Abstract
In the past two decades, remarkable advances have been made in the development of technologies used to engineer new aptamers and ribozymes. This has encouraged interest among researchers who seek to create new types of gene control systems that can be made to respond specifically to small molecule signals. Validation that RNA molecules can exhibit the characteristics needed to serve as precision genetic switches has come from the discovery of numerous classes of natural ligand-sensing RNAs called riboswitches. While a great deal of progress has been made towards engineering useful designer riboswitches, considerable advances are needed before the performance characteristics of these RNAs match those of protein systems that have been co-opted to regulate gene expression. In this review, we will evaluate the potential for engineered RNAs to regulate gene expression and lay out possible paths to designer riboswitches based upon currently available technologies. Furthermore, we will discuss some technical advances that would empower RNA engineers who seek to make routine the production of designer riboswitches that can function in eukaryotes.
Keywords: allosteric, aptamer, directed evolution, riboswitch, ribozyme, RNA switch
Introduction
Soon after researchers established methods for engineering RNA aptamers that selectively bind small molecule ligands,1–3 the prospects for engineering sensors and switches made from RNA began to be explored. Although a variety of useful applications for ligand-binding RNAs can be envisioned4–6, the ability to create user-defined gene control elements remains one of the most promising goals.7–11 Questions regarding whether RNA molecules can be made to perform adequately as molecular sensors and switches will be addressed in some detail in the following paragraphs. But perhaps the most compelling reason to pursue the development of RNA-based gene control mechanisms is that they promise to supply simple cis-acting, modular, and non-immunogenic systems for use in future gene therapy applications. Engineered RNA switches could be used to tune the expression levels of targeted genes using drug like molecules, thereby facilitating levels of transgene expression that are optimal for each individual.
At first glance, proposals to use RNA to create designer gene control elements that respond to small ligands might seem far-fetched. Some RNA experts have proposed12, 13 that RNA is a fundamentally deficient medium for forming biological receptors and catalysts when compared to proteins. In part, such views are reinforced by the fact that gene control in most organisms appears to be dominated by protein factors14. This finding suggests that evolution has chosen its champion, and that RNA has been soundly beaten. From among the diversity of protein-based gene control systems known, several already have been co-opted for use as gene control systems in eubacteria15 and in eukaryotes16–18. For most of these protein-based systems to function optimally, the expression level of the protein factor has to be tightly regulated because over expression or under expression could result in a decreased dynamic range of the genetic switch. For example, over expression of a protein gene-control factor could result in higher background activation of the gene being controlled. Furthermore, most of the protein-based switches used in eukaryotes are engineered versions of non-human proteins, which could prove to be immunogenic.
In stark contrast, riboswitches that respond to small molecules and that have been validated as functionally equivalent to protein gene control systems are far more rare19. However, since these RNA domains are embedded within the mRNA whose expression is under control, there should be an equivalent number of RNA switches expressed relative to the number of adjoining coding regions. Furthermore, an engineered genetic switch composed of RNA is expected to be far less likely to elucidate an immune response.
Even the discovery of numerous microRNAs in plants20 and animals21 does not offer support for the hypothesis that RNA is a robust medium for forming genetic sensors and switches. Although microRNAs appear to influence the expression of thousands of genes in some eukaryotes22, 23, they recognize their mRNA targets via Watson/Crick base pairing and require a multi-protein complex called RISC24 to carry out their gene control actions. To eliminate the need for protein factors and to permit the development of gene control elements that respond to various ligands, RNAs must be able to form complex-folded structures that selectively respond to the binding of ligands.
In this review, we will highlight some of the recent advances made in RNA switch engineering and point out some discoveries of natural metabolite-sensing RNAs that provide precedents of RNAs that serve as proficient genetic switches. Furthermore, we will discuss some of the challenges needed to be addressed if the prospects for this technology are to be fully realized.
Engineered RNA switches that respond to small molecules
Despite initial concerns regarding the functional capability of RNA compared to proteins, numerous examples of ligand-binding RNA switches have been created in the laboratory. Creating the initial RNA switch constructs25, 26 involved the simple fusion of a ligand-binding aptamer with a ribozyme. If joined appropriately, ligand binding to the aptamer causes a shape change in the adjoining ribozyme domain, and thereby results in allosteric control4–6. Finding the right nucleotides and structures to create a functional bridge between aptamer and ribozyme could be achieved by testing a few variant designs25, or could harness the power of directed evolution to find a few functional constructs in large random-sequence populations26.
From a narrow perspective, engineering these switches is surprisingly easy. RNA structural domains tend to be modular27 and functional domains such as aptamers and ribozymes frequently retain their activities when repositioned into new constructs. Likewise, finding bridges between aptamers and ribozymes that confer 10- to 100,000-fold allosteric regulation can be as simple as testing a dozen rationally-designed constructs, or turning a few rounds of evolution in a test tube. Entirely new aptamers with novel ligand specificities can be isolated by exploiting allosteric ribozymes28, 29, and RNA switches with diverse ligand specificities have even been made to function as features of array-based analyte sensors30, 31.
Several early reports involving the grafting of aptamers onto ribozymes32 or messenger RNAs33, 34 revealed some of the potential of engineered RNAs to serve as ligand-modulated gene control systems. Unfortunately, a number of factors intervene to prevent many engineered RNA switches from becoming useful genetic switches. For example, the functions of most aptamers have not been validated in cells, the folding of RNA constructs might differ between test tube and cell, or the ribozyme chosen for RNA switch construction might not be appropriate for controlling gene expression. Some of these factors are discussed in greater detail below.
Riboswitches: natural metabolite-sensing RNAs
Although many challenges face RNA engineers who see to create designer gene control elements, they can proceed with absolute confidence that RNA switches can exhibit the performance characteristics needed to find widespread utility in gene control. Nature long ago has validated RNA switch technology, and numerous examples of metabolite-sensing RNA switches exist in modern cells35–38. These natural ligand-responsive genetic switches are called riboswitches39, and are usually found in certain mRNAs of bacteria where they commonly regulate the expression of enzymes that catalyze key chemical transformations.
As with engineered RNA switches, each metabolite-responsive riboswitch carries at least one aptamer that selectively binds its target ligand. A few riboswitches carry two aptamers in tandem of either identical or distinct ligand specificities40, 41. Such arrangements allow the resulting genetic switch to respond to two different metabolites42, or to provide greater responsiveness to small changes in metabolite concentration43, 44. A cooperative RNA switch that requires two different ligands for activation has been engineered previously45, demonstrating that even the more complex architectures of natural riboswitches can be emulated by engineered constructs.
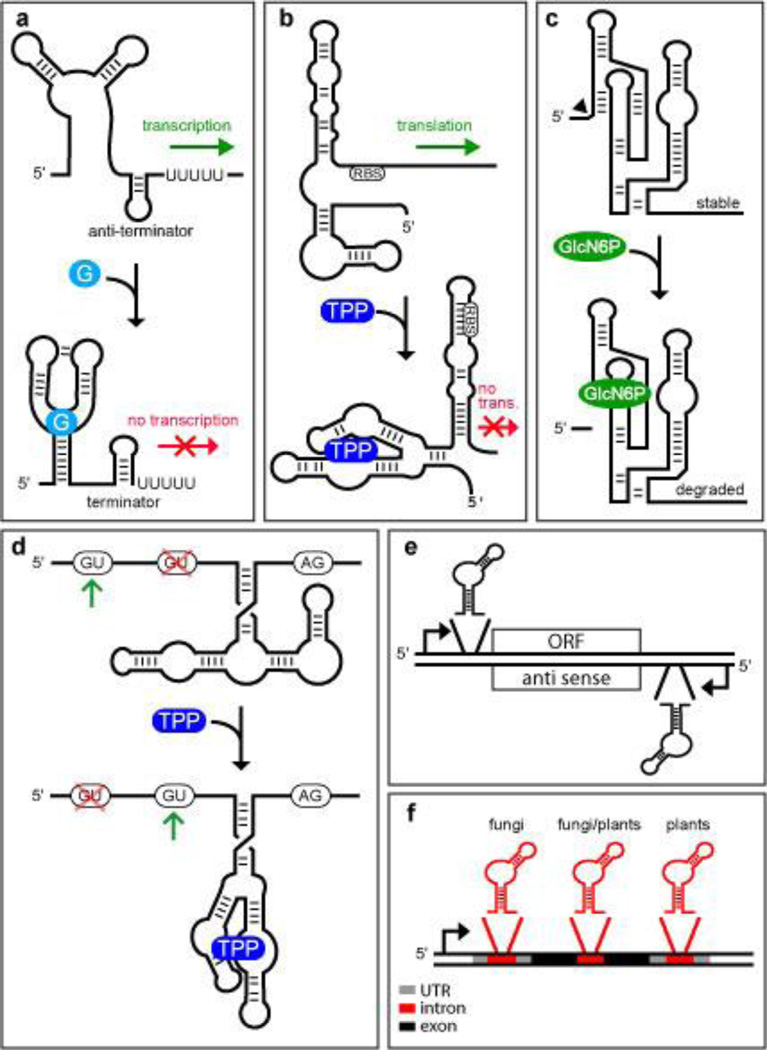
Examples of known riboswitch classes all carry additional nucleotides adjacent to the aptamer that control gene expression. In many cases, these additional nucleotides comprising the expression platform can be directed by ligand binding to adopt an alternative structure that somehow controls expression of the adjoining coding region (Figure 1a-d). If riboswitches naturally have explored numerous mechanisms for gene control, then reverse engineering the mechanisms used in modern cells could provide a powerful predictor of the most effective designs to be used by RNA switch engineers.
Figure 1.
Natural Riboswitch Locations and Mechanisms. (a) Transcription regulation based on a guanine-sensing riboswitch.121 When guanine (G) binds, the folding pathway favors the formation of a terminator stem at the expense of antiterminator stem formation. This architecture yields ‘off’ switch function because guanine causes transcription to terminate before the coding region of the mRNA is synthesized. Less common are examples of ‘on’ switch function where ligand binding favors antiterminator stem formation, as is observed with a related adenine-sensing riboswitch.122 (b) Translational regulation based on a TPP riboswitch.51 In the absence of the riboswitch ligand, expression of the down stream gene is permitted because translation can be initiated at the ribosomal binding site (RBS). However, in the presence of the ligand, the RNA adopts an alternate conformation that does not permit translation to be initiated at the RBS.123 (c) Metabolite-triggered ribozyme regulation of gene expression by GlcN6P riboswitches. When the ribozyme cofactor GlcN6P is low in concentration, the ribozyme does not undergo efficient self-cleavage and the stable mRNA can be translated. When GlcN6P is bound by the RNA, the ribozyme undergoes efficient self-cleavage, and the 3′ cleavage fragment including the ORF is rapidly degraded by a nuclease.57 (d) Control of alternative splicing by a TPP-sensing riboswitch in eukaryotes. When TPP concentrations are low, nucleotides from the unoccupied aptamer base pair near the second 5′ splice site, forcing the spliceosome to use the first 5′ splice site. When TPP is bound, the nucleotides formerly blocking the splice site are now involved in binding the ligand. This allows the spliceosome to choose the second 5′ splice site to yield an alternatively spliced mRNA. In fungi, ligand binding yields an alternatively spliced mRNA lacking upstream open reading frames (uORFs) that otherwise would decoy the ribosome from initiating translation at the main ORF.61 (e) Eubacterial riboswitch placement. Most riboswitches found in eubacteria are present within the 5′ untranslated regions (UTR) of mRNAs and directly control expression of a downstream open reading frame (ORF). There is bioinformatics evidence that at least one riboswitch controls the expression of an antisense RNA to regulate protein expression from a separate mRNA indirectly.53 (f) Eukaryotic riboswitch placement. Riboswitches have been found in introns located within the 5′ UTRs, coding regions, and 3′ UTRs of eukaryotic mRNAs.
In bacteria, the majority of riboswitches appear to regulate gene expression by influencing transcription termination or translation initiation.38 For transcription termination control, usually a portion of the aptamer sequence required to bind the ligand also can base pair with nucleotides in the downstream expression platform that are required to form an intrinsic terminator46, 47 or antiterminator stem (Figure 1a). This establishes a mutually-exclusive folding pathway for the riboswitch whose pathway choice is dictated by the availability of the metabolite target48, 49. If the riboswitch permits formation of the transcription terminator stem, then RNA polymerase will cease building the nascent mRNA, and gene expression is inhibited because the adjoining open reading frame (ORF) of the mRNA is not generated.
Similarly, riboswitches that control translation initiation commonly exploit ligand binding to control expression platform folding near the ribosome binding site of the adjoining open reading frame (Figure 1b).50–52 Theoretically, riboswitches that control translation initiation in bacteria could control the expression of full-length mRNAs. However, it also seems possible that the failure of ribosomes to bind to a nascent mRNA could cause the transcription termination protein Rho to cause premature transcription termination38. In other words, translation control by bacterial riboswitches might actually be a stealth form of transcription control.
Some of the more obscure mechanisms for riboswitch gene control might be well suited for future gene therapy applications. For example, there is bioinformatics evidence53 that a bacterial riboswitch for the coenzyme S-adenosylmethionine likely controls the expression of a separate mRNA via an antisense mechanism (Figure 1e). However, the riboswitch RNA probably uses a conventional transcription termination mechanism to control the production of its antisense domain, so this riboswitch mechanism would not be directly applicable in eukaryotes.
A rare but very effective riboswitch mechanism involves the control of self-cleavage of the mRNA for the GlmS protein in many Gram-positive bacteria (Figure 1c).54 This riboswitch class exploits a self-cleaving ribozyme that promotes glmS mRNA destruction when bound to the amino sugar compound glucosamine-6-phosphate (GlcN6P). Unlike the engineered allosteric ribozymes mentioned above, GlcN6P riboswitches use the ligand as a cofactor55, 56 to promote RNA cleavage by internal phosphoester transfer54. In this example, the ribozyme self-cleaves in the 5′ untranslated region (UTR) of the mRNA, and therefore does not directly inhibit gene expression. Expression is rapidly down regulated because the cleaved ribozyme product is targeted for mRNA decay by a specific nuclease enzyme57. Similarly, ribozymes designed to regulate gene expression through RNA cleavage will need to interfere with key steps of the gene expression pathway unless they cleave within an ORF. Unfortunately, engineering ribozymes that require specific ligands to function as cofactors for RNA transesterification might be too challenging to make this mechanism a routine choice for designer gene control systems.
To date, riboswitches that bind to the coenzyme thiamin pyrophosphate (TPP)51, 58 are the most widespread known. Unlike their typical use of transcription and translation gene control mechanisms in bacteria, TPP riboswitches in fungi59–61 and plants62–64 control pre-mRNA splicing (Figure 1d), and are found in several locations within mRNAs (Figure 1f). In some instances61, 62, TPP binding within an intron controls alternative splicing of the segment carrying the aptamer. This results because ligand binding sequesters nucleotides of the aptamer that otherwise would base pair with a 5′ splice site and prevent access by the spliceosome.
Given that a few TPP riboswitches are the only validated metabolite-binding riboswitches in eukaryotes, it is not yet clear whether splicing control is a preferential mechanism for the control of gene expression by RNA switches. It is interesting to note that self-splicing group I ribozymes65 are naturally responsive to guanosine compounds, and may represent a very widespread type of riboswitch that could modulate gene expression if its guanosine substrates vary sufficiently in concentration. This ribozyme class also can be made allosteric for other ligands32, and this offers the possibility that engineered group I ribozymes could be harnessed to provide a gene control system that would work in organisms from all three domains of life.
Engineering RNA aptamers for genetic switch applications
Any RNA switch that is engineered to respond to a small molecule ligand will only be as good as the aptamer it is built upon. Obvious characteristics such as ligand specificity and affinity are just two factors that must be considered. Perhaps the most overlooked component of engineering novel genetic switches made of RNA is that not all aptamers will have the characteristics needed to function well inside cells. For example, will an aptamer maintain sufficient selectivity and affinity when grafted to its regulatory setting? Will these characteristics, as well as its allosteric interplay with flanking sequences be retained when presented in a complex cellular milieu?
Riboswitch aptamers and their adjoining expression platforms are functionally validated in the organisms from which they have been isolated. However, riboswitches respond to natural compounds, which may not be suitable for use as ligands for designer gene control elements. If natural aptamers are excluded as modules for RNA switch construction, this leaves only the collection of engineered aptamers to choose from. Unfortunately, despite nearly two decades of aptamer engineering research, only a few aptamers that respond to small molecules have been validated to function inside cells32–34, 66–69, and some of these are not suitable for use as gene regulators in humans. For example, one of the most commonly used aptamers for constructing prototype RNA switches senses theophylline, which in bacteria has both poor bioavailability and toxic effects at high concentrations32, 70. Furthermore, it is known that the affinities exhibited by independently-folding aptamers can be eroded when they are integrated with accessory domains25, 29.
The types of problems noted above could be better overcome if new aptamers with a variety of functional characteristics could be easily made. Of particular interest would be the validation of a larger collection of aptamers capable of recognizing non-toxic small molecules that are also readily bioavailable. Access to aptamers that bind compounds known to have desirable pharmacokinetics would allow RNA switch engineers to choose from a collection of validated aptamers that sense compounds ideal for use as genetic switch triggers.
Many existing aptamers for small molecules have been made using a method of directed evolution called SELEX (systematic evolution of ligands by exponential enrichment)1–3. SELEX methods typically require that the target ligand be immobilized on a solid support. This arrangement creates several problems for those who seek to rapidly generate large numbers of aptamers for a diverse set of ligands. In addition to the costs of the reagents, the specialized skills needed to prepare and characterize each solid support reduce the enthusiasm that many have for generating their own aptamers. Not every compound of interest will be compatible with the chosen immobilization chemistry, and ligand immobilization reduces the number of potential contact surfaces that are available to interact with the binding pocket of the aptamer.
Some of these limitations can be avoided by using a directed evolution strategy that does not require target immobilization. A process called allosteric selection has been developed whereby novel aptamers can be isolated without using ligand-derivatized solid supports. One variation of this strategy makes use of a population of RNAs carrying a self-cleaving ribozyme fused to a random-sequence domain71–74. The RNAs are subjected to selection and amplification procedures75 that allow the isolation of RNA variants that function as allosteric self-cleaving ribozymes.
More recently, a method for allosteric selection has been developed for the isolation of DNA76 and RNA aptamers77 in which the population of random-sequence RNAs flanked by primer binding sites is immobilized, rather than their small molecule targets. In this allosteric selection strategy, RNAs are immobilized onto streptavidin-coated magnetic beads via a biotinylated DNA oligonucleotide complementary to a portion of one of the two primer binding sites. Immobilized RNAs are exposed to the target molecule resulting in the elution of a subset of the RNAs due to ligand-induced refolding and release from the tethered oligonucleotide.
Allosteric selection methods have been used to isolate multiple classes of aptamers for ligands that are added to the selection reactions simultaneously.28 This demonstrated another advantage of the allosteric selection strategy: the parallel selection of aptamers from a single directed evolution lineage. Despite distinct advantages, existing allosteric selection strategies are still burdened by methodological constraints that hinder conventional SELEX methods. Functional RNAs must be isolated, usually by time-consuming chromatography or electrophoresis procedures. The selected RNAs must be amplified, and typical methods for amplification using PCR can give artifacts that require troubleshooting attention to overcome. Until methodological advances are made in aptamer selection technology, RNA engineers might only slowly add to the limited collection of aptamers with validated function in cells.
Engineering aptamer-based genetic control systems
There is a growing list of reports demonstrating engineered aptamer-based RNA switches (or ‘synthetic riboswitches’), and there are several recent reviews7–11, 78, 79 that already discuss these studies in considerable detail. In the following paragraphs, we focus our discussion on how synthetic riboswitches compare with their natural counterparts and on some of the strategies and methods used to create gene control constructs based on aptamers. We have organized our discussion by classifying synthetic riboswitches according to their proposed mechanism of action.
Aptamers as steric blocks to ribosome scanning
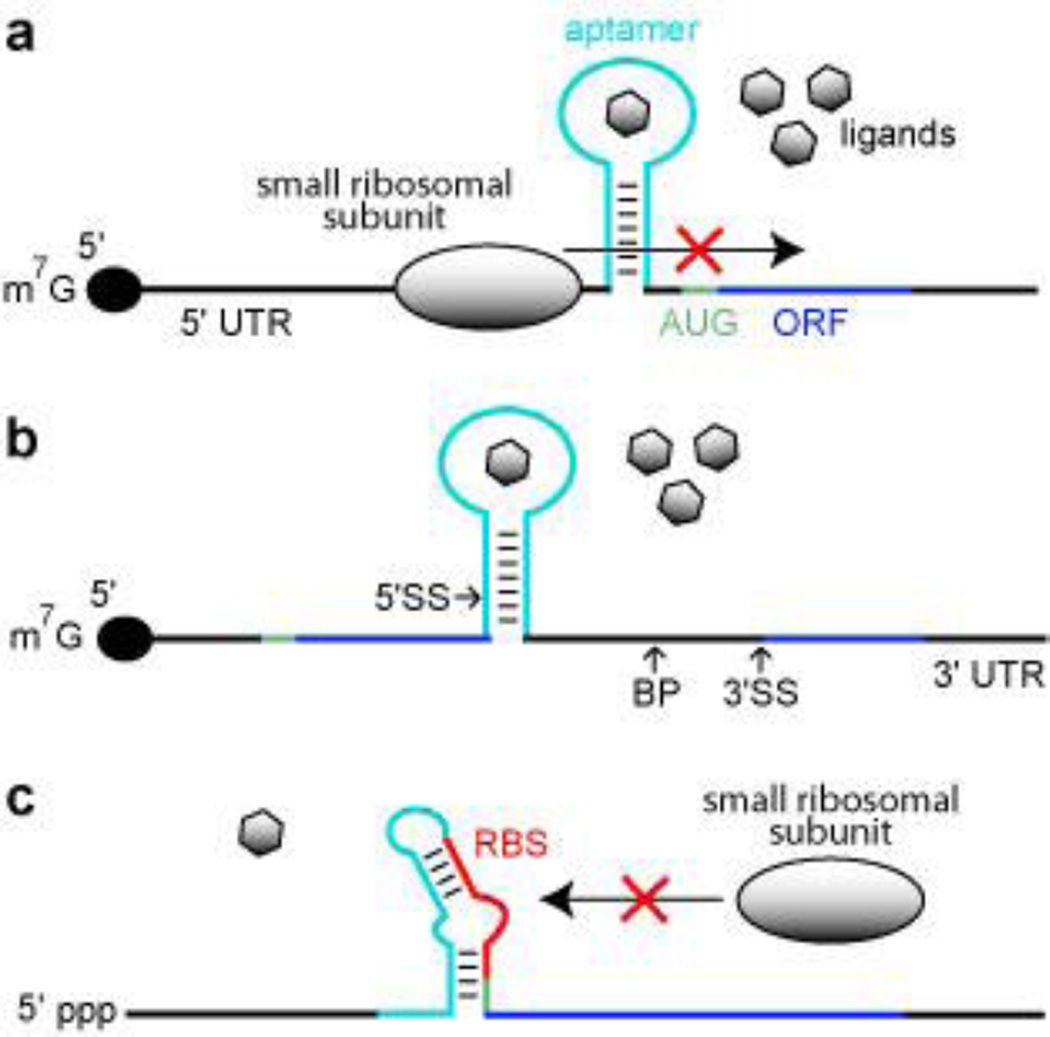
One of the simplest conceivable mechanisms by which an aptamer can be used to regulate gene expression in eukaryotes is to harness a ligand-bound aptamer as a molecular roadblock. Since eukaryotic ribosomes commonly recognize 5′ cap structures of mRNAs and scan along the single-stranded 5′ UTR for a start codon,80 the placement of an aptamer in this region that forms a thermodynamically stable complex with a ligand should permit translation modulation. A mechanism involving either the ligand-mediated disruption of ribosome assembly or scanning has been invoked for gene regulation effects observed using aptamers for antibiotics33 and for dye compounds33, 34 (Figure 2a).
Figure 2.
Aptamer-based synthetic riboswitches. (a) Aptamer-mediated inhibition of ribosomal scanning in S. cerevisiae. Ligand binding by the aptamer stabilizes a structure within the 5′ UTR that precludes the ribosome from reaching the AUG start codon. Black lines represent UTRs, green line represents the start codon, dark blue line identifies the ORF, and the light blue line represents the aptamer domain. (b) Aptamer mediated inhibition of mRNA splicing in S. cerevisiae. Ligand binding stabilizes a structure that sequesters the 5′ splice site (5′ SS), which precludes efficient splicing. BP and 3′ SS designate the branch point and the 3′ splice site, respectively. Additional annotations are as described in a. (c) Aptamer mediated inhibition of ribosome binding in E. coli. The absence of ligand binding allows the aptamer sequence to base pair with the ribosome binding site (RBS), which inhibits ribosome binding. Annotations are as described in a.
A more recent study using engineered yeast transcripts81, 82 indicates that locating the aptamer close to the 5′ cap favors ligand control of ribosome loading, while locating the aptamer close to the start codon favors ligand control of ribosome scanning. Therefore, even when aptamers are used without integration with an expression platform, there is variability in both mechanism and in the level of gene control that is observed when the aptamer is placed at different locations in the 5′ UTR.
Despite the simplicity of such a mechanism, there are no known natural examples of this mechanism in bacteria or in eukaryotes. Since bacterial ribosomes initiate translation by binding to a ribosome binding site located a few nucleotides upstream of a translation start codon83, it is not surprising that roadblock aptamers have not been identified in these organisms. In contrast, eukaryotes seem far more likely to carry roadblock aptamers. However, since so few examples of metabolite-binding aptamers have been found in eukaryotes, it is not clear how frequently used this mechanism might be.
Rational design of aptamer-regulated mRNA splicing
All of the TPP sensing riboswitches examined in any detail in eukaryotes reside in introns located in the 5′ UTRs59–61, the coding region61, 64, or the 3′ UTRs59, 62, 63 of eukaryotic mRNAs. These riboswitches control gene expression via regulation of pre-mRNA splicing, which suggests that this mechanism could be a favorable one to exploit for engineering gene control systems.
There have been several recent reports describing the successful harnessing of nuclear pre-mRNA splicing as a mechanism for engineered riboswitches. For example, Gaur and colleagues have created theophylline-dependent splicing constructs wherein ligand binding to the aptamer blocks the 3′ splice site84, 85, or the branch site69. Similarly, Suess and colleagues have created a collection of tetracycline-responsive gene control constructs in yeast69. For example, one “off” switch construct exhibited 16-fold modulation of reporter gene expression was shown using in-line probing29, 86 to sequester the 5′ splice site when ligand binds. Therefore this construct most likely represses gene expression by controlling spliceosome access to the 5′ splice site (Figure 2b).
These studies demonstrate that all the main features of nuclear introns can be exploited to control gene expression by inserted aptamers. Unfortunately, not every rationally-designed construct will function as a robust genetic switch, and so a certain amount of in vitro or in vivo screening is required to find constructs with the desired ‘on’ or ‘off” switch function with the dynamic range necessary for an application. For the yeast example69, researchers placed introns of the yeast actin and U3 genes directly downstream of the start codon of a reporter gene. The reporter-intron fusion constructs were further modified to contain a single tetracycline aptamer at various locations within the intron close to either the 5′ splice site (5′ SS) or the branch point. In vivo reporter assays were then used to screen for constructs that exhibited robust responses to the addition of tetracycline. Since a no-fail process for engineering riboswitches based purely on rational design strategies is unlikely to be created soon, in vivo screening for the performance characteristics desired will be an essential part of future engineering efforts.
As is found with tandem riboswitches in nature41–44, multiple engineered riboswitches could be used to control a single gene to yield greater regulatory complexity or to increase the dynamic range of gene control. To demonstrate the latter principle, Suess and colleagues69 integrated two tetracycline-modulated splicing domains into a single ORF to generate a construct that exhibited as much as a 32-fold modulation of gene expression. A similar effect was observed when they integrated synthetic riboswitches for tetracycline that independently block scanning ribosomes or that control splicing. While tandem synthetic riboswitches for the same ligand could be used to increase the dynamic range for gene expression, tandem arrangements of synthetic riboswitches with different ligand specificities could be used to set gene expression at specific levels within this dynamic range using different ligands as opposed to using different concentrations of the same ligand.
Synthetic riboswitches for use in bacteria
Many bacteria make extensive use of metabolite-sensing riboswitches, and in this regard they should be ideal organisms for use in engineering and applying synthetic riboswitches. However, a few aspects of bacterial biochemistry can confound engineering efforts. For example, mechanisms such as aptamer roadblocks and aptamer control of nuclear splicing that have been validated in eukaryotes69, 84, 85 are not transferable to bacteria.
For the same reason, bacteria might not always make good test sites for synthetic riboswitches that are intended to be used in eukaryotes. Synthetic riboswitches that control transcription termination or translation initiation in bacteria will be unlikely to do the same in eukaryotes. However, a synthetic riboswitch that forms a strong local structure that controls transcription termination or translation initiation in bacteria may also form a strong local structure when moved to a eukaryotic mRNA. If this structure is positioned as a roadblock or is designed to include a sequence element for nuclear pre-mRNA splicing, then the same synthetic riboswitch sequence might me made to use different mechanisms to function in organisms from different domains of life.
One of the most challenging factors restricting the design of synthetic riboswitches in bacteria is the speed at which the riboswitch must function. The turnover of mRNA transcripts in bacteria can be orders of magnitude faster than that of eukaryotic mRNAs, with some bacterial mRNAs having a half life measured in seconds. This can place severe kinetic constraints on the function of aptamers and expression platforms, as the aptamer and its flanking control region must fold rapidly and influence mRNA transcription or translation before the mRNA is naturally destroyed.
It is known that some natural bacterial riboswitches operate under kinetic41, 48, 49 rather than thermodynamic87 constraints. For example, if a riboswitch fails to bind its ligand before RNA polymerase moves beyond the influence of the transcription terminator in an expression platform, then the opportunity for the riboswitch to control expression is lost. Some riboswitch aptamers simply do not have enough time to reach thermodynamic equilibrium with their ligands, and this causes them to trigger gene control only when the ligand is present at concentrations much higher than the dissociation constant (KD) for the aptamer-ligand complex48, 49, 88, 89.
Given these constraints, it is likely that the development of synthetic riboswitches for use in bacteria would benefit greatly from employing in vivo screening for function. Recently, Gallivan and coworkers have reported the use of in vivo screening to identify numerous theophylline-dependent riboswitches. 90 Variable-length random-sequence linkers ranging from 4 to 8 random-sequence nucleotides were placed between a theophylline aptamer and the ribosome binding site of a β-galactosidase reporter gene. Functional synthetic riboswitches were identified by screening Escherichia coli colonies for theophylline-induced reporter gene expression. The vast majority of sequences from the population of constructs were unresponsive to theophylline, but several dozen constructs were confirmed to function as synthetic riboswitches.
One engineered riboswitch (clone 8.1) identified from the random N8 population exhibits 36-fold activation of gene expression in the presence of theophylline. This RNA controls gene expression via sequestering the ribosome binding site (Figure 2c), and thus mimics the mechanism used by many naturally occurring riboswitches.38 In addition, the authors provide some evidence suggesting that the construct could be thermodynamically controlled. If true, this synthetic riboswitch mimics the function of a natural adenine-sensing riboswitch that is able to equilibrate between ligand bound and unbound states on a timescale that would permit translation to be controlled even after transcription is completed.
In a striking demonstration of biological function, the clone 8.1 riboswitch was used to create bacterial cells whose motility is controlled by theophylline.91 This was achieved simply by grafting the synthetic riboswitch to an mRNA encoding the CheZ protein of E. coli. CheZ is a phosphatase whose action is required for E. coli to rotate their flagellar motor in a counterclockwise fashion to generate directional motion of the cell. Since the clone 8.1 riboswitch activates gene expression when bound to its ligand, E. coli cells lacking the wild-type cheZ gene and carrying only the riboswitch-cheZ mRNA fusion actively colonize trails of theophylline that have been applied to solid media.
This series of experiments demonstrates the promise of applying genetic screening protocols to identify synthetic riboswitches that have the performance characteristics needed for cellular function (Figure 3). A recent report by Suess and colleagues describes the utility of genetic screening in eukaryotes as well.68 Such methods should be useful for creating additional synthetic riboswitches that also can be moved to different mRNAs retention of gene control activity. To forward this goal, Gallivan and coworkers have recently reported the harnessing of the cell motility phenotype to directly select for variant theophylline-sensing constructs that control gene expression.92 Instead of screening colonies for ligand-dependent reporter gene activity, they select for changes in cell motility upon theophylline addition. This rapid and inexpensive cell-based selection method should be useful for isolating a variety of synthetic riboswitches based on aptamer constructs that are validated to function in cells.
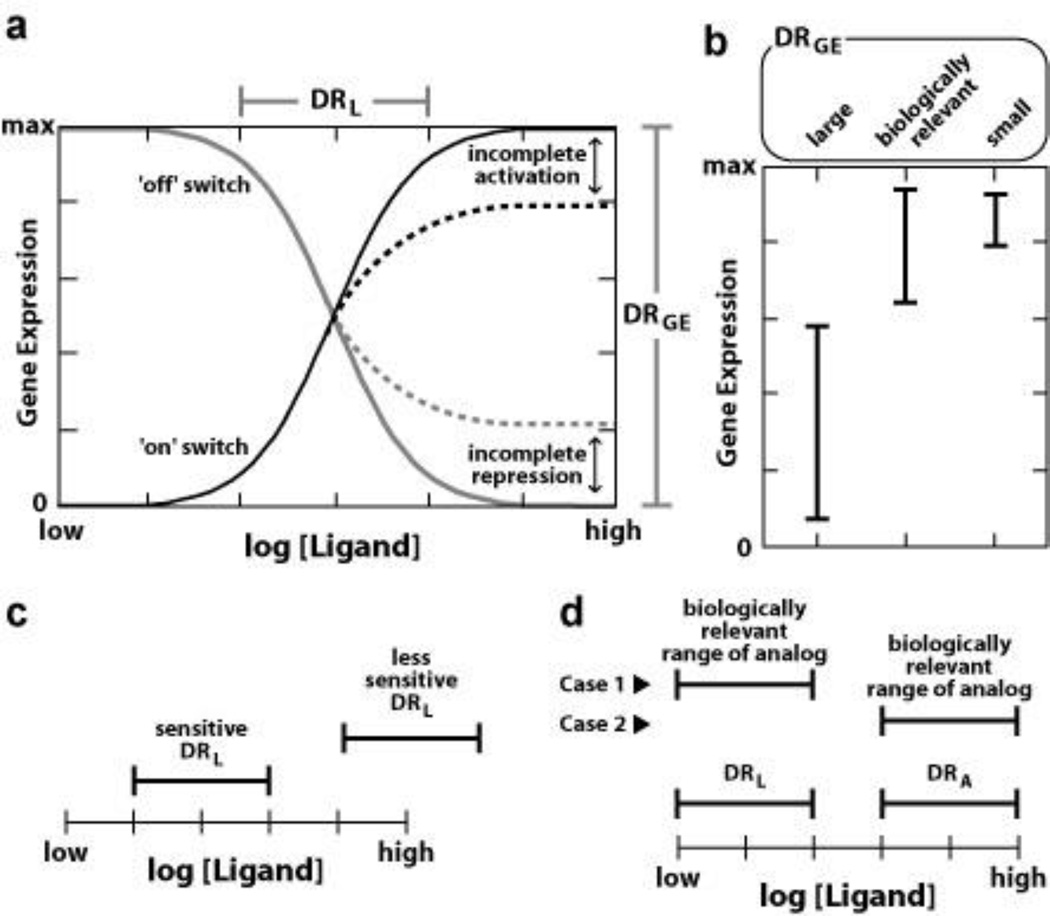
Figure 3.
Functional parameters for aptamer- and allosteric ribozyme-based gene control elements. (a) Each RNA switch will exhibit a dynamic range for ligand binding (DRL) and for gene expression (DRGE). The DRGE will vary widely among synthetic or natural riboswitches. The DRL should exhibit an 88-fold change in ligand concentration required to range from 10% to 90% gene expression for a riboswitch that carries a single aptamer domain,43 unless other factors are in play (e.g. non-linear cellular uptake of ligand; RNA folding problems). Dashed lines indicate the effects of imperfect gene control function, observed when the riboswitch RNA misfolds or otherwise fails to perfectly activate or repress gene expression even when folded correctly. (b) Comparison of theoretical DRGE ranges reveals that there will be a biologically relevant DRGE that natural or synthetic riboswitches need to encompass for their action to exhibit meaningful gene control. It is possible that synthetic riboswitches that exhibit the best DRGE (large) might be less useful than a construct that has a far poorer DRGE (small) that overlaps the biologically relevant range. (c) Sensitivity of ligand binding is critical for synthetic riboswitch utility. Less sensitive constructs might exhibit a DRL that is outside the range of ligand concentrations that can be attained in cells. (d) Specificity of ligand recognition is critical for synthetic riboswitch utility. If the aptamer has a DRL that is orders of magnitude better that that for a close analog (DRA) then gene control should be selectively triggered by the desired ligand if the analog is present in similar concentrations. However, if the analog naturally is present at concentrations that are orders of magnitude higher in concentration than the ligand, inappropriate regulation of gene expression may result.
Engineering aptamer-ribozyme fusions for gene control
As noted above, natural glmS ribozymes use ligand-triggered RNA cleavage to control gene expression. Since the GlcN6P ligand is a cofactor for the cleavage reaction, and not just a passive allosteric ligand, exploiting the precise mechanism of glmS ribozymes with synthetic riboswitches would severely restrict ligand choices to those that could function as cofactors. The development of allosteric ribozymes26 as synthetic riboswitches would permit a broader range of ligands to be used.
Challenges faced by ribozyme engineers
Although some advances have been made that bring allosteric ribozymes closer to utility as designer gene control elements, there remain some substantial barriers to their widespread use as synthetic riboswitches. Initially, a ribozyme with the reactivity necessary to affect gene expression must be identified for use as a catalytic platform. This ribozyme must have a structural feature whose folding is important for active site function, and where the bio-compatible aptamer can be grafted to efficiently control ribozyme action. Even if the independent ribozyme and aptamer modules chosen are ideal for in vivo use, the rate constants for ribozyme activity and the KD values for ligand binding to the aptamer frequently are diminished when the two domains are linked to form an allosteric construct. If the parental modules have rate constants or KD values that are barely sufficient for in vivo utility, any erosion of functional characteristics when allosteric ribozymes are made could render them useless for gene control applications. Furthermore, if an allosteric ribozyme exhibits adequate functional characteristics for in vivo use, any propensity for misfolding will limit the dynamic range for gene control due to incomplete gene repression or activation.
Self-cleaving ribozymes93 are attractive starting points for engineering synthetic riboswitches. Indeed, self-cleaving hammerhead ribozymes are commonly used as catalytic platforms for engineering allosteric ribozymes. The first examples of allosteric hammerhead ribozymes included only the catalytic core25, 26, and lacked accessory domains that were more recently found to substantially improve the rate constant for RNA cleavage.94–96 Some full-length hammerhead ribozymes are capable of regulating gene expression in mammals,97–99 and therefore it seems possible that allosteric ribozymes based upon the this self-cleaving motif can be engineered to function as designer ligand-responsive gene control elements.
Designing allosteric ribozymes for gene control
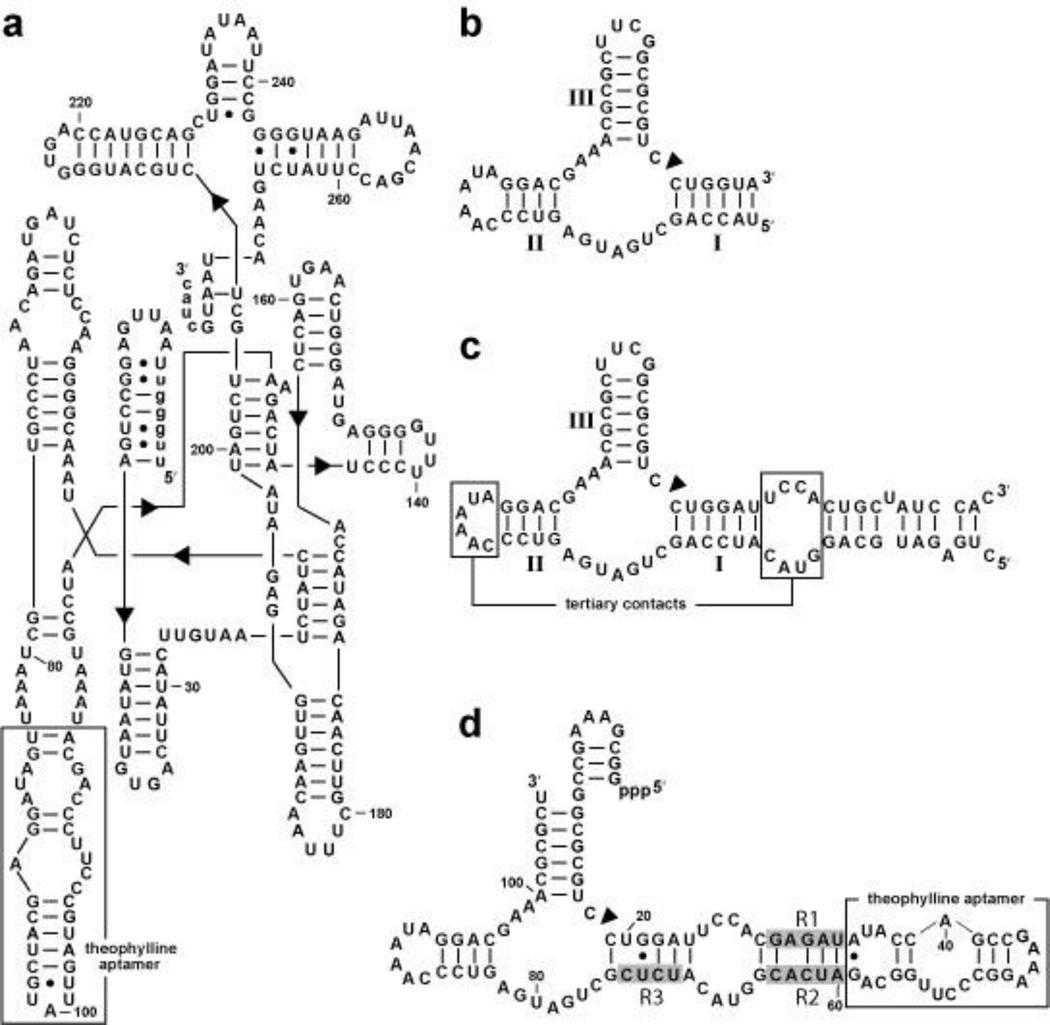
Allosteric ribozymes that modulate gene expression have been generated by using the Group I self-splicing intron as a catalytic platform.32 Eight constructs were created by grafting the theophylline aptamer to the group I self-splicing intron via P5 or P6. One of these constructs, Th2P6 (Figure 4a), exhibited a 4-fold increase in the observed rate constant for splicing in the presence of theophylline. DNA encoding this ribozyme was cloned into a thymidylate synthase gene and the vectors were introduced into a strain of E. coli otherwise lacking thymidylate synthase. Cells encoding the fusion construct were unable to grow on minimal medium lacking thymidine, unless supplemented with theophylline. This assay demonstrating biological function of the RNA switch presumably could be employed to select for functional gene control elements from large population of allosteric ribozyme candidates.
Figure 4.
Allosteric ribozymes as catalytic platforms for synthetic riboswitches. (a) Sequence and secondary structure model of an allosteric group I intron (Th2P6).32 Uppercase and lowercase letters identify intron and exon sequences, respectively. (b) A sequence encompassing only the minimal catalytic core of a hammerhead ribozyme with stems I through III identified. (c) Sequence and secondary structural model of the full-length Schistosoma mansoni hammerhead ribozyme.124 A construct based on this ribozyme is known to function in vivo.98 (d) Sequence and secondary structural model of a high-speed allosteric hammerhead ribozyme derived from the parental ribozyme in c. R1 through R3 (shaded nucleotides) were derived by in vitro selection from random-sequence domains. Theophylline binding is expected to permit the RNA to form tertiary contacts between the accessory domains in stems I and II and thereby exhibit high ribozyme activity.
In some instances, rapid switching between structural states will be required to efficiently control gene expression.48, 49 Therefore, subtle structural alterations involving modest thermodynamic differences could be a desirable feature of allosteric ribozyme mechanisms. Some allosteric ribozymes are believed to function via a slip-structure conformational change, wherein realignment of base pairs in a ‘communication module’ allows ligand binding to trigger changes in the adjoining ribozyme active site.25, 26, 100 Since communication modules are small, both rational design and computer-aided design strategies have been successfully used to create aptamer-ribozyme fusion constructs that exhibit robust allosteric activities. For example, an algorithm was used to computationally select 23 allosteric ribozyme candidates from a pool of 4096 sequences.101 Three of these constructs, made by fusing a FMN aptamer to a hammerhead ribozyme, exhibited 4- to 60-fold activation in the presence of FMN.
More recently, researchers appear to have created several allosteric ribozymes that function in vivo.102 Aptamers for theophylline or tetracycline were grafted to hammerhead ribozymes with the expectation that some might function in yeast cells to modulate the expression of a reporter gene when the appropriate ligands are introduced. The most responsive constructs identified exhibit no more than three-fold modulation of reporter gene expression in response to ligand addition, which demonstrates that engineering robust RNA switches for in vivo application is nontrivial. Similarly, these investigators have created numerous additional allosteric ribozyme constructs that frequently alter gene expression by less than two fold.103 The dynamic ranges of gene expression exhibited by these cis acting RNA switches102, 103 are far lower than the dynamic ranges for the functions of many allosteric ribozyme reported previously.26, 28–31, 74, 81, 90, 104, 105 However, it is not yet clear whether these constructs exhibit poor ON/OFF gene expression ratios because the RNA constructs simply are poor allosteric ribozymes or because the allosteric ribozyme activities are poorly manifest in a cellular environment. Additional experiments are needed to assess whether in vivo function requires self-cleavage for these102, 103 and other examples106, 107 of allosteric ribozymes, and whether the proposed mechanisms for gene control are correct. Regardless, even modest RNA switch function, with or without ribozyme cleavage, could be used to generate therapeutically useful levels of a protein product if only small changes in gene expression are needed.
A greater understanding of the mechanisms of gene control used by existing engineered hammerhead ribozyme constructs should also help assess whether these constructs based on the full-length hammerhead construct have a rate constant for self cleavage that is sufficient for in vivo application. In addition, definitive establishment of mechanism could facilitate the engineering of allosteric ribozymes that exhibit even greater dynamic range for gene control. Allosteric ribozymes based on the minimal hammerhead ribozyme usually incorporate the aptamer in stem II of the ribozyme (Figure 4b). However, this arrangement would disrupt the tertiary contacts used by full-length hammerhead ribozymes to achieve their high rate constants for RNA cleavage (Figure 4c). Therefore, a different attachment point is required. We have employed in vitro selection to isolate allosteric hammerhead ribozymes that retain the natural accessory domains, but integrate a theophylline aptamer in the stem I region of the RNA.74
A construct carrying a theophylline aptamer was fused to a full-length hammerhead ribozyme carrying three random-sequence domains that link to the aptamer (Figure 4d). Ligand-mediated alteration of the structure of stem I should modulate ribozyme activity by permitting or disrupting tertiary structure formation between stems I and II. In vitro selection yielded variant RNAs with observed rate constants (kobs) as high as 8 min−1 in the presence of theophylline and increases in rate constants (kobs) for RNA cleavage of up to 285 fold compared to those measured in the absence of ligand. Although these kobs values are predicted to be sufficient for gene control applications, these ribozymes do not display robust allosteric modulation of gene expression in mammalian cells.
Since there are many possible reasons for why these allosteric ribozymes selected in vitro do not control gene expression in cells (Figure 3), in vivo selection protocols for allosteric ribozyme function may be useful to bypass these problems. Indeed, Wieland and Hartig have recently described the use of in vivo selection to isolate an aptamer-hammerhead fusion that exhibits robust gene control activity in E. coli.104 These researchers fused a theophylline aptamer to stem III of a full-length hammerhead ribozyme to create an allosteric ribozyme wherein ligand binding activates ribozyme activity. The synthetic riboswitch is designed to work by ribozyme cleavage of the 5′ portion of the mRNA, which otherwise would block access to the ribosome binding site for the adjacent ORF. Similar allosteric hammerhead ribozyme designs also could be used in eukaryotes where ribozyme cleavage would destabilize mRNA by removing the 5′ cap, as has recently been demonstrated in apicomplexan parasites.108
Future directions
Surveying the landscape of natural and synthetic riboswitches reveals that RNA switches that use aptamer-only and aptamer-ribozyme mechanisms for gene control are possible. Both modes of riboswitch-mediated gene control have advantages and disadvantages, and therefore developing both technologies seems prudent. The main advantages of engineering aptamer-based RNA switches are that there are relatively simple methods available for their isolation, and there is no need for the aptamer to be subsequently grafted onto ribozymes. However, it is not clear that aptamer roadblocks or other structures stabilized by ligand binding will be sufficiently stable in all genetic contexts to maximally modulate gene expression. In contrast, ribozymes that splice or cleave mRNA would make irreversible changes to their substrate, and therefore they could modulate expression of an ORF even if the ligand-induced shape change is transient. Of course, integrating two RNA domains to work in concert and exhibit robust ribozyme activity in vivo is more challenging.
The use of aptamer or ribozyme fusions with mRNAs to control gene expression mimics a mode of regulation that is commonly found with riboswitches. However, there are intermolecular interactions between regulatory RNAs and gene expression machinery that also could be exploited to create designer gene control elements made of RNA. Aptamers or ribozymes could be grafted to microRNAs to control their processing and subsequent use in gene control.109, 110 RNAs such as 6S in bacteria111, 112 and B2 RNA in eukaryotes113, 114 naturally bind to RNA polymerase and more globally regulate transcription.115 The number of trans-acting non-coding RNAs in bacteria and in eukaryotes is far greater.116, 117 The functions of these RNAs likewise could be engineered to exhibit control by small-molecule ligand binding.
The greater diversity of engineered RNA gene control options is already being explored. For example, Liu and coworkers have reported the in vivo selection of an RNA that functions as a trans-acting ligand-dependent transcriptional activator in S. cerevisiae.67, 118 They created their own trans-acting RNA activator of transcription via in vivo selection.117 This was accomplished by recruiting a random-sequence population of RNAs to the His3 gene using MS2 RNA hairpins and a LexA-MS2 fusion protein. Members of the RNA population with specific structures that can activate His3 expression were selected by growing cells in medium lacking histidine. This in vivo selection resulted in the isolation of an RNA that activates transcription 53 fold. In a subsequent in vivo selection, the RNA-based transcriptional activator was converted into a ligand-dependent RNA-based transcriptional activator67 fusing a tetramethylrosamine aptamer119 to the RNA-based transcription activator via a random N7 domain. In vivo selection again was used to isolate an RNA transcriptional activator that is 10-fold more active in the presence of the ligand.
The vast majority of designer riboswitches engineered to date have been generated using rational design or in vivo screening. These methods limit the sequence space that can be explored when engineered riboswitches and therefore might not be well suited to identify gene control elements that match the performance characteristics of naturally occurring riboswitches. Increasingly, new methods use in vivo selection techniques are being developed for the isolation of engineered riboswitches.92, 120 These techniques more closely mimic the natural process of evolution by allowing more sequence space to be explored than current in vivo screening methods, but with far time constraints than nature demands.
Current cell-based selection methods are usually restricted to sample sizes of no more than a few million variants. Perhaps RNA switches with the desired properties can be successfully isolated from populations of this size if the variants are derived from pre-existing aptamer and ribozyme domains that are validated to function in cells. If novel aptamer or ribozyme domains are desired, then an in vitro selection method must be employed that samples far greater sequence space. Therefore, the best strategy may be to use in vitro selection and in vivo selection in series to isolate distinct classes of synthetic riboswitches. In vitro selection allows one to reach deep in to sequence space for millions of novel variants of allosteric constructs. In vivo selection is then used to sift through the enriched population for those that retain the desired function when expressed in cells. Such methods should yield synthetic riboswitches that have far greater ligand selectivity and mechanistic diversity than the natural metabolite-responsive riboswitches known to date.
Acknowledgments
We thank members of the Breaker lab for helpful discussions. This work was supported by the Howard Hughes Medical Institute, and by grants from the NSF and the NIH.
References
- 1.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 2.Osborne SE, Ellington AD. Nucleic acid selection and the challenge of combinatorial chemistry. Chem Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 4.Breaker RR. Engineered allosteric ribozymes as biosensor components. Curr Opin Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- 5.Silverman SK. Rube Goldberg goes (ribo)nuclear? Molecular switches and sensors made from RNA. RNA. 2003;9:377–383. doi: 10.1261/rna.2200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breaker RR. Natural and engineered nucleic acids as tools to explore biology. Nature. 2004;432:838–845. doi: 10.1038/nature03195. [DOI] [PubMed] [Google Scholar]
- 7.Davidson EA, Ellington AD. Engineering regulatory RNAs. Trends Biotechnol. 2005;23:109–112. doi: 10.1016/j.tibtech.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 9.Bauer G, Suess B. Engineered riboswitches as novel tools in molecular biology. J Biotechnol. 2006;124:4–11. doi: 10.1016/j.jbiotec.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Gallivan JP. Toward reprogramming bacteria with small molecules and RNA. Curr Opin Chem Biol. 2007;11:612–619. doi: 10.1016/j.cbpa.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Win MN, Smolke CD. RNA as a versatile and powerful platform for engineering genetic regulatory tools. Biotechnol Genet Eng Rev. 2007;24:311–346. doi: 10.1080/02648725.2007.10648106. [DOI] [PubMed] [Google Scholar]
- 12.Ellington A. What’s so great about RNA? ACS Chem. Biol. 2007;2:445–448. doi: 10.1021/cb700139t. [DOI] [PubMed] [Google Scholar]
- 13.Doudna JA, Lorsch JR. Ribozyme catalysis: not different, just worse. Nat Struct Mol Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- 14.Ptashne M, Gann A. Genes & Signals. New York: CSHL Press, Cold Spring Harbor; 2002. [Google Scholar]
- 15.Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 16.Toniatti C, Bujard H, Cortese R, Ciliberto G. Gene therapy progress and prospects: transcription regulatory systems. Gene Therapy. 2004;11:649–657. doi: 10.1038/sj.gt.3302251. [DOI] [PubMed] [Google Scholar]
- 17.Goverdhana S, Puntel M, Xiong W, Zirger JM, Barcia C, Curtin JF, et al. Regulatable gene therapy expression systems for gene therapy applications: progress and future challenges. Molecular Therapy. 2005;12:189–211. doi: 10.1016/j.ymthe.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore I, Samalova M, Kurup S. Transactivated and chemically inducible gene expression in plants. Plant J. 2006;45:651–683. doi: 10.1111/j.1365-313X.2006.02660.x. [DOI] [PubMed] [Google Scholar]
- 19.Phan TT, Schumann W. Development of a glycine-inducible expression system for Bacillus subtilis. J Biotechnol. 2007;128:486–499. doi: 10.1016/j.jbiotec.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Dugas DV, Bartel B. MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol. 2004;7:512–520. doi: 10.1016/j.pbi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 21.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 22.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 24.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 25.Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 26.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaeger L, Chworos A. The architectonics of programmable RNA and DNA nanostructures. Curr Opin Struct Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Koizumi M, Soukup GA, Kerr JNQ, Breaker RR. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat Struct Biol. 1999;6:1062–1071. doi: 10.1038/14947. [DOI] [PubMed] [Google Scholar]
- 29.Soukup GA, DeRose EC, Koizumi M, Breaker RR. Generating new ligand-binding RNAs by affinity maturation and disintegration of allosteric ribozymes. RNA. 2001;7:524–536. doi: 10.1017/s1355838201002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seetharaman S, Zivarts M, Sudarsan N, Breaker RR. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat Biotechnol. 2001;19:336–341. doi: 10.1038/86723. [DOI] [PubMed] [Google Scholar]
- 31.Hesselberth JR, Robertson MP, Knudsen SM, Ellington AD. Simultaneous detection of diverse analytes with an aptazyme ligase array. Anal Biochem. 2003;312:106–112. doi: 10.1016/s0003-2697(02)00441-4. [DOI] [PubMed] [Google Scholar]
- 32.Thompson KM, Syrett HA, Knudsen SM, Ellington AD. Group I aptazymes as genetic regulatory switches. BMC Biotechnol. 2002;2:21–33. doi: 10.1186/1472-6750-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 34.Grate D, Wilson C. Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg Med Chem. 2001;9:2565–2570. doi: 10.1016/s0968-0896(01)00031-1. [DOI] [PubMed] [Google Scholar]
- 35.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 36.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 37.Coppins RL, Hall KB, Groisman EA. The intricate world of riboswitches. Curr Opin Microbiol. 2007;10:176–181. doi: 10.1016/j.mib.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nahvi A, Sudarsan N, Ebert MS, Zoy X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 40.Stoddard CD, Batey RT. Mix-and-match riboswitches. ACS Chem Biol. 2006;1:751–754. doi: 10.1021/cb600458w. [DOI] [PubMed] [Google Scholar]
- 41.Breaker RR. Complex riboswitches. Science. 2008;319:1795–1797. doi: 10.1126/science.1152621. [DOI] [PubMed] [Google Scholar]
- 42.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, et al. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 43.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 45.Jose AM, Soukup GA, Breaker RR. Cooperative binding of effectors by an allosteric ribozyme. Nucleic Acids Res. 2001;29:1631–1637. doi: 10.1093/nar/29.7.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 47.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 48.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 49.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 50.Nou XW, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci USA. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 52.Fuchs RT, Grundy FJ, Henkin TM. S-adnosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proc Natl Acad Sci USA. 2007;104:4876–4880. doi: 10.1073/pnas.0609956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004;32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 55.Klein DJ, Ferré-D’Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 56.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the glmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 21:3356–3368. doi: 10.1101/gad.1605307. 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 59.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, et al. Thiamine-regulated gene expression of spergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 61.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 62.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woodson SA. Structure and assembly of group I introns. Curr Opin Struct Biol. 2005;15:324–330. doi: 10.1016/j.sbi.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 66.Harvey I, Garneau P, Pelletier J. Inhibition of translation by RNA-small molecule interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 68.Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schroeder R, Suess B. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007;35:4179–4185. doi: 10.1093/nar/gkm425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J Am Chem Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 71.Koizumi M, Soukup GA, Kerr JN, Breaker RR. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat Struct Biol. 1999;6:1062–1071. doi: 10.1038/14947. [DOI] [PubMed] [Google Scholar]
- 72.Soukup GA, Emilsson GA, Breaker RR. Altering molecular recognition of RNA aptamers by allosteric selection. J Mol Biol. 2000;298:623–632. doi: 10.1006/jmbi.2000.3704. [DOI] [PubMed] [Google Scholar]
- 73.Zivarts M, Liu Y, Breaker RR. Engineered allosteric ribozymes that respond to specific divalent metal ions. Nucleic Acids Res. 2005;33:622–631. doi: 10.1093/nar/gki182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Link KH, Guo L, Ames TD, Yen L, Mulligan RC, Breaker RR. Engineering high-speed allosteric hammerhead ribozymes. Biol Chem. 2007;388:776–786. doi: 10.1515/BC.2007.105. [DOI] [PubMed] [Google Scholar]
- 75.Roth A, Breaker RR. Selection in vitro of allosteric ribozymes. Methods Mol Biol. 2004;252:145–164. doi: 10.1385/1-59259-746-7:145. [DOI] [PubMed] [Google Scholar]
- 76.Nutiu R, Li YF. In vitro selection of structure-switching signaling aptamers. Angew Chem Int Ed Engl. 2005;44:1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 77.Morse DP. Direct selection of RNA beacon aptamers. Biochem Biophys Res Commun. 2007;359:94–101. doi: 10.1016/j.bbrc.2007.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suess B, Weigand JE. Engineered riboswitches. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 79.Wieland M, Hartig J. Artificial riboswitches: synthetic mRNA-based regulators of gene expression. ChemBioChem. 2008;9:1873–1878. doi: 10.1002/cbic.200800154. [DOI] [PubMed] [Google Scholar]
- 80.Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;98:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 81.Hanson S, Berthelot K, Fink B, McCarthy JEG, Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol Microbiol. 2003;49:1627–1637. doi: 10.1046/j.1365-2958.2003.03656.x. [DOI] [PubMed] [Google Scholar]
- 82.Hanson S, Bauer G, Fink B, Suess B. Molecular analysis of a synthetic tetracycline-binding riboswitch. RNA. 2005;11:503–511. doi: 10.1261/rna.7251305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaberdin VR, Bläsi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 84.Kim D-S, Gusti V, Pillai SG, Gaur RK. An artificial riboswitch for controlling pre-mRNA splicing. RNA. 2005;11:1667–1677. doi: 10.1261/rna.2162205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gusti V, Kim DS, Gaur RK. Sequestering of the 3′ splice site in a theophylline-responsive riboswitch allows ligand-dependent control of alternative splicing. Oligonucleotides. 2008;18:93–99. doi: 10.1089/oli.2007.0107. [DOI] [PubMed] [Google Scholar]
- 86.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 88.Wang JX, Breaker RR. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol. 2008;86:157–168. doi: 10.1139/O08-008. [DOI] [PubMed] [Google Scholar]
- 89.Tomšič J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenoslymethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insight into their function. Chem Biol. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J Am Chem Soc. 2007;129:6807–6811. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Topp S, Gallivan JP. Random walks to synthetic riboswitches – A high-throughput selection based on cell motility. Chembiochem. 2008;9:210–213. doi: 10.1002/cbic.200700546. [DOI] [PubMed] [Google Scholar]
- 93.Cochrane JC, Strobel SA. Catalytic strategies of self-cleaving ribozymes. Acc Chem Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 94.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 95.De la Pena M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shepotinovskaya IV, Uhlenbeck OC. Catalytic diversity of extended hammerhead ribozymes. Biochemistry. 2008;47:7034–7042. doi: 10.1021/bi7025358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, et al. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- 98.Yen L, Magnier M, Weissleder R, Stockwell BR, Mulligan RC. Identification of inhibitors of ribozyme self-cleavage in mammalian cells via high-throughput screening of chemical libraries. RNA. 2006;12:797–806. doi: 10.1261/rna.2300406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–903. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kertsburg A, Soukup GA. A versatile communication module for controlling RNA folding and catalysis. Nucleic Acids Res. 2002;30:4599–4606. doi: 10.1093/nar/gkf596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hall B, Hesselberth JR, Ellington AD. Computational selection of nucleic acid biosensors via a slip structure model. Biosens Bioelectron. 2007;22:1939–1947. doi: 10.1016/j.bios.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 102.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wieland M, Hartig J. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew Chem Int Ed Engl. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- 105.Topp A, Gallivan JP. Riboswitches in unexpected places – A synthetic riboswitch in a protein coding region. RNA. doi: 10.1261/rna.1269008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ogawa A, Maeda M. An artificial aptazyme-based riboswitch and its cascading system in E. col. Chembiochem. 2008;9:206–209. doi: 10.1002/cbic.200700478. [DOI] [PubMed] [Google Scholar]
- 107.Ogawa A, Maeda M. Aptazyme-based riboswitches as label-free and detector-free sensors for cofactors. Bioorg Med Chem Lett. 2007;17:3156–3160. doi: 10.1016/j.bmcl.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 108.Agop-Nersesian C, Pfahler J, lanzer M, Meissnerr M. Functional expression of ribozymes in Apicomplexa: Towards exogenous control of gene expression by inducible RNA-cleavage. Int J Parasitol. 2008;38:673–681. doi: 10.1016/j.ijpara.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 109.An C, Trinh VB, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tuleuova N, An C, Ramanculov E, Revzin A, Yokobayashi Y. Modulating endogenous gene expression of mammalian cells via RNA-small molecule interaction. Biochem Biophys Res Commun. 2008 doi: 10.1016/j.bbrc.2008.08.112. in press. [DOI] [PubMed] [Google Scholar]
- 111.Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–784. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol. 2005;12:313–319. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- 113.Allen TA, Von Kaenel S, Goodrich JA, Jugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 114.Espinosa CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 115.Barrandon C, Spiluttini B, Bensaude O. Non-coding RNAs regulating the transcriptional machinery. Biol Cell. 2008;100:83–95. doi: 10.1042/BC20070090. [DOI] [PubMed] [Google Scholar]
- 116.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 117.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 118.Buskirk AR, Kehayova PD, Landrigan A, Liu DR. In vivo evolution of an RNA-based transcriptional activator. Chem Biol. 2003;10:533–540. doi: 10.1016/s1074-5521(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 119.Grate D, Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc Natl Acad Sci USA. 1999;96:6131–6136. doi: 10.1073/pnas.96.11.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nomura Y, Yokobayashi Y. Reengineering a natural riboswitch by dual genetic selection. J Am Chem Soc. 2007;129:13814–13815. doi: 10.1021/ja076298b. [DOI] [PubMed] [Google Scholar]
- 121.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 122.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 123.Rentmeister A, Mayer G, Kuhn K, Famulok M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007;35:3713–3722. doi: 10.1093/nar/gkm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]