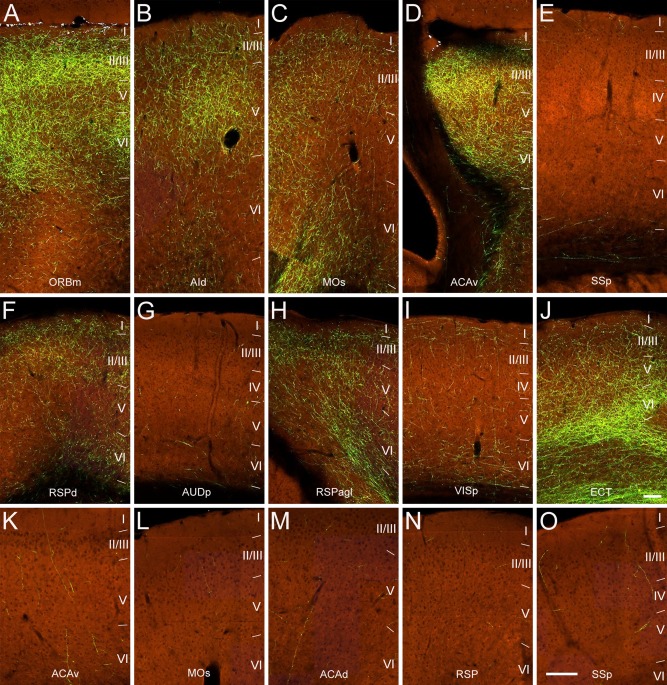
Higher‐magnification images of projections from the AAV‐GFP injection in the claustrum of the right hemisphere in a Gnb4‐IRES2‐Cre mouse (485903475) reveal laminar distribution of axon terminals in different cortical regions. Regions shown correspond to the large (ipsilateral) and small (contralateral) boxes in Figure 11: ORBm (A), AId (B), MOs (C), ACAv (D), SSp (E), RSPd (F), AUDp (G), RSPagl (H), VISp (I), ECT (J), contralateral ACAv (K), contralateral MOs (L), contralateral ACAd (M), contralateral RSP (N), and contralateral SSp (O). Images are oriented with pial surface at top and white matter at bottom. I–VI represent cortical layers. Lines indicate borders between cortical layers. Scale bars = 100 μm in J (applies to A–J); 180 μm in O (applies to K–O).
Projections from the claustrum were much sparser in contralateral cortical areas than in ipsilateral areas. They were found in the contralateral ORBm, ORBvl, PL, ILA, MOs, ACAv, ACAd, AId, AIp, RSPd, and SSp (Fig. 12K–O) and were barely observed in visual and auditory areas. Because labeling was too sparse, laminar distribution patterns of labeled axons in these contralateral cortical areas were not well defined.
In addition to the neocortex, claustral projections were also found in the olfactory areas and retrohippocampal regions. Olfactory areas receiving claustral inputs include the anterior olfactory nucleus (Fig. 11A), taenia tecta (Fig. 11A), PIR (Fig. 11B–F), nucleus of the lateral olfactory tract (Fig. 11D), cortical amygdala area (Fig. 11D–F), piriform‐amygdala area (Fig. 11E), and postpiriform transition area (Fig. 11F). Retrohippocampal regions receiving claustral inputs include ENTm, ENTl, PRE, POST, PAR, and SUB (Fig. 11G,H). Labeled axons in ENTm and ENTl terminated densely in layer 6 and moderately in layers 2/3 and 5, and very sparsely in layer 1; those in PRE, POST, and PAR terminated densely in layer 2/3 and very sparsely in layer 1, and those in SUB terminated sparsely in the pyramidal layer (Fig. 11G,H).
The centers of the four claustrum injection sites are located 2.6 mm (Ntng2, 513773998), 3.1 mm Ntng2, 485846989), 3.5 mm (Gnb4, 485903475), and 3.8 mm (wild‐type, 180436360) from the anterior edge of the frontal pole (Fig. 13). Although the locations of these injection sites varied, overall projection patterns from the claustrum to cortex were similar among these cases. The additional projections observed in the wild‐type injection likely were due to injection site contamination in gustatory, visceral, and insular cortices, as well as endopiriform nucleus, which was confirmed by specific patterns of subcortical projections (Fig. 13, insets). In this case, strong projections were seen in the contralateral insular cortex, which otherwise only received very sparse projections in the three Cre mouse injections. This likely is due to the less restricted injection in the wild‐type mouse, which contaminated the neighboring agranular insular cortex that sends strong projections to its contralateral side (Reep and Winans, 1982).
Figure 13.
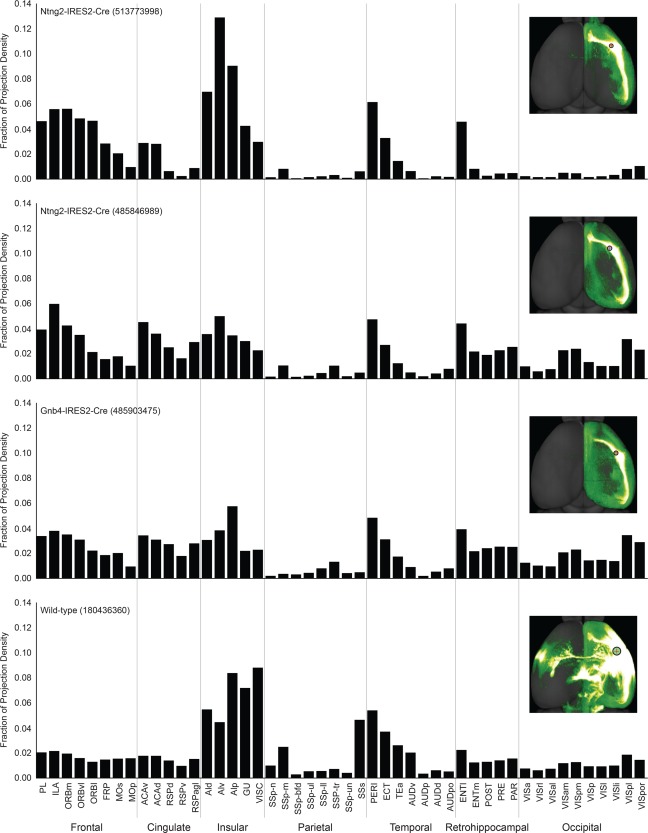
Histograms showing fractions of projection densities from the claustrum to different neocortical and allocortical areas in four AAV‐GFP injection experiments in three Cre mice and one wild‐type mouse. Each inset reveals the location of the injection site and projections in a dorsal view. The four injections are ordered from anterior to posterior.
In the four claustral injections, no projection to the contralateral claustrum was detected (Fig. 11), with the exception of a couple of nearby axon fibers. Within the ipsilateral claustrum, axons travel from the injection sites forward or backward along the ipsilateral claustrum toward the prefrontal or retrohippocampal regions (Figs. 10, 11). Although this is not entirely certain from our data because of the nearly saturated signals, these axons appeared to send collaterals and possibly form connections with other claustral neurons.
As described above, marker genes Ntng2 and Gnb4 are not exclusive to the claustrum; they are also expressed in nearby endopiriform nucleus and deep layers of the gustatory area and visceral area, albeit more sparsely (Fig. 2). Consequently, the Ntng2‐IRES2‐Cre and Gnb4‐IRES2‐Cre lines can also drive expression in these structures. However, specific targeting of the AAV tracer into each structure (sometimes fortuitously) can further enhance the specificity. For example, the Ntng2 injection 485846989 (Fig. 10C) was clearly centered in the claustrum, although it did encroach into the endopiriform nucleus as was confirmed by axonal labeling in the anterior olfactory nucleus. The Gnb4 injection 485903475 (Fig. 10D) was also clearly centered in the claustrum, although it did have contamination in deep layers of the agranular insular cortex, posterior part, which was confirmed by weak labeling in the midline thalamic nuclei. For both Gnb4 and Ntng2 injections, a few axon segments were found in the magnocellular and parvocellular parts of the ventral posteromedial thalamic nucleus, suggesting that the injections had minimal involvement of the visceral and gustatory areas. Note that the projection patterns from these injections that were relatively specific to the claustrum differed greatly from those injections in the same Cre lines but relatively specific to adjacent structures (e.g., Ntng2 injection 485847695, deep layers of the agranular area; Gnb4 injection 485875903, endopiriform nucleus; Gnb4 injection 485902743, deep layers of the visceral area), supporting the specificity of the claustrocortical projection patterns that we observed.
Claustral projections were observed in subcortical structures, in particular in the amygdala and striatum in all four injections (Fig. 11B,E). However, projections to the midline thalamic nuclei, hypothalamic nuclei, and brainstem nuclei were minor and inconsistent across the four injections. This difference could be due to cell‐type‐specific projections or contamination of the injection sites. Further investigation of claustrosubcortical projections is needed (Mathur, 2014).
Quantitative analysis of the claustrocortical and corticoclaustral connections
To reveal the trend of projection strengths from the claustrum to the neocortex and allocortex, we computed the fractional density of projection signal in each cortical area. Projection density in a target structure/area was defined as the ratio of projection signal volume in that structure relative to the total volume of that structure. Projection density in each structure was then converted to a fraction of total density within each experiment, and the sum of all fractional projection densities would therefore add up to 1 (Fig. 13). Fractional density is relative to each experiment, compensating for the difference in the size of each injection site. This quantitative analysis confirmed the qualitative observation described above that the claustrum projects to all cortical areas and revealed a crude topographic organization (Fig. 13). The anterior claustrum injection had more projections to anterior cortical areas and less to occipital areas and retrohippocampal regions compared with the middle and posterior claustrum injections, which showed an opposite trend. It should be noted that the high values in the insular areas for all four cases were due largely to fluorescence signal spillover from the injection sites.
To compare ascending and descending projection strengths among areas in different experiments, we computed normalized projection density (projection signal volume divided by both injection site volume and target structure volume) in representative injection cases for each cortical area (Table 3) and the claustrum (Ntng2‐IRES2‐Cre, 485846989). Figure 14 shows the normalized projection density for both ascending projections from the claustrum to cortical areas (color‐coded in blue) and descending projections from cortical areas to the claustrum (in red). The width of the line is proportional to the normalized projection density. This quantitative result confirms that the claustrum has reciprocal connections with nearly all ipsilateral and many contralateral cortical areas. This analysis also shows that the claustrum receives strong inputs from cortical areas PL, ILA, ORB, ACA, PERI, and ENT and in turn sends strong outputs back to these areas (Fig. 14A). The claustrum receives weak inputs from sensory areas and sends weak outputs back to these areas (Fig. 14A). The claustrum receives strong contralateral inputs from cortical areas PL, ILA, ORBm, ORBvl, and ACAv, but these cortical areas receive much weaker contralateral projections from the claustrum (Fig. 14B).
Figure 14.
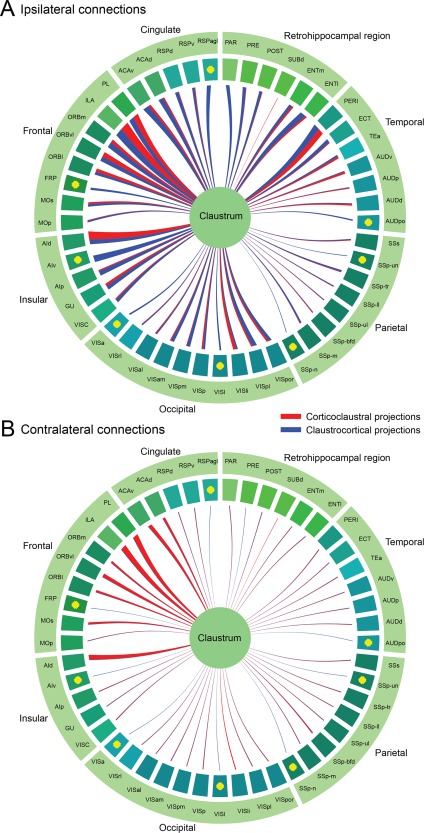
Circle diagrams showing the normalized projection densities from different neocortical and allocortical areas to claustrum and vice versa. A: Ipsilateral connections. B: Contralateral connections. The width of the line is proportional to the normalized projection density. Corticoclaustral projections are color‐coded in red and claustrocortical projections in blue. Yellow dots indicate no injections or lack of good injections. Cortical areas are color‐coded as in the Allen mouse reference atlas.
DISCUSSION
The present study began by defining the boundary of the claustrum in mouse by means of cytoarchitecture, chemoarchitecture, genoarchitecture, and connectivity. This allowed us to reconstruct the 3D claustrum, with a volume of 0.275 mm3, more than 200 times smaller than the neocortex. Using both qualitative and quantitative analyses of the connections between claustrum and cortex we have demonstrated that almost all cortical areas send inputs to both ipsi‐ and contralateral claustrum in a crude topographic manner. The claustrum, in turn, sends projections back to all ipsilateral cortical areas and to several contralateral cortical areas that mirror ipsilateral areas that receive strong or moderate claustral inputs (Fig. 14).
Delineation of the boundary of the claustrum
Our primary understanding of brain organization derives from its structure, as defined by unique characteristics in cytoarchitecture, myeloarchitecture, chemoarchitecture, topography, connectivity, and function. More than 20,000 genes have been mapped onto the entire mouse brain (Lein et al., 2007; Ng et al., 2009). Genoarchitecture, the differential topographic patterning of expressed genes, has been used to delineate brain structures and their evolutionary relations across species (Puelles and Ferran, 2012; Zeng et al., 2012). With the claustral molecular maker (Gng2) combined with tract‐tracing, the shape, size and location of the claustrum were modified from what was shown in a well‐established rat atlas (Paxinos and Watson, 1998), revealing a relatively smaller claustrum, especially in its anterior extent (Mathur et al., 2009; Mathur, 2014). Using the same gene and other claustral makers, we made similar findings for the mouse claustrum. Located beneath the agranular insular area, the claustrum is smaller than previously indicated in the mouse atlases (Paxinos and Franklin, 2001; Dong, 2008) and in some studies (Smith and Alloway, 2014; Zingg et al., 2014) but is similar to that in other studies (Druga et al., 1993; Real et al., 2006). In addition to Gng2, we found a large set of genes with enriched expression in the mouse claustrum (Table 2). Among these, Cux2, Lxn, and Ntng2 were also found to be differentially expressed in both mouse and monkey claustrum (Watakabe et al., 2014). Furthermore, Gng2 and Ntng2 were found in human claustrum (Pirone et al., 2012), and Oprk1 (kappa opioid receptor) expression is the strongest in both mouse and human claustrum (Peckys and Landwehrmeyer, 1999). It seems likely that these genes are well‐conserved in the claustrum across species and that they serve as molecular markers of anatomical delineation.
In previous rodent studies (Druga et al., 2014; Smith and Alloway, 2014), the claustrum was divided into an oval, neuron‐dense “ventral claustrum” (not to be confused with the outdated terminology of “ventral claustrum,” a structure that is now commonly referred to as the endopiriform nucleus), and a moon‐shaped, neuron‐sparse “dorsal claustrum” located beneath the gustatory and visceral areas. In the present study, we found the connectivity to be markedly different between these two subdivisions, in addition to other molecular and cytoarchitectonic differences. The lack of projections from the “dorsal claustrum” compared with the “ventral claustrum,” which projects to the entire ipsilateral cortex, suggests that the “dorsal claustrum” should not be considered as a part of the claustrum but rather as a part of layer 6 of the gustatory and visceral areas. In addition, we also compared some of the genes negatively expressed in the claustrum with the “shell/core” zones immunostained with VGLUT2 and calretinin (Real et al., 2003, 2006). Our results indicate that the oval “core” is the true claustrum and the “shell” more reasonably belongs to layer 6 of the agranular insular area. We therefore conclude that the claustrum is a single nucleus of gray matter beneath the agranular insular area. This subcortical nucleus has crude topographical and reciprocal connections with almost all cortical areas.
Comparison with previous connectivity studies
Claustrocortical and corticoclaustral projections have been extensively studied in mammals, especially in monkeys and cats (Edelstein and Denaro, 2004; Druga, 2014). These studies demonstrate that the claustrum connects with frontal, parietal, temporal, and occipital areas. Recent human and nonhuman primate studies with diffusion tensor imaging or constrained spherical deconvolution tractography confirmed that the claustrum connects through fiber bundles with frontal, parietal, temporal, and occipital cortices (Park et al., 2012; Milardi et al., 2015; Torgerson et al., 2015) and extends to contralateral claustrum through interclaustral connections (Milardi et al., 2015). In these studies, low imaging resolution could not reveal the directionality of the connections between the claustrum and the cortex. Using a sensitive anterograde viral tracing method, we found that the claustrum projects to all ipsilateral cortical areas with varying strengths and receives inputs from almost all ipsilateral cortical areas as well. In addition to ipsilateral claustrocortical connections, we found weak projections of the claustrum to many contralateral cortical areas. However, we did not find any direct connection between the ipsilateral and the contralateral claustra, contrary to the human study (Milardi et al., 2015). Our finding of strong connections of the claustrum with the prefrontal and cingulate areas is consistent with that reported in humans (Torgerson et al., 2015). Although the total volumes and numbers of cortical areas differ greatly among these species, the consistency of widespread distribution of connections and selective strong connections suggests that the fundamental organization of the claustrum is similar.
Topography is one of the organizational principles for information processing in brains. It applies also to the connections between the claustrum and the cortex. Retrograde tracer studies in monkeys and cats have revealed that claustral projection neurons are organized topographically (Pearson et al., 1982; Minciacchi et al., 1985; Tanne‐Gariepy et al., 2002; Gattass et al., 2014). Retrogradely labeled neurons within the claustrum form anteroposterior elongated zones: an anterior zone to frontal cortex, a middle zone to parietal cortex, and a posterior zone to occipital cortex. These cortically related zones are not strictly segregated from each other but overlap to a certain extent (Clasca et al., 1992; Tanne‐Gariepy et al., 2002). Compared with that in monkeys and cats, the claustrum in rats contains only two considerably overlapping cortically related zones, the anterodorsal zone with intermingled neurons projecting to motor and somatosensory areas and the posteroventral zone with intermingled neurons projecting to visual and auditory areas (Sloniewski et al., 1986; Sadowski et al., 1997; Kowianski et al., 2000). Consistent with retrograde tracing studies, our anterograde tracing also shows a crude topography of claustrocortical projections; the anterior part of the claustrum sends more projections to anterior cortical areas than the posterior part of the claustrum, whereas the posterior part of the claustrum does the opposite. For descending pathways, our findings that the primary visual area projects to the posterior part of the claustrum and that the higher visual areas project to almost the entire anteroposterior extent of the claustrum are consistent with previous studies in monkeys, cats, rabbits, and rats as well as guinea pigs and ferrets (LeVay and Sherk, 1981; Pearson et al., 1982; Sloniewski et al., 1986; Jakubowska‐Sadowska et al., 1998; Patzke et al., 2014). Therefore, like claustrocortical projections, the corticoclaustral projections in mice are also organized in a crude dorsoventral and anteroposterior topographic fashion. Descending axons from various cortical areas form anteroposterior elongated terminal fields in the claustrum with considerable overlap. Substantial overlapping of cortical inputs, particularly from frontal, cingulate, insular, temporal, and entorhinal cortices, is reminiscent of prefrontostriatal projections (Mailly et al., 2013), suggesting that multimodal information processing may occur within the claustrum (Hadjikhani and Roland, 1998).
Reciprocal claustrocortical connections have been shown in different species with bidirectional transport tracers (e.g., WGA‐HRP or BDA) which reveal retrograde neurons as well as axon terminals in targets (Neal et al., 1986; Beneyto and Prieto, 2001; Patzke et al., 2014). Similarly, we found that the claustrum has reciprocal connections with all of the cortical areas studied and that there is a generally proportional relationship between strengths of descending projections from cortical areas to claustrum and ascending projections from claustrum to cortical areas (Fig. 14). We also found reciprocal connections between claustrum and allocortical areas such as the entorhinal cortex and unidirectional connections from the claustrum to regions around the subiculum, consistent with previous studies in rats (van Groen and Wyss, 1990a, 1990b).
Bilateral claustrocortical and corticoclaustral projections have been described for cats and rats (Norita, 1977; Squatrito et al., 1980; Macchi et al., 1981; Minciacchi et al., 1985; Li et al., 1986; Sloniewski et al., 1986; Sadowski et al., 1997; Beneyto and Prieto, 2001). However, some studies showed only ipsilateral corticoclaustral projections after injections in primary sensory areas and portions of primary motor area (Olson and Graybiel, 1980; Alloway et al., 2009; Patzke et al., 2014). With sensitive viral tracer injected into the claustrum, our results show bilateral claustrocortical projections with ipsilateral dominance (Fig. 10C,D). The discrepancy between the studies may be due to the extremely weak claustrocortical projections to contralateral primary sensory and motor areas. For the descending pathways, we found that most of the cortical areas send stronger projections to ipsilateral claustrum than to contralateral, except that two areas (cingulate and secondary motor areas) send stronger projections to the contralateral claustrum than the ipsilateral. Why these differences exist compared with the rest of the cortex is not known. Through its bilateral connections with cortex, the claustrum is capable of maintaining or coordinating hemispheric functional cortical connectivity.
Laminar distribution of inputs to cortex may indicate the flow of information in the brain. Anterograde tracing in cats and tree shrews demonstrated that claustral projections preferentially terminate in layers 4 and 6 of the primary visual area (Carey et al., 1980; Olson and Graybiel, 1980; LeVay and Sherk, 1981; LeVay, 1986). This laminar distribution pattern is referred to as a “feedforward connection,” which extends from lower to higher hierarchical areas (Felleman and Van Essen, 1991). However, a study in cats investigating synaptic organization of claustral projections to the primary visual cortex using light and electron microscopy demonstrates synapses in all cortical layers, suggestive of a “lateral connection” (da Costa et al., 2010). Our results likewise demonstrate that the claustrocortical projections are distributed, sparsely, across all layers of VISp, SSp, and AUDp, without layer 4 dominance (Fig. 12). In contrast to the primary sensory areas, more projections terminated in layers 2/3 than in layers 5 and 6 of the midline cortical areas, without any apparent relation to feedforward, feedback, or lateral connections (Felleman and Van Essen, 1991). This is also the case for claustral projections to the allocortical areas. In ENT, for example, projections are terminated strongly in layer 6, moderately in layers 5 and 2/3, and weakly in layer 1 (Fig. 11). The functional significance of these connections remains unknown. Nevertheless, our anterograde viral tracing results are consistent with previous studies in rats and mice that used using conventional anterograde tracing methods (Zhang et al., 2001; Zingg et al., 2014).
Functional implications
Recent large‐scale tracing studies have resulted in the construction of complex interareal, corticocortical connectomes in monkeys and rodents (Markov et al., 2014; Oh et al., 2014; Zingg et al., 2014; Bota et al., 2015), revealing modules or networks specialized for processing certain brain functions. The present study shows that the claustrum has strong reciprocal and bilateral connections with the midline cortical areas, including prelimbic, infralimbic, orbital, anterior cingulate, and retrosplenial areas, as well as strong reciprocal connections with the ipsilateral perirhinal and entorhinal areas. Midline and temporal cortical areas and retrohippocampal areas have been associated with a broad range of emotional, cognitive, and attentional processes.
Our quantitative analysis reveals that the claustrum is preferentially connected in a topographic, reciprocal, and bilateral manner to most, but curiously not all, cortical areas. In particular, only sparse and/or weak ascending and descending connections are apparent between the claustrum and the primary motor cortex, primary and secondary somatosensory cortex, primary auditory cortex, and primary visual cortex. The claustrum may therefore act as a superhub to coordinate a far‐flung network of cortical regions.
One important question left unanswered by our analysis is the extent to which any individual claustrum neuron projects widely to many cortical regions or whether, depending on the anterior–posterior location of its cell body within the claustrum, it has a well‐focused cortical axonal target zone.
The functional implications of such a widespread claustrocortical network remain open. As alluded to above, the claustrum has been implicated in mediating the integrated character of any one conscious experience (Crick and Koch, 2005), in amplifying cortical oscillations (Smythies et al., 2012, 2014), in detecting salient events (Remedios et al., 2010, 2014), in allocating selective attention (Mathur, 2014; Goll et al., 2015), and in controlling and/or switching between the resting state network and the central executive network (Reser et al., 2014). Claustrum‐specific transgenic mice, in combination with optogenetics and modern physiological techniques, will make probing such functions possible.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
ROLE OF AUTHORS
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: QW, LN, JAH, SWO, HZ. Acquisition of data: JAH, SMS, AB. Analysis and interpretation of data: QW, LN, DF, YL, JJR, CK, HZ. Drafting of the manuscript: QW, CK, HZ. Critical revision of the manuscript for important intellectual content: QW, CK, HZ. Study supervision: CK, HZ.
DATA ACCESSIBILITY
All connectivity, ISH, and reference data for the Allen Mouse Brain Connectivity Atlas are available through the data portal (http://mouse.brain-map.org).
ACKNOWLEDGMENTS
We thank Linda Madisen, Hong Gu, and Maya Mills for generating the new Cre lines; Marty Mortrud, Phil Bohn, and Ben Ouellette for stereotaxic injections; Anh Ho for TissueCyte imaging; Karla Hirokawa for confocal imaging; and the Transgenic Colony Management team, especially James Harrington, for mouse husbandry. The authors thank the Allen Institute founders, Paul G. Allen and Jody Allen, for their vision, encouragement, and support.
This article was published online on 27 June 2012. An error was subsequently identified. This notice is included in the online and print versions to indicate that both have been corrected 01 July 2016.
LITERATURE CITED
- Alloway KD, Smith JB, Beauchemin KJ, Olson ML. 2009. Bilateral projections from rat MI whisker cortex to the neostriatum, thalamus, and claustrum: forebrain circuits for modulating whisking behavior. J Comp Neurol 515:548–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudin SA, Singh T, Agster KL, Burwell RD. 2013. Borders and comparative cytoarchitecture of the perirhinal and postrhinal cortices in an F1 hybrid mouse. Cereb Cortex 23:460–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneyto M, Prieto JJ. 2001. Connections of the auditory cortex with the claustrum and the endopiriform nucleus in the cat. Brain Res Bull 54:485–498. [DOI] [PubMed] [Google Scholar]
- Bota M, Sporns O, Swanson LW. 2015. Architecture of the cerebral cortical association connectome underlying cognition. Proc Natl Acad Sci U S A (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, Sawchenko PE. 2007. Time course and distribution of inflammatory and neurodegenerative events suggest structural bases for the pathogenesis of experimental autoimmune encephalomyelitis. J Comp Neurol 502:236–260. [DOI] [PubMed] [Google Scholar]
- Bunce JG, Zikopoulos B, Feinberg M, Barbas H. 2013. Parallel prefrontal pathways reach distinct excitatory and inhibitory systems in memory‐related rhinal cortices. J Comp Neurol 521:4260–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwell RD. 2001. Borders and cytoarchitecture of the perirhinal and postrhinal cortices in the rat. J Comp Neurol 437:17–41. [DOI] [PubMed] [Google Scholar]
- Cai Y, Zhang Q, Wang C, Zhang Y, Ma T, Zhou , Tian M, Rubenstein JL, Yang Z. 2013. Nuclear receptor COUP‐TFII‐expressing neocortical interneurons are derived from the medial and lateral/caudal ganglionic eminence and define specific subsets of mature interneurons. J Comp Neurol 521:479–497. [DOI] [PubMed] [Google Scholar]
- Carey RG, Bear MF, Diamond IT. 1980. The laminar organization of the reciprocal projections between the claustrum and striate cortex in the tree shrew, Tupaia glis . Brain Res 184:193–198. [DOI] [PubMed] [Google Scholar]
- Chorazyna H, Stepien L, Sychowa B. 1965. Changes in auditory conditioning in dogs after lesions of the claustrum. Proc XXIII Int Cong Physiol Sci Tokyo 13:456–457. [Google Scholar]
- Clarey JC, Irvine DR. 1986. Auditory response properties of neurons in the claustrum and putamen of the cat. Exp Brain Res 61:432–437. [DOI] [PubMed] [Google Scholar]
- Clasca F, Avendano C, Roman‐Guindo A, Llamas A, Reinoso‐Suarez F. 1992. Innervation from the claustrum of the frontal association and motor areas: axonal transport studies in the cat. J Comp Neurol 326:402–422. [DOI] [PubMed] [Google Scholar]
- Cortimiglia R, Crescimanno G, Salerno MT, Amato G. 1991. The role of the claustrum in the bilateral control of frontal oculomotor neurons in the cat. Exp Brain Res 84:471–477. [DOI] [PubMed] [Google Scholar]
- Crick FC, Koch C. 2005. What is the function of the claustrum? Philos Trans R Soc Lond B Biol Sci 360:1271–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa NM, Fursinger D, Martin KA. 2010. The synaptic organization of the claustral projection to the cat's visual cortex. J Neurosci 30:13166–13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Li B, Liu Y, Iqbal K, Grundke‐Iqbal I, Gong C. 2009. Dysregulation of insulin signaling, glucose transporters, O‐GlcN acylation, and phosphorylation of tau and neurofilaments in the brain: Implication for Alzheimer's disease. Am J Pathol 175:2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HW. 2008. The allen reference atlas: a digital color brain atlas of the C57BL/6J male mouse. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- Druga R. 2014. The structure and connection of the claustrum In: Smythies JR, Edelstein LR, Ramachandran VS, editors. The claustrum: structural, functional, and clinical neuroscience. San Diego: Academic Press; p 29–84. [Google Scholar]
- Druga R, Chen S, Bentivoglio M. 1993. Parvalbumin and calbindin in the rat claustrum: an immunocytochemical study combined with retrograde tracing frontoparietal cortex. J Chem Neuroanat 6:399–406. [DOI] [PubMed] [Google Scholar]
- Druga R, Salaj M, Barinka F, Edelstein L, Kubova H. 2014. Calretinin immunoreactivity in the claustrum of the rat. Front Neuroanat 8:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein LR, Denaro FJ. 2004. The claustrum: a historical review of its anatomy, physiology, cytochemistry and functional significance. Cell Mol Biol 50:675–702. [PubMed] [Google Scholar]
- Eskilsson A, Tachikawa M, Hosoya K, Blomqvist A. 2014. Distribution of microsomal prostaglandin E synthase‐1 in the mouse brain. J Comp Neurol 522:3229–3244. [DOI] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. 1991. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex 1:1–47. [DOI] [PubMed] [Google Scholar]
- Frontera JG, Stiehl WL. 1963. Further results on stimulation of insula and claustrum in monkeys (Macaca mulatta). Anat Rec 145:319–320. [Google Scholar]
- Gabor AJ, Peele TL. 1964. Alterations of behavior following stimulation of the claustrum of the cat. Electroencephalogr Clin Neurophysiol 17:513–519. [DOI] [PubMed] [Google Scholar]
- Garrett ME, Nauhaus I, Marshel JH, Callaway EM. 2014. Topography and areal organization of mouse visual cortex. J Neurosci 34:12587–12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattass R, Soares JG, Desimone R, Ungerleider LG. 2014. Connectional subdivision of the claustrum: two visuotopic subdivisions in the macaque. Front Syst Neurosci 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll Y, Atlan G, Citri A. 2015. Attention: the claustrum. Trends Neurosci 38:486–495. [DOI] [PubMed] [Google Scholar]
- Hadjikhani N, Roland PE. 1998. Cross‐modal transfer of information between the tactile and the visual representations in the human brain: a positron emission tomographic study. J Neurosci 18:1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JA, Oh SW, Zeng H. 2012. Adeno‐associated viral vectors for anterograde axonal tracing with fluorescent proteins in nontransgenic and cre driver mice. Curr Protoc Neurosci Chapter 1, Unit 1, 20 21–18. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Zeng H. 2013. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci 36:183–215. [DOI] [PubMed] [Google Scholar]
- Ishii K, Tsuji H, Tamaoka A. 2011. Mumps virus encephalitis with symmetric claustrum lesions. AJNR 32:E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowska‐Sadowska K, Moryś J, Sadowski M, Kowiański P, Karwacki Z, Narkiewicz O. 1998. Visual zone of the claustrum shows localizational and organizational differences among rat, guinea pig, rabbit and cat. Anat Embryol 198:63–72. [DOI] [PubMed] [Google Scholar]
- Koubeissi MZ, Bartolomei F, Beltagy A, Picard F. 2014. Electrical stimulation of a small brain area reversibly disrupts consciousness. Epilepsy Behav 37:32–35. [DOI] [PubMed] [Google Scholar]
- Kowianski P, Morys J, Sadowski M, Dziewiatkowski J. 2000. Qualitative and quantitative differences in the motor and somatosensory cortical projections of the rat claustrum‐‐combined retrograde transport and stereological studies. Fol Morphol 59:111–119. [PubMed] [Google Scholar]
- Kuan L, Li Y, Lau C, Feng D, Bernard A, Sunkin SM, Zeng H, Dang C, Hawrylycz M, Ng L. 2015. Neuroinformatics of the Allen Mouse Brain Connectivity Atlas . Methods 73:4–17. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. 2007. Genome‐wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. [DOI] [PubMed] [Google Scholar]
- LeVay S. 1986. Synaptic organization of claustral and geniculate afferents to the visual cortex of the cat. J Neurosci 6 3564–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVay S, Sherk H. 1981. The visual claustrum of the cat. I. Structure and connections. J Neurosci 1:956–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZK, Takada M, Hattori T. 1986. Topographic organization and collateralization of claustrocortical projections in the rat. Brain Res Bull 17:529–532. [DOI] [PubMed] [Google Scholar]
- Lim DH, Ledue J, Mohajerani MH, Vanni MP, Murphy TH. 2013. Optogenetic approaches for functional mouse brain mapping. Front Neurosci 7:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi G, Bentivoglio M, Minciacchi D, Molinari M. 1981. The organization of the claustroneocortical projections in the cat studied by means of the HRP retrograde axonal transport. J Comp Neurol 195:681–695. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. 2010. A robust and high‐throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, Gu H, Mills M, Cheng A, Tasic B, Nguyen TN, Sunkin SM, Benucci A, Nagy A, Miyawaki A, Helmchen F, Empson RM, Knopfel T, Boyden ES, Reid RC, Carandini M, Zeng H. 2015. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85:942–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailly P, Aliane V, Groenewegen HJ, Haber SN, Deniau JM. 2013. The rat prefrontostriatal system analyzed in 3D: evidence for multiple interacting functional units. J Neurosci 33:5718–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov NT, Ercsey‐Ravasz MM, Ribeiro Gomes AR, Lamy C, Magrou L, Vezoli J, Misery P, Falchier A, Quilodran R, Gariel MA, Sallet J, Gamanut R, Huissoud C, Clavagnier S, Giroud P, Sappey‐Marinier D, Barone P, Dehay C, Toroczkai Z, Knoblauch K, Van Essen DC, Kennedy H. 2014. A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cereb Cortex 24:17–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Garrett ME, Nauhaus I, Callaway EM. 2011. Functional specialization of seven mouse visual cortical areas. Neuron 72:1040–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur BN. 2014. The claustrum in review. Front Syst Neurosci 8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur BN, Caprioli RM, Deutch AY. 2009. Proteomic analysis illuminates a novel structural definition of the claustrum and insula. Cereb Cortex 19:2372–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milardi D, Bramanti P, Milazzo C, Finocchio G, Arrigo A, Santoro G, Trimarchi F, Quartarone A, Anastasi G, Gaeta M. 2015. Cortical and subcortical connections of the human claustrum revealed in vivo by constrained spherical deconvolution tractography. Cereb Cortex 25:406–414. [DOI] [PubMed] [Google Scholar]
- Minciacchi D, Molinari M, Bentivoglio M, Macchi G. 1985. The organization of the ipsi‐ and contralateral claustrocortical system in rat with notes on the bilateral claustrocortical projections in cat. Neuroscience 16:557–576. [DOI] [PubMed] [Google Scholar]
- Neal JW, Pearson RC, Powell TP. 1986. The relationship between the auditory cortex and the claustrum in the cat. Brain Res 366:145–151. [DOI] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong HW, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, Bokil H, Mitra PP, Puelles L, Hohmann J, Anderson DJ, Lein ES, Jones AR, Hawrylycz M. 2009. An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci 12:356–362. [DOI] [PubMed] [Google Scholar]
- Norita M. 1977. Demonstration of bilateral claustro‐cortical connections in the cat with the method of retrograde axonal transport of horseradish peroxidase. Arch Histol Jpn 40:1–10. [DOI] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, Mortrud MT, Ouellette B, Nguyen TN, Sorensen SA, Slaughterbeck CR, Wakeman W, Li Y, Feng D, Ho A, Nicholas E, Hirokawa KE, Bohn P, Joines KM, Peng H, Hawrylycz MJ, Phillips JW, Hohmann JG, Wohnoutka P, Gerfen CR, Koch C, Bernard A, Dang C, Jones AR, Zeng H. 2014. A mesoscale connectome of the mouse brain. Nature 508:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CR, Graybiel AM. 1980. Sensory maps in the claustrum of the cat. Nature 288:479–481. [DOI] [PubMed] [Google Scholar]
- Park S, Tyszka JM, Allman JM. 2012. The claustrum and insula in Microcebus murinus: a high resolution diffusion imaging study. Front Neuroanat 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzke N, Innocenti GM, Manger PR. 2014. The claustrum of the ferret: afferent and efferent connections to lower and higher order visual cortical areas. Front Syst Neurosci 8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. 2001. The mouse brain in stereotaxic coordinates, 2nd ed. New York: Academic Press. [Google Scholar]
- Paxinos G, Watson C. 1998. The rat brain in stereotaxic coordinates, 4th ed. San Diego: Academic Press . [Google Scholar]
- Pearson RC, Brodal P, Gatter KC, Powell TP. 1982. The organization of the connections between the cortex and the claustrum in the monkey. Brain Res 234:435–441. [DOI] [PubMed] [Google Scholar]
- Peckys D, Landwehrmeyer GB. 1999. Expression of mu, kappa, and delta opioid receptor messenger RNA in the human CNS: a 33P in situ hybridization study. Neuroscience 88:1093–1135. [DOI] [PubMed] [Google Scholar]
- Pirone A, Cozzi B, Edelstein L, Peruffo A, Lenzi C, Quilici F, Antonini R, Castagna M. 2012. Topography of Gng2‐ and NetrinG2‐expression suggests an insular origin of the human claustrum. PLoS One 7:e44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptito M, Lassonde MC. 1981. Effects of claustral stimulation on the properties of visual cortex neurons in the cat. Exp Neurol 73:315–320. [DOI] [PubMed] [Google Scholar]
- Puelles L, Ferran JL. 2012. Concept of neural genoarchitecture and its genomic fundament. Front Neuroanat 6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman FE, Baizer JS. 2007. Neurochemically defined cell types in the claustrum of the cat. Brain Res 1159:94–111. [DOI] [PubMed] [Google Scholar]
- Real MA, Davila JC, Guirado S. 2003. Expression of calcium‐binding proteins in the mouse claustrum. J Chem Neuroanat 25:151–160. [DOI] [PubMed] [Google Scholar]
- Real MA, Davila JC, Guirado S. 2006. Immunohistochemical localization of the vesicular glutamate transporter VGLUT2 in the developing and adult mouse claustrum. J Chem Neuroanat 31:169–177. [DOI] [PubMed] [Google Scholar]
- Reep RL, Winans SS. 1982. Efferent connections of dorsal and ventral agranular insular cortex in the hamster, Mesocricetus auratus . Neuroscience 7:2609–2635. [DOI] [PubMed] [Google Scholar]
- Remedios R, Logothetis NK, Kayser C. 2010. Unimodal responses prevail within the multisensory claustrum. J Neurosci 30:12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remedios R, Logothetis NK, Kayser C. 2014. A role of the claustrum in auditory scene analysis by reflecting sensory change. Front Syst Neurosci 8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reser DH, Richardson KE, Montibeller MO, Zhao S, Chan JM, Soares JG, Chaplin TA, Gattass R, Rosa MG. 2014. Claustrum projections to prefrontal cortex in the capuchin monkey (Cebus apella). Front Syst Neurosci 8:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz AO. 1960. Vias de degeneracion a partir de estereotaxis del claustrum cerebral (analisis en el gato). Ann Anat 9:283–307. [Google Scholar]
- Sadowski M, Morys J, Jakubowska‐Sadowska K, Narkiewicz O. 1997. Rat's claustrum shows two main cortico‐related zones. Brain Res 756:147–152. [DOI] [PubMed] [Google Scholar]
- Salerno MT, Cortimiglia R, Crescimanno G. 1984. [Effects of stimulation of the claustrum on the spontaneous activity of pyramidal neurons of the contralateral motor areas 4 and 6 in the cat]. Boll Soc Ital Biol Sper 60:1975–1981. [PubMed] [Google Scholar]
- Segundo JP, Machne . 1956. Unitary responses to afferent volleys in lenticular nucleus and claustrum. J Neurophysiol 19:325–339. [DOI] [PubMed] [Google Scholar]
- Sherk H. 1986. The claustrum and the cerebral cortex In: Jones EG, Peters A, editors. cerebral cortex, vol 5, sensorimotor areas and aspects of cortical connectivity. New York: Plenum Press. [Google Scholar]
- Sherk H, LeVay S. 1981. The visual claustrum of the cat. III. Receptive field properties. J Neurosci 1:993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloniewski P, Usunoff KG, Pilgrim C. 1986. Retrograde transport of fluorescent tracers reveals extensive ipsi‐ and contralateral claustrocortical connections in the rat. J Comp Neurol 246:467–477. [DOI] [PubMed] [Google Scholar]
- Smith JB, Alloway KD. 2010. Functional specificity of claustrum connections in the rat: interhemispheric communication between specific parts of motor cortex. J Neurosci 30:16832–16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Alloway KD. 2014. Interhemispheric claustral circuits coordinate sensory and motor cortical areas that regulate exploratory behaviors. Front Syst Neurosci 8:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies J, Edelstein L, Ramachandran V. 2012. Hypotheses relating to the function of the claustrum. Front Integr Neurosci 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies J, Edelstein L, Ramachandran V. 2014. Hypotheses relating to the function of the claustrum II: does the claustrum use frequency codes? Front Integr Neurosci 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I, Hassmannova Y, Albe‐Fessard D. 1970. A macrophysiological study of functional organization of the claustrum. Exp Neurol 29:31–51. [DOI] [PubMed] [Google Scholar]
- Sperner J, Sander B, Lau S, Krude H, Scheffner D. 1996. Severe transitory encephalopathy with reversible lesions of the claustrum. Pediatr Radiol 26:769–771. [DOI] [PubMed] [Google Scholar]
- Squatrito S, Battaglini PP, Galletti C, Riva Sanseverino E. 1980. Projections from the visual cortex to the contralateral claustrum of the cat revealed by an anterograde axonal transport method. Neurosci Lett 19:271–275. [DOI] [PubMed] [Google Scholar]
- Stegeman S, Jolly LA, Premarathne S, Gecz J, Richards LJ, Mackay‐Sim A, Wood SA. 2013. Loss of Usp9x disrupts cortical architecture, hippocampal development and TGFbeta‐mediated axonogenesis. PLoS One 8:e68287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman AA, Krsnik Z, Sun J, Rasin MR, State MW, Sestan N, Louvi A. 2009. Developmentally regulated and evolutionarily conserved expression of SLITRK1 in brain circuits implicated in Tourette syndrome. J Comp Neurol 513:21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanne‐Gariepy J, Boussaoud D, Rouiller EM. 2002. Projections of the claustrum to the primary motor, premotor, and prefrontal cortices in the macaque monkey. J Comp Neurol 454:140–157. [DOI] [PubMed] [Google Scholar]
- Torgerson CM, Irimia A, Goh SY, Van Horn JD. 2015. The DTI connectivity of the human claustrum. Hum Brain Mapp 36:827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Groen T, Wyss JM. 1990a. The connections of presubiculum and parasubiculum in the rat. Brain Res 518:227–243. [DOI] [PubMed] [Google Scholar]
- van Groen T, Wyss JM. 1990b. The postsubicular cortex in the rat: characterization of the fourth region of the subicular cortex and its connections. Brain Res 529:165–177. [DOI] [PubMed] [Google Scholar]
- Wang Q, Burkhalter A. 2007. Area map of mouse visual cortex. J Comp Neurol 502:339–357. [DOI] [PubMed] [Google Scholar]
- Wang Q, Henry AM, Harris JA, Oh SW, Joines KM, Nyhus J, Hirokawa KE, Dee N, Mortrud M, Parry S, Ouellette B, Caldejon S, Bernard A, Jones AR, Zeng H, Hohmann JG. 2014. Systematic comparison of adeno‐associated virus and biotinylated dextran amine reveals equivalent sensitivity between tracers and novel projection targets in the mouse brain. J Comp Neurol 522:1989–2012. [DOI] [PubMed] [Google Scholar]
- Wang TW, Stromberg GP, Whitney JT, Brower NW, Klymkowsky MW, Parent JM. 2006. Sox3 expression identifies neural progenitors in persistent neonatal and adult mouse forebrain germinative zones. J Comp Neurol 497:88–100. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Ohsawa S, Ichinohe N, Rockland KS, Yamamori T. 2014. Characterization of claustral neurons by comparative gene expression profiling and dye‐injection analyses. Front Syst Neurosci 8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet‐Bongaarts AL, Smith KA, Ebbert AJ, Swanson B, Kuan L, Page DT, Overly CC, Lein ES, Hawrylycz MJ, Hof PR, Hyde TM, Kleinman JE, Jones AR. 2012. Large‐scale cellular‐resolution gene profiling in human neocortex reveals species‐specific molecular signatures. Cell 149:483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xu M, Kamigaki T, Hoang Do JP, Chang WC, Jenvay S, Miyamichi K, Luo L, Dan Y. 2014. Selective attention. Long‐range and local circuits for top‐down modulation of visual cortex processing. Science 345:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang , Hannesson DK, Saucier DM, Wallace AE, Howland J, Corcoran ME. 2001. Susceptibility to kindling and neuronal connections of the anterior claustrum. J Neurosci 21:3674–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg B, Hintiryan H, Gou L, Song MY, Bay M, Bienkowski MS, Foster NN, Yamashita S, Bowman I, Toga AW, Dong HW. 2014. Neural networks of the mouse neocortex. Cell 156:1096–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All connectivity, ISH, and reference data for the Allen Mouse Brain Connectivity Atlas are available through the data portal (http://mouse.brain-map.org).


