ABSTRACT
NMD is a highly conserved pathway that degrades specific subsets of RNAs. There is increasing evidence for roles of NMD in development. In this commentary, we focus on spermatogenesis, a process dramatically impeded upon loss or disruption of NMD. NMD requires strict regulation for normal spermatogenesis, as loss of a newly discovered NMD repressor, UPF3A, also causes spermatogenic defects, most prominently during meiosis. We discuss the unusual evolution of UPF3A, whose paralog, UPF3B, has the opposite biochemical function and acts in brain development. We also discuss the regulation of NMD during germ cell development, including in chromatoid bodies, which are specifically found in haploid germ cells. The ability of NMD to coordinately degrade batteries of RNAs in a regulated fashion during development is akin to the action of transcriptional pathways, yet has the advantage of driving rapid changes in gene expression.
KEYWORDS: RNA decay, NMD, spermatogenesis, neofunctionalization, subfunctionalization
NMD
NMD is a highly conserved RNA degradation pathway that operates in all eukaryotes examined so far, from yeast to humans.31 NMD has two known roles. First, it is a RNA surveillance pathway that degrades aberrant mRNAs harboring premature termination codons (PTCs) generated as a result of mutation or biosynthetic errors; e.g., aberrant RNA splicing. While the functional significance of this RNA surveillance role is not known, it is likely to help protect cells from the deleterious effects of dominant-negative truncated proteins translated from PTC-bearing mRNAs.10,41 Second, NMD serves as a regulator of normal gene expression through its ability to degrade a subset of functional wild-type RNAs.16 NMD magnitude is regulated in a stage-, tissue-, and cell type-specific manner, allowing it to coordinately alter the levels of these natural target mRNAs.18,22,59 Thus, NMD acts much like a transcriptional pathway, but rather than regulating the synthesis of groups of RNAs, it regulates their decay. This ability raises the exciting possibility that NMD controls or influences specific biological processes. Indeed, increasing evidence—from organisms across the phylogenetic scale—support the notion that NMD is critical for specific developmental events.19,55,60
NMD is a translation-dependent mechanism that degrades RNAs harboring in-frame stop codons in specific contexts. The most well understood context that triggers NMD is an exon-exon junction downstream of the stop codon terminating the main open reading frame (ORF). Exon-exon junctions serve as nucleation points that recruit the exon junction complex (EJC), a set of proteins deposited on RNAs after splicing that mediate several functions, including NMD activation.17 The EJC contains a highly stable tetramer core comprised of the RNA-binding proteins eIF4AIII, MLN51, Y14, and MAGOH. In the current consensus model of NMD, RNA decay is triggered when the EJC interacts with the NMD factor UPF3B (Fig. 1). UPF3B is part of a “NMD core complex,” which also contains the RNA helicase, UPF1, and the adaptor protein UPF2. This NMD core complex is thought to form at sites of translation termination, which limits this complex to only interacting with EJCs recruited downstream of the main ORF stop codon, as upstream EJCs are ejected by the pioneer round of translation.24 The consequence of this is that, only stop codons in middle exons, not the final exon, elicit EJC-dependent NMD. This provides a simple mechanism by which PTCs are distinguished from normal stop codons, as the main ORF ends in the final exon in most normal mRNAs, making them immune to NMD. However, an interesting subset of normal (including alternatively spliced) mRNAs terminate their main ORF in a middle exon, allowing these particular mRNAs to be degraded by NMD in a regulated manner. As described below, other contexts can also elicit NMD (e.g., long 3′ untranslated regions [UTRs]), further expanding the repertoire of normal mRNAs subject to regulated decay by NMD.
Figure 1.
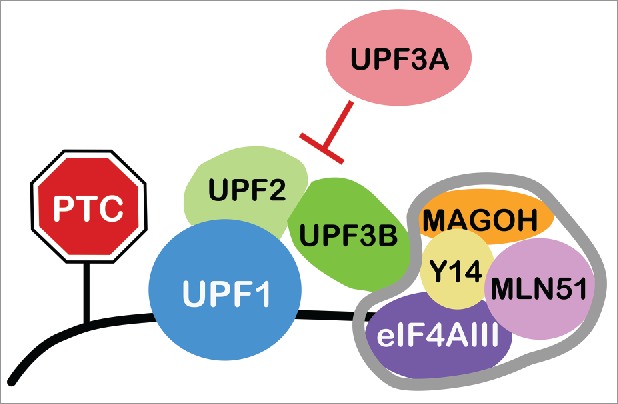
Triggering and Regulating NMD. RNA degradation elicited by the NMD pathway depends on UPF proteins and is enhanced by the exon junction complex (EJC), whose core components are outlined in gray. UPF3B is thought to be critical for NMD because it drives the interaction of the EJC with the NMD adaptor protein, UPF2, and the RNA helicase UPF1. UPF3A has the opposite function – it represses NMD.45
The UPF3 gene paralogs
Invertebrates harbor a single copy of the NMD gene, UPF3, whereas vertebrates have two: the autosomal gene, UPF3A, and the X-linked gene, UPF3B. Thus, the duplication event that gave rise to these gene paralogs likely occurred at the dawn of the vertebrate lineage ∼400 million years ago.45
Why do two UPF3 paralogs exist?
UPF3B encodes a well-studied protein with well-defined biochemical functions required for a branch of the NMD pathway.8,23 In the most commonly accepted model of NMD, UPF3B serves as an adaptor protein that forms a large molecular complex that drives RNA decay.7 In higher eukaryotic species, UPF3B accomplishes this by directly interacting with both the EJC and NMD core protein UPF2.6 Mutations in the human UPF3B gene cause intellectual disability and are highly associated with neuro-developmental disorders, including schizophrenia and autism.42,49,50 While the underlying basis for its role in neural development and cognition are not known, in vitro studies have shown that UPF3B promotes neural differentiation and it is regulated by neurally expressed microRNAs that influence neural self-renewal vs. differentiation decisions.20,29 UPF3B also shapes the unfolded protein response, a pathway critical for normal neural development.21
Until recently, the functional role of UPF3B's paralog partner—UPF3A—has been unclear. Gain-of-function studies conducted before the emergence of the RNA interference (RNAi) technique showed that UPF3A had little or no NMD activity when tethered downstream of a stop codon in reporter RNAs.30 This was perplexing given that the UPF3A gene has existed since the first vertebrates emerged. What functional value did UPF3A provide to avoid initial extinction at the dawn of the vertebrate lineage? How did it later become fixed in vertebrate genomes? One possibility is that UPF3A served as a back-up gene for UPF3B, a notion supported by the discovery that the UPF3A protein is dramatically upregulated in response to loss or depletion of UFP3B.10,50 However, since natural selection acts on benefits provided in the present, not potential future applications, it is not obvious how such functional redundancy could have been selected for. Furthermore, it is paradoxical as to why UPF3A has only weak NMD activity when it has had ∼400 million years to perfect this activity. This is particularly perplexing when one considers that a single amino-acid substitution (mimicking that of UPF3B) is sufficient to convert UPF3A into a strong NMD factor.26 If UPF3A functions in NMD, why would this factor avoid acquiring strong NMD-promoting activity over evolutionary time?
UPF3A is a NMD repressor
To begin to address these questions, Shum et al. elected to re-examine the function of UPF3A by taking a loss-of-function approach.45 Using RNAi, these investigators depleted UPF3A in several cell lines, both mouse and human, and made the surprising discovery that this led to downregulation of most NMD substrate mRNAs tested. As a control, they depleted well-established NMD factors and found that this upregulated NMD substrates, as expected. This opposing activity of UPF3A raised the possibility that it is a NMD repressor (Fig. 1). As further support for this, UPF3A depletion increased NMD activity as measured with a NMD reporter. To assess whether UPF3A serves as a global repressor of the NMD pathway, RNA sequencing (RNA-seq) half-life analysis was employed in cells depleted of UPF3A. This revealed that many more transcripts were destabilized than stabilized upon depletion of UPF3A, providing support for the idea that UPF3A acts as a broadly acting NMD repressor. As described below, the ability of UPF3A to repress NMD is likely physiologically significant, as it provides a mechanism to coordinately stabilize batteries of mRNAs critical for specific biological events.
Shum et al. next investigated the underlying mechanism by which UPF3A represses NMD.45 To elucidate whether the UPF2-interacting domain in UPF3A is required, they examined an alternative isoform of UPF3A lacking a functional UPF2-interaction domain as a result of a naturally occurring alternative splicing event. This UPF3A variant failed to inhibit NMD and did not rescue NMD repression in cells that had been depleted of UPF3A, suggesting that the UPF2-binding activity of UPF3A is essential for NMD repression. To elucidate the role of the EJC-interaction domain of UPF3A, they tested a UPF3A mutant lacking this domain and found that it retained NMD repressive activity. This indicated that the EJC-interaction domain of UPF3A is dispensable for UPF3A's ability to repress NMD. This raised the possibility that UPF3A's EJC-interacting domain is largely inactive and that UPF3A represses NMD by sequestering the essential NMD factor, UPF2, away from the EJC and perhaps other components of the NMD machinery. In support of this model, the C-terminal half of UPF3A (which houses the EJC-interaction domain) was found to be both poorly conserved and highly divergent in sequence from the equivalent region in UPF3B. Furthermore, previous studies had suggested that UPF3A interacts more weakly with the EJC than does UPF3B.9 If indeed UPF3A serves as NMD repressor by virtue of a weak EJC-interacting domain, this predicts that substituting it with UPF3B's strong EJC interacting domain would convert UPF3A into an NMD-promoting factor. Consistent with this prediction, this swap caused UPF3A to acquire modest, but significant, NMD-promoting activity. Shum et al. also performed the converse experiment: they replaced UPF3B's strong EJC-interaction domain with UPF3A's weak EJC interaction domain, and found that this converted UPF3B into an NMD inhibitor.
Together, these findings supported a model in which UPF3A acts as a molecular decoy that discourages UPF2 from creating a complex with the EJC, thereby inhibiting transcripts from being degraded by NMD (Fig. 1). Other experiments performed by Shum et al. also supported this model. For example, co-immunoprecipitation experiments showed that depletion of UPF3A increased the interaction of endogenous EJC components with the central NMD factor UPF1. They also found—through an in vivo tethering assay—that depleting UPF3A did not affect the ability of tethered UPF2 (bound downstream of a stop codon) to elicit NMD. This suggests that UFP3A does not inhibit UPF2s ability to recruit other NMD factors, but rather it acts by sequestering UPF2 from the NMD machinery.
The role of UPF3A in germ cells in vivo
In order for UPF3A to modulate the magnitude of NMD in a biologically meaningful way, it is critical that UPF3A expression itself is regulated. High levels of UPF3A in a given tissue would presumably strongly repress NMD and thus robustly stabilize NMD target transcripts, a subset of which could encode proteins critical for that tissue. Conversely, a tissue with low levels of UFP3A would be expected to have strong NMD, which would robustly destabilize NMD substrates that might otherwise cause aberrant events in that tissue. While early studies showed that UPF3A is broadly expressed in all tissues, there were hints that it is regulated. For example, Upf3a mRNA levels were found to differ in adult tissues, with particularly high expression in the testis.44,57 Evidence suggested that UPF3A might also be regulated at the protein level. Loss or depletion of UPF3B was found to dramatically stabilize UPF3A protein through a mechanism involving competition of these two proteins with UPF2.9 This raised the possibility that tissue- or stage-specific alterations in UPF3B level could, in turn, lead to complementary changes in UPF3A level. In their recent study, Shum et al. examined UPF3A expression in more detail and made three key observations: (i) UPF3A protein is highly expressed in early mouse embryos; (ii) UPF3A protein is also abundant in the adult testis but is largely undetectable in other adult tissues; and (iii) Upf3a mRNA and UPF3A protein are both expressed in a stage-specific manner during spermatogenesis, with expression highest in spermatocytes, the stage of germ cell development in which meiosis occurs.45 Together, these results suggested that UPF3A expression is regulated at both the mRNA and protein level in a manner that largely restricts its expression to the early embryo and the male germline.
To test whether UPF3A functions in these contexts, Shum et al. generated and phenotypically analyzed Upf3a-mutant mice.45 They found that global knockout (KO) of Upf3a caused early embryonic lethality (before embryonic day 8.5), with embryo defects observable even at the pre-implantation stage. This result clearly indicated that UPF3A functions in early embryogenesis, perhaps because it represses NMD to stabilize key transcripts important for pre-implantation development.
To specifically examine the role of UPF3A in male germline, Shum et al. crossed Upf3a-floxed mice with the Stra8-Cre line to generate conditional (c) KO mice that ablated Upf3a in germ cells just prior to the initiation of spermatogenesis. They found these mice exhibited a dramatic reduction in the number of spermatocytes at the postnatal stage, indicative of delayed or inhibited progression through meiosis. Consistent with this, adult Upf3a-cKO mice had reduced numbers of more advanced germ cells, including sperm. Histological staining showed abundant vacuoles in the seminiferous tubule sections of these mutant mice, further indicative of spermatogenic failure.
These results supported a model in which UPF3A must be highly expressed in spermatocytes to repress NMD and thereby stabilize key NMD substrate mRNAs. In support of this model, Shum et al. found that purified spermatocytes from Upf3a-cKO mice had lower levels of NMD substrates (indicative of increased NMD) compared with control mice spermatocytes. Some of these NMD substrates may be critical for meiotic progression, which would explain why normal spermatogenesis was perturbed in Upf3a-cKO mice. Indeed, RNA-seq half-life analysis identified NMD substrates encoding proteins known to function in meiosis and related events, including EXO1, SGOL2, and TERF.
Intriguingly, Shum et al. found that even Upf3a haploinsufficiency in male germ cells was sufficient to activate NMD and cause spermatogenic defects in the testis. The finding that only a 50% decrease in UPF3A perturbed meiotic progression suggested that one or more of its target RNAs must be tightly regulated for normal spermatogenesis. Together, these findings strongly suggested that NMD must be suppressed to allow for normal spermatogenesis, particularly during the meiotic stage of male germ cell development.
Evolutionary implications
The finding that UPF3A is a NMD repressor, while UPF3B is an NMD activator, leads to an interesting evolutionary question. What evolutionary forces led to the evolution of these antagonistic paralogs? Previous studies have shown that gene paralogs can evolve in a variety of ways. In “neofunctionalization,” duplicated genes accumulate mutations that result in a new function.2 In “subfunctionalization” a new function is not acquired, instead the ancestral function(s) of the original single-copy gene is divided among the daughter genes, either through their different expression patterns or differential retention of specific ancestral functions.11 Which of these evolution modes apply to the UPF3A/UPF3B paralogs?
Neofunctionalization: Acquisition of NMD suppression
Let us first consider neofunctionalization, which is the most intuitively obvious model for UPF3A/UPF3B evolution. According to this model, the ancestral UPF3B gene encoded a NMD factor and the duplicated UPF3A gene copy acquired – through evolutionary time – the ability to repress NMD (Fig. 2, upper). This scenario predicts that modern day organisms with a single copy of the UPF3 gene express an NMD-promoting version of the UPF3 protein. Indeed, all invertebrates that have been examined (such as S. cerevisiae, C. elegans, A. thaliana, and D. melanogaster) harbor a single UPF3 gene that encodes an NMD-promoting factor.
Figure 2.
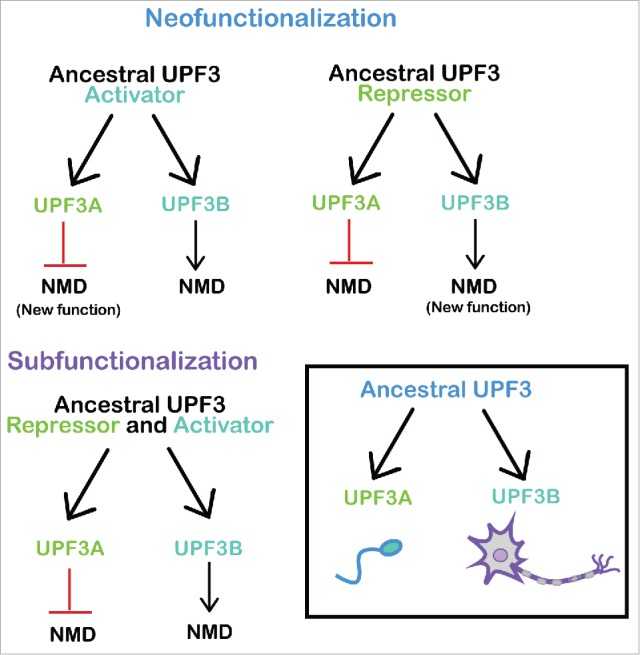
UPF3 Evolution. (Upper) Neofunctionalization models in which UPF3A acquired the ability to repress NMD (left) or UPF3B acquired the ability to activate NMD (right). (Lower left) Subfunctionalization model in which a putative ancestral bi-functional UPF3 gene segregated its NMD repressor and NMD stimulatory activities to UPF3A and UPF3B, respectively. (Lower right) UPF3A and UPF3B evolved to function in spermatogenesis and neurogenesis, respectively. These NMD regulators also act in other biological systems.
The mechanism by which UPF3A suppresses NMD (described above) provides a clue as to how the new function—NMD suppression—could quickly evolve. A simple mutation that inactivated the EJC-interaction domain would have been sufficient to convert an ancestral UPF3 protein with NMD activity into a NMD repressor. Indeed, Shum et al. showed that mutational inactivation of the EJC-interaction domain was sufficient to convert the modern day NMD factor, UPF3B, into a NMD repressor.45 The ability to rapidly acquire this new function also provides a simple explanation for how the newly duplicated UPF3A was rapidly selected for after it first arose at the dawn of vertebrate lineage. If, instead, UPF3A's new function required a lengthy stepwise process involving multiple mutation events, it is not obvious how it could have avoided extinction. We suggest that new functions requiring single or few mutations have commonly been responsible for the fixation of gene paralogs over evolutionary time. In some cases, gene paralogs could have lost a functional domain and generated dominant-negative proteins, as exemplified by UPF3A. In other cases, simple mutations may have created new activities.
Why did UPF3B maintain the ability to promote NMD, while UPF3A acquired the ability to repress NMD? The available evidence suggests that this allowed these two paralogs to confer unique functions in different tissues (Fig. 2, upper). In modern day organisms, UPF3B is required for a specific branch of the NMD pathway, where it functions during brain development.8,35,50 UPF3B promotes the differentiation of neural progenitor cells and may be involved in the ability of NMD to influence neural proliferation versus differentiation decisions by virtue of its ability to degrade specific transcripts.20,29 Thus, by subtly controlling the stabilities of batteries of mRNAs, the UPF3B branch of NMD may have played an important role in the evolution of the vertebrate brain.
In contrast to UPF3B, UPF3A functions in the testis, as described above.45 Perhaps this divide in UPF3 function occurred because the early vertebrate germline uniquely required NMD repression rather than NMD activation. Selective forces may have acted to use UPF3A to drive transient stabilization of key stage-specific transcripts, a notion supported by the finding that UPF3A is expressed in a stage-specific manner in modern day male germ cells. In summary, we suggest that the different expression patterns and functions of UPF3A and UPF3B are likely to have evolved from unique requirements of different cell types in evolving vertebrates.
Neofunctionalization: Acquisition of NMD-promoting activity
Another possibility is that the ancestral UPF3 gene encoded a NMD repressor rather than a NMD factor (Fig 2, upper right). While intuitively less obvious than the model described above, this alternative neofunctionalization model has some merits. A key point in its favor is that, in principal, the ancestral UPF3 protein would only require having a UPF2-interaction domain, not an EJC-interaction domain. Because this is a simpler molecule than a NMD activator, it is a reasonable possibility that UPF3 only evolved to this extent prior to the emergence of vertebrates. If so, it could have served the same role then as UPF3A does now — to suppress NMD in order to stabilize critical transcripts. It may have had an ancient role in the early embryo and then later acquired a germline expression pattern to function in vertebrate spermatogenesis (Fig. 2, lower right). In contrast to UPF3A, which maintained its function, UPF3B acquired a new activity—NMD-promoting activity—by gaining the ability to interact with the EJC. This could have occurred in a slow step-wise fashion or in a more rapid manner through recombination events that allowed it to acquire the EJC-interaction domain from another gene. Once created, the newly minted UPF3B protein likely evolved to promote mRNA decay in tissues and stages where this was favorable. The modern day role of UPF3B in nervous system development suggests that UPF3B was selected to function in developing neurons, where it served to promote the decay of key transcripts critical for the development of these cells (Fig. 2, lower right). A candidate is the mRNA encoding the pro-neural differentiation protein, SMAD7, which must be degraded by NMD to maintain the neural stem cell state.29
Subfunctionalization: Distribution of NMD-promoting and NMD-repressive activities
A third model for how the UPF3A/UPF3B paralogs arose is subfunctionalization. In subfunctionalization, only the function(s) of the ancestral gene are used in the duplicated daughter genes.32 If the ancestral gene has a single function, it can be “sub-functionalized” through different expression patterns of the daughter genes, thereby distributing the function to different cell types or tissues. Alternatively, if the ancestral gene has different functions, these can be distributed to different daughter paralogs, as this provides certain benefits, such as the ability to be independently optimized. For the subfunctionalization model to apply to the UPF3A/UPF3B paralogs, one must hypothesize that the ancestral UPF3 protein had both NMD-promoting and NMD-inhibitory activities (Fig. 2, lower left). This possibility is supported by the characteristics of the modern-day UPF3A protein. Shum et al. showed that while UPF3A is primarily a NMD inhibitor, it acts as a NMD factor for some NMD substrates in some cell types.45 While the underlying mechanism was not explored, a reasonable possibility is that UPF3A's weak EJC-interacting domain is sufficient to allow UPF3A to promote the decay transcripts that are “on the verge” of being degraded by NMD.
Primordial organisms may have had a form of UPF3 with both NMD-promoting and –repressing activities because insufficient time had passed to perfect either one. In these early organisms, transcripts may have co-evolved with UPF3; those that needed to be degraded evolved to be sensitive to its pro-decay function and those that required stabilization evolved to respond to its anti-decay function. Once the UPF3 gene duplicated, this provided an opportunity to separate its two functions, with UPF3B losing NMD suppression activity, and UPF3A largely losing NMD stimulatory activity. As these singular properties appeared, UPF3A and UPF3B may have optimized them and acquired expression patterns that permitted them to act in the appropriate cell types and stages of development (Fig. 2, lower).
If indeed the subfunctionalization model applies to UPF3A/UPF3B evolution, it would be a novel example of this mode of evolution. Rather than a single function being sub-divided in different tissues, it would be an example of opposing functions undergoing such sub-division.52 Other examples of gene paralogs with opposing functions have been discovered,48,58 and thus it will be interesting to determine whether the subfunctionalization or neofunctionalization models apply to these cases as well.
Evidence that NMD is critical for spermatogenesis
As described above, Shum et al. provided several lines of evidence that NMD must be suppressed for normal spermatogenesis.45 Is this because NMD requires tight regulation? Or, instead, might NMD have no role at all in spermatogenesis? To address whether NMD functions in spermatogenesis, Bao et al. conditionally knocked out the essential NMD factor gene, Upf2, in male germ cells.4 They first used Vasa-Cre mice to conditionally knock out Upf2 in fetal germ cells (specifically at the prospermatogonia stage). These Upf2-cKO mice had small testes with few germ cells and were infertile. Histological analyses showed that young (postnatal day 3 [P3]) Upf2-cKO mice had reduced numbers of undifferentiated spermatogonia, suggesting that NMD is required for very early stages of germ cell development. To examine a later stage of germ cell development, Bao et al. conditionally ablated Upf2 in male germ cells postnatally. Like mice with embryonic ablation of Upf2, these cKO mice had small testes and were infertile. Histological analysis at P14 showed these postnatal cKO mice had few spermatocytes, raising the possibility of a block in meiotic progression. This phenotype, along with vacuoles in seminiferous tubules, were also observed in “hyper NMD” Upf3a-cKO mice,45 raising the possibility that a carefully titrated amount of NMD is critical for meiotic progression and the normal morphology of seminiferous tubules.
NMD factors also have functions in other pathways and thus defects in Upf2 knockout mice do not necessarily result entirely from perturbed NMD. While it is not known whether human UPF2 has non-NMD functions, yeast UPF2 has been shown to function in telomeres and chromatin silencing.1,12,27 Some mammalian NMD factors, including UPF1, have been shown to have non-NMD functions, including in the Staufen-mediated mRNA decay pathway.38 Thus, it is possible that mammalian UPF2 has a non-NMD function and that disruption of this function has a role in the spermatogenic defects observed by Bao et al. However, it is worth noting that studies in Drosophila, zebrafish and mice have shown that loss or depletion of several different NMD factors, including UPF2, lead to very similar defects, suggesting that perturbed NMD is responsible.3,33,45,51,54,55
Additional recent support for the notion that NMD is critical for spermatogenesis comes from a recent study by Fanourgakis et al.14 These investigators studied mutant mice lacking the tudor domain protein, TDRD6, that were previously shown to exhibit developmental arrest in haploid germ cells (at the round-to-elongated spermatid stage) and are infertile.53 TDRD6 is a component of the chromatoid body (CB), a cytoplasmic entity present exclusively in round spermatids. CBs harbors concentrated amounts of specific proteins and RNAs involved in post-transcriptional gene regulation.34 Tdrd6-KO mice have highly disrupted “ghost”-like CBs, suggesting that TDRD6 is critical for CB formation and/or maintenance.53 Because the CBs in Tdrd6-KO mice are largely defective, these mutant mice are valuable models for understanding the in vivo roles of the CB. Fanourgakis et al. hypothesized that CBs are critical for NMD, based on the previous finding that the NMD protein, UPF1, is normally highly concentrated in CBs.34 In support of their hypothesis, Fanourgakis et al. found that the NMD protein, UPF1, was largely absent from CBs in Tdrd6-KO mice. Furthermore, they found that round spermatids from Tdrd6-KO mice exhibited a profound defect in the interaction of UPF1 with UPF2. Because the RNA helicase activity of UPF1 requires the presence of UPF2, this implied that the round spermatids in Tdrd6-KO mice have deficient NMD. As further support, these investigators found that NMD substrates were elevated in these mutant mice, as described below. Together with the findings of Bao et al., these results suggested that NMD has critical roles in spermatogenesis.
NMD regulation during germ cell development
In order for NMD to play roles in specific stages of spermatogenesis, it is critical that its magnitude is regulated in a stage-specific manner. Downregulation of NMD would lead to stabilization of its many target mRNAs, while NMD upregulation would have the converse effect. Both would likely exert major effects on spermatogenesis, particularly if NMD's target transcripts encode proteins with important roles in germ cells, including broad cellular roles such as proliferation and differentiation, and germ cell-specific events such as meiosis.
Evidence that NMD is upregulated as germ cell development proceeds comes from Bao et al., who found that many NMD genes, including Upf1 and Upf2, are upregulated in enriched spermatocytes and round spermatids compare with enriched immature germ cells (spermatogonia)4 (Fig. 3A, Model I). Time-course analysis of the first wave of spermatogenesis confirmed that Upf1 and Upf2 are upregulated at the spermatocyte stage, with sustained or elevated expression in round spermatids.4,14 However, there is also evidence that NMD is instead downregulated in spermatocytes (Fig. 3B, Model II). Shum et al. found that the NMD repressor protein, UPF3A, is present at highest levels in spermatocytes45 (Fig. 3B). They also found that the X-linked NMD gene, Upf3b, is transcriptional silenced by the meiotic sex chromosome inactivation mechanism at this stage (Fig. 3B). In the future, it will be important to determine whether these alterations in UPF1, UPF2, UPF3A, and UPF3B levels lead to a significant changes in NMD magnitude. This will depend, in part, on which of these factors are rate limiting for NMD in developing germ cells. With regard to UPF3A, it is worth noting that while it serves as a broadly acting NMD repressor in both mouse and human cells, a small subset of NMD substrates are destabilized by UPF3A in a stem cell line.45 It is not yet known if UPF3A destabilizes some NMD substrates in germ cells.
Figure 3.
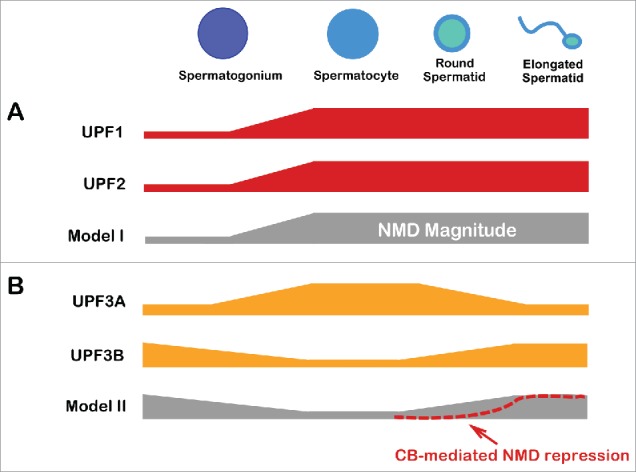
NMD modulation during male germ cell development. (A) UPF1 and UPF2 levels increase when dividing male germ cells (spermatogonium) differentiate into meiotic germ cells (spermatocytes), leading to the hypothesis that NMD magnitude is upregulated at this transition point (Model I). Sustained high expression of these NMD factors in early differentiating haploid germ cells (round spermatids) is consistent with high NMD magnitude being maintained in these cells. (B) The inverse expression pattern of the NMD repressor, UPF3A, and the NMD factor, UPF3B, in spermatocytes leads to the hypothesis that NMD magnitude is suppressed in these cells to stabilize key mRNAs important for meiosis (Model II). The additional finding that NMD factors are sequestered in chromatoid bodies (CBs) in round spermatids raises the possibility that NMD is also suppressed at this stage of development. This could provide a means to stabilize mRNAs for translation at the next developmental stage – mature haploid germ cells (elongated spermatids).
The role of chromatoid bodies in NMD
An intriguing line of evidence that NMD is regulated at the haploid stage of germ cell development comes from Meikar et al. and Fanourgakis et al.14,34 These studies found that several NMD factors are highly concentrated in CBs (with little or no UPF1 in the cytosol), raising the possibility that CBs are sites of highly active NMD. This hypothesis is also supported by studies on processing (P) bodies, which have several characteristics in common with CBs,25 and have been shown to support NMD.39 Together, these observations lead to the intriguing model that CBs are, in some respects, the germ cell version of P bodies and that both are sites of active NMD.
While an intriguing idea, it is important to keep in mind that it has proven difficult to elucidate the functional activities of cytoplasmic bodies. Indeed, it has been shown that P body formation can be a consequence, rather than a cause, of RNA-mediated decay and repression events.13 In the case of CBs, in order for them to have a causal role in NMD, it is critical that they support active translation, as NMD is translation dependent, based on its requirement of a stop codon, as well as empirical studies showing that impairment of translation blocks NMD.5 Do CBs support translation? While a definitive answer to this question is not known, there are several lines of evidence suggesting that the answer is “no.” For example, CBs lack detectable ribosomal proteins and rRNA.61 They also harbor several RNAs that are translationally repressed.36,43 This raises the possibility that rather than being sites of active NMD, CBs are actually sites of inactive NMD (Fig. 3B, Model 2).
What purpose might this serve? It is well established that some genes (most notably the protamine genes) are transcribed at the round spermatid stage but are not translated until later, at the elongated spermatid stage.15 Thus, it is critical that a subset of mRNAs is stabilized in round spermatids so that they remain intact for later translation. Perhaps NMD repression is one means by which this is achieved. Repression of NMD in round spermatids could also stabilize mRNAs that are translated and function directly in these cells.
To definitively assess how NMD is regulated during germ cell development, it will be important, in the future, to use assays that directly measure NMD magnitude in different germ cell stages. Given that germ cells tend to change their characteristics in primary culture (indeed, post-proliferative stages tend to die in culture), methods that measure NMD magnitude in individual cells in vivo, such as NMD reporter mice, hold perhaps the greatest future promise.
Recognition of NMD substrates in germ cells
The NMD pathway recognizes specific features in RNA that leads to their decay. Many of these features only elicit decay when in specific contexts, many of which are undefined. Thus, it is often not possible to predict if a given transcript will be degraded by NMD. As described in the Introduction, the most well-established and reliable NMD-inducing feature is an exon-exon junction downstream of the stop codon terminating the main ORF.37 Both Bao et al. and Fanourourgakis et al. examined whether this feature triggers NMD in round spermatids. Bao et al. used RNAseq analysis to define alternatively spliced transcripts that contain a stop codon in this context. These investigators found that surprisingly few transcripts with a stop codon in this context were upregulated in Upf2-cKO mice testes relative to control mice testes. While, at face value, this suggested that NMD does not degrade most PTC-bearing transcripts, this analysis was performed on whole testes from adult Upf2-cKO mice, which are largely depleted of both meiotic and haploid germ cells. Thus, it is difficult to distinguish between effects of Upf2 on NMD target transcript levels vs. germ cell subsets. To address this, the authors purified round spermatids from adult Upf2-cKO mice and compared them with purified round spermatids from control mice. RNAseq analysis did not reveal significant upregulation of PTC-bearing transcripts in the enriched Upf2-cKO round spermatids, providing evidence that the EJC-dependent branch of NMD is absent or weak in round spermatids. The notion that EJC-dependent branch of NMD is not measurably active in round spermatids was confirmed by Fanourgakis et al., who found that purified KO spermatids did not exhibit a significant difference in the expression of dEJ-bearing mRNA, as compare with control spermatids, based on RNAseq analysis. They confirmed this in selected cases by PCR analysis.
The possibility that round spermatids do not have EJC-dependent NMD is exciting, as no other instance of cell type-specific immunity to NMD had previously been reported. How could this occur? As described above, round spermatids are unique in having NMD factors highly concentrated in cytoplasmic bodies—CBs—that some evidence suggest are translationally inactive and thus might not support NMD. If so, round spermatids would be expected to largely lack EJC-dependent NMD.
Fanourgakis et al. and Bao et al. considered another possibility – that round spermatids do have active NMD, but that these germ cells respond to a non-EJC feature. In particular, these investigators examined long 3′ UTRs, which have been shown to induce NMD in some cases. 31 Both groups obtained evidence that long 3′ UTRs elicit NMD in round spermatids. Bao et al. observed an accumulation of alternatively spliced transcripts with longer 3′ UTRs in purified Upf2-cKO spermatids as compared with control spermatids. Fanourgakis et al. found that more long 3′ UTR transcripts were upregulated than downregulated in Tdrd6-KO mice. They also examined RNAs with multiple isoforms and found that, in most cases, only the long 3′ UTR isoform was upregulated in the KO mice, providing very strong evidence that NMD is impacted. These authors also dealt with the possibility that the alterations in the levels of mRNAs with long 3′ UTRs were indirectly affected by NMD. Using RIP analysis, they found that the Tdrd6-KO mice round spermatids exhibited impaired UPF1 binding with most mRNAs with long 3′ UTRs that they tested, providing evidence that loss of Tdrd6 upregulates these mRNAs through the direct action of NMD. Together, these findings from Fanourgakis et al. and Bao et al. strongly suggest that NMD is active in round spermatids and that this pathway targets a subset of transcripts with long 3′ UTRs.
This data potentially impacts past work showing that round spermatid transcripts tend to have short 3′ UTRs.28,40 This phenomenon was previously interpreted as the result of a shift in the relative efficiency of upstream vs. downstream polyadenylation. However, it could instead result from an increased NMD magnitude in round spermatids, leading to reduced numbers of RNAs with long 3′ UTRs in these meiotic cells.
If indeed round spermatids use the NMD pathway to degrade transcripts with long 3′ UTRs, how does one reconcile this with the data suggesting that the EJC-dependent branch of NMD is not active in these cells? Both Bao et al. and Fanourgakis et al. favor the notion that germ cells are unique in recognizing one NMD-inducing feature but not another. If true, it will be interesting in the future to determine how this accomplished. For example, do round spermatids have sub-optimal levels or an inactive form of an EJC component or downstream factor? Another outstanding question is how could round spermatids engage in 3′ UTR-dependent NMD if the NMD factors are sequestered in translationally silent CBs in round spermatids? One possibility is that while most NMD factors are sequestered in CBs, a modest amount of NMD factors are also present in the cytosol where translation is active. If low levels of NMD factors are sufficient for 3′UTR-dependent NMD but not EJC-dependent NMD, this would explain why round spermatids are unique in having branch-specific NMD.
While the notion of cell type- and branch-specific NMD is an intriguing explanation for the data from Bao et al. and Fanourgakis et al, there are others. One alternative possibility is that EJC-dependent NMD and long 3′UTR-dependent NMD occur in different germ cell subsets. The round spermatids analyzed by Fanourgakis et al. are reported to be ∼70% pure and thus the 30% contaminating cells could be responsible for long 3′ UTR-elicited NMD.
Bao et al. also examined how NMD impacts spermatocytes. They compared purified spermatocytes from Upf2-cKO and control mice, and obtained evidence that, like round spermatids, spermatocytes have long 3′UTR-dependent NMD, but no detectable EJC-dependent NMD. However, there are several caveats to consider. The KO spermatocytes they used for their analysis were from Upf2-cKO adults that had suffered from massive germ cell defects and loss since the postnatal period (at least since P14). Thus could have adversely influenced results. For example, the broad testicular defects and disorganization might have caused aberrant gene expression in the few remaining live spermatocytes, which could have obscured effects on NMD targets. Presuming that spermatocyte death is an ongoing and stochastic process in Upf2-cKO mice, RNAseq analysis of spermatocytes from these mice would likely reflect gene expression in both healthy and dying spermatocyes. Finally, the data provided by Bao et al. suggest that spermatocyte death in Upf2-cKO mice first occur in mid-meiosis, which predicts that purified Upf2-cKO spermatocytes would be enriched for early spermatocytes. Thus, comparison with control spermatocytes would likely enrich for early meiosis genes and dilute the ability to identify NMD substrates.
Thus, we consider that “the jury is still out” with regard to the nature of NMD substrates in germ cells. If EJC-dependent NMD does not occur in these cells, this will add to an already long list of unusual qualities of male germ cells.46 If instead, EJC-dependent NMD does occur in germ cells, but it is restricted to particular stages of germ cell development, this will be an equally interesting possibility that will lead to future research to uncover the underlying molecular mechanism responsible.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Addinall SG, Holstein EM, Lawless C, Yu M, Chapman K, Banks AP, Ngo HP, Maringele L, Taschuk M, Young A, et al.. Quantitative fitness analysis shows that NMD proteins and many other protein complexes suppress or enhance distinct telomere cap defects. PLoS Genet 2011; 7(4):e1001362; PMID:21490951; http://dx.doi.org/ 10.1371/journal.pgen.1001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assis R, Bachtrog D. Neofunctionalization of young duplicate genes in Drosophila. Proc Nat Acad Sci U S A 2013; 110(43):17409-14; PMID:24101476; http://dx.doi.org/ 10.1073/pnas.1313759110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avery P, Vicente-Crespo M, Francis D, Nashchekina O, Alonso CR, Palacios IM. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA (New York, NY) 2011; 17(4):624-638; PMID:21317294; http://dx.doi.org/ 10.1261/rna.2404211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao J, Vitting-Seerup K, Waage J, Tang C, Ge Y, Porse BT, Yan W. UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3′UTR transcripts. PLoS Genet 2016; 12(5):e1005863; PMID:27149259; http://dx.doi.org/ 10.1371/journal.pgen.1005863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 1995; 270(48):28995-29003; PMID:7499432; http://dx.doi.org/ 10.1074/jbc.270.48.28995 [DOI] [PubMed] [Google Scholar]
- 6.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 2008; 15(1):85-93; PMID:18066079; http://dx.doi.org/ 10.1038/nsmb1330 [DOI] [PubMed] [Google Scholar]
- 7.Celik A, Kervestin S, Jacobson A. NMD: At the crossroads between translation termination and ribosome recycling. Biochimie 2015; 114:2-9; PMID:25446649; http://dx.doi.org/ 10.1016/j.biochi.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA 2nd, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. EMBO J 2007; 26:1820-1830; PMID:17363904; http://dx.doi.org/ 10.1038/sj.emboj.7601628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan W-K, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gécz J, Wilkinson MF. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol 2009; 16(7):747-753; PMID:19503078; http://dx.doi.org/ 10.1038/nsmb.1612 [DOI] [PubMed] [Google Scholar]
- 10.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 2007; 76:51-74; PMID:17352659; http://dx.doi.org/ 10.1146/annurev.biochem.76.050106.093909 [DOI] [PubMed] [Google Scholar]
- 11.Cusack BP, Wolfe KH. When gene marriages don't work out: divorce by subfunctionalization. Trend Genet 2007; 23(6):270-272; PMID:17418444; http://dx.doi.org/ 10.1016/j.tig.2007.03.010 [DOI] [PubMed] [Google Scholar]
- 12.Dahlseid JN, Lew-Smith J, Lelivelt MJ, Enomoto S, Ford A, Desruisseaux M, McClellan M, Lue N, Culbertson MR, Berman J. mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae. Eukaryotic Cell 2003; 2(1):134-142; PMID:12582130; http://dx.doi.org/ 10.1128/EC.2.1.134-142.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 2007; 8(1):9-22; PMID:17183357; http://dx.doi.org/ 10.1038/nrm2080 [DOI] [PubMed] [Google Scholar]
- 14.Fanourgakis G, Lesche M, Akpinar M, Dahl A, Jessberger R. Chromatoid body protein TDRD6 supports long 3′ UTR triggered nonsense mediated mRNA decay. PLoS Genet 2016; 12(5):e1005857; PMID:27149095; http://dx.doi.org/ 10.1371/journal.pgen.1005857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fajardo MA, Haugen HS, Clegg CH, Braun RE. Separate elements in the 3′ untranslated region of the mouse protamine 1 mRNA regulate translational repression and activation during murine spermatogenesis. Dev Biol 1997; 191(1):42-52; PMID:9356170; http://dx.doi.org/ 10.1006/dbio.1997.8705 [DOI] [PubMed] [Google Scholar]
- 16.He F, Jacobson A. Nonsense-mediated mRNA Decay: Degradation of defective transcripts is only part of the story. Annu Rev Genet 2015; 49:339-366; PMID:26436458; http://dx.doi.org/ 10.1146/annurev-genet-112414-054639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hir HL, Sauliere J, Wang Z. The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol 2016; 17(1):41-54; PMID:26670016; http://dx.doi.org/ 10.1038/nrm.2015.7 [DOI] [PubMed] [Google Scholar]
- 18.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdisciplinary Reviews: RNA 2012; 3(6):807-828; PMID: 26397022; http://dx.doi.org/23821644 10.1002/wrna.1137 [DOI] [PubMed] [Google Scholar]
- 19.Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: To die or not to die, that is the question. Curr Opin Genet Devt 2011; 21(4):422-430; PMID: 21550797; http://dx.coi.org/23821644 10.1016/j.gde.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet 2013; 22(23):4673-4687; PMID:23821644; http://dx.doi.org/ 10.1093/hmg/ddt315 [DOI] [PubMed] [Google Scholar]
- 21.Karam R, Lou C-H, Kroeger H, Huang L, Lin JH, Wilkinson MF. The unfolded protein response is shaped by the NMD pathway. EMBO Rep 2015; 16(5):599-609; PMID:25807986; http://dx.doi.org/ 10.15252/embr.201439696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karam R, Wengrod J, Gardner LB, Wilkinson MF. Biochimica et biophysica acta regulation of nonsense-mediated mRNA decay: Implications for physiology and disease. BBA - Gene Regulat Mech 2013; 1829(6-7):624-633; PMID:23500037; http://dx.doi.org/ 10.1016/j.bbagrm.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karousis ED, Nasif S, Mühlemann O. Nonsense-mediated mRNA decay: Novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA 2016; 7(5):661-82; PMID:27173476; http://dx.doi.org/ 10.1002/wrna.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J 2001; 20(8):2062-2068; PMID:11296238; http://dx.doi.org/ 10.1093/emboj/20.8.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A 2006; 103(8):2647-52; PMID:16477042; http://dx.doi.org/ 10.1073/pnas.0509333103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA (New York, NY) 2006; 12(6):1015-1022; PMID:16601204; http://dx.doi.org/ 10.1261/rna.12506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew JE, Enomoto S, Berman J. Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol Cell Biol 1998; 18(10):6121-30; PMID:9742129; http://dx.doi.org/ 10.1128/MCB.18.10.6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, Graber JH. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucl Acid Res 2007; 35(1):234-246; PMID:17158511; http://dx.doi.org/ 10.1093/nar/gkl919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep 2014; 6(4):748-764; PMID:24529710; http://dx.doi.org/ 10.1016/j.celrep.2014.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lykke-Andersen J, Shu MD, Steitz J. Human Upf proteins target an mRNA for nonsense-mediated decay when downstream of a termination codon. Cell 2000; 103(7):1121-1131; PMID:11163187; http://dx.doi.org/ 10.1016/S0092-8674(00)00214-2 [DOI] [PubMed] [Google Scholar]
- 31.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 2015; 16(11):665-677; PMID:26397022; http://dx.doi.org/ 10.1038/nrm4063 [DOI] [PubMed] [Google Scholar]
- 32.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics 2000; 154(1):459-473; PMID:10629003; http://dx.doi.org/ 10.1371/journal.pgen.0040029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 2001; 10(2):99-105; PMID:11152657; http://dx.doi.org/ 10.1093/hmg/10.2.99 [DOI] [PubMed] [Google Scholar]
- 34.Meikar O, Vagin VV, Chalmel F, Sostar K, Lardenois A, Hammell M, Jin Y, Da Ros M, Wasik KA, Toppari J, et al.. An atlas of chromatoid body components. RNA 2014; 20(4):483-95; PMID:24554440; http://dx.doi.org/ 10.1261/rna.043729.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen LS, Jolly L, Shoubridge C, Chan WK, Huang L, Laumonnier F, Raynaud M, Hackett A, Field M, Rodriguez J, et al.. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry 2012; 17(11):1103-15; PMID:22182939; http://dx.doi.org/ 10.1038/mp.2011.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen Chi M, Chalmel F, Agius E, Vanzo N, Khabar KSA, Jégou B, Morello D. Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. PLoS ONE 2009; 4(3):e4900; PMID:19333380; http://dx.doi.org/ 10.1371/journal.pone.0004900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholson P, Mühlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Bioch Soc Transact 2010; 38(6):1615-1620; PMID:21118136; http://dx.doi.org/ 10.1042/BST0381615 [DOI] [PubMed] [Google Scholar]
- 38.Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA 2013; 4(4):423-435; PMID:23681777; http://dx.doi.org/ 10.1002/wrna.1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell 2007. [DOI] [PubMed] [Google Scholar]
- 40.Pero R, Lembo F, Chieffi P, Del Pozzo G, Fedele M, Fusco A, Bruni CB, Chiariotti L. Translational regulation of a novel testis-specific RNF4 transcript. Mol Reprod Dev 2003; 66(1):1-7; PMID:12874792; http://dx.doi.org/ 10.1002/mrd.10322 [DOI] [PubMed] [Google Scholar]
- 41.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol 2009; 21(3):394-402; PMID:19359157; http://dx.doi.org/11113196 10.1016/j.ceb.2009.02.007 [DOI] [PubMed] [Google Scholar]
- 42.Rujescu D, Ingason A. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet 2009; 18(5):988-996; PMID: 18945720; http://dx.doi.org/11113196 10.1093/hmg/ddn351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato H, Tsai-Morris CH, Dufau ML. Relevance of gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) in the structural integrity of the chromatoid body during spermatogenesis. Biochim Biophy Acta Mol Cell Res 2010; 1803(5):534-543; PMID: 20176059; http://dx.doi.org/11113196 10.1016/j.bbamcr.2010.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to saccharomyces cerevisiae Upf2 protein and Upf3 protein. Mol Cell Biol 2001; 21:209-223; PMID:11113196; http://dx.doi.org/ 10.1128/MCB.21.1.209-223.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shum EY, Jones SH, Shao A, Dumdie J, Krause MD, Chan WK, Lou CH, Espinoza JL, Song HW, Phan MH, et al.. The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell 2016; 165(2):382-395; PMID:27040500; http://dx.doi.org/ 10.1016/j.cell.2016.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A, et al.. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Reports 2013; 3(6):2179-2190; PMID:23791531; http://dx.doi.org/ 10.1016/j.celrep.2013.05.031 [DOI] [PubMed] [Google Scholar]
- 47.Stalder L, Mühlemann O. The meaning of nonsense. Trend Cell Biol 2016; 18(7):315-321; PMID:18524595; http://dx.doi.org/17185322 10.1016/j.tcb.2008.04.005 [DOI] [PubMed] [Google Scholar]
- 48.Stoick-Cooper C, Weidinger G, Riehle K, Hubbert C, Major M, Fausto N, Moon R. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 2007; 134:479-489; PMID:17185322; http://dx.doi.org/ 10.1242/dev.001123 [DOI] [PubMed] [Google Scholar]
- 49.Swerdlow N, Weber M, Qu Y, Light GA, Braff DL. Realistic expectations of prepulse inhibition in translational models for schizophrenia research. Psychopharmacology (Berl) 2008; 199(3):331-388; PMID: 18568339; http://dx.doi.org/17704778 10.1007/s00213-008-1072-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarpey P, Raymond F, Nguyen L, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, et al.. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 2007; 39:1127-33; PMID:17704778; http://dx.doi.org/ 10.1038/ng2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thoren LA, Nørgaard GA, Weischenfeldt J, Waage J, Jakobsen JS, Damgaard I, Bergström FC, Blom AM, Borup R, Bisgaard HC, et al.. UPF2 is a critical regulator of liver development, function and regeneration. PLoS ONE 2010; 5(7); PMID:20657840; http://dx.doi.org/ 10.1371/journal.pone.0011650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.VanderSluis B, Bellay J, Musso G, Costanzo M, Papp B, Vizeacoumar FJ, Baryshnikova A, Andrews B, Boone C, Myers CL. Genetic interactions reveal the evolutionary trajectories of duplicate genes. Mol Syst Biol 2010; 6:429; PMID:21081923; http://dx.doi.org/ 10.1038/msb.2010.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasileva A, Tiedau D, Firooznia A, Müller-Reichert T, Jessberger R. Tdrd6 Is Required for Spermiogenesis, Chromatoid Body Architecture, and Regulation of miRNA Expression. Curr Biol 2009; 19(8); PMID:19345099; http://dx.doi.org/ 10.1016/j.cub.2009.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Mönch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Gen Dev 2008; 22(10):1381-1396; PMID:18483223; http://dx.doi.org/ 10.1101/gad.468808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wittkopp N. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol 2009; 29:3517-28; PMID:19414594; http://dx.doi.org/ 10.1128/MCB.00177-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian non- sense-mediated mRNA decay. Mol Cell Biol 2006; 26(4):1272-87; PMID:16449641; http://dx.doi.org/ 10.1128/MCB.26.4.1272-1287.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zetoune A, Fontaniere S, Magnin D, Anczukow O, Buisson M, Zhang C, Mazoyer S. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet 2008; 9:83; PMID:19061508; http://dx.doi.org/ 10.1186/1471-2156-9-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Duc A-C, Rao S, Sun X-L, Bilbee A, Rhodes M, Li Q, Kappes D, Rhodes J, Wiest D. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev Cell 2013; 24:411-425; PMID:23449473; http://dx.doi.org/ 10.1016/j.devcel.2013.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lou C-H, Dumdie J, Goetz A, et al. Nonsense-Mediated RNA Decay Influences Human Embryonic Stem Cell Fate. Stem cell reports. 2016;6(6):844-857 . doi: 10.1016/j.stemcr.2016.05.008. PMID: 27304915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stalder L, Mühlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18(7):315-321 . doi: 10.1016/j.tcb.2008.04.005. PMID: 18524595 [DOI] [PubMed] [Google Scholar]
- 61.Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. NatRevMolCell Biol. 2007;8:85-90. doi: 10.1038/nrm2081. PMID: 17183363 [DOI] [PubMed] [Google Scholar]


