Abstract
We detected the mcr-1 gene in 21 (14.8%) Salmonella isolates from pigs at slaughter; 19 were serovar Typhimurium sequence type 34. The gene was located on IncHI2-like plasmids that also harbored IncF replicons and lacked a conjugative transfer region. These findings highlight the need to prevent further spread of colistin resistance in animals and humans.
Keywords: mcr-1 gene, colistin, Salmonella enterica serovar Typhimurium ST34, bacteria, antimicrobial resistance, clonal spread, sequence type, pigs, slaughter, food safety, zoonoses, China
Since our report of emergence of a plasmid-mediated colistin resistance mechanism involving the mcr-1 gene in Escherichia coli (1), mcr-1 has been found in >30 countries in 5 continents in <1 year (2). E. coli is the main host of mcr-1, although several other Enterobacteriaceae, including Salmonella, have also been implicated as carriers of this gene (3,4). We screened pigs at slaughter for Salmonella isolates to elucidate the epidemiology of mcr-1 in this major foodborne pathogen, which a serious public health problem.
The Study
During July 2013–May 2014, a total of 1,780 cecum samples were obtained from pigs at 5 slaughter houses in southern and central China. Samples were incubated in buffered peptone water for 20 h and then inoculated into chromogenic medium selective for Salmonella spp. (CHROMagar Microbiology, Paris, France) for 24 h. Suspected Salmonella colonies were selected (1 isolate was selected from each sample) and identified by using PCR for detection of the invA gene.
We obtained 142 Salmonella isolates and screened them for mcr-1 by using PCR and sequencing with primers described (1); 21 (14.8%) were positive for mcr-1. These isolates were serotyped as described (5). We determined the clonal relationship of mcr-1–positive isolates by using pulsed-field gel electrophoresis (PFGE) with XbalI restriction enzyme and identified sequence types (STs) of these isolates by using multilocus sequence typing (http://mlst.warwick.ac.uk/mlst/dbs/Senterica).
Most of the isolates had indistinguishable PFGE patterns and were clonally related to serovar Typhimurium ST34 (n = 19), which was the dominant type (Table). ST34 is common in isolates from humans in China and other countries and has been linked to food-producing animals (6,7).
Table. Characteristics of mcr-1–positive Salmonella isolates from pigs at slaughter, China, 2013–2014*.
| Strain | Serovar | PFGE type | Sequence type | S1-PFGE, plasmid, kb | Colistin MIC, mg/L | Polymyxin B MIC, mg/L | Other antimicrobial drug resistance† | Resistance gene‡ |
|---|---|---|---|---|---|---|---|---|
| SH149 | Typhimurium | A5 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH143 | Typhimurium | A1 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH138 | Typhimurium | A1 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH93 | Typhimurium | A1 | 34 | ≈180 | 2 | 8 | FFC, SXT, TET | floR, oqxAB |
| SH33 | Typhimurium | A1 | 34 | ≈110 | 2 | 4 | AMP, STR, SXT, TET | oqxAB |
| SH137 | Typhimurium | A1 | 34 | ≈180 | 2 | 8 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SA316 | Typhimurium | A1 | 34 | ≈180 | 2 | 8 | AMP, STR, GEN, FFC, SXT, TET | floR |
| SH127 | Typhimurium | A1 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH128 | Typhimurium | A1 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH133 | Typhimurium | A1 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH134 | Typhimurium | A1 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH17 | Typhimurium | A1 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH239 | Typhimurium | A1 | 34 | ≈180, ≈138 | 2 | 8 | AMP, STR, GEN, FFC, SXT, TET | floR |
| SH271 | Typhimurium | A1 | 34 | ≈180 | 2 | 8 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH83 | Typhimurium | A1 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH205 | Typhimurium | A1 | 34 | ≈180 | 2 | 8 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH39 | Typhimurium | A2 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH178 | Typhimurium | A1 | 34 | ≈180 | 2 | 4 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH175§ | Typhimurium | A4 | 34 | ≈180 | 2 | 8 | AMP, STR, GEN, FFC, SXT, TET | floR, oqxAB |
| SH36§ | Heidelberg | B | ND | ≈180 | 2 | 8 | STR, FFC, SXT | floR, oqxAB |
| Z4P319S§ | London | C | ND | ≈180 | 1 | 2 | AMP, STR, GEN, FFC,CIP, SXT, TET | floR, oqxAB |
*According to recommendations of the joint Clinical and Laboratory Standards Institute (CLSI)–European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Polymyxin Breakpoints Working Group (http://www.eucast.org/), the International Organization for Standardization standard broth microdilution method (20776–1) was used to determine colistin MICs for these strains. Antimicrobial susceptibility was determined and evaluated according to CLSI document no. M100-S25 (http://shop.clsi.org/site/Sample_pdf/M100S25_sample.pdf). Resistance to florfenicol was interpreted according to EUCAST clinical breakpoints and epidemiological cutoff values (>16 mg/L) (http://mic.eucast.org/Eucast2/). MICs of olaquindox for all isolates, except SH36, were >64 mg/L. AMP, ampicillin; CIP, ciprofloxacin; FFC, florfenicol; GEN, gentamicin; ND, not determined; STR, streptomycin; SXT, sulfamethoxazole/trimethoprim; TET, tetracycline. †Boldface indicates resistance phenotypes transferred to a recipient by transformation. ‡Boldface indicates resistance genes transferred to a recipient by transformation. §Original strains of transformants.
We determined MICs for 12 antimicrobial drugs for all mcr-1–positive isolates by using agar dilution methods or a broth microdilution method. Colistin showed MICs of 1–2 mg/L for these isolates (Table). More than 80% of the isolates were also resistant to ampicillin, streptomycin, florfenicol, tetracycline, sulfamethoxazole/trimethoprim, and gentamicin; 76.2% showed reduced susceptibility (MIC >0.06 mg/L) to ciprofloxacin.
We identified 2 lipopolysaccharide regulatory genes (pmrA and pmrB) by using and PCR and sequencing (8). Plasmid-mediated resistance genes floR (florfenicol resistance) and oqxAB (olaquindox and ciprofloxacin resistance) were detected by using PCR. None of the 21 isolates had mutations in the pmrA and pmrB genes, which are associated with colistin resistance in Salmonella isolates. A total of 20 and 19 isolates had the floR and oqxAB genes, respectively. Salmonella Typhimurium ST34 has also been associated with the spread of the oqxAB gene in human clinical isolates in China (9), and Salmonella Typhimurium ST34 coproducing oqxAB or floR genes has also been isolated from humans in Europe and Canada (4,10).
To determine the genetic context of mcr-1, we used PCR mapping with primers ISAP-F (5′-CGAAGCACCAAGACATCA-3′) and MCR-R (5′-CCACAAGAACAAA CGGACT-3′). Insertion sequence ISApl1 was found upstream of mcr-1 for all isolates.
We determined transferability and location of mcr-1 by using conjugation, transformation, and hybridization (with S1-PFGE nuclease). These procedures showed that mcr-1 genes were located on an ≈180-kb plasmid, except for those in strains SH33 (≈110-kb plasmid) and SH239 (≈180-kb plasmid and ≈138-kb plasmid) (Table). However, the mcr-1 gene could not be transferred to E. coli C600 by conjugation.
We then randomly selected 6 mcr-1–positive Salmonella isolates (SH138, SH149, SH175, SH36, SH39, and Z4P319S) for chemical transformation, which was successful for SH175, SH36, and Z4P319S. S1-PFGE confirmed that transformants harbored single plasmids and had 8–16-fold higher MICs for colistin than the recipient E. coli DH5α. The floR and oqxAB genes were also transferred with the mcr-1 gene.
We used Hiseq Technology (Illumina, San Diego, CA, USA) to sequence plasmid DNA purified from transformants of Salmonella Heidelberg SH36 and Salmonella London Z4P319S and genomic DNA extracted from the original isolate (Salmonella Typhimurium SH138, which was selected as a representative ST34 strain). We assembled sequence reads were assembled into contigs by using SOAPdenovo version 2.04 (http://soap.genomics.org.cn/soapdenovo.html) and the separated plasmid contigs of the 3 mcr-1–carrying plasmids from chromosomal contigs and compared them with pHNSHP45–2 by using blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and BRIG (11). We then analyzed replicon sequence types of these plasmids by using the Plasmid MLST Database (http://pubmlst.org/plasmid/) and performed analysis and annotation of the partial sequence of mcr-1–carrying plasmids by using the RAST Server (12), ISfinder (https://www-is.biotoul.fr), BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and Gene Construction Kit 4.0 (Textco BioSoftware, Inc., West Lebanon, NH, USA).
For the 3 isolates, we found mcr-1 on derivatives of IncHI2-like plasmids, which also harbored IncF (F4) and IncFIB (B53) replicons. The 3 plasmids were named pHNSH36 (from strain SH36), pHNZ319S (from strain Z4P319S), and pHNSH138 (from strain SH138). IncHI2 plasmids, such as pHNSHP45–2, have been frequently associated with global spread of mcr-1 genes (3,13,14).
Compared with pHNSHP45–2 (in E. coli isolated from a pig in China [14]), the 3 plasmids all had a typical IncHI2-type backbone coding replication, transfer, maintenance, and stability functions, and a multiresistance region. However, we did not identify conjugative transfer region 1 of IncHI2 plasmids, including traJGIH and trhRYXFHG, in the 3 plasmids (Figure 1, panel A), which might be the reason why these plasmids were not transferred by conjugation.
Figure 1.
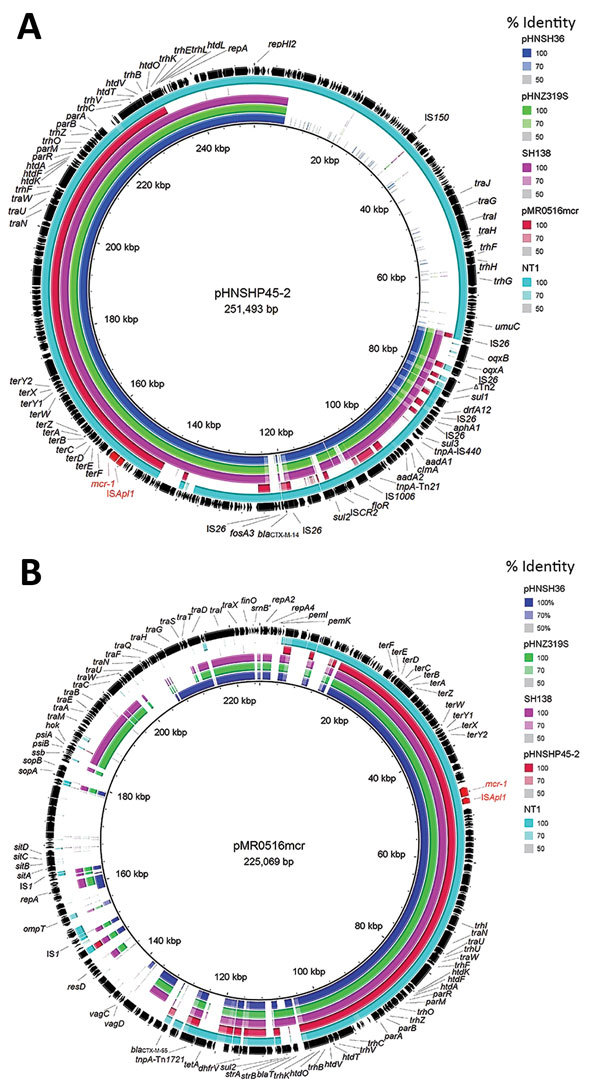
Sequence comparison of scaffolds (portions of genome sequences reconstructed from end-sequenced whole-genome clones) identified in mcr-1–positive plasmids pHNSH36, pHNZ319S, and pHNSH138 with 2 mcr-1–bearing plasmids pHNSHP45-2 (GenBank accession no. KU341381) and pMR0516mcr (GenBank accession no. KX276657), and contigs identified in mcr-1–positive genomes of Escherichia coli strain NT1 in BRIG (11) (GenBank accession LSBW01000090.1) obtained during analysis of mcr-1–positive Salmonella isolates from pigs at slaughter, China, 2013–2014. Arrows indicate positions and direction of transcription of genes. Reference plasmids pHNSHP45–2 (A) and pMR0516mcr (B) are indicated in black in the outer circles.
The multiresistance region contained numerous resistance genes, such as oqxAB, floR, sul1, cmlA, aadA2, and complete or truncated insertion sequences and transposons (IS26, ΔTn2, Tn21, IS1006, and ISCR2), which was similar to that of pHNSHP45-2, except that pHNSH36, pHNZ319S, and pHNSH138 were missing fosA3 and blaCTX-M-14 (Figure 1, panel A). The 2,635-bp module (mcr-1-ΔISApl1) was similar to that of pHNSHP45-2, but different in some aspects, such as insertion sites and orientation, which were identical to those of the novel IncHI2-IncF plasmid pMR0516mcr (GenBank accession no. KX276657) found in a clinical E. coli isolate from the United States (15), and human E. coli strain TN1 from Vietnam (GenBank accession no. LSBW01000090.1) (Figure 2).
Figure 2.
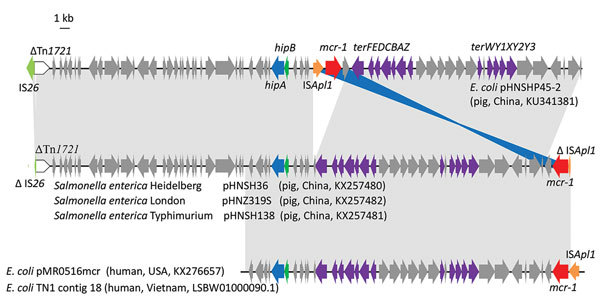
Genetic organization of scaffolds (portions of genome sequences reconstructed from end-sequenced whole-genome clones) containing mcr-1 harbored by plasmids pHNSH36, pHNZ319S, and pHNSH138 obtained during analysis of mcr-1–positive Salmonella isolates from pigs at slaughter, China, 2013–2014, and structural comparison with plasmids pHNSHP45–2, pMR0516mcr, and Escherichia coli TN1 contig 18. Arrows indicate positions and direction of transcription of genes. Regions with >99% homology are indicated in gray or blue. Triangles indicate truncated genes. Information in parentheses after isolates indicate source, location, and GenBank accession number.
Similar to pMR0516mcr (IncHI2-F18:A-:B1) (Figure 1, panel B), the 3 plasmids also contained an IncF-type backbone, including regions coding replication and partial regions coding transfer, maintenance, and stability functions. Similar to pHNSHP45-2, the plasmid carrying mcr-1 in strain NT1 belonged to the ST3-IncHI2 plasmid group. However, pMR0516mcr and the 3 plasmids described in this study were not typeable because there were no smr0199 loci in the IncHI2 plasmid (Figure 1, panel B). These data suggest that the mcr-1-ISApl1 module might have first been inserted into IncHI2 plasmids and that recombination between IncF- and IncHI2-type plasmids might have occurred subsequently, resulting in acquisition of the IncF-backbone fragment and deletion of the IncHI2 transfer region 1 or combination with the replication region in some instances.
Conclusions
We found that spread of mcr-1 in pigs at slaughter in China was associated with clonal dissemination of Salmonella Typhimurium ST34. This organism has also been detected in Portugal and England (3,4). The presence of indistinguishable IncHI2-F4:A-:B5 plasmids in different Salmonella serovars indicates that horizontal transfer of mcr-1–harboring plasmids might have also contributed to spread of mcr-1 in Salmonella spp.
Other drug-resistance genes, such as floR and oqxAB, were always transferred with mcr-1 by IncHI2-F4:A-:B5 plasmids, which led to successful flow of mcr-1–harboring Salmonella isolates under various selective pressures because florfenicol and olaquindox are widely used in farm animals in China. In addition, spread of similar IncHI2-like plasmids and Salmonella Typhimurium ST34 clones carrying mcr-1 in different countries highlights the need for internationally coordinated intervention strategies to limit its further dissemination.
Acknowledgments
This study was supported in part by grants from the National Key Basic Research Program of China (no. 2013CB127200) and the National Natural Science Foundation of China (no. 31272610 and no. 31625026).
Biographies
Y.D. has served on advisory boards for Shionogi, Meiji Seika Pharma, Tetraphase Pharmaceuticals, and Merck, and has received research funding from Merck and The Medicines Company for studies not related to this study.
Ms. Yi is a researcher at the College of Veterinary Medicine, South China Agricultural University, Guangdong, China. Her primary research interest is microbiology, including antimicrobial resistance genes and their means of dissemination.
Footnotes
Suggested citation for this article: Yi L, Wang J, Gao Y, Liu Y, Doi Y, Wu R, et al. mcr-1−harboringSalmonella enterica serovar Typhimurium sequence type 34 in pigs, China. Emerg Infect Dis. 2017 Feb [date cited].
These authors contributed equally to this article.
References
- 1.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 2.Schwarz S, Johnson AP. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother. 2016;71:2066–70. 10.1093/jac/dkw274 [DOI] [PubMed] [Google Scholar]
- 3.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. 2016;71:2300–5. 10.1093/jac/dkw093 [DOI] [PubMed] [Google Scholar]
- 4.Campos J, Cristino L, Peixe L, Antunes P. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. 2016;21:30270. 10.2807/1560-7917.ES.2016.21.26.30270 [DOI] [PubMed] [Google Scholar]
- 5.Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galán JE, Ginocchio C, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–9. 10.1016/0890-8508(92)90002-F [DOI] [PubMed] [Google Scholar]
- 6.Sun J, Ke B, Huang Y, He D, Li X, Liang Z, et al. The molecular epidemiological characteristics and genetic diversity of Salmonella Typhimurium in Guangdong, China, 2007-2011. PLoS One. 2014;9:e113145. 10.1371/journal.pone.0113145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antunes P, Mourão J, Pestana N, Peixe L. Leakage of emerging clinically relevant multidrug-resistant Salmonella clones from pig farms. J Antimicrob Chemother. 2011;66:2028–32. 10.1093/jac/dkr228 [DOI] [PubMed] [Google Scholar]
- 8.Quesada A, Porrero MC, Téllez S, Palomo G, García M, Domínguez L. Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enterica isolated from poultry and swine. J Antimicrob Chemother. 2015;70:71–4. 10.1093/jac/dku320 [DOI] [PubMed] [Google Scholar]
- 9.Wong MH, Yan M, Chan EW, Liu LZ, Kan B, Chen S. Expansion of Salmonella Typhimurium ST34 clone carrying multiple resistance determinants in China. Antimicrob Agents Chemother. 2013;57:4599–601. 10.1128/AAC.01174-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulvey MR, Finley R, Allen V, Ang L, Bekal S, El Bailey S, et al. Emergence of multidrug-resistant Salmonella enterica serotype 4,[5],12:i:- involving human cases in Canada: results from the Canadian Integrated Program on Antimicrobial Resistance Surveillance (CIPARS), 2003-10. J Antimicrob Chemother. 2013;68:1982–6. 10.1093/jac/dkt149 [DOI] [PubMed] [Google Scholar]
- 11.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhi C, Lv L, Yu LF, Doi Y, Liu JH. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16:292–3. 10.1016/S1473-3099(16)00063-3 [DOI] [PubMed] [Google Scholar]
- 14.Brennan E, Martins M, McCusker MP, Wang J, Alves BM, Hurley D, et al. Multidrug-resistant Escherichia coli in bovine animals, Europe. Emerg Infect Dis. 2016;22:1650–2. 10.3201/eid2209.160140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, et al. Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: First Report of mcr-1 in the United States. Antimicrob Agents Chemother. 2016;60:4420–1. 10.1128/AAC.01103-16 [DOI] [PMC free article] [PubMed] [Google Scholar]


