Abstract
Antiviral defenses are inappropriately activated in systemic lupus erythematosus (SLE) and association between SLE and the antiviral helicase gene, IFIH1, is well established. We sought to extend the previously reported association of pathogenic soluble mediators and autoantibodies with mouse Mda5 to its human ortholog, IFIH1. To better understand the role this gene plays in human lupus, we assessed association of IFIH1 variants with soluble mediators and autoantibodies in 357 European-American SLE patients, first-degree relatives, and unrelated, unaffected healthy controls. Association between each of 135 genotyped SNPs in IFIH1 and four lupus-associated plasma mediators, IL-6, TNF-α, IFN-β, and IP-10, were investigated via linear regression. No significant associations were found to SNPs orthologous to those identified in exon 13 of the mouse. However, outside of this region there were significant associations between IL-6 and rs76162067 (p = 0.008), as well as IP-10 and rs79711023 (p = 0.003), located in a region of IFIH1 previously shown to directly influence MDA-5 mediated IP-10 and IL-6 secretion. SLE patients and FDRs carrying the minor allele for rs79711023 demonstrated lower levels of IP-10, while only FDRs carrying the minor allele for rs76162067 demonstrated an increased level of IL-6. This would suggest that the change in IP-10 is genotypically driven, while the change in IL-6 may be reflective of SLE transition status. These data suggest that IFIH1 may contribute to SLE pathogenesis via altered inflammatory mechanisms.
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease marked by immune dysregulation and chronic inflammation resulting from reduced immunologic tolerance to nuclear self-antigens. One mechanism by which autoimmunity may develop is through an innate immune response to viral infection that results in cross-reactive recognition of self-antigen via molecular mimicry [1]. One component of antiviral immune defense is the family of retinoic acid-inducible gene-I-like receptors (RIG-I like receptors [RLRs]). RLRs are encoded by a number of IFN-inducible genes [2]. One such IFN-inducible gene, Interferon induced with helicase C domain 1 (IFIH1), has previously been associated with multiple autoimmune conditions, including SLE [3, 4]. IFIH1 has been associated in SLE with increased sensitivity to serum Type I IFN and the presence of anti-dsDNA autoantibodies [5].
In a recent study, melanoma-differentiation-associated gene 5 (Mda5), the mouse ortholog to IFIH1, was shown to be associated with IFN-β, IL-6, CXCL10 (IP-10), ISg56, TNF-α, anti-nuclear antibodies (ANA), and anti-dsDNA antibodies [2]. The Mda5 mutant mice carrying a single missense mutation within exon 13 had significantly higher mRNA expression levels of IFN-β, IL-6, IP-10, ISg56, and TNF-α within their kidneys relative to congenic wild-type (WT) mice. There were also significant increases in serum ANA and anti-dsDNA antibodies in Mda5 mutant mice compared to WT mice. Coinciding with an increase in these pro-inflammatory mediators, Mda5 mutant mice spontaneously developed lupus-like nephritis and systemic autoimmune symptoms without viral infection.
The findings of Funabiki et al. link Mda5 with dysregulated immune function resulting in lupus-like disease [2]. As these associations have yet to be investigated in human SLE, we sought to determine if IFIH1, the human ortholog of Mda5, is associated with altered soluble mediators and autoantibodies in SLE patients compared to unaffected first-degree relatives of SLE patients (FDRs) and unrelated, unaffected controls with no family history of SLE. We have recently demonstrated immunologic differences between FDRs who remained unaffected during a follow-up period of over six years vs. those who have transitioned to classified SLE and unrelated, unaffected healthy individuals [6].
Materials and methods
Study population
Experiments were performed in accordance with the Helsinki Declaration and approved by the Institutional Review Board (IRB) of the Oklahoma Medical Research Foundation (OMRF). Appropriately consented, retrospectively collected, and de-identified clinical and genotyping data, as well as serum and plasma samples, were utilized for the study; the study was approved by the OMRF IRB under exemption 45 CFR 46.101(b) (4). A total of 357 EA study participants, including SLE patients meeting >4 cumulative ACR criteria [7], unaffected first degree relatives of SLE patients (FDRs) not included in the current study, and unrelated, unaffected controls with no family history of SLE, were retrospectively selected from the Oklahoma Rheumatic Disease Research Core Center (ORDRCC) (Table 1). Individuals donating samples to the ORDRCC completed IRB-approved, written consent forms prior to sample procurement. Written consent was recorded into the electronic, de-identified ORDRCC database; a copy of the signed consent form was given to each ORDRCC participant and the original document retained by the clinical coordinator. These individuals were selected based on the availability of retrospectively collected, appropriately consented, genotyping data, as well as serum and plasma samples, to assess levels of soluble mediators and the presence of lupus-associated autoantibody specificities. The inclusion of FDRs in addition to cases and controls boosted the power of our analysis from 0.25 (cases only) to 0.80 (cases + FDRs + controls), f2 effect size = 0.05. For the assessment of association between genetic variants and a quantitative trait (as presented here), the classification of an individual as diseased or not is only relevant in that it 1) helps to identify individuals that may be on treatment affecting levels of the quantitative trait being examined and 2) it allows for assurance that the distribution of the trait being examined is as inclusive as possible. Thus, for our purposes, FDRs, which are known to have elevated levels of many autoantibodies and soluble mediators [6, 8, 9], were included to boost the power, represent a larger range of the trait distribution and allow us to assess if treatment was influencing the values of the cytokines or autoantibodies being studied.
Table 1. European-American Study Participants.
| Disease Status | SLEb | FDRc | Ctld |
|---|---|---|---|
| n (%a) | 128 (36) | 144 (40) | 85 (24) |
| Gender: n (%) | |||
| • Female | 121 (95) | 130 (90) | 82 (96) |
| • Male | 7 (5) | 14 (10) | 3 (4) |
| Age: Median (Interquartile Range) | 48 (34–54) | 49 (40–58) | 53 (52–61) |
a Percent of 357 total participants.
b SLE = patients meeting ≥ 4 ACR criteria.
c FDR = first degree relatives of SLE patients meeting < 4 ACR criteria.
d Ctl = unrelated, unaffected controls with no family history of SLE.
Genotyping
Samples were genotyped on the Illumina ImmunoChip array on an Illumina iScan according to the manufacturers protocols [10]. Genotypes were called using Opticall [11] using the default options with the addition of -nointcutoff option in order to manually remove intensity outliers. Genotype clusters were determined using Evoker [12]. Standard quality control procedures were as follows: removal of samples with mean intensity values <0.25 or >3; removal of SNPs and samples with call rates <90%; removal of samples with heterozygosity >3 sigma; and removal of related samples. Principal component analysis was performed using EIGENSTRAT and genetic outliers were removed as described in [13]. Insertion/deletions were also removed prior to analyzing the data. After quality control, 135 observed SNPs remained and no imputation was performed.
Detection of plasma soluble mediators
Plasma levels of IL-6, TNF-α, IFN-β, and IP-10 (CXCL10) were assessed by xMAP multiplex assay (eBioscience/Affymetrix, Santa Clara, CA), utilizing a single lot of assay plates to limit lot-specific assay variability [14]. In addition, SLE patient, FDR, and control samples were included on the same assay plate; a known control serum was included on each plate (Cellgro human AB serum, Cat#2931949, L/N#M1016) to limit inter-plate variability. Data were obtained using the Bio-Rad BioPlex 200® array system (Bio-Rad Technologies, Hercules, CA), with a lower boundary of 100 beads per analyte per sample. Analyte concentrations were interpolated from 5-parameter logistic nonlinear regression standard curves. Analytes below the detection limit were assigned a value of 0.001 pg/ml. Well-specific validity was assessed by AssayCheX™ QC microspheres (Radix BioSolutions, Georgetown, TX, USA) to evaluate non-specific binding. Mean inter-assay coefficient of variance (CV) of multiplexed bead-based assays for cytokine detection has previously been shown to be 10–14% [15, 16], and a similar average CV (11%) was obtained across the analytes in this assay using healthy control serum. Intra-assay precision of duplicate wells averaged <10% CV in each multiplex assay.
Detection of SLE-associated autoantibodies
Serum and plasma samples were screened for autoantibody specificities using the BioPlex 2200 multiplex system (Bio-Rad Technologies). The BioPlex 2200 ANA kit uses fluorescently dyed magnetic beads for simultaneous detection of 11 autoantibody specificities, including SLE-associated autoantibodies against dsDNA, chromatin, Ro/SSA, La/SSB, Sm, the Sm/RNP complex, and RNP[9]. Anti-dsDNA (IU/mL) has a previously determined positive cutoff of 10 IU/mL; an Antibody Index (AI) value (range 0–8) is reported by the manufacturer to reflect the fluorescence intensity of each of the other autoantibody specificities with a positive cutoff as AI = 1.0. The AI scale is standardized relative to calibrators and control samples provided by the manufacturer.
Statistical methods
Power calculations were completed using the powr.f2.test() function in the “pwr” package in R (version 1.1–3) [17]. IFIH1 association analyses of soluble mediators (IL-6, TNF-α, IFN-β, and IP-10) and lupus-associated autoantibody (anti-dsDNA and ANA [positive for any of the following autoantibody specificities: dsDNA, chromatin, Ro/SSA, La/SSB, Sm, SmRNP, nRNP) data obtained via xMAP multiplex assays were restricted to those previously investigated by Funabiki et al. [2]. Soluble mediator and autoantibody data were transformed via Box-Cox transformation and linear regression analysis was used to assess associations between transformed values and each individual SNP. Age and gender were included as covariates and an additive model of inheritance was assumed in the linear regression analyses.
Association analysis of soluble mediator and autoantibody levels, as well as the number of detected SLE-associated autoantibody specificities, with SNPs in exon 13 of IFIH1 was subsequently expanded to include the 135 quality controlled SNPs within the IFIH1 gene. Linkage disequilibrium between novel significant SNPs and previously identified SLE-associated SNPs was analyzed using the LDmatrix module in LDlink with all European populations [18]. Differences in levels of soluble mediators between major and minor alleles were determined by unpaired t-test (transformed data) and Mann-Whitney test (untransformed concentration data). Differences in levels of soluble mediators between SLE patients (cases), unaffected FDRs, and unrelated, unaffected controls were determined by ANOVA with Tukey’s multiple comparison (transformed data) and Kruskal-Wallis with Dunn’s multiple comparison (untransformed concentration data). Differences in levels of soluble mediators between SLE patients (cases), unaffected FDRs, and unrelated, unaffected controls carrying either major (CC) or minor (TC) alleles of rs7316162067 or rs79711023 for IL-6 or IP-10 levels, respectively, were determined by Kruskal-Wallis with Dunn’s multiple comparison (untransformed concentration data).
Results
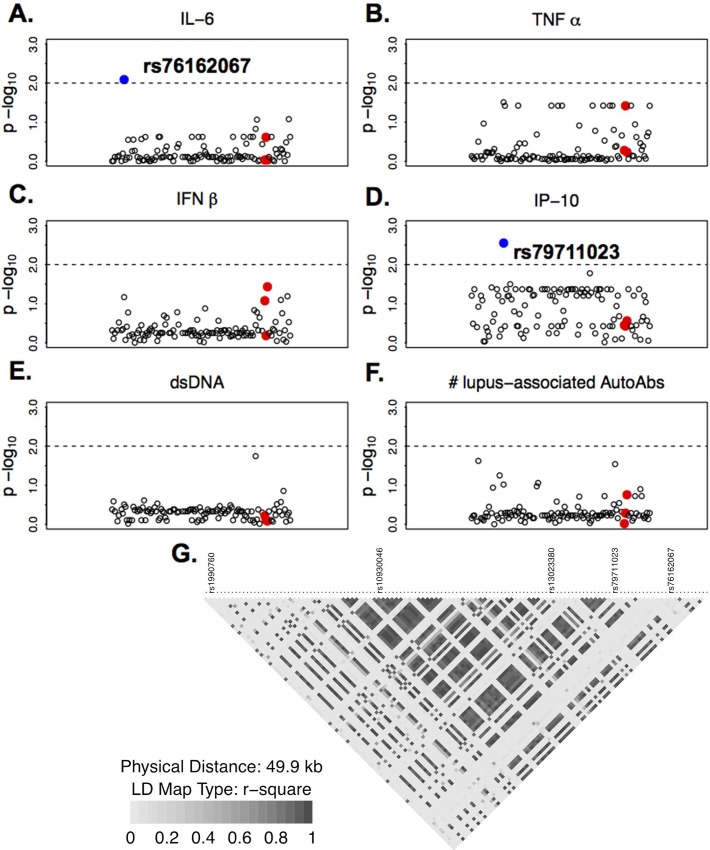
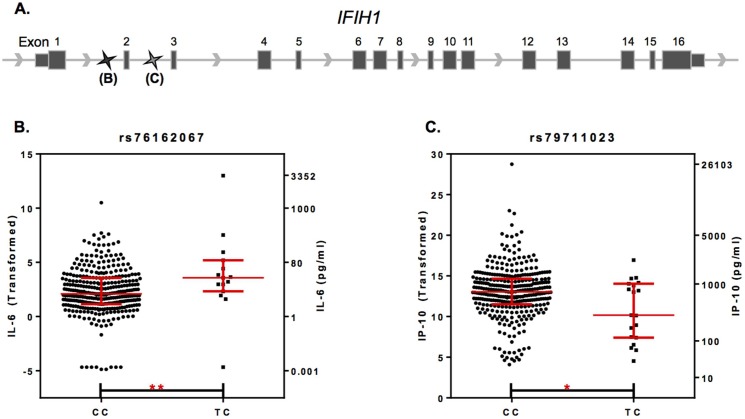
We sought to determine if the soluble mediators that significantly associated with mutant Mda5 [2], also associated with the human ortholog, IFIH1. Plasma levels of select soluble mediators and the presence of lupus-associated autoantibody specificities in 357 individuals (SLE patients, FDRs of SLE patients not included in this study, and unrelated, unaffected controls with no family history of SLE were analyzed along with 135 quality controlled SNPs in the region of IFIH1 (Fig 1). The zoom plots presented demonstrate no significant associations (p<0.01) between variants within IFIH1 exon 13 (red dots) and plasma levels of IL-6, TNF-α, IFN-β, IP-10, or anti-dsDNA (Fig 1A–1E). The number of positive lupus-associated autoantibody specificities was also not significantly associated with variants in exon 13 (Fig 1F). However, significant associations were found with SNPs outside of exon 13 (blue dots). IL-6 was significantly associated with rs76162067 (p = 0.008; Fig 1A), while IP-10 was significantly associated with rs79711023 (p = 0.003; Fig 1D), both of which are upstream of previously identified SLE-associated SNPs, rs13023380, rs10930046, and rs1990760 ([4, 5], Fig 1G). Results from LDlink show that neither rs76162067 nor rs79711023 are in strong linkage disequilibrium with any of the previously identified SLE-associated SNPs mentioned above (S1 Table). Of the 357 individuals assessed, 14 carried the minor (TC) allele for rs73162067, an intronic SNP located between exons 1 and 2, while 19 individuals carried the minor (TC) allele for rs79711023, an intronic SNP located between exon 2 and 3 (Fig 2A). Those individuals carrying the minor allele for rs76162067 and rs79711023 exhibited higher plasma levels of IL-6 (p = 0.0086) and lower plasma levels of IP-10 (p = 0.0250), respectively (Fig 2B and 2C). These data confirm in humans the presence of significant associations between IFIH1 and distinct pro-inflammatory mediators.
Fig 1. Genotypic Associations between IFIH1 and Immune Mediators.
Zoom plots of associations between SNPs in IFIH1 and box-cox transformed plasma levels of (A) IL-6, (B) TNF-α, (C) IFN-β, (D) IP-10, (E) anti-dsDNA, and (F) total number of positive lupus-associated autoantibody specificities (dsDNA, chromatin, Ro/SSA, La/SSB, Sm, SmRNP, and nRNP, as described in Materials and Methods), displayed as negative log transformed p-values (y-axis). Red dots indicate SNPs within Exon 13. LD map of IFIH1 SNPs is presented (G).
Fig 2. IFIH1 effect on plasma levels of IL-6 and IP-10.
Exon map of IFIH1 is presented (A) containing labeled SNPs rs76162067 and rs79711023. IL-6 (B) and IP-10 (C) levels in study participants expressing the major (CC) or minor (CT) allele of SNP rs76162067 or rs79711023, respectively, are expressed as box-cox transformed (left y-axis) and concentration (right y-axis) values. Exon order is displayed L-R for ease of interpretation, although IFIH1 is transcribed R-L from the negative strand. Data in B-C are presented as median ± interquartile range. *p≤0.05, **p<0.01 Mann-Whitney test.
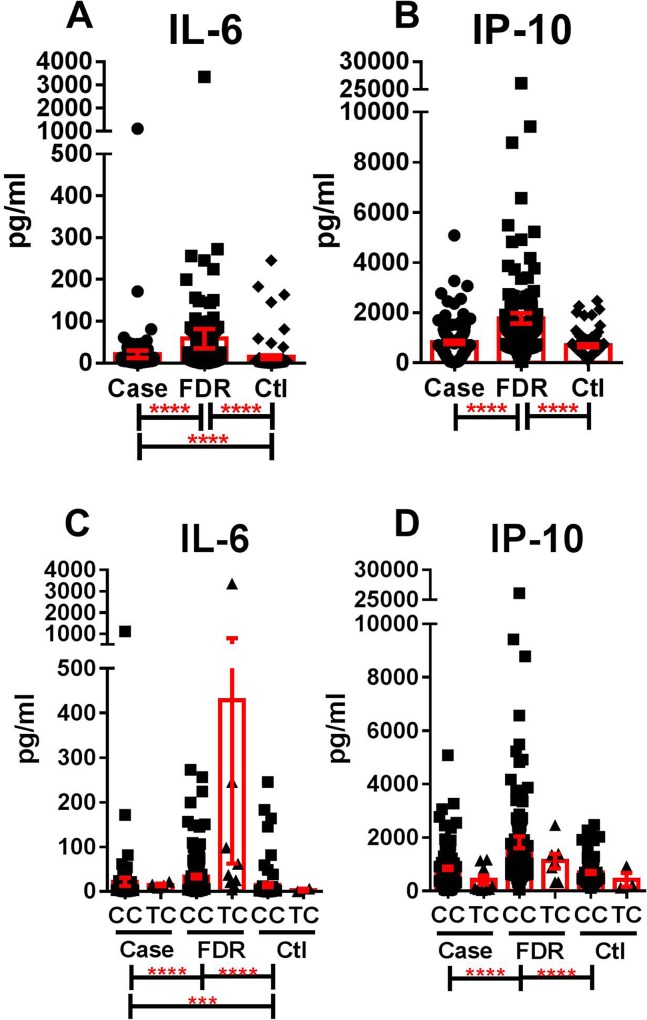
We next evaluated IL-6 and IP-10 levels by affectation status in SLE patients compared to unaffected FDRs and unrelated controls (Fig 3). FDRs had the highest plasma levels of both IL-6 (Fig 3A) and IP-10 (Fig 3B), significantly higher than both cases and unrelated controls (p<0.0001). Cases had significantly higher plasma levels of IL-6 than unrelated controls (p<0.0001). This pattern was unchanged by the presence of major (CC) or minor (TC) alleles of rs7316162067 (Fig 3C) or rs79711023 (Fig 3D) for IL-6 or IP-10 levels, respectively. Although SLE patients were more likely than FDRs and Ctls to have a history of prednisone, hydroxychloroquine, or immunosuppressant medication usage (p<0.0001, S2 Table), neither the presence nor absence of these medications altered the pattern of IL-6 and IP-10 levels in FDRs, SLE patients, and controls, such that FDRs continued to have the highest levels of IL-6 and IP-10, followed by cases and controls (S1 Fig). Although immunosuppressent use decreased IL-6 and IP-10 levels, while prednisone use increased these cytokine levels in cases alone (S1 Fig), correcting for the presence of treatment did not alter the significance of the previous association analysis (rs76162067, p = 0.016; rs76162067, p = 0.024). Finally, other soluble mediators that were not found to be associated with SNPs evaluated within IFIH1, including TNF-α (Fig 1B) and IFN-β (Fig 1C), were also highest in FDRs, followed by SLE patients (cases) and controls (S2A and S2B Fig). In addition to neither anti-dsDNA levels (Fig 1E), nor number of SLE-associated autoantibody specificities (Fig 1F) associated with IFIH1 (Fig 1E), there were no difference in anti-dsDNA levels between SLE patients, FDRs, and controls (S2C Fig). However, cases did have significant accumulation of SLE-associated autoantibody specificities compared to FDRs (p = 0.004) and controls (p = 0.012) (S2D Fig). These data suggest that IFIH1-independent effects are present that contribute to altered pro-inflammatory mediator levels and the presence of SLE-associated autoantibodies.
Fig 3. Altered IL-6 and IP-10 levels in SLE patients and lupus relatives.
Plasma levels of IL-6 (A, C) and IP-10 (B, D) were assessed in SLE patients (case), unaffected first-degree relatives of SLE patients (FDR), and unrelated, unaffected controls with no family history of SLE (Ctl). IL-6 (C) and IP-10 (D) levels are compared by genotype of associated SNPs rs76162067 and rs79711023, respectively. Data are presented as mean + SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 by Kruskal-Wallis test with Dunn’s multiple comparison for all groups assessed (Case/FDR/Ctl [A-B]; Case/FDR/Ctl carrying major (CC) or minor (TC) alleles [C-D]). Only comparisons found to be statistically significant were marked.
Discussion
We sought to determine if pro-inflammatory mediators previously shown to be associated with Mda5, resulting in a lupus-like phenotype in mice [2], also associated with IFIH1 in humans. Contrary to the results of the motivating study in mice, we see no association of soluble mediators with SNPs in exon 13 of IFIH1. When we expanded our analyses, however, to include 135 SNPs across IFIH1 we found two promising effects associated with IL-6 and IP-10. Although previous investigations have shown IFIH1, IL-6, and IP-10 to be associated with SLE [4, 19, 20], none have shown a direct connection between human IFIH1 and these two soluble mediators. SNPs significantly associated with IL-6 (rs76162067) and IP-10 (rs79711023) are located within the region of exons 1–3 that encodes the CARD1/CARD2 domain of the MDA5 protein [4]. The CARD1/CARD2 domain of IFIH1-encoded MDA5 has been shown to signal through the adaptor protein, interferon-β promoter stimulator (IPS-1) [21], necessary for MDA-5 mediated secretion of IL-6 (via NFκB) and IP-10 (via NFκB and IRF3) [22].
We have recently demonstrated that IL-6 [23] and IP-10 [24] are altered early in pre-clinical SLE pathogenesis, prior to the onset of clinical disease, and additional alterations occur again prior to imminent clinical disease flare in patients with classified SLE [14]. In addition to the two genetic variants associated with IL-6 and IP-10 in the current study, SNP rs13023380 has recently been associated with the CARD1/CARD2 encoding region of IFIH1 in SLE patients [4] and lies closest to the IL-6 and IP-10 associated SNPs in our IFIH1 LD map. These findings suggest that the IPS-1 binding region MDA5 is significantly associated with SLE and may contribute to altered IL-6 and IP-10 levels. We observed that those subjects with the minor allele for rs76162067 tended to have higher levels of IL-6, while those with the minor allele for rs79711023 tended to have lower levels of IP-10, including both SLE patients and FDRs of SLE patients, who are at increased risk of transitioning to classified disease [6, 25]. These results suggest that genetic variants within IFIH1 may directly contribute to alterations in the levels of inflammatory mediators associated with SLE pathogenesis and will be the subject of future study.
When we stratified the soluble mediator data by disease status, as mentioned previously, unaffected FDRs of SLE patients not included in the current study had the highest plasma levels of IL-6 and IP-10, significantly higher than both cases and unrelated, unaffected controls, independent of medication usage. These data suggest that FDRs have some level of immune dysregulation that may be partially genetically driven by IFIH1, as has been previously shown for SLE patients [4, 5]. We have recently shown that these unaffected FDRs also displayed the highest plasma levels of the regulatory mediator IL-10 compared to FDRs who transitioned to classified SLE and unrelated, unaffected healthy controls, independent of medication usage [6]. This suggests that although FDRs of SLE patients exhibit pro-inflammatory immune dysregulation compared to unrelated controls, they have heightened regulatory activity that may allow them to remain unaffected, as these FDRs have been followed for an average of 6.7 years without transition to classified SLE [6].
The lack of direct replication between mouse and human genetic associations can occur for a number of reasons. First, differences in linkage disequilibrium (LD) may explain the absence of a significant association within exon 13 as we are not directly observing all possible variation in human IFIH1. LD is roughly a measure of how often alleles are inherited together. If, for example, certain alleles are commonly inherited together in mice at a different frequency than they are in humans, we would expect to see differences between species. Although conservation of haplotype structure across mammals has previously been reported on a genome-wide scale, LD block size and structure at the gene/exon level is indeed species-dependent [26]. For instance, the significant exon 13 variants previously reported in Mda5 may be in LD with a causal variant in the mouse model but not so with the human model. A limitation of the current study is that only 135 of the 1824 IFIH1 variants identified in dbSNP [27] were available for analysis. It is possible that some additional, potentially significant SNPs around exon 13 were not sequenced or that a discrepancy in LD structure failed to capture the association signal. Previously identified SNPs shown to be significantly associated with SLE [4, 5] were found to lie downstream of rs76162067 and rs79711023 on our LD map. This is not surprising given that those SNPs were pinpointed using case-control analyses, rather than analysis based on immune mediator phenotype in the current study. That being said, it is quite exciting that the two SNPs we identified in the current study are located in the region of IFIH1 known to encode the CARD1/CARD2 domain that drives MDA5-mediated IL-6 and IP-10 production, as well as lying in proximity of a previously identified SNP associated with SLE that also lies within the CARD1/CARD2 domain encoding region of IFIH1 [4].
Despite differences between our findings and those of Funabiki et al., associations of genetic variants with a common physiological function have been noted in mouse and human models containing modest phenotypic differences between the two species [28–30]. As such, the results of both our investigation and the Funabiki study support the role of IFIH1 in autoimmune disease through its potential to influence dysregulation of inflammatory pathways. Future studies will build upon our exciting findings that SLE-associated mediators IL-6 and IP-10 are associated with genetic variants of IFIH1 that directly impact MDA5-mediated secretion of these soluble mediators in SLE patients and their blood relatives. Of particular interest will be the potential effects of these IFIH1 variants on the ability of MDA5 to respond to dsRNA and changes in signaling pathways that lead to IL-6 and IP-10 secretion that contribute to altered risk of either developing SLE or having increased morbidity due to a predisposition for altered disease activity in patients with classified SLE.
Supporting information
Comparison of IL-6 and IP-10 levels between each study group assessed by medication usage.
(PPTX)
Comparison of mediators and autoantibodies between each study group.
(PPTX)
R2 values generated from LDmatrix module in the LDlink program referenced from all European populations in 1000 Genomes.
(XLSX)
Counts and percentages of medication use within each study group.
(XLSX)
Acknowledgments
This study was supported by the National Institute of Allergy, Immunology and Infectious Diseases, Office of Research on Women’s Health, National Institute of General Medical Sciences, and the National Institute of Arthritis, Musculoskeletal and Skin Diseases under award numbers U01AI101934, R01AI024717, U19AI082714, U54GM104938, P30GM103510, P30GM110766, P30AR053483, RC1AR058554, U34AR067392, and HHSN266200500026C. This publication is the sole responsibility of the authors and does not represent the views of the National Institutes of Health.
Data Availability
A portion of these data were obtained using clinical diagnostic assays under the purview of the FDA and thus are not available to us. However, all data used in the analyses presented in this manuscript are available upon request from the corresponding author, Courtney G. Montgomery, at Courtney-Montgomery@omrf.org. Data will be available for researchers who meet criteria for access to confidential, anonymous data.
Funding Statement
This study was supported by the National Institute of Allergy, Immunology and Infectious Diseases, Office of Research on Women’s Health, National Institute of General Medical Sciences, and the National Institute of Arthritis, Musculoskeletal and Skin Diseases under award numbers U01AI101934, R01AI024717, U19AI082714, U54GM104938, P30GM103510, P30GM110766, P30AR053483, RC1AR058554, U34AR067392, and HHSN266200500026C. This publication is the sole responsibility of the authors and does not represent the views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fujinami RS, Oldstone MB. Amino acid homology between the encephalitogenic site of myelin basic protein and virus: mechanism for autoimmunity. Science. 1985;230(4729):1043–5. [DOI] [PubMed] [Google Scholar]
- 2.Funabiki M, Kato H, Miyachi Y, Toki H, Motegi H, Inoue M, et al. Autoimmune Disorders Associated with Gain of Function of the Intracellular Sensor MDA5. Immunity. 2014;40(2):199–212. 10.1016/j.immuni.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 3.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324(5925):387–9. PubMed Central PMCID: PMC2707798. 10.1126/science.1167728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molineros JE, Maiti AK, Sun C, Looger LL, Han S, Kim-Howard X, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS genetics. 2013;9(2):e1003222 PubMed Central PMCID: PMC3575474. 10.1371/journal.pgen.1003222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. J Immunol. 2011;187(3):1298–303. PubMed Central PMCID: PMC3304466. 10.4049/jimmunol.1100857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munroe ME, Young KA, Kamen DL, Guthridge JM, Niewold TB, Costenbader KH, et al. Soluble Mediators and Clinical Features Discern Risk of Transitioning to Classified Disease in Relatives of Systemic Lupus Erythematosus Patients. Arthritis & rheumatology. 2016;In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. Epub 1997/10/27. [DOI] [PubMed] [Google Scholar]
- 8.Laustrup H, Heegaard NH, Voss A, Green A, Lillevang ST, Junker P. Autoantibodies and self-reported health complaints in relatives of systemic lupus erythematosus patients: a community based approach. Lupus. 2004;13(10):792–9. 10.1191/0961203304lu2015oa [DOI] [PubMed] [Google Scholar]
- 9.Bruner BF, Guthridge JM, Lu R, Vidal G, Kelly JA, Robertson JM, et al. Comparison of autoantibody specificities between traditional and bead-based assays in a large, diverse collection of patients with systemic lupus erythematosus and family members. Arthritis Rheum. 2012;64(11):3677–86. Epub 2012/11/01. PubMed Central PMCID: PMC3490432. 10.1002/art.34651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13(1):101 Epub 2011/02/25. 10.1186/ar3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah T, Liu J, Floyd J, Morris JA, Wirth N, Barrett JC, et al. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics. 2012;28(12):1598–603. 10.1093/bioinformatics/bts180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris JA, Randall JC, Maller JB, Barrett JC. Evoker: a visualization tool for genotype intensity data. Bioinformatics. 2010;26(14):1786–7. 10.1093/bioinformatics/btq280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–9. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 14.Munroe ME, Vista ES, Guthridge JM, Thompson LF, Merrill JT, James JA. Pro-inflammatory adaptive cytokines and shed tumor necrosis factor receptors are elevated preceding systemic lupus erythematosus disease flare. Arthritis & rheumatology. 2014;66(7):1888–99. PubMed Central PMCID: PMC4128244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupont NC, Wang K, Wadhwa PD, Culhane JF, Nelson EL. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J Reprod Immunol. 2005;66(2):175–91. Epub 2005/07/21. 10.1016/j.jri.2005.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dossus L, Becker S, Achaintre D, Kaaks R, Rinaldi S. Validity of multiplex-based assays for cytokine measurements in serum and plasma from "non-diseased" subjects: comparison with ELISA. J Immunol Methods. 2009;350(1–2):125–32. Epub 2009/09/15. 10.1016/j.jim.2009.09.001 [DOI] [PubMed] [Google Scholar]
- 17.Champely S, Ekstrom C, Dalgaard P, Gill J, Weibelzahl S, De Rosario H. Basic Functions for Power Analysis. 2016.
- 18.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–7. PubMed Central PMCID: PMCPMC4626747. 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong KO, Tan AW, Thong BY, Lian TY, Cheng YK, Teh CL, et al. Enhanced expression of interferon-inducible protein-10 correlates with disease activity and clinical manifestations in systemic lupus erythematosus. Clin Exp Immunol. 2009;156(1):134–40. Epub 2009/01/30. PubMed Central PMCID: PMC2673751. 10.1111/j.1365-2249.2009.03880.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swaak AJ, van Rooyen A, Aarden LA. Interleukin-6 (IL-6) and acute phase proteins in the disease course of patients with systemic lupus erythematosus. Rheumatology international. 1989;8(6):263–8. [DOI] [PubMed] [Google Scholar]
- 21.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–8. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- 22.Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203(7):1795–803. PubMed Central PMCID: PMCPMC2118350. 10.1084/jem.20060792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R, Munroe ME, Guthridge JM, Bean KM, Fife DA, Chen H, et al. Dysregulation of Innate and Adaptive Serum Mediators Precedes Systemic Lupus Erythematosus Classification and Improves Prognostic Accuracy of Autoantibodies J Autoimmun. 2016;Epub Ahead of Print. [DOI] [PMC free article] [PubMed]
- 24.Munroe ME, Lu R, Zhao YD, Fife DA, Robertson JM, Guthridge JM, et al. Altered type II interferon precedes autoantibody accrual and elevated type I interferon activity prior to systemic lupus erythematosus classification. Ann Rheum Dis. 2016;Epub Ahead of Print. Epub 25 Jan 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flesher DL, Sun X, Behrens TW, Graham RR, Criswell LA. Recent advances in the genetics of systemic lupus erythematosus. Expert review of clinical immunology. 2010;6(3):461–79. PubMed Central PMCID: PMC2897739. 10.1586/eci.10.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guryev V, Smits BM, van de Belt J, Verheul M, Hubner N, Cuppen E. Haplotype block structure is conserved across mammals. PLoS genetics. 2006;2(7):e121 PubMed Central PMCID: PMC1523234. 10.1371/journal.pgen.0020121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29(1):308–11. PubMed Central PMCID: PMC29783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung CL, Chan V, Kung AW. A differential association of ALOX15 polymorphisms with bone mineral density in pre- and post-menopausal women. Human heredity. 2008;65(1):1–8. 10.1159/000106057 [DOI] [PubMed] [Google Scholar]
- 29.Beaulieu LM, Lin E, Mick E, Koupenova M, Weinberg EO, Kramer CD, et al. Interleukin 1 receptor 1 and interleukin 1beta regulate megakaryocyte maturation, platelet activation, and transcript profile during inflammation in mice and humans. Arteriosclerosis, thrombosis, and vascular biology. 2014;34(3):552–64. PubMed Central PMCID: PMC4070375. 10.1161/ATVBAHA.113.302700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehlers CL, Walter NA, Dick DM, Buck KJ, Crabbe JC. A comparison of selected quantitative trait loci associated with alcohol use phenotypes in humans and mouse models. Addiction biology. 2010;15(2):185–99. PubMed Central PMCID: PMC2848508. 10.1111/j.1369-1600.2009.00195.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of IL-6 and IP-10 levels between each study group assessed by medication usage.
(PPTX)
Comparison of mediators and autoantibodies between each study group.
(PPTX)
R2 values generated from LDmatrix module in the LDlink program referenced from all European populations in 1000 Genomes.
(XLSX)
Counts and percentages of medication use within each study group.
(XLSX)
Data Availability Statement
A portion of these data were obtained using clinical diagnostic assays under the purview of the FDA and thus are not available to us. However, all data used in the analyses presented in this manuscript are available upon request from the corresponding author, Courtney G. Montgomery, at Courtney-Montgomery@omrf.org. Data will be available for researchers who meet criteria for access to confidential, anonymous data.