Abstract
Small nucleolar RNAs (snoRNAs) are conserved noncoding RNAs best studied as ribonucleoprotein (RNP) guides in RNA modification1,2. To explore their role in cancer, we compared 5,473 tumor-normal genome pairs to identify snoRNAs with frequent copy number loss. The SNORD50A-SNORD50B snoRNA locus was deleted in 10–40% of 12 common cancers, where its loss was associated with reduced survival. A human protein microarray screen identified direct SNORD50A and SNORD50B RNA binding to K-Ras. Loss of SNORD50A and SNORD50B increased the amount of GTP-bound, active K-Ras and hyperactivated Ras-ERK1/ERK2 signaling. Loss of these snoRNAs also increased binding by farnesyltransferase to K-Ras and increased K-Ras prenylation, suggesting that KRAS mutation might synergize with SNORD50A and SNORD50B loss in cancer. In agreement with this hypothesis, CRISPR-mediated deletion of SNORD50A and SNORD50B in KRAS-mutant tumor cells enhanced tumorigenesis, and SNORD50A and SNORD50B deletion and oncogenic KRAS mutation co-occurred significantly in multiple human tumor types. SNORD50A and SNORD50B snoRNAs thus directly bind and inhibit K-Ras and are recurrently deleted in human cancer.
The two major classes of snoRNAs, C/D-box and H/ACA-box snoRNAs, modify rRNAs, tRNAs and small nuclear RNAs (snRNAs) to assist in the production of functional ribosomes3 in association with proteins that can include fibrillarin and dyskerin4,5. Recent studies, however, suggest that snoRNAs may have broader roles, including in genetic disorders6, human variation7, hematopoiesis8, metabolism9 and neoplasia10,11.
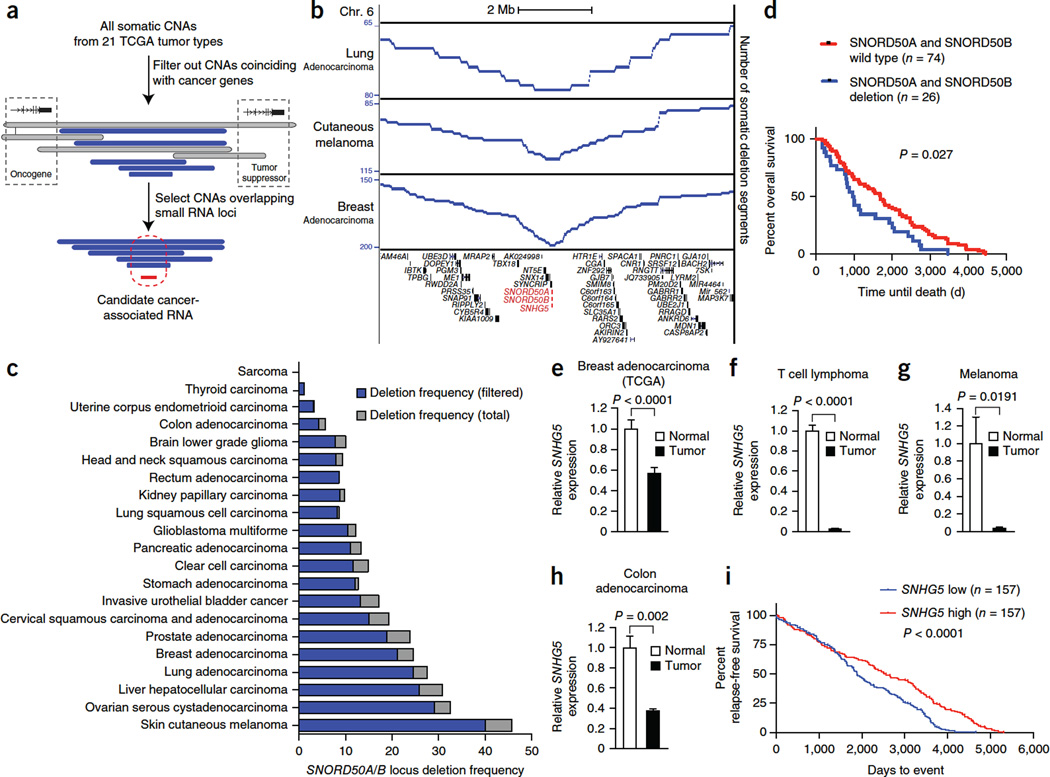
To screen for snoRNAs recurrently altered in cancer, we analyzed copy number alterations (CNAs) in 5,473 pairs of tumor and matching normal genomes in 21 human cancer types in The Cancer Genome Atlas (TCGA) data set, focusing on snoRNA locus alterations distant from known cancer-associated genes (Fig. 1a–c). Somatic loss of the adjacent SNORD50A and SNORD50B (SNORD50A/B) snoRNA genes occurred at a frequency of >10% in tumors for 12 of 21 TCGA tumor types (Fig. 1b,c), as determined using conservative CNA thresholds (Supplementary Fig. 1a–c). By comparison, publicly available TCGA analysis with the GISTIC algorithm places the overall incidence of SNORD50A/B deletion across all tumor types at 24.9%, with SNORD50A/B in significant deletion peaks. Our analysis observed somatic SNORD50A/B deletions in at least 20% of melanomas as well as ovarian, liver, lung and breast tissue malignancies, suggesting a role for SNORD50A and SNORD50B loss in cancer.
Figure 1.
Frequent deletion of SNORD50A/B in human cancers, SNORD50A/B expression and patient survival. (a) Schematic of the approach to identify altered snoRNA-encoding genomic loci in cancer using TCGA data. All somatic deletion segments, including those spanning contiguous oncogenes or tumor-suppressor genes, are shown in gray, whereas filtered deletions that do not also span known cancer-associated genes are shown in blue. (b) Somatic SNORD50A/B deletion in individual cancer types. (c) SNORD50A/B locus deletion frequency in 21 TCGA tumor types unfiltered with CNAs involving cancer-associating genes (gray) or filtered (blue). (d) Overall patient survival in the TCGA invasive breast cancer cohort as a function of filtered SNORD50A/B deletion. Analysis of statistical significance was performed using the log-rank (Mantel-Cox) test. (e–h) Expression of the SNORD50A/B host gene, SNHG5, in TCGA breast cancer data (62 tumor-matched normal breast tissue pairs) (e), published lymphoma data (10 tumor and 28 normal T cell samples)29 (f), published melanoma data (2 tumor and 2 normal melanocyte samples)30 (g) and published colon cancer data (45 tumor and 39 normal colon tissue samples)31 (h). Statistical significance was determined using an unpaired two-sided t test, except in the case of breast adenocarcinoma, where the samples are patient matched and a paired t test was used. In all cases, data represent means ± s.d. (i) Overall survival as a function of SNHG5 expression from a breast cancer cohort annotated in GEO series GSE6532.
Consistent with this hypothesis, SNORD50A/B loss was associated with decreased overall survival in the TCGA cohort of breast adenocarcinoma (Fig. 1d). Additionally, levels of RNA transcripts from the host gene for SNORD50A and SNORD50B, SNHG5, were decreased in the multiple cancer types where its expression has been measured (Fig. 1e–h). Analysis of other annotated snoRNA host genes demonstrated that the downregulation observed for SNHG5 is not a general phenomenon in cancer (Supplementary Fig. 1d). Decreases in SNHG5 expression were in some cases more profound than the incidence of genomic deletion for a given tumor, suggesting that additional mechanisms might exist to downregulate its expression in cancer. Analysis of transcription factors binding near the SNHG5 promoter was performed using Encyclopedia of DNA Elements (ENCODE) chromatin immunoprecipitation and sequencing (ChIPseq) data and TCGA melanoma RNA sequencing (RNA-seq) data, identifying HMGN5 and MYC among the transcription factors whose expression correlated most with SNHG5 expression (Supplementary Fig. 1e–g). Methylation at the CpG island nearest to SNHG5 did not correlate with SNHG5 expression (Supplementary Fig. 1h). Decreased SNHG5 expression was associated with reduced survival in both the breast cancer and cutaneous melanoma patient cohorts (Fig. 1i and Supplementary Fig. 1i). Therefore, the SNORD50A/B locus is commonly deleted in multiple human cancer types, and SNORD50A/B loss correlates with poorer clinical outcome.
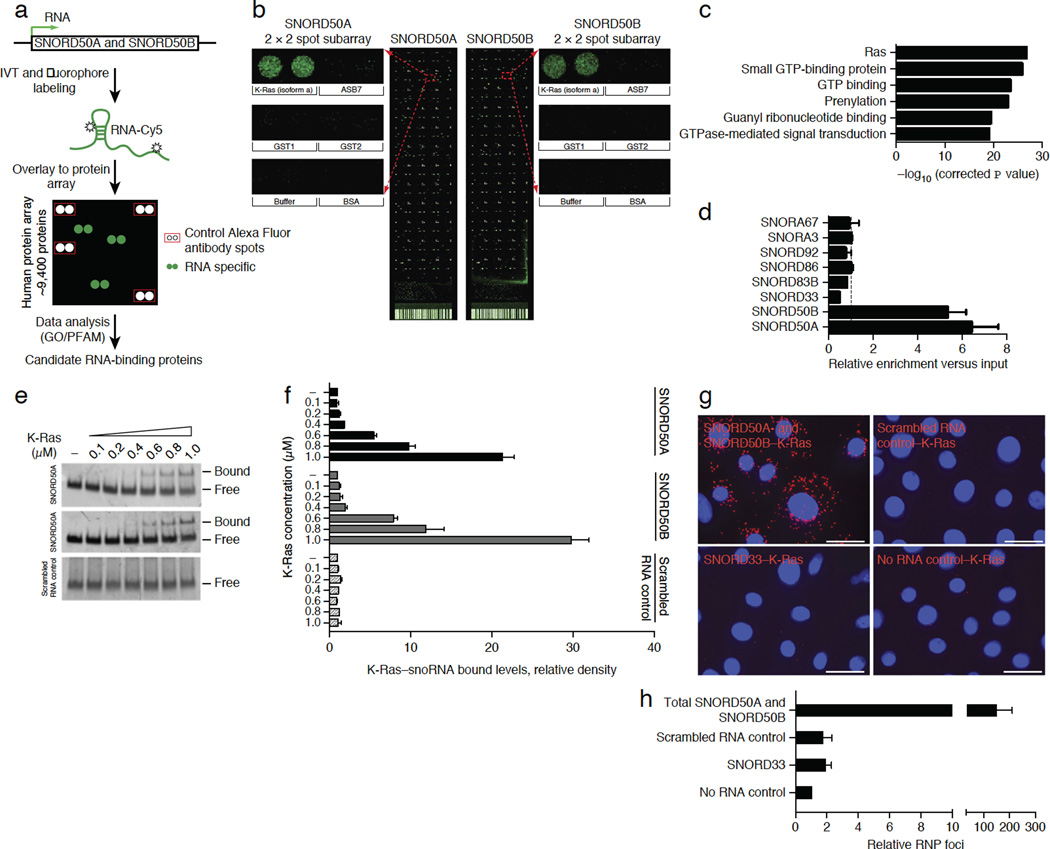
SNORD50A and SNORD50B are co-located on chromosome 6q14.3. RNA-seq of polyadenylated RNA by the ENCODE Project did not detect a transcript spanning SNORD50A and SNORD50B, indicating that they do not function as nuclear long noncoding RNA (lncRNA) caps12. SNORD50A and SNORD50B encode two C/D box–containing snoRNAs that specify sites for 2′-O-ribose methylation on target RNAs, such as 28S rRNA13. Because such housekeeping functions do not suggest obvious links to carcinogenesis and because RNA-protein interactions are integral to snoRNA action, we undertook a screen to identify cancer-relevant proteins that bind SNORD50A and SNORD50B. We modified a recently developed approach for binding of long RNA to high-density protein microarrays of 9,125 recombinant human proteins14,15 to facilitate small RNA hybridization (SR-PMA) (Fig. 2a). Hybridization of 75-nucleotide (nt) SNORD50A and 71-nt SNORD50B RNAs identified specific binding to independently spotted recombinant K-Ras4A and K-Ras4B proteins (Fig. 2b and Supplementary Fig. 2a–e). Although SNORD50A and SNORD50B bound K-Ras most strongly, they also bound additional GTPases (Fig. 2c, Supplementary Fig. 2c,f and Supplementary Table 1). In contrast, hybridization with 30 other noncoding and coding RNAs, including ten additional snoRNAs, failed to detect binding to even a single GTPase (Supplementary Fig. 2g and Supplementary Table 2). We confirmed SNORD50A and SNORD50B binding to K-Ras using ultraviolet light (UV) cross-linking and immunoprecipitation (CLIP), which identified K-Ras association with SNORD50A and SNORD50B but not with control C/D-box or H/ACA-box snoRNAs (Fig. 2d). Moreover, CLIP demonstrated that other Ras isoforms also bind SNORD50A and SNORD50B (Supplementary Fig. 2h). Further confirmation of this interaction was obtained by electromobility shift assay with purified recombinant K-Ras protein (Fig. 2e,f). SNORD50A and SNORD50B therefore bind Ras superfamily GTPases.
Figure 2.
SNORD50A and SNORD50B directly bind K-Ras. (a) Schematic of hybridization to microarrays containing 9,125 recombinant human proteins. (b) Protein microarray signal at the K-Ras4A protein isoform for SNORD50A and SNORD50B. Enlarged duplicate spotted K-Ras signals plus adjacent protein controls are shown. (c) Top gene ontology (GO) terms for SNORD50Aand SNORD50B-binding proteins. The significance of gene term enrichment calculated with a modified Fisher’s exact test using control spot subtracted 9,125 proteins on ProtoArray as a background universe. (d) CLIP assay demonstrates K-Ras association with SNORD50A and SNORD50B in vivo. The dotted line represents a value of 1.0, of no enrichment over input (n = 3, mean of three biological replicates ± s.d.). (e) Electromobility gel shift assay of SNORD50A and SNORD50B versus scrambled RNA control with increasing amounts of purified recombinant human K-Ras protein. (f) Quantification of the electromobility gel shift assay. Data are represented as the means ± s.d. of four independent experiments. (g) Intracellular RP-PLA in normal human epidermal keratinocytes identifying extranuclear association of SNORD50A and SNORD50B with endogenous K-Ras protein (orange); K-Ras binding to the SNORD33 snoRNA, scrambled RNA and no RNA represent controls. Scale bars, 100 µm. (h) Quantification of PLA (means ± s.d., n = 3).
To identify the sequences responsible for the K-Ras–snoRNA interaction, a series of KRAS and SNORD50A mutants were generated (Supplementary Fig. 3a–d). Deletion mutants were generated in the K-Ras nucleotide-binding region, the switch I region, the switch II region and the C-terminal region. Deletions within both switch regions did not reduce K-Ras binding to SNORD50A; however, deletions within the N-terminal nucleotide-binding region and C-terminal region reduced binding by 30% and 50%, respectively, suggesting that residues involved in SNORD50A interaction are widely distributed across K-Ras (Supplementary Fig. 3a,b). To identify these residues, we superimposed the K-Ras crystal structure onto the SRP54-SRP complex with 7S RNA; this predicted that the positively charged surface residues—Lys5, Lys42, Arg149 and Arg161—of K-Ras might interact with RNA. Consistent with this notion, mutagenesis of these residues further reduced K-Ras binding to SNORD50A by 65% (Supplementary Fig. 3a,b). To map the SNORD50A nucleotide sequences required for K-Ras binding, we made short deletions within the C, C′, D and D′ boxes of SNORD50A and then assessed binding to K-Ras (Supplementary Fig. 3c,d). Deletions within the C′, D and D′ boxes modestly reduced K-Ras binding; however, a 7-nt deletion within the C box largely abolished it (Supplementary Fig. 3c,d). Because K-Ras protein has been largely observed in extranuclear cell compartments and snoRNAs are primarily, but not exclusively, found in the nucleolus and Cajal bodies, we next examined where the SNORD50A and SNORD50B RNA interactions with K-Ras proteins occur. To do this, we modified proximity ligation analysis (PLA), which can detect protein-protein interactions in intact cells16, to identify RNA-protein interactions within cells (RP-PLA). Exogenously delivered, biotin-labeled SNORD50A and SNORD50B distributed to the nucleolus and other parts of the cell as expected (Supplementary Fig. 3e) and identified SNORD50A and SNORD50B interacting with K-Ras protein outside the nucleus (Fig. 2g,h).
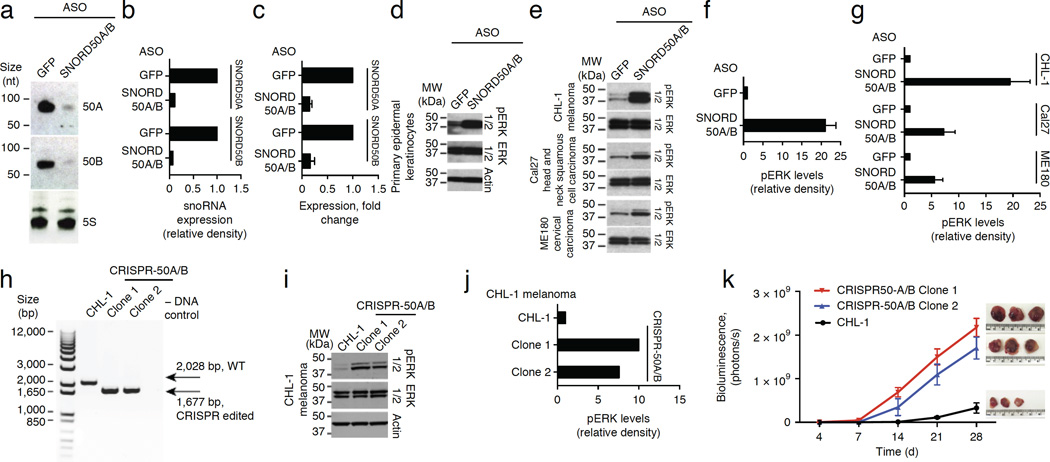
The frequency of SNORD50A/B deletion in cancer and the direct binding of SNORD50A and SNORD50B to K-Ras protein raised the possibility that SNORD50A/B loss might contribute to activation of downstream K-Ras targets. To test this possibility, we first depleted SNORD50A and SNORD50B expression, using antisense oligonucleotides (ASOs) because the small size of these snoRNAs constrained conventional RNA interference techniques. Decreased SNORD50A and SNORD50B levels were verified by both quantitative RT-PCR (qRT-PCR) and RNA blot analysis (Fig. 3a–c), with the latter demonstrating single bands of full length, indicating that these snoRNAs are not processed to shorter snoRNA-derived RNA fragment (sdRNAs), including processed snoRNAs (psnoRNAs)3. Among K-Ras–activated targets is the ERK1/2 mitogen-activated protein kinase (MAPK) pathway, one of the signaling cascades most commonly upregulated in human cancers17,18. Consistent with an impact on K-Ras action, SNORD50A and SNORD50B depletion led to ERK1/2 MAPK activation in both normal epithelial cells and cancer cells with intact Ras-ERK1/2-MAPK pathway genes (Fig. 3d–g). MAPK cascade activation was due to loss of SNORD50A/B as opposed to its host gene, SNHG5, as SNHG5 depletion showed no effects on the MAPK pathway (Supplementary Fig. 4a–c).
Figure 3.
Impact of SNORD50A and SNORD50B loss on K-Ras. (a) RNA blot demonstrating depletion of cellular SNORD50A and SNORD50B RNA by delivery of separate ASO specific to SNORD50A and SNORD50B (SNORD50A/B). (b) Quantification of RNA blot analysis. (c) Quantitative PCR demonstrating depletion of cellular SNORD50A and SNORD50B (means ± s.d., n = 4). (d) SNORD50A and SNORD50B depletion in normal human epidermal keratinocytes increases levels of active phosphorylated ERK1/2 MAPKs. (e) SNORD50A and SNORD50B depletion in multiple cancer cell lines with intact Ras-ERK1/2 MAPK signaling components increases levels of active ERK1/2. (f) Quantification of active, phosphorylated ERK1/2 as a function of SNORD50A and SNORD50B depletion in primary epidermal keratinocytes. (g) Quantification of active phosphorylated ERK1/2 as a function of SNORD50A and SNORD50B depletion in cancer cell lines. Data in f and g are represented as means ± s.d. of three independent experiments. (h) PCR analysis of genomic DNA from CRISPR-edited CHL-1 melanoma cells (CRISPR-50A/B) demonstrating deletion of the adjacent SNORD50A and SNORD50B genes in duplicate independent CHL-1 clones. (i) CRISPR-mediated SNORD50A/B deletion increases levels of active ERK1/2. (j) Quantification of active, phosphorylated ERK1/2 as a function of SNORD50A and SNORD50B depletion. (k) Tumor growth kinetics in vivo assessed by bioluminescent imaging of wild-type CHL-1 parental melanoma cells and two independent SNORD50A/B-deleted clones (n = 3 mice/group, means ± s.d.). Explanted tumor xenografts at day 28 are shown at right.
Strong activation of wild-type Ras, as well as active oncogenic Ras, reduces cell proliferation in vitro, in a process with features of oncogene-induced senescence; in epithelial cells, the resulting growth arrest can be partially rescued by CDK4 (refs. 19,20). Depletion of SNORD50A and SNORD50B reduced proliferation in vitro comparably to overexpression of oncogenic KRAS encoding a p.Gln61Leu substitution (Supplementary Fig. 4d–i). Moreover, much as in oncogenic Ras-induced growth inhibition, this effect was partially rescued by CDK4 (Supplementary Fig. 4h,i), indicating that the effects of SNORD50A and SNORD50B loss recapitulate these aspects of Ras activation.
To evaluate the consequences of SNORD50A/B loss on tumorigenesis, we undertook CRISPR-mediated gene deletion in CHL-1 melanoma cells (Fig. 3h and Supplementary Fig. 5a); these cells retain intact and inducible Ras-MAPK pathway function. Parallel studies were performed in A549 and NCI-H23 lung cancer cells with oncogenic KRAS mutations (Supplementary Fig. 5). As in ASO-mediated knockdown, SNORD50A/B-deleted clones displayed increased levels of active, phosphorylated ERK1/2 in CHL-1 (Fig. 3i,j), A549 (Supplementary Fig. 5e,f,h,i) and NCI-H23 (Supplementary Fig. 5k,l,n,o) cells. This effect was not associated with signs of increased ribosomal stress, as assessed by ribosome profiling/Riboseq (Supplementary Fig. 5b,c). SNORD50A/B deletion enhanced tumor growth in vivo in all three tumor cell types (Fig. 3k and Supplementary Fig. 6a–c). SNORD50A/B gene loss thus accelerates tumor growth in vivo in the context of both wild-type and oncogenic mutant KRAS.
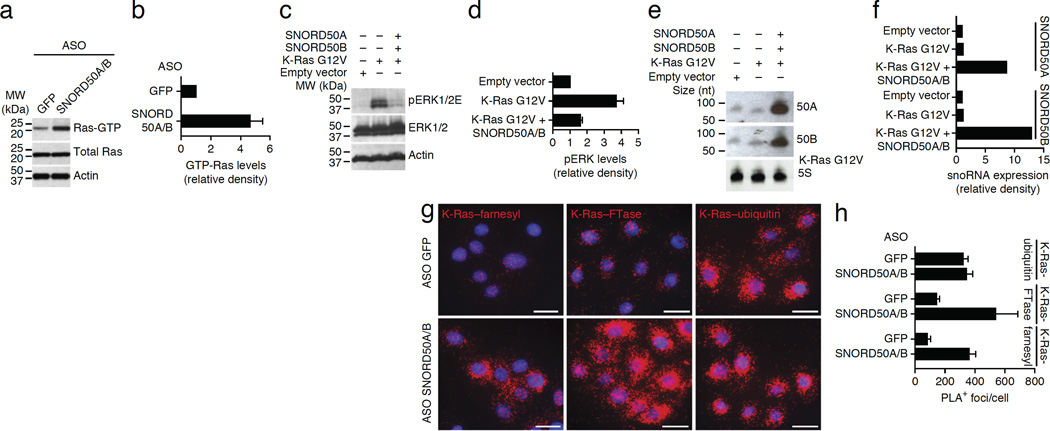
Depletion of SNORD50A and SNORD50B increased levels of active GTP-bound K-Ras (Fig. 4a,b). Overexpression of SNORD50A and SNORD50B in vitro acutely diminished the levels of active ERK1/2 induced by oncogenic K-Ras (Fig. 4c–f), indicating that SNORD50A and SNORD50B oppose oncogenic signaling by K-Ras. Consistent with this hypothesis, tumor cells overexpressing SNORD50A and SNORD50B did not show long-term persistence (Supplementary Fig. 7a,b).
Figure 4.
Impact of SNORD50A and SNORD50B on K-Ras function. (a) Depletion of SNORD50A and SNORD50B increases cellular levels of active, GTP-bound K-Ras protein. (b) Quantification of GTP-bound K-Ras protein. (c) Enforced expression of SNORD50A and SNORD50B suppresses the increased levels of phosphorylated ERK1/2 induced by mutant oncogenic K-Ras Gly12Val. (d) Quantification of phosphorylated ERK1/2 as a function of combined SNORD50A and SNORD50B (SNORD50A/B) overexpression. (e) RNA blot demonstrating enforced expression of exogenous SNORD50A and SNORD50B RNA. (f) Quantification of RNA blot analysis. (g) The effects of SNORD50A and SNORD50B depletion on K-Ras protein farnesylation and association with FTase. Orange shows PLA signal for farnesylated K-Ras (left column), FTase-K-Ras proximity (middle column) and ubiquitinated K-Ras (right column). Scale bars, 100 µm. (h) PLA quantification. Data are representative of two (b,d) and at least three (h) independent experiments and are shown as means ± s.d.
For full biological activity, both wild-type and oncogenic mutant Ras family proteins require prenylation by enzymes such as the CAAX farnesyltransferase (FTase), which can place lipid moieties on the C terminus of K-Ras. We thus next studied K-Ras farnesylation as a function of SNORD50A and SNORD50B. There are no antibodies that specifically recognize farnesylation of K-Ras; however, PLA can detect protein post-translational modifications (PTMs) by combining an antibody to a target protein with a PTM-specific antibody, thereby only showing signal when the PTM is present on the protein of interest21. We combined antibodies to K-Ras with those specific for the farnesyl moiety to detect K-Ras farnesylation within cells. Depletion of SNORD50A and SNORD50B quantitatively increased K-Ras farnesylation within cells without altering the signal for another K-Ras PTM, namely ubiquitination (Fig. 4g,h). Consistent with this finding, SNORD50A/B-knockout cells displayed increased amounts of prenylated K-Ras protein (Supplementary Fig. 7c–h). Loss of SNORD50A and SNORD50B therefore increases the levels of active, prenylated K-Ras.
To probe the basis for the enhanced K-Ras farnesylation seen with loss of SNORD50A and SNORD50B, we next asked whether SNORD50A and SNORD50B serve as endogenous inhibitors of FTase binding to K-Ras. Within cells, loss of SNORD50A and SNORD50B enhanced the interaction between K-Ras and FTase (Fig. 4g,h). We confirmed this result using recombinant K-Ras and FTase proteins with in vitro–transcribed SNORD50A and SNORD50B. Consistent with observations in cells, SNORD50A and SNORD50B reduced binding of recombinant K-Ras protein to FTase in far western blotting (Supplementary Fig. 7i,j), suggesting that SNORD50A and SNORD50B inhibit activating K-Ras farnesylation by impairing FTase binding to K-Ras.
These data indicate that SNORD50A/B snoRNAs are recurrently lost in cancer and that the mature snoRNAs bind and suppress the activity of the Ras oncoproteins. Loss of these snoRNAs enhances K-Ras association with FTase as well as causing hyperactivation of the downstream ERK1/2 MAPK pathway. Increased tumorigenesis occurs with combined oncogenic KRAS mutation and SNORD50A/B deletion, suggesting that frequent SNORD50A/B deletion might cooperate with oncogenic KRAS mutations in cancer to drive Ras-MAPK hyperactivation. Consistent with this hypothesis, we observed that SNORD50A/B deletion displayed statistically significant co-occurrence up to P = 1.1 × 10−4 with KRAS, HRAS and NRAS mutations in multiple cancer types (Supplementary Tables 3 and 4). Of interest, recent work found that mutational co-occurrence is more common than mutual exclusivity by identifying 148 co-occurring significantly mutated gene (SMG) combinations in comparison to 14 mutually exclusive SMGs; co-occurrence was observed between genes residing in the same signaling pathway, including the Ras and phosphoinositide 3-kinase (PI3K) pathways22. The co-occurrence of KRAS mutations with SNORD50A/B deletion in human cancers is consistent with our observation that SNORD50A/B deletion enhances the tumorigenesis of cells expressing oncogenic K-Ras mutants and supports the premise that mutational cooperativity occurs among genes that act within specific cancer-relevant pathways.
snoRNAs have recently been implicated in multiple processes, including neoplasia. For example, SNORA42 was found to be amplified in some non–small cell lung cancer tumors23. SNORD50A/B itself was previously observed to map to translocations of uncertain functional impact in B cell lymphoma24 and to also display point mutations in a small subset of prostate tumor cell lines and tumors, although with unclear pathogenic consequences, as similar alterations were observed in germline DNA25. The observation here that SNORD50A and SNORD50B bind multiple GTPases may reflect a conserved class of interactions. Consistent with this possibility, in yeast, the Bms1 ribosomal GTPase protein was found to bind the yeast U3 snoRNA directly during biosynthesis of the 40S ribosomal subunit26. Additionally, 23S rRNA may regulate the function of ribosomal GTPases27,28. In spite of the association of snoRNAs with the nucleolus, roles for them in the cytoplasm have been recently appreciated, including in metabolic stress9. SNORD50A and SNORD50B snoRNAs therefore represent GTPase-binding snoRNAs with effects on K-Ras regulation and tumorigenesis.
ONLINE METHODS
Copy number variation analysis
Level 3, segmented SNP6 copy number data mapped to hg19 for each cancer type were downloaded from the TCGA Data Portal on 24 August 2012. Tumor samples with matched normal samples were exclusively used for downstream analysis to determine somatic CNAs. The numbers of analyzed normal-tumor pairs were as follows: sarcoma, 17; thyroid carcinoma, 256; uterine endometrioid carcinoma, 428; colon adenocarcinoma, 371; glioma, 140; head and neck carcinoma, 279; rectum adenocarcinoma, 139; kidney papillary carcinoma, 471; lung squamous cell carcinoma, 278; glioblastoma multiforme, 484; pancreatic adenocarcinoma, 45; clear cell carcinoma, 102; stomach adenocarcinoma, 157; bladder cancer, 76; cervical carcinoma, 93; prostate adenocarcinoma, 138; breast adenocarcinoma, 824; lung adenocarcinoma, 297; hepatocellular carcinoma, 81; ovarian cystadenocarcinoma, 550; and melanoma, 247. The working list of cancer-associated genes from the Cancer Gene Census was downloaded from the Wellcome Trust Sanger Institute Cancer Genome Project, and their genomic coordinates were mapped to hg19. snoRNA coordinates were downloaded from GENCODE Genes v12 within the UCSC Genome Browser Table Browser. Tumor sample segmented intervals that overlapped a cancer-associated gene were filtered out from analysis. Somatic CNA for each snoRNA was determined by subtracting the copy number value for the segment containing the snoRNA in the tumor sample from the copy number in the normal sample. A somatic CNA cutoff greater than an absolute value of 0.25 was used.
Survival curve analysis
Annotated clinical data for TCGA breast cancer samples were downloaded in matrix format from the UCSC Cancer Genome Browser. Additional survival data associated with SNHG5 gene expression were obtained from GEO under accession GSE6532. ‘High’ and ‘low’ values correspond to the upper and lower half of the expression values within the data set. Statistical significance was analyzed using the log-rank (Mantel-Cox) test. DNA sequencing data from somatic mutations of paired samples from TCGA were downloaded to compare the mutual exclusivity or co-occurrence of SNORD50A and SNORD50B loss with that of somatic mutation and copy number variants of cancer-associated genes. Significant co-occurrence or mutual exclusivity was determined using Fisher’s exact test.
Expression analysis
Expression of the SNORD50A and SNORD50B host gene SNHG5 was determined from TCGA data for breast cancer. Expression data were analyzed using probe A_23_P361087 from the custom Agilent array. Fragments per kilobase of transcript per million mapped reads (FPKM) values for SNHG5 were used from primary melanoma cultures. Statistical significance was determined using an unpaired two-sided t test, except in the case of breast adenocarcinoma, where the samples are patient matched and a paired t test was used. Tumor-normal sample sizes were as follows: colon, 45 and 39; breast, 62 and 62; T cell lymphoma, 10 and 28; and melanoma, 2 and 2. For analysis of snoRNA host gene expression, TCGA breast invasive carcinoma gene expression data (PANCAN normalized) were downloaded from the UCSC Cancer Genome Browser (version dated 24 February 2015). Somatic gene expression changes of all snoRNA host genes with the gene prefix “SNHG” were calculated by subtracting expression values for a matched normal sample for those from a primary tumor sample in the same patient. The aggregate log2-tranformed gene expression changes are plotted, and only matched tumor-normal samples were included, resulting in n = 113.
Transcription factor and methylation analyses
The SNHG5 promoter region (±500 bp with respect to the transcription start site (TSS)) was overlapped with all called peaks from ENCODE ChIP-seq data, and 68 different transcription factors were identified as bound. For each of these transcription factors, the Pearson correlation was calculated between expression of the transcription factor and expression of SNHG5 in the TCGA melanoma RNA-seq data. Methylation values from the CpG island probe nearest to the SNHG5 promoter (cg10705909) were obtained from TCGA data for melanoma, correlated against SNHG5 expression in matched samples and plotted as a scatterplot. Statistical significance was determined using an unpaired two-tailed t test.
Mutational analysis of co-occurrence
All TCGA Pan-Cancer gene-level non-silent somatic mutations and TCGA Pan-Cancer gene-level copy number (GISTIC2) data were downloaded from the UCSC Cancer Genome Browser. In total, 4,224 samples with both mutation and copy number data were analyzed. Gene-level copy number alterations >0.25 (log2) for KRAS, HRAS and NRAS were classified as amplifications, whereas gene-level copy number alterations <−0.25 (log2) for SNHG5 were classified as deletions. Fisher’s exact tests were performed between SNORD50A/B loss and RAS amplification or mutation to determine significant co-occurrence or mutual exclusivity.
Protein microarray analyses
DNA encoding SNORD50A and SNORD50B was synthesized by overlapping PCR of a construct with an SP6 promoter at the 5′ end, the resulting fragment was cloned into the pCR2.1TOPO vector (Life Technologies), and RNAs were produced and labeled with Cy5 dye with a ratio of RNA:Label IT Cy5 reagent of 1:3 and a reaction time of not more then 30 min at 37 °C to achieve between 0.5 and 1 Cy5 dye unit covalently attached to each RNA used. For RNA incubation, the ProtoArray Human Protein Microarray v5.0 (Life Technologies) was used. GO analysis and PFAM domain analysis of RNA-binding proteins were performed with Benjamini-Hochberg correction of the P value. The P value for the Venn diagram illustrating overlap of two independent microarray incubations was calculated using Fisher’s exact test.
snoRNA mutagenesis
pCR2.1TOPO-SNORD50A vector was used as a template for inverse PCR using oligonucleotides targeting the snoRNA C, D, C′ and D′ boxes and containing compatible infusion arms with reverse oligonucleotide. The following mutant forms of SNORD50A were generated and used for RNA in vitro transcription and gel shifts: pCR2.1TOPO-SNORD50A′C (SNORD50A deletion of 6–12 bp), pCR2.1TOPO-SNORD50A′D (SNORD50A deletion of 69–72 bp), pCR2.1TOPO-SNORD50A′C′ (SNORD50A deletion of 35–40 bp) and pCR2.1TOPO-SNORD50A′D′ (SNORD50A deletion of 25–28 bp).
Mutant K-Ras proteins
Deletions in the KRAS (4B) gene coding sequence were introduced by overlapping PCR amplification, and resulting sequences were cloned into the pLEX lentiviral vector containing a sequence encoding a FLAG-HA-HIS (FHH) tag in frame. For controls, wild-type KRAS coding sequence and eGFP were engineered to express the same tag and subcloned into the pLEX lentiviral vector. To match expression of delivered wild-type K-Ras protein with endogenous protein levels, the uORF-K-Ras (wild type) construct was generated containing a short upstream ORF (uORF) 5′ to the coding sequence. Using these approaches, the following constructs were generated: pLEX-FHH-K-Ras (wild type), pLEX-uORF-FHH-K-Ras (wild type), pLEX-uORF-FHH-GFP, pLEX-FHH-K-Ras′10–18 (targeting the K-Ras nucleotide-binding site), pLEX-FHH-K-Ras′28–39 (targeting the switch I region), pLEX-FHH-K-Ras′57–63 (targeting the switch II region) and pLEX-FHH-K-Ras′165–183 (targeting the C-terminal region). To predict the K-Ras residues that potentially interact with SNORD50A and SNORD50B snoRNA for point mutagenesis, the crystal structure of wild-type K-Ras (Protein Data Bank (PDB), 4LPK) was overlaid onto the crystal structure of the SRP54-SRP19 (PDB, 2V3C) 7S.S-SRP RNA complex, which contains a Ras GTPase-like domain, using PyMOL software. Site-directed mutagenesis was used to change all candidate residues to isoleucine, and the resulting sequences were cloned to generate the pLEX-FHH-K-Ras-K5I, pLEX-FHH-K-Ras-K42I, pLEX-FHH-K-Ras-R149I and pLEX-FHH-K-Ras-R161I constructs used for CLIP.
Gel-shift assays
For wild-type and mutant snoRNAs, pCR2.1TOPO vector containing snoRNAs under the control of the SP6 promoter and flanked with an SspI site at the 3′ end was used. Plasmid DNA was digested with SspI, and 4 pg of linear DNA was then in vitro transcribed in a 50-µl total volume consisting of 1× Transcription buffer (Promega), 10 mM DTT (Promega), 1 mM NTPs (Invitrogen), 40 U RNaseOUT (Invitrogen) and 60 U SP6 RNA polymerase at 37 °C for 4 h, after which DNA was digested by the addition of 2 U DNase I at 37 °C for 15 min. For gel-shift assays, 0.2 µM of the full-length or deletion-mutant forms of snoRNAs SNORD50A, SNORD50B or scrambled 74-bp (USCR) RNAs was used with the indicated amount or 0.6 µM of purified recombinant K-Ras protein (Abcam, ab96817). snoRNAs were denatured at 90 °C for 30 s in 10 µl of water, and samples were immediately transferred to ice; after adding an equal volume of 40 mM HEPES (pH 7.4) with 2 mM DTT, the sample volume was brought up to room temperature for 5 min to allow equilibration. Next, K-Ras protein was added, and samples were incubated an additional 15 min and then analyzed using a 10% TBE polyacrylamide gel; gels were scanned using a Typhoon 9410 scanner (GE Healthcare), with band intensity quantified using ImageJ software.
UV cross-linking and immunoprecipitation
Human kidney fibroblast 293 cells were transfected with pLEX-FHH-K-Ras (wild type), uORFFHH-K-Ras (wild type) or mutant forms and selected in 0.5 µg/ml puromycin for 48 h. For CLIP assessment of Ras isoform activity, primary human keratinocytes were infected with VSVG pseudotyped pLEX-FHH-uORF-EGFP, pLEX-FHH-uORF-K-Ras, pLEX-FHH-uORF-N-Ras or pLEX-FHH-uORF-H-Ras isoforms and selected in 0.5 mg/ml puromycin. Cells were subjected to 0.3 J/cm2 UV-C cross-linking on ice, lysed directly in the plate in CLIP lysis buffer (50 mM Tris, pH 7.5, 10% glycerol, 200 mM NaCl, 5 mM EDTA, 0.5% sarkosyl, 0.2% Tween and 0.1% Igepal) and briefly sonicated. Lysate (2 mg) was incubated with a 30-µl bead bed of anti-FLAG-M2 agarose (Sigma). Complexes were then eluted twice with 0.5 ml of CLIP lysis buffer containing 0.5 mg/ml FLAG-M2 peptide (MDYKDDDDK). cDNA synthesis was performed using the iScript cDNA synthesis kit (Bio-Rad) with reactions spiked with 0.2 pmol of 3′-end target-specific oligonucleotides, and qRT-PCR was performed.
RNA-protein proximity ligation assays
For PLA, Duolink In Situ regents were used (Sigma) in accordance with the manufacturer’s instructions. Primary antibodies were as follows: polyclonal antibody to biotin (A150-109, Bethyl Laboratories), monoclonal antibody to Ras (C-4) (sc-166691, Santa Cruz Biotechnology), polyclonal antibody to farnesyl (AB4073, Millipore), antibody to farnesyl transferase FTβ (X-28) (sc-137. Santa Cruz Biotechnology) and polyclonal antibody to ubiquitin (FL-76) (sc- 9133, Santa Cruz Biotechnology). For RP-PLA, primary keratinocytes plated in eight-well chamber slides were transfected with 1.5 nM chemically synthesized 5′–biotin labeled snoRNAs or scrambled 74-bp RNA using Lipofectamine RNAiMAX reagent (Life Technologies). Forty-eight hours after transfection, slides were washed with PBS and fixed in fresh 3.7% formaldehyde for 10 min at room temperature, followed by two PBS washes and incubation for 10 min with 70% ethanol. Signal was detected using the DeltaVision wide-field Deconvolution System (Applied Precision), and particles were quantified using the ImageJ software particle analysis tool. Localization of transfected SNORD50A and SNORD50B within the nucleolus was performed in live cells using the Nucleolar-ID Green detection kit (Enzo Life Sciences). The specificity of nucleolar costaining was confirmed by nucleolar depletion after treatment with 5 mM actinomycin D for 4 h at 37 °C.
snoRNA depletion
All ASOs were synthesized as 5-10-5 RNA-DNA chimeric oligonucleotides linked with phosphorothioate backbones, with ten deoxyribonucleotides flanked by five 2′-O-MOE–modified ribonucleotides at both sides (z, 2′-O-methoxyethyl; x, phosphorothioate backbone) and are listed in Supplementary Table 5. American Type Culture Collection (ATCC)-verified cell lines were purchased directly from ATCC and subjected to mycoplasma testing. For knockdown of targeted snoRNAs, subconfluent (40–60%) primary keratinocytes, CHL-1 melanoma cells, Cal27 head and neck squamous cell carcinoma cells or ME180 cervical carcinoma cells were treated with the ASOs at a final concentration of 50 nM with 5 µl of RNAiMAX.
SNHG5 depletion
Small interfering RNA (siRNA) oligonucleotide duplexes were synthesized by Ambion (Life Technologies). Human keratinocytes (1 × 106) were electroporated with 1 nmol of siRNA nucleotides using the Amaxa neonatal keratinocyte nucleofection kit (Lonza). The efficiency of knockdown was evaluated using 5 µg of total RNA isolated with TRIzol reagent (Ambion) by RNA blot analyses using antisense probes to SNORD50A and SNORD50B and the Gene Images AlkPhos Direct labeling and detection system (GE Healthcare) or by qRT-PCR.
SNORD50A/B deletion
CRISPR-based genome editing was performed in CHL-1, A549 and NCI-H23 cells. Two target sequences specific to SNORD50B and SNORD50A were designed and fused to sequence encoding guide RNA (gRNA) whose expression was driven by the U6 promoter cloned in pBluescriptII(+)KS vector. In total, 2.5 pg of CRISPR-50B and CRISPR-50A targeting plasmids together with pcDNA3.3TOPO-Cas98 was nucleofected (Lonza) into CHL-1 cells or transfected into the lung cancer cell lines (A549 and NCI-H23) using TransIT-X2 (Mirus). After 5 d, cells received two nucleofections and transfections, and the population was subjected to serial dilution and clonally selected from single cells. Genomic PCR was used to verify loss of wild-type alleles, confirmed by Sanger sequencing of 24 clones. No wild-type alleles were present in the analyzed reads, demonstrating that the two alleles differed by one nucleotide (T) at the CRISPR-edited junction (Supplementary Fig. 5).
SNORD50A and SNORD50B overexpression
Sequences from the SNHG5 gene were amplified using primers flanking both SNORD50B and SNORD50A and cloned into the pSPARTA vector upstream of the gene encoding DsRed Express 2 in HindIII and BamHI sites, with their expression under the control of the human PGK promoter. The final vector, pSPARTA-HGU50ABDsRedExpress2 was verified by sequencing and used for transfection of 293 human fibroblasts. For long-term SNORD50A and SNORD50B expression, cells were selected in 0.35 mg/ml puromycin: DsRed Express 2 expression was monitored with fluorescence microscopy and snoRNA expression levels were evaluated by quantitative PCR. To evaluate the impact of transient enforced SNORD50A and SNORD50B expression on oncogenic K-Ras, the pSPARTA-HGU50AB-DsRedExpress2 vector was transfected together with pBABE-uORF-K-Ras-G12V plasmid containing the sequence for the oncogenic K-Ras Gly12Val mutant fused at the 5′ end with an upstream ORF to control oncogenic K-Ras expression. Cells were collected 48 h after transfection, and phosphorylated ERK levels were detected by immunoblot analysis.
Far western blotting
The indicated amounts of recombinant FTase (PR-361, Jena Biosciences) and MBP control in 4 µl of phosphate buffer were spotted in triplicate onto nitrocellulose membrane 0.5 cm apart, and membranes were blocked in TBST in 5% milk blocking buffer for 1 h at room temperature. Membranes were rinsed in binding buffer (50 mM Tris (pH 7.4), 1 mM DTT and 1% BSA), and, after supplementing with 500 U/ml RNaseOUT, each membrane received 1 pg/ml recombinant K-Ras protein (Abcam, ab96817). Additionally, 1.5 pg/ml of in vitro–transcribed SNORD50A and SNORD50B was added to membrane 2, while 1.5 pg/ml in vitro–transcribed scrambled RNA control of similar size and nucleotide composition was incubated with membrane 3. After washing, membranes were incubated with pan-Ras monoclonal antibody (C-4, Santa Cruz Biotechnology) in blocking buffer for 1 h at room temperature. Membranes were washed again, incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature and then washed; signal was developed using enhanced chemiluminescent (ECL) reagent (Thermo).
Ras prenylation analyses
K-Ras prenylation was evaluated by increased electrophoretic mobility of K-Ras protein due to post-translational modification involving attachment of the farnesyl or geranyl lipid residues to the Ras CAAX box. Two 15-cm plates of parental NCI-H23, A549 and CHL-1, as well as CRISPR-edited clones, were grown to 80% confluence and, after washing twice with ice-cold PBS, collected in 1.5 ml of modified RIPA buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.5% Igepal and 0.1% sodium deoxycholate) on ice. The lysate was centrifuged 5 min at 13,000 rpm to remove cell debris, and total protein concentration was then measured using the BCA protein assay kit (Thermo). The lysate was divided evenly into two tubes containing 2 mg of total protein each; 3 µg of monoclonal antibody to Ras (C-4) or IgG was added to each tube, and tubes were incubated overnight on a rotating wheel at 4 °C. The next day, 20 µl of protein A/G beads (Thermo) was added to each tube, and tubes were incubated for an additional 2 h at 4 °C on a rotating wheel. Beads were washed at least three times for 10 min each wash using modified RIPA buffer, and samples were eluted in SDS-PAGE loading dye. Proteins were analyzed with SDS-PAGE and, after transfer to a membrane, probed with primary monoclonal antibody to Ras (C-4, Santa Cruz Biotechnology) followed by goat anti-mouse secondary antibody conjugated to IRD 680 (LI-COR). Signal was detected using the Li-COR Odyssey Clx imager, and the ratio between unprocessed and prenylated forms of Ras protein was quantified using Image Studio 3.1.4. Software.
Immunoblot analyses
The following antibodies were used: polyclonal antibody to phosphorylated ERK1/2 (197G2) (4377, Cell Signaling Technology), monoclonal antibody to phosphorylated MEK1/2 (41G9) (9154, Cell Signaling Technology), polyclonal antibody to ERK1/2 (9102, Cell Signaling Technology), polyclonal antibody to MEK1/2 (9122, Cell Signaling Technology) and monoclonal antibody to Ras (C-4) (sc-166691, Santa Cruz Biotechnology).
Cell lines
CHL-1 human melanoma, NCI-H23 lung adenocarcinoma and A549 lung carcinoma cells were purchased from ATCC. The CHL-1 and A549 lines were authenticated by genetic profiling using polymorphic short tandem repeat (STR) loci by Promega. Primary human keratinocytes were isolated from fresh biopsy samples approved by the Stanford University institutional review board and were propagated for up to five passages. All cell lines and primary cells were confirmed to be free from mycoplasma using the MycoAlert mycoplasma detection kit (Lonza).
Ribosome profiling
As a positive control for ribosome stress, CHL-1 control cells were treated with a combination of the amino acid analog l-azetidine-2-carboxylic-acid (AZC; 10 mM) and proteasome inhibitor MG132 (20 µM) for 20 min. Cells were washed with ice-cold PBS and lysed on ice with cyclohexamide-containing buffer; deep sequencing libraries for ribosome profiling were prepared with rRNA removed before reverse transcription using Ribo-Zero (Epicenter, MRZH116). Sequencing reads were mapped to the human genome (hg19) using TopHat2. To further illustrate 5′ ribosome pausing, we derived the ribosome pausing index (PI)13 of individual transcripts by calculating the normalized ribosome density within a ten-codon window from the start codon (5′ PI) or stop codon (3′ PI). The ribosome pausing index was calculated by dividing the 5′ PI by the 3′ PI.
K-Ras activation assay
Subconfluent (40–60%) primary keratinocytes were treated with ASOs at a final concentration of 50 nM and 5 µl of RNAiMAX transfection reagent in complete growth medium, followed by collection of cells 24 h after treatment. GTP-bound K-Ras pulldown was performed using the Ras activation assay kit (Millipore).
Cell proliferation assays
Cell proliferation assays were performed using the Click-iT EdU kit according to the manufacturer’s instructions (Life Technologies). Cells were analyzed 24 h after SNORD50A and SNORD50B knockdown using 50 nM ASO1 and ASO2 mixtures. For rescue experiments, primary keratinocytes were transfected with the LZRS-CDK4 retroviral vector 48 h before ASO-mediated snoRNA depletion or infection with the pLEXK-Ras-Q61L lentiviral vector.
Tumor xenografts and bioluminescence imaging
Mouse and human tissue studies were approved by the Stanford institutional review boards. Parental or CRISPR-edited, SNORD50A/B-deleted CHL-1, A549 or NCI-H23 tumor cells (1 × 106) that were also stably infected with a puromycin-selectable retroviral vector containing firefly luciferase were suspended in a 1:1 solution of PBS and Matrigel (BD Biosciences) and injected without blinding to construct identity into the subcutaneous space of hairless 8-week-old immunodeficient NOD SCID gamma female mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG), Jackson Laboratory). Tumor viability was measured using the in vivo imaging system IVIS-200 (Xenogen). Images representing in vivo bioluminescent signal were superimposed on grayscale reference images of the mice obtained under weak illumination. Total photon emission (in units of photons per second) from defined regions of interest was quantified using LivingImage 4.4 (Caliper LifeSciences). Animal cohort numbers were chosen based on results from xenograft pilot studies in mice. The sample exclusion criteria were determined before experiments for technical failures such as misinjection of cells in tumor xenograft studies. Physical randomization was performed using animal tag number.
Statistical tests
Statistical analyses were performed as described in the figure legends and relevant sections of the Online Methods. Data are presented as means ± s.d. For survival analysis, analysis of statistical significance was performed using the log-rank (Mantel-Cox) test. Mutual exclusivity or co-occurrence of SNORD50A/B deletion with somatic mutation of cancer-associated genes was determined using Fisher’s exact test. Statistical significance of SNHG5 expression in different tumor types was determined using an unpaired two-sided t test, except in the case of breast adenocarcinoma, where the samples are patient matched and a paired t test was used. P < 0.05 was considered to indicate significance.
Supplementary Material
Acknowledgments
We thank J. Crabtree, J. Ferrell, S. Artandi, A. Oro, H. Chang and members of the Khavari laboratory for presubmission review and advice. This work was supported by the US Veterans Affairs Office of Research and Development and by US National Institutes of Health/National Cancer Institute grant CA142635 and by US National Institutes of Health/National Institute of Arthritis Musculoskeletal and Skin Diseases grant AR49737 to P.A.K.
Footnotes
Accession codes. RNA binding to protein microarray data and Riboseq data have been deposited in the Gene Expression Omnibus (GEO) under accessions GSE51817 and GSE71763.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Z.S. designed and executed experiments, analyzed data and wrote the manuscript. D.E.W., D.J., A.J.U., A.B., R.F., B.J.Z., Y.C. and F.M. executed experiments, analyzed data and contributed to design of experiments. D.J. and R.M.S. executed experiments. J.D.P. helped design experiments and analyzed data. P.A.K. designed experiments, analyzed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 2.Kiss T. Small nucleolar RNA–guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falaleeva M, Stamm S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays. 2013;35:46–54. doi: 10.1002/bies.201200117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, Meier UT. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reichow SL, Hamma T, Ferre-D’Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahoo T, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhartiya D, Talwar J, Hasija Y, Scaria V. Systematic curation and analysis of genomic variations and their potential functional consequences in snoRNA loci. Hum. Mutat. 2012;33:E2367–E2374. doi: 10.1002/humu.22158. [DOI] [PubMed] [Google Scholar]
- 8.Bellodi C, et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Rep. 2013;3:1493–1502. doi: 10.1016/j.celrep.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel CI, et al. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011;14:33–44. doi: 10.1016/j.cmet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mannoor K, Liao J, Jiang F. Small nucleolar RNAs in cancer. Biochim. Biophys. Acta. 2012;1826:121–128. doi: 10.1016/j.bbcan.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer. 2012;12:84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 12.Yin QF, et al. Long noncoding RNAs with snoRNA ends. Mol. Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 13.Rebane A, Roomere H, Metspalu A. Locations of several novel 2′-O-methylated nucleotides in human 28S rRNA. BMC Mol. Biol. 2002;3:1. doi: 10.1186/1471-2199-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siprashvili Z, et al. Identification of proteins binding coding and non-coding human RNAs using protein microarrays. BMC Genomics. 2012;13:633. doi: 10.1186/1471-2164-13-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 17.Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard AL, Hayward NK. Molecular pathways: mitogen-activated protein kinase pathway mutations and drug resistance. Clin. Cancer Res. 2013;19:2301–2309. doi: 10.1158/1078-0432.CCR-12-0383. [DOI] [PubMed] [Google Scholar]
- 19.Lazarov M, et al. CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat. Med. 2002;8:1105–1114. doi: 10.1038/nm779. [DOI] [PubMed] [Google Scholar]
- 20.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 21.Weibrecht I, et al. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev. Proteomics. 2010;7:401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- 22.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei YP, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31:2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka R, et al. Intronic U50 small-nucleolar-RNA (snoRNA) host gene of no protein-coding potential is mapped at the chromosome breakpoint t(3;6)(q27;q15) of human B-cell lymphoma. Genes Cells. 2000;5:277–287. doi: 10.1046/j.1365-2443.2000.00325.x. [DOI] [PubMed] [Google Scholar]
- 25.Dong XY, et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum. Mol. Genet. 2008;17:1031–1042. doi: 10.1093/hmg/ddm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karbstein K, Jonas S, Doudna JA. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell. 2005;20:633–643. doi: 10.1016/j.molcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Clementi N, Polacek N. Ribosome-associated GTPases: the role of RNA for GTPase activation. RNA Biol. 2010;7:521–527. doi: 10.4161/rna.7.5.12467. [DOI] [PubMed] [Google Scholar]
- 28.Clementi N, Chirkova A, Puffer B, Micura R, Polacek N. Atomic mutagenesis reveals A2660 of 23S ribosomal RNA as key to EF-G GTPase activation. Nat. Chem. Biol. 2010;6:344–351. doi: 10.1038/nchembio.341. [DOI] [PubMed] [Google Scholar]
- 29.Piccaluga PP, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J. Clin. Invest. 2007;117:823–834. doi: 10.1172/JCI26833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flockhart RJ, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skrzypczak M, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE. 2010 Oct 1; doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.