Abstract
The rumen is lined on the luminal side by a stratified squamous epithelium that is responsible for not only absorption, but also transport, extensive short-chain fatty acid (SCFA) metabolism and protection. Butyrate has been demonstrated to initiate the differentiation of the tissue following introduction of solid feed to the weaning neonate as well as affecting the metabolism of other nutrients and absorption of nutrients in in vitro experiments. The objective of the present study was to validate expression stability of eight putative reference genes bovine rumen, considering the intrinsic heterogeneity of bovine rumen with regard to different luminal characteristics due to direct infusion of butyrate to double the intra-ruminal content of the rumen liquor. Our focus was on identifying stable reference genes which are suitable to normalize real-time RT-qPCR experiments from rumen samples collected from clinical assays, irrespective of localization within the organ and the across physiological state. The most stably expressed genes included: ACTB, UXT, DBNDD2, RPS9, DDX54 and HMBS. Their high stability values suggest these reference genes will facilitate better evaluation of variation of across an array of conditions including: localization within the rumen, differences among cattle fed an array of rations, as well as response to development in the weaning animal. Moreover, we anticipate these reference genes may be useful for expression studies in other ruminants.
Introduction
Given the unique placement of the organ in the gastrointestinal tract of the ruminant animal, the ruminal epithelium plays a vital and unique role in its health and productivity. Moreover, the ruminal epithelium is not simply a barrier to nutrient diffusion, but through the metabolism of short chain fatty acids (SCFA), primarily butyrate, acts to maintain the integrity of metabolite concentration gradients. In addition, ruminal SCFA metabolism protects against potentially detrimental decreases in blood pH [1,2]. Acetate, propionate and butyrate are metabolized to different extents by the ruminal epithelium. The impact of butyrate on the tissue is actively being investigated as a putative on-farm additive to enhance rumen development in weanling animals. Factors contributing to variation in feed efficiency among dairy cattle are not well understood, but likely include differences in basal energy requirements, levels of tissue metabolism, stress, and physical activity, and differences in nutrient digestibility or metabolism (e.g., during rumen microbial fermentation: [3]). Furthermore, these physiological factors likely have a genetic component [4]. Investigations into the molecular and genetic/epigenetic aspects of feed efficiency are needed to develop management strategies and selective tools for enhancing nutrient use efficiency by dairy cattle.
Visceral tissues represent 8–12% of body mass, yet account for a highly variable, and disproportionate amount of energy and nutrient use in ruminants (50–60% of total body nutrient use [5]). To date, our understanding of regulation of development of these organs is limited, in part, due to the inability to assess temporal changes in the gastrointestinal (GI) tract in vivo. Use of biopsy approaches can at a minimum provide invaluable information regarding the expression of genes under the influence of an array of luminal conditions.
Existing research on this subject was conducted in vitro using cell lines and short-term culture [6,7,8], or in vivo using experimental treatments and slaughter [8,9], combined with cell culture. Using biopsy procedures developed previously permits examination of the timeline of intestinal adaptive responses in vivo and enables critical sample collection without slaughter.
We sought to evaluate different reference genes for their potential use as internal normalization controls in gene expression measurements of rumen epithelial tissue from ontogenic studies in vivo to assess potential for luminal factors to impact tissue gene expression. In order to reduce the likelihood that our candidate genes exhibited regulated covariation, we chose a group of eight references with varied roles in cellular metabolism and molecular functions such as cytoskeletal structure (β-actin, ACTB), protein interactions (dystrobrevin binding protein, DBNDD2), ribosome biogenesis (DEAD box polypeptide 54, DDX54), glucose metabolism (glyceraldehyde-3-phosphate dehydrogenase, GAPDH), general metabolism (hydroxymethylbilane synthase, HMBS), matrix organization and collagen formation (cyclophilin B, PPIB), ribosomal structure (ribosomal protein S9, RPS9), and signaling pathways (prefoldin-like chaperone, UXT).
We attempted to (1) address each critical issue that needs to be carefully considered in a qPCR assay, and (2) identify a highly stable set of reference genes for butyrate-rumen-based experiments. Our results demonstrate highly stable values thus they may serve as useful guidelines or as a foundation point for reference gene selection for expression studies in other ruminants.
Material and methods
All procedures involving animals were approved by the ARS Northeast Area Beltsville Location Institutional Animal Care and Use Committee protocol number 12–008.
Animals and treatments
Holstein dry cows (n = 6) were surgically fitted with rumen fistulae using established methods. After recovery, cows were housed in tie stalls with ad libitum access to water and were maintained on the standard dry cow ration for the entirety of the experiment. Cows were allowed exercise daily, which was limited to 1 h/d during the infusion portion of the experiment. Ruminal infusion of butyrate was initiated immediately following 0h sampling (baseline controls) and thereafter continued for 168h at a rate of 5.0L/d of a solution representing > 10% of daily anticipated metabolizable energy intake in a buffered saliva solution (pH 7.0; 3.8% KHCO3, 7.3%NaHCO3) as a continuous infusion. Following the 168h infusion, infusion was terminated and cows were maintained on the dry cow ration for an additional 168h for sampling. Rumen epithelial samples were serially collected via biopsy through rumen fistulae at 0, 24, 72 and 168h of infusion, and 168h post infusion. Rumen epithelial samples were saved in RNAlater (Life Technologies, Grand Island, NY) and stored in −80C until RNA extraction.
Sampling
Rumen biopate (papillae) were collected by grab biopsy from the ventral cranial sac of the rumen and immediately processed for RNA collection. Briefly, rumen papillae samples were washed in large volumes of physiological saline to remove visible debris before being stabilized in RNAlater according to the manufacturer’s instructions, and stored at −80°C until RNA extraction. Tissues were homogenized in Qiazol Lysis Reagent (Qiagen, Valencia, CA) and total RNA was extracted using the RNeasy Mini Kit (Qiagen) with on-column DNase digestion.
RNA concentration was determined by measuring the optical density at 260 nm using a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA quality was assessed by combining information from several control steps. First, purity was inferred from the absorption ratios using the NanoDrop. Only the samples with A260/A280 absorption ratios from 1.8–2.2 and A260/A230 > 1.5 were used in the analysis. Then RNA integrity was assessed by microcapillary electrophoresis with an Experian RNA StdSens Chip and Experion Bioanalyzer (Bio-Rad Laboratories, Hercules, CA), showing rRNA subunits with 18S/28S peaks on the virtual gel and electropherograms. Only RNA samples with electropherograms of high quality and RQI ≥ 8 were used for further study. All samples were pure and free from protein and organic pollutants derived from the RNA extraction. After performing RNA quality controls, RNA was deemed as suitable for qPCR analysis.
First strand cDNA synthesis and genomic DNA contamination
RNA samples were adjusted to the same concentration, measured on NanoDrop and adjusted again in order to equalize the RNA input in the subsequent reverse-transcription reaction. Then, RNA (1 μg) was reversed-transcribed with a blend of oligo(dT) and random hexamers primers using the iScript cDNA synthesis kit (Bio-Rad), according to the manufacturer’s instructions. The cDNAs were diluted to a final volume of 200 μl. Absence of genomic DNA contamination in RNA samples was tested by PCR using primers designed to amplify a 716-bp fragment (spanning exons 4, 5 and 6) of the β-actin gene (AC_000182exon4: 5’-GGCTACAGCTTCACCACCAC-3’, AC_000182exon6: 5’-ACTCCTGCTTGCTGATCCAC-3’). A quantity equivalent to the cDNA used as template in the subsequent amplification PCR (i.e., 5 ng of gDNA) was used as positive control. Clear distinguishable 496-bp bands were found over the cDNA samples. No cDNA showed the 716-bp fragment. As second way to test the gDNA contamination, we designed (if possible) and checked reference genes primers to generate amplicons with different length from gDNA or cDNA (Table 1). S1 Fig. shows those primer pairs positions mapped to their respective gene sequence using Bioconductor and the IRanges package [10,11]. After performing gDNA control, the cDNA samples were deemed as suitable for qPCR analysis.
Table 1. Description of reference genes and primer sequences.
Primer PCR efficiency and PCR Tm product data represent mean values ± SE (n = 30). PCR efficiencies (E) calculated according to the equation (1 + E) = 10slope. Free energy for potential secondary structures in the amplicon is shown (ΔG, kcal/mole).
| Gen Id | Symbol | Name | Primer Sequence (5'-3') | Exon | ΔG (kcal/mole) | PCR E | PCR product cDNA/gDNA | PCR product Tm (°C) |
|---|---|---|---|---|---|---|---|---|
| NM_174152 | PPIB | cyclophilin B | 4806CGGCAAAGTTCTAGAGGGCA | 4 | 0.55 | 1.06±0.08 | 85/830 | 83.18±0.25 |
| 5636CACGTCCTTCAGAGGCTTGT | 5 | |||||||
| NM_173979 | ACTB | β-actin | 2714AGTACTCCGTGTGGATTGGC | 5 | 0.59 | 1.00±0.04 | 78 | 81.22±0.28 |
| 2791ACTCCTGCTTGCTGATCCAC | 5 | |||||||
| NM_001046207 | HMBS | hydroxymethylbilane synthase | 3468ACCGCGCTCTCTAAGATTGG | 4–5 | 0.34 | 1.01±0.06 | 66/155 | 80.50±0.15 |
| 3622CCTCTCCAAAGCATGCTCCA | 5 | |||||||
| NM_001034034 | GAPDH | glyceraldehyde-3-phosphate dehydrogenase | 3374GTCATCCCTGAGCTCAACGG | 7 | 0.07 | 1.00±0.05 | 69/155 | 79.57±0.22 |
| 3528AACAGACACGTTGGGAGTGG | 8 | |||||||
| NM_001037471 | UXT | prefoldin-like chaperone | 6090CACATGTTGCTAGAGGGGCT | 5–6 | 0.5 | 1.01±0.06 | 66/281 | 81.18±0.28 |
| 6370TCAGTGCTGAGTCTCTGGGA | 6 | |||||||
| NM_001130748 | DBNDD2 | dystrobrevin binding protein | 2270GTGGAGCTTATCGACCTGGG | 2 | 0.82 | 0.99±0.07 | 74 | 82.00±0.19 |
| 2343GGAGTTGGTGGAGGGTCTTC | 2 | |||||||
| XM_002694516 | DDX54 | DEAD box polypeptide 54 | 15818AAGAAGCGGTTTGTGGGACA | 18 | 0.9 | 0.99±0.06 | 74 | 84.43±0.17 |
| 15891CTGATGTAGCGGCCACTCTC | 18 | |||||||
| BC148016 | RPS9 | ribosomal protein S9 | 6113TGCTGGATGAGGGCAAGATG | 3 | 0.98 | 1.02±0.03 | 75 | 84.37±0.22 |
| 6187GCAGGCGTCTCTCCAAGAAA | 3 |
Selection of internal control genes
We selected several candidate reference genes based on the following criteria: (i) sequences should encode for proteins that perform varied roles in cellular metabolism with different molecular functions in order to minimize the effect of co-regulation, (ii) sequences which had previous been assessed for stability in, at least, slightly similar biological contexts, and (iii) because we aimed to perform several in silico controls on the primer pairs and amplicons, sequences with reliable and accessible prior information. However, we designed the primers de novo to meet our quality control criteria. Thus, qPCR primers were designed with the following criteria: lengths of 20 nucleotides, Tm of 60±1°C and GC content of 50–60% yielding PCR amplicon lengths of 60–90 nucleotides. We chose candidate sequences from studies aimed to measure mRNA abundance of rumen epithelial genes [12], genes involved in ruminal development [13], glucose transporters in the mammary gland [14] or orthologs of stable references previously used in different tissues in the phylogenetically bovine closely related Capra hircus [15,16]. All qPCR primers were tested for specificity using NCBI’s BLAST software [17]. For the prediction of secondary structure of the amplicons, we used the mfold Web Server with default settings of minimal free energy, 50 mM Na+, 3mM Mg2+, and annealing temperature of 60°C [18]. We chose primers that would yield amplicons with minimal secondary structures and melting temperatures that would not hamper annealing. We ultimately selected 8 primer pairs that met our quality controls. Designed primers were synthesized by Integrated DNA Technologies (Coralville, IA). Table 1 shows the overall mean real-time qPCR amplification efficiency of each primer pair (E) estimated from the data obtained from the exponential phase of each individual amplification plot and the equation (1+E) = 10slope using LinReg software and the criteria of including three to five fluorescent data points with R2≥0.995 to define a linear regression line [19]
Real-time qPCR assays
PCR reactions were carried out in an IQ5 (Bio-Rad) thermal cycler using SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad) to monitor dsDNA synthesis. Reactions contained 1 μl of the diluted cDNA as a template and 0.150 μM of each primer in a total volume reaction of 20 μl. Master mix was prepared and dispensed into individual wells using electronic Eppendorf Xplorer® multipipettes (Eppendorf AG, Hamburg, Germany). The following standard thermal profile was used for all qPCRs: polymerase activation (95°C for 3 min), amplification and quantification cycles repeated 40 times (95°C for 30 sec, 60°C for 1 min). The specificity of the primer pairs was checked by melting-curve analysis performed by the PCR machine after 40 amplification cycles (60 to 95°C) and is shown in S2 Fig. Fluorescence was analyzed using iQ5 2.1 standard optical system analysis software v2.1 (Bio-Rad). All amplification plots were analysed using a base line threshold of 30 relative fluorescence units (RFU) to obtain Cq (quantification cycle) values for each gene-cDNA combination.
Data analysis
To determine which reference genes were best suited for transcript normalization in bovine rumen, we first used the statistical algorithm geNorm [20]. In a second approach, the coefficient of variation of normalized relative expression levels was calculated based on the formulas (formula 11, 13, 15, 17, 18, 19 and 20) described in the qBase software [21]. In short, Cq values were incorporated into an in-house developed R script [22] and transformed into relative quantities (RQs) using the efficiency of each primer pair and the sample with the lowest Cq as a calibrator. Then, a sample-specific normalization factor (NF) was estimated as a geometric mean of RQs for the candidate genes. Finally, the mean coefficient of variation (CV) for all reference genes was calculated as the arithmetic mean of the CV estimated from the different reference genes.
Assessment of normalization
To validate the reliability of the results we addressed the influence of the choice of one reference gene on the interpretation of output data. First, we estimated two normalization factors (determined by calculating the geometric mean of the two best-scored and the two worst-scored reference genes) and we plotted the normalized relative quantities for two genes over the time-course experiment and for all samples. As a second approach to validate the stable expression of the references, the expression level of a gene of interest encoding a regulator of chromosome condensation (RCC2, NM_001101911F: 5’-CCTGTGGGGCTGAATTCAGT-3’; NM_001101911FRv: 5’-GTTGTGTCCCAGCTGACCAT-3’) was quantified at two different time points (0–14 days) and was normalized to those normalization factors. Finally, we addressed the normalization based on the use of one single reference gene. Expression level ratios for RCC2 were quantified at days 0 and 14 normalized to each of the reference genes using the delta-delta method modified by the efficiency correction as described by [23].
Results and discussion
Previously, in lactating cattle, similar infusion approaches elicited changes in a wide array of gene families [24]. Prior experiments, in lactating cattle [24,25], demonstrated that butyrate concentration in the rumen attained pleateau by 168 h, and had largely returned to pre-infusion concentrations by 168 post removal. Thus, changes reflecting new expression levels are best assessed at these time points while changes occurring in 24 and 48 h are more likely to reflect gene expression changes in response to the luminal environment and thus, will likely lend insight into mechanism of action accounting for the response.
Quality controls and overall gene expression
Due to the difficulty in comparing various data sets related to perturbation of rumen SCFA concentrations (e.g., sub acute ruminal acidosis models, direct infusion, physiological status[8]) and putative power of RNAseq as a tool for assessment of transcriptomic changes in response, a validated set of reference sequences will enhance future interpretations. We anticipated a need for a solid validation of a select set of potential reference genes using two bioinformatics algorithms: geNorm and qBase. Factors known to affect the reliability of gene expression data such as RNA quality, DNase treatment, two-step RT-qPCR, the use of the same RT master mix that generated one cDNA batch, primer design keeping in mind the presence of secondary structures in the primer as well as in the amplicon, PCR efficiency correction, and non-specific amplification were closely controlled during the experiment. High-quality total RNA was obtained and evaluated by microcapillary electrophoresis and absorbance ratios. To exclude genomic DNA contamination, a fragment of ACTB cDNA (GenBank accession number NM_173979) was amplified with primers designed to span two introns to generate bands clearly distinguishable between cDNA and gDNA. No cDNA showed the 716-bp fragment characteristic from gDNA. Also, for some sequences was possible to design assays with primers spanning an intron. Primer position on the sequence, number of exon and different sizes for amplicons that we used to confirm that our material was free of gDNA contamination is shown in Table 1. After performing the in silico quality controls on primer pairs and amplicon sequences described in the Materials and Methods, 8 primer pairs from these genes were retained for the experimental analysis. Amplification specificity of all SYBR Green assays was confirmed by running a dissociation protocol using incremental temperatures to 95°C. The melting curve analysis showed that each of the 8 primer pairs amplified a single product.
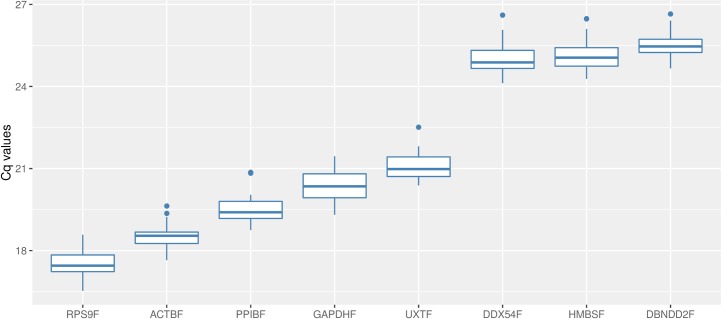
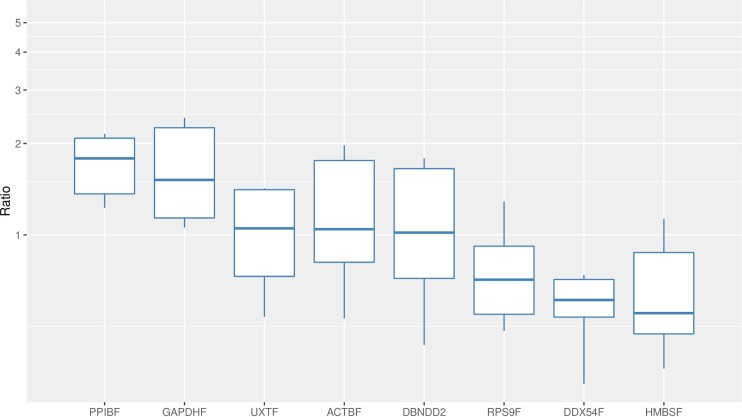
Real-time qPCR reactions provided data within variable Cq value ranges for the 8 references. RPS9F had the lowest mean Cq value (17.51), while DBNDD2F had the highest one (25.52), showing an expression level approximately >250-fold lower than RPS9F (Fig 1). Preliminary analysis of data showed that individual reference genes had similar Cq values variation across all studied samples (~2 cycles). The smallest variation was observed for ACTBF (1.98 cycles), while DDX54F was the gene with the widest range (2.49 cycles). All assays were found to have high PCR efficiency of amplification. Amplification efficiencies varied from 0.988 for DDX54F to 1.057 for PPIBF indicating comparable amplification in the 30 cDNAs tested (Table 1).
Fig 1. Boxplot of Cq values.
Variations of the quantification cycle are shown for all examined reference genes sorted by expression range (n = 30). Each box represents the 50% quartile; the median is figured as the line in each box; whiskers depict the 25% minimum and maximum range of the Cq value.
Stability analysis
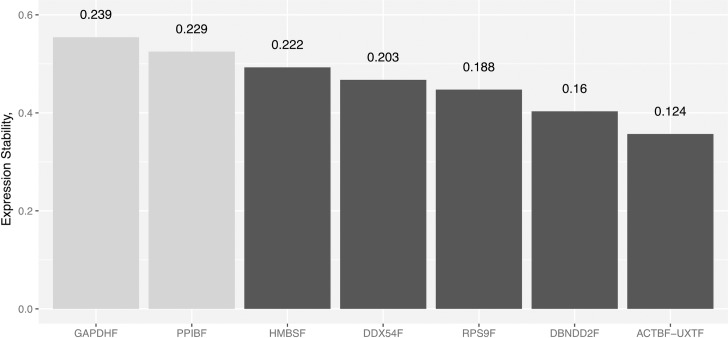
To analyze the stability of the expression and identify suitable reference genes, we used the statistical algorithm geNorm [20]. The geNorm software tool represents the most commonly used algorithm to evaluate candidate reference genes and has established itself as the de facto standard method [26,27]. The program defines a stability measure (M) as the average pairwise variation between a gene and all other reference genes. The stability measurement relies on the principle that the expression ratios of two ideal internal genes are identical in all samples, regardless of the experimental condition. The lower the M value, the more stably expressed is the gene. In a second approach, the CV for all reference genes were determined based on the equations defined by the qBase framework [21]. Fig 2 shows the ranking of the genes tested in our samples according to their M and CV values. In our material, without exception, stability values were much lower than the default threshold of 1.5 defined in the algorithm. In fact, none of the tested candidate reference genes exceed the M value = 0.6. In addition, 6 out the 8 references show M values < 0.5 indicating that their stable expression makes them highly suitable as internal references in rumen gene expression analysis. geNorm algorithm indicated that ACTBF and UXTF are the most stable genes (M = 0.36), whereas the lowest stability value was recorded for the GAPDH gene (M = 0.55). Genes with the values inside the optimal range for homogeneous sample panels (M and CV values lower than 0.5 and 25% respectively) according to [21] were the most stably expressed and included: ACTBF, UXTF, DBNDD2F, RPS9F, DDX54F and HMBSF (M = 0.49 and CV = 0.222).
Fig 2. geNorm ranking of 8 reference genes from bovine rumen samples.
Vertical numbers at the top indicate the CV values of the reference genes involved in the normalization. References showing highly stable expression (M values < 0.5) are represented as black bars.
Validation of reference genes
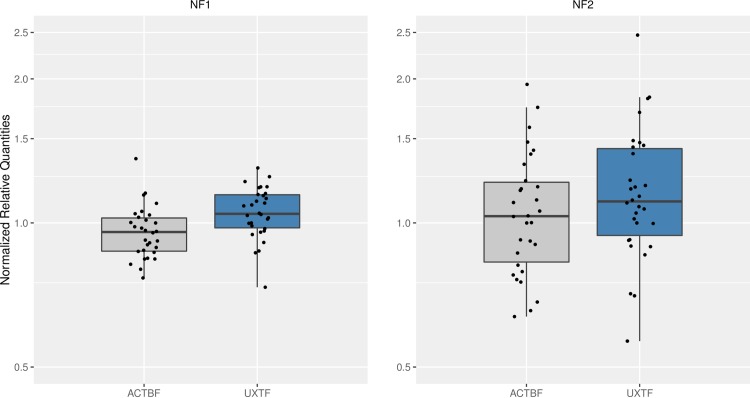
To test the putative reference genes identified above, we estimated the normalized relative quantities of the two best reference set across the samples compared to their overall geometric mean. The values were closely distributed around 1-fold and only a small difference between samples could be detected (1.77-fold on average) when we used the two best-scored references as calibrators (Fig 3). When we normalized the expression of the same genes against the geometric mean of the references with the highest M value, the values were also closely distributed around 1-fold. However, we clearly observed average differences of over 3-fold between samples (3.70-fold). This illustrates that the selection of the references matters and how in the second case, by using a references pair that it is not the optimal, we add technical variation during the normalization step instead of removing variation (Fig 3).
Fig 3. Evaluation of reference genes for bovine rumen.
The relative quantities of genes ACTBF and UXTF were normalized for each sample against the 2 best-scored (NF1) and the 2 worst-scored references (NF2). Y-axis is shown in logarithmic scale.
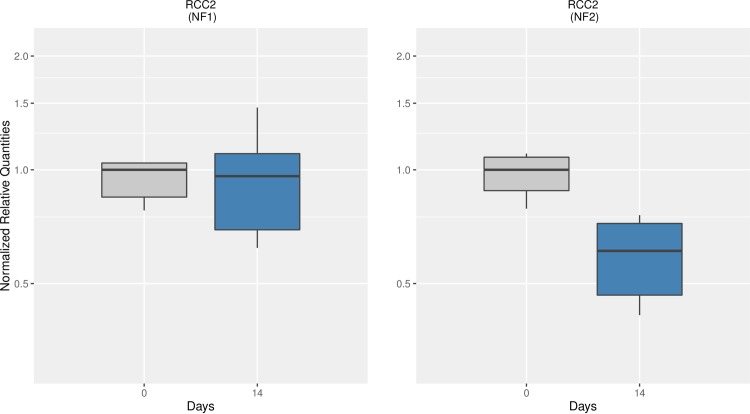
In a second attempt to validate the stability of the references, we monitored the mean expression of one target gene at days 0 and 14. Normalized relative quantities of the regulator of chromosome condensation RCC2 were obtained using two normalization factors: again, the use of the 2 best-scored references (NF1) and the use of the 2 worst-scored references (NF2). Fig 4 shows the relationship of the transcript levels between those days. Fourteen days after butyrate infusion, we did not observe any difference in the amount of RCC2 transcripts in the rumen epithelium when the normalization factor NF1 was used. However, when we measured the transcriptional activity of the same gene, on the same subjects, at the same time-points but using the NF2, the mRNA expression levels of RCC2 decreased about ~ 2-fold. The shift in the average expression levels is due to solely the selection of the reference genes to build the normalization factor. Therefore, selection of references without previous knowledge of their expression stability values are likely to create technical artifacts due to their intrinsic variation.
Fig 4. Normalized relative quantities of RCC2 over days 0 and 14.
Data were normalized for each sample against the 2 best-scored (NF1) and the 2 worst-scored references (NF2). NRQ were rescaled to day 0 that was arbitrarily set as 1.
Single reference-based normalization
One of the most intriguing issues in the field of qPCR relates to normalization using only one single non-validated reference gene. Despite the vast number of publications showing that the use of a single reference yields unreliable data, this approach is still routinely used in animal, plant and biomedical research. After the publication of the MIQE guidelines, Short [28] noted that only 10.5% of qPCR analyses published in three leading high-impact journals used more than one single reference gene. The high stability expression levels found in our study (low M and CV values) might lead one to think that our results are not compromise by the use of one single reference gene during the normalization. In order to test this hypothesis, we quantified again the ratio of RCC2 at days 0 and 14, but this time we normalized that level relative to each of the 8 stable reference genes identified in the study. Normalization of RCC2 mRNA with the various references showed that the ratios obtained were highly different depending on the reference gene used. It is noteworthy to mention that we found even contradictory results, as some ratios suggest up-regulation of RCC2 (ratio > 1-fold) whereas some ratios indicate down-regulation over the same period of time (ratio < 1-fold). The average RCC2 mRNA ratio (0–14 days) was 1.31-fold but it ranged from being down-regulated (1.75-fold) to being up-regulated (3.17-fold), depending only on the normalizer chosen (Fig 5). Thus, even working with highly stable genes across a given dataset, as in our case here, we found that differences in expression levels of ~5.6-fold can be generated based on only the choice of the single reference gene. It is likely that those differences will be greater if the reference has not been validated or worse, if it is a non-stable reference gene. With this analysis we demonstrate that the use of one non-validated reference gene is also a source of introduction of technical variation, may create confounding variation and may lead to misinterpretation of our conclusions. This also points out the highly improbable chance that small differences in gene expression, such as those a priori expected for a transcription factor or other regulators, could be detected based on the sole use of one reference gene.
Fig 5. Normalization of RCC2 mRNA expression at 0 and 14 days after butyrate infusion using 1 single reference gene.
The X-axis shows the internal gene that was used for estimating each ratio. Y-axis is shown in logarithmic scale.
Conclusions
Increasingly there is interest in understanding the impact of visceral organ impact, including gastrointestinal tissues, on animal biology across agricultural species. Therefore identifying a stable set of quality reference genes which are suitable to normalize real-time RT-qPCR experiments from rumen epithelial tissue samples is needed. We have found that the most stably expressed genes were ACTB, UXT, DBNDD2, RPS9, DDX54 and HMBS and their high stability indicate that these reference genes will facilitate better evaluation of variation across an array of physiological conditions including: localization within the rumen, differences among cattle fed an array of rations, as well as response to development in the weaning animal. Moreover, we anticipate these reference genes may be useful for expression studies in other ruminants.
Supporting information
X-axis represent fragment length in bp. Bottom row on X-axis shows number and position of exons. On top of exons, primer sequences forward and reverse are shown. Red color denotates forward primer spanning an intron.
(TIF)
(TIF)
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the Fulbright/Ministry of Education of Spain Visiting Scholarship PRX14/00601 to Dr. María José Ranilla. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stevens, CE. Fatty acid transport through the rumen epithelium. In: Phillipson, AT editor. Physiology of Digestion and Metabolism in the RuminantOriel Press, Ltd., Newcastle upon Tyne, England;1969. pp. 101–112.
- 2.Galfi, P, S Neogrady and T Sakata. Effects of volatile fatty acids on the epithelial cell proliferation of the digestive tract and its hormonal mediation. In:Tsuda, T., Y. Sasaki and R. Kawashima editors. Physiological Aspects of Digestion and Metabolism in Ruminants:Proceedings of the Seventh International Symposium on Ruminant PhysiologyAcademic Press, Inc., San Diego. 1991. pp. 49-59.
- 3.Greenfield TL, Baldwin RL, Erdman RA, McLeod KR. Ruminal fermentation and intestinal flow of nutrients by lactating cows consuming brown midrib corn silages Journal of dairy science 2001;84 (11): 2469–2477. 10.3168/jds.S0022-0302(01)74697-8 [DOI] [PubMed] [Google Scholar]
- 4.Crews DH Jr. Genetics of efficient feed utilization and national cattle evaluation: A review. Genet. Mol. Res. 2005;4:152–165. [PubMed] [Google Scholar]
- 5.McLeod KR, Baldwin RL. Effects of diet forage: concentrate ratio and metabolizable energy intake on visceral organ growth and in vitro oxidative capacity of gut tissues in sheep. Journal of Animal Science 2000;78 (3):, 760–770. [DOI] [PubMed] [Google Scholar]
- 6.Wang L-Q, Baldwin RL VI and Jesse BW. Identification of two cDNA clones encoding small proline-rich proteins expressed in sheep ruminal epithelium. Biochem. J. 1996:317: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane MA, Baldwin RL VI, Jesse BW Developmental changes in ketogenic enzyme gene expression during sheep rumen development. J Anim Sci 2002; 80:1538–1544. [DOI] [PubMed] [Google Scholar]
- 8.Penner GB, Steele MA, Aschenbach JR, McBride BW. Ruminant nutrition symposium: molecular adaptation of ruminal epithelia to highly fermentable diets. J Anim Sci 2011; 89:1108–1119. 10.2527/jas.2010-3378 [DOI] [PubMed] [Google Scholar]
- 9.Steele MA, Vandervoort G, AlZahal O, Hook SE, Matthews JC, McBride BW. Rumen epithelial adaptation to high-grain diets involves the coordinated regulation of genes involved in cholesterol homeostasis. Physiol Genomics. 2011; 43:308–316. 10.1152/physiolgenomics.00117.2010 [DOI] [PubMed] [Google Scholar]
- 10.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, et al. Software for Computing and Annotating Genomic Ranges. Prlic A, editor. PLOS Comput Biol. Public Library of Science; 2013;9: e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. BioMed Central Ltd; 2004;5: R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yohe TT. Performance and Development of the Rumen in Holstein Bull Calves Fed an Aspergillus Oryzae Fermentation Extract. 2014. Available from: https://etd.ohiolink.edu/!etd.send_file?accession=osu1397769968&disposition=inline [DOI] [PubMed]
- 13.Naeem A, Drackley JK, Stamey J, Loor JJ. Role of metabolic and cellular proliferation genes in ruminal development in response to enhanced plane of nutrition in neonatal Holstein calves. J Dairy Sci 2012;95:1807–1820. 10.3168/jds.2011-4709 [DOI] [PubMed] [Google Scholar]
- 14.Gross JJ, Dorland HA, Wellnitz O, Bruckmaier RM. Glucose transport and milk secretion during manipulated plasma insulin and glucose concentrations and during LPS‐induced mastitis in dairy cows. Journal of Animal Physiology and Animal Nutrition. 2015;99: 747–756. 10.1111/jpn.12259 [DOI] [PubMed] [Google Scholar]
- 15.Tao H, Chang G, Xu T, Zhao H, Zhang K, Shen X. Feeding a High Concentrate Diet Down-Regulates Expression of ACACA, LPL and SCD and Modifies Milk Composition in Lactating Goats. PLoS ONE. 2015;10: e0130525 10.1371/journal.pone.0130525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu W, Lin Y, Liao H, Wang Y. Selection of Reference Genes for Gene Expression Studies Related to Intramuscular Fat Deposition in Capra hircus Skeletal Muscle. PLoS ONE. 2015;10: e0121280 10.1371/journal.pone.0121280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990; 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 18.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters. 2003;339: 62–66. [DOI] [PubMed] [Google Scholar]
- 20.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. BioMed Central Ltd; 2002;3: research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology. BioMed Central Ltd; 2007;8: R19 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2015. URL https://www.R-project.org/. [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29: e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldwin RL VI, Wu S, Li W, Li C. Bequette B J and Li RW. Quantification of transcriptome responses of the rumen epithelium to butyrate infusion using RNA-seq technology. Gene Regulation and Systems Biology. 2012; 6:67–80. 10.4137/GRSB.S9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C Li RW, Baldwin RL, Blomberg L, Wu S, Li W. Transcriptomic sequencing reveals a set of unique genes activated by butyrate-induced histone modification. Gene Regulation and Systems Biology. 2016;10: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227–230. 10.1016/j.ymeth.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 27.Jacob F, Guertler R, Naim S, Nixdorf S, Fedier A, Hacker NF, et al. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS ONE. Public Library of Science; 2013;8: e59180 10.1371/journal.pone.0059180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huggett JF, Bustin SA. Standardisation and reporting for nucleic acid quantification. Accred Qual Assur. Springer-Verlag; 2011;16: 399–405. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
X-axis represent fragment length in bp. Bottom row on X-axis shows number and position of exons. On top of exons, primer sequences forward and reverse are shown. Red color denotates forward primer spanning an intron.
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper.