Abstract
Breastfeeding is associated with a lower incidence of obesity, diabetes, and cardiovascular disease later in life. While macrosomic infants have a higher risk of developing obesity and other metabolic disorders. Breast milk may contain special nutrients to meet the different growth needs of different infants. Whether mothers make breast milk different to meet the requirement of macrosomic infants is still unknown. Here, we conducted a comparison between mothers delivering macrosomic and non-macrosomic infants in colostrum endogenous peptides. More than 400 peptides, originating from at least 34 protein precursors, were identified by Liquid Chromatography/Mass Spectrometry (LC/MS). Out of these, 29 peptides found to be significant differently expressed (|fold change| ≥ 3, P < 0.01). Blastp analysis revealed 41 peptides may have established biological activities, which exhibit immunomodulating, antibacterial action, antioxidation, opioid agonist and antihypertensive activity. Furthermore, we found that peptide located at β-Casein 24-38 AA has antimicrobial effect against E. coli, Y. enterocolitica and S. aureus. While, κ-Casein 89-109 AA-derived peptide plays as a regulator of preadipocyte proliferation. The profile of endogenous peptides from macrosomic term infants is different from non-macrosomic terms. This different peptide expression potentially has specific physiological function to benefit macrosomic infants. Finally, we believe that our research is a meaningfull finding which may add to the understanding of milk peptide physiological action.
Keywords: human milk, macrosomic fetuses, peptidomics, antibacterial activity, adipocyte proliferation
INTRODUCTION
Pregnancies with a macrosomic infants gradually comprise a subgroup of high-risk pregnancies. As a common pregnant complication, macrosomia seriously threatens the health of pregnant women [1, 2]. These abnormal weight gain affects both healthy of the fetus and the mother. It is always accompanied with prolonged labour and abnormal haemorrhage during delivery, leading to perineal laceration and even cesarean section (CS) [3]. Meanwhile, these fetuses frequently bring with shoulder dystocia and brachial plexus palsy, even that such as severe birth asphyxia and neonatal mortality [4]. Macrosomic fetuses are at risk for metabolic abnormalities complications like electrolyte disturbances, glycopenia and hyperbilirubinaemia during neonatal period [5]. Among the long-term, obesity and metabolic disorders, cardiovascular disease and childhood cancers have to be mentioned [6–8].
As the most suitable food for the newborn, milk is characterized by its good for nutrition, easier to digest, immune regulating properties and anti-infective function, which all provide nutrition and protection for the infant during early life [6, 9, 10]. Recent research finds that breastfeeding helps to prevent the incidence of obesity, diabetes as well as cardiovascular disease later in life [11–13]. Hence, human milk is especially important for fetal macrosomias who are more likely than other infants to be obese later in life. Nowdays, scientists belive that the composition of breast milk is various according to different growth needs of different infants, such as preterm human milk is more suitable for babies born too early. So we suppose mothers may produce different milk for macrosomic and non-macrosomic infants, also these infants respond differently to the breast milk they drink.
Despite being known and studied for years, the low molecular weight (LMW) portion of proteome have never before attracted enough attention. In fact, this protein/peptide fraction is key factor that regulates many genes, participates in various pathways of disease and now deserves more and more attention these days [14–15]. Recent researchs have found a large number of functional endogenous peptides in the milk thus making human milk a carrier of biochemical messages [16–20]. Over 300 naturally produced peptides were identified originating from the protein composition of breast milk and antimicrobial activity of the peptide mixture was firstly considered [16]. David et al. (2014) did a comparison on the entire endogenous peptides from human milk and the milk gastric juice of term infants to investigate their componet difference [19]. The recognition of specific roles performed by milk endogenous peptides raises questions about whether women delivering macrosomic fetuses bring more benefits to macrosomic infants? Elucidation of the significance of the naturally occurring peptides in the milk to development in the human newborn remains to be further studied.
Here, we carried out a quantitative study on human milk endogenous peptides from macrosomic and non-macrosomic terms. The broad aim of this research was to develop an effective ultrafiltration method to purify naturally occurring human milk peptides for Liquid Chromatography/Mass Spectrometry (LC/MS) analysis. A main objective was to quantitatively compare the naturally occurring peptide profile of human milks collected from mothers delivering macrosomic and non-macrosomic infants. Another goal was to identify known peptide hormones in human milk of these different expression peptides. We further revealed that peptide located at β-Casein 24-38 AA (NQELLLNPTHQIYPV, henceforth called Casein 24) shows antimicrobial activities against E. coli, Y. enterocolitica and S. aureus. Peptide located at κ-Casein 89-109 AA (PHAQIPQRQYLPNSHPPTVVR, henceforth called Casein 89) exhibits growth inhibiting activity in human preadipocytes.
RESULT
Maternal and neonatal characteristics
Table 1 shows the basic characteristics of maternity and infant involved in this test. All recruited women belong to the same age group and have similar body mass index (P > 0.05). The macrosomic infants' body weight exhibits significant difference when compared with control group (P < 0.05). Further more, both 1 h and 2 h oral glucose tolerance test (OGTT) levels were significantly higher in macrosomic infants group.
Table 1. Maternal and neonatal clinical characteristics.
| Clinical Features | Macrosomia (n= 6) | Control (n = 6) |
|---|---|---|
| Maternal age (mean ± SD) | 30.8 ± 1.4 | 29.7 ± 1.8 |
| Maternal BMI (mean ± SD) | 31.3 ± 0.6 | 26.3 ± 1.9 |
| Week gestation (mean ± SD) | 36.2 ± 0.8 | 36.4 ± 0.5 |
| Mode of devliviery CS (%) | 100 % | 100 % |
| Brith weight (g) (mean ±SD) | 4120.5 ± 54.3a | 3220.5 ± 26.9 |
| OGTT fasting | 5.7 ± 0.3 | 4.7 ± 0.2 |
| 1 h | 9.5 ± 0.3a | 5.3 ± 0.3 |
| 2 h | 7.2 ± 0.4a | 4.9 ± 0.5 |
P < 0.05 when compared with control group.
BMI, body mass index.
SD, standard deviation.
Sample preparation
Mucins, caseins, and whey proteins are the leading member of the human milk total protein contents, the peptides account for only a small fraction. Therefore, ultrafiltration using molecular weight cut-off (MWCO) filters was used to enrich the peptide part. Studies on plasma and cerebrospinal fluid reveal that peptides always adsorbed on proteins surface and may therefore reduce the efficiency of ultrafiltration separation [21, 22]. Denaturing and reducing solution (7 M urea, 2 M thiourea, and 20 mM DTT) was used in order to disrupt the protein–peptides interactions. Mass spectrum analysis confirmed that the method can identify more peptides than prior studies on endogenous peptides [16–18].
Peptide identification and quantitative analysis
Peptides compostion of the pooled human milk from mothers delivering macrosomic and non-macrosomic infants were directly analyzed by LC-MS after isotopomeric dimethyl labeled. More than 400 peptides, originating from at least 34 protein precursors, were identified from the six individuals (Table S1). Hierarchical clustering analysis showed a remarkable peptide expression profile change between the two compared groups (Figure S1). Of these peptide, 15 peptides presented high levels (a fold change ≥ 3.0, P < 0.05) in the milk from mothers delivering macrosomic infants, while 14 peptides accounted for the main composition of peptides in the milk from mothers delivering non-macrosomic infants (a fold change ≤−3.0, P < 0.05) (Table 2).
Table 2. Part peptides that are differentially expressed in human milk from women delivering macrosomic infants.
| Sequence | Mass | Protein Names | log2(M/H) | P-value |
|---|---|---|---|---|
| Peptides down-regulated in human milk from women delivering macrosomic infants (> 3 folds) | ||||
| LGSAMQNTQNLLQMPY | 1807.9 | Alpha-2-macroglobulin | −5.40 | 9.82E-16 |
| LWSVPQPKVLPIPQQV | 1828.1 | Beta-casein | −4.62 | 2.82E-02 |
| LPIPQQVVPYPQRAVPVQ | 2028.2 | Beta-casein | −3.87 | 5.24E-05 |
| VPQPIPQTLALPPQP | 1594.9 | Beta-casein | −3.68 | 1.24E-05 |
| DLENLHLPL | 1062.6 | Beta-casein | −3.50 | 4.83E-17 |
| LALPPQP | 734.4 | Beta-casein | −3.48 | 2.53E-05 |
| PHAQIPQRQYLPNSHPPTVVR | 1745.9 | Kappa-casein | −3.44 | 4.92E-02 |
| GRVMPVL | 770.4 | Beta-casein | −3.27 | 3.24E-05 |
| WRKMCRKLLDMTFSSKTNTLVVR | 2869.5 | vWF-cleaving protease | −3.26 | 4.81E-11 |
| MSRLEVVFTALMNSK | 1724.9 | Cholesteryl ester transfer protein | −3.19 | 3.24E-05 |
| DTVYTKGRVMPVL | 1477.8 | Beta-casein | −3.18 | 5.40E-05 |
| LLLNPTHQIYPVTQPLAPVH | 2250.3 | Beta-casein | −3.18 | 5.30E-04 |
| LLLNPTHQIYPVTQPLAPVHNPISV | 2760.5 | Beta-casein | −3.06 | 2.62E-02 |
| VLPIPQQVVPYPQRAVPVQALL | 2424.4 | Beta-casein | −3.03 | 1.53E-05 |
| Peptides up-regulated in human milk from women delivering macrosomic infants (> 3 folds) | ||||
| LALPPQPLWSVPQPK | 1670.0 | Beta-casein | 3.11 | 1.54E-06 |
| LQPLMQQVPQPIP | 1487.8 | Beta-casein | 3.54 | 1.53E-05 |
| NQELLLNPTHQIYPV | 1063.6 | Beta-casein | 3.72 | 2.52E-02 |
| VMPVLKSPTIPFFDPQIPKLTDLEN | 2838.5 | Beta-casein | 3.93 | 4.83E-07 |
| LWSVPQPKVLPIPQQVVPYP | 2284.3 | Beta-casein | 4.06 | 5.44E-05 |
| LLNPTHQIYPVT | 1394.8 | Beta-casein | 4.17 | 2.54E-06 |
| SPTIPFFDPQIPKLTDLEN | 2171.1 | Beta-casein | 4.19 | 2.53E-05 |
| ILPLAQPAVVLPVPQPEIMEVPKA | 2548.5 | Beta-casein | 4.26 | 2.52E-02 |
| PHAQIPQRQYLPNSHPPTVVR | 1745.9 | Kappa-casein | 4.44 | 4.92E-02 |
| SIQLPTTVRDIMNRW | 1829.0 | Membrane alanine aminopeptidase variant | 4.54 | 5.27E-03 |
| ASQLMGENRTMTIHNGMFFST | 2372.1 | Fibrinogen beta chain | 5.09 | 4.92E-02 |
| EDLIDEDDIPVRSFFP | 1905.9 | C4A variant protein | 5.23 | 2.74E-03 |
| ATSSLCSVTNTSMMTSE | 1805.7 | Ascites sialoglycoprotein | 5.31 | 4.51E-03 |
| LAQPAVVLPVPQPEIMEVPK | 2154.2 | Beta-casein | 6.29 | 1.12E-09 |
| MKFISTSLLLMLLVSSLS | 1982.1 | B cell-attracting chemokine 1 | 7.46 | 1.13E-12 |
M = Peptide Intensity (Human milk from women delivering macrosomic infants).
H = Peptide Intensity (Human milk from women delivering no-macrosomic infants).
Characteristics of peptides identified from human milk
Fristly, we analyzed the general features of the milk peptides identified from human milk. Both the molecular weight (MW) and isoelectric point (PI) of these peptides spread over a wide range, but we can observed that most of the peptides focused between 14–22 KDa and a acidic PI range (4.0–7.0), as shown in Figure 1A and 1B. Moreover, the relative distribution of PI versus MW also has a certain unique. Points of these identified peptides mainly gather into three groups, which distributed around PI 4, PI 6 and PI 10 (Figure 1C).
Figure 1. Characteristics of Peptides identified by Liquid Chromatography/Mass Spectrometry (LC/MS).
(A) Distribution of molecular weight (MW) of peptides identified of colostrums samples milk. (B) Distribution of molecular pI of peptides identified of colostrums samples milk. (C) Scatter plot of MW versus isoelectric point (PI) distribution of the peptides. (D) Peptide numbers for each milk protein precursors. (E) Selected peptide ladder sequence and the locations of the 17 ladders identified in κ-casein. (F) Distribution of the four cleavage sites in the identified peptides.
Cleavage site patterns of identified endogenous peptides
A typical characteristic of these identified peptides was 279 peptides broken from protein of β-casein, occupied the highest proportion of the endogenous peptides from human milk (Figure 1D). Other peptides mostly derive from human milk proteins such as α-casein (CASA), κ-casein (CSN3), Polymeric immunoglobulin receptor (PIGR), Complement C4-A (C4A), Bile salt-activated lipase (BAL), Mucin-4 (MUC4), A disintegrin and metalloproteinase with thrombospondin motifs 13 (ADAM-TS 13), Perilipin-2 (ADFP), Alpha-2-macroglobulin (Alpha-2-M), Mothers against decapentaplegic homolog 1 (BSP II), Cytotactin (TNC), Fibrinopeptide B (FGB) and Fibrinopeptide A (FGA) (Figure 1D). In the further analysis, we observed that many of the peptides come from the same segment of one protein, which was in consistent with the observation in the serum peptidome analysis [23]. Another easily cleaved protein by the endogenous enzymes, κ-casein, generated 17 peptides in our case. However, the distribution of these peptides sequences were hardly a random, which arrayed like a ladder and came from 1 segments of κ-casein. The locations of the 17 peptide ladders identified in κ-casein were marked in Figure 1E.
We further combined peptidome analysis of human milk by LC/MS with bioinformatic analysis of the cleavage site distribution at the amino terminal end (N-terminal) and carboxyl terminal end (C-terminal) of the observed endogenous peptides. Briefly, all these four cutting sites of all the identified peptides were used to investigate the nature of proteolytic enzymes within human milk. Results showed that leucine (L) was the most popular C-terminal post-cleavage site of the preceding peptide, while proline (P) dominated the cleavage site of C terminus of the identified peptide. Leucine (L) is the most cleavaged site of N terminus of the identified peptide and N-terminal pre-cleavage site (Figure 1F).
Putative functional peptides
Previous literatures showed the known human milk functional peptides have various biological effects such as immunomodulating (QPTIPFFDPQIPK) [24], antibacterial (QELLLNPTHQYPVTQPL/APVHNPISV) [25], antioxidant (WSVPQPK) [26], opioid agonist (YPFVEPI/YPFVE) [27], antihypertensive (HLPLP) [26]. Table 2 summarizes some peptides in human milk which had been report with potential beneficial effect on human health. We are surprised that 41 of the endogenous milk peptides shared > 50 % homology with known functional peptides, promising similar beneficial effects (Table 3). Of these peptides twelve matched known immunomodulatory sequences, eighteen matched antibacterial sequences, four matched antioxidant sequences, four matched opioid agonist sequences and three matched antihypertensive sequence (sequences shown in Table 3). It's worth noting that all these 41 peptides are primarily from β-casein. More research is needed for the remaining unknown function peptides.
Table 3. Putative functional peptides derived from human milk.
| Identified peptides | Precursor protein | Fragment | Functional peptides | Known activity |
|---|---|---|---|---|
| QPTIPFFDPQIPK | Immunomodulating | |||
| FDPQIPKLTDLEN | Beta-casein | 125−137 | ||
| FDPQIPKLTDLENL | Beta-casein | 125−138 | ||
| GRVMPVLKSPTIPFFDPQIP | Beta-casein | 111−130 | ||
| GRVMPVLKSPTIPFFDPQIPK | Beta-casein | 111−131 | ||
| IPFFDPQIPKLTDLEN | Beta-casein | 122−137 | ||
| MPVLKSPTIPFFDPQIP | Beta-casein | 114−130 | ||
| MPVLKSPTIPFFDPQIPK | Beta-casein | 114−131 | ||
| SPTIPFFDPQIPK | Beta-casein | 119−131 | ||
| SPTIPFFDPQIPKLT | Beta-casein | 119−133 | ||
| SPTIPFFDPQIPKLTDLEN | Beta-casein | 119−137 | ||
| VLKSPTIPFFDPQIPK | Beta-casein | 116−131 | ||
| VMPVLKSPTIPFFDPQIPK | Beta-casein | 113−131 | ||
| QELLLNPTHQYPVTQPL APVHNPISV | Antibacterial | |||
| ELLLNPTHQIYPVTQPLAPV | Beta-casein | 41−60 | ||
| ELLLNPTHQIYPVTQPLAPVHNPIS | Beta-casein | 41−65 | ||
| LLLNPTHQIYPVTQPLAPVHNPIS | Beta-casein | 42−65 | ||
| LLLNPTHQIYPVTQPLAPVHNPISV | Beta-casein | 42−66 | ||
| LLLNQELLLNPTHQIYPVTQPLAP | Beta-casein | 36−59 | ||
| LLLNQELLLNPTHQIYPVTQPLAPV | Beta-casein | 36−60 | ||
| LLNPTHQIYPVTQPLAPVHNPIS | Beta-casein | 43−65 | ||
| LLNPTHQIYPVTQPLAPVHNPISV | Beta-casein | 43−66 | ||
| LLNQELLLNPTHQIYP | Beta-casein | 22−37 | ||
| LLNQELLLNPTHQIYPV | Beta-casein | 22−38 | ||
| LNPTHQIYPVTQPLAPVHNPIS | Beta-casein | 44−65 | ||
| NPTHQIYPVTQPLAPVHNPIS | Beta-casein | 45−65 | ||
| NPTHQIYPVTQPLAPVHNPISV | Beta-casein | 44−66 | ||
| NQELLLNPTHQIYPV | Beta-casein | 24−38 | ||
| QIYPVTQPLAPVHNPISV | Beta-casein | 49−66 | ||
| VTQPLAPVHNPISV | Beta-casein | 53−66 | ||
| YPVTQPLAPVHNPIS | Beta-casein | 51−65 | ||
| YPVTQPLAPVHNPISV | Beta-casein | 51−66 | ||
| LWSVPQPK | Beta-casein | 8−15 | WSVPQPK | Antioxidant |
| LWSVPQPKVLPIP | Beta-casein | 8−20 | ||
| LWSVPQPKVLPIPQQ | Beta-casein | 8−22 | ||
| LWSVPQPKVLPIPQQV | Beta-casein | 8−23 | ||
| YPFVEPI/YPFVE | Opioid agonist | |||
| IYPFVEPIPYGFLPQN | Beta-casein | 64−79 | ||
| QPQPLIYPFVEPIP | Beta-casein | 59−72 | ||
| YPFVEPIPYGFL | Beta-casein | 65−76 | ||
| YPFVEPIPYGFLP | Beta-casein | 65−77 | ||
| HLPLP | Antihypertensive | |||
| DLENLHLPL | Beta-casein | 46−54 | ||
| DLENLHLPLP | Beta-casein | 46−55 | ||
| DLENLHLPLPLL | Beta-casein | 46−57 |
Antimicrobial activity of Casein 24
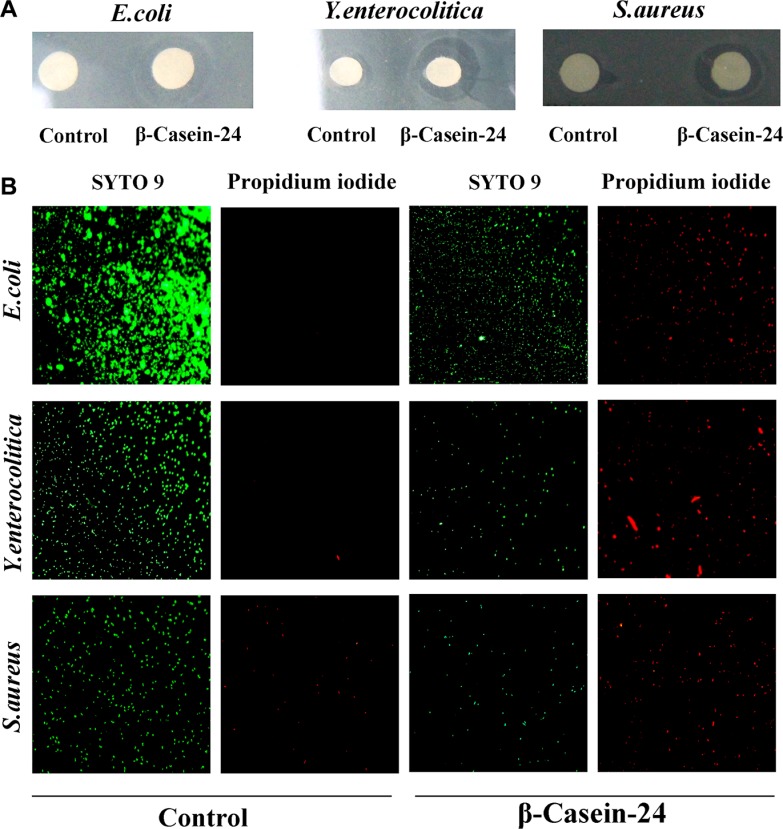
Minimal inhibitory concentration (MIC), agar well diffusion and microscopic assessment experiments were employed for the investigation of Casein 24 antimicrobial activity against E. coli, Y. enterocolitica and S. aureus. Casein 24 exhibited antimicrobial activity against all the test strains (Figure 2A). The MIC values of Casein 41 against the bacterial strains are summarized in Table 4. The microscopic assessment of Casein 24 against the bacterial strains were showed in Figure 2B. Of the test stain, E. coli and Y. enterocolitica were more sensitive to Casein 24. Intensive research will be carried out in the future.
Figure 2. Casein 24 antimicrobial activity against E. coli, Y. enterocolitica and S. aureus.
(A) Agar well diffusion assay. (B) Microscopic assessment of Casein 24 antimicrobial activity. Live cells emit green fluorescence and dead cells appear red (due to propidium iodide uptake).
Table 4. The minimal inhibitory concentration of Casein 41 to selected microorganisms.
| Microorganisms | |||
|---|---|---|---|
| S. aureus | K. pneumoniae | E. coli | |
| Minimum inhibitory concentration (μM) | 12.5−25 | 25 | 12.5−25 |
Effect of Casein 89 on human preadipocyte proliferation
Study on multifunctional peptides from human milk promots κ-casein derived peptides exhibit growth-promoting effect on 3T3-L1 cell (Mouse embryonic fibroblast cell line) [28]. Also, our peptide quantitative analysis revealed κ-casein derived peptide Casein 89 specially high-expressed in macrosomic group. To gain more experimental result supports, we investigated whether Casein 89 has a stimulating or inhibiting activity in human preadipocytes. We firstly provened that FITC-labelled Casein 89 could successfully get into the preadipocytes (Figure 3A). Its further effect was examined by incubating human preadipocytes with Casein 89 at different concentrations (0.05 μmol/ml, 0.5 μmol/ml and 5 μmol/ml), and measuring viability by the CCK-8 assay and xCELLigence® impedance-based cell analysis system. CCK-8 assay and xCELLigence assays revealed that the absolute numbers of living cells were reduced in Casein 89 treated cultures compared to control cultures at 72 h (Figure 3B). We observed a dose dependent moderate inhibition to Casein 89 treatment (Figure 3C).
Figure 3. Cell Proliferation Assays of Casein 89.
(A) FITC-labelled Casein 89 could successfully get into the preadipocytes. (B) Real-Time Monitoring of Casein 89 effect on human preadipocytes. (C) CCK-8 cell proliferation assay for Casein 89 treatment. Data was expressed as mean ± SD. *P < 0.05, when compared with control group.
DISCUSSION
A subfield of proteomics named peptidomics [29], which studies endogenous peptides that occur in body fluids, tissues, and cell lines, has recently increased. In present study, we conducted a comparison between mothers delivering macrosomic and non-macrosomic infants in colostrum endogenous peptides. To efficiently identify these naturally occurring peptides from human milk, a novel comprehensive approach was invented. Centrifugal ultrafiltration strategy is a well-established method based on a size-exclusion mechanism for concentrating and purifying peptide [30, 31]. Finally, more than 400 peptides were identified in both groups. Compared with previous studies on endogenous milk peptides [16, 18], more peptides were identified in this study (over 400 peptides vs 248 peptides, over 400 peptides vs over 300 peptides). This prompts ultrafiltration could be a more effective approach for endogenous peptides preparation.
Previous studies on endogenous peptides mainly focused on the proteolytic enzymes cleaving profile of the hydrolysed milk protein product [16, 18, 19], but lack a quantitative analysis of the complete protein-released peptidome, especially its correlation with disease progression. In fact, the quantitative description of peptides levels and their alterations under various physiological or pathological conditions, may reflect the biological processes occurring in body, are inevitably related to the proteolytic enzymes activities, and hus might be served as diagnostic biomarkers of pathological state. In this study, stable isotope dimethyl labeling was applied to quantitative the peptide composition of human milk from mothers delivering macrosomic and non-macrosomic infants [20], to uncover whether the human milk peptides amounts ranging from colostrum stage to mature milk stage. Results showed that 28 peptide were present at a statistically significant changes, accounting for about 7% of the total identified peptides.
Proteolytic activities of proteases in milk contribute to the production of smaller fragments such as peptides in milk [31]. Such peptides could become active only after releasing from their parent protein and this could attribute to gastric and pancreatic enzymes digestion process [32, 33]. The beneficial health effects of milk protein-derived bioactive peptides have been proved in many articles, which exhibit antimicrobial, antihypertensive, antioxidative, antithrombotic, mineral binding, immunomodulatory and opioid activities, among others [32, 34–36]. The function of these peptides is based on their amino acid composition and sequence signature. The high sequence homology between the naturally produced milk peptides with those reviewed peptides in previous articles suggests the likely function of these peptides. In this study, blastp analysis revealed high homology to query sequence with identities more than 50%. Of these peptides, twelve matched known immunomodulatory sequences, eighteen matched antibacterial sequences, four matched antioxidant sequences, four matched opioid agonist sequences and three matched antihypertensive sequence. The functional peptides may be specifically releasing to benefit macrosomic infants health and ensures safe growth and development during the early stages of their life.
Infection of newborns, as a major health care issue, is the leading cause of infants mortality during birth, while breast milk fed infants are at lower risk for infections [37, 38]. Early human milk feedings could provide protection for the newborn against these infection. However, the biological function which peptides widely dispay is its antibacterial effect. Combined with previous bioinformatic analysis, we are pretty sure milk endogenous peptides could play an important role in anti-infection. Experiments in our study confirmed that Casein 24 has a bacteria-fighting powers against E. coli, Y. enterocolitica and S. aureus. A previous study on pathogens inveterate that E. coli, Y. enterocolitica and S. aureus were the major strains in our neonatal intensive care unit (NICU) [39]. Nowdays, many gram negative pathogens show multidrug-resistant, including organisms that express extendedspectrum beta-lactamases such as K. pneumoniae, K. oxytoca, and other enterobacteriaceae such as E. coli. With the abusive application of a large number of antibiotics, antibiotics are subjected to certain constraints for bacterial resistance and screening bottleneck of new antibiotic. In this context, antimicrobial peptides promise to be the most ideal alternative medicine.
Also, current evidences indicate that the bacterial species of gut microbiota has been proposed to be a contributory factor in the development and maintenance of children and adolescent obesity [40, 41]. Abundant species of endogenous antimicrobial peptides were revealed in human milk of our study, especially the antimicrobial effect of Casein 24 was confirmed. So we speculate that these nature occurring peptides has a significant effect on gut microbiota composition, then influences host metabolism, adiposity and obesity complications. Perhaps, these effects of peptides could provide a protection for macrosomia.
Obesity and related metabolic diseases has become an epidemic [42]. In general, the offspring of obese women are more likely to be macrosomic than these offspring from their lean counterparts [43]. So inhibiting the proliferation of human preadipocyte may be a better regulation of the fetal macrosomia weight gain. During the last decade, peptides have been shown to exert a variety of metabolic effects. Natriuretic peptides could effective degrade lipids in adipose tissue [44], it has been proved that its plasma levels are negatively related to the development of insulin resistance and metabolic syndrome [45]. Several researches have revealed the potential therapeutic role of Angiotensin-(1–7) on treating and preventing metabolic disorders including both lipid and glucose disorders [46]. In consistent with the above studies, our studied showed that macrosomic group specially high-expressed peptide Casein 89 exhibit growth inhibiting activity in human preadipocytes.
It should be noted that this study also had some limitations. First, macrosomia occurs mainly in pregnancies complicated by Type-1 diabetes, Type-2 diabetes and gestational diabetes (GDM) [47, 48]. But during sample collection, the influence of diabetes and GDM dieases were excluded. Second, protection effects of milk endogenous peptides in macrosomia must be work on a whole, while integrated research on these peptides are lacking. Third, human milk will inevitably experience a process of digestion in gastrointestinal tract after taking, which peptides could escape digestion and present in blood stream are still unknown. Further more, the effects of endogenous peptides must be varied and we merely revealed its function from antimicrobial activity and inhibiting adipocyte hyperplasia sides.
In summary, we investigated the peptides profile in human milk from mothers delivering macrosomic infants, as well as detecting abundance changes of these peptides. Knowledge gained from these peptides in human milk will provide some insights into the regulatory mechanism involved in immunomodulating, antibacterial, antioxidant, opioid agonist, and developmental advantages on macrosomic infants from breastfeeding. It has been shown for the first time, that naturally occurring peptides in milk from macrosomic group, which show inhibiting properties on adipocytes. We believe that our research is a meaningfull finding which may add to the understanding of milk peptide physiological action for interested researchers.
MATERIALS AND METHODS
Sample collection
Colostrums from women delivering macrosomic infants (n = 6) and their matched controls (n = 6) were obtained at Nanjing Maternal and Child Health Hospital, China. Patients members were fully informed of the research and signed medical informed consent and this study was approved by Nanjing Medical University, Human Research Ethics Committee and Nanjing Maternal and Child Health Hospital. 10–20 ml of milk was collected using a mechanical breast pump (Ameda Egnell, Basel, Switzerland) by each lactating mother in the first 72 hours after birth. The milk was immediately stored on dry ice during transport to the laboratory (up to 1 h), and then centrifuged (1000 g, 20 min, 4°C) to remove the lipid layer and cell debris pellet. The aqueous phase (skim milk) was then added aliquot of protease inhibitor mixture (Complete Mini EDTA-free, Roche, Basel, Switzerland) and stored at −80°C.
Sample treatment
Samples were then centrifuged at 120,00 g at 4°C for 30 min after thawing on the ice, and the supernatant was collected. Protein concentrations of all the samples were determined by the bicinchoninic acid (BCA) method (Pierce, Rochford, USA), using BSA as a standard. In the ultrafiltration method, 50 μL of milk samples were two-fold diluted in a denaturing and reducing solution (7 M urea, 2 M thiourea, and 20 mM DTT), and transferred to centrifugal filter devices. Molecular weight cut-off filters (Millipore, Billerica, MA, USA) of 10 kDa were washed with 0.5 ml H2O prior to use. The milk samples were centrifuged through the filters according to the manufacturer's recommendations. The filtrates were then desalted and concentrated by C18 solid phase extraction (SPE) (Strata C18-E, 55 μm, 760A, 100 mg/mL, Phenomenex, Torrance, CA, USA), and finally lyophilized. The derived peptides of the different samples are then labeled with isotopomeric dimethyl labels [20]. The labeled samples were mixed and simultaneously analyzed by LC-MS/MS whereby the mass difference of the dimethyl labels was used to compare the peptide abundance in the different samples.
Liquid chromatography/mass spectrometry (LC/MS)
The freeze dried peptides was dissolved in 0.1% formic acid and filtered through a 0.45 μm membrane before injection. Reverse-phase chromatography was performed using a LC Packings C18 trap column (Acclaim PepMap100, 75 μm × 20 mm,) and separated with a LC packings C18 column (Acclaim PepMap, 75 μm × 150 mm) coupled to an Ultimate 3000 nano-LC system (Eksigent Technologies, Dublin, CA). The mobile phase was composed of: (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. A linear gradient from 0% to 20% B delivered at 200 μl/min over 50 min was used. The eluate was monitored by absorbance at 280 nm and five fractions were collected. The samples were directly injected into a MALDI TOF/TOF (Ultraflextreme, Bruker Daltonics, Bremen, Germany) instrument operated in the positive ion mode, following the previous strategy [49]. Full scan analysis was performed over the m/z range 400–5000 at 3 spectra/s. The capillary voltage and cone voltage were maintained at 3.9 kV and 40 V, respectively. For MS analysis, 2,000 single shot spectra were accumulated from 10 random positions on each sample, irradiating each position with 200 laser pulses.
The MS data were searched using the Mascot database search (http://www.matrixscience.com) against the SwissProt sequence database (http://www.expasy.org/tools/) considering the following variable modifications: phosphorylation, carbamylation, deamidation, methionine oxidation, acetylation, sulfation, and oxidized as well as reduced cysteines [15, 50]. The mass tolerance for precursor ion masses was set to 100 ppm and for miss cleavage, a single amino acid. All included peptides in the search were at least 6 amino acids long. PEAKS software (version 7.0, Bioinformatics Solutions) was also used to search the databases using MS/MS spectral data.
Search for potential bioactive peptides
In order to further uncover the function of the specific peptides, we refered to the method as David et al. (2013) provided [16]. We first search for the known functional peptide in previous studies, then compared these identified peptides to each breast milk peptide using DNAman software. We also considered 50% indentity of the query sequence as a retaining minimum standard, so as to remove false positives.
Antimicrobial activity assays of Casein 24
Bioinformatics analysis prompted Casein 41 has potential antimicrobial activity. The chemical synthetic peptide Casein 41 was purchased from Science Peptide Biological Technology CO.LTD (Shanghai, China). Then, the antimicrobial action of Casein 41 was tested against S. aureus, K. pneumoniae and E. coli using minimal inhibitory concentration, agar well diffusion assay and microscopic assessment experiment. Briefly, these three microorganisms were cultured overnight, so as to enhance the activity of bacteria. MIC was determined in 96-well polypropylene microtitre plates by concentration-dependent reducing Casein 41 binding. Sterile water was used as positive controls, while wells containing peptide without bacterial suspension were served as negative controls. The active strains were coated on a petri dish, then filter paper with 25 uM Casein 41 were covered on it and incubated overnight. The filter paper with sterile water was added as the negative control. For the microscopic assessment of Casein 24 activity, bacteria sample was treated with SYTO 9 and propidium iodide stains mixture (live/dead® Baclight™ bacterial viability kit, Thermofisher, USA) following the manufacturer's instruction. Microscope images were captured by using a fluorescence microscope (Zeiss, Imager.A2, Germany). Bacterium suspension without any disposition were served as negative controls. All experiments were repeated three times with nearly identical results.
Cell proliferation assays of Casein 89
Human preadipocytes were proliferated in Preadipocyte Medium (PAM, ScienCell Research Laboratories) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. FITC-labelled Casein 89 synthesised from Science Peptide Biological Technology CO.LTD was added into the medium at 60% confluence, fluorescent was detected after 6 h culture using a fluorescence microscope. The Cell Counting Kit-8 (CCK-8, Dojindo, Tokyo, Japan) experiment was carried out in 96-well plate to determine the proliferation promotion effect of peptide Casein 89. Moreover, a real-time cell proliferation assay was performed using the xCELLigence® impedance-based, label-free, real time cell analysis system (ACEA Biosciences, San Diego CA, USA). Cell indices were plotted against cultured time as growth curves. Cells were plated in 10 % FBS media at the indicated density in 16 microtiter untreated E-plates (ACEA Biosciences, San Diego CA, USA). Peptide Casein 89 treatments were initiated after 4 h incubation. Cell state changes were continuously monitored every hour for a period of 72 h. The peptide solvent phosphate buffer saline was used as a positive control.
Statistical analysis
Data from the experiments were analyzed by Student's unpaired or paired t-tests where appropriate with a significance of P < 0.05. The threshold value we used to screen differentially expressed peptides is a fold change ≥ 3.0 or ≤ −3.0 (P < 0.01). All data were expressed as the mean ± standard deviation (SD) in this study.
SUPPLEMENTARY MATERIALS FIGURE AND TABLE
Abbreviations
- CS
cesarean section
- LMW
low molecular weight
- MWCO
molecular weight cut-off-LC/MS liquid chromatography/mass spectrometry
- Casein 24
peptide located at β-Casein 24-38 AA
- Casein 89
peptide located at κ-Casein 89-109 AA
- MW
molecular weight
- PI
isoelectric point
- CASA
α-casein
- CSN3
κ-casein
- PIGR
polymeric immunoglobulin receptor
- C4A
complement C4-A
- BAL
bile salt-activated lipase
- MUC4
mucin-4
- ADAM-TS 13
a disintegrin and metalloproteinase with thrombospondin motifs 13
- ADFP
perilipin-2
- Alpha-2-M
alpha-2-macroglobulin
- BSP II
mothers against decapentaplegic homolog 1
- TNC
cytotactin
- FGB
fibrinopeptide B
- FGA
fibrinopeptide A
- N-terminal
amino terminal end
- C-terminal
carboxyl terminal end
- L
leucine
- P
proline
- MIC
minimal inhibitory concentration
- NICU
neonatal intensive care unit
- DM1
Type-1 diabetes
- DM2
Type-2 diabetes
- GDM
gestational diabetes mellitus
- SPE
solid phase extraction
- FBS
fetal bovine serum
- CCK-8
cell counting kit-8
- SD
standard deviation
- OGTT
oral glucose tolerance test
Footnotes
CONFLICTS OF INTEREST
All authors read and approved the final manuscript. No conflicts of interest were reported.
GRANT SUPPORT
This work was funded by a grant from the National Key Basic Research Program of China(2013CB530604), the Key project of the National Natural Science Foundation of China (81330067), the National Natural Science Foundation of China (Grant No. 81300683, 81471086 and 81200642), Nanjing Medical Science and Technique Development Foundation (JQX13012).
Authors' contributions
Chenbo Ji and Xirong Guo designed the research; Yun Li, Lianghui You and Chunmei Shi conducted the research; Xing Wang performed the statistical analysis; Xianwei Cui and Lei Yang prepared the initial manuscript draft and revised subsequent drafts.
REFERENCES
- 1.Maayan-Metzger A, Lubin D, Kuint J. Hypoglycemia rates in the first days of life among term infants born to diabetic mothers. Neonatology. 2009;96:80–85. doi: 10.1159/000203337. [DOI] [PubMed] [Google Scholar]
- 2.Lu Y, Zhang J, Lu X, Xi W, Li Z. Secular trends of macrosomia in southeast China, 1994-2005. BMC Public Health. 2011;11:818. doi: 10.1186/1471-2458-11-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Decker A, Platt RW, Kramer MS. How big is too big? The perinatal consequences of fetal macrosomia. Am J Obstet Gynecol. 2008;198:517. doi: 10.1016/j.ajog.2007.12.005. e1–6. [DOI] [PubMed] [Google Scholar]
- 4.Lipscomb KR, Gregory K, Shaw K. The outcome of macrosomic infants weighing at least 4500 grams: Los Angeles County + University of Southern California experience. Obstet Gynecol. 1995;85:558–564. doi: 10.1016/0029-7844(95)00005-C. [DOI] [PubMed] [Google Scholar]
- 5.Walsh JM, McAuliffe FM. Prediction and prevention of the macrosomic fetus. Eur J Obstet Gynecol Reprod Biol. 2012;162:125–130. doi: 10.1016/j.ejogrb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics. 2005;115:e290–296. doi: 10.1542/peds.2004-1808. [DOI] [PubMed] [Google Scholar]
- 7.Hermann GM, Dallas LM, Haskell SE, Roghair RD. Neonatal macrosomia is an independent risk factor for adult metabolic syndrome. Neonatology. 2010;98:238–244. doi: 10.1159/000285629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ornoy A. Prenatal origin of obesity and their complications: Gestational diabetes, maternal overweight and the paradoxical effects of fetal growth restriction and macrosomia. Reprod Toxicol. 2011;32:205–212. doi: 10.1016/j.reprotox.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Lis J, Orczyk-Pawiłowicz M, Kątnik-Prastowska I. Proteins of human milk involved in immunological processes. Postepy Hig Med Dosw (Online) 2013;67:529–547. doi: 10.5604/17322693.1051648. [DOI] [PubMed] [Google Scholar]
- 10.Molinari CE, Casadio YS, Hartmann BT, Livk A, Bringans S, Arthur PG, Hartmann PE. Proteome mapping of human skim milk proteins in term and preterm milk. J Proteome Res. 2012;11:1696–1714. doi: 10.1021/pr2008797. [DOI] [PubMed] [Google Scholar]
- 11.Schack-Nielsen L, Michaelsen KF. Breast feeding and future health. Curr Opin Clin Nutr Metab Care. 2006;9:289–296. doi: 10.1097/01.mco.0000222114.84159.79. [DOI] [PubMed] [Google Scholar]
- 12.Gouveri E, Papanas N, Hatzitolios AI, Maltezos E. Breastfeeding and diabetes. Curr Diabetes Rev. 2011;7:135–142. doi: 10.2174/157339911794940684. [DOI] [PubMed] [Google Scholar]
- 13.Owen CG, Whincup PH, Cook DG. Breast-feeding and cardiovascular risk factors and outcomes in later life: evidence from epidemiological studies. Proc Nutr Soc. 2011;70:478–484. doi: 10.1017/S0029665111000590. [DOI] [PubMed] [Google Scholar]
- 14.Hu L, Li X, Jiang X, Zhou H, Jiang X, Kong L, Ye M, Zou H. Comprehensive peptidome analysis of mouse livers by size exclusion chromatography prefractionation and nanoLC-MS/MS identification. J Proteome Res. 2007;6:801–808. doi: 10.1021/pr060469e. [DOI] [PubMed] [Google Scholar]
- 15.Hölttä M, Zetterberg H, Mirgorodskaya E, Mattsson N, Blennow K, Gobom J. Peptidome analysis of cerebrospinal fluid by LC-MALDI MS. PLoS One. 2012;7:e42555. doi: 10.1371/journal.pone.0042555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dallas DC, Guerrero A, Khaldi N, Castillo PA, Martin WF, Smilowitz JT, Bevins CL, Barile D, German JB, Lebrilla CB. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res. 2013;12:2295–2304. doi: 10.1021/pr400212z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nonaka A, Nakamura T, Hirota T, Matsushita A, Asakura M, Ohki K, Kitakaze M. The milk-derived peptides Val-Pro-Pro and Ile-Pro-Pro attenuate arterial dysfunction in L-NAME-treated rats. Hypertens Res. 2014;37:703–707. doi: 10.1038/hr.2014.72. [DOI] [PubMed] [Google Scholar]
- 18.Baum F, Fedorova M, Ebner J, Hoffmann R, Pischetsrieder M. Analysis of the endogenous peptide profile of milk: identification of 248 mainly casein-derived peptides. J Proteome Res. 2013;12:5447–5462. doi: 10.1021/pr4003273. [DOI] [PubMed] [Google Scholar]
- 19.Dallas DC, Guerrero A, Khaldi N, Borghese R, Bhandari A, Underwood MA, Lebrilla CB, German JB, Barile D. A peptidomic analysis of human milk digestion in the infant stomach reveals protein-specific degradation patterns. J Nutr. 2014;144:815–820. doi: 10.3945/jn.113.185793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 21.Robbins RJ, Villanueva J, Tempst P. Distilling cancer biomarkers from the serum peptidome: high technology reading of tea leaves or an insight to clinical systems biology? J Clin Oncol. 2005;23:4835–4837. doi: 10.1200/JCO.2005.02.912. [DOI] [PubMed] [Google Scholar]
- 22.Shores KS, Knapp DR. Assessment approach for evaluating high abundance protein depletion methods for cerebrospinal fluid (CSF) proteomic analysis. J Proteome Res. 2007;6:3739–3751. doi: 10.1021/pr070293w. [DOI] [PubMed] [Google Scholar]
- 23.Hong WX, Liu W, Zhang Y, Huang P, Yang X, Ren X, Ye J, Huang H, Tang H, Zhou G, Huang X, Zhuang Z, Liu J. Identification of serum biomarkers for occupational medicamentosa-like dermatitis induced by trichloroethylene using mass spectrometry. Toxicol Appl Pharmacol. 2013;273:121–129. doi: 10.1016/j.taap.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Azuma N, Nagaune S, Ishino Y, Mori H, Kaminogawa S, Yamauchi K. DNA-synthesis stimulating peptides from human β-casein. Agric Biol Chem. 1989;53:2631–2634. [Google Scholar]
- 25.Hayes M, Stanton C, Fitzgerald GF, Ross RP. Putting microbes to work: dairy fermentation, cell factories and bioactive peptides. Part II: bioactive peptide functions. Biotechnol J. 2007;2:435–449. doi: 10.1002/biot.200700045. [DOI] [PubMed] [Google Scholar]
- 26.Hernández-Ledesma B, Q. A, Amigo L, Recio I. Identification of bioactive peptides after digestion of human milk and infant formula with pepsin and pancreatin. Int Dairy J. 2007;17:42–49. [Google Scholar]
- 27.Jarmolowska B, S.-S. E, Kostyra E, Kostyra H, Mierzejewska D, Darmochwal-Marcinkiewicz K. Opioid activity of Humana formula for newborns. J Sci Food Agric. 2007;87:2247–2250. [Google Scholar]
- 28.Mandal SM, Bharti R, Porto WF, Gauri SS, Mandal M, Franco OL, Ghosh AK. Identification of multifunctional peptides from human milk. Peptides. 2014;56:84–93. doi: 10.1016/j.peptides.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Schrader M, Schulz-Knappe P. Peptidomics technologies for human body fluids. Trends Biotechnol. 2001;19:S55–60. doi: 10.1016/S0167-7799(01)01800-5. [DOI] [PubMed] [Google Scholar]
- 30.Soloviev M, Finch P. Peptidomics: bridging the gap between proteome and metabolome. Proteomics. 2006;6:744–747. doi: 10.1002/pmic.200500878. [DOI] [PubMed] [Google Scholar]
- 31.Dallas DC, Murray NM, Gan J. Proteolytic Systems in Milk: Perspectives on the Evolutionary Function within the Mammary Gland and the Infant. J Mammary Gland Biol Neoplasia. 2015;20:133–147. doi: 10.1007/s10911-015-9334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raikos V, Dassios T. Health-promoting properties of bioactive peptides derived from milk proteins in infant food: a review. Dairy Sci Technol. 2014;94:91–101. doi: 10.1007/s13594-013-0152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.HK Milk-derived bioactive peptides: from science to applications. J Funct Foods. 2009;1:177–187. [Google Scholar]
- 34.Nagpal R, Behare P, Rana R, Kumar A, Kumar M, Arora S, Morotta F, Jain S, Yadav H. Bioactive peptides derived from milk proteins and their health beneficial potentials: an update. Food Funct. 2011;2:18–27. doi: 10.1039/c0fo00016g. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto N, Ejiri M, Mizuno S. Biogenic peptides and their potential use. Curr Pharm Des. 2003;9:1345–1355. doi: 10.2174/1381612033454801. [DOI] [PubMed] [Google Scholar]
- 36.Fekete áA, Givens DI, Lovegrove JA. The impact of milk proteins and peptides on blood pressure and vascular function: a review of evidence from human intervention studies. Nutr Res Rev. 2013;26:177–190. doi: 10.1017/S0954422413000139. [DOI] [PubMed] [Google Scholar]
- 37.Lawn JE, Cousens S, Zupan J, Lancet Neonatal Survival Steering Team 4 million neonatal deaths: when? Where? Why? Lancet. 2005;365:891–900. doi: 10.1016/S0140-6736(05)71048-5. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Schmitz G, Levy O. Development of newborn and infant vaccines. Sci Transl Med. 2011;3:90ps27. doi: 10.1126/scitranslmed.3001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen T, Li Y, Liu H, Lu Q. Distribution and drug resistance of pathogens in aneonatal intensive care unit. Chin J Infect Control. 2013;12:300–303. [Google Scholar]
- 40.Harley IT, Karp CL. Obesity and the gut microbiome: Striving for causality. Mol Metab. 2012;1:21–31. doi: 10.1016/j.molmet.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu P, Li M, Zhang J, Zhang T. Correlation of intestinal microbiota with overweight and obesity in Kazakh school children. BMC Microbiol. 2012;12:283. doi: 10.1186/1471-2180-12-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popkin BM, Kim S, Rusev ER, Du S, Zizza C. Measuring the full economic costs of diet, physical activity and obesity-related chronic diseases. Obes Rev. 2006;7:271–293. doi: 10.1111/j.1467-789X.2006.00230.x. [DOI] [PubMed] [Google Scholar]
- 43.Higgins L, Greenwood SL, Wareing M, Sibley CP, Mills TA. Obesity and the placenta: A consideration of nutrient exchange mechanisms in relation to aberrant fetal growth. Placenta. 2011;32:1–7. doi: 10.1016/j.placenta.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 44.Beleigoli AM1, Diniz MF, Ribeiro AL. Natriuretic peptides: linking heart and adipose tissue in obesity and related conditions—a systematic review. Obes Rev. 2009;10:617–626. doi: 10.1111/j.1467-789X.2009.00624.x. [DOI] [PubMed] [Google Scholar]
- 45.Hsieh JC, Wang JH, Lee CJ, Chen YC, Liou HH, Hsu BG. Low serum long-acting natriuretic peptide level correlates with metabolic syndrome in hypertensive patients: a cross-sectional study. Arch Med Res. 2013;44:215–220. doi: 10.1016/j.arcmed.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Fujimoto M, Masuzaki H, Tanaka T, Yasue S, Tomita T, Okazawa K, Fujikura J, Chusho H, Ebihara K, Hayashi T, Hosoda K, Nakao K. An angiotensin II AT1 receptor antagonist, telmisartan augments glucose uptake and GLUT4 protein expression in 3T3-L1 adipocytes. FEBS Lett. 2004;576:492–497. doi: 10.1016/j.febslet.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 47.Verheijen EC, Tuffnell DJ. Outcomes of pregnancies in women with Type 1 diabetes in Scotland: a national population-based study. BJOG. 2003;110:966. doi: 10.1111/j.1471-0528.2003.03021.x. [DOI] [PubMed] [Google Scholar]
- 48.Clausen TD, Mathiesen E, Ekbom P, Hellmuth E, Mandrup-Poulsen T, Damm P. Poor pregnancy outcome in women with type 2 diabetes. Diabetes Care. 2005;28:323–328. doi: 10.2337/diacare.28.2.323. [DOI] [PubMed] [Google Scholar]
- 49.Liu F, Zhao C, Liu L, Ding H, Huo R, Shi Z. Peptidome profiling of umbilical cord plasma associated with gestational diabetes-induced fetal macrosomia. J Proteomics. 2016;139:38–44. doi: 10.1016/j.jprot.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Hölttä M, Minthon L, Hansson O, Holmén-Larsson J, Pike I, Ward M, Kuhn K, Rüetschi U, Zetterberg H, Blennow K, Gobom J. An integrated workflow for multiplex CSF proteomics and peptidomics-identification of candidate cerebrospinal fluid biomarkers of Alzheimer's disease. J Proteome Res. 2015;14:654–663. doi: 10.1021/pr501076j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.