Abstract
We enrolled a total of 277 patients who received nephrectomy due to clear cell renal cell carcinoma (ccRCC) in Zhongshan Hospital from Jan 2005 to Jun 2007. Immunohistochemistry was performed to evaluate the impact of CLEC-2 positive cell infiltration on the overall survival (OS) and recurrence-free survival (RFS) of patients with ccRCC. Kaplan-Meier analysis showed that high CLEC-2 positive cell infiltration in tumor tissue indicated poorer OS and RFS (OS, p < 0.001; RFS, p = 0.002). High CLEC-2 positive cell infiltration is also an independent risk factor for OS and RFS in multivariate analyses (OS, p = 0.004; RFS, p = 0.009). CLEC-2 positive cell infiltration could also stratify ccRCC patients' survival with University of California Integrated Staging System (UISS) stratum in the mediate-risk and high-risk groups. We constructed two nomograms incorporating parameters derived from multivariate analyses to predict patients' OS and RFS (OS, c-index 0.813; RFS, c-index 0.716). In conclusion, high CLEC-2 positive cell infiltration in ccRCC is an independent adverse prognostic factor for patients, and established nomograms based on this information could help predict ccRCC patients' OS and RFS.
Keywords: c-type lectin-like receptor 2, clear cell renal cell carcinoma, prognostic factor, overall survival, recurrence-free survival
INTRODUCTION
Renal cell carcinoma (RCC) is the most common malignant cancer in the kidney, accounting for 2% to 3% of all adult malignancies [1], and clear cell renal cell carcinoma (ccRCC) is the most common histological subtype, responsible for most deaths. There are approximately 16.6 women and 37.7 men diagnosed with RCC per 100,000 people every year in China [2]. The natural history of renal cell carcinoma is very complicated and about 20-40% patients would develop recurrences or metastasis even after undergoing curative nephrectomy [3]. Currently, TNM stage, Fuhrman grade and several integrated models like University of California Integrated Staging System (UISS), and Mayo Clinic stage, size, grade and necrosis (SSIGN) score are being used to predict the clinical outcome of RCC. However, these models may not be enough due to the genetic complexity and heterogeneity of the disease [4]. A more accurate prediction model is needed and combining some important molecular biomarkers with current models is probably a new and effective way.
The c-type lectin-like receptor 2 (CLEC-2) known as an emerging pattern recognition receptors for the activation of innate immunity, is a type II membrane protein with a c-type lectin like domain and a single hemITAM motif. CLEC-2 was first identified in a bioinformatic screen in search of c-type lectin receptors and CLEC-2 mRNA was found in the liver and myeloid cells including monocytes, dendritic cells, NK cells, and granulocytes [5]. CLEC-2 signaling modulates toll-like receptor agonists and promotes induction of IL-10 [6]. Later on after systematically analyzing, CLEC-2 was recognized as a platelet activating receptor for the snake venom toxin rhodocytin inducing platelet aggregation. CLEC-2 also possesses an endogenous ligand, the mucin-like glycoprotein podoplanin found on lymphatic endothelium, stromal of secondary lymphoid organs and some cancer cells [7]. Ligation of CLEC-2 with podoplanin elicits strong platelet activation, and it is identified that platelet activation is known to promote tumor metastasis, which may be triggered by podoplanin up-regulation [8]. All these evidence suggests a potential role of CLEC-2 in cancer immunomodulation and metastasis.
However, no researchers have assessed the correlation between clec-2 positive cell infiltration and clinical outcomes of ccRCC patients before. We wondered whether CLEC-2 could become a potential prognostic marker for patients with RCC. Thus we looked into CLEC-2 positive cell infiltration in a large set of clear cell RCC patients by means of immunohistochemistry. The impact of CLEC-2 positive cell infiltration on patients' overall survival (OS) and recurrence-free survival (RFS) were analyzed.
RESULTS
Associations between CLEC-2 positive cell infiltration and clinicopathological characteristics
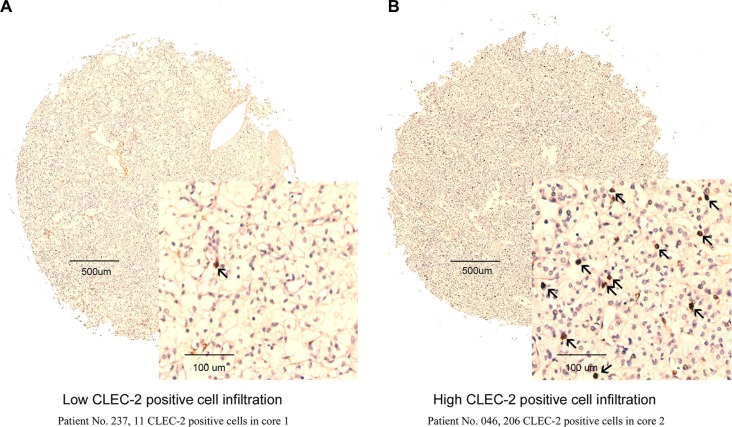
CLEC-2 was mainly expressed in stromal cells compared with tumor tissues (Figure 1A, 1B). A specimen tissue was considered high CLEC-2 positive cell infiltration if it contains more than 53 CLEC-2 positive cells, otherwise low CLEC-2 positive cell infiltration. As we can see from Table S1, CLEC-2 positive cell infiltration is apparently associated with tumor size (p = 0.015), pathological T stage (p = 0.042), TNM stage (p = 0.016) and necrosis (p = 0.038). Other clinicopathological parameters of the ccRCC patients were not associated with CLEC-2 positive cell infiltration.
Figure 1. CLEC-2 positive cell infiltration in clear-cell renal cell carcinoma (ccRCC) tissues.
Representative CLEC-2 immunohistochemical (IHC) images of ccRCC tumor tissues with low CLEC-2 positive cell infiltration (Patient No. 237, 11 CLEC-2 positive cells in core 1) (A) and high CLEC-2 positive cell infiltration (Patient No. 046, 206 CLEC-2 positive cells in core 2) (B). Arrows indicate CLEC-2 positive cells.
Association between CLEC-2 positive cell infiltration and clinical outcomes
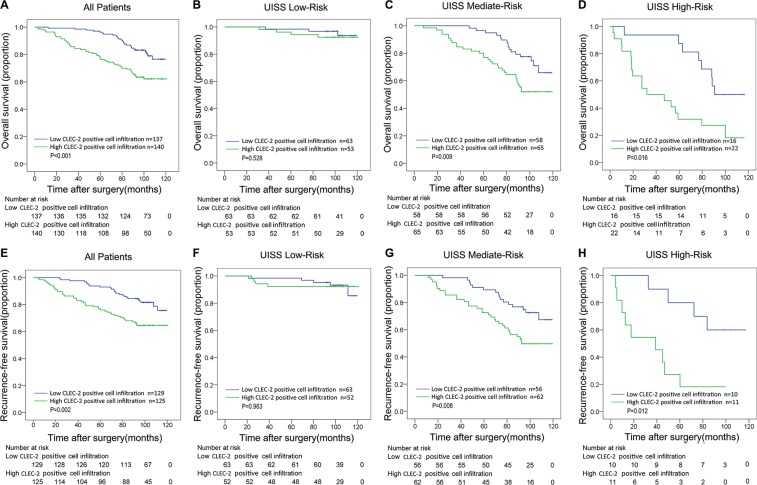
The median follow-up time for all available patients was 98.63 months (range 2.63–120.47). The mean follow-up time was 91.06 months. 79 in 277 patients (28.5%) died during the follow up and 68 in 254 patients (26.8%) experienced disease relapse. We compared overall survival and recurrence-free survival according to CLEC-2 positive cell infiltration in order to further investigate the prognostic value of CLEC-2 positive cell infiltration. Obviously, ccRCC patients with high CLEC-2 positive cell infiltration had a poorer OS and RFS compared to those with low CLEC-2 positive cell infiltration. Kaplan–Meier survival analysis was used and as was shown in Figure 2A and 2E, high CLEC-2 positive cell infiltration was a significant negative prognostic predictor for patients included in the study (OS, p < 0.001; RFS, p = 0.002).
Figure 2. Overall survival (OS) and Recurrence-free survival (RFS) analyses of patients with ccRCC based on CLEC-2 positive cell infiltration.
Kaplan-Meier analysis of OS in All Patients group (n = 277) (A); and in UISS Low-Risk group (n = 116) (B) in UISS Mediate-Risk group (n = 67) (C) and in UISS High-Risk group (n = 74) (D); Kaplan-Meier analysis of RFS in All Patients group (n = 254) (E); in UISS Low-Risk group (n = 115) (F) in UISS Mediate-Risk group (n = 118) (G) and in UISS High-Risk group (n = 21) (H). P value was calculated by log-rank test.
We first conducted univariate analyses using number of CLEC-2 positive cells as a continuous variable. It was a risk factor for OS and RFS (OS, p < 0.001; RFS, p = 0.001) (Table S2). CLEC-2 positive cell infiltration as a dichotomous variable was also a risk factor for OS and RFS (OS, p = 0.001; RFS, p = 0.002) (Table S2), indicating an important impact of CLEC-2 positive cell infiltration on clinical outcome. To evaluate the robustness of the prognostic value of CLEC-2 positive cell infiltration and control for confounders, we performed Cox multivariate regression analyses and found that high CLEC-2 positive cell infiltration was still an independent risk factor for both OS and RFS (OS, p = 0.004; RFS, p = 0.009) (Table 1), together with other clinicopathological parameters like pathological T stage, Fuhrman grade, Necrosis, ECOG PS and distant metastasis. We also found that CLEC-2 positive cell infiltration could stratify ccRCC patients' survival in the UISS mediate-risk and high-risk groups, in which high CLEC-2 positive cell infiltration turned out to be an independent risk factor in both OS and RFS analyses (OS, p = 0.009, RFS, p = 0.008 in mediate-risk groups; OS, p = 0.016, RFS, p = 0.012 in high-risk groups) (Figure 2C and 2G, Figure 2D and 2H), while in the low-risk groups it is not statistically significant (Figure 2B and 2E). This probably indicates that CLEC-2 positive cells may function more in patients with higher pathological T stage, Fuhrman grade and/or ECOG PS.
Table 1. Proportional hazard model for overall survival and recurrence free survival prediction.
| Variables | OS (n = 277) | RFS(n = 254) | ||
|---|---|---|---|---|
| HR (95%CI) | P-value† | HR (95%CI) | P-value† | |
| Pathological T stage | 0.001 | <0.001 | ||
| pT1 | Reference | Reference | ||
| pT2 | 2.468 (1.217–5.004) | 0.012 | 2.144 (0.947–4.855) | 0.068 |
| pT3 | 2.968 (1.732–5.088) | < 0.001 | 3.181 (1.806–5.602) | < 0.001 |
| pT4 | 4.116 (1.080–15.681) | 0.038 | 11.160 (3.285–37.913) | < 0.001 |
| Distant metastasis | ||||
| Yes vs No | 2.804 (1.421–5.533) | 0.003 | ||
| Fuhrman grade | < 0.001 | < 0.001 | ||
| 1 | Reference | Reference | ||
| 2 | 1.641 (0.585–4.600) | 0.346 | 1.297(0.504–3.339) | 0.589 |
| 3 | 3.711 (1.231–11.189) | 0.020 | 3.868 (1.376–10.871) | 0.010 |
| 4 | 15.148 (3.117–73.613) | 0.001 | 13.530 (2.909–62.929) | 0.001 |
| Necrosis | ||||
| Present vs Absent | 1.871 (1.023–3.425) | 0.042 | 1.853(1.010–3.486) | 0.046 |
| ECOG PS | 0.001 | 0.001 | ||
| 0 | Reference | Reference | ||
| 1 | 2.392 (1.451–3.970) | 0.001 | 2.180 (1.231–3.861) | 0.008 |
| 2 | 2.914 (1.079–7.870) | 0.035 | 6.776 (2.547–18.028) | < 0.001 |
| 3 | 4.200 (1.212–14.550) | 0.024 | 5.792 (1.848–18.148) | 0.003 |
| CLEC-2 positive cell infiltration | ||||
| High vs Low | 2.065 (1.258–3.390) | 0.004 | 2.057 (1.200–3.524) | 0.009 |
ECOG PS = Eastern Cooperative Oncology Group performance status; HR = hazard ratio; CI = confidence interval; OS = overall survival; RFS = recurrence free survival;
Data obtained from the Cox proportional hazards model, P-value < 0.05 was regarded as statistically significant.
Extension of current prognostic model with CLEC-2 positive cell infiltration
The sensitivity of the predictive system calculated by C-index could be increased if we combine CLEC-2 positive cell infiltration information into the SSIGN and UISS score system. The SSIGN/UISS simplified three risk groups have been used in this procedure. CLEC-2 positive cell infiltration together with SSIGN score had increased predictive power compared to SSIGN score alone in both OS (C-index 0.744 vs. 0.725) and RFS (C-index 0.672 vs. 0.631) The situation was the same for UISS score, as adding CLEC-2 positive cell infiltration information into the UISS model could also improve its predictive power for both OS (C-index 0.763 vs. 0.743) and RFS (C-index 0.682 vs. 0.638) prediction (Table 2).
Table 2. Comparison of the predictive accuracy of the prognostic models.
| Models | Overall survival | Recurrence free survival | ||
|---|---|---|---|---|
| C-index | AIC | C-index | AIC | |
| CLEC-2 | 0.615 | 845.59 | 0.615 | 600.31 |
| TNM | 0.706 | 811.82 | 0.608 | 605.91 |
| TNM + CLEC-2 | 0.738 | 804.29 | 0.664 | 597.78 |
| SSIGN | 0.725 | 809.12 | 0.631 | 603.6 |
| SSIGN + CLEC-2 | 0.744 | 801.30 | 0.672 | 595.93 |
| UISS | 0.743 | 804.84 | 0.638 | 608.98 |
| UISS + CLEC-2 | 0.763 | 794.91 | 0.682 | 600.00 |
| Nomogram | 0.813 | 758.85 | 0.716 | 521.21 |
C-index, concordance index; AIC, Akaike information criterion; SSIGN, Mayo clinic stage, size, grade, and necrosis score; UISS, UCLA Integrated Staging System. C-index and AIC were calculated from 1000 bootstrap sample.
Prognostic nomograms for OS and RFS
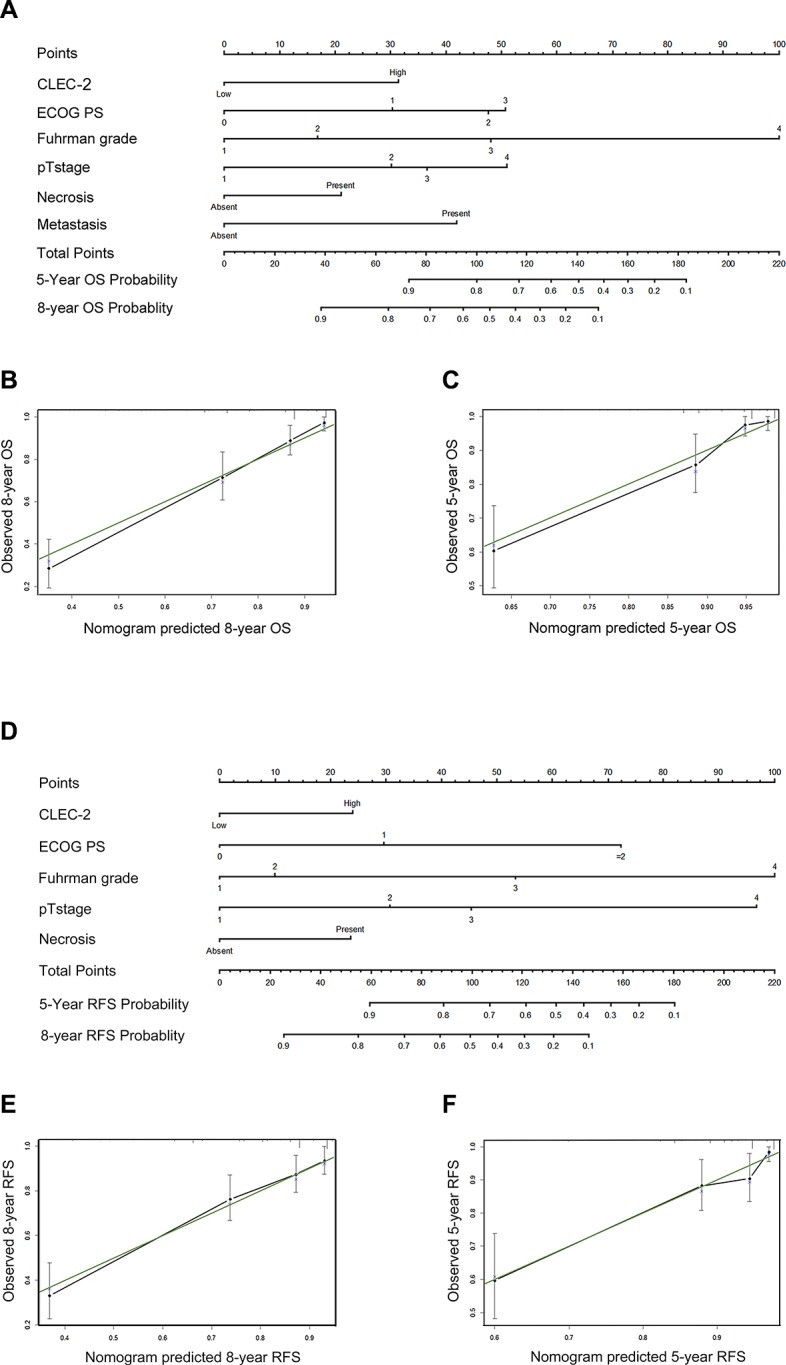
After incorporating significant prognostic factors concluded from Cox multivariate analyses, we established two nomograms to predict 5-year and 8-year ccRCC patients' OS and RFS (Figure 3). Pathological T stage, distant metastasis, Fuhrman grade, necrosis status, ECOG PS and CLEC-2 positive cell infiltration were included. A score was assigned to each level of the variables, and the total score could be used to predict the probability of survival. After performing Bootstrap validations we found that the calibration plots showed good consistency between the predicted and actual observation (Figure 3B, 3C; Figure 3E, 3F). The C-index indicated a good predictive accuracy for nomograms in both OS and RFS (OS, C-index 0.813; RFS, C-index 0.716).
Figure 3. Prognostic nomograms and calibration plots for OS and RFS prediction.
(A) Six independent prognostic factors including CLEC-2 positive cell infiltration, ECOG PS, Fuhrman grade, pathological T stage, necrosis and metastasis were identified and entered into the nomogram. (B) Calibration curves for predicting 8-year OS of ccRCC patients. (C) Calibration curves for predicting 5-year OS of ccRCC patients. (D) Five independent prognostic factors including CLEC-2 positive cell infiltration, ECOG PS, Fuhrman grade, pathological T stage and necrosis were identified and entered into the nomogram. (E) Calibration curves for predicting 8-year RFS of ccRCC patients. (F) Calibration curves for predicting 5-year RFS of ccRCC patients.
DISCUSSION
In this study, we detected the infiltration of CLEC-2 positive cell in ccRCC using 277 specimen tissues with immunohistochemistry, and it turned out that high CLEC-2 positive cell infiltration in tumor tissue is correlated with a poor prognosis. As an independent poor prognostic factor for OS and RFS of ccRCC patients, the infiltration level of CLEC-2 positive cells can be added to current prognostic models like TNM stage, UISS and SSIGN in order to improve the predictive accuracy. Moreover, we constructed two nomograms incorporating CLEC-2 positive cell infiltration with other significant parameters derived from multivariate analysis to predict patients' OS and RFS. C-indexes indicated that the two nomograms performed better than current prognostic models.
In this study, the CLEC-2 positive staining was mainly found in tumor stromal cells and it is known that CLEC-2 is mainly expressed on myeloid cells. Some myeloid cells like tumor associated macrophages (TAMs), neutrophils and myeloid-derived suppressor cells (MDSC) could help tumor angiogenesis, invasion, metastasis, and meditate certain immune-suppressive function under specific circumstances. This indicated that those CLEC-2 positive cells in this study might have a role inimmunomodulation and help cancer immunologic escape [6, 9]. According to previous studies, CLEC-2 positive cells could mediate tumor progression mainly in two ways, suppressive immunoregulation and platelet activation. Diego Mourão-Sa et al. found that CLEC-2 could signal via Syk, Ca21 and NFAT, leading to Syk phosphorylation, calcium signaling and NFAT activation in myeloid cells. Notably, by activating NFAT, CLEC-2 can modulate the effect of signals induced by other innate receptors such as toll-like receptors, resulting in selectively increased production of cytokine IL-10 [6]. The strong immune suppressive effects of IL-10 might indicate a role for CLEC-2 positive myeloid cells in immune suppression and tumor progression [10]. Different from other CLRs, receptor crosslinking by anti-CLEC-2 mAb was unable to activate CARD9/NF-κB pathway pathway, which is an important downstream signal of CLRs in inducing pro-inflammatory response [11, 12]. Thus CLEC-2 activation cannot increase serum levels of some pro-inflammatory cytokines like TNF, IL-6, IL1b, IL-1a or IL-12/IL-23p40, which might further prove its immune suppressive role. Therefore, ligation of CLEC-2 might promote cancer progression by creating an immunosuppressive environment and help cancer cell escape from immune surveillance.
On the other hand, CLEC-2 ligation with podoplanin could elicit strong platelets activation [7], which protects them from shear stress and NK cells in the blood stream and serves for tumor cell nestling [13]. Activated platelets release growth factors, promoting tumor angiogenesis and growth. The endogenous ligand of CLEC-2, podoplanin, could increase tumor cells motility by remodeling actin in the cytoskeleton and correlated with the onset of epithelial to mensenchymal transition (EMT), a key role in tumor metastasis [14, 15]. Interaction between podoplanin and CLEC-2 may regulate tumor invasion and metastasis and might be a potential target for therapy of metastasis.
In conclusion, we have identified that CLEC-2 positive cell infiltration correlates with ccRCC patients' survival and can be used as a novel prognostic factor in predicting patients OS and RFS. However, there are still some limitations. This is a retrospective study in nature and the number of patients enrolled is limited. Besides, CLEC-2 may be expressed in varying amounts in different areas of the tumor and the way we chose cutoff point with X-tile might lead to potential overfitting bias. A mutli-centered prospective external validation is needed. Moreover, further experimental studies are also required to identify the detailed role of CLEC-2 positive cells in ccRCC.
MATERIALS AND METHODS
Patient selection
The study included a total of 277 patients who received radical or partial nephrectomy due to clear cell RCC in Zhongshan Hospital, Fudan University from Jan 2005 to Jun 2007. All the patients were consecutively included if they met the criterion of having pathologically proven ccRCC, having received partial or radical nephrectomy and possessing available Formalin Fixed Paraffin Embedded (FFPE) specimen of tumor mass (≥ 1 cm3). Patients were excluded if they had other malignant tumor before, or histories of adjuvant or neo-adjuvant therapies including targeted therapies. Samples with over 80% necrotic or hemorrhagic area and patients with bilateral tumors were also excluded. Fudan University, Zhongshan Hospital research medical ethics committee approved this study and informed consent was given for the use of clinical specimens in this study.
Data collection
The primary outcome was OS, which was calculated from the time of operation to the time of death. RFS was defined as the time from nephrectomy to the time of first recurrence. During the first five years, the interval of follow up was three months and one year later then. Data were censored when the patient died or was alive at Jan 30, 2015, the last follow up time. The analysis of recurrence-free survival excluded fifteen patients with metastasis at surgery and eight patients with missing recurrence state.
Baseline clinical characteristics and complete follow-up outcome included in the database were re-examined. Two pathologists (Yuan J. and Jun H.) reviewed the H&E slides to reconfirm histological subtype, stage, and Fuhrman grade. One urologist reassessed all the MRI and CT scans. Histological subtype of ccRCC were reconfirmed according to 2014 EAU guidelines [16]. TNM stage and Fuhrman grade were based on the 2010 AJCC TNM classification and 2012 ISUP consensus, respectively [17, 18]. Patient risks are stratified according to the SSIGN, UISS and SSIGN localized (Leibovich) score according to original scoring algorithm [19–21].
Immunochemistry
Tissue microarrays were constructed as previously described [22]. This was a different cohort but we used the same method. Immunohistochemical staining was performed on tissue microarrays and primary antibodies against human CLEC-2 (Anti-CLEC-2 antibody, orbr3344, Biobirt, diluted 1/100) was used. Antibody specificity was confirmed by immunochemistry and western blot. The staining results were scanned by a microscopy system (Leica DM6000 B, Leica Microsystems GmbH, Mannheim, Germany). We recorded images with Leica CV-M2CL camera and analyzed them with Leica Ariol 4.0 software. Specimens were considered as high CLEC-2 positive cell infiltration if there were over 53 CLEC-2 positive cells in a tissue core, otherwise low CLEC-2 positive cell infiltration. Each patient had two tissue cores, and the number of CLEC-2 positive cells calculated as the average of them. The cutoff point was selected according to optimal p value with X-tile, version 3.6.1 (Yale University, New Haven, Connecticut) [23].
Statistical analysis
The relationship between CLEC-2 positive cell infiltration and clinicopathological parameters of the patients was assessed by χ2 test, Fisher's exact method and Cochran-Mantel-Haenszel χ2 test. Survival curves were established with Kaplan-Meier method and compared with log-rank test. Univariate and multivariate Cox proportional hazard models were used to evaluate the HR (Hazard ratio) and 95% CI (confidence interval). Two nomograms were formed to predict the OS and RFS. We calculated concordance index to compare the prognostic or predictive accuracy of different models. Statistical analyses were performed using SPSS Statistics 21.0 (SPSS Inc., Chicago, IL), R software version 3.0.2 with the “rms” package (R Foundation for Statistical Computing, Vienna, Austria) and Stata (version 12.1; StataCorp LP, TX, USA). All statistical tests were 2-sided and P < .05 was regarded as statistically significant.
SUPPLEMENTARY MATERIALS TABLES
Acknowledgments
Y. Xiong for acquisition of data, analysis and interpretation of data, statistical analysis and drafting of the manuscript; L. Liu, Y. Xia, J. Wang, W. Xi, Q. Bai,Y. Qu and Q. Long for technical and material support; J. Xu and J. Guo for study concept and design, analysis and interpretation of data, drafting of the manuscript, obtained funding and study supervision. All authors read and approved the final manuscript. The authors would like to thank Dr. Yuan Ji, Dr. Jun Hou and Ms. Haiying Zeng (Department of Pathology, Zhongshan Hospital of Fudan University) for diagnosis confirmation and technical assistance, respectively.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
GRANT SUPPORT
This study was funded by grants from National Key Projects for Infectious Diseases of China (2012ZX10002012-007, 2016ZX10002018-008), National Natural Science Foundation of China (31100629, 31270863, 81372755, 31470794, 81401988, 81402082, 81402085, 81471621, 81472227, 81472376, 31570803, 81501999 and 81572352), Science and Technology Commission of Shanghai Municipality (14ZR 1406300) and Program for New Century Excellent Talents in University (NCET-13-0146). All these study sponsors have no roles in the study design, in the collection, analysis, and interpretation of data.
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, Parkin DM, Ferlay J, Li L, Chen Y. Estimates of cancer incidence in China for 2000 and projections for 2005. Cancer epidemiology, biomarkers & prevention. 2005;14:243–250. [PubMed] [Google Scholar]
- 3.Stewart GD, O'Mahony FC, Powles T, Riddick AC, Harrison DJ, Faratian D. What can molecular pathology contribute to the management of renal cell carcinoma? Nature reviews Urology. 2011;8:255–265. doi: 10.1038/nrurol.2011.43. [DOI] [PubMed] [Google Scholar]
- 4.Volpe A, Patard JJ. Prognostic factors in renal cell carcinoma. World journal of urology. 2010;28:319–327. doi: 10.1007/s00345-010-0540-8. [DOI] [PubMed] [Google Scholar]
- 5.Colonna M, Samaridis J, Angman L. Molecular characterization of two novel C-type lectin-like receptors, one of which is selectively expressed in human dendritic cells. European journal of immunology. 2000;30:697–704. doi: 10.1002/1521-4141(200002)30:2<697::AID-IMMU697>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 6.Mourao-Sa D, Robinson MJ, Zelenay S, Sancho D, Chakravarty P, Larsen R, Plantinga M, Van Rooijen N, Soares MP, Lambrecht B, Reis e Sousa C. CLEC-2 signaling via Syk in myeloid cells can regulate inflammatory responses. European journal of immunology. 2011;41:3040–3053. doi: 10.1002/eji.201141641. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki-Inoue K, Kato Y, Inoue O, Kaneko MK, Mishima K, Yatomi Y, Yamazaki Y, Narimatsu H, Ozaki Y. Involvement of the snake toxin receptor CLEC-2, in podoplanin-mediated platelet activation, by cancer cells. The Journal of biological chemistry. 2007;282:25993–26001. doi: 10.1074/jbc.M702327200. [DOI] [PubMed] [Google Scholar]
- 8.Lowe KL, Navarro-Nunez L, Watson SP. Platelet CLEC-2 and podoplanin in cancer metastasis. Thrombosis research. 2012;129:S30–37. doi: 10.1016/S0049-3848(12)70013-0. [DOI] [PubMed] [Google Scholar]
- 9.Kerrigan AM, Dennehy KM, Mourao-Sa D, Faro-Trindade I, Willment JA, Taylor PR, Eble JA, Reis e Sousa C, Brown GD. CLEC-2 is a phagocytic activation receptor expressed on murine peripheral blood neutrophils. Journal of immunology. 2009;182:4150–4157. doi: 10.4049/jimmunol.0802808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annual review of immunology. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 11.Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C, Forster I, Ruland J. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 12.Goodridge HS, Shimada T, Wolf AJ, Hsu YM, Becker CA, Lin X, Underhill DM. Differential use of CARD9 by dectin-1 in macrophages and dendritic cells. Journal of immunology. 2009;182:1146–1154. doi: 10.4049/jimmunol.182.2.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schacht V, Dadras SS, Johnson LA, Jackson DG, Hong YK, Detmar M. Up-regulation of the lymphatic marker podoplanin, a mucin-type transmembrane glycoprotein, in human squamous cell carcinomas and germ cell tumors. The American journal of pathology. 2005;166:913–921. doi: 10.1016/S0002-9440(10)62311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer science. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin-Villar E, Megias D, Castel S, Yurrita MM, Vilaro S, Quintanilla M. Podoplanin binds ERM proteins to activate RhoA and promote epithelial-mesenchymal transition. Journal of cell science. 2006;119(Pt 21):4541–4553. doi: 10.1242/jcs.03218. [DOI] [PubMed] [Google Scholar]
- 16.Ljungberg B, Bensalah K, Canfield S, Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L, Merseburger AS, Mulders P, Powles T, Staehler M, et al. EAU guidelines on renal cell carcinoma: 2014 update. European urology. 2015;67:913–924. doi: 10.1016/j.eururo.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Kim SP, Alt AL, Weight CJ, Costello BA, Cheville JC, Lohse C, Allmer C, Leibovich BC. Independent validation of the 2010 American Joint Committee on Cancer TNM classification for renal cell carcinoma: results from a large, single institution cohort. The Journal of urology. 2011;185:2035–2039. doi: 10.1016/j.juro.2011.02.059. [DOI] [PubMed] [Google Scholar]
- 18.Delahunt B, Cheville JC, Martignoni G, Humphrey PA, Magi-Galluzzi C, McKenney J, Egevad L, Algaba F, Moch H, Grignon DJ, Montironi R, Srigley JR, Members of the IRTP The International Society of Urological Pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. The American journal of surgical pathology. 2013;37:1490–1504. doi: 10.1097/PAS.0b013e318299f0fb. [DOI] [PubMed] [Google Scholar]
- 19.Leibovich BC, Blute ML, Cheville JC, Lohse CM, Frank I, Kwon ED, Weaver AL, Parker AS, Zincke H. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: a stratification tool for prospective clinical trials. Cancer. 2003;97:1663–1671. doi: 10.1002/cncr.11234. [DOI] [PubMed] [Google Scholar]
- 20.Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. The Journal of urology. 2002;168:2395–2400. doi: 10.1016/S0022-5347(05)64153-5. [DOI] [PubMed] [Google Scholar]
- 21.Zisman A, Pantuck AJ, Wieder J, Chao DH, Dorey F, Said JW, deKernion JB, Figlin RA, Belldegrun AS. Risk group assessment and clinical outcome algorithm to predict the natural history of patients with surgically resected renal cell carcinoma. Journal of clinical oncology. 2002;20:4559–4566. doi: 10.1200/JCO.2002.05.111. [DOI] [PubMed] [Google Scholar]
- 22.Pan D, Xu L, Liu H, Zhang W, Zhu Y, Xu J, Gu J. Interleukin-11 receptor predicts post-operative clinical outcome in patients with early-stage clear-cell renal cell carcinoma. Japanese journal of clinical oncology. 2015;45:202–209. doi: 10.1093/jjco/hyu194. [DOI] [PubMed] [Google Scholar]
- 23.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clinical cancer research. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.