“It is not in the stars to hold our destiny but in ourselves”
- William Shakespeare
Population-based cohort studies have established a consistent link between obesity and atrial fibrillation (AF)1–3. Whether this relationship is causal, though, remains a question. Associations observed in epidemiological studies could arise due to confounding by behavioral or socioeconomic factors, or as a result of obesity’s common co-occurrence with hypertension, diabetes, and other established risk factors for AF. Although observational studies can adjust for known confounders using statistical techniques, the existence of unknown or unmeasured confounders and lack of precision in the measured variables can lead to residual confounding. Randomized controlled trials (RCTs) remain the gold standard for establishing the direct, causal effect of risk factor modification on the development of a disease related to that risk factor. However, RCTs are not always feasible due to the long follow-up time required to observe sufficient numbers of outcomes and the difficulties of maintaining complex intervention protocols. Indeed, no large-scale RCT of weight interventions for the prevention of AF have been reported to date. In this issue of Circulation, Chatterjee et al.4 use the random assignment of genetic variants – an approach termed “Mendelian randomization” 5,6 – to provide support for a causal relationship between obesity and AF.
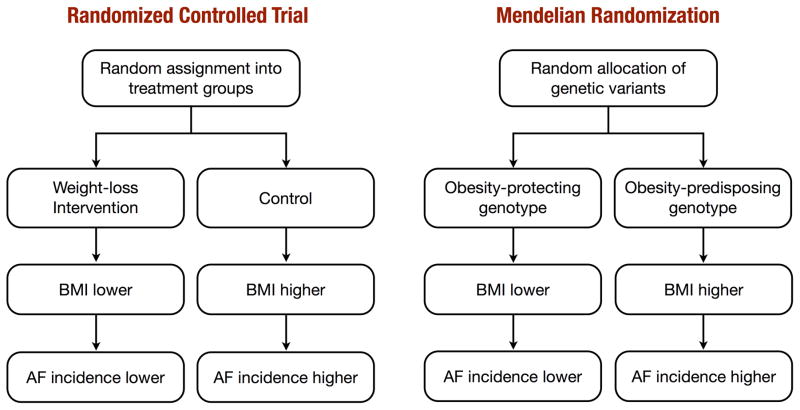
This method is founded on the principle that an individual’s genotype is determined randomly at conception from his/her parental genotypes. Furthermore, according to Mendel’s law of independent assortment, genetic variants governing variation in one trait are inherited independently of those influencing another trait. Therefore, genotype will in most cases be unrelated to behavioral, dietary, and other factors that could confound the association between obesity and AF. Unlike observational studies, where obese individuals are likely to be different from those with normal weight with respect to many confounding factors, those with and without an obesity predisposing genotype are expected to be balanced with regard to measured and unmeasured confounders. For these reasons, Mendelian randomization studies have often been likened to natural randomized trials, in which genotype plays the role of random treatment assignment (Figure 1). The fact that genotype is determined before birth and stays constant throughout life also rules out the possibility of reverse causation. Hence, a difference in the outcome (AF) between the genotype groups is taken to be evidence of the causal effect of the exposure (obesity).
Figure 1.
Comparison of a randomized controlled trial and Mendelian randomization study
To perform a successful Mendelian randomization study, one needs to find a genetic variant that can reliably predict the exposure. Recent genome-wide association studies have identified multiple single nucleotide polymorphisms (SNPs) in dozens of genetic loci that are associated with increased body-mass index (BMI) and adiposity7,8. Because each of these variants typically exerts only a small effect on BMI, to obtain a more powerful predictor, one common approach is to combine multiple variants into a single genetic risk score, whereby all obesity predisposing alleles that a person carries are added up in a weighted sum, with weights proportional to their effect sizes. Chatterjee and colleagues used two genetic predictors of BMI: a variant in the fat mass and obesity-associated (FTO) gene and a BMI gene score comprised of 39 BMI-associated SNPs identified by previous genome-wide association studies7,8. They first confirmed that both genetic instruments were associated with increased BMI. In a meta-analysis of several prospective studies including 51,646 individuals of European ancestry, they showed that each unit increase in the genetic risk score was associated with an average increase in BMI of 1.05 kg/m2. They next showed that the same one-unit increase in the genetic risk score was associated with an 11% increase in the risk of incident AF, lending support to the hypothesis that obesity is causally related to incident AF.
Although Mendelian randomization provides a powerful approach to strengthen the conclusions of observational studies, it has several limitations. First, as noted above, the validity of the method relies on the assumption that genotype is unrelated to any factors that could confound the exposure-outcome relationship. While this assumption is often untestable, empirical evidence suggests that it is plausible in many situations9. For example, a previous study10 found no association between BMI-associated variants and several traditional risk factors of cardiovascular disease. On the other hand, an earlier study showed that a closely linked FTO variant was associated with blood pressure, and concluded that obesity was causally related to hypertension11. This alone does not invalidate the use of FTO variants as genetic instruments for testing the causal relationship between BMI and AF. On the contrary, this suggests that if hypertension is a direct consequence of obesity, rather than a correlate, adjusting for it could obscure the true causal role of obesity. Similar arguments may be made for other AF risk factors such as sleep apnea. Nevertheless, adjustment for hypertension and other risk factors did not alter the associations between genotype and AF in the current study. Conversely, the association between genetic variants and AF was completely attenuated after accounting for measured BMI, further supporting the notion that the relationship was causally mediated by BMI, rather than through alternate pathways, due to pleiotropy (the effect of a particular gene on multiple risk factors) or linkage disequilibrium (statistical association between different genetic variants induced by the tendency of alleles that are close together on a chromosome to be inherited together). Second, weak genetic instruments, that explain too little variation in the exposure, could bias causal estimates or result in failure to establish causal relationships due do a lack of power. A recent article described this situation as analogous to interpreting the effects of a lipid lowering trial on coronary heart disease when the therapy studied (such as niacin or fibrates) has a weak effect on lowering LDL-cholesterol12. The large sample size and the use of a genetic score including multiple SNPs robustly associated with BMI partially alleviate this concern.
What are the implications of these findings? While the study provides strong support for the causal role of obesity in the development of AF, the estimate of causal effect size should be interpreted with caution and should not be extrapolated to other situations. Estimates from Mendelian randomization studies are likely to be different than the effects of weight-loss interventions for several reasons. First of all, Mendelian randomization estimates represent the effect of life-long differences in BMI, whereas most interventions are applied to individuals for a limited duration of time. In this respect, it is interesting to note that the causal effect of genetically determined BMI on AF (~10% increase in AF risk for a one-unit increase in genetically determined BMI) was larger in magnitude (but not statistically different) than the observational association between measured BMI and incident AF (5% increase in AF risk for a one-unit increase in measured BMI), suggesting that genotype may better reflect the effects of cumulative exposure to increased BMI compared with cross-sectional assessments. Second, causal effect estimates from Mendelian randomization studies can be thought of as a population-average effect (i.e., as if the intervention was applied to the entire population), and could be different than the effect of interventions applied to specific subgroups. For example, a recent analysis of the Swedish Obese Subjects (SOS) trial demonstrated that an average weight loss of 18% or nearly 50 pounds through bariatric surgery reduced the risk of AF by 29%13. Based on the current study, however, a similar reduction in risk should be achieved with only a ~3-point reduction in BMI, corresponding to a weight loss of 15–25 pounds (depending on one’s height). Furthermore, it is not known whether obesity-mediated risk for AF is affected by body fat distribution, which has recently been shown to be a strong determinant of cardiovascular risk, independent of BMI14, or if it might be modified by behavioral factors, such as nutrition and physical activity. Only future RCTs can establish the true magnitude of AF risk reduction resulting from weight loss in different patient groups. Nevertheless, the data provided by Chatterjee and colleagues suggest that early prevention of obesity is likely to have a greater impact in reducing the risk of AF, and should be a focus of public health interventions.
It is important to note that while Mendelian randomization studies use genetic variants as a proxy for an exposure, their main goal is not to identify genetic determinants of disease, which might be used for genetic screening, but to provide an insight about the role of modifiable (non-genetic) exposures in disease etiology5,6. As has recently been demonstrated, improving one’s environment with lifestyle behaviors such as increased physical activity and a healthy diet can decrease risk of heart disease regardless of one’s inherent genetic risk15. Whereas genetic variants can contribute to inter-individual differences in BMI, they cannot explain the expanding obesity epidemic over recent decades, as the genetic makeup of the population could not have changed over a span of two or three generations. Thus, lifestyle interventions directed at primary prevention of obesity may be the best way to reduce the growing prevalence of AF. Although our genetic predispositions for obesity are pre-determined at birth, they do not have to define our destiny for disease - it remains within ourselves to alter that destiny through a healthy lifestyle and avoidance of weight gain.
Footnotes
Conflict/Disclosure Statement: Dr. Neeland is supported by grant K23DK106520 from the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institute of Health and by the Dedman Family Scholarship in Clinical Care from UT Southwestern. Dr. Kozlitina reports no disclosures related to this paper.
References
- 1.Wang TJ, Parise H, Levy D, D’Agostino RB, Sr, Wolf PA, Vasan RS, Benjamin EJ. Obesity and the risk of new-onset atrial fibrillation. JAMA. 2004;292:2471–2477. doi: 10.1001/jama.292.20.2471. [DOI] [PubMed] [Google Scholar]
- 2.Frost L, Hune LJ, Vestergaard P. Overweight and obesity as risk factors for atrial fibrillation or flutter: the Danish Diet, Cancer, and Health Study. Am J Med. 2005;118:489–495. doi: 10.1016/j.amjmed.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 3.Wanahita N, Messerli FH, Bangalore S, Gami AS, Somers VK, Steinberg JS. Atrial fibrillation and obesity - results of a meta-analysis. Am Heart J. 2008;155:310–315. doi: 10.1016/j.ahj.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee NA, Giulianini F, Geelhoed B, Lunetta KL, Misialek JR, Niemeijer MN, Rienstra M, Rose LM, Smith AV, Arking DE, Ellinor PT, Heeringa J, Lin H, Lubitz SA, Soliman EZ, Verweij N, Alonso A, Benjamin EJ, Gudnason V, Stricker BH, van der Harst P, Chasman DI, Albert CM. Genetic obesity and the risk of atrial fibrillation - causal estimates from Mendelian Randomization. Circulation. 2016 doi: 10.1161/CIRCULATIONAHA.116.024921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davey Smith G, Ebrahim S. ‘Mendelian randomisation’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 6.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 7.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Magic, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Procardis C, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berndt SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, Esko T, Fall T, Ferreira T, Gentilini D, Jackson AU, Luan J, Randall JC, Vedantam S, Willer CJ, Winkler TW, Wood AR, Workalemahu T, Hu YJ, Lee SH, Liang L, Lin DY, Min JL, Neale BM, Thorleifsson G, Yang J, Albrecht E, Amin N, Bragg-Gresham JL, Cadby G, den Heijer M, Eklund N, Fischer K, Goel A, Hottenga JJ, Huffman JE, Jarick I, Johansson Å, Johnson T, Kanoni S, Kleber ME, König IR, Kristiansson K, Kutalik Z, Lamina C, Lecoeur C, Li G, Mangino M, McArdle WL, Medina-Gomez C, Müller-Nurasyid M, Ngwa JS, Nolte IM, Paternoster L, Pechlivanis S, Perola M, Peters MJ, Preuss M, Rose LM, Shi J, Shungin D, Smith AV, Strawbridge RJ, Surakka I, Teumer A, Trip MD, Tyrer J, Van Vliet-Ostaptchouk JV, Vandenput L, Waite LL, Zhao JH, Absher D, Asselbergs FW, Atalay M, Attwood AP, Balmforth AJ, Basart H, Beilby J, Bonnycastle LL, Brambilla P, Bruinenberg M, Campbell H, Chasman DI, Chines PS, Collins FS, Connell JM, Cookson WO, de Faire U, de Vegt F, Dei M, Dimitriou M, Edkins S, Estrada K, Evans DM, Farrall M, Ferrario MM, Ferrières J, Franke L, Frau F, Gejman PV, Grallert H, Grönberg H, Gudnason V, Hall AS, Hall P, Hartikainen AL, Hayward C, Heard-Costa NL, Heath AC, Hebebrand J, Homuth G, Hu FB, Hunt SE, Hyppönen E, Iribarren C, Jacobs KB, Jansson JO, Jula A, Kähönen M, Kathiresan S, Kee F, Khaw KT, Kivimäki M, Koenig W, Kraja AT, Kumari M, Kuulasmaa K, Kuusisto J, Laitinen JH, Lakka TA, Langenberg C, Launer LJ, Lind L, Lindström J, Liu J, Liuzzi A, Lokki ML, Lorentzon M, Madden PA, Magnusson PK, Manunta P, Marek D, März W, Mateo Leach I, McKnight B, Medland SE, Mihailov E, Milani L, Montgomery GW, Mooser V, Mühleisen TW, Munroe PB, Musk AW, Narisu N, Navis G, Nicholson G, Nohr EA, Ong KK, Oostra BA, Palmer CN, Palotie A, Peden JF, Pedersen N, Peters A, Polasek O, Pouta A, Pramstaller PP, Prokopenko I, Pütter C, Radhakrishnan A, Raitakari O, Rendon A, Rivadeneira F, Rudan I, Saaristo TE, Sambrook JG, Sanders AR, Sanna S, Saramies J, Schipf S, Schreiber S, Schunkert H, Shin SY, Signorini S, Sinisalo J, Skrobek B, Soranzo N, Stančáková A, Stark K, Stephens JC, Stirrups K, Stolk RP, Stumvoll M, Swift AJ, Theodoraki EV, Thorand B, Tregouet DA, Tremoli E, Van der Klauw MM, van Meurs JB, Vermeulen SH, Viikari J, Virtamo J, Vitart V, Waeber G, Wang Z, Widén E, Wild SH, Willemsen G, Winkelmann BR, Witteman JC, Wolffenbuttel BH, Wong A, Wright AF, Zillikens MC, Amouyel P, Boehm BO, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Cupples LA, Cusi D, Dedoussis GV, Erdmann J, Eriksson JG, Franks PW, Froguel P, Gieger C, Gyllensten U, Hamsten A, Harris TB, Hengstenberg C, Hicks AA, Hingorani A, Hinney A, Hofman A, Hovingh KG, Hveem K, Illig T, Jarvelin MR, Jöckel KH, Keinanen-Kiukaanniemi SM, Kiemeney LA, Kuh D, Laakso M, Lehtimäki T, Levinson DF, Martin NG, Metspalu A, Morris AD, Nieminen MS, Njølstad I, Ohlsson C, Oldehinkel AJ, Ouwehand WH, Palmer LJ, Penninx B, Power C, Province MA, Psaty BM, Qi L, Rauramaa R, Ridker PM, Ripatti S, Salomaa V, Samani NJ, Snieder H, Sørensen TI, Spector TD, Stefansson K, Tönjes A, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Vollenweider P, Wallaschofski H, Wareham NJ, Watkins H, Wichmann HE, Wilson JF, Abecasis GR, Assimes TL, Barroso I, Boehnke M, Borecki IB, Deloukas P, Fox CS, Frayling T, Groop LC, Haritunian T, Heid IM, Hunter D, Kaplan RC, Karpe F, Moffatt MF, Mohlke KL, O’Connell JR, Pawitan Y, Schadt EE, Schlessinger D, Steinthorsdottir V, Strachan DP, Thorsteinsdottir U, van Duijn CM, Visscher PM, Di Blasio AM, Hirschhorn JN, Lindgren CM, Morris AP, Meyre D, Scherag A, McCarthy MI, Speliotes EK, North KE, Loos RJ, Ingelsson E. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey Smith G, Lawlor D, Harbord R, Timpson N, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2008;4:1985–1992. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordestgaard BG, Palmer TM, Benn M, Zacho J, Tybjaerg-Hansen A, Davey Smith G, Timpson NJ. The effect of elevated body mass index on ischemic heart disease risk: causal estimates from a Mendelian randomisation approach. PLoS Med. 2012;9:e1001212. doi: 10.1371/journal.pmed.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timpson NJ, Harbord R, Davey Smith G, Zacho J, Tybjaerg-Hansen A, Nordestgaard BG. Does greater adiposity increase blood pressure and hypertension risk? Mendelian randomisation using FTO/MC4R genotype. Hypertension. 2009;54:84–90. doi: 10.1161/HYPERTENSIONAHA.109.130005. [DOI] [PubMed] [Google Scholar]
- 12.Ference BA. Mendelian randomization studies: Nature’s randomized trials. [Accessed January 10, 2017];American College of Cardiology website. 2015 Jun 12; www.acc.org. http://www.acc.org/latest-in-cardiology/articles/2015/06/11/13/17/mendelian-randomization-studies.
- 13.Jamaly S, Carlsson L, Peltonen M, Jacobson P, Sjostrom L, Karason K. Bariatric surgery and the risk of new-onset atrial fibrillation in swedish obese subjects. J Am Coll Cardiol. 2016;68:2497–2504. doi: 10.1016/j.jacc.2016.09.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neeland IJ, Turer AT, Ayers CR, Berry JD, Rohatgi A, Das SR, Khera A, Vega GL, McGuire DK, Grundy SM, de Lemos JA. Body fat distribution and incident cardiovascular disease in obese adults. J Am Coll Cardiol. 2015;65:2150–2151. doi: 10.1016/j.jacc.2015.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, Chasman DI, Baber U, Mehran R, Rader DJ, Fuster V, Boerwinkle E, Melander O, Orho-Melander M, Ridker PM, Kathiresan S. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]