Abstract
Activity-based protein profiling (ABPP) serves as a chemical proteomic platform to discover and characterize functional amino acids in proteins on the basis of their enhanced reactivity towards small-molecule probes. This approach, to date, has mainly targeted nucleophilic functional groups, such as the side chains of serine and cysteine, using electrophilic probes. We show here that "reverse-polarity" (RP)-ABPP using clickable, nucleophilic hydrazine probes can capture and identify protein-bound electrophiles in cells, including the pyruvoyl cofactor of S-adenosyl-l-methionine decarboxylase (AMD1), which we find is dynamically controlled by intracellular methionine concentrations, and a heretofore unknown modification – an N-terminally bound glyoxylyl group – in the poorly characterized protein secernin-3. RP-ABPP thus provides a versatile method to monitor the metabolic regulation of electrophilic cofactors and discover novel types of electrophilic modifications on proteins in human cells.
Summary
A chemical proteomic strategy is described for the discovery of protein-bound electrophilic groups in human cells and used to characterize dynamic regulation of the pyruvoyl catalytic cofactor in S-adenosyl-l-methionine decarboxylase and to discover an N-terminal glyoxylyl modification on Secernin proteins.
Post-translational modifications (PTMs) serve to diversify protein structure and function in important ways, including, but not limited to the regulation of protein activity, protein-protein interactions, and protein localization and stability in cells1. Proteins are subject to a wide array of chemically diverse PTMs, which have historically been discovered on a case-by-case basis by in-depth investigations of individual proteins2. Advances in mass spectrometry (MS) technologies have, however, made it possible to discover novel PTMs with remarkable scope, sensitivity, and structural resolution, leading to an expanded understanding of diverse modification states, such as the N-acetylation/acylation of lysines3 and electrophilic/oxidative modification of cysteines4,5. It is likely, however, that many functionally important PTMs remain to be discovered because they are, as-of-yet, structurally unpredicted and/or occur on proteins of unknown function3.
Various approaches have been used to discover and characterize PTMs, including exploiting their often atypical chemical reactivity for covalent tagging and enrichment6–8. Activity-based protein profiling (ABPP) also uses chemical probes that react with large classes of proteins based on shared functional and/or structural properties9. Such aberrant ‘reactivity’ can originate from conserved amino acid residues that confer special activities to proteins, such as enzymatic catalysis, and, accordingly, ABPP has been applied to characterize diverse enzyme classes in native biological systems9. To date, ABPP has principally focused on targeting nucleophilic functional groups10, largely because the main twenty proteinogenic amino acids are replete with nucleophilic side chains, but devoid of reactive electrophiles. Proteins, however, also use electrophilic groups for function, which are typically acquired through installation as covalent PTMs1,2,11 or by binding of exogenous cofactors12.
Because electrophilic PTMs define reactive centers that are uncommon and not easily predicted from the primary structures of proteins, it seems likely that additional uncharacterized electrophilic functional groups exist in the human proteome. We reasoned that, by reversing the polarity of activity probes from electrophilic to nucleophilic, ABPP could potentially be adapted to globally discover protein-bound electrophiles in native biological systems. Here, we show that “reverse polarity” (RP)-ABPP with clickable hydrazine probes can identify functional electrophilic PTMs on proteins in living cells (in situ) and reveal dynamic changes in protein-bound electrophile status in response to metabolic perturbations. We also show that RP-ABPP uncovers structurally novel electrophilic PTMs that occur on conserved residues in proteins of uncharacterized function.
Results
In situ profiling with hydrazine probes in human cells
Previous studies have used nucleophilic probes to characterize PTMs such as N-linked glycosylation13 and ADP-ribosylation14, but, in these cases, additional chemistry is required (oxidation and exogenous catalysts, respectively) to promote reactions. Nucleophilic probes have also been applied to characterize various protein modifications that are caused by oxidative stress and aging, including direct oxidation of amino acids (e.g., cysteine sulfenylation15; protein carbonylation16) and aspartyl and asparaginyl cyclized amino-succinimide modifications17, 18, respectively. Here, we aimed to complement and advance this past work by developing a chemical proteomic method to discover electrophilic PTMs that are installed into proteins for primary functional purposes. To bias our profiles toward functional electrophilic sites, we elected to capture these sites in living cells (in situ) without exposure to exogenous oxidative stressors and to focus on proteins that showed strong (near-complete) reactivity with nucleophile probes in this endogenous setting.
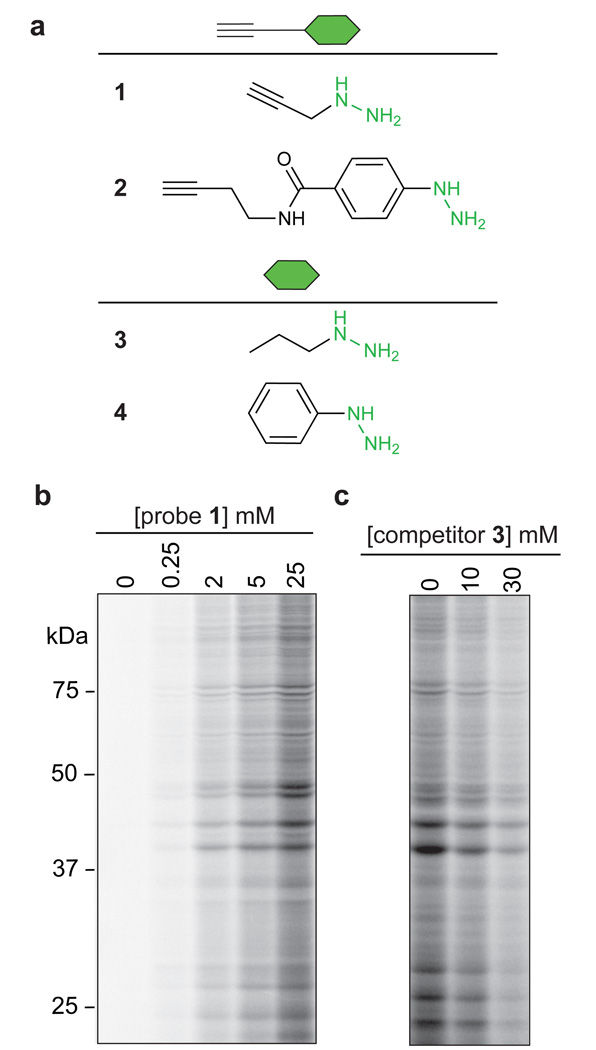
For our initial RP-ABPP studies, we selected propargyl hydrazine (probe 1; Fig. 1a) as a prototype nucleophilic probe to capture protein-bound electrophiles in cells, where the hydrazine was intended to serve as a reactive nucleophile and the alkyne as a latent affinity handle for conjugation by Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC, or ‘click’ chemistry)19 to azide reporter tags for protein detection, enrichment, and identification20. We also employed a phenyl hydrazine derivative (probe 2; Fig. 1a) as a second probe for analysis. The selection of hydrazines for RP-ABPP builds on knowledge that this class of nucleophiles can covalently inhibit enzymes that use both oxidative21 and electrophilic cofactors11, 22 (Supplementary Fig. 1). We treated HEK293T cells with varying concentrations of probe 1 (30 min, 37 °C), lysed cells, and conjugated cell proteomes to a rhodamine-azide (Rh-N3) reporter tag. Clear concentration-dependent protein labeling was observed (Fig. 1b and Supplementary Fig. 2) and this labeling was suppressed (Fig. 1c and Supplementary Fig. 2) by increasing concentrations of co-administered non-clickable propyl hydrazine competitor (3; Fig. 1a). Protein staining did not reveal any obvious changes in the expression or abundance of proteins in cells treated with probe 1 (Supplementary Fig. 2), and cell viability assays confirmed that the hydrazine probes were not toxic to cells under these conditions (Supplementary Methods).
Fig 1. Reverse polarity (RP)-ABPP with hydrazine probes in human cells.
a, Structures of hydrazine probes (alkyl probe 1 and aryl probe 2) and corresponding non-clickable analogs or competitors (3 and 4). b, SDS-PAGE analysis of the soluble proteome of HEK293T cells treated with probe 1 (30 min), revealing the concentration-dependent labeling of proteins by 1 as measured by CuAAC to a rhodamine-azide tag (fluorescent gel shown in grayscale). c, Treatment of HEK293T cells with competitor 3 blocks in a concentration-dependent manner the labeling of proteins by 1 (3 mM, 30 min).
High-reactivity targets of hydrazine probes in human cells
We next set out to identify proteins modified by probe 1 using multidimensional liquid chromatography-tandem mass spectrometry (LC/LC-MS/MS)23 combined with SILAC (Stable Isotopic Labeling of Amino acids in Cell culture) methodology24 (Fig. 2a), where cells were grown in medium containing either natural abundance lysine and arginine (‘light’) or 13C- and 15N-enriched amino acid isotopologues (‘heavy’) and subject to different probe treatment conditions followed by lysis, combination, conjugation to a biotin-azide reporter tag, enrichment by streptavidin chromatography, and quantitative proteomic analysis. MS1 and MS2 data provided information on the relative quantity and identity of enriched proteins, respectively25.
Fig 2. Identification of protein targets of hydrazine probes.
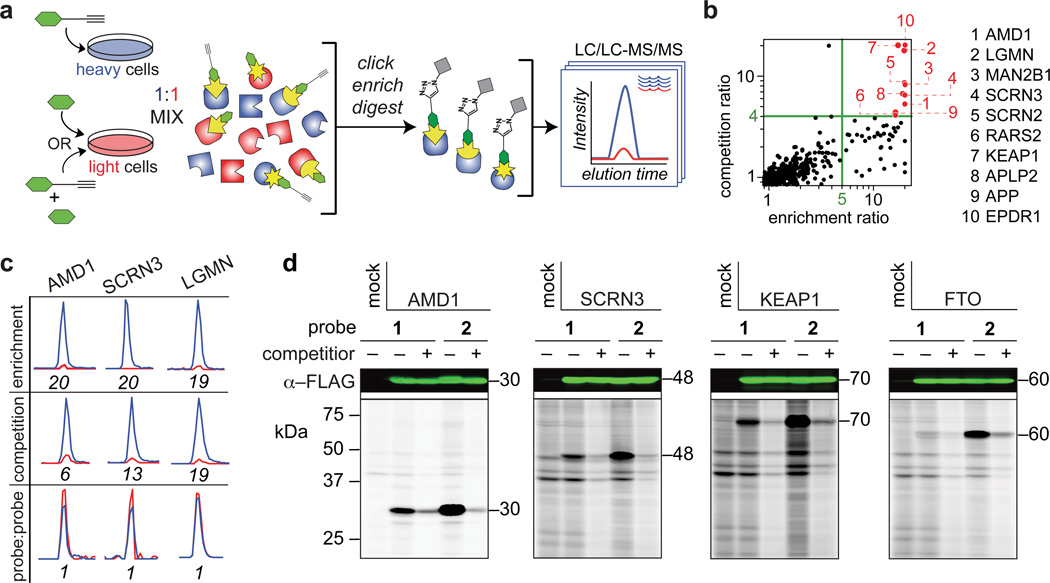
a, Schematic for MS-based quantitative (SILAC) proteomics experiments (enrichment and competition) as described in the text. Heavy (H) and light (L) cells, proteomes, and peptides are depicted in blue and red, respectively. b, Quadrant plot of average competition versus enrichment SILAC ratios from quantitative proteomics experiments (left). Probe-1 targeted proteins (upper right quadrant) are highlighted in red and listed to the right of the plot. c, Extracted parent ion chromatograms and corresponding H/L ratios for representative tryptic peptides of three protein targets of probe 1 quantified in enrichment, competition, and probe vs. probe control experiments. d, Western blots (upper) and RP-ABPP data (lower) for hydrazine probe-treated transfected cells expressing the indicated protein targets. The first lane in each panel corresponds to a control transfection (‘mock’) with the appropriate empty expression vector as described in Supplementary Methods. Molecular weights (kDa) are indicated.
Two types of experiments were performed – 1) direct enrichment of probe 1-labeled proteins from heavy cells in comparison to light control cells treated with the non-clickable hydrazine (3) at the same concentration as 1 (3 mM for 0.5 h); and 2) competition experiments where both heavy and light cells were treated probe 1, but light cells were also treated with 10X competitor 3 (Fig. 2a) – in two different human cell lines (HEK293T cells and the human breast cancer cell line MDA-MB-231). Proteins both substantially enriched (MS1heavy:light ratios > 5) and competed by 3 (MS1heavy:light ratios > 4) were considered candidate targets of probe 1. We reasoned that, by requiring both enrichment and competition, we would focus our interpretation on proteins that possess electrophilic modifications capable of near-complete reaction with probe 1 and thereby avoid following up on lower stoichiometry adducts that may originate from minor side reactions with weaker electrophilic groups (e.g., sulfenylated cysteines26, 27 or esterified carboxylate side-chains or C-termini17). The target list was further refined by requiring that each protein was detected with a minimum of three unique quantified peptides per experiment and by averaging protein ratios across three or more biological replicates per cell line (Supplementary Table 1).
We plotted our competition versus enrichment data (Fig. 2b and Supplementary Table 2), which illustrated four categories of proteins – 1) a lower left quadrant of unenriched, background proteins; 2) a nearly unoccupied (as expected) upper left quadrant that would designate proteins competed by 3, but not enriched by 1; 3) a lower right quadrant populated by a substantial number of proteins that were enriched by 1, but not competed by 3; and 4) an upper right quadrant, which housed ten proteins that showed strong enrichment and competition and were therefore designated as high-reactivity targets of hydrazine probes (Fig. 2b and Table 1). Representative peptide ratios for three of these high-reactivity proteins from enrichment, competition, and control (where heavy and light cells were treated with equal concentrations of 1) experiments are shown in Fig. 2c. We also performed analogous experiments with the aryl hydrazine probe 2 (1 mM, 0.5 h), which furnished a larger list of high-reactivity proteins (33 in total; Supplementary Fig. 3 and Supplementary Table 3) that contained several of the targets of probe 1 (compared in Supplementary Table 4), as well as additional proteins that may reflect an expanded chemical reactivity for probe 2.
Table 1.
High-reactivity protein targets of probe 1.
| Probe targets | Probe reactivity |
Function | Electrophile | |
|---|---|---|---|---|
| 1. |
S-adenosylmethionine (AdoMet) decarboxylase (AMD1/AdoMetDC) |
1, 2 | polyamine biosynthesis (cancer) | pyruvamide* |
| 2. | Legumain (LGMN) | 1, 2 | cysteine protease (cancer) | aspartimide† |
| 3. | α-mannosidase (MAN2B1) | 1, 2 | glycohydrolase (Mannosidosis) | unknown |
| 4. | Secernin-3 (SCRN3) | 1, 2 |
N-terminal nucleophile (Ntn) hydrolase‡ |
unknown |
| 5. | Secernin-2 (SCRN2) | 1, 2 | ||
| 6. | probable arginine tRNA ligase (RARS2) | 1 | protein synthesis (Pontocerebellar hypoplasia 6)‡ |
unknown |
| 7. | Kelch-like ECH-associated protein-1 (KEAP1) | 1, 2 | antioxidant response regulator (cancer) |
unknown |
| 8. | β-amyloid-like precursor protein-2 (APLP2) | 1 | Aβ precursor: amyloid plaques (Alzheimer's disease) |
unknown |
| 9. | β-amyloid precursor protein (APP) | 1 | ||
| 10. | ependymin-related protein-1 (EPDR1) | 1, 2 | neuron regeneration and memory‡ | unknown |
| 11. | Fat mass and obesity-associated Fe(II)- and 2-oxoglutarate (2OG)-dependent dioxygenase (FTO) |
2 | RNA demethylation [obesity and growth retardation, developmental delay, and facial dysmorphism (GDFD)] |
unknown |
Unable to predict the chemical properties of all potential probe adducts, we considered the possibility that targets may be lost due to potentially unstable hydrazone adducts that could be generated via capture of certain carbonyl electrophiles. Inspired by previous methods that use reductants to convert labile hydrazones to more stable hydrazides16, we performed an enrichment profiling experiment with probe 1 in the presence of 50 mM NaCNBH3 added during cell lysis. We did not, however, observe a change or increase in 1-enriched proteins under these conditions, suggesting that targets reacting with 1 via hydrazone formation were stable to our streptavidin enrichment and proteomic analysis protocol (Supplementary Table 1).
We next attempted to validate a representative subset of high-reactivity targets by recombinant expression. Four proteins were selected, of which three (AMD1, SCRN3 and KEAP1) reacted with both probes 1 and 2 and one (FTO) was preferentially targeted by probe 2. We confirmed the recombinant expression of each protein in transfected HEK293T cells by western blotting [Fig. 2d (upper blots)] and found that, in each case, treatment of transfected cells with probe 1 or 2, followed by conjugation to azide-rhodamine, furnished a strong fluorescent band at the appropriate molecular weight that was absent in mock-transfected control cells [Fig. 2d (lower gels)]. Probe labeling of each protein was blocked by treatment with excess non-clickable agents 3 or 4 [Fig. 2d (lower gels)]. For FTO, we observed selective reactivity with the aryl probe 2 over the alkyl probe 1 (Fig. 2d), matching the proteomic data obtained for endogenously expressed FTO in cells (Supplementary Fig. 3). These data indicate that hydrazine reactivity is an intrinsic property of the protein targets of probes 1 and 2 that is shared by both the endogenous and recombinant forms of these proteins.
Monitoring electrophilic cofactor in AMD1 in cells
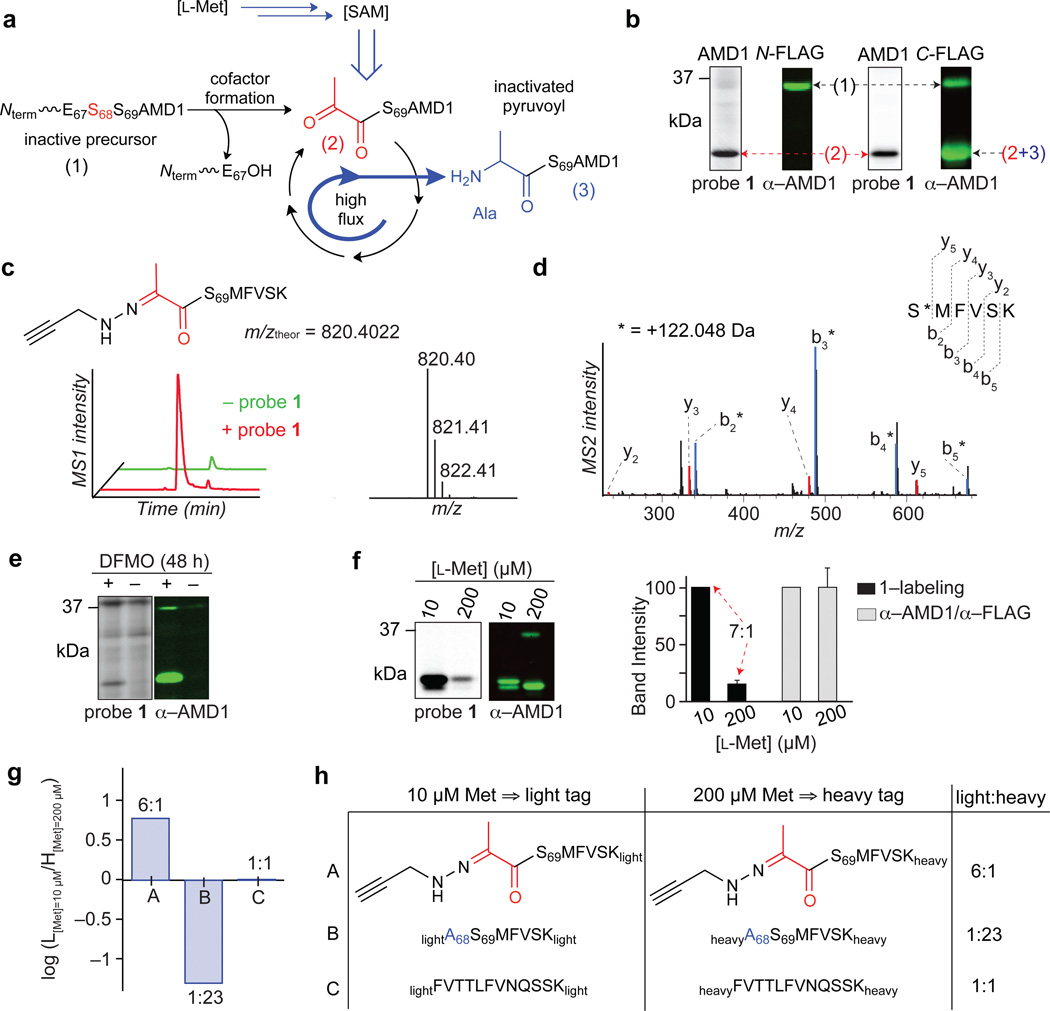
Literature searches revealed that some of the probe targets are known to possess electrophilic PTMs (Table 1). Prominent among these was S-adenosyl-l-methionine (SAM) decarboxylase (AMD1 or AdoMetDC), which employs an N-terminally bound pyruvoyl group generated from serine to catalyze the rate-limiting step in polyamine biosynthesis28. AMD1 can be inhibited by hydrazines and related nucleophiles that target the enzyme’s pyruvoyl cofactor22, 29, which is installed by a putrescine-induced auto-cleavage of the inactive proenzyme (38 kDa) to generate a processed, catalytically competent enzyme (30 kDa) (Fig. 3a). Blotting with an anti-FLAG antibody detected both the full-length pro- and processed active forms of AMD1 when expressed as a C-terminal FLAG-tagged protein in transfected HEK293T cells, whereas only the larger pro-form was detected for an AMD1 protein bearing the FLAG tag on its N-terminus (Fig. 3b), consistent with the expected processing event that cleaves an N-terminal portion of AMD1. Importantly, for either N- or C-terminally tagged AMD1, strong labeling with probe 1 was observed exclusively for the processed form of these proteins (Fig. 3b). We next treated active human AMD1 expressed and purified from E. coli with probe 1, followed by tryptic digestion of the protein and LC-MS/MS analysis, which identified the N-terminal pyruvoyl modification as the site of 1 reactivity (Fig. 3c, d and Supplementary Fig. 4a). We also observed loss of the unmodified N-terminal pyruvoyl tryptic peptide in 1-treated AMD1 samples (Supplementary Fig. 4b), indicating that the reaction between 1 and the pyruvoyl group of AMD1 proceeded to near-completion. We did not observe evidence of modification of any other tryptic peptides from AMD1 (Supplementary Fig. 4c, d), supporting that probe 1 reacted specifically with the N-terminal pyuvoyl group.
Fig. 3. Functional profiling of the pyruvoyl cofactor of AMD1 by hydrazine probes.
a, Shown are three forms of AMD1 – inactive proenzyme (38 kDa) (1), catalytically competent N-terminal pyruvoyl-containing enzyme (30 kDa) expected to be probe-reactive (2), and inactivated enzyme bearing an alanine form of the cofactor (3). b, Expression and probe 1-labeling profiles of N- (left) versus C-terminal (right) FLAG-tagged AMD1 in transfected HEK293T cells. Forms (1)–(3) are indicated. c, Extracted parent ion chromatograms (left) and corresponding isotopic envelope (right) for the N-terminal pyruvoyl peptide of AMD1 labeled by probe 1. d, MS2 spectra of the N-terminal pyruvoyl peptide of AMD1 labeled by probe 1. The b- and y-ions assign the labeled site (*) to the N-terminal Ser69 residue. e, Expression and probe 1-labeling profiles of endogenous AMD1 in HEK293T cells following treatment with difluoromethylornithine (DFMO). f, Expression and probe 1-labeling profiles of AMD1-transfected cells cultured in media containing 10 versus 200 µM l-methionine (left) and corresponding band intensities for each (right). Results represent average values from three biological replicates, and standard deviations are represented by error bars for the low methionine condition, where band intensities were normalized to the high methionine condition. g, From cells generated as in f, ratios for AMD1 peptides – probe 1-labeled pyruvoyl (A) and alanine (B) forms of N-terminal peptide, as well as an internal peptide (C) – in experiments comparing cells grown in low (10 µM) versus high (200 µM) methionine and compared by modifying corresponding peptides with light and heavy formaldehyde, respectively. Results are from a single experiment representative of two independent biological replicates. h, Summary table of data in g. Results shown are from a single experiment representative of two independently performed experiments.
Difluoromethylornithine (DFMO), an inhibitor of putrescine biosynthesis, has been shown to increase the expression of AMD130, and we confirmed that probe 1 could detect DMFO-induced changes in the endogenous concentration of active AMD1 in cells (Fig. 3e). We also found that lowering the concentration of l-methionine, a biosynthetic precursor for SAM, from that found in standard culture media (200 µM) to physiological serum (10 µM)31, increased probe 1 labeling of recombinant AMD1 in cells by ~seven-fold with negligible changes in AMD1 expression (Fig. 3f).
A mechanism to explain the regulation of AMD1 activity by SAM concentration has been described for the purified enzyme in vitro32, where each catalytic event is partitioned such that a small fraction of the pyruvoyl cofactor is inactivated irreversibly to alanine (Ala) instead of being regenerated (Fig. 3a and Supplementary Fig. 5). Consistent with this model being operational in cells, the differences observed in probe 1-labeling of AMD1 in low versus high methionine could reflect changes in the N-terminal structure of AMD1. We tested this premise by culturing AMD1-transfected HEK293T cells in high versus low methionine, treating cells with probe 1, and then enriching AMD1 protein from each cell preparation with anti-FLAG antibodies. The AMD1 protein samples were further purified by SDS-PAGE, digested in gel with trypsin, and the resulting peptides modified with isotopically heavy or light formaldehyde. Combining heavy and light samples (corresponding to cells grown in high and low methionine), followed by LC-MS/MS analysis, revealed much greater signals for the probe 1-labeled pyruvoyl N-terminal tryptic peptide of AMD1 in low methionine-treated cells (A, Fig. 3g, h and Supplementary Table 5). Conversely, the alanine form of the N-terminal tryptic peptide of AMD1 was dramatically increased in the high methionine-exposed cells (B, Fig. 3g, h and Supplementary Table 5). We also measured an internal tryptic peptide from AMD1, which revealed no changes in protein expression caused by low versus high methionine exposure (C, Fig. 3g, h and Supplementary Table 5). These data, taken together, indicate that changes in methionine content dramatically alter the fraction of active, N-terminal pyruvoyl-modified AMD1 in cells, such that persistent metabolic flux under high methionine conditions leads to progressive loss of the cofactor and catalytic activity. That probe 1 monitors the post-translational regulation of AMD1 in cells indicates hydrazines can serve not only as inhibitors22, but also activity-based probes for this enzyme.
Discovery of an N-terminal glyoxylyl modification on SCRN3
Our studies with AMD1 confirmed that the hydrazine probes react with electrophilic cofactors of established functionality in enzymes. We therefore extrapolated that additional targets of probes 1 and 2 might possess as-of-yet structurally uncharacterized electrophilic modifications. To address this question, we adapted our chemical proteomic approach to identify the peptide, rather than the whole protein, harboring the probe-reactive electrophile. This site-specific profiling method, termed isoTOP-ABPP33, leverages isotopically differentiated, protease-cleavable biotin-azide tags to enrich and release probe-labeled peptides as mass-differentiated pairs (Fig. 4a). While this approach has been used successfully to identify the sites of reactivity for several probes34, in these past instances, the sites all represented natural amino acids. Here, we faced the additional challenge that the sites of probe 1/2 reactivity were expected to be non-natural (e.g., PTM-modified amino acids) and, in most cases, not predictable from the sequences of protein targets.
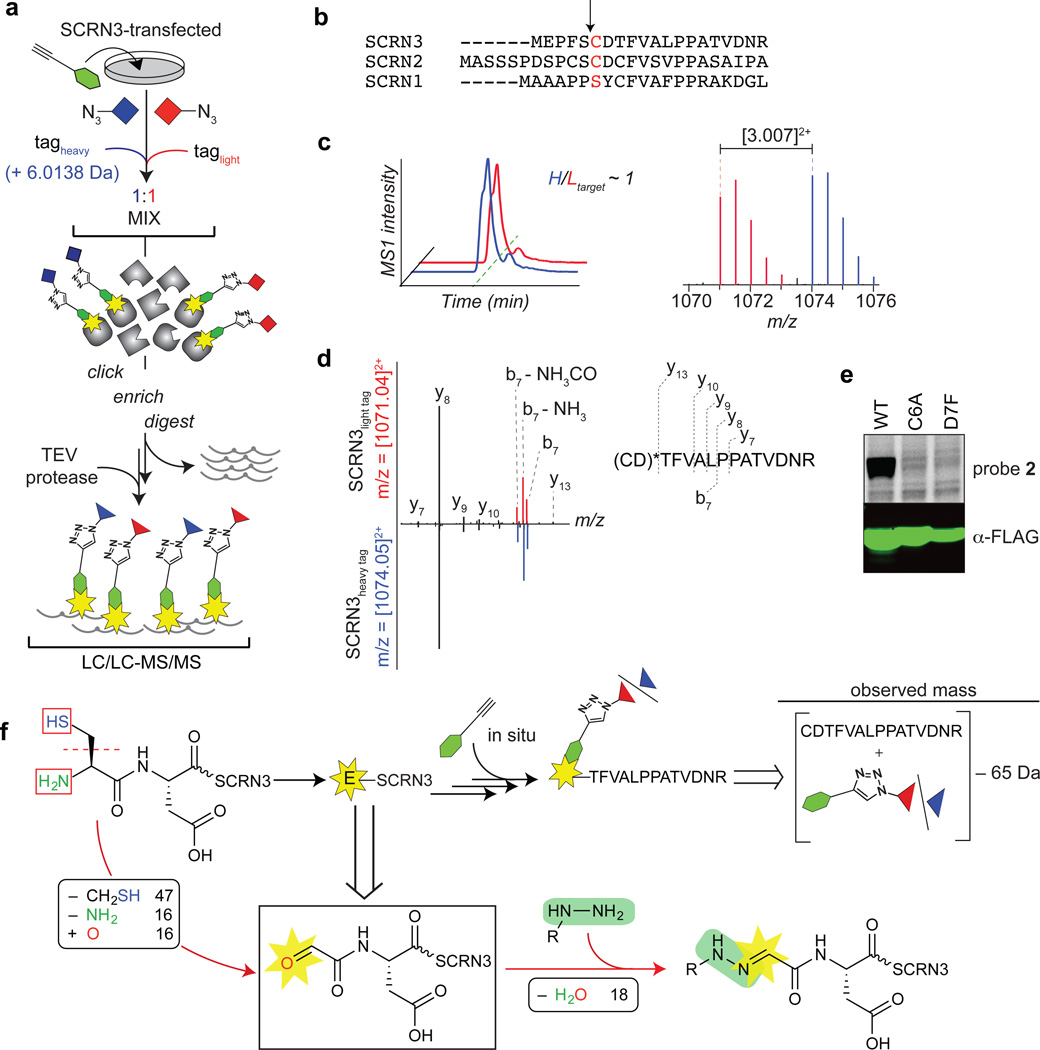
Fig. 4. Identification of a hydrazine-reactive site in secernin-3 (SCRN3).
a, Characterization of probe labeled peptides using the isoTOP-ABPP method as described in the text. b, N-terminal sequence alignment of human secernin proteins. The predicted catalytic residues for the putative Ntn hydrolase activities of secernins are highlighted (in red) and the associated cleavage sites of predicted proforms are indicated by the arrow. c, Extracted double-charged MS1 ion chromatograms (left) and corresponding isotopic envelopes (right) for co-eluting heavy- and light-tagged peptides labeled by probe 2 (in blue and red, respectively). d, Comparison of high-resolution MS2 spectra generated from light- versus heavy-tagged parent ions (left). The y-ions resolve the modified site (*) to the N-terminal cysteine and/or adjacent aspartate (right). e, Probe 2-labeling and expression profiles of Cys6-to-Ala6 (C6A) and Asp7-to-Phe7 (D7F) mutant SCRN3 proteins compared to wild-type (WT) SCRN3. f, Reaction scheme (upper schematic) and MS results-based prediction of the structure of the probe 2-SCRN3 adduct (lower schematic).
We first confirmed that isoTOP-ABPP correctly assigned the site of probe 1 (and 2) reactivity in AMD1 as the N-terminal pyruvoyl modification using both recombinant and endogenous sources of the enzyme (Supplementary Figs. 6 and 7). We next selected for analysis Secernin-3 (SCRN3), a poorly characterized protein from the target list that reacted with both probes 1 and 2 (Table 1) and is predicted on the basis of sequence to be an N-terminal cysteine nucleophile (Ntn) hydrolase (Fig. 4b)35. Based on these predictions, Cys6 of SCRN3 would serve as the N-terminal nucleophile responsible for autoproteolytic processing via a mechanism that is shared by other Ntn hydrolases36, 37 (Supplementary Fig. 8); however, to our knowledge, SCRN3 has not been experimentally verified to undergo this type of N-terminal processing. SCRN3-transfected HEK293T cells were treated with probe 2 (1 mM, 0.5 h), lysed, conjugated to a 1:1 ratio of isotopically heavy and light biotin-azide tags, and enriched on streptavidin beads. Enriched proteins were then digested on-bead by sequential proteolysis, first with trypsin to remove unlabeled peptides, followed by TEV protease to release probe-labeled peptides, which were analyzed by LC-MS/MS, where authentic probe-labeled peptides were expected to migrate as isotopically differentiated mass pairs with a heavy:light ratio of 1. The most abundant probe-labeled peptide pair that conformed to these specifications (Fig. 4c and Supplementary Table 6) was both manually and computationally identified as a half-tryptic peptide spanning from Cys6 to Arg20 of SCRN3 based on the pattern of y-ions in the high-resolution tandem mass spectra (Fig. 4d, left). Importantly, the y13-ion assigned the probe modification site to one of the first two residues (Fig. 4d, right), which was supported by mutation of either residue – Cys6 or Asp7 – which abolished probe 2 labeling (Fig. 3e). Similar data were obtained for SCRN3-transfected cells treated with probe 1 (Supplementary Fig. 9).
The data accumulated thus far indicated that i) SCRN3 contains a hydrazine-reactive group at or near Cys6, which represented the presumed N-terminus of the protein, as predicted by the activation mechanism for Ntn proteins (Supplementary Fig. 8); and ii) electrophile formation and subsequent reaction with probe 1 or 2 yielded a net loss of 65 Da from the summated mass of an unmodified N-terminal Cys6-Arg-20 peptide combined with the masses of the hydrazine probes (Fig. 4f, upper). The most logical structure conceived to fit these criteria was the hydrazone product of a reaction between hydrazine probes and an N-terminal glyoxylyl group originating from Cys6 (Fig. 4f, lower and Supplementary Fig. 10a, b).
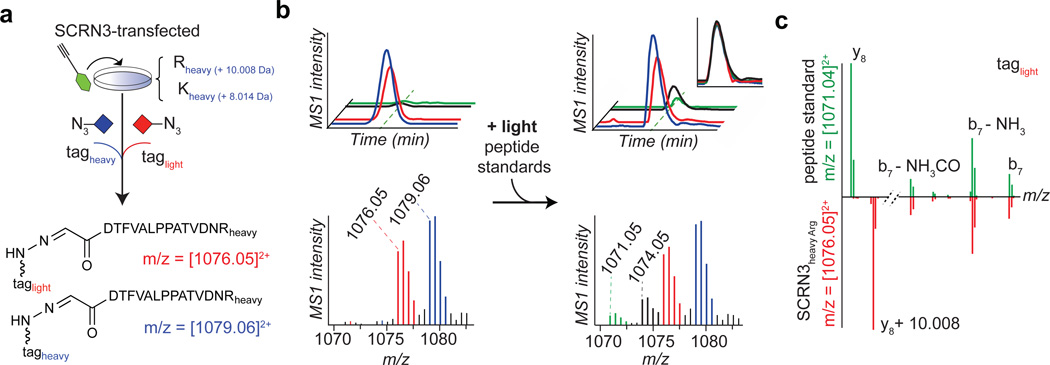
To test our structural predictions, we synthesized a peptide standard for the hydrazone product of a probe 2-Glyoxylyl6-Arg20 reaction and then combined the standard with proteomic lysates prepared from probe 2-treated SCRN3-transfected HEK293T cells that had been grown in heavy arginine/lysine, such that the peptide standard and probe 2-labeled endogenous SCRN3 peptide would be isotopically distinguishable (Fig. 5a). Note that these experiments were performed with probe 2 rather than probe 1 because probe 2 appeared to produce higher product yields with SCRN3 (Fig. 2d). The peptide standard and the 2-labeled endogenous SCRN3 peptide co-eluted by LC (Fig. 5b) and displayed tandem mass spectra that differed by the predicted 10 Da in the y8-ion containing the heavy C-terminal Arg residue, but were identical across the b7-ion series lacking this residue (Fig. 5c). These data supported the structural assignment that SCRN3 possesses an N-terminal glyoxylyl group originating from the conserved Cys6 residue.
Fig. 5. Evidence supporting the structural assignment of an N-terminal glyoxylyl group in SCRN3.
a–c, Heavy-Arg/Lys-labeled SCRN3-transfected cells treated with probe 2, followed by processing by isoTOP-ABPP, furnishes an isotopically differentiated probe 2-labeled SCRN3 peptide pair (red and blue) (a), which co-elutes with a light amino acid-labeled probe 2-Glyoxylyl6-Arg20 standard (also an isotopically differentiated peptide pair; black and green). Inset chromatogram shows all four traces scaled to the same intensity (upper right, inset plot) to show co-elution of endogenous and standard 2-Glyoxylyl6-Arg20 SCRN3 peptides. Corresponding isotopic envelopes are shown below the chromatograms. c, Comparison of high-resolution MS2 spectra for light-tagged standard versus endogenous 2-Glyoxylyl6-Arg20 SCRN3 peptides (inverted y-axis) distinguished by the expected 10 Da mass shift for y8-ions containing the C-terminal Arg residue.
We obtained additional confirmation for the N-terminal glyoxylyl modification of SCRN3 by subjecting SCRN3-transfected cell lysates to reductive amination with (NH4)2SO4, which furnished the predicted N-terminal glycine peptide (Gly6-Arg20) for the trypsin-digested, affinity-purified SCRN3 protein (Supplementary Fig. 11). The parent mass for this peptide (Supplementary Fig. 11, left) and its corresponding b-, but not y-ions (Supplementary Fig. 11, right) also shifted in mass by 1 Da when (NH4)2SO4 was substituted with 15N-enriched (NH4)2SO4.
Further analysis of the MS data from SCRN3-transfected cells (Supplementary Table 6) identified another prominent probe 2-enriched product that matched the predicted mass for an N-terminal pyruvoyl modification of SCRN3 (Supplementary Table 7). This product was detected with ~50% of the ion intensity of the N-terminal glyoxylyl modified SCRN3 (Supplementary Fig. 12a). Both the N-terminal glyoxylyl and pyruvoyl forms of recombinant SCRN3 were also observed with alkyl probe 1 (Supplementary Table 7), and their relative intensities preserved under different sample preparation conditions (Supplementary Fig. 12b), suggesting that both electrophilic modification states occur for SCRN3 in situ.
In support of the pyruvoyl structure assignment and a model for glyoxylyl formation that involves an apparent 2e− oxidation of the N-terminal Cys residue of SCRN3 via Cα–Cβ bond cleavage and oxidative deamination, we found that SCRN3-transfected cells grown in media supplemented with l-[15N13C3]cysteine shifted the parent mass and b7-ion series, but not y8-ion for the probe 2-labeled pyruvoyl peptide by the expected 3 Da (Supplementary Fig. 12c) and the corresponding glyoxylyl peptide by the expected 2 Da (Supplementary Fig. 12d). In contrast, internal peptides that contained a Cys residue shifted the expected 4 Da in parent mass, accounting for all 4 heavy atoms of the Cys residue (Supplementary Table 8).
As an initial attempt to estimate the fraction of SCRN3 protein bearing the N-terminal glyoxylyl modification, we reacted lysates from probe 2-treated SCRN3-transfected (and heavy amino acid-labeled) HEK293T cells with aniline to generate an iminium adduct that could be reduced with NaCNBH3 to a more stable amine product (Supplementary Fig. 13a, b), following the reactivity trends that have been exploited in bioconjugation approaches38, 39. We also synthesized an aniline-modified and reduced glyoxylyl N-terminal SCRN3 peptide standard, along with a control standard for an internal SCRN3 tryptic peptide, and compared the ratios of heavy parent ion peak intensities for the aniline-modified N-terminal and internal peptides with near-equivalent amounts of the corresponding peptide standards (Supplementary Fig. 13c, d). The ratios of the aniline-appended N-terminal and internal peptides compared to their respective standards provided an estimate of 10 ± 3% for the N-terminal glyoxylyl-modified SCRN3. We view this estimate as a lower limit for the fraction of glyoxylyl-modified SCRN3 in cells, since the probe 2 and aniline reactions with this form of SCRN3 may proceed with less than 100% efficiency in cell lysates, and it is also possible that, by overexpressing SCRN3 in transfected cells, we may have saturated the putative endogenous processing system that installs the glyoxylyl modification. At least partly supporting these hypotheses, we estimated that only 15 ± 3% (n=4) of the recombinant SCRN3 possessed an intact Cys6 side chain in transfected cell preparations (Supplementary Fig. 14).
Finally, in initial attempts to assess features required for SCRN3’s N-terminal glyoxylyl/pyruvoyl modifications, we evaluated a variant of SCRN3 where amino acids 2–5 were deleted and found that this mutant protein was still converted to the glyoxylyl/pyruvoyl form, albeit with an apparently lower efficiency than the wild-type SCRN3 protein (Supplementary Fig. 15 and Supplementary Table 7). This result indicates that an N-terminal leader sequence is not required for SCRN3 processing or installation of its N-terminal electrophilic modifications. We also found noted that SCRN3 reacts with hydrazine probes when expressed in mammalian cells, but not E. coli (Supplementary Fig. 16). This contrasts with the profile of AMD1, which reacts well with probe 1 or 2 when expressed in E. coli (Supplementary Fig. 16). These data are consistent with the established autonomous mechanism of installation for AMD1’s pyruvoyl modification and suggest further that the N-terminal glyoxylyl/pyruvoyl groups in SCRN3 may require enzymatic machinery of the mammalian host cell for generation. In support of this hypothesis, LC/LC-MS/MS characterization of SCRN3 expressed and purified from bacteria failed to detect the N-terminal glyoxylyl/pyruvoyl states of the protein, instead revealing only the unprocessed N-terminus containing the initiator methionine residue and the Cys6 N-terminus presumably generated by autocleavage (Supplementary Table 9).
Discussion
Our results indicate that both known and novel electrophilic modifications on proteins can be profiled in cells using simple hydrazine probes. That at least some of the sites of hydrazine reactivity represent electrophilic cofactors important for enzyme catalysis indicates that 1 and 2 can be considered authentic ABPP probes. The profiling of dynamic changes in the pyruvoyl modification state of AMD1 by 1 provides a compelling example of how RP-ABPP can be used to evaluate protein function in cellular systems. In this regard, previous studies have shown that heightened methionine concentrations in cells leads to proteasomal degradation of AMD1 by a mechanism postulated to involve conversion of the N-terminal pyruvoyl cofactor to alanine40. We did not observe alterations in AMD1 levels in our studies of high versus low methionine, despite substantial changes in the fraction of N-terminal pyruvoyl versus alanine forms of the enzyme, suggesting that catalytic flux-mediated decreases in AMD1 activity can precede, or even occur without effects on AMD1 degradation in cells.
Our further discovery of an N-terminal glyoxylyl modification on the poorly characterized protein SCRN3 underscores the potential of RP-ABPP to illuminate novel structural features of potential functionality in the human proteome, of which we speculate there are others still awaiting excavation. For nucleophilic probes, like the simple hydrazines used herein, however, which have the potential to react with a diversity of electrophilic modifications, structural characterization of these modifications remains a major challenge. In some cases, the electrophilic modification could be inferred from the literature. The lysosomal protease legumain (LGMN), for instance, harbors an active site aspartimide (succinimide) that catalyzes peptide ligation reactions in vitro41. Considering that legumain was a high-reactivity target of probes 1 and 2 and further that succinimide electrophiles are known to react with hydrazines18 (see Supplementary Fig. 10c for structure and reaction), our findings suggest that the aspartimide cofactor observed previously on purified legumain in vitro41 also exists endogenously, raising the possibility that legumain could function as both a protease and ligase in cells. For proteins like SCRN3 that possess structurally novel electrophilic modifications, we acknowledge that elucidating their source of reactivity with hydrazine probes may require considerable effort and further methodological advances. We believe that such endeavors are worthwhile, however, especially given that some of the other proteins displaying high reactivity with hydrazine probes have compelling disease relationships (e.g., AMD142 and KEAP143 with cancer; APPs with Alzheimer’s disease44; FTO with obesity45).
We are unsure why the aryl hydrazine 2 showed broader proteomic reactivity compared to alkyl hydrazine 1, but we note that, from a chemical perspective, the aryl hydrazine derivative is likely to form hydrazones of greater stability with respect to hydrolysis46, enabling the capture and identification of proteins that possess electrophilic modifications that form otherwise unstable adducts with alkyl hydrazines11, 47. Probe 2 might also have expanded reactivity towards proteins that deviate from electrophilic chemistry and instead involve, for instance, radical-based mechanisms21.
The SCRN proteins, conserved from bacteria to higher mammals, have three members in humans – SCRN1–3 – however, only SCRN2 and SCRN3 were identified as targets of the hydrazine probes herein. Notably, both of these proteins, as well as their orthologues in other organisms, but not SCRN1, share an ‘SCD’ motif that includes the glyoxylyl/pyruvoyl modification site discovered for SCRN3, suggesting that the N-terminal processing and modification of SCRN3 may be conserved across SCRN2/3 proteins, but differ for SCRN1 (Supplementary Fig. 17). Despite being superficially classified as Ntn hydrolases, SCRN2/3, based on our data, appear more likely to operate by a distinct catalytic mechanism that involves electrophilic modification of their N-termini. In considering potential functions for SCRN2/3, we note that formylglycine (fGly)-dependent enzymes, which possess a structurally related aldehyde modification, exploit the hydrated form of this group for nucleophilic attack on substrates to catalyze reactions such as sulfate ester hydrolysis48. fGly cofactors, however, are not generated at autoproteolyzed N-termini of proteins, but rather installed by 2e− oxidation of internal cysteine (or serine) residues catalyzed by the enzyme sulfatase-modifying factor 1 (SUMF1)48. A similar installation mechanism seems incompatible with generating the N-terminal glyoxylyl and pyruvoyl modifications for SCRN3.
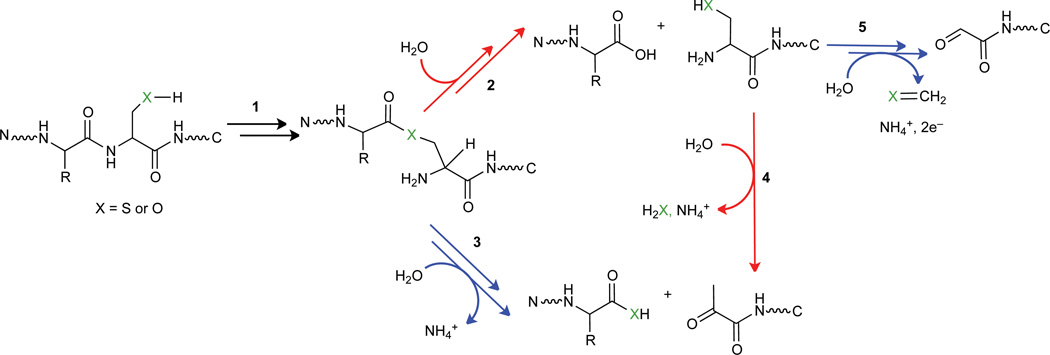
Some common mechanistic features can be found in the activation chemistries of Ntn hydrolases and pyruvoyl-dependent decarboxylases1,49. Autoproteolysis occurs via an N→O or N→S acyl shift where nucleophilic side chains attack upstream peptide bonds to form cyclic intermediates that reopen as oxo/thioesters via C–N bond cleavage (step 1, Fig. 6). Next, their respective active sites direct divergent outcomes (hydrolysis versus β-elimination) to yield catalytic machinery of opposite polarity (steps 2 versus 3, respectively, Fig. 6). N-terminal cysteine-derived pyruvoyl groups have been proposed to act as the likely cofactor for bacterial reductases50, 51 and can form chemically at engineered N-termini with pyridoxal-5’-phosphate (PLP)52, 53. However, detection of a cysteine N-terminus, if autoactivated as predicted for Ntn hydrolases, might favor pyruvoyl formation via β-elimination uncoupled from proteolysis (step 4). Self-processing of this type, however, is unlikely to produce the glyoxylyl form of SCRN3, and, there is no report, to our knowledge, of a cysteine-to-glyoxylyl transformation. This modification can be described as an aldol cleavage coupled to oxidative deamination, perhaps occurring in single or sequential steps (step 5). Both products could emerge, in principle, from transformations catalyzed by PLP-dependent enzymes12.
Fig. 6. Possible routes for N-terminal processing of SCRN3 in comparison to the established mechanisms of N-terminal maturation for other protein classes.
The mechanisms for Ntn hydrolases and pyruvoyl-dependent decarboxylases diverge at the transient ester intermediate formed during autoproteolysis (step 1). Hydrolysis yields a catalytic nucleophile for Ntn hydrolases (step 2), whereas active site-directed β-elimination generates an electrophilic pyruvoyl cofactor for decarboxylases (step 3). Detection of a new N-terminus containing an intact side chain, shown as a product of step 2, suggests that formation of pyruvoyl and glyoxylyl groups may occur via 4 and 5, respectively, in the N-terminal processing of SCRN3.
The mechanism of formation and consequent function of the N-terminus glyoxylyl/pyruvoyl modification on SCRN3 represent important areas for future investigation. That SCRN3, with its N-terminal modified forms, shares features in common with a subclass of bacterial reductases51 points to one potential catalytic function that should be further explored. Alternatively, at this stage, we cannot exclude the possibility that the glyoxylyl group, rather than imparting a catalytic activity to SCRN3, might instead represent a non-catalytic form of the protein that serves a different function. Regardless, the strong conservation of the N-terminal cysteine residue across the SCRN2/3 protein family argues for a functional role for the modified electrophilic form of this residue.
In summary, our findings demonstrate that the principles of ABPP can be extended to the functional and structural characterization of electrophilic modifications on proteins. We note that proteinaceous electrophilic cofactors, such as pyruvoyl and glyoxylyl PTMs, cannot be easily predicted by analysis of protein sequences, underscoring the value of chemical proteomic methods like RP-ABPP for their de novo discovery in native biological systems. That our initial studies, which only profiled two human cell lines, succeeded in identifying a novel N-terminal glyoxylyl modification that appears to be a conserved feature of a poorly characterized class of proteins (the secernins) leads us to speculate that many other proteins in the human proteome may harbor electrophilic groups. We anticipate that the characterization of these groups would benefit from the use of additional types of nucleophilic probes, akin to the diverse sets of electrophilic probes employed in more classical ABPP studies of proteins that use different nucleophilic residues for function9. Of course, mapping electrophilic groups will confront the added challenge of structural characterization of these PTMs, but our studies show how RP-ABPP probes can facilitate this effort (by both enriching these modifications and generating stable adducts with predictable shifts in mass values). Finally, we recognize that, ultimately, one of the most important questions is – what types of functions do electrophilic modifications impart on proteins? To the extent that RP-ABPP probes have the potential to not only characterize, but also inhibit the function of proteins that possess electrophilic modifications, these probes could serve as launching points for more advanced and selective small-molecule inhibitors that modulate electrophile-dependent function for basic and translational research purposes.
Methods
See Supplementary Information for a detailed Methods section.
Supplementary Material
Acknowledgments
We acknowledge Steven Ealick and Leslie Kinsland for the AdoMetDC bacterial expression plasmid, Keriann Backus for the cleavable tags, Gonzalo González-Páez and Dennis Wolan for TEV protease, Melissa Dix and Jim Moresco for instrument assistance, Armand Cognetta and Kenneth Lum for software and data assistance, Liron Bar-Peled and Stephan Hacker for plasmids and Silvia Ortega-Gutiérrez for helpful discussions. This work was supported by the National Institutes of Health Grants CA132630 (B.F.C.), P41 GM103533 and U54 GM114833-02 (J.R.Y.), an NSF Graduate Research Fellowship DGE-1346837 (E.J.O.) and a Helen Hay Whitney Postdoctoral Fellowship sponsored by Merck (M.L.M).
Footnotes
Author Contributions
M.L.M and B.F.C. conceived the project, designed experiments and composed the manuscript. M.L.M performed all experiments. L.H. wrote software and J.R.Y. provided advice. M.L.M., L.H., B.E.C. and B.F.C. analyzed data. M.L.M., E.J.O. and P.E.D. designed peptide standards. M.L.M. and E.J.O. synthesized peptides. M.L.M. and B.D.H. synthesized probes.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Okeley NM, van der Donk WA. Novel cofactors via post-translational modifications of enzyme active sites. Chem. Biol. 2000;7:159–171. doi: 10.1016/s1074-5521(00)00140-x. [DOI] [PubMed] [Google Scholar]
- 3.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baez NO, Reisz JA, Furdui CM. Mass spectrometry in studies of protein thiol chemistry and signaling: opportunities and caveats. Free Radical Biol. Med. 2015;80:191–211. doi: 10.1016/j.freeradbiomed.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furdui CM, Poole LB. Chemical approaches to detect and analyze protein sulfenic acids. Mass Spectrom. Rev. 2014;33:126–146. doi: 10.1002/mas.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuh KN, Pratt MR. Chemical methods for the proteome-wide identification of posttranslationally modified proteins. Curr. Opin. Chem. Biol. 2015;24:27–37. doi: 10.1016/j.cbpa.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tate EW, Kalesh KA, Lanyon-Hogg T, Storck EM, Thinon E. Global profiling of protein lipidation using chemical proteomic technologies. Curr. Opin. Chem. Biol. 2015;24:48–57. doi: 10.1016/j.cbpa.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Li XD. Chemical proteomics approaches to examine novel histone posttranslational modifications. Curr. Opin. Chem. Biol. 2015;24:80–90. doi: 10.1016/j.cbpa.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 10.Shannon DA, Weerapana E. Covalent protein modification: the current landscape of residue-specific electrophiles. Curr. Opin. Chem. Biol. 2015;24:18–26. doi: 10.1016/j.cbpa.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Klinman JP, Bonnot F. Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ. Chem. Rev. 2014;114:4343–4365. doi: 10.1021/cr400475g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips RS. Chemistry and diversity of pyridoxal-5'-phosphate dependent enzymes. Biochim. Biophys. Acta. 2015;1854:1167–1174. doi: 10.1016/j.bbapap.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 14.Morgan RK, Cohen MS. A clickable aminooxy probe for monitoring cellular ADP-ribosylation. ACS Chem. Biol. 2015;10:1778–1784. doi: 10.1021/acschembio.5b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta V, Carroll KS. Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta. 2014;1840:847–875. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madian AG, Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J. Proteome Res. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alfaro JF, et al. Chemo-enzymatic detection of protein isoaspartate using protein isoaspartate methyltransferase and hydrazine trapping. Anal. Chem. 2008;80:3882–3889. doi: 10.1021/ac800251q. [DOI] [PubMed] [Google Scholar]
- 18.Klaene JJ, Ni W, Alfaro JF, Zhou ZS. Detection and quantitation of succinimide in intact protein via hydrazine trapping and chemical derivatization. J. Pharm. Sci. 2014;103:3033–3042. doi: 10.1002/jps.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 21.Binda C, et al. Structural and mechanistic studies of arylalkylhydrazine inhibition of human monoamine oxidases A and B. Biochemistry. 2008;47:5616–5625. doi: 10.1021/bi8002814. [DOI] [PubMed] [Google Scholar]
- 22.Shantz LM, Stanley BA, Secrist JA, 3rd, Pegg AE. Purification of human S-adenosylmethionine decarboxylase expressed in Escherichia coli and use of this protein to investigate the mechanism of inhibition by the irreversible inhibitors, 5'-deoxy-5'-[(3-hydrazinopropyl)methylamino]adenosine and 5'-([(Z)-4-amino-2-butenyl]methylamino)-5'-deoxyadenosine. Biochemistry. 1992;31:6848–6855. doi: 10.1021/bi00144a027. [DOI] [PubMed] [Google Scholar]
- 23.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 24.Mann M. Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 25.Adibekian A, et al. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat. Chem. Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalle-Donne I, et al. Protein carbonylation: 2,4-dinitrophenylhydrazine reacts with both aldehydes/ketones and sulfenic acids. Free Radical Biol. Med. 2009;46:1411–1419. doi: 10.1016/j.freeradbiomed.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 27.Gupta V, Paritala H, Carroll KS. Reactivity, selectivity and stability in sulfenic acid detection: A comparative study of nucleophilic and electropilic probes. Bioconjugate Chem. 2016 doi: 10.1021/acs.bioconjchem.6b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolbert WD, et al. The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase. Biochemistry. 2001;40:9484–9494. doi: 10.1021/bi010735w. [DOI] [PubMed] [Google Scholar]
- 29.McCloskey DE, et al. New insights into the design of inhibitors of human S-adenosylmethionine decarboxylase: studies of adenine C8 substitution in structural analogues of S-adenosylmethionine. J. Med. Chem. 2009;52:1388–1407. doi: 10.1021/jm801126a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shantz LM, Holm I, Janne OA, Pegg AE. Regulation of S-adenosylmethionine decarboxylase activity by alterations in the intracellular polyamine content. Biochem. J. 1992;288:511–518. doi: 10.1042/bj2880511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zinellu A, et al. Plasma methionine determination by capillary electrophoresis-UV assay: application on patients affected by retinal venous occlusive disease. Anal. Biochem. 2007;363:91–96. doi: 10.1016/j.ab.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Xiong H, Stanley BA, Pegg AE. Role of cysteine-82 in the catalytic mechanism of human S-adenosylmethionine decarboxylase. Biochemistry. 1999;38:2462–2470. doi: 10.1021/bi9825201. [DOI] [PubMed] [Google Scholar]
- 33.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)–a general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 35.Pei J, Grishin NV. Peptidase family U34 belongs to the superfamily of N-terminal nucleophile hydrolases. Protein Sci. 2003;12:1131–1135. doi: 10.1110/ps.0240803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brannigan JA, et al. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 37.Xiong H, Pegg AE. Mechanistic studies of the processing of human S-adenosylmethionine decarboxylase proenzyme. Isolation of an ester intermediate. J. Biol. Chem. 1999;274:35059–35066. doi: 10.1074/jbc.274.49.35059. [DOI] [PubMed] [Google Scholar]
- 38.Dirksen A, Dirksen S, Hackeng TM, Dawson PE. Nucleophilic catalysis of hydrazone formation and transimination: implications for dynamic covalent chemistry. J. Am. Chem. Soc. 2006;128:15602–15603. doi: 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]
- 39.Dirksen A, Dawson PE. Rapid oxime and hydrazone ligations with aromatic aldehydes for biomolecular labeling. Bioconjug. Chem. 2008;19:2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yerlikaya A, Stanley BA. S-adenosylmethionine decarboxylase degradation by the 26S proteasome is accelerated by substrate-mediated transamination. J. Biol. Chem. 2004;279:12469–12478. doi: 10.1074/jbc.M312625200. [DOI] [PubMed] [Google Scholar]
- 41.Dall E, Fegg JC, Briza P, Brandstetter H. Structure and mechanism of an aspartimide-dependent peptide ligase in human legumain. Angew. Chem. Int. Ed. 2015;54:2917–2921. doi: 10.1002/anie.201409135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 43.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blennow K, Mattsson N, Scholl M, Hansson O, Zetterberg H. Amyloid biomarkers in Alzheimer's disease. Trends Pharmacol. Sci. 2015;36:297–309. doi: 10.1016/j.tips.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490:267–272. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buckingham J. The chemistry of arylhydrazones. Q. Rev. Chem. Soc. 1969;23:37–56. [Google Scholar]
- 47.Huizinga EG, et al. Active site structure of methylamine dehydrogenase: hydrazines identify C6 as the reactive site of the tryptophan-derived quinone cofactor. Biochemistry. 1992;31:9789–9795. doi: 10.1021/bi00155a036. [DOI] [PubMed] [Google Scholar]
- 48.Appel MJ, Bertozzi CR. Formylglycine, a post-translationally generated residue with unique catalytic capabilities and biotechnology applications. ACS Chem. Biol. 2015;10:72–84. doi: 10.1021/cb500897w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Q, Buckley D, Guan C, Guo HC. Structural insights into the mechanism of intramolecular proteolysis. Cell. 1999;98:651–661. doi: 10.1016/s0092-8674(00)80052-5. [DOI] [PubMed] [Google Scholar]
- 50.Kabisch UC, et al. Identification of D-proline reductase from Clostridium sticklandii as a selenoenzyme and indications for a catalytically active pyruvoyl group derived from a cysteine residue by cleavage of a proprotein. J. Biol. Chem. 1999;274:8445–8454. doi: 10.1074/jbc.274.13.8445. [DOI] [PubMed] [Google Scholar]
- 51.Andreesen JR. Glycine reductase mechanism. Curr. Opin. Chem. Biol. 2004;8:454–461. doi: 10.1016/j.cbpa.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl. Microbiol. Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- 53.Scheck RA, Dedeo MT, Iavarone AT, Francis MB. Optimization of a biomimetic transamination reaction. J. Am. Chem. Soc. 2008;130:11762–11770. doi: 10.1021/ja802495w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.