Abstract
Peptide‐based hydrogels have attracted significant interest in recent years as these soft, highly hydrated materials can be engineered to mimic the cell niche with significant potential applications in the biomedical field. Their potential use in vivo in particular is dependent on their biocompatibility, including their potential to cause an inflammatory response. In this work, we investigated in vitro the inflammatory potential of a β‐sheet forming peptide (FEFEFKFK; F: phenylalanine, E: glutamic acid; K: lysine) hydrogel by encapsulating murine monocytes within it (3D culture) and using the production of cytokines, IL‐β, IL‐6 and TNFα, as markers of inflammatory response. No statistically significant release of cytokines in our test sample (media + gel + cells) was observed after 48 or 72 h of culture showing that our hydrogels do not incite a pro‐inflammatory response in vitro. These results show the potential biocompatibility of these hydrogels and therefore their potential for in vivo use. The work also highlighted the difference in monocyte behaviour, proliferation and morphology changes when cultured in 2D vs. 3D. © 2016 The Authors Journal of Peptide Science published by European Peptide Society and John Wiley & Sons Ltd.
Keywords: peptide, hydrogel, monocytes, cell culture, inflammatory response
Introduction
In the past two decades, significant efforts have been made to develop novel biomaterials for a variety of biomedical applications such as regenerative medicine and cell therapy. One such class of material, which has attracted significant interest, is hydrogel as this soft, highly hydrated material can be engineered to mimic the cell niche to promote in vitro and in vivo tissue regeneration 1, 2, 3, 4, 5 or can be used as drug and cell in vivo delivery platforms. 6, 7, 8, 9 A variety of approaches can be used to design hydrogels, one such approach is self‐assembly of small molecules (low molecular weight hydrogelators – LMWH). One such class of LMWH are short synthetic peptides, which are of significant interest as they can be synthesised using standard chemical routes and therefore be obtained with high definition and high purity. In addition, being built out of natural amino acids, they can be designed to be biocompatible and biodegradable and can be metabolised by the body.10, 11, 12
A number of molecular designs have been developed for the synthesis of self‐assembling peptide LMWHs with the four main families being amphiphilic peptides, 13, 14, 15 short peptide derivatives, 16, 17, 18, 19, 20 α‐helix/coil‐coil peptides 21, 22 and β‐sheet peptides. 4, 23, 24, 25, 26, 27, 28, 29 β‐sheet peptides are of particular interest as they allow the fabrication of very stable hydrogels with properties that can be tailored through peptide design, media properties and processing. We have recently investigated the self‐assembly and gelation properties of a family of β‐sheet peptides 30, 31, 32, 33, 34 based on the design developed by Zhang and co‐workers. 23, 24, 25, 35 This design, which is based on the alternation of hydrophilic and hydrophobic residues, allows the synthesis of peptides that self‐assemble into anti‐parallel β‐sheet rich fibres. Above a critical gelation concentration, these fibres entangle and/or associate to form three‐dimensional networks that have the ability to trap water, i.e.: form hydrogels (Figure 1).
Figure 1.
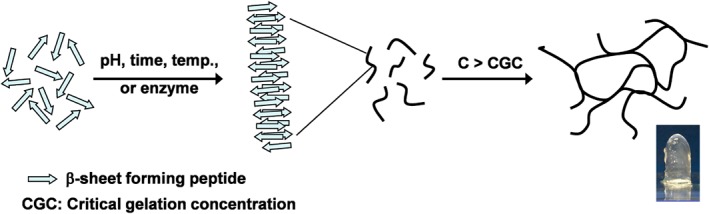
Schematic representation of the self‐assembly and gelation process of β‐sheet forming peptides.
Biomaterial biocompatibility covers a variety of aspects including toxicity and biodegradability as well as inflammatory potential. In this work, we decided to focus on the investigation in vitro of the inflammatory properties of our hydrogel using monocytes. Monocytes are inflammatory cells that circulate within the bloodstream. In response to injury events, such as the implantation of a biomaterial, they migrate into the tissue/biomaterial and become macrophages. There are four subtypes of macrophages: M1, which have a pro‐inflammatory function; M2a, which have an anti‐inflammatory function; M2b, which are involved in immune‐regulation and M2c, which are responsible for tissue remodelling. 36 The presence of M1 promotes the necessary inflammatory response to protect the body initially after surgical implantation. 37, 38, 39 However, a prolonged M1 response ultimately leads to failure of the biomaterial to integrate. 40, 41 M1 macrophages respond to the pro‐inflammatory cytokine interferon‐γ and lipopolysaccharides (LPS), an integral part of gram‐negative bacterial membranes. In the presence of these signals, macrophages produce interlukin‐1β (IL‐1β), 6 (IL‐6), 12, 15, 18 and 23; tumour necrosis factor‐α (TNFα); chemokine ligand 15, 20 and α‐chemokine ligands 9, 10, 11 and 13. Monocytes can therefore be used as a marker of inflammatory response in vitro. Haines‐Butterick et al. tested the inflammatory properties of a 20 amino acid peptide hydrogel in 2D by seeding murine monocytes on the surface of the hydrogels and quantifying the production of TNFα. 42 They found minimal amounts of TNFα produced in response to their hydrogel scaffold. In our study, we decided to look at cytokine production after 3D encapsulation of monocytes in the hydrogel as this configuration is more representative of the in vivo situation, where the monocytes invade biomaterials. In our case, the production of three cytokines IL‐1β, IL‐6 and TNFα were quantified.
Materials and Methods
Monocyte Culture
Frozen murine monocytes (J774.2 Cell Line, Sigma–Aldrich, Dorset, UK) were cultured in DMEM (D6546, Sigma–Aldrich, Dorset, UK) containing 2 mM L‐glutamine (GE Healthcare Life Sciences, Buckinghamshire, UK) and 10% fetal bovine serum (FBS) (GE Healthcare Life Sciences, Buckinghamshire, UK). All cells were grown in an atmosphere of 5% CO2/95% air at 37 °C. Cells were dislodged into the media using a cell scraper prior to passaging. Cells were used at passage 5 in all experiments.
Hydrogel Preparation
Hydrogels were prepared by dissolving ultraviolet sterilised FEFEFKFK peptide (Biomatik, Ontario, Canada; Purity 95% TFA salt) to a concentration of 20 mg ml−1 using a 30 : 70 mixture of sterile phosphate buffered saline solution (GE Healthcare Life Sciences, Buckinghamshire, UK) and sterile distilled water. After vortexing and overnight incubation at 80 °C, the peptide solution (pH 2–3) formed a hydrogel upon cooling at room temperature.
Cell Encapsulation
To form the hydrogel within a ThinCert well, insert with 1.0‐µm pore size (Greiner Bio‐One Ltd, Gloucestershire, UK), 0.5 ml of hydrogel, prepared as described previously, was pipetted into the insert and placed within a 12‐well plate (Greiner Bio‐One Ltd, Gloucestershire, UK). The hydrogel pH was adjusted to 7 by adding 50 µl of a 0.5 M NaOH solution to the top of the hydrogel drop by drop and then gently mixing the NaOH in with the pipette tip (shear modulus of hydrogel after pH adjustment ~10 kPa.2). Cells were added to the top of the gel in 100 µl of cell suspension and mixed into the gel using the tip of the pipette to give a final encapsulated cell concentration of 2 × 105 cells ml−1. Finally, 1 ml of complete media was added to the well plate around the insert containing the hydrogel before being placed in the incubator.
Study Design
Table 1 describes the composition of the samples used in our study. The third negative control (NC3) was prepared as described previously, but media alone was added to the gel compared with media with cells in the test sample (TEST). For the positive controls, 100 µl of a 20 µg ml−1 LPS (Sigma–Aldrich) solution was mixed with the cells prior to plating the cells (PC1) or encapsulating the cells in the hydrogel (PC2). At 48 and 72 h samples' supernatant was removed and frozen at −80 °C for further analysis. The supernatant that was removed was replaced with an equivalent amount of complete media.
Table 1.
Sample composition
| Sample | Abbreviation | Cell number | Gel volume (ml) | Amount of LPS added (µg) | Volume of media added (ml) |
|---|---|---|---|---|---|
| Media only Negative control | NC1 | No cells | No gel | 0 | 1 |
| Media + cell Negative control | NC2 | 2 × 105 a | No gel | 0 | 1 |
| Media + gel Negative control | NC3 | No cells | 0.5 | 0 | 1 |
| Media + cell + LPS Positive control | PC1 | 2 × 105 a | No gel | 2 | 1 |
| Media + gel + LPS Positive control | PC2 | 2 × 105 b | 0.5 | 2 | 1 |
| Media + gel + cells Test sample | TEST | 2 × 105 b | 0.5 | 0 | 1 |
per 0.5 ml of hydrogel;
added to well surface.
Detection of IL‐1β, TNFα and IL6 by Enzyme‐linked Immunoabsorbent Assay
Samples were tested for the presence of pro‐inflammatory cytokines with enzyme‐linked immunoabsorbent assay (ELISA). All ELISA components were purchased from R&D systems (Abingdon, UK). Details are collated in Table 2. A seven‐point standard curve was generated for each cytokine tested using the supplied recombinant mouse cytokine in twofold serial dilutions and a standard blank of phosphate buffered saline.
Table 2.
Specification of ELISA tests used
| Cytokine | Capture antibody | Detection antibody | High standard (pg ml−1) |
|---|---|---|---|
| IL‐1β | Rat anti‐mouse IL‐1β | Biotinylated goat anti‐mouse IL‐1β | 1 000 |
| TNFα | Goat anti‐mouse TNFα | Biotinylated goat anti‐mouse TNFα | 2 000 |
| IL6 | Rat anti‐mouse IL‐6 | Biotinylated goat anti‐mouse IL‐6 | 1 000 |
Statistical Analysis
A one‐way analysis of variance to determine statistical significance with a 95% (p < 0.05) and 99.5% (p < 0.005) confidence interval and post hoc Tukey tests were performed using ezANOVA software.
Results and Discussion
In Figure 2, the experimental design used for this study is presented. In order to confirm the validity of the approach, a set of two negative (media and media + cells) and one positive (media + cells + LPS) controls with no hydrogels were prepared. These samples allowed us to confirm that the monocytes did respond to the presence of LPS. For the testing of the hydrogel in addition to the test sample (gel + media + cells), a negative (gel + media) control and a positive (gel + media + cells + LPS) control were prepared.
Figure 2.
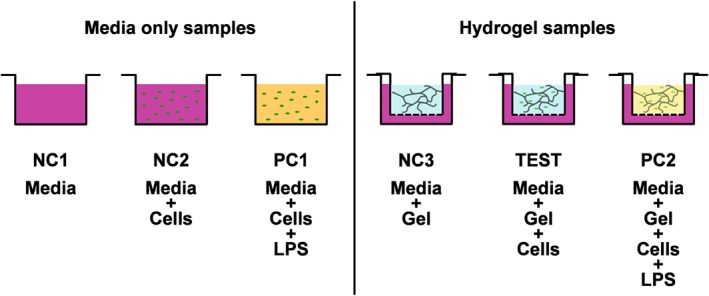
Experimental design. NC1: negative control 1 (media); NC2: negative control 2 (media + cells); PC1: positive control 1 (media + cells + LPS); NC3: negative control 3 (gel + media); TEST: test sample (gel + media + cells); PC2: positive control 2 (gel + media + cells + LPS).
When dealing with hydrogels, ingress and egress via diffusion of large molecules such as LPS and proteins needs to be carefully considered. Diffusion of large molecules will depend on the hydrogel network properties (e.g. mesh size and charge) and the molecules' properties (e.g. aggregation propensity and hydrodynamic radius). In previous work, we have shown that the mesh size of our hydrogel is in the range of 20–30 nm.31 LPS are amphipathic molecules (~10–20 kDa), which in solution aggregate to form large structures (spherical and/or cylindrical micelles) with large hydrodynamic radii (~60–100 nm). 43, 44 On the other hand, interleukins and TNFα are found in solution in mono, di and/or trimeric states (~10–50 kDa) 45 and adopt compact conformations resulting in small hydrodynamic radii (~2–10 nm). We therefore expected LPS to be unable to diffuse into the hydrogels, while interleukins and TNFα were expected to be able to diffuse out. Preliminary tests indeed confirmed these assumptions. Neither interleukins nor TNFα were detected when LPS was simply added to the surrounding media of a hydrogel with monocytes encapsulated suggesting that the monocytes did not come into contact with LPS. On the other hand, when LPS was added to the cell solution prior to encapsulation into the hydrogels, interleukins and TNFα were detected in the surrounding media from 48 h onwards showing that the interleukins and TNFα were indeed able to diffuse out of the hydrogels. As a consequence of these findings, LPS was mixed into the hydrogels with the cells for the positive control sample, PC2. The production of interleukins and TNFα was measured by collecting the surrounding media and subsequently using ELISA as described in the Materials and Methods section. The detailed diffusive properties of these hydrogels have been the subject of a number of articles from our previous work8, 34 and work of other groups6, 46 and are beyond the scope of this article.
In Figure 3, light microscopy images obtained immediately after seeding the monocytes and after 48 and 72 h of cell culture are presented. As can be seen for the samples prepared without gel (2D culture), significant differences were observed between the negative control, NC2 (cell + media), and the positive control, PC1 (cell + media + LPS). For NC2, the monocytes were seen to proliferate rapidly, and after 48 h, they were already confluent, covering the full surface of the culture well. When LPS was added (PC1), the number of monocytes visibly decreased after 48 and 72 h of culture, and their morphology changed. Cells appeared larger and more extended suggesting that the monocytes responded to the LPS and differentiated into macrophages. When the monocytes were seeded in the hydrogel, the difference in morphology between the sample with LPS (PC2) and without LPS (TEST) was less obvious. Close inspection of the images seems to suggest a slight decrease in monocyte number, but no significant change in cell morphology was observed. As will be discussed next, for PC1 and PC2, a significant increase in cytokine level was detected suggesting that the monocytes were activated by the LPS in PC2 sample too. The difference observed in cell morphologies between 2D and 3D culture is not unexpected, indeed significant work in the literature has shown that cell behaviour and morphology can differ substantially when cultured in 2D or 3D. In our case, although the monocytes responded to the supplemented LPS even in the hydrogel, as will be shown next, there was no significant change in cell morphology probably because of the cells being encapsulated within a 3D matrix.
Figure 3.
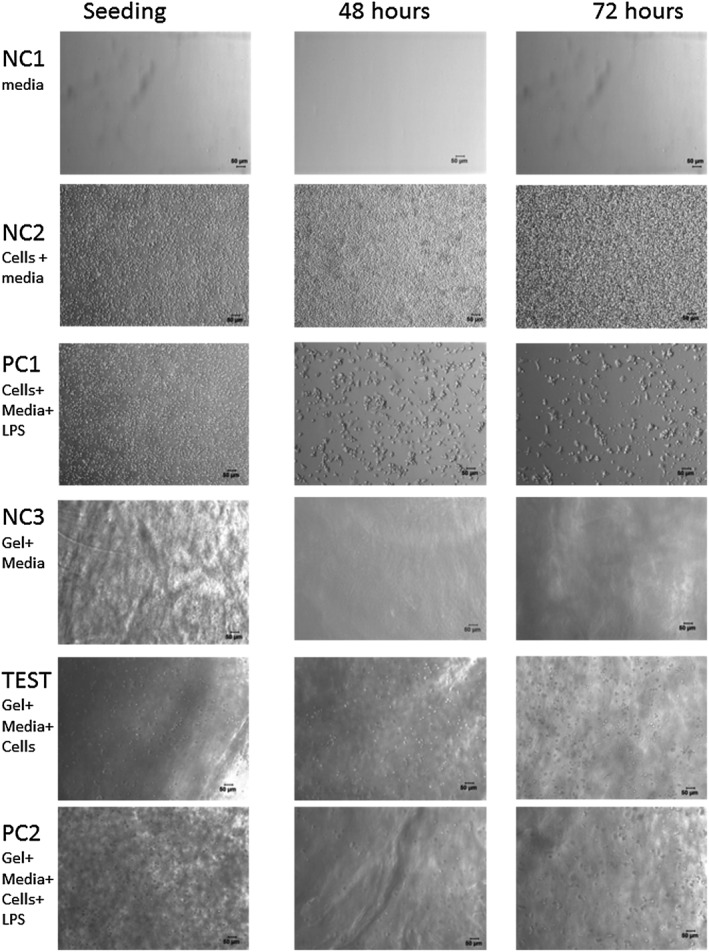
Light microscope images of samples at the time of seeding and after 48 and 72 h of culture. Scale bar represents 50 µm.
In Figure 4, the amount of IL‐1β, TNFα and IL‐6 detected for each sample after 48 and 72 h of culture are shown. For IL‐6 and TNFα, significant and statistically meaningful levels of cytokines were detected only for the positive controls, PC1 and PC2. For IL‐1β, no statistical difference in interleukin produced between NC1 and NC3, and the TEST sample were observed. As NC1 and NC3 did not contain cells, this suggests that no statistically significant production of IL‐1β was detected in the TEST sample. On the other hand, a statistically significant amount of IL‐1β was detected in the two positive controls, PC1 and PC2. In this case though, statistically significant amounts of IL‐1β were detected in the negative control NC2, which contained cells. The increase of detectable IL‐1β in this sample can be explained by the rapid growth of these cells in a limited space, as shown by the light microscopy images, causing cell death and release of IL‐1β. This effect has been noted in another study where a lower seeding density was used. 42 Similar results to those seen for IL‐1β were observed for IL‐6 and TNFα. There was significantly more cytokine production in the two positive control samples, PC1 and PC2. There was no significant cytokine production detected in any of the negative controls; NC1, NC2 or NC3, or TEST samples. The lack of cytokines detected when the monocytes were encapsulated within our hydrogels suggests that they do not incite a pro‐inflammatory response in vitro making them a good candidate for use in vivo.
Figure 4.
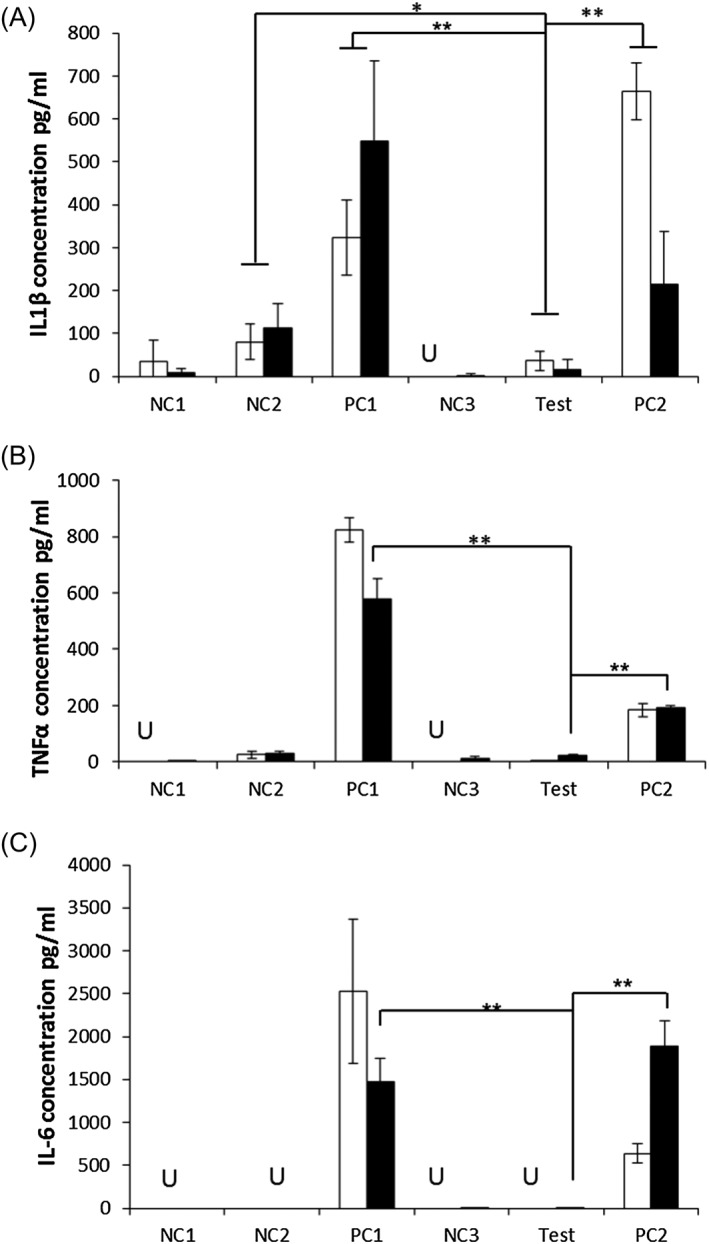
Amount of (A) IL‐1β, (B) TNFα, (C) IL‐6 detected in the different samples (see Figure 2 for details) in response to octapeptide hydrogel at 48 (white bars) and 72 h (black bars). Data points are mean of 6 replicates. Error bars represent standard deviation. U = undetectable; * p < 0.05; **p < 0.005.
Conclusions
We investigated in vitro the inflammatory potential of our peptide hydrogels by encapsulating murine monocytes within them (3D culture) and using the production of cytokines, IL‐β, IL‐6 and TNFα, as markers of inflammatory response. No statistically significant release of cytokines in our test sample was observed after 48 or 72 h of culture showing that our hydrogels do not incite a pro‐inflammatory response in vitro. These results show the potential biocompatibility of these hydrogels and therefore their potential for use in vivo. Our work also showed that 2D and 3D culture of monocytes in the presence or absence of LPS yielded very different cell behaviour. In the absence of LPS, the monocytes were found to proliferate rapidly in 2D while in 3D limited proliferation was observed. In both cases, the cell morphology remained unchanged. When LPS was added in 2D, limited cell proliferation and significant changes in cell morphology were observed, while in 3D, no significant changes in cell behaviour could be observed; although in both cases, an increase in cytokine production was observed confirming that in both cases, the monocytes were responding to the presence of LPS.
Acknowledgements
A. S., A. F. M. and V. L. W. are grateful to the EPSRC (fellowship no. EP/K016210/1) and the EU FP7 NMP programme (BIOSCENT) for financial support. All research data supporting this publication are directly available within this publication.
Markey, A. , Workman, V. L. , Bruce, I. A. , Woolford, T. J. , Derby, B. , Miller, A. F. , Cartmell, S. H. , and Saiani, A. (2017) Peptide hydrogel in vitro non‐inflammatory potential. J. Pept. Sci., 23: 148–154. doi: 10.1002/psc.2940.
References
- 1. Mujeeb A, Miller AF, Saiani A, Gough JE. Self‐assembled octapeptide scaffolds for in vitro chondrocyte culture. Acta Biomater. 2013; 9(1): 4609–4617. [DOI] [PubMed] [Google Scholar]
- 2. Castillo Diaz LA, Saiani A, Gough JE, Miller AF. Human osteoblasts within soft peptide hydrogels promote mineralisation in vitro. Journal of Tissue Engineering 2014; 5 2041731414539344: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Worthington P, Pochan DJ, Langhans SA. Peptide hydrogels–versatile matrices for 3D cell culture in cancer medicine. Front. Oncol. 2015; 5: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gelain F, Horii A, Zhang SG. Designer self‐assembling peptide scaffolds for 3‐D tissue cell cultures and regenerative medicine. Macromol. Biosci. 2007; 7(5): 544–551. [DOI] [PubMed] [Google Scholar]
- 5. Saracino GAA, Cigognini D, Silva D, Caprini A, Gelain F. Nanomaterials design and tests for neural tissue engineering. Chem. Soc. Rev. 2013; 42(1): 225–262. [DOI] [PubMed] [Google Scholar]
- 6. Altunbas A, Pochan DJ. Peptide‐based and polypeptide‐based hydrogels for drug delivery and tissue engineering. In Peptide‐Based Materials 2012; 310: 135–167. [DOI] [PubMed] [Google Scholar]
- 7. Leite DM, Barbu E, Pilkington GJ, Lalatsa A. Peptide self‐assemblies for drug delivery. Curr. Top. Med. Chem. 2015; 15(22): 2277–2289. [DOI] [PubMed] [Google Scholar]
- 8. Tang C, Miller AF, Saiani A. Peptide hydrogels as mucoadhesives for local drug delivery. Int. J. Pharm. 2014; 465: 427–435. [DOI] [PubMed] [Google Scholar]
- 9. Yan C, Mackay ME, Czymmek K, Nagarkar RP, Schneider JP, Pochan DJ. Injectable solid peptide hydrogel as a cell carrier: effects of shear flow on hydrogels and cell payload. Langmuir 2012; 28(14): 6076–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Du X, Zhou J, Shi J, Xu B. Supramolecular hydrogelators and hydrogels: from soft matter to molecular biomaterials. Chem. Rev. 2015; 115(24): 13165–13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fichman G, Gazit E. Self‐assembly of short peptides to form hydrogels: design of building blocks, physical properties and technological applications. Acta Biomater. 2014; 10(4): 1671–1682. [DOI] [PubMed] [Google Scholar]
- 12. Acar H, Srivastava S, Chung EJ, Schnorenberg MR, Barrett JC, LaBelle JL, Tirrell M. Self‐assembling peptide‐based building blocks in medical applications. Adv. Drug Deliv. Rev. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beniash E, Hartgerink JD, Storrie H, Stendahl JC, Stupp SI. Self‐assembling peptide amphiphile nanofiber matrices for cell entrapment. Acta Biomater. 2005; 1(4): 387–397. [DOI] [PubMed] [Google Scholar]
- 14. Hartgerink JD, Beniash E, Stupp SI. Peptide‐amphiphile nanofibers: a versatile scaffold for the preparation of self‐assembling materials. Proc. Natl. Acad. Sci. U. S. A. 2002; 99(8): 5133–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matson JB, Stupp SI. Self‐assembling peptide scaffolds for regenerative medicine. Chem. Commun. 2012; 48(1): 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jayawarna V, Ali M, Jowitt TA, Miller AE, Saiani A, Gough JE, Ulijn RV. Nanostructured hydrogels for three‐dimensional cell culture through self‐assembly of fluorenylmethoxycarbonyl‐dipeptides. Adv. Mater. 2006; 18(5): 611–614. [Google Scholar]
- 17. Jayawarna V, Richardson SM, Hirst AR, Hodson NW, Saiani A, Gough JE, Ulijn RV. Introducing chemical functionality in Fmoc‐peptide gels for cell culture. Acta Biomater. 2009; 5(3): 934–943. [DOI] [PubMed] [Google Scholar]
- 18. Tang C, Smith AM, Collins RF, Ulijn RV, Saiani A. Fmoc‐diphenylalanine self‐assembly mechanism induces apparent pK(a) shifts. Langmuir 2009; 25(16): 9447–9453. [DOI] [PubMed] [Google Scholar]
- 19. Cardoso AZ, Alvarez Alvarez AE, Cattoz BN, Griffiths PC, King SM, Frith WJ, Adams DJ. The influence of the kinetics of self‐assembly on the properties of dipeptide hydrogels. Faraday Discuss. 2013; 166: 101–116. [DOI] [PubMed] [Google Scholar]
- 20. Cheng G, Castelletto V, Moulton CM, Newby GE, Hamley IW. Hydrogelation and self‐assembly of Fmoc‐tripeptides: unexpected influence of sequence on self‐assembled fibril structure, and hydrogel modulus and anisotropy. Langmuir 2010; 26(7): 4990–4998. [DOI] [PubMed] [Google Scholar]
- 21. Banwell EF, Abelardo ES, Adams DJ, Birchall MA, Corrigan A, Donald AM, Kirkland M, Serpell LC, Butler MF, Woolfson DN. Rational design and application of responsive alpha‐helical peptide hydrogels. Nat. Mater. 2009; 8(7): 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walshaw J, Woolfson DN. Open‐and‐shut cases in coiled‐coil assembly: alpha‐sheets and alpha‐cylinders. Protein Sci. 2001; 10(3): 668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caplan MR, Moore PN, Zhang SG, Kamm RD, Lauffenburger DA. Self‐assembly of a beta‐sheet protein governed by relief of electrostatic repulsion relative to van der Waals attraction. Biomacromolecules 2000; 1(4): 627–631. [DOI] [PubMed] [Google Scholar]
- 24. Caplan MR, Schwartzfarb EM, Zhang SG, Kamm RD, Lauffenburger DA. Effects of systematic variation of amino acid sequence on the mechanical properties of a self‐assembling, oligopeptide biomaterial. Journal of Biomaterials Science‐Polymer Edition 2002; 13(3): 225–236. [DOI] [PubMed] [Google Scholar]
- 25. Caplan MR, Schwartzfarb EM, Zhang SG, Kamm RD, Lauffenburger DA. Control of self‐assembling oligopeptide matrix formation through systematic variation of amino acid sequence. Biomaterials 2002; 23(1): 219–227. [DOI] [PubMed] [Google Scholar]
- 26. Leon EJ, Verma N, Zhang SG, Lauffenburger DA, Kamm RD. Mechanical properties of a self‐assembling oligopeptide matrix. Journal of Biomaterials Science‐Polymer Edition 1998; 9(3): 297–312. [DOI] [PubMed] [Google Scholar]
- 27. Cui HG, Krikorian V, Thompson J, Nowak AP, Deming TJ, Pochan DJ. Preparation and characterization of synthetic polypeptide single crystals with controlled thickness. Macromolecules 2005; 38(17): 7371–7377. [Google Scholar]
- 28. Ozbas B, Rajagopal K, Schneider JP, Pochan DJ. Semiflexible chain networks formed via self‐assembly of beta‐hairpin molecules. Phys. Rev. Lett. 2004; 93(26): DOI: 10.1103/PhysRevLett.93.268106. [DOI] [PubMed] [Google Scholar]
- 29. Yucel T, Micklitsch CM, Schneider JP, Pochan DJ. Direct observation of early‐time hydrogelation in beta‐hairpin peptide self‐assembly. Macromolecules 2008; 41(15): 5763–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boothroyd S, Saiani A, Miller AF. Formation of mixed ionic complementary peptide fibrils. Macromolecular Symposia 2008; 273: 139–145. [Google Scholar]
- 31. Saiani A, Mohammed A, Frielinghaus H, Collins R, Hodson N, Kielty CM, Sherratt MJ, Miller AF. Self‐assembly and gelation properties of alpha‐helix versus beta‐sheet forming peptides. Soft Matter 2009; 5(1): 193–202. [Google Scholar]
- 32. Boothroyd S, Miller AF, Saiani A. From fibres to networks using self‐assembling peptides. Faraday Discuss. 2013; 166: 195–207. [DOI] [PubMed] [Google Scholar]
- 33. Boothroyd S, Saiani A, Miller AF. Controlling network topology and mechanical properties of co‐assembling peptide hydrogels. Biopolymers 2014; 101(6): 669–680. [DOI] [PubMed] [Google Scholar]
- 34. Roberts D, Rochas C, Saiani A, Miller AF. Effect of peptide and guest charge on the structural, mechanical and release properties of beta‐sheet forming peptides. Langmuir 2012; 28(46): 16196–16206. [DOI] [PubMed] [Google Scholar]
- 35. Hong YS, Legge RL, Zhang S, Chen P. Effect of amino acid sequence and pH on nanofiber formation of self‐assembling peptides EAK16‐II and EAK16‐IV. Biomacromolecules 2003; 4(5): 1433–1442. [DOI] [PubMed] [Google Scholar]
- 36. Sridharan R, Cameron AR, Kelly DJ, Kearney CJ, O'Brien FJ. Biomaterial based modulation of macrophage polarization: a review and suggested design principles. Materials Today 2015; 18(6): 313–325. [Google Scholar]
- 37. Anderson JM, Rodriguez A, Chang DT. Foreign body reaction to biomaterials. Semin. Immunol. 2008; 20(2): 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Franz S, Rammelt S, Scharnweber D, Simon JC. Immune responses to implants – a review of the implications for the design of immunomodulatory biomaterials. Biomaterials 2011; 32(28): 6692–709. [DOI] [PubMed] [Google Scholar]
- 39. Gardner AB, Lee SK, Woods EC, Acharya AP. Biomaterials‐based modulation of the immune system. Biomed. Res. Int. 2013; 2013: 732182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004; 25(12): 677–86. [DOI] [PubMed] [Google Scholar]
- 41. Rees AJ. Monocyte and macrophage biology: an overview. Semin. Nephrol. 2010; 30(3): 216–33. [DOI] [PubMed] [Google Scholar]
- 42. Haines‐Butterick LA, Salick DA, Pochan DJ, Schneider JP. In vitro assessment of the pro‐inflammatory potential of beta‐hairpin peptide hydrogels. Biomaterials 2008; 29(31): 4164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hayter JB, Rivera M, McGroarty EJ. Neutron scattering analysis of bacterial lipopolysaccharide phase structure. Changes at high pH. J. Biol. Chem. 1987; 262(11): 5100–5. [PubMed] [Google Scholar]
- 44. Santos NC, Silva AC, Castanho MA, Martins‐Silva J, Saldanha C. Evaluation of lipopolysaccharide aggregation by light scattering spectroscopy. Chembiochem 2003; 4(1): 96–100. [DOI] [PubMed] [Google Scholar]
- 45. Lewit‐Bentley A, Fourme R, Kahn R, Prange T, Vachette P, Tavernier J, Hauquier G, Niers W. Structure of tumour necrosis factor by X‐ray solution scattering and preliminary studies by single crystal X‐ray diffraction. J. Mol. Biol. 1988; 199(2): 389–92. [DOI] [PubMed] [Google Scholar]
- 46. Branco MC, Pochan DJ, Wagner NJ, Schneider JP. Macromolecular diffusion and release from self‐assembled beta‐hairpin peptide hydrogels. Biomaterials 2009; 30(7): 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]


