ABSTRACT
RNA modifications have long been known to be central in the proper function of tRNA and rRNA. While chemical modifications in mRNA were discovered decades ago, their function has remained largely mysterious until recently. Using enrichment strategies coupled to next generation sequencing, multiple modifications have now been mapped on a transcriptome-wide scale in a variety of contexts. We now know that RNA modifications influence cell biology by many different mechanisms — by influencing RNA structure, by tuning interactions within the ribosome, and by recruiting specific binding proteins that intersect with other signaling pathways. They are also dynamic, changing in distribution or level in response to stresses such as heat shock and nutrient deprivation. Here, we provide an overview of recent themes that have emerged from the substantial progress that has been made in our understanding of chemical modifications across many major RNA classes in eukaryotes.
KEYWORDS: Epitranscriptome, gene expression, post-transcriptional regulation, protein translation, RNA modification
Introduction
It has been known for decades that RNA from all kingdoms of life can be post-transcriptionally modified with over 100 chemical moieties. Initial studies were largely limited to more abundant modifications: N1-methyladenosine (m1A), pseudouridine (Ψ), and 2′-O-methylation (2′OMe) in tRNA and rRNA, and N6-methyladenosine (m6A), 5-methylcytidine (m5C), and 2′O methylation (2′OMe) in mRNA and viral RNA.1-3 This early research not only began the work of exhaustively cataloging these modifications, but also began to uncover the mechanisms by which these small chemical additions influence RNA folding and function. This field has experienced a recent resurgence, particularly with respect to mRNA modifications, driven in part by technological advances in high throughput sequencing and mass spectrometry that have allowed deeper analysis of less prevalent modifications on less abundant RNA species. With these tools in hand, the mechanisms by which RNA modifications influence cell biology are beginning to be elucidated.
The discovery and characterization of FTO as a mammalian mRNA m6A demethylase highlighted the idea that chemical modifications on RNA could represent a reversible and dynamic mode of post-transcriptional regulation.4 Using an antibody-based enrichment strategy followed by high throughput sequencing, the distribution of m6A sites was mapped throughout the mammalian transcriptome, revealing the characteristic topology in the 3′UTR that suggested its function.5,6 As we learn more about m6A and other mRNA modifications, their distributions in the transcriptome, and their functions in biology, it is becoming clear that different modifications in different sequence contexts provide an additional layer of complexity to the post-transcriptional regulation of gene expression. Ongoing work has also revealed new facets of the structural and functional roles that chemical modifications play on tRNA and rRNA. As we learn more about the networks that control their installation, interplay between mRNA, tRNA, and rRNA modification machinery has also been uncovered, adding a complex layer of regulation to protein translation. Rather than providing an exhaustive overview of our current understanding of RNA modifications, here we will focus primarily on recent studies that highlight emerging themes in the biology of chemical modifications in eukaryotic RNA.
mRNA
The presence of chemical modifications on mRNA was uncovered in the 1970s, when poly(A) tail-based purification techniques first allowed for sufficiently pure mRNA preparations to rule out contamination with other RNA species.1-3 These studies revealed the presence of internal m6A, m5C, and ribose 2′OMe (Fig. 1). The functional significance of these modifications in mRNA remained mysterious for decades, but recent studies have begun to shed light on how they influence cellular processes. The discovery of the first mRNA m6A demethylase FTO,4 quickly followed by a second demethylase ALKBH5,7 and the METTL3/METTL14 methyltransferase complex,8 brought mRNA modifications back to the forefront. The major implication of these discoveries was that mRNA modifications are likely reversible and regulated, renewing interest in uncovering their biological functions. Mapping of m6A sites using an antibody-based enrichment strategy followed by high throughput sequencing uncovered the transcriptome-wide topology of this modification, revealing enrichment around the stop codon and in the 3′UTR of thousands of transcripts.5,6 The exact numbers vary between studies, particularly between individual experiments, but the numbers of reproducible peaks seem to converge on approximately 13,000 sites in 5000–7000 genes, highlighting the need for consistent analysis of replicates. YTHDF2, the first m6A-binding protein to be characterized in detail, was shown to mediate an m6A-dependent reduction in half-life of its mRNA targets.5,9 Recent work has now revealed a molecular mechanism for this effect: YTHDF2 binds m6A and recruits the CCR4-NOT deadenylase complex to mRNA, resulting in CAF1- and CCR4-dependent deadenylation and subsequent degradation of m6A-containing transcripts.10
Figure 1.
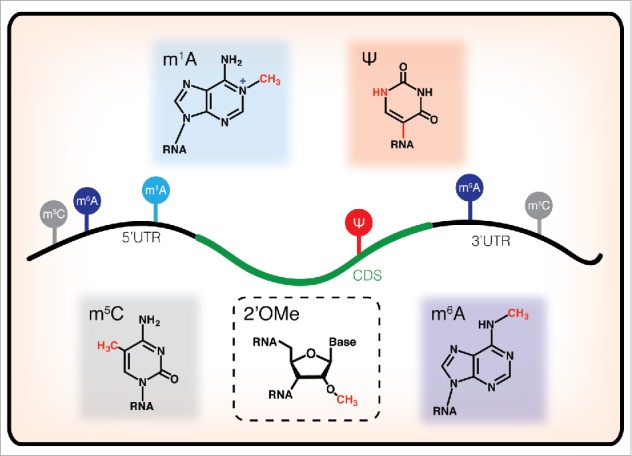
Chemical structures of RNA modifications currently characterized in mRNA, schematized with their reported localization along transcripts. m6A has known functional roles at both the 5′UTR and the 3′UTR, regulating mRNA translation and decay, respectively. m1A is localized primarily in the 5′UTR and early coding sequence, m5C is found primarily in untranslated regions, and Ψ is distributed throughout protein coding sequences, but their functions remain mysterious. While 2′OMe has been detected in mRNA samples, its distribution in the transcriptome has not yet been mapped.
Though initial reports highlighted its 3′ localization, m6A is also found in the 5′UTR, a region of mRNA closely tied to the regulation of translation. In fact, m6A in the 5′UTR mediates cap-independent translation through eIF3,11,12 a strikingly different effect from the decay pathway described above. Interestingly, this process also involves YTHDF2, so while m6A may mediate very different mRNA processing events depending on its context, specific components may be shared between pathways. In this second example, YTHDF2 is a protective factor against demethylation of m6A sites in the 5′UTR by FTO, rather than a direct link to processing machinery.12 This protection of 5′ m6A sites provides a route to selective mRNA translation during heat shock stress. Dynamic modification of RNAs in all major classes during stress responses is an emerging theme, which we will highlight throughout this review. It will also be interesting to see whether other YTH domain-containing proteins show the same versatility: YTHDF1 binds m6A in mRNA and promotes translation initiation,13 and recently YTHDC1 was shown to affect splicing as a nuclear m6A reader protein.14,15 It remains to be seen what other functions these proteins may have in cells.
At this stage, m6A is the most extensively characterized mRNA modification — we know the machinery that installs and removes it, m6A-specific binding proteins have been identified, and multiple biological systems in which it plays a role have been described.16 However, numerous other modifications have been added to the mRNA repertoire. m1A has been described in tRNA and rRNA,17,18 but we now know that it is also present in mRNA, albeit at levels approximately 10-fold lower than m6A.19,20 In the studies that first described its distribution in mRNA, one identified 7,154 peaks in 4,151 coding genes,19 while the other identified 887 peaks in 600 genes.20 The discrepancy is primarily due to methodological differences that inherently yield different levels of stringency, and that minimize spontaneous isomerization of m1A to m6A to different degrees. Li et al. employed an approach based on antibody enrichment followed by demethylation to identify demethylation-sensitive sites. The study by Dominissini et al., on the other hand, relied on antibody enrichment of freshly purified mRNA to call peaks. While this study did utilize the Dimroth rearrangement, which converts m1A to m6A, to reverse the stops and mutations introduced by reverse transcriptase upon encountering an m1A site, the rearrangement signature was not a requirement for calling an m1A peak. While its function in mRNA remains unclear, it is dynamic in response to nutrient starvation and heat shock, it has a characteristic localization on the 5′ end of transcripts, and it positively correlates with protein level. These features are all suggestive of a role in translation that has yet to be uncovered. While m1A tends to appear in relatively more structured and higher GC-content regions, NMR studies recently demonstrated its propensity to locally disrupt duplex formation, suggesting that structural changes in m1A-containing 5′UTRs may influence translation.21 This is consistent with the fact that m1A carries a positive charge at physiological pH, and that m1A sites have no clearly identifiable sequence motif.19,20
The most abundant RNA modification in cells overall, pseudouridine (Ψ), is generated through enzyme-mediated isomerization of uridine. On the order of 250 to 300 sites have now been mapped in yeast mRNA at single base resolution.22-24 In human, the reported number of mRNA Ψ sites varies widely from 96 in one study,23 to 353 in a second,22 and up to 2084 in a third study.25 This variation is likely the result of technical differences in experimental design and data analysis. While all three studies relied on the chemical reactivity of Ψ with N-cyclohexyl-N′-β-(4-methylmorpholinium) ethylcarbodiimide (CMC), Li et al. synthesized a biotinylated CMC derivative for an enrichment step, likely allowing them to identify low abundance sites that the other methods might miss. Though many of these sites have been attributed to specific enzyme activities, the biological function of Ψ has yet to be elucidated. In keeping with an emerging theme among mRNA modifications, however, Ψ is dynamic in heat shock, nutrient deprivation, and other stresses.22,23,25
While 5-methylcytidine (m5C) is best known for its central role as an epigenetic mark in DNA, it was also described in early studies of RNA methylation.3 Mapping of m5C in RNA was initially attempted by adapting the bisulfite sequencing technology used for m5C in DNA, leading to the identification of approximately 10,000 m5C sites in mRNA.26,27 Subsequently, new methods were developed that leverage m5C methyltransferases to enrich m5C-containing RNA, either by incorporating 5-azacytidine into RNA,28 or by mutating the methyltransferase29 to enable modified RNA capture. These methods identified far fewer m5C sites in mRNA, on the order of a few hundred to one thousand, respectively.30 It remains unclear whether this discrepancy is the result of a biological difference (since the latter two studies looked at only a single m5C methyltransferase, NSUN2), or if it is the result of technical differences. While its biological function remains unclear, some of the machinery that regulates m5C in different RNA classes has been identified: Trm4p (in yeast),31 and DNMT2 and NSUN2 (in human) have been identified as m5C methyltransferases.27,32 While the roles and specificities of these enzymes is controversial, their functional characterization will likely aid in uncovering the roles of m5C in the future.33-35 For instance, a recent study suggested functional roles for m5C oxidation to 5-hydroxymethylcytidine in D. melanogaster mRNA.33
Finally, while the majority of RNA modifications occur on the base moiety, methylation on the ribose 2′ hydroxyl to form 2′OMe has been found on all four ribonucleosides in many RNA classes. In eukaryotic rRNA, its installation is mediated by small nucleolar RNAs (snoRNAs) through a guide-RNA mediated process.36 Though its presence in mRNA was first described at the same time as m6A in 1974 (Fig. 1),1,2 its distribution in mRNA has not yet been reported. Recently, however, new methods have been developed to map 2′OMe utilizing either chemical strategies,37 or methylation-sensitive reverse transcriptase enzyme.38 Thus far these approaches have identified 2′OMe sites only in abundant RNA species, but future technical advances may eventually reveal the 2′OMe distribution in mRNA.
miRNA and lncRNA
N6-methylation of mRNA is thought to be a cotranscriptional process, leaving open the possibility that other RNA Polymerase II products also contain m6A. Indeed, a search for over-represented sequence motifs in miRNA-containing regions revealed that the primary consensus sequence for the m6A methyltransferase METTL3 is prevalent in these regions.39 METTL3 depletion and overexpression resulted in diminished and increased mature miRNA levels, respectively. This effect was recapitulated in in vitro processing reactions; methylated pre-let-7e was processed more efficiently than its unmethylated counterpart, indicating that methylation status influences processing.
The lncRNA XIST is also m6A-methylated, carrying a number of m6A residues that appear to be critical for its gene silencing activity.40 XIST is m6A-methylated by METTL3, which is recruited to XIST through the proteins RBM15 and RBM15B. In an inducible XIST expression system, loss of METTL3 allows for induction of XIST, but its gene silencing function is impaired. However, loss of m6A on XIST can be rescued by tethering the m6A-binding protein YTHDC1 directly to XIST, suggesting that this is another case in which the recruitment of a specific m6A-binding protein mediates the effect of m6A on a central cellular process.
While these are only two recent examples, they highlight how RNA modification status can influence both RNA processing and function through common machinery. In the context of mRNA, the m6A-binding protein YTHDC1 has been implicated in splicing,15 so it could represent another case in which an m6A-binding protein coordinates different cellular processes in different contexts.
Viral infection
The RNA species discussed in the previous sections are primarily products of RNA Polymerase II. Upon viral infection, host cell machinery is often hijacked to enable viral infection and replication, at the expense of normal cellular function. Late in their life cycle, for instance, retroviruses use RNA Polymerase II to transcribe viral DNA.41 Some RNA modifications, such as m6A, are thought to be installed contranscriptionally, perhaps one of the many processes coordinated by the RNA Polymerase II C-terminal repeat domain. The presence of m6A in viral RNA has, indeed, been known for decades, having been observed in B77 avian sarcoma virus, Rous sarcoma virus, simian virus 40, influenza, and adenoviruses in the 1970s.42-46 However, akin to the case with mRNA, the functions and regulation of RNA modifications in host-virus interactions have only recently begun to be unveiled. HIV-1 is a very illustrative example of this, as it exploits multiple RNA modifications to its advantage. In order to replicate, HIV-1 reverse transcriptase (RT) must synthesize a DNA intermediate using tRNALys3 as a primer for minus-strand strong-stop synthesis. During plus-strand strong-stop synthesis, however, N1-methylation at adenosine 58 (A58) in tRNALys3 is required to properly terminate DNA synthesis.47,48 Replacement of A58 with U, which is not methylated to produce the block, allows HIV-1 RT to read past this point and inhibits HIV-1 replication.
A trio of recent studies has now revealed that HIV-1 viral RNA itself is also m6A-methylated, and that proteins involved in m6A regulation influence viral infection.49-51 Though each study took different approaches and reaches different conclusions in some cases, they do converge in many respects. They find that HIV-1 RNA carries multiple m6A peaks that appear to be important in infection, and that the cellular host responds with infection-specific methylation of multiple transcripts potentially involved in the response to viral infection. Knockdown of the METTL3/METTL14 methyltransferase complex inhibits replication and viral release. Lichinchi et al. demonstrate that methylation of a specific site in the Rev response element (RRE) of the env gene increases its affinity for Rev, facilitating Rev's export function. Reducing methylation at this site by knocking down METTL3/METTL14 or by mutating the site substantially inhibits viral RNA export into the cytoplasm, an essential step in the HIV-1 replication cycle.50 Interestingly, Tirumuru et al. report a binding site for the m6A-binding protein YTHDF1 in this region. The proteins YTHDF1, YTHDF2 and YTHDF3, all established m6A-binding proteins, appear to negatively regulate HIV-1 viral reverse transcription and mRNA transcription in this study.51 It remains to be seen whether the YTH proteins play a role in the Rev-mediated export model described by Lichinchi et al. or whether they represent an independent effect. In the third study, however, the opposite effect was observed with respect to the YTHDF proteins, HIV-1 mRNA expression, and viral replication.49 The specific m6A sites reported in the three studies also differ, perhaps reflecting different cell lines and viral clones used in each study. Ongoing work will clarify the effect of RNA modifications on HIV-1 and explore their effects other viruses.
tRNA
Of all major RNA classes, tRNAs carry the largest complement of chemical modifications. Each molecule carries, on average, 14 modifications that contribute to its function.18,52 While a detailed description of all the known modifications on tRNA, their respective functions, and their regulatory machineries is beyond the scope of this overview, decades of work have revealed some interesting paradigms by which modifications influence tRNA function.
In some cases, tRNA modifications directly influence folding, and thereby, function. A classic example is m1A58, a highly conserved modification in tRNAs from all three domains of life. Detailed studies in S. cerevisiae reveal that it is present in 23 out of 34 tRNAs and is installed by the Trm6p/Trm61p methyltransferase complex.53 Disruption of Trm6p/Trm61p enzymatic activity results in a slow growth phenotype, driven largely by the degradation of unmethylated tRNAiMet. The situation in human is strikingly similar, and the human homologs TRMT6 and TRMT61A show 20% and 30% similarity to the yeast enzymes at the amino acid level, respectively.17,54 Supporting this conserved role, ALKBH1 was recently reported to demethylate m1A58 in human tRNAiMet as well as several other tRNA species.55 Loss of ALKBH1 increases the cellular tRNAiMet level, corroborating the idea that m1A58 methylation is essential for tRNAiMet stability. ALKBH1-mediated tRNA demethylation also attenuates the rate of protein synthesis. While m1A in tRNAiMet represents one of the few cases where a single tRNA modification is absolutely essential, there are many other cases where loss of one or more modifications results in misfolding and subsequent degradation.56,57
Since protein production relies on the availability of tRNA pools, tRNA maturation and processing are key factors in translation regulation. tRNA maturation is a complex process that involves multiple changes in subcellular localization, throughout which modifications are added and can act as markers of maturation. In the anti-codon loop, modified nucleosides can affect protein translation directly, both as a fundamental quality control mechanism and in response to cellular signaling or stress. Chemical modifications in the anti-codon stem loop region influence translation speed and fidelity by altering codon-anti-codon interactions and by controlling reading frame.58-61 In S. cerevisiae, Trm9p-mediated methylation to form 5-methylcarboxymethyluridine at the wobble positions of tRNAARG(UCU) and tRNAGLU(UUC) enhances translation of transcripts with higher levels of these codons upon DNA damage.62 In this case, the higher levels of these codons in ribonucleotide reductase 1 and 3 (Rnr1p and Rnr3p) allow for enhanced production of these key players in the DNA damage response. Thus the ability of modifications to influence translation directly can be implemented to bias the cell toward efficiently translating proteins required for a robust cellular response.
In addition to regulating the translation of specific mRNAs during stress responses, tRNAs can also be cleaved into second messenger signaling molecules. This process is also influenced by modifications — yeast Trm9p, and mammalian NSUN2 and DNMT2 have all been implicated in regulating cleavage through their methyltransferase activity.32,63-65 The resulting fragments have been demonstrated to bind and inhibit translation machinery,66 as well as bind and destabilize mRNAs directly.67 In mouse epidermal stem cells, the expression level of the RNA m5C methyltransferase NSUN2 is low but increases over the course of differentiation. This occurs with a concomitant increase in protein translation that is, counter intuitively, not correlated with increased proliferation. Deletion of NSUN2 results in tRNA hypomethylation, accumulation of tRNA fragments, and reduced translation, favoring the stem cell state.67 By this mechanism, loss of NSUN2 enhances the self-renewal of stem cells in a tumor setting as well, but it renders them sensitive to stress because they are unable to activate stress response pathways that require tRNA methylation. How tRNA fragments influence cell biology more broadly remains to be worked out. However, the fact that their accumulation has already been connected to neuronal loss,65 cancer progression,67 and stem cell differentiation68 suggests that this may represent a general mechanism by which tRNA modifications can influence cell signaling processes in both normal and disease states.
As more modification machinery is characterized and as sequencing technology continues to improve, the contributions of individual and groups of modifications will continue to be uncovered.69,70 Here we have laid out three mechanisms by which modifications influence tRNA biology — through effects on folding, by regulating mRNA decoding, and by influencing tRNA cleavage into potential signaling messengers — but there are likely many more. Given the evolutionary conservation of these modifications and the 1–2% of the genome that generates the proteins devoted to this task,71 we have likely only scratched the surface of tRNA biology and how it is influenced by the broad diversity of chemical modifications.
rRNA
In contrast to tRNA, rRNA carries a more limited repertoire of modifications. Given the ribosome's central role in all domains of life, some have speculated that perhaps these specific modifications were strongly selected for throughout evolution.71 Indeed, they tend to occur on highly conserved residues in areas of the ribosome central to its function. The majority of these modifications are Ψ and ribose 2′OMe, but m1A, N6,N6-dimethyladenosine (m6,6A), m5C, 3-methyluridine (m3U), N7-methylguanosine (m7G), and N4-acetylcytidine (ac4C), among others, have also been found in eukaryotic rRNAs.18 Despite the smaller variety, rRNA is heavily modified, with human rRNAs estimated to carry 94 2′OMe and 95 Ψ.
Mapping rRNA modifications onto the structure of the yeast ribosome has yielded hints as to their function in this complex machinery. In the small subunit, two m6,6A and an ac4C surround the decoding site where tRNAs read out mRNA sequence, while in the large subunit, an m1A, m5C, and m3U sit near the catalytic peptidyl transferase center.36 This structural information also suggests that intersubunit contacts involving rRNA modifications and ribosomal proteins may be involved in the complex motion required for successful mRNA translation.
While many RNA modifications occur in highly conserved areas, there is heterogeneity at some sites that alter ribosome composition. With the development of RiboMeth-seq, we can now estimate the stoichiometry of 2′OMe in rRNA in S. cerevisiae.37 While the majority of sites are fully methylated, a small proportion (8 out of 54) is methylated at less than 90%. Knockdown of specific guide RNAs unexpectedly changes the level of not only the modification specified by that guide, but of others as well, implying interdependent and potentially co-regulated processes. Hypermethylation of at least one 28S rRNA site in an aggressive breast cancer cell line is correlated with lower IRES-dependent translation and disregulated codon-anti-codon recognition.73 Similarly, loss of the pseuodouridine synthase Cbf5p in yeast also results in decreased tRNA binding, loss of translational fidelity, and reduction in IRES-mediated translation, phenotypes that are also evident in mouse and human cells upon loss of DKC1.74,75
While there is mounting evidence that chemical modifications in rRNA are crucial to ribosome biogenesis and to maintenance of accurate and efficient protein synthesis, not the least of which is its strong conservation throughout evolution, many questions remain as to their mechanisms of action. As more details regarding rRNA modifications and the cellular machinery that controls them are revealed, however, we must take care in attributing function to the modifications themselves versus the enzymes that install them. It has now been well established both in yeast and human systems that, in some cases, enzymes bind to rRNA early in biogenesis, but do not actually catalyze a reaction until a later phase of maturation.76,77 Similarly, in some cases enzymes modify RNAs in multiple classes,78 so loss of function phenotypes must be analyzed with care. Thus it is crucial that, as researchers delve deeper into the functions of modifications in all RNA classes, the roles of the enzymes and the modifications themselves are disentangled.79
Discussion
RNA modifications have been a fascinating mystery since they were discovered decades ago, and despite extensive progress in characterizing their distributions and functional roles, we are only scratching the surface of how they influence cell biology. In this overview, we have tried to highlight recent discoveries that illustrate themes in the emerging biology of RNA modifications. While modifications have long been studied in the context of tRNA and rRNA, over the last 5–6 y there has been a renewed focus on mRNA modifications. The most well characterized, m6A, has now been shown to play a role in mRNA decay,9 translation,11,13 splicing,5,15 and alternative polyadenylation,80 the circadian clock,81 stem cell differentiation,82 and heat shock response,12 among others. The transcriptome-wide distributions of other mRNA modifications such as Ψ, m1A, and m5C have recently been mapped, and we will likely see more in the near future. While the biological functions of these more recently characterized modifications remain unclear, a unifying theme among them all is that their levels and/or distribution change in response to stresses and nutrient deprivation. Moreover, tRNA and rRNA modifications change in response to stress as well, suggesting that RNA modifications may represent a rapid way to regulate protein translation in response to cellular signals. There are now reported examples where the protein machinery involved in regulating RNA modification levels affects multiple classes of RNA, so it is possible that this crosstalk could help to coordinate mRNA, tRNA and rRNA function simultaneously.78 It has also become clear that depending on the context of the modification and its localization, for instance in the 5′UTR or 3′UTR of a transcript, the same protein can coordinate with different cellular machinery and result in very different outcomes.10,11
While substantial progress has been made in mapping RNA modification sites with high throughput sequencing-based approaches, some major challenges remain. First, in the majority of cases, even if single base resolution can be achieved, we cannot determine the absolute stoichiometry of modification at a given site. Second, while there is overlap in transcripts reported with multiple different modifications, we cannot dissect whether different pools of the same transcript are differentially modified, let alone parse out how combinations of modifications influence function. Third, current approaches all require a relatively large amount of input material, preventing studies of rare cell populations and clinical samples. The future of the field will likely involve progress on two fronts: on the one hand, developing techniques that will allow us to map modifications at single base resolution with stoichiometric information, and on the other, uncovering the mechanisms by which modifications regulate additional biological processes.
Disclosure of potential conflicts of interest
The authors declare no competing interests.
Acknowledgments
The authors apologize to colleagues whose work was not cited owing to space limitations. This work was supported by National Institutes of Health HG008688 and GM071440 (C.H.). C.H. is an investigator of the Howard Hughes Medical Institute (HHMI). S.N. is an HHMI fellow of the Damon Runyon Cancer Research Foundation (DRG-2215–15).
References
- 1.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A 1974; 71:3971-5; PMID:4372599; http://dx.doi.org/ 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry RP, Kelley DE. Existence of Methylated Messenger RNA in Mouse L Cells. Cell 1974; 1:37-42; http://dx.doi.org/ 10.1016/0092-8674(74)90153-6 [DOI] [Google Scholar]
- 3.Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res 1975; 2:1653-68; PMID:1187339; http://dx.doi.org/ 10.1093/nar/2.10.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011; 7:885-7; PMID:22002720; http://dx.doi.org/ 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201-6; PMID:22575960; http://dx.doi.org/ 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 6.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012; 149:1635-46; PMID:22608085; http://dx.doi.org/ 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al.. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013; 49:18-29; PMID:23177736; http://dx.doi.org/ 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014; 10:93-5; PMID:24316715; http://dx.doi.org/ 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014; 505:117-20; PMID:24284625; http://dx.doi.org/ 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun 2016; 7:12626; PMID:27558897; http://dx.doi.org/ 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015; 163:999-1010; PMID:26593424; http://dx.doi.org/ 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015; 526:591-4; PMID:26458103; http://dx.doi.org/ 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015; 161:1388-99; PMID:26046440; http://dx.doi.org/ 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 2014; 10:927-9; PMID:25242552; http://dx.doi.org/ 10.1038/nchembio.1654 [DOI] [PubMed] [Google Scholar]
- 15.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al.. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell 2016; 61:507-19; PMID:26876937; http://dx.doi.org/ 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 16.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 2014; 15:293-306; PMID:24662220; http://dx.doi.org/ 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 17.Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 2005; 11:1281-90; PMID:16043508; http://dx.doi.org/ 10.1261/rna.5040605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al.. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262-7; PMID:23118484; http://dx.doi.org/ 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al.. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016; 530:441-6; PMID:26863196; http://dx.doi.org/ 10.1038/nature16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol 2016; 12:311-6; PMID:26863410; http://dx.doi.org/ 10.1038/nchembio.2040 [DOI] [PubMed] [Google Scholar]
- 21.Zhou H, Kimsey IJ, Nikolova EN, Sathyamoorthy B, Grazioli G, McSally J, Bai T, Wunderlich CH, Kreutz C, Andricioaei I, et al.. m(1)A and m(1)G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat Struct Mol Biol 2016; 23:803-10; PMID:27478929; http://dx.doi.org/ 10.1038/nsmb.3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al.. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014; 159:148-62; PMID:25219674; http://dx.doi.org/ 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014; 515:143-6; PMID:25192136; http://dx.doi.org/ 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 2014; 9:e110799; PMID:25353621; http://dx.doi.org/ 10.1371/journal.pone.0110799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 2015; 11:592-7; PMID:26075521; http://dx.doi.org/ 10.1038/nchembio.1836 [DOI] [PubMed] [Google Scholar]
- 26.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 2009; 37:e12; PMID:19059995; http://dx.doi.org/ 10.1093/nar/gkn954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 2012; 40:5023-33; PMID:22344696; http://dx.doi.org/ 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 2013; 31:458-64; PMID:23604283; http://dx.doi.org/ 10.1038/nbt.2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et al.. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 2013; 4:255-61; PMID:23871666; http://dx.doi.org/ 10.1016/j.celrep.2013.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol 2013; 14:215; PMID:24286375; http://dx.doi.org/ 10.1186/gb4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA 1999; 5:1105-18; PMID:10445884; http://dx.doi.org/ 10.1017/S1355838299982201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010; 24:1590-5; PMID:20679393; http://dx.doi.org/ 10.1101/gad.586710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006; 311:395-8; PMID:16424344; http://dx.doi.org/ 10.1126/science.1120976 [DOI] [PubMed] [Google Scholar]
- 34.Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA 2008; 14:1663-70; PMID:18567810; http://dx.doi.org/ 10.1261/rna.970408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev 2007; 21:261-6; PMID:17289917; http://dx.doi.org/ 10.1101/gad.1472907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma S, Lafontaine DL. ‘View From A Bridge’: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem Sci 2015; 40:560-75; PMID:26410597; http://dx.doi.org/ 10.1016/j.tibs.2015.07.008 [DOI] [PubMed] [Google Scholar]
- 37.Birkedal U, Christensen-Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H. Profiling of Ribose Methylations in RNA by High-Throughput Sequencing. Angew Chem Int Edit 2015; 54:451-5; PMID:25417815; http://dx.doi.org/ 10.1002/anie.201408362 [DOI] [PubMed] [Google Scholar]
- 38.Aschenbrenner J, Marx A. Direct and site-specific quantification of RNA 2′-O-methylation by PCR with an engineered DNA polymerase. Nucleic Acids Res 2016; 44:3495-502; PMID:27016740; http://dx.doi.org/ 10.1093/nar/gkw200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N-6-methyladenosine marks primary microRNAs for processing. Nature 2015; 519:482-+; PMID:25799998; http://dx.doi.org/ 10.1038/nature14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016; 537:369-73; PMID:27602518; http://dx.doi.org/ 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goff SP. Host factors exploited by retroviruses. Nat Rev Microbiol 2007; 5:253-63; PMID:17325726; http://dx.doi.org/ 10.1038/nrmicro1541 [DOI] [PubMed] [Google Scholar]
- 42.Beemon K, Keith J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol 1977; 113:165-79; PMID:196091; http://dx.doi.org/ 10.1016/0022-2836(77)90047-X [DOI] [PubMed] [Google Scholar]
- 43.Canaani D, Kahana C, Lavi S, Groner Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res 1979; 6:2879-99; PMID:223130; http://dx.doi.org/ 10.1093/nar/6.8.2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dimock K, Stoltzfus CM. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry 1977; 16:471-8; PMID:189800; http://dx.doi.org/ 10.1021/bi00622a021 [DOI] [PubMed] [Google Scholar]
- 45.Hashimoto SI, Green M. Multiple methylated cap sequences in adenovirus type 2 early mRNA. J Virol 1976; 20:425-35; PMID:978798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J Virol 1976; 20:45-53; PMID:1086370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell 1979; 18:93-100; PMID:509527; http://dx.doi.org/ 10.1016/0092-8674(79)90357-X [DOI] [PubMed] [Google Scholar]
- 48.Renda MJ, Rosenblatt JD, Klimatcheva E, Demeter LM, Bambara RA, Planelles V. Mutation of the methylated tRNA(Lys)(3) residue A58 disrupts reverse transcription and inhibits replication of human immunodeficiency virus type 1. J Virol 2001; 75:9671-8; PMID:11559799; http://dx.doi.org/ 10.1128/JVI.75.20.9671-9678.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al.. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 2016; 19:675-85; PMID:27117054; http://dx.doi.org/ 10.1016/j.chom.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 2016; 1:16011; PMID:27572442; http://dx.doi.org/ 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 2016; 5:e15528.; PMID:27371828; http://dx.doi.org/ 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010; 24:1832-60; PMID:20810645; http://dx.doi.org/ 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 1998; 12:3650-62; PMID:9851972; http://dx.doi.org/ 10.1101/gad.12.23.3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozanick SG, Bujnicki JM, Sem DS, Anderson JT. Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 Mtase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res 2007; 35:6808-19; PMID:17932071; http://dx.doi.org/ 10.1093/nar/gkm574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016; 167:816-28; PMID:27745969; http://dx.doi.org/ 10.1016/j.cell.2016.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexandrov A, Chernyakov I, Gu WF, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Molecular Cell 2006; 21:87-96; PMID:16387656; http://dx.doi.org/ 10.1016/j.molcel.2005.10.036 [DOI] [PubMed] [Google Scholar]
- 57.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5 ′–3 ′ exonucleases Rat1 and Xrn1. Gene Dev 2008; 22:1369-80; PMID:18443146; http://dx.doi.org/ 10.1101/gad.1654308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rezgui VA, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PG. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci U S A 2013; 110:12289-94; PMID:23836657; http://dx.doi.org/ 10.1073/pnas.1300781110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tukenmez H, Xu H, Esberg A, Bystrom AS. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res 2015; 43:9489-99; PMID:26283182; http://dx.doi.org/ 10.1093/nar/gkv832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hori H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front Genet 2014; 5:144; PMID:24904644; http://dx.doi.org/ 10.3389/fgene.2014.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gustilo EM, Vendeix FA, Agris PF. tRNA's modifications bring order to gene expression. Curr Opin Microbiol 2008; 11:134-40; PMID:18378185; http://dx.doi.org/ 10.1016/j.mib.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 2007; 28:860-70; PMID:18082610; http://dx.doi.org/ 10.1016/j.molcel.2007.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klassen R, Paluszynski JP, Wemhoff S, Pfeiffer A, Fricke J, Meinhardt F. The primary target of the killer toxin from Pichia acaciae is tRNA(Gln). Mol Microbiol 2008; 69:681-97; PMID:18532979; http://dx.doi.org/ 10.1111/j.1365-2958.2008.06319.x [DOI] [PubMed] [Google Scholar]
- 64.Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem 2012; 287:42708-25; PMID:23086926; http://dx.doi.org/ 10.1074/jbc.M112.371799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al.. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 2014; 33:2020-39; PMID:25063673; http://dx.doi.org/ 10.15252/embj.201489282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011; 43:613-23; PMID:21855800; http://dx.doi.org/ 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015; 161:790-802; PMID:25957686; http://dx.doi.org/ 10.1016/j.cell.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N, et al.. Stem cell function and stress response are controlled by protein synthesis. Nature 2016; 534:335-40; PMID:27306184; http://dx.doi.org/ 10.1038/nature18282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 2015; 12:835-7; PMID:26214130; http://dx.doi.org/ 10.1038/nmeth.3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 2015; 12:879-84; PMID:26237225; http://dx.doi.org/ 10.1038/nmeth.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bjork GR. Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog Nucleic Acid Res Mol Biol 1995; 50:263-338; PMID:7538683; http://dx.doi.org/ 10.1016/S0079-6603(08)60817-X [DOI] [PubMed] [Google Scholar]
- 72.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2011; 2:611-31; PMID:21823225; http://dx.doi.org/ 10.1002/wrna.79 [DOI] [PubMed] [Google Scholar]
- 73.Belin S, Beghin A, Solano-Gonzalez E, Bezin L, Brunet-Manquat S, Textoris J, Prats AC, Mertani HC, Dumontet C, Diaz JJ. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One 2009; 4:e7147; PMID:19779612; http://dx.doi.org/ 10.1371/journal.pone.0007147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, et al.. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 2011; 44:660-6; PMID:22099312; http://dx.doi.org/ 10.1016/j.molcel.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 2006; 312:902-6; PMID:16690864; http://dx.doi.org/ 10.1126/science.1123835 [DOI] [PubMed] [Google Scholar]
- 76.Letoquart J, Huvelle E, Wacheul L, Bourgeois G, Zorbas C, Graille M, Heurgue-Hamard V, Lafontaine DL. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc Natl Acad Sci U S A 2014; 111:E5518-26; PMID:25489090; http://dx.doi.org/ 10.1073/pnas.1413089111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lafontaine D, Vandenhaute J, Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev 1995; 9:2470-81; PMID:7590228; http://dx.doi.org/ 10.1101/gad.9.20.2470 [DOI] [PubMed] [Google Scholar]
- 78.Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, Blumberg A, Schlesinger O, Bieri P, Greber B, et al.. Mitochondrial 16S rRNA Is Methylated by tRNA Methyltransferase TRMT61B in All Vertebrates. PLoS Biol 2016; 14:e1002557; PMID:27631568; http://dx.doi.org/ 10.1371/journal.pbio.1002557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zorbas C, Nicolas E, Wacheul L, Huvelle E, Heurgue-Hamard V, Lafontaine DL. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol Biol Cell 2015; 26:2080-95; PMID:25851604; http://dx.doi.org/ 10.1091/mbc.E15-02-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al.. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev 2015; 29:2037-53; PMID:26404942; http://dx.doi.org/ 10.1101/gad.269415.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al.. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013; 155:793-806; PMID:24209618; http://dx.doi.org/ 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- 82.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al.. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015; 347:1002-6; PMID:25569111; http://dx.doi.org/ 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]


