SUMMARY
Genetic variation drives phenotypic diversity and influences the predisposition to metabolic disease. Here, we characterize the metabolic phenotypes of eight genetically distinct inbred mouse strains in response to a high-fat/high-sucrose diet. We found significant variation in diabetes-related phenotypes and gut microbiota composition among the different mouse strains in response to the dietary challenge and identified taxa associated with these traits. Follow-up microbiota transplant experiments showed that altering the composition of the gut microbiota modifies strain-specific susceptibility to diet-induced metabolic disease. Animals harboring microbial communities with enhanced capacity for processing dietary sugars and for generating hydrophobic bile acids showed increased susceptibility to metabolic disease. Notably, differences in glucose-stimulated insulin secretion between different mouse strains were partially recapitulated via gut microbiota transfer. Our results suggest that the gut microbiome contributes to the genetic and phenotypic diversity observed among mouse strains and provide a link between the gut microbiome and insulin secretion.
Keywords: Metabolic disease, gut microbiome, pancreatic islets, insulin secretion
eTOC blurb
Host genetics modulates the development of metabolic disease and shapes the composition of the gut microbiota. Kreznar et al. demonstrate that the gut microbiota contributes to strain-specific susceptibility to diet-induced metabolic disease and identify links between microbial metabolism and insulin secretion.
INTRODUCTION
The intestinal microbiota exerts a profound influence on development, physiology and health (Clemente et al. 2012; Sommer & Bäckhed 2013; Tremaroli & Bäckhed 2012). Although there is substantial interpersonal variation in the composition of the gut microbiota among unrelated healthy subjects, sequencing studies have revealed distal gut community patterns associated with different pathological states, including obesity and diabetes (Ridaura et al. 2013; Qin et al. 2012; Karlsson et al. 2013). Remarkably, alterations in the intestinal microbiota composition have been shown to modulate insulin sensitivity (Vrieze et al. 2010) —a key feature in metabolic disease and type 2 diabetes (T2D), and thus play a role in diabetes susceptibility. s
Dietary components that are not efficiently absorbed in the proximal intestine reach the distal gut where they are metabolized by gut microbes. Intestinal microbes impact our health in part by generating numerous metabolites from our diet. Short-chain fatty acids (SCFA), mainly acetate, propionate and butyrate, are produced through bacterial fermentation of dietary carbohydrates. SCFA serve as energy and signaling molecules in the intestine and peripheral organs (Besten et al. 2013). Specifically, SCFA are important regulators of both energy and glucose homeostasis (Besten et al. 2013; Koh et al. 2016). For example, butyrate improves insulin sensitivity (Gao et al. 2009; Hartstra et al. 2015) and T2D patients have reduced levels of butyrate-producing bacteria (Qin et al. 2012). Additionally, acetate modulates insulin secretion from β-cells (Priyadarshini et al. 2015; Perry et al. 2016). While primarily associated with metabolic benefits, increased concentrations of butyrate and acetate have been found in the cecum of obese mice, suggesting an increased ability of the microbiome to harvest energy from the diet (Turnbaugh et al. 2006).
Gut microbes also impact host physiology by modifying bile acids (BA) synthesized by the host (Houten et al. 2006; Kuipers et al. 2014; Ryan et al. 2014; Sayin et al. 2013). In addition to their role in emulsifying lipids, BA function as hormones through their ability to activate nuclear hormone receptors (D. J. Parks et al. 1999) and G-coupled protein receptors (Kawamata et al. 2003). They modulate glucose homeostasis, lipid metabolism, energy expenditure, and intestinal motility (Kuipers et al. 2014). Primary BA are synthesized from cholesterol in the liver (Russell 2009), stored in the gallbladder, and secreted into the duodenum upon ingestion of a meal. The gut microbiota catalyzes the production of secondary BA via deconjugation, dehydrogenation, epimerization, and dehydroxylation of primary BA (Ridlon et al. 2006). BA with different modifications vary in their ability to activate receptors and affect host physiology (Makishima et al. 1999; Kuipers et al. 2014). Subjects with T2D have altered circulating BA profiles. Treatment of T2D subjects with compounds that increase fecal excretion of BA and modify BA composition improves their glycemic status (Handelsman 2011).
Mouse genetics can be employed to explore the relationships between diet, host genetics, and metabolic responses (O’Connor et al. 2014; B. W. Parks et al. 2013; Ussar et al. 2015). The Collaborative Cross (CC) is a systems genetics mouse resource that consists of a panel of recombinant inbred lines and an outbred stock derived from eight genetically diverse founder strains. These include five classical inbred strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ and NZO/HILtJ), and three wild-derived strains (CAST/EiJ, PWK/PhJ, WSB/EiJ) (Churchill et al. 2004; Roberts et al. 2007; Aylor et al. 2011).
We examined the metabolic phenotypes and gut microbiota composition of the eight CC founder strains in response to chronic consumption of two defined diets: a high-fat/high-sucrose diet (HF/HS) and a control diet. We found remarkable variation in diabetes-related phenotypes and gut microbiota composition as a function of host genotype and diet, and we identified bacterial taxa that correlate with metabolic traits, including body weight, glucose, and insulin levels. Germ-free (GF) mice were colonized with microbiota derived from two founder strains that exhibited divergent metabotypes, C57BL/6J and CAST/EiJ. The transplanted animals were maintained on the HF/HS diet and then subjected to metabolomic and metagenomic analyses. We identified functional differences attributable to the two transplanted microbial communities, including insulin secretion responses and susceptibility to diet-induced metabolic disease.
RESULTS
Host metabolic responses to diet are influenced by genetic background
We assessed the variability of diet-induced metabolic responses of the eight genetically diverse CC founder strains: A/J, C57BL/6J (B6), 129S1/SvImJ (129), NOD/ShiLtJ (NOD), NZO/HILtJ (NZO), CAST/EiJ (CAST), PWK/PhJ (PWK), WSB/EiJ (WSB). All mice were obtained from the Jackson Laboratory, maintained in the same vivarium and fed the same diet, so that the only known difference among the strains is genetics. We placed four-week-old male mice from each strain on either a control or a high-fat high-sucrose (HF/HS) diet for 22 weeks (Table S1).
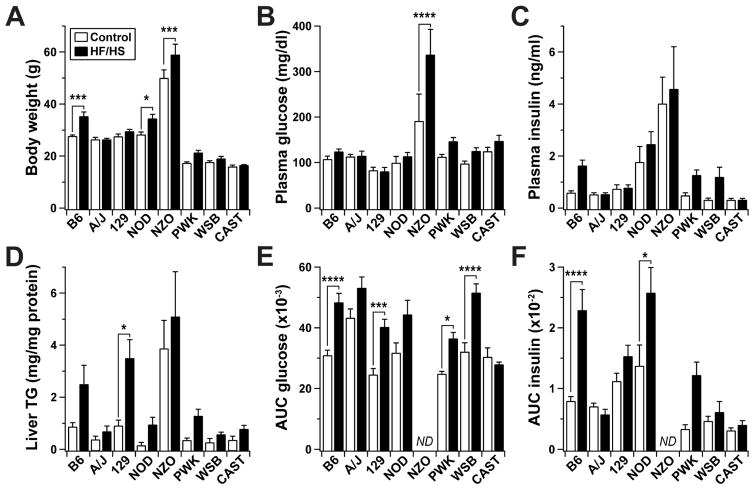
The CC founder strains displayed a wide range of body weight and metabolic responses to the dietary challenge (Figure 1 and S1). Two-way ANOVA analysis of the clinical traits revealed a significant strain effect for fasting insulin (F = 14.94, p < 0.0001). We also observed significant strain-diet interactions for body weight (F = 3.19, p < 0.01) and fasting glucose (F = 2.81, p < 0.01). Significant strain and diet effects were also seen for hepatic triglyceride content (F = 10.96, p < 0.0001; F = 11.92, p < 0.001, respectively) effects. Liver triglyceride content showed high inter-strain variation, with 129 having the most significant response to diet (p < 0.05) (Figure 1D). NZO mice were the only strain to become overtly diabetic (glucose levels >300 mg/dl) as a consequence of HF/HS feeding. With the exception of NZO mice, which did not survive past 18 weeks on the HF/HS diet, B6 mice were the most responsive to diet. HF/HS-fed B6 mice became obese (p < 0.01) and developed insulin resistance and glucose intolerance after ~8 weeks (Figure 1A and S1A-C). In addition to differences in diet responsiveness, the strains varied in both absolute levels of insulin and change in insulin levels over time, suggesting a significant divergence in insulin sensitivity among the strains (Figure S1B).
Figure 1. Segregation of metabolic syndrome among CC founder mice.
Male mice were maintained on the high-fat/high-sucrose (HF/HS) or a control diet for 22 weeks beginning at 4 weeks of age. (A) Body weight, (B) fasting plasma glucose and (C) insulin, and (D) hepatic triglyceride content determined for all mice at 26 weeks of age. Areas under the curve (AUC) for (E) glucose and (F) insulin during oral glucose tolerance test (oGTT) conducted at 22 weeks of age. Insulin and glucose values were determined from plasma following a 4 hour fast. No data (ND) were collected for NZO mice during oGTT. In all panels, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by two-way ANOVA (diet and strain) with Bonferroni’s multiple comparisons test to assess within-strain differences. Data are mean ± SEM, n ≥ 9 mice/genotype/diet.
To assess whole-body glucose homeostasis and more directly evaluate the underlying role of the pancreatic islets in the control of plasma insulin, we measured plasma glucose and insulin during an oral glucose tolerance test (oGTT). Both plasma glucose and insulin during the oGTT varied dramatically between the strains. We computed the area under the curve (AUC) for each trait to determine the overall excursion in glucose and insulin that occurred during the oGTT (Figure 1E–F and S2). We observed a wide inter-strain range of responses in plasma insulin during the oGTT (F = 12.84, p < 0.0001) (Figure 1F and 2B). Changes in plasma insulin may reflect altered insulin secretion from β-cells, peripheral insulin resistance, reduced insulin clearance, or any combination thereof. 129 and WSB showed diet-induced glucose intolerance, but minimal changes in their insulin response during the oGTT (Figure 1E–F and S2A), suggesting that their glucose intolerance may be driven by altered insulin secretion and/or enhanced insulin clearance. Remarkably, insulin secretion and glucose tolerance were completely unaffected by the HF/HS diet in CAST. Furthermore, the kinetics of the glucose and insulin responses were more rapid in CAST than in all other strains (Figure S2), suggesting that CAST mice may employ different pathways underlying glucose-stimulated insulin secretion and whole-body glucose disposal.
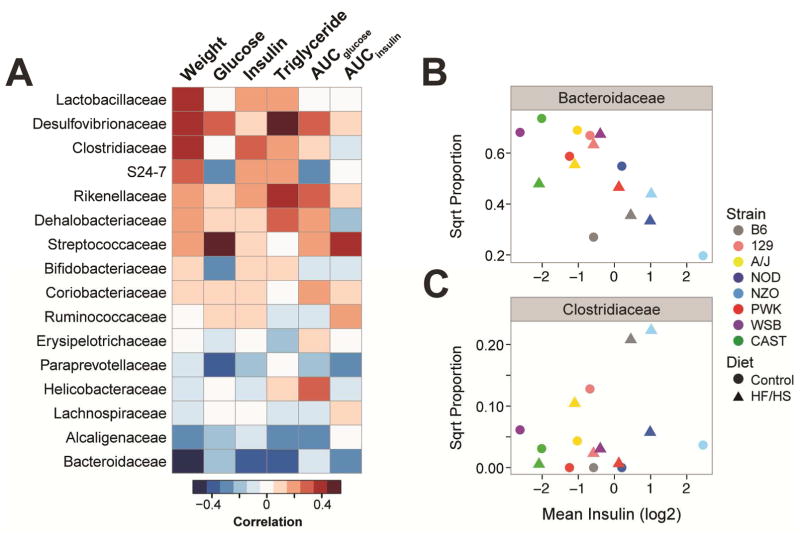
Figure 2. Gut microbial taxa correlate with metabolic phenotypes.
(A) Heat map illustrates Pearson’s pair-wise correlation between microbial families and diabetes-related clinical traits measured in the 8 CC founder mice (n ≥ 9 mice/genotype/diet). Microbial families are ordered by their correlation to body weight. Red, positive correlation; blue, negative. Area under the curve (AUC) values for insulin and glucose were computed from oGTT conducted at 22 weeks; other metrics were collected at 26 weeks. Correlation coefficients and p-values found in Table S2. Contributions of strain and diet on the correlations observed between fasting insulin and (B) the Bacteroidaceae family, and (C) the Clostridiaceae family.
Diet and host genotype influence microbiota composition
Gut microbes influence the development of metabolic disease. We characterized the cecal microbiomes of the eight CC founder strains by 16S rRNA sequencing. We compared the cecal microbiomes employing UniFrac, a phylogenetic distance metric used to measure differences in bacterial community structure (Lozupone & Knight 2005). Principal coordinates analysis (PCoA) of 16S rRNA unweighted UniFrac distances revealed a strong influence of strain (PERMANOVA, p < 0.001) and diet (PERMANOVA, p < 0.001) on microbial community composition (Figure S3A). Consistent with previous studies, the effect of diet on gut microbial composition varied among the strains (O’Connor et al. 2014; B. W. Parks et al. 2013; Carmody et al. 2015), where B6, CAST and NOD mice showed the greatest microbiome response to diet (Figure S3A).
We detected eight bacterial phyla among the mice (Figure S3B). Bacteroidetes and Firmicutes dominated the gut of all strains on either diet, accounting for >90% of the sequenced reads. As reported by other studies, we observed a decrease in the Bacteroidetes:Firmicutes ratio and an increase in Proteobacteria in the HF/HS-fed mice (Ley et al. 2005; Hildebrandt et al. 2009). In fact, Proteobacteria showed the greatest fold change in abundance in response to diet: HF/HS feeding caused an average 5.4-fold change (p < 0.0001), although the relative increase varied among strains.
Microbial taxa correlate with metabolic phenotypes
To determine whether strain-dependent variability in microbiota composition was associated with the dramatic differences in the diabetes-related clinical traits, we computed Pearson’s correlations between abundance of family-level taxa and the metabolic traits among the 8 CC founder mice (Figure 2A). We focused our analysis on families that were present in at least 7 of the founder strains. Bacteroidaceae was among the most negatively correlated with several metabolic phenotypes, including body weight, fasting plasma insulin and AUCinsulin during the oGTT. The Bacteroidaceae family belongs to the Bacteroidetes phylum and is typically found at higher levels in fecal samples of lean vs. obese individuals (Ley et al. 2005; Turnbaugh et al. 2009). Conversely, Clostridiaceae and Rikenellaceae showed the strongest positive correlations with plasma insulin levels. Our analysis also identified strong positive correlations between fasting plasma glucose and the Streptococcaceae and Desulfovibrionaceae families. Members of these families have previously been shown to be enriched in the fecal microbiome of patients with T2D (Qin et al. 2012; Karlsson et al. 2013).
Some of the correlations mentioned above varied significantly as a function of host diet and strain (Table S2). For example, the negative correlation observed between fasting insulin levels and Bacteroidaceae had a significant strain effect (p < 0.0001 ). We also observed a slight diet effect (p < 0.001), which is likely driven by the low abundance and high fasting insulin levels in the chow-fed NZO mice (Figure 2B). We also observed a significant diet effect for the relationship between Clostridiaceae and fasting insulin levels (p < 0.05), but there was also a strain difference that seems to be driven by NZO on chow diet (p < 0.001) (Figure 2C).
These results suggest that diet and genetic background are major determinants of gut microbial composition and metabolic disease. However, the relative contributions of host genetic variance vs. microbial-derived genetic variation across different mouse strains in the development of diet-induced metabolic phenotypes remain largely unknown.
The gut microbiome is a source of genetic variation that influences host-associated differences in diet-induced metabotypes
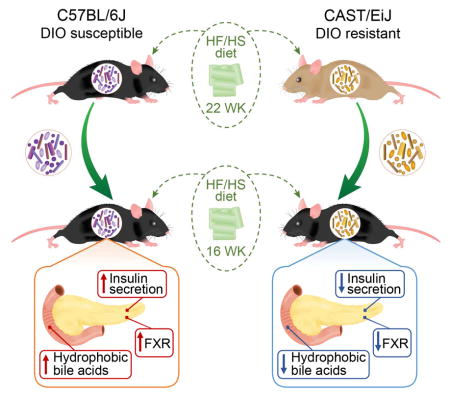
To directly test the influence of gut microbes on the metabolic phenotypes observed among the founder strains, we performed cecal transplants into germ-free B6 (B6-GF) hosts, leveraging two CC founder strains that showed disparate responsiveness to the HF/HS diet. The B6 strain became obese, insulin resistant, and glucose intolerant, whereas the CAST strain remained lean and insulin-sensitive despite HF/HS feeding (Figure 1).
As mentioned above, B6 and CAST mice had significantly different intestinal microbiota (PERMANOVA, F = 4.86, p < 0.001) (Figure S3A). B6 mice harbored a significantly greater abundance of microbial families with strong positive correlations with metabolic traits, such as weight and insulin (i.e. Clostridiaceae, p < 0.05), while CAST mice had a greater representation of families with significant negative correlations (i.e. Bacteroidaceae, p < 0.01) (Figure 2A and S3C).
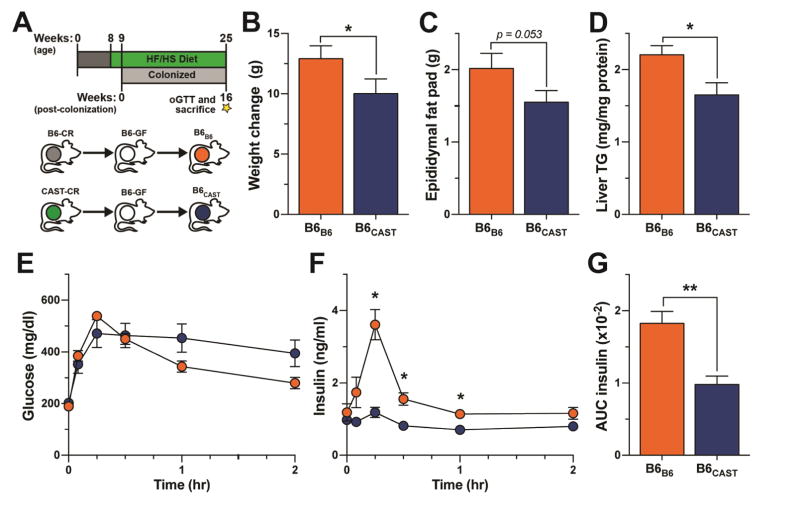
We transplanted cecal microbiota from either conventionally-raised B6 (B6-CR) or CAST (CAST-CR) donor mice into 9-week-old B6-GF recipient mice, to yield B6B6 or B6CAST mice, respectively. Transplanted animals were housed by treatment group in separate vinyl gnotobiotic isolators and maintained on a HF/HS diet for 16 weeks following colonization (Figure 3A). A dietary treatment of 16 weeks allows robust development of metabolic phenotypes associated with consumption of HF/HS diet.
Figure 3. Divergent effects of B6 and CAST microbiomes on diet-induced metabolic phenotypes.
(A) Transplant experimental design. (B) Total weight change, (C) epididymal fat pad mass and (D) quantification of hepatic triglyceride (TG) contents. (E and F) Glucose and insulin values during oGTT and (G) AUC insulin in B6B6 and B6CAST mice. All measurements shown collected 16-weeks post-colonization. *p < 0.05, **p < 0.01 by Student’s t-test. Data are mean ± SEM, n = 7 for B6B6 and n = 6 for B6CAST mice.
Recipient mice recapitulated microbial and metabolic phenotypes observed in the respective donor strains (Figure 3 and 4). B6B6 mice gained ~25% more weight, had larger epididymal fat pad mass and showed greater hepatic triglyceride accumulation than B6CAST mice (Figure 3). Additionally, oGTT revealed that while the plasma glucose levels resulting from an orally administered bolus of glucose did not significantly differ between the two groups of transplanted mice (Figure 3E), the insulin responses were dramatically different (Figure 3F). The glucose challenge evoked a much larger insulin response in B6B6 mice than in B6CAST mice. The low insulin response in B6CAST mice resembled the insulin response of the CAST-CR donors (Figure 1F and S1F). These results suggest that the effectiveness of insulin to maintain euglycemia was greater in the mice receiving the CAST microbiota than in mice receiving the B6 microbiota (Figure 3E–F).
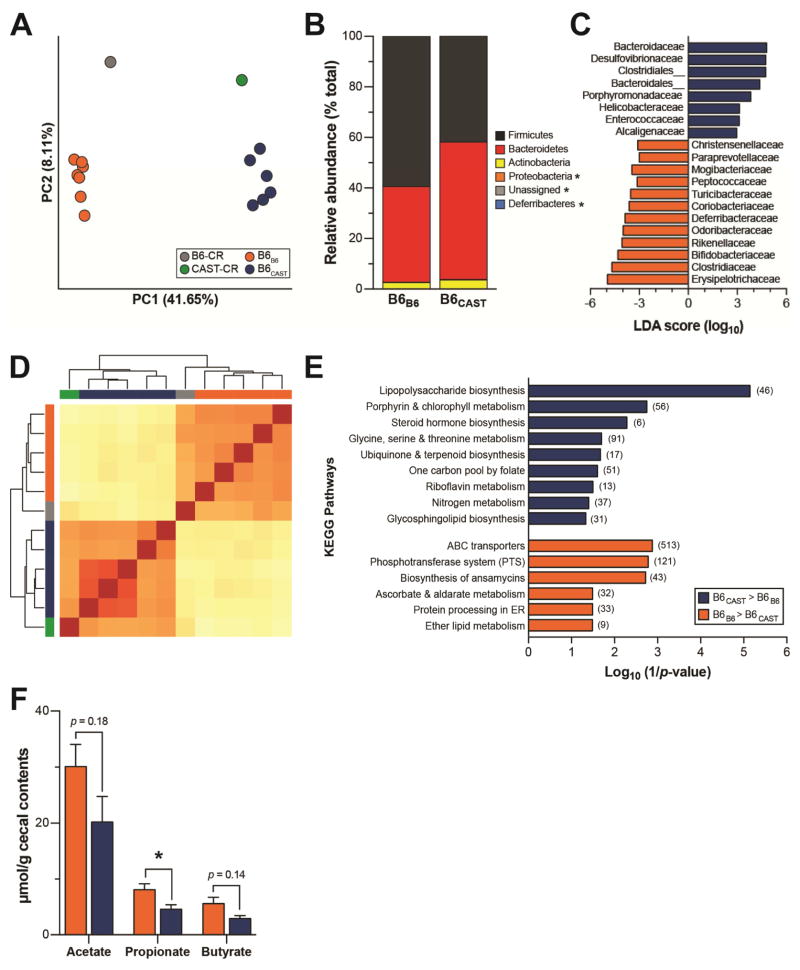
Figure 4. Gut microbiota composition and function of transplant recipients.
(A) Principal coordinate analysis (PCoA) of unweighted UniFrac distances for the fecal microbiota of transplant donors and recipients at sacrifice. Each circle represents an individual mouse. Percent variation explained by each PC is shown in parentheses. (B) Relative abundance of major microbial phyla ordered by increasing mean abundance; * denotes mean phyla abundance <1%. (C) Microbial families differentially enriched in either B6CAST (blue) or B6B6 (orange) as determined by linear discriminant analysis (LDA) with effect size (LEfSe). (D) Clustering of mice based on relative abundance of KEGG metabolic pathways using euclidian distance measurement with complete linkage hierarchical clustering; B6-CR (grey), CAST-CR (green), B6B6 (orange), B6CAST (blue). (E) KEGG categories enriched in either CAST (blue) or B6 (orange) transplanted microbiomes. (F) Targeted GC-MS analysis of cecal short-chain fatty acids; *p < 0.05 by Student’s t-test. Data are mean ± SEM, n = 6–7 mice/recipient group and n = 2–3 mice/donor group. For metagenomics analysis n=5 mice/recipient group.
16S rRNA gene profiling of the donor cecal inoculum and transplant recipient fecal samples show that recipient mice were successfully colonized with the donor’s microbiota. B6B6 and B6CAST mice assumed a phylogenetically similar composition to that of their respective donors as confirmed by PCoA of unweighted UniFrac distances (Figure 4A). As seen in the founders, Bacteroidetes and Firmicutes comprised ~90% of the microbiome, although the abundance of Firmicutes was higher in B6B6 (p < 0.05) (Figure 4B). We identified taxonomic differences in the microbiota composition between the two recipient groups using linear discriminant analysis (LDA) effect size (LEfSe) with LDA score >2 (Segata et al. 2011). We found 20 microbial families that were differentially enriched in the fecal microbiota of B6B6 versus B6CAST mice. There were 12 microbial families that were enriched in B6B6, of which 7 belonged to the Firmicutes phyla (Figure 4C). Some of the families differentially represented in the transplanted animals overlap with taxa that are significantly correlated with metabolic phenotypes in the founder strains (Figure 2). Notably, B6B6 mice exhibited higher levels of Clostridiaceae (p < 0.01), which is positively associated with insulin secretion in the founder strains (Figure 2), whereas B6CAST mice had higher levels of Bacteroidaceae (p < 0.01), which is negatively associated with body weight and insulin secretion (Figure 2). These results are concordant with the metabolic phenotypes observed in the transplanted mice and suggest that the distinct microbial gut communities influence metabolic changes evoked by HF/HS feeding, including insulin secretion,
We characterized the functional potential of transplanted communities by sequencing and analyzing their metagenomes. Metagenomic analysis of the same samples further validated that the B6 and CAST-derived microbiota were distinct from one another, with donors clustering with their respective transplant recipients (Figure 4D). We identified several thousand genes differentially represented between the B6 and CAST microbiota (Table S4). This metagenomic analysis also revealed microbial functions that were enriched in each transplanted microbial community (Table S5). The most enriched microbial pathways in B6B6 mice included genes involved in membrane transport, and carbohydrate and lipid metabolism (Figure 4E). For example, the ABC transporters and phosphotransferase system (PTS) pathways were enriched in mice colonized with the B6 microbiota (p < 0.01). PTS are a class of transport systems involved in the uptake and phosphorylation of a variety of carbohydrates that can be subsequently fermented to SCFA (Deutscher et al. 2006). It has been previously reported that diet-induced obese mice have a concomitant enrichment of microbial pathways involved in PTS and elevated concentrations of SCFA (Turnbaugh et al. 2008), reflecting an increased capacity for energy harvest. Consistent with these results, targeted GC/MS analysis of SCFA in cecal contents disclosed that B6B6 mice had an increased concentration of the major fermentation end-products, compared with B6CAST (Figure 4F). Conversely, B6CAST microbiota were enriched in genes related to the vitamin B12 (cobalamin) biosynthetic pathway (Figure S4A), synthesis of other B vitamins and enzyme co-factors, as well as lipopolysaccharide (LPS) biosynthesis (Figure 4E and S4B). A difference in LPS biosynthetic potential may reflect the composition of the B6CAST microbiota, which has a significantly higher relative abundance of gram negative Bacteroidetes than the B6B6 microbiota (Figure S3B). Our findings mirror those described previously in T2D patients relative to diabetes-free control patients (Qin et al. 2012; Karlsson et al. 2013)—both the microbiota of T2D patients and our metabolically-diseased mice with B6 microbiota show enrichment in KEGG pathways involved in membrane transport, while diabetes-free patients and mice with the CAST microbiota exhibit enrichment in vitamin and cofactor biosynthesis.
B6 and CAST microbiota produce divergent bile acid profiles
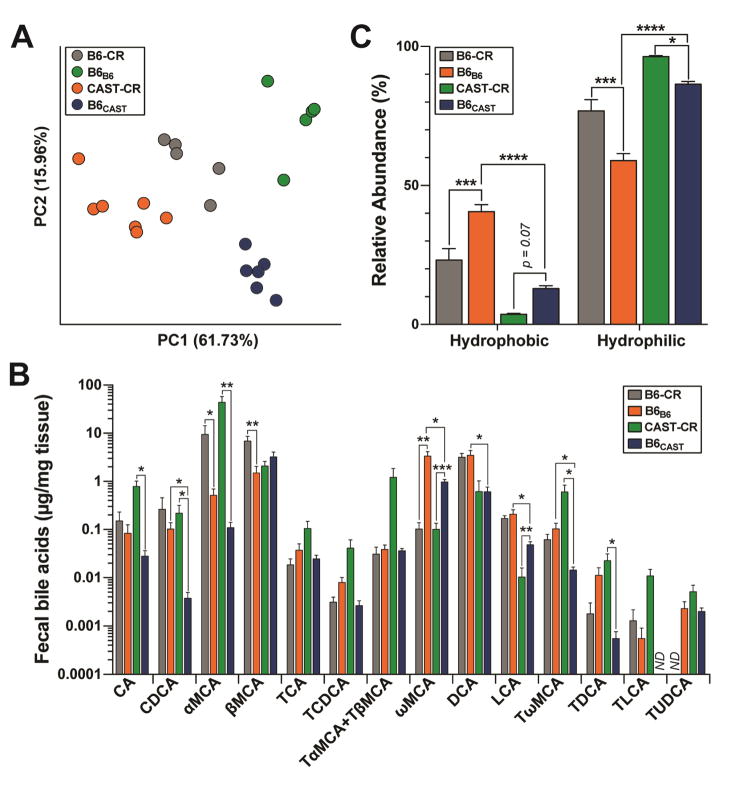
Gut microbes impact host physiology in part by modulating the composition of the BA pool. We determined fecal BA profiles of the transplanted mice and HF/HS-fed B6-CR and CAST-CR mice by UPLC/MS-based quantification of primary and the most abundant secondary BA. The BA composition of B6B6 mice closely resembled that of B6-CR donor mice, whereas B6CAST exhibited a BA profile that was intermediate between CAST-CR and B6-CR mice (Figure 5A). Microbiota composition was also a significant predictor of BA composition. Bray-Curtis dissimilarity-based PCA revealed clustering of the BA profiles by microbiota composition.
Figure 5. B6 and CAST microbiota produce different bile acid profiles.
(A) Principal component analysis of the square root proportion of 14 major bile acid species (ng/mg). Each dot represents the bile acid profile of an individual mouse. Percent variation explained by each PC is shown in parentheses. (B) Abundance of fecal bile acids, and (C) relative abundance of hydrophobic and hydrophilic BA species determined by UPLC-MS/MS from fecal samples collected at 12-weeks post-colonization. No data (ND). *p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA with Bonferroni’s multiple comparisons test. Data are mean ± SEM, n= 6–7 for transplant recipients, and n= 5 for CR mice.
Although the B6CAST microbiota composition resembled that of CAST-CR (Figure 4A), there were significant differences in BA profiles between these groups, suggesting that variation in circulating BA is under the control of both host genetics and gut microbiota. For example, the primary BA cholic acid (CA), chenodeoxycolic acid (CDCA) and α-muricholic acid (α-MCA) were significantly higher in CAST-CR mice compared to B6CAST mice (p < 0.01, p < 0.05, p < 0.01, respectively) (Figure 5B). Moreover, taurine-conjugated muricholic acids (MCAs) were significantly higher in CAST-CR mice compared with B6CAST mice. In contrast, these differences in taurine conjugation were not present between B6-CR and B6B6 mice. Taurine conjugation of MCAs is a host process (Ridlon et al. 2006), further highlighting the interaction of host genetics and microbiome in modulating host BA profiles.
B6-CR and B6B6 mice had a significantly greater representation of hydrophobic BA species (e.g., deoxycholic acid, lithocholic acid (Figure 5B–C)), which are elevated in humans and mice with insulin resistance (Ryan et al. 2014; Prawitt et al. 2011). Microbial metabolism of bile acids generally leads to a more hydrophobic bile acid pool, which facilitates fecal elimination of bile acids. Bile salt hydrolases (BSH) are involved in the hydrolysis of conjugated BA, a necessary step for the production of secondary BA. Consistent with the results presented above, there were a higher number of distinct BSH genes in the B6 microbiota relative to CAST microbiota (13 annotated BSH genes highly abundant in the B6 microbiota relative to CAST vs. two annotated BSH genes highly abundant in the CAST microbiota relative to B6, Table S4). Furthermore, the two groups of recipient mice had vastly different fecal BA profiles. Chenodeoxycholic acid (CDCA; p < 0.05), deoxycholic acid (DCA; p < 0.01), lithocholic acid (LCA; p < 0.01), ω-muricholic acid (ωMCA; p < 0.05), and tauro-ω-muricholic acid (TωMCA; p < 0.05) were all significantly higher in B6B6 than in B6CAST (Figure 5B). DCA was the most abundant BA species in B6B6 mice, and was also ~5-fold more abundant in B6-CR vs. CAST-CR mice. DCA contributes to microbial dysbiosis, a hallmark of metabolic disease, and is positively associated with higher levels of Firmicutes (Islam et al. 2011). Tauroursodeoxycholic acid (TUDCA) was >2-fold higher in CAST-CR mice compared to the transplanted animals, but was not detected in B6-CR mice. Interestingly, administration of TUDCA has been shown to decrease hepatic steatosis and improve insulin resistance in genetically obese mice (Kars et al. 2010; Ozcan et al. 2006), suggesting a potential protective role. These results reveal differences in BA profiles linked to both host genotype and gut microbial composition. They also suggest that the differential responses to prolonged HF/HS diet consumption between B6 and CAST mice could be mediated at least in part by differences in microbial BA metabolism.
Gut microbiota influences insulin secretion
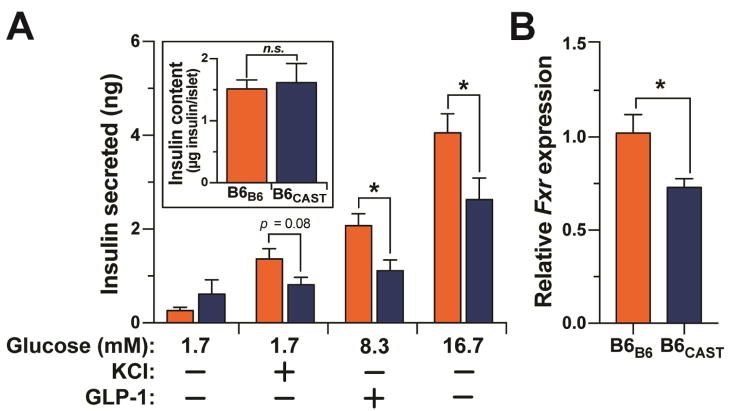
The most dramatic phenotype difference we observed between B6B6 and B6CAST mice was in insulin secretion, where B6CAST mice had a blunted insulin response during the oGTT (Figure 3E). This attenuated response in B6CAST mice may also reflect low insulin secretion from β-cells and/or increased insulin clearance. To determine whether the differential insulin response during the oGTT in the B6B6 vs. B6CAST mice resulted from altered insulin secretion, we performed ex vivo insulin secretion assays on isolated islets. Islets were harvested from B6-GF mice 1 month after successful colonization with either CAST-CR or B6-CR cecum-derived microbiota (Figure S5).
The isolated islets partially recapitulated the reduced insulin secretion observed in the CAST-colonized mice in vivo (Figure 3E). The comparison between the B6-GF mice receiving B6 vs. CAST microbiota allowed us to estimate the contribution of the microbiota to the strain difference in insulin secretion (Figure 6A). Accordingly, the reduction in insulin secretion caused by CAST microbiota colonization in B6 mice was ~33%.
Figure 6. CAST and B6 microbiomes differentially regulate insulin secretion and Fxr expression in pancreatic islets.
(A) Total islet insulin content and glucose-stimulated insulin secretion in response to low glucose (3.3 mM), low glucose plus KCl (40 mM), high glucose (16.7), and high glucose plus GLP-1 (100 mM) from islets isolated from B6B6 and B6CAST mice. The number of islets and the insulin content per islet were not different between the groups. (B) Relative expression of Fxr mRNA from isolated islets. Figure S6 shows microbiota composition for donor and transplanted communities. *p < 0.05 by Student’s t-test. Data are mean ± SEM, n = 5.
Circulating acetate is capable of modulating insulin secretion from pancreatic islets. Specifically, recent studies have shown that acetate directly enhances glucose-stimulated insulin secretion through activation of free fatty acid receptors on β-cells (Priyadarshini et al. 2015) and the parasympathetic nervous system (Perry et al. 2016). Therefore, we measured concentrations of SCFA in plasma and cecum, but found no differences in levels of acetate between B6B6 and B6CAST mice (Figure S6A–B), suggesting that the divergent effects of the B6 and CAST microbiota on insulin secretion are unlikely to stem from differences in acetate.
Recent in vitro studies have also identified BA as important regulators of islet function (Düfer et al. 2012; Renga et al. 2010). We investigated the plasma BA profiles in the B6B6 and B6CAST mice used for insulin secretion studies (Figure S6C–D). B6CAST BA profiles were composed of a significantly higher percentage of hydrophilic BA (Figure S6C). Consistent with a previous report (Sayin et al. 2013), BA profiles were dominated by taurine-conjugated species, with TωMCA and TβMCA being the two most abundant in both groups of animals (Figure S6D). In B6B6 mice, the hydrophobic secondary BA DCA and LCA were significantly higher than in B6CAST mice (Figure S6D).
BA regulate insulin secretion through the activation of specific receptors in islets. For instance, BA can directly increase insulin secretion and production through activation of farnesoid X receptor (Fxr) in β-cells (Düfer et al. 2012; Renga et al. 2010). Expression of Fxr is increased in an agonist-dependent manner (Lee et al. 2006). Remarkably, we found that expression of Fxr was significantly higher in B6B6 islets compared with B6CAST islets (Figure 6B). These results suggest that the gut microbiota modulate BA-dependent signaling in pancreatic islets.
DISCUSSION
The collective genetic variance of the eight CC strains is roughly equivalent to that of the entire human population, with the three wild-derived strains (WSB, CAST and PWK) accounting for ~75% of the genetic diversity within the cohort (Roberts et al. 2007). Remarkably, these three wild-derived strains captured the full scope of dietary responsiveness observed across the panel (Figure 1 and S1). HF/HS feeding had no effect on any of the phenotypes measured in CAST mice, whereas it resulted in weight gain, glucose intolerance and insulin resistance in B6 mice. Additionally, the diet caused a simultaneous increase in weight and glucose in NZO mice. We also identified significant differences in the gut microbiota composition among strains and between diets. All animals were obtained from the same facility, and subject to the same environmental conditions throughout the study, and genetic differences among the mice is the only known variable. Together, these results support a role for host genetics to regulate the composition of the microbiota. However, it’s important to note that although large population studies have identified highly heritable taxa, the genetic architecture underlying these taxa is highly complex with relatively small effect sizes that are difficult to replicate (Benson 2016).
From the CC founder panel, we identified B6 and CAST as the two strains with the most divergent phenotypes. Previous studies have exploited the differential response to diet-induced metabolic disease between B6 and CAST to identify genetic loci associated with metabolic disease (Mehrabian et al. 2000; Mehrabian et al. 1998). In these studies, the gut microbiome was not may have contributed to the metabolic differences between strains.
In order to dissect the contribution of the microbiome of B6 and CAST to their contrasting metabolic profiles, we resorted to fecal transplantation experiments. B6-GF mice colonized with the CAST microbiota were less affected by chronic HF/HS feeding relative to B6-GF mice colonized with the B6 microbiota. The mice receiving the CAST microbiota secreted far less insulin in response to a glucose challenge, but were still able to maintain normal blood glucose levels.
We consistently identified microbial taxa in both the CC founders and transplant recipients associated with metabolic traits. Clostridiaceae showed the strongest positive correlation with plasma insulin levels and weight gain (Figure 2A). Clostridiaceae also had a strong positive correlation with AUC insulin, a proxy for pancreas function. OTUs within the Clostridiaceae family have previously been both positively and negatively associated with metabolic traits (Ussar et al. 2015; Karlsson et al. 2013), and a recent study showed a positive correlation between an increase in BMI and an increase of SCFA-producing Closdiria species in Danish infants (Bergström et al. 2014). In contrast to the elevated Clostridiaceae in mice with a B6 microbiota, Bacteroidaceae was significantly higher in CAST-CR and B6CAST mice (Figure S3C and 4C). Bacteroidaceae was negatively correlated with body weight, circulating insulin and AUCinsulin (Figure 2A). A previous report found that daily oral administration of Bacteroides uniformis, a member of the Bacteroidaceae family, ameliorated metabolic dysfunction resulting from a high-fat diet (Gauffin Cano et al. 2012). This species also evoked a reduction in hepatic triglyceride levels, consistent with our observations that B6CAST mice have lower hepatic lipid levels compared to B6B6 mice. Fecal abundance of members of the Bacteroidaceae family, including Bacteroides vulgatus, has also been reported to be lower in humans with T2D (X. Wu et al. 2010). Despite the high abundance of Bacteroidaceae in B6CAST mice, we did not observe complete protection from diet-induced metabolic disease that we observed in CAST-CR mice, suggesting that host factors, or taxa that failed to colonize transplanted mice (e.g., Verrucomicrobiaceae), contribute to the metabotype differences.
Vitamin B12 is exclusively produced by microbes (Martens et al. 2002) and several members of the Bacteroidaceae family transport, metabolize and produce vitamin B12 analogs (Goodman et al. 2009; Degnan et al. 2014; M. Wu et al. 2015). Metagenomic analysis of the microbial communities from mice with the CAST microbiota revealed microbial functional enrichment for pathways involved in the biosynthesis of vitamin B12 (Figure S4A), which is necessary for DNA synthesis, neurological function, hematopoiesis, epigenetic modifications, and propionate metabolism (Kibirige & Mwebaze 2013). Importantly, deficiencies in vitamin B12 are commonly observed in individuals with T2D and gestational diabetes (Kibirige & Mwebaze 2013; Krishnaveni et al. 2009), and B12 therapy improves insulin resistance and endothelial function in patients with metabolic syndrome by mechanisms that are not fully elucidated (Setola et al. 2004).
Our metagenomic analysis also revealed that genes involved in LPS production are enriched in the CAST-transplanted microbiome (Figure 4E and SF4B). This finding was surprising given that increased levels of LPS have been causally linked to the development of metabolic disease, yet B6CAST mice are partially protected from the effects of HF/HS feeding relative to B6B6 animals (Figure 3). Taxonomic evaluation of the metagenomic data indicated that the Bacteroidetes phylum is the major contributor to the increased abundance of genes from this pathway (Table S4). This is relevant because unrelated bacteria generate structurally distinct LPS molecules with varying capacity to elicit an innate immune response (Whitfield & Trent 2014). Notably, a recent study showed that LPS derived from E. coli generates a strong inflammatory signal, whereas LPS derived from members of the Bacteroidetes phylum inhibited the host immune response (Vatanen et al. 2016). The differential ability of LPS sub-types to modify host physiology may explain why LPS has been shown to both stimulate (Nguyen et al. 2014) and attenuate (Amyot et al. 2012) insulin secretion. Studies aimed at testing the roles of LPS derived from phylogenetically diverse taxa on metabolic disease and insulin secretion are warranted to further clarify how structural differences in this molecule affect host metabolism.
In addition to LPS, gut microbes produce SCFA, which are important energy and signaling molecules implicated in metabolic disease. For instance, butyrate has been shown to improve whole-body insulin sensitivity (Gao et al. 2009) and patients with T2D have reduced levels of butyrate-producing bacteria (Qin et al. 2010). SCFA are also elevated in individuals with diet-induced obesity, which is consistent with the elevated cecal SCFA levels in B6B6 mice (Turnbaugh et al. 2008). Interestingly, SCFA are also known regulators of insulin sensitivity and secretion. Acetate can modulate insulin secretion from β-cells either directly through FFAR2 or via parasympathetic activation (Priyadarshini et al. 2015; Perry et al. 2016). However, we did not observe differences in concentrations of plasma or cecal acetate in the transplanted animals (Figure 4F and S6A–B). Therefore, it is unlikely that the differences in insulin secretion could be attributed to SCFA and consequentially implies there are multiple pathways through which the gut microbiota can module insulin secretion from β-cells.
Gut microbes are responsible for the production of the highly hydrophobic secondary BA DCA and LCA through the dehydroxylation of the primary BA, CA and CDCA, in the colon. Removal of glycine/taurine BA conjugates via BSH enzymes is a prerequisite for 7α/β-dehydroxylation of primary BA into secondary BA (Batta et al. 1990). Interestingly, there were 13 predicted BSH genes that were more abundant in the B6 metagenome but only two in the CAST metagenome. One possible interpretation of this result is that there may be more bacterial species present in the B6 microbiome that are able to deconjugate BA. Consistent with this, B6B6 mice had significantly higher levels of secondary BA as well as hydrophobic BA species than B6CAST mice (Figure 5B–C and S6C–D), both of which are elevated in humans and mice with insulin resistance (Ryan et al. 2014; Prawitt et al. 2011). Furthermore, DCA has been positively associated with higher levels of Firmicutes (Islam et al. 2011). This is consistent with our findings as B6-CR founders and B6B6 had a significantly greater relative abundance of Firmicutes and fecal DCA than CAST-CR and B6CAST (Figure S3B and 5B). Conversely, B6CAST had a higher abundance of hydrophilic BA and the majority of the BA pool was comprised of the mouse primary BA, βMCA (Figure 5B–C).
The BA receptor Fxr is expressed in pancreatic β-cells and its activation via BA enhances insulin secretion (Kumar et al. 2012; Renga et al. 2010). Hydrophobic BA such as CDCA, DCA, LCA, and their taurine conjugates are known ligands of Fxr. The hydrophobic TCDCA increases insulin production and secretion through an FXR-dependent regulation of KATP channels (Düfer et al. 2012). Moreover, β-cell FXR activation in diabetic leptin receptor deficient (db/db) mice and NOD mice increases insulin secretion and delays the development of diabetes (Renga et al. 2010; Zhang et al. 2006). We detected higher levels of LCA and DCA in the feces and plasma of B6B6 mice relative to B6CAST mice (Figure 5B and S6C–D), along with increased expression of Fxr in pancreatic islets from B6B6 mice (Figure 6B). Altogether, this suggests that the gut microbiota and BA composition could modulate pancreatic function and insulin secretion.
We have highlighted four examples of microbial-derived products, vitamin B12, SCFAs, LPS, and BA, as plausible mediators of the microbiome effect on insulin secretion. However, there are thousands of other metabolites that were not characterized in our study and could also play an important role in regulating host metabolism. Future experiments using gnotobiotic mice colonized with defined communities that have different metabolic capabilities will provide mechanistic insights into the communication between gut microbes and the host.
EXPERIMENTAL PROCEDURES
Mouse husbandry
Animal care and study protocols were approved by the University of Wisconsin - Madison Animal Care and Use Committee.
Collaborative Cross (CC) mouse husbandry
Mice were housed on a 12 h-light:dark cycle. CC founder strains were obtained from The Jackson Laboratory (Bar Harbor, ME, USA) and were bred at University of Wisconsin, Madison. Mice were group housed by strain (2 mice/cage) and diet under a temperature- and humidity-controlled conditions, and received ad libitum access to water and food. After 4 weeks of age, mice were maintained on either a control (TD.08810, Envigo Teklad, 16.8%-kcal fat, 60.9% carbohydrate, 22.3% protein) or a high-fat high-sucrose diet (TD.08811, Envigo Teklad, 44.6%-kcal fat, 40.6% carbohydrate, 14.8% protein) (Table S1). Strains were housed within the same vivarium throughout the duration of the study.
Gnotobiotic mouse husbandry
C57BL/6J germ-free mice were bred and housed in the Microbial Sciences Building vivarium at University of Wisconsin-Madison to generate mice used in this study. B6-CR and B6-GF mice were housed in separate plastic flexible vinyl gnotobiotic isolators under temperature- and humidity-controlled conditions (12 hr light:dark). Fresh cecal contents were collected from 15-week old conventional B6-CR and CAST-CR mice maintained on the HF/HS diet (n = 2 to 3 mice per donor cecal microbiota samples). Cecal contents from B6 and CAST donor mice were resuspended in rich medium (1:100 w/vol) inside an anaerobic chamber. Suspensions were transferred into anaerobic sealed tubes and moved into gnotobiotic isolators. 9-week-old B6-GF male mice were inoculated via a single oral gavage with ~0.2 ml of cecal incoula (Turnbaugh et al. 2009). Each group of mice was housed in a controlled environment in separate plastic flexible vinyl gnotobiotic isolators under standard conditions. Recipient mice received sterilized water and HF/HS diet (TD.0.8811) ad libitum beginning one week before colonization.
Metabolic phenotypes
Mice were fasted for 4 h before blood collection. Fasting levels of glucose, insulin and triglycerides were quantified at regular intervals, along with body weight. Glucose tolerance tests were performed by administering an oral dose of glucose (2 g/kg body weight) after a 4 h fast. Blood was collected at 0, 5, 15, 30, 60 and 120 minutes to assess glucose and insulin levels. Hepatic triglycerides were extracted following the Bligh and Dyer method.
Microbiome sequencing and analysis
DNA was isolated from cecal contents and feces by extraction using a bead-beating protocol (Turnbaugh et al. 2009). The V4 region of the 16S rRNA gene was amplified using barcoded primers and sequencing was performed using the Illumina MiSeq platform. 16S rRNA sequences were analyzed using QIIME (Quantitative Insights Into Microbial Ecology) software package (Caporaso et al. 2010). Linear discriminant analysis (LDA) effect size (LEfSe) analysis was performed using standard parameters (p < 0.05 and LDA score 2.0) (Segata et al. 2011). For the metagenomic analysis, DNA fragments ~350–400 bp were sequenced using Illumina Rapid HiSeq 2000. Details about metagenomic sequence analysis, 16S rRNA sequencing and analyses are provided in Supplemental Experimental Procedures.
Statistical analysis
The data are expressed as mean ± SEM and analyzed using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA). Multiple groups were analyzed by one-way or two-way ANOVA followed by Bonferroni’s multiple comparisons test. Significant differences between two groups were evaluated by two-tailed unpaired Student’s t-test or Mann-Whitney U test for samples that were not normally distributed. Pearson’s correlations between microbiota and phenotypes and association testing were performed in R. The level of significance was set at p < 0.05; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Highlights.
Host genotype affects the abundance of taxa associated with metabolic disease.
Gut microbiota affect susceptibility to diet induced metabolic disease.
The gut microbiome modulates insulin secretion.
Acknowledgments
The authors thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing sequencing and support services, and the University of Wisconsin Center for High Throughput Computing (CHTC) in the Department of Computer Sciences for providing computational resources, support, and assistance. We also thank the University of Wisconsin Mass Spectrometry Facility for technical assistance. Finally, we are grateful to Dr. Jeffrey Gordon for his support on this project.
This work was supported in part by grants NIH DK1018259-01 (F.E.R.), DK101573 (A.D.A), RR021940 (B.H.) and GM077336 (B.H.) and by the Clinical and Translational Science Award (CTSA) program, through the NIH National Center for Advancing Translational Sciences (NCATS) grants UL1TR000427 (A.T.B) and KL2TR000428 (F.E.R.). L.L.T. is supported by a NLM Computation and Informatics in Biology and Medicine Postdoctoral Fellowship (5T15LM007359). J.H.K. is supported by the NIH National Institute of Allergy and Infectious Diseases grant T32AI55397.
Footnotes
See Supplemental Information for detailed description of methods related to (i) plasma and liver measurements, (ii) HPLC/MS and GC/MS assays of plasma and cecal contents, (iii) microbiome analyses (iv) islet isolation and GSIS, and (v) qRT-PCR assays.
AUTHOR CONTRIBUTIONS
F.E.R., A.D.A., M.P.K. and J.H.K. conceived project; J.H.K., E.I.V., M.E.R., D.S.S., K.L.S., B.S.Y., A.T.B. and W.Z. performed experiments and interpreted results; J.H.K., M.P.K., L.L.T. prepared figures; J.H.K., M.P.K., A.D.A. and F.E.R. wrote the manuscript; W.Z., L.L.T., M.E.R, and B.H. provided critical feedback. All authors read and agreed on the final version of the manuscript.
ACCESSION NUMBERS
The data reported in this paper are accessible in the European Nucleotide Archive (ENA) database under accession ID PRJEB15120.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amyot J, et al. Lipopolysaccharides impair insulin gene expression in isolated islets of Langerhans via Toll-Like Receptor-4 and NF-κB signalling. O. S. Shirihai, ed. PloS one. 2012;7(4):e36200. doi: 10.1371/journal.pone.0036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylor DL, et al. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome research. 2011;21(8):1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta AK, et al. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. The Journal of biological chemistry. 1990;265(19):10925–10928. [PubMed] [Google Scholar]
- Benson AK. The gut microbiome-an emerging complex trait. Nature genetics. 2016;48(11):1301–1302. doi: 10.1038/ng.3707. [DOI] [PubMed] [Google Scholar]
- Bergström A, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Applied and environmental microbiology. 2014;80(9):2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besten den G, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. Journal of lipid research. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell host & microbe. 2015;17(1):72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nature genetics. 2004;36(11):1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Clemente JC, et al. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a modulator of gut microbial ecology. Cell metabolism. 2014;20(5):769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiology and molecular biology reviews: MMBR. 2006;70(4):939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düfer M, et al. Bile acids acutely stimulate insulin secretion of mouse β-cells via farnesoid X receptor activation and K(ATP) channel inhibition. Diabetes. 2012;61(6):1479–1489. doi: 10.2337/db11-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58(7):1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauffin Cano P, et al. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. S. Bereswill, ed. PloS one. 2012;7(7):e41079. doi: 10.1371/journal.pone.0041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell host & microbe. 2009;6(3):279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman Y. Role of bile acid sequestrants in the treatment of type 2 diabetes. Diabetes care. 2011;34(Suppl 2) Supplement_2:S244–50. doi: 10.2337/dc11-s237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartstra AV, et al. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes care. 2015;38(1):159–165. doi: 10.2337/dc14-0769. [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–24. e1–2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. The EMBO journal. 2006;25(7):1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam KBMS, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141(5):1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Kars M, et al. Tauroursodeoxycholic Acid may improve liver and muscle but not adipose tissue insulin sensitivity in obese men and women. Diabetes. 2010;59(8):1899–1905. doi: 10.2337/db10-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. The Journal of biological chemistry. 2003;278(11):9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- Kibirige D, Mwebaze R. Vitamin B12 deficiency among patients with diabetes mellitus: is routine screening and supplementation justified? Journal of diabetes and metabolic disorders. 2013;12(1):17. doi: 10.1186/2251-6581-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, et al. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Krishnaveni GV, et al. Low plasma vitamin B12 in pregnancy is associated with gestational “diabesity” and later diabetes. Diabetologia. 2009;52(11):2350–2358. doi: 10.1007/s00125-009-1499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap--bile acids in metabolic control. Nature reviews. Endocrinology. 2014;10(8):488–498. doi: 10.1038/nrendo.2014.60. [DOI] [PubMed] [Google Scholar]
- Kumar DP, et al. Activation of transmembrane bile acid receptor TGR5 stimulates insulin secretion in pancreatic β cells. Biochemical and biophysical research communications. 2012;427(3):600–605. doi: 10.1016/j.bbrc.2012.09.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FY, et al. FXR, a multipurpose nuclear receptor. Trends in biochemical sciences. 2006;31(10):572–580. doi: 10.1016/j.tibs.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Ley RE, et al. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M, et al. Identification of a nuclear receptor for bile acids. Science (New York, NY) 1999;284(5418):1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Martens JH, et al. Microbial production of vitamin B12. Applied microbiology and biotechnology. 2002;58(3):275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- Mehrabian M, et al. Genetic control of HDL levels and composition in an interspecific mouse cross (CAST/Ei x C57BL/6J) Journal of lipid research. 2000;41(12):1936–1946. [PubMed] [Google Scholar]
- Mehrabian M, et al. Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. The Journal of clinical investigation. 1998;101(11):2485–2496. doi: 10.1172/JCI1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, et al. Lipopolysaccharides-mediated increase in glucose-stimulated insulin secretion: involvement of the GLP-1 pathway. Diabetes. 2014;63(2):471–482. doi: 10.2337/db13-0903. [DOI] [PubMed] [Google Scholar]
- O’Connor A, et al. Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics. Mammalian genome: official journal of the International Mammalian Genome Society. 2014;25(11–12):583–599. doi: 10.1007/s00335-014-9540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science (New York, NY) 2006;313(5790):1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell metabolism. 2013;17(1):141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DJ, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science (New York, NY) 1999;284(5418):1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Perry RJ, et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature. 2016;534(7606):213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt J, Caron S, Staels B. Bile acid metabolism and the pathogenesis of type 2 diabetes. Current diabetes reports. 2011;11(3):160–166. doi: 10.1007/s11892-011-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini M, et al. An Acetate-Specific GPCR, FFAR2, Regulates Insulin Secretion. Molecular endocrinology (Baltimore, Md) 2015;29(7):1055–1066. doi: 10.1210/me.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Renga B, et al. The bile acid sensor FXR regulates insulin transcription and secretion. Biochimica et biophysica acta. 2010;1802(3):363–372. doi: 10.1016/j.bbadis.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (New York, NY) 2013;341(6150):1241214–1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. Journal of lipid research. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Roberts A, et al. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mammalian genome: official journal of the International Mammalian Genome Society. 2007;18(6–7):473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW. Fifty years of advances in bile acid synthesis and metabolism. Journal of lipid research. 2009;50(Suppl) Supplement:S120–5. doi: 10.1194/jlr.R800026-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KK, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509(7499):183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell metabolism. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Segata N, et al. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setola E, et al. Insulin resistance and endothelial function are improved after folate and vitamin B12 therapy in patients with metabolic syndrome: relationship between homocysteine levels and hyperinsulinemia. European Journal of Endocrinology. 2004;151(4):483–489. doi: 10.1530/eje.0.1510483. [DOI] [PubMed] [Google Scholar]
- Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nature reviews Microbiology. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, et al. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell host & microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussar S, et al. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell metabolism. 2015;22(3):516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165(4):842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A, et al. The environment within: how gut microbiota may influence metabolism and body composition. Diabetologia. 2010;53(4):606–613. doi: 10.1007/s00125-010-1662-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annual review of biochemistry. 2014;83(1):99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- Wu M, et al. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science (New York, NY) 2015;350(6256):aac5992–aac5992. doi: 10.1126/science.aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Current microbiology. 2010;61(1):69–78. doi: 10.1007/s00284-010-9582-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(4):1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.