Summary
Protein function originates from a cooperation of structural rigidity, dynamics at different timescales and allostery. However, how these three pillars of protein function are integrated is still only poorly understood. Here we show how these pillars are connected in Protein Tyrosine Phosphatase 1B (PTP1B), a drug target for diabetes and cancer that catalyzes the dephosphorylation of numerous substrates in essential signaling pathways. By combining new experimental and computational data on wt-PTP1B and ≥10 PTP1B variants in multiple states, we discovered a fundamental and evolutionarily conserved CH/π switch that is critical for positioning the catalytically important WPD loop. Furthermore, our data show that PTP1B uses conformational and dynamic allostery to regulate its activity. This shows that both conformational rigidity and dynamics are essential for controlling protein activity. This connection between rigidity and dynamics at different timescales is likely a hallmark of all enzyme function.
Keywords: protein tyrosine phosphatase, PTP1B, enzyme, NMR spectroscopy, X-ray crystallography, allostery
Graphical abstract
Introduction
Enzymes require conformational and dynamic events to perform chemistry. Some of these events are required for catalysis and some, for a subset of enzymes, are required for the allosteric control of the catalytic cycle. Investigations on the nature of the conformations, including balance between rigidity and flexibility, and the type of motions that control enzymes have only recently been initiated (Butterwick et al., 2004; Mandel et al., 1996). Protein tyrosine phosphatase 1B (PTP1B, PTPN1), the first non-receptor bound protein tyrosine phosphatase (PTP) isolated, is the best studied member of the human PTP family (Alonso et al., 2004). Since its discovery more than 25 years ago (Tonks et al., 1988), PTP1B has been shown to play a central role in multiple cellular processes, especially glucose uptake, body mass regulation, motility and proliferation. It does so by dephosphorylating multiple substrates including scaffolding proteins and receptor tyrosine kinases. Consequently, PTP1B is a validated target for multiple diseases, especially diabetes and cancer (Feldhammer et al., 2013).
Like all members of this family, PTP1B catalyzes the hydrolysis of phosphorylated tyrosine residues using a phosphocysteine intermediate (Pannifer et al., 1998). The catalytic site is defined by: (1) the PTP loop ([I/V]HCxxGxxR[S/T]G), which includes the catalytic cysteine, C215; (2) the WPD loop, 179WPD181, which contains the D181 that functions as the proton donor and acceptor during phosphoryl transfer; and (3) the substrate binding loop (SBL), which restricts dephosphorylation to tyrosine residues (Figures 1A and S1A)(Andersen et al., 2001). Substrate binding is accompanied by a rotation of the WPD loop from an open (hydrolysis incompetent) to a closed (hydrolysis competent) position (Figure 1B). Most human PTPs, including PTP1B, have catalytic hydrolysis rates (kcat) of ~15-60 s−1 and it was shown that in PTP1B the motion of the WPD-loop is the rate limiting step for hydrolysis (Whittier et al., 2013).
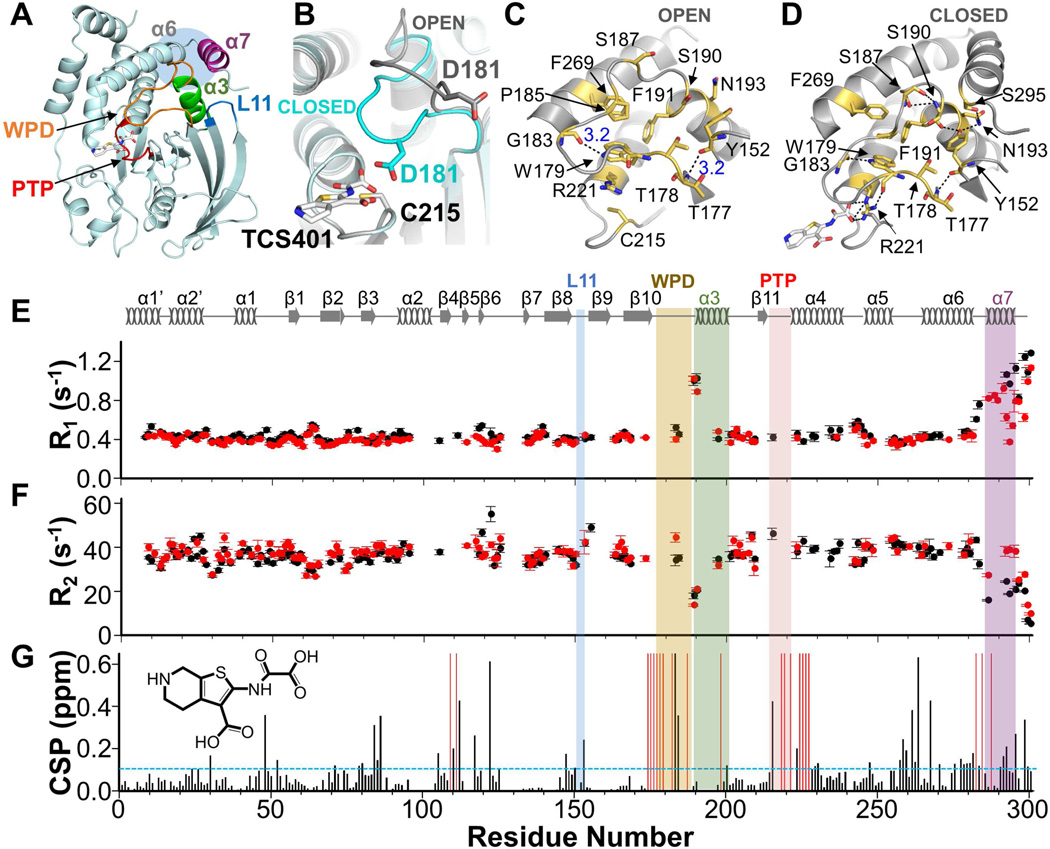
Figure 1. Dynamics and CSPs of PTP1B vs PTP1B:TCS401.
(A) PTP1B (PDB 5K9W); helices: α3 (green), α7 (purple); loops: WPD (orange), PTP (red), L11 (blue). Active site inhibitor TCS401 is shown as sticks. Blue shaded circle highlights the allosteric binding pocket.
(B) Open (gray, PDB 5K9V) and closed (cyan, PDB 5K9W) conformations of the WPD loop. C215, catalytic nucleophile, D181, catalytic acid/base and TCS401 are shown as sticks.
(C) Open state with key residues shown as sticks and labeled; polar interactions, black dotted lines.
(D) Same as (C), except closed state.
(E) 15N longitudinal R1 relaxation rates for PTP1B (black) and PTP1B:TCS401 (red); 850 MHz; 298 K. Secondary structures are shown.
(F) Same as (E) except 15N transverse R2 relaxation rates for PTP1B (black) and PTP1B:TCS401 (red) are shown.
(G) CSPs for PTP1B:TCS401 (1:3 molar ratio). Residues with significant CSPs are above the dashed line (σ= 0.11 ppm, standard deviation, sd). Residues broadened beyond detectability upon TCS401 binding are shown as red bars. Labeling as in (A). Chemical structure of TCS401 is shown.
See also Figures S1, S2.
Substrate binding initiates WPD loop closure. The PTP loop residue R221 then rotates about its γ-carbon to form a salt bridge with the bound phosphate (Pannifer et al., 1998) (Figures 1C and 1D). This rotation allows the WPD loop to close about the active site and positions D181 for hydrolysis. Although the structural changes that occur upon loop closure have been well characterized (Jia et al., 1995), a comprehensive understanding of the residues that are essential for WPD loop closure and, in turn PTP1B activity, is still missing.
Nearly a decade ago, PTP1B was discovered to have an allosteric binding pocket ~20 Å away from the catalytic site, at the intersection of helices α3-α6-α7 (Figure 1A) (Wiesmann et al., 2004). When benzofuran compounds bind this pocket, the activity of PTP1B is potently inhibited. Although initial molecular dynamics (MD) studies suggested that truncating and/or mutating helix α7 affects WPD loop closure (Li et al., 2014; Olmez and Alakent, 2011; Shinde and Sobhia, 2013), there is still no molecular model that explains how perturbations from the allosteric site are propagated to the active site. Furthermore, the role of dynamics and/or conformational changes in this process remain essential, open questions. To address these gaps, we employed a comprehensive experimental approach integrating NMR spectroscopy, X-ray crystallography, molecular dynamics (MD) simulations and biochemistry. Our data enabled us to define the complete regulatory network of PTP1B, which shows that PTP1B uses both conformational and dynamic allostery to communicate between its allosteric and active sites. We also discovered that WPD loop rigidity is essential for PTP1B function. Finally, we identified a chemically inert, distal residue (>15 Å from the catalytic center) that, when mutated, results in a folded but, remarkably, catalytically dead enzyme. Together, this work lays the foundation for a comprehensive understanding of conformation and dynamics in the entire PTP family specifically, and enzyme function and protein allostery generally.
Results
PTP1B helices α3 and α7, but not the WPD loop, exhibit fast timescale dynamics
We previously reported the sequence-specific backbone assignment of PTP1B (~35 kDa; residues 1-301; hereafter referred to as PTP1B) (Krishnan et al., 2014). While the sequence-specific backbone assignment is ~45% complete in helix α3 and ~50% complete in the E-loop (which contains the 100% conserved glutamic acid residue that coordinates the PTP loop and R221), the assignments for all other biologically important regions of PTP1B (PTP, WPD, Q, SBL, helices α6 and α7; Figures 1A and S1A) are highly complete. Missing NMR assignments in large proteins such as PTP1B are either due to slow H/D back exchange after protein expression in D2O-based medium or intermediate conformational exchange, which broadens peaks beyond detection (Peti and Page, 2016). As loops and helices often have fast H/D back exchange, it is most likely that residues in helix α3 and the E-loop are missing because of intermediate conformational exchange.
The chemical shift index (CSI; 13Cα and 13Cβ chemical shifts), which reports on protein secondary structural elements, correlates well with our PTP1B crystal structures (PTP1B; residues 1-301) for helices α1-α6. However, the lower than average CSI values for helix α7 show that this helix is not fully populated in solution (Figure S1B), an observation that correlates with a lack of density for helix α7 in multiple PTP1B crystal structures (Barford et al., 1994). Helix α7 is also not fully populated in longer PTP1B constructs (Krishnan et al., 2014), showing that this is not an artifact from working with only the PTP1B catalytic domain.
To detect PTP1B motions, we measured 15N NMR relaxation data (R1, R2) that probe fast timescale backbone dynamics. The data show that PTP1B largely exhibits uniform dynamics throughout its backbone with the expected increases in loop regions. However, three regions exhibited unexpected behavior: (1) the WPD loop, (2) helix α3 and (3) helix α7. First, in spite of the fact that the WPD loop oscillates between an open and closed conformation at an intermediate timescale (µs; Figures 1B-1F), its fast timescale dynamics mirrored the minimal overall dynamics of the PTP1B backbone (also clearly demonstrated by the relaxation data of PTP1B variants; see below). Second, and different from the WPD loop, the residues in helix α3, which are distal from the catalytic site and C-terminal to the WPD loop, were either broadened beyond detectability or had increased fast timescale dynamics. Third, residues in helix α7 exhibited much higher dynamics than the rest of PTP1B. A Bracken plot (R1*R2 vs R2/R1), which indirectly reports conformational exchange, showed that ~25 residues have significant chemical exchange or motional anisotropy, many of which belong to residues in helices α3 and α7 (Figure S1C) (Kneller et al., 2002). Over the past decades it has become clear that motions in enzymes are often coordinated with function (Boehr et al., 2006; Henzler-Wildman and Kern, 2007; Lisi and Loria, 2016; Mittermaier and Kay, 2009). Thus, these data suggest that helices α3 and α7 play key roles in PTP1B activity.
Active site inhibitor binding perturbs the allosteric pocket
We then investigated the consequences of active site inhibitor binding, which results in a closed conformation of the WPD loop, on PTP1B chemical shifts and dynamics. TCS401 is a small (306 Da) active site inhibitor that binds PTP1B with a KD of ~26 ± 2 µM (Table S1) (Iversen et al., 2000). As expected, chemical shift perturbations (CSPs, reflecting local and global changes in PTP1B structure) and the broadening of peak line widths beyond detectability were observed for residues at the TCS401 binding site, including the PTP-, WPD-loops and helix α4 (Figures 1G and S2A). However, CSPs were also detected in distal residues, including residues from loop L11 (150-153) and at the allosteric site (helices α3, α6 and α7), both ~20-25 Å away from catalytic cysteine, C215 (Figure 1A and 1B). These data show that active site inhibitor binding and WPD loop closure alter the chemical environment of allosteric site residues and also suggest that these changes are mediated by an allosteric path that includes residues from L11.
Inhibitor binding quenches the dynamics of helix α7
To see if altered dynamics accompanies active site inhibitor binding, we repeated the NMR dynamics measurements with TCS401-saturated PTP1B. The most significant changes were observed in helix α7, where the dynamics detected in the TCS401-free state were quenched in the TCS401-saturated state (Figures 1E and 1F). Additionally, residues in the WPD loop, the PTP loop and helix α4 broadened beyond detectability, likely due to intermediate exchange. The Bracken plot for TCS401-saturated PTP1B shows nearly no difference to that obtained for TCS401-free PTP1B (Figure S1D). Thus, for those residues that are accessible using NMR measurements, i.e. not broadened beyond detection, the data show that the change in dynamics in helix α7 relies primarily on fast timescale dynamics.
We also determined the crystal structures of PTP1B (residues 1-301) in both the free and TCS401-bound states. Free PTP1B lacks electron density for helix α7 (Barford et al., 1994), suggesting that this helix is flexible and/or adopts multiple states in the crystal, consistent with the intermediate CSI values obtained by NMR for this region. In contrast, in the PTP1B:TCS401 complex, the electron density for helix α7 is present, allowing helix α7 to be readily modeled (Figure S2B and S2C). This shows that helix α7 transitions from a flexible to an ordered conformation between the free and inhibitor bound complexes, correlating perfectly with the observed quenched dynamics for this helix upon TCS401 binding. Together, these results show that both altered dynamics and CSP detected conformational changes contribute to PTP1B allostery. The results also suggest that helix α7 and its intrinsic dynamics are central elements of the allosteric network of PTP1B.
Helix α7 is the hub of the PTP1B allosteric network
To define the role of helix α7 in PTP1B activity and allostery, we studied a variant of PTP1B in which helix α7 is deleted, PTP1BΔ7 (residues 1-284). As previously observed (Wiesmann et al., 2004), helix α7 is important for PTP1B function as the catalytic activity of PTP1BΔ7 is ~40% lower than that of PTP1B (Figures 2A; Table S1). However, this is not due to a change in structure, as the structures of PTP1BΔ7 and PTP1B are identical (RMSD = 0.19 Å; RMSDs are for residues 1-280; Figure 2B; Table S2). To determine if the deletion of helix α7 alters PTP1B conformation and dynamics in solution, we used NMR spectroscopy. The quality of the 2D [1H,15N] TROSY spectrum of PTP1BΔ7 is excellent, with peak intensities that are more uniform than those observed for PTP1B, allowing for the sequence-specific backbone assignment to be readily completed. The residues affected by the absence of helix α7 were then determined using CSP mapping. As expected, CSPs were observed for residues abutting helix α7 (helices α1’, α2’ and α6). However, CSPs were also observed for A189 and S190 in helix α3 (Figure 2C). Furthermore, 15N backbone dynamics showed that PTP1BΔ7 exhibits remarkably uniform dynamics throughout its backbone (Figures S3A and S3B). Consistent with this, the Bracken plot showed only ~10 residues, about 15 fewer than in PTP1B, with chemical exchange or motional anisotropy (Figure S3C). Together, these data show that helix α7 influences helices α3 and α6 and that this interaction is strictly required for the majority of the intermediate timescale dynamics observed in PTP1B, explaining the reduction of activity in PTP1BΔ7.
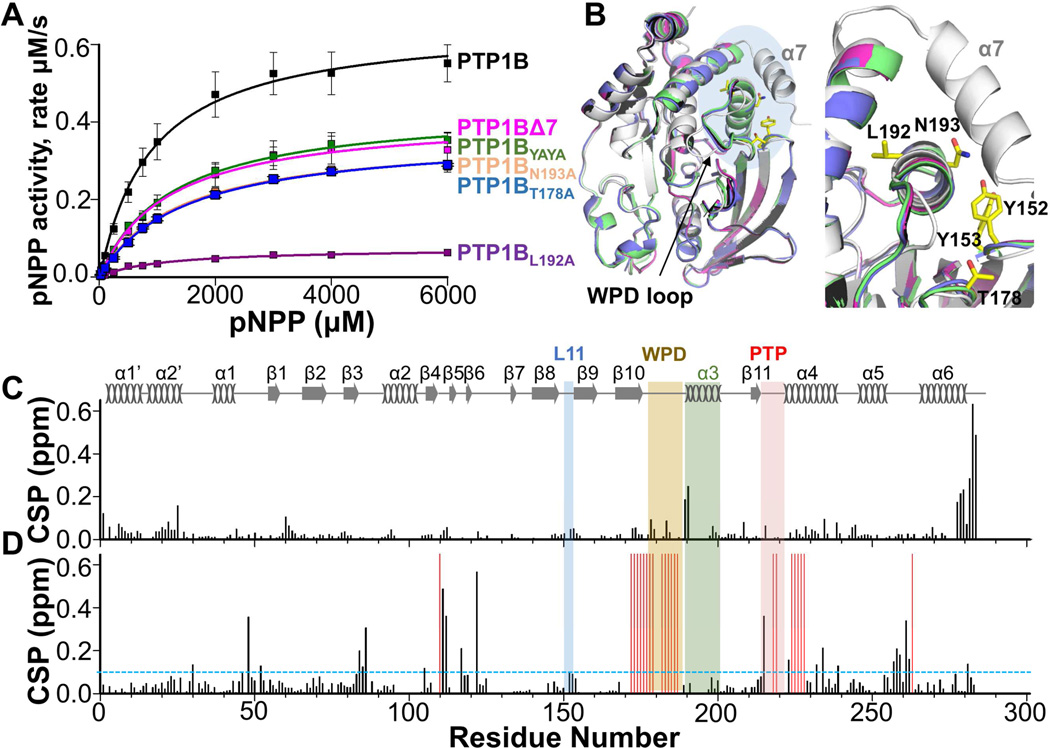
Figure 2. Activity of PTP1B variants.
(A) Enzymatic activity of PTP1B and variants (pNPP assays; ± SE, n = 3-9).
(B) Overlay of PTP1B (black, PDB 5K9V), PTP1B:TCS401 (gray, PDB 5K9W), PTP1BΔ7 (pink, PDB 5KA0), PTP1BYAYA (green, PDB 5KA2) and PTP1BT178A (blue, PDB 5KA4). Blue shade shows close-up region on right; L192, N193, Y152, Y153 and T178 (yellow sticks).
(C) CSPs of PTP1BΔ7 vs PTP1B.
(D) CSPs of PTP1BΔ7 vs PTP1BΔ7:TCS401 (1:3 molar ratio). Residues with significant CSPs are above the dashed line (= 0.09 ppm, sd). Residues broadened beyond detectability upon TCS401 binding are shown as red bars. Secondary structures are indicated; labeling as in Figure 1A.
See also Figure S3.
We then examined the closed state of PTP1BΔ7 by studying TCS401-saturated PTP1BΔ7. PTP1BΔ7 binds TCS401 with the same binding affinity (KD = 33 ± 6 µM) and in the same conformation as PTP1B (Table S1). Likewise, the CSP analysis between the free and TCS401-saturated PTP1BΔ7 was highly similar to that observed for free and TCS401- saturated PTP1B (Figure 2D); namely, CSPs were largely confined to active site residues, including a complete loss of WPD-loop resonances due to line broadening. However, there were unexpected differences. Unlike TCS401-saturated PTP1B, TCS401-saturated PTP1BΔ7 lacked CSPs in loop L11 and in helices α3 and α6 (L267 broadens beyond detectability, G277 has a small CSP). The lack of these CSPs show that TCS401 binding does not directly perturb the chemical shifts of residues in helices α3 and α6, but instead results from a shifted conformational ensemble that requires helix α7 to communicate between the PTP1B active and allosteric sites. As expected, fast timescale 15N backbone dynamics of TCS401-saturated PTP1BΔ7 showed no differences when compared to unbound PTP1BΔ7, supporting the CSP results (Figures S3A, S3B and S3D). Together, these data provide additional evidence that the protein dynamics of helix α7 constitute a critical and necessary component of the communication pathway between the PTP1B active and allosteric sites.
The PTP1B allosteric pathway includes L11
TCS401 binding to PTP1B resulted in CSPs in L11, a loop distal from the active site (Figure 1G). To determine the role of L11 in the PTP1B allosteric pathway, we generated different L11 variants and tested their activities (Table S1). While the activities of all variants were reduced, the variant with the lowest activity was Y152AY153A (PTP1BYAYA) (Figures 2A; Table S1). The structure of PTP1BYAYA is identical to PTP1B (RMSD = 0.37 Å) and its 2D [1H,15N] TROSY spectrum is of excellent quality. A CSP analysis of PTP1BYAYA with PTP1B showed that most CSPs are local to the L11 mutations (residues 147-155). However, we also identified CSPs for WPD loop residues (Y176, T177, T178 and W179; Figure 3A) and throughout helix α7. 15N backbone dynamics of PTP1BYAYA mirrored that of PTP1B (Figure 3A) as did the Bracken plot (Figure S4A and S4B). Thus, the CSPs revealed the presence of a communication network between the WPD loop and helix α7 that is mediated by residues in L11; i.e., L11 constitutes a key component of the allosteric pathway.
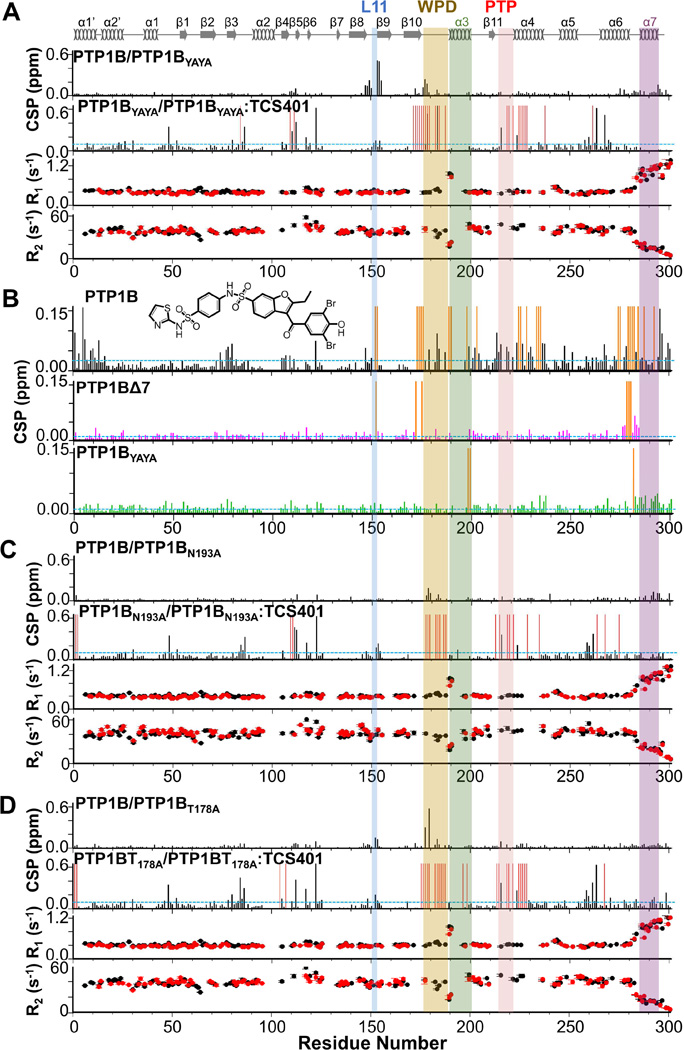
Figure 3. 15N dynamics and CSPs of PTP1B variants.
(A) Top, CSPs of PTP1BYAYA vs PTP1B and PTP1BYAYA vs PTP1BYAYA:TCS401 (1:3 molar ratio). Residues with significant CSPs are above the dashed line (σ= 0.1 ppm, sd). Bottom, 15N R1 and R2 relaxation rates for PTP1BYAYA (black) and PTP1BYAYA:TCS401 (red).
(B) CSPs of PTP1B (top), PTP1BΔ7 (middle) and PTP1BYAYA (bottom) with the allosteric inhibitor (1:3 molar ratio). Residues with significant CSPs are above the dashed line (PTP1B, σ = 0.02 ppm; PTP1BΔ7, σ = 0.01 ppm; PTP1BYAYA, σ = 0.01 ppm; sd). Residues broadened beyond detectability upon allosteric inhibitor binding are shown as orange bars. Chemical structure of allosteric inhibitor is shown.
(C) Top, CSPs of PTP1BN193A vs PTP1B and PTP1BN193A vs PTP1BN193A:TCS401 (1:3 molar ratio). Residues with significant CSPs are above the blue dashed line (σ = 0.1 ppm, sd). Bottom, 15N R1 and R2 relaxation rates for PTP1BN193A (black) and PTP1BN193A:TCS401 (red).
(D) Top, CSPs of PTP1BT178A vs PTP1B and PTP1BT178A vs PTP1BT178A:TCS401 (1:3 molar ratio) complex. Residues with significant CSPs are above the dashed line (σ = 0.09 ppm, sd). Bottom, 15N R1 and R2 relaxation rates for PTP1BT178A (black) and PTP1BT178A:TCS401 (red).
For (A, B, C) residues broadened beyond detectability upon inhibitor binding are shown as red bars. Secondary structures are indicated; labeling as in Figure 1A.
See also Figure S4.
To probe the closed state, we studied TCS401-saturated PTP1BYAYA. PTP1BYAYA binds TCS401 with an identical affinity and conformation as PTP1B (KD = 26 ± 3 µM; RMSD = 0.18 Å). The CSPs were largely unchanged from those observed for PTP1B. Namely, TCS401 binding resulted in CSPs around the active site and a complete loss of WPD-loop resonances due to line broadening. However, and very different from PTP1B, no CSPs were detected in helices α3 or α7 (CSPs were observed for helix α6 residue L267; Figure 3A); i.e., PTP1BYAYA is more similar to PTP1BΔ7 than PTP1B. The 15N backbone dynamics of TCS401-saturated PTP1BYAYA mirrored those of TCS401-saturated PTP1B, with the notable exception that helix α7 remained flexible in the inhibitor-bound state (Figure 3A). Thus, these data show that the altered dynamics accompanying allosteric signaling is communicated by L11 residues Y152 and Y153. Interestingly, these residues were previously reported to be phosphorylated by the insulin receptor tyrosine kinase (Bandyopadhyay et al., 1997; Dadke et al., 2000; Li et al., 2005), suggesting they might form a feedback signaling module within the insulin pathway via this allosteric network.
To confirm that these residues are critical for allostery, we measured CSPs of different PTP1B variants (PTP1B, PTP1BΔ7, PTP1BYAYA) in the presence and absence of an allosteric inhibitor, 3-(3,5-Dibromo-4-hydroxy-benzoyl)-2-ethyl-benzofuran-6-sulfonicacid-(4-(thiazol-2-ylsulfamyl)-phenyl)-amide, hereafter referred to as the allosteric inhibitor (Wiesmann et al., 2004). This inhibitor binds PTP1B with a KD of 7.8 ± 0.8 µM, while it binds PTP1B variants with 2- to 4-fold lower affinities (Table S1). As expected, a CSP analysis of allosteric inhibitor saturated PTP1B showed CSPs and peak broadening beyond detectability for all peaks in the allosteric inhibitor binding pocket. However, CSPs and peak broadening were also detected for residues outside this site, including L11 residues Y152, Y153, residues in the WPD loop, residues in helices α4 and α6 as well as A189, S190 and V198 (helix α3). In contrast, these same CSP measurements with PTP1BΔ7 and PTP1BYAYA showed far fewer (or nearly no) CSPs, other than in the allosteric inhibitor binding pocket, as well as far fewer peaks that broadened beyond detectability (Figure 3B). This confirms that helix α7 and L11 residues Y152 and Y153 define the key elements of the allosteric pathway in PTP1B.
L11 connects the allosteric binding pocket to the WPD loop via N193 and T178
Our crystals structures show that a hydrogen bond network is formed between Y152 (L11), S295 (α7) and N193 (α3) when PTP1B binds TCS401. This suggests that N193 is also part of the allosteric network. Like PTP1BYAYA, PTP1BN193A has reduced activity (Figures 2A; Table S1). This was not due to a change in conformation, as the CSI (data not shown) and stability analysis of PTP1BN193A (Tables S2 and S3) showed that the mutation did not change the overall fold. A CSP analysis with PTP1B revealed CSPs throughout helix α7 and in the WPD loop (Figure 3C), confirming that helix α3, along with L11, the WPD loop and helix α7, defines the allosteric pathway between the active and allosteric binding sites. 15N backbone dynamics of PTP1BN193A mirrors the dynamics of PTP1B (Figure 3C), as does the Bracken plot (Figure S4C and S4D).
PTP1BN193A binds TCS401 with an affinity and conformation similar to that observed for PTP1B (KD = 37 ± 4 µM, Table S1; the increased RMSD, 0.88 Å, is due to changes in loops away from the active site as, in this variant, helix α7 is not ordered in the closed state). The CSPs observed between free and TCS401-saturated PTP1BN193A mirrored the PTP1B results. Namely, CSPs were localized to the active site, including the hallmark loss of peak intensities corresponding to WPD-loop residues. Similarly, L11 residues also showed CSPs, with Y152 showing a larger CSP in PTP1BN193A than PTP1B, possibly due to its inability to form a hydrogen bond with N193. In contrast, PTP1BN193A helices α3 and α7 were much less affected by TCS401 binding than they were in PTP1B. Rather, the CSP pattern was similar to that observed for PTP1BYAYA (Figure 3C). The 15N backbone dynamics of TCS401- saturated PTP1BN193A were also similar to PTP1BYAYA. Namely, helix α7 stayed fully flexible in the TCS401-bound state (Figure 3C). Together, the data demonstrate that N193 (α3), via Y152 (L11) and helix α7, is an essential component of the communication pathway between the active and allosteric sites.
We then set out to identify the residues from the WPD loop that participate in this pathway. We generated multiple WPD variants and tested their effect on PTP1B activity. PTP1BT178A exhibited significantly reduced activity (Figure 2A; Table S1). The PTP1BT178A crystal structure was identical to PTP1B (RMSD = 0.46 Å), with the exception of an unusually long distance between the side chains of N193 and Y152 (7.9 Å compared to 4.0 Å in PTP1B). Furthermore, the 2D [1H,15N] TROSY spectrum of PTP1BT178A allowed for NMR analysis. An overlay of the PTP1BT178A and PTP1B spectra showed only minor changes in chemical shifts, with the exception of the expected CSPs localized about T178 in the WPD loop. Additional CSPs were identified for Y152 and Y153, demonstrating that the mutation of T178 to an alanine alters the chemical environment of L11 residues (Figure 3D). 15N backbone dynamics of PTP1BT178A mirrors the dynamics of PTP1B (Figure 3D), as does the Bracken plot (Figure S4E and S4F).
PTP1BT178A binds TCS401 with a slightly lower affinity, but with a conformation identical to that observed for PTP1B (KD = 45 ± 3.5 µM, Table S1; RMSD = 0.12 Å). A CSP analysis between free and TCS401-saturated PTP1BT178A shows CSPs in the active site and a loss of peaks of WPD loop residues due to line broadening. CSPs were also observed in peaks corresponding to E loop residues, demonstrating that changes in the WPD loop directly affect the E loop environment. Furthermore, CSPs were observed in peaks corresponding to L11 residues Y152 and Y153. Correlating well with the results of other PTP1B variants, fewer residues in helices α3 (only F196, V198), α6 (L267, S270, G277) and α7 (Q290, W291) showed CSPs or line broadening (Figure 3D). We also performed 15N backbone dynamics of TCS401-saturated PTP1BT178A; helix α7 stayed flexible in the inhibitor-bound state. This confirms a role for residue T178 in the altered dynamics that supplements the allosteric signaling in PTP1B (Figure 3D).
Finally, while PTP1BN193A, PTP1BYAYA and PTP1BΔ7 have catalytic activities that are lower than PTP1B, their affinities for the active site inhibitor TCS401 are essentially identical. This suggests that while phosphate binding is comparable, the phosphate release step is slower in these variants, as confirmed by the higher Ki values (competition between substrate and TCS401) measured for PTP1BN193A, PTP1BYAYA and PTP1BΔ7 (Figure S4G, Table S3). Thus, these data show that the allosteric network likely enhances the phosphate release step in the PTP1B catalytic cycle.
PTP1B activity requires intermediate timescale dynamics and rigidity in the WPD loop
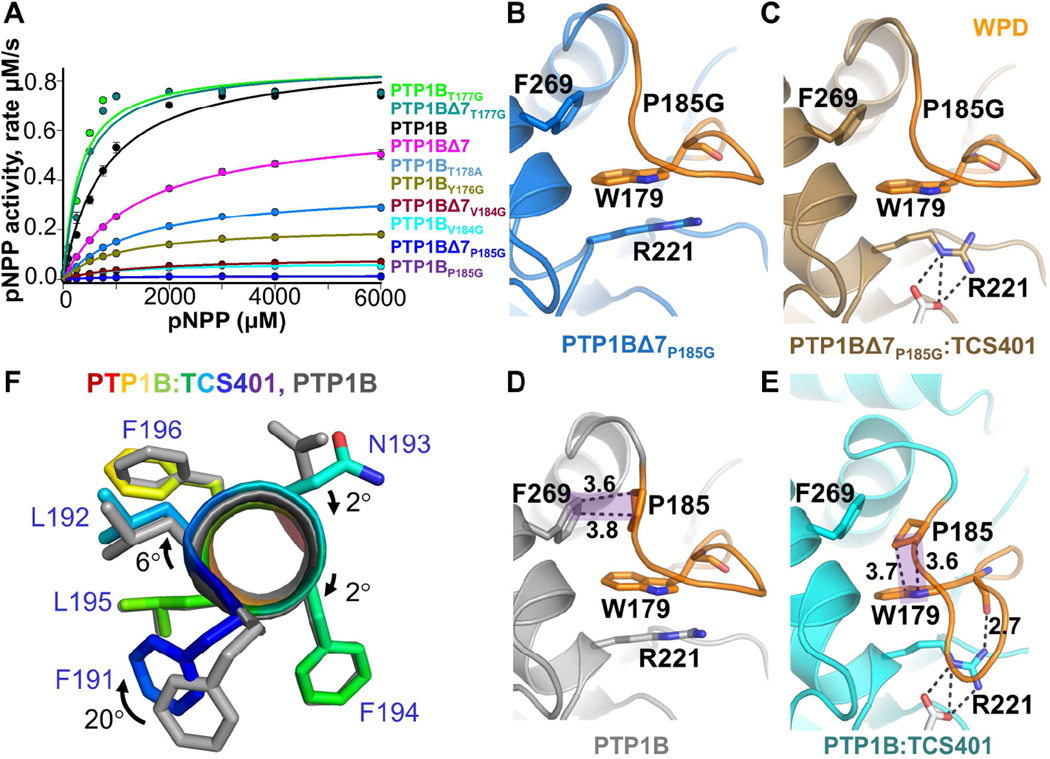
Our unexpected discovery that the WPD loop is rigid on the ps-ns timescale led us to test the role of WPD loop residues that border the 179WPD181 sequence on PTP1B function. Previous studies showed that intermediate timescale WPD loop dynamics play an important role in PTP1B activity (Brandao et al., 2012; Whittier et al., 2013). We tested this by studying variants designed to increase loop dynamics; namely by inserting flexible glycine residues adjacent to the WPD sequence. Surprisingly, only a single variant, T177G, increased the activity of PTP1B (~5 fold; Figure 4A; Table S1); all others either reduced (Y176G, V184G) or abolished (P185G; catalytically dead) catalysis. The activities of the variants (T177G, V184G and P185G) in allosterically dead PTP1BΔ7 were identical to the PTP1B variants, confirming that these changes in activity were due solely to changes in WPD loop conformation and dynamics.
Figure 4. P185 provides the rigidity that mediates the opening and closing of the WPD loop.
(A) Activities of PTP1B and WPD loop variants (pNPP assays; ±SE, n=3-6).
(B) Structure of PTP1BΔ7P185G (blue, PDB 5KAA).
(C) Structure of PTP1BΔ7P185G:TCS401 (brown, PDB 5KAB).
(D) Structure of PTP1B (gray, PDB 5K9V).
(E) Structure of PTP1B:TCS401 (cyan, PDB 5K9W). In (D) and (E), P185 forms a CH-π stacking interaction (shaded purple) between F269 (helix α6) and W179 (WPD loop) in the open and closed states, respectively. Hydrogen bond (2.7 Å) between the side chain of R221 and backbone HN of W179 is detected in PTP1B:TCS401 but not in PTP1BΔ7P185G:TCS401.
(F) Overlay of PTP1B and PTP1B:TCS401, showing the rotation of helix α3 between the open and closed state; F191 (dark blue) rotates by 20°.
To evaluate the closed state of these mutants, we tested their ability to bind to TCS401. All variants bound TCS401 with affinities identical to PTP1B; the only exception was PTP1BP185G, the catalytically dead variant. To understand how this variant binds TCS401 more tightly (KD = 20 ± 0.6 µM; Table S1) yet is still catalytically dead, we determined its structure. Unlike PTP1B, the WPD loop in PTP1BΔ7P185G:TCS401 is not closed, but instead adopts an open conformation (Figures 4B and 4C). The first step of WPD loop closure is the rotation of the PTP loop R221 sidechain, which creates the space needed for the WPD loop to close about the active site. Although this rotation of R221 is observed in PTP1BΔ7P185G:TCS401, the WPD loop fails to close. Our PTP1B structures reveal why this is the case. In the open state, P185 forms a CH/π interaction with F269 from helix α6 and a hydrophobic interaction with P180 from the WPD loop (Figure S4H). In contrast, in the closed state, P180 rotates downwards towards the R221 sidechain, breaking its hydrophobic interaction with P185. This allows P185 to rotate by ~50° and form a second CH/π interaction with W179 (Figures 4D and 4E). A movement of both P185 and W179 deeper into the central hydrophobic pocket of PTP1B accompanies this interaction. This causes both F269 and especially F191 to rotate out of the pocket away from the catalytic site and, in the case of F191, towards helix α6 (Figure 4F). The P185 rotation and the movement of F191 allows the C-terminal portion of the WPD loop to collapse towards helix α3. This results in the formation of one additional helical turn accompanied by a rotation of the helix itself, both of which allow the sidechain of S187 to bind and cap the HN backbone of S190.
In the absence of P185, however, none of these changes occur (Figures 4B-4F). This suggests that the CH/π switch is indispensable for WPD loop function, as, in its absence, PTP1BP185G does not close and, consequently, cannot mediate catalysis. The importance of this interaction is consistent with the finding that the energy of prolinetryptophan stacked interactions, like that observed between P185 and W179, can be large (~7 kcal/mol) (Biedermannova et al., 2008); i.e., without this stabilization energy, the loop remains open. Finally, because this interaction results in the movement of F191 towards helix α6 upon inhibitor binding, this suggests that the distance between helices α3 and α6 is of critical importance to allow for these changes, and as a consequence catalysis, to occur.
Chemical-shift-restrained molecular dynamics (MD) simulations capture the key elements of the PTP1B allosteric network
We performed MD simulations to derive an atomic-level description of the dynamics in PTP1B that underlies the experimentally identified allosteric network. To improve accuracy and sampling, we performed experimentally-guided simulations, where the NMR chemical shifts were used as a system-specific force field correction (Camilloni et al., 2012; Tiberti et al., 2015). Importantly, because the NMR chemical shifts are time- and ensemble-averaged over multiple, interconverting conformations, the experimental restraints were enforced such that individual conformations do not have to satisfy all data, as long as the entire ensemble of conformations is in agreement with experiment. In practice, this was achieved by replica-averaged simulations, in which we simulated multiple copies of the protein in parallel.
We used the crystal structure of PTP1B bound to TCS401 (PDB 5K9W; ligand was removed in silico) with a closed WPD loop as starting point for our simulations and applied chemical shift restraints over four copies. Two of the four simulations result in a WPD loop in an open-like conformation, while the other two result in a closed WPD loop. In contrast, control simulations that did not include experimental restraints showed only marginal changes in the WPD loop conformations (closed or open) over the timescale of the simulation (~2 µs; Figure S5A and S5B). Thus, the use of chemical shift restraints allows us to follow the closed-to-open transition of the WPD loop and therefore provides an independent approach to determine the motions and interactions that accompany loop closure.
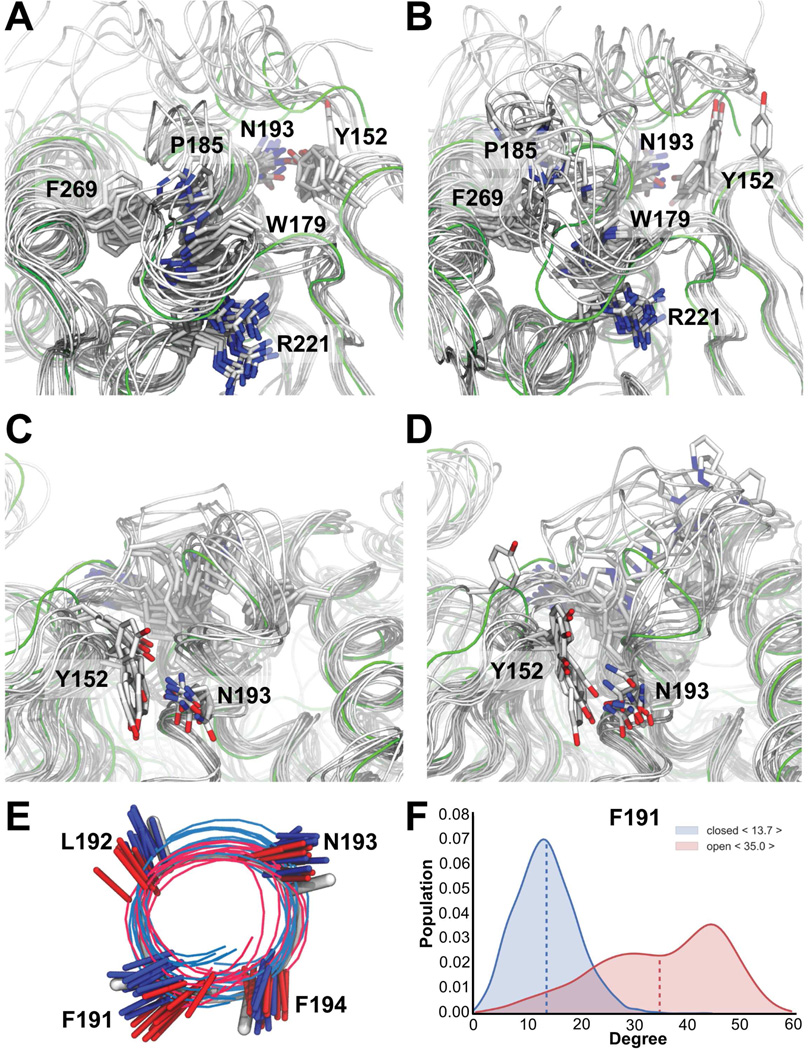
The simulations confirm the crystallographic results which show that WPD loop residue P185 forms CH/π stacking interactions with either F269 or W179. When the WPD loop is closed, P185 interacts with W179 (Figure 5A), whereas the two replicas with an open WPD loop conformation show increased dynamics of P185, now mainly interacting with F269 (Figure 5B). Interestingly, we also identified a correlation between the conformation of the WPD loop and an interaction between Y152 (L11) and N193 (helix α3). Namely, these residues interact primarily when the WPD loop is closed (Figures 5C and S5D). This agrees with the crystallographic and NMR data, which show that when the WPD loop is closed, helix α3 typically forms an extra turn. In contrast, when the WPD loop is open, helix α3 is shorter and twisted (Figures 5E, 5F, S5C and S5D). As also suggested by the crystallographic data, these structural and dynamic changes are linked to a weakened interaction between Y152 and N193 (Figure 5D). As expected, the simulations confirm that the interaction between R221 and W179 is more often observed when the WPD loop is in a closed conformation and less when it is in an open conformation.
Figure 5. Chemical shift restrained MD captures the opening and closing mechanism of the WPD loop.
(A) Superposition of 10 structures from 2 of the 4 simulations sampling the closed state of the WPD loop. The backbone of each structure is shown as a gray tube and key residues are highlighted as sticks. Crystal structure of PTP1B:TCS401 (green; PDB 5K9W). The P185 interaction with F269 and W179 is present in the majority of frames. Furthermore, R221 anchors W179 via the expected sidechain:backbone interactions.
(B) Superposition of 10 structures from the 2 simulations sampling the WPD-loop open conformation.
(C) Same as (A) but showing the interaction between Y152 and N193; present in the majority of frames.
(D) Same as (B), but showing the interaction between Y152 and N193; mostly absent in the open conformation.
(E) MD simulations highlight conformational differences in helix α3 between the open (red) and closed states (blue).
(F) The most notable difference is the ‘F191 push’. See also Figure S5.
Finally, we see a weaker, but a clearly observable correlation between the conformation of the WPD loop and that of helix α7. In particular, we find less well-defined packing between helices α3 and α7 when the WPD loop is open. The chemical shift restraints ensured that we observe partial unfolding of helix α7 (~40% formed; Figures S5C-S5F). Nevertheless, we note that on the timescales accessed here, as well as due to the well-known bias in current force fields to form compact structures (Piana et al., 2014), we do not expect our simulations to capture fully the disordered nature of helix α7. Despite these issues and the difficulty obtaining converged ensembles, our simulations capture many of the key events in the allosteric communication network that links the active site with helix α7.
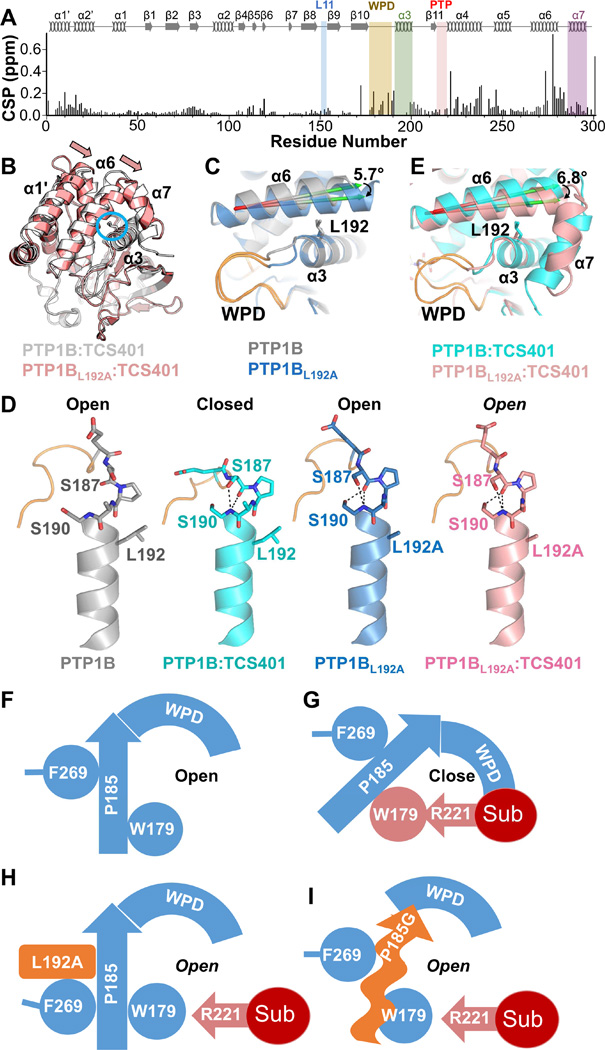
L192 is not allosteric but instead functions as a wedge that facilitates WPD loop closure
The most obvious candidate for defining the distance between helices α3 and α6 is L192. PTP1BL192A has ~10-fold reduced activity (Figure 2A; Table S1). Furthermore, a CSP analysis of PTP1BL192A vs PTP1B revealed major changes in every region of the spectrum, including the ability to assign additional residues in helix α3. CSPs were observed in helices α1’, α2’, α3, α4 and α6, and also in the WPD- and PTP-loops (Figure 6A). This suggests that there are significant changes in the L192A conformation. This was confirmed by its structure (RMSD of 0.80 Å with PTP1B), which showed that helices α1’ and α6 collapse towards helix α3 by ~6°, filling the pocket created by the missing L192 sidechain (Figures 6B and 6C). As a result, the WPD loop adopts a unique conformation, in which residues 178-185 adopt an open conformation while loop residues 186-190 adopt a closed conformation (Figure 6C). Furthermore, S187 forms the hydrogen bond with the HN backbone of S190 that is otherwise only seen when the WPD loop is closed (Figure 6D).
Figure 6. PTP1BL192A.
(A) CSPs of PTP1BL192A vs PTP1B. Secondary structures are indicated; labeling as in Figure 1A.
(B) Overlay of PTP1B:TCS401 (gray; PDB 5K9W) and PTP1BL192A:TCS401 (pink; PDB 5KA9); L192 (sticks) is circled. Arrows highlight the shift of helix α6.
(C) Overlay of PTP1B (grey; PDB 5K9V) and PTP1BL192A (blue; PDB 5KA8).
(D) The position of the WPD loop (residues 186-190, orange) in PTP1B (gray; PDB 5K9V), PTP1B:TCS401 (cyan, PDB 5K9W), PTP1BL192A (blue, PDB 5KA8) and PTP1BL192A:TCS401 (pink, PDB 5KA9); hydrogen bonds are shown as black dotted lines.
(E) Overlay of PTP1B:TCS401 (cyan; PDB 5K9W) with PTP1BL192A:TCS401 (pink; PDB 5KA9). The conformational change in helix α6 displaces the helix α7 from its closed conformation.
(F) Model of the PTP1B WPD loop in the open state.
(G) Substrate binding (red) causes R221 to engage W179, initiating a rotation of P185 and the closure of WPD loop.
(H) The L192A mutation shifts helix α6 closer to helix α3, restricting F191 rotation and preventing WPD loop closure.
(I) The P185G mutation prevents the closure of the WPD loop.
In spite of these changes, PTP1BL192A still binds TCS401 (KD = 39 ± 6 µM; Table S1). However, the structure of the complex shows that, like the P185G mutant, the WPD loop of the L192A mutant does not close but instead stays open (Figure 6E). This is because the pocket into which F191 normally binds to permit WPD loop closure is occupied by A273 from the collapsed helix α6. This constrains F191 to its open state conformation, which inhibits the movement of W179 and, in turn, the W179-P185 CH/π stacking interaction. Thus, L192 functions as a ‘wedge’ that props helix α6 away from F191 binding pocket, ensuring that F191 can rotate upon substrate binding and, in turn, facilitate WPD loop closure Figures 6F-6I). Thus, we have identified a single point variant of PTP1B (PTP1BL192A) that is more than 15 Å away from the PTP1B active site and not a member of the catalytically important loops, yet, remarkably, still results in a catalytically dead enzyme.
Discussion
Protein dynamics are essential for protein function and regulation (Akimoto et al., 2013; Henzler-Wildman and Kern, 2007; Lisi and Loria, 2016; Masterson et al., 2010; Tzeng and Kalodimos, 2012). Most metabolic enzymes turnover quickly, and thus are highly efficient catalysts that generate large quantities of product. In contrast, eukaryotic posttranslational modification enzymes, especially phosphatases such as PTP1B, are fundamentally different as they function to fine-tune biological signaling pathways and activities. Thus, these posttranslational modification enzymes are often slower and subject to multiple levels of regulation. By combining experimental PTP1B structural and dynamics data with mutational studies and molecular dynamics simulations, we show that the essential steps in PTP1B catalysis, including product hydrolysis, require both protein rigidity and slow dynamics, whereas its allosteric regulation requires fast dynamics.
In PTP1B, the first step of the catalytic cycle is WPD loop closure. This occurs (or the population of closed WPD state increases) when the negatively charged oxygens of the substrate phosphate bind R221. To mediate this coordination, the sidechain of R221 rotates by ~90°. This leads to the formation of a hydrogen bond with the HN backbone of W179 in the WPD loop and establishes a key connection between substrate binding and the WPD-loop (Figures 7A and 7B). Here, we discovered that P185 is essential for this process. Namely, the R221-W179 hydrogen bond results in the movement of the P180 sidechain away from the W179 aromatic sidechain, which results in the formation of a CH/π interaction between W179 and P185. In the open conformation, P185 is stabilized by a CH/π interaction with F269 and a hydrophobic interaction with P180 (Figures 7C and 7D). Thus, the switch (~50° rotation of the P185) of the π interaction partner between F269 (open) and W179 (closed) is the joint that allows for WPD loop closure. This explains the surprising rigidity of the WPD loop in fast timescale motion experiments (15N R1) as well as the observation that the P185G mutation leads to a catalytically dead enzyme (although rare, reduced activity upon glycine substitutions have been identified in other enzymes, i.e., triosephosphate isomerase (Xiang et al., 2004))). The movement of the W179 sidechain and the P185 CH/π rearrangement necessitates a concurrent movement of other residues, most importantly F191, which becomes wedged between helices α3 and α6 (the ‘F191 push’) (Figure 7E and 7F). This P185-dependent WPD loop movement is the basic requirement for PTP function and is independent of allostery as it also functions in PTP1BΔ7. Strikingly, P185 is 100% conserved in the PTP family and thus this mode of action of the WPD loop is likely conserved in all PTPs (Table S4).
Figure 7. The activity cycle of PTP1B.
The PTP1B catalytic cycle and its allosteric control (free/open, outer circle; inhibitor bound/closed, inner circle).
(A→B) Binding of the active site inhibitor TCS401 pushes the W179 aromatic side chain towards P185 (P185 makes a CH/π switch from F269 to W179).
(C→D) This allows for the closing of the WPD loop.
(E→F) The resulting ‘F191 push’ causes helix α3 to rotate.
(G→H) This allows S190 to hydrogen bond with S187, extending and stabilizing helix α3.
(I→J) This allows N193 to interact with Y152 and E295.
In turn, the ‘F191 push’ causes helix α3 to rotate (~20° at F191). This leads to: (1) the formation of a hydrogen bond between residues S187 and the NH backbone of S190, which stabilizes the closed state of the WPD loop (Figure 7G and 7H) and (2) a new rotamer conformation of N193 that results in a hydrogen bond network with S295 (α7) and Y152 (L11) (Figure 7I and 7J). This latter network, which is only observed in the closed state, enhances substrate/phosphate release, as demonstrated by the higher Ki values for allosterically dead PTP1B variants. It also stabilizes helix α7, which, as observed in the NMR measurements, otherwise has increased dynamics. This helix α7 stabilization both increases the activity of PTP1B by ~50%, as demonstrated by the mutational studies, and allows for its allosteric modulation and inhibition. This differs significantly from the mechanism of allostery previously described, based only on crystal structures, in which allosteric inhibitor binding was hypothesized to sterically block WPD closure. We show that allosteric inhibitor binding prevents helix α3 from rotating and, in turn, prevents PTP1B from forming the key hydrogen bond interaction network needed for allostery. Furthermore, we also identified all residues that define the PTP1B allosteric pathway (T178, Y152, Y153 and residues in helix α7). PTP1B variants of these residues will be effective screening tools for high-throughput allosteric inhibitor screening, as these variants decouple allostery from activity in PTP1B. Equally exciting, Y152 and Y153 are phosphorylated in vivo and this is likely an endogenous mechanism for allosterically controlling PTP1B activity (Bandyopadhyay et al., 1997; Dadke et al., 2000; Li et al., 2005).
Finally, this analysis shows that both protein rigidity and dynamics are essential for PTP1B function. It also shows that distinct dynamic timescales underlie different aspects of PTP1B function. This result is novel as previous studies of adenylate kinase (AdK) (Henzler-Wildman et al., 2007) suggested that fast timescale motions are coupled with slower timescale motions and, together, regulate the catalytic cycle. Our results show that PTP1B is different. In PTP1B, rigidity and slow timescale motions are required for catalysis, while fast timescale motions allow for its allosteric control. Thus, dynamics at different timescales are not coupled as they are in AdK, but are decoupled in PTP1B. This demonstrates that the molecular mechanisms that govern enzyme function differ for distinct enzymes. However, this behavior of PTP1B is logical from a chemical point of view: chemistry requires that the reaction components are precisely positioned; slow dynamics ensure this is achieved. Thus, in PTP1B, slow dynamics is important for substrate binding and product release while fast dynamics, in elements distal from the chemical reaction, is important for the allosteric control of the overall function.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by Wolfgang Peti (wolfgangpeti@email.arizona.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Expression plasmids contained cDNA sequences from Homo sapiens. Recombinant proteins were overexpressed in E. coli BL21 (DE3) RIL cells (Agilent) in LB broth or D20- based M9, minimal salt media containing 15NH4Cl and/or [2H,13C]-D-glucose (CIL or Isotec). Cells were grown at 37°C at 250 rpm in 2 L shaking flasks until A600 reached ~0.6, cooled, then induced with isopropyl b-D-1-thiogalactopyranoside (IPTG, GoldBIO) to a final concentration of 1 mM. After induction, cells were grown for 20 hrs at 18°C.
METHOD DETAILS
Protein Expression and Purification
DNA coding the human PTP1B catalytic domain PTP1B 1-301 (residues 1-301) was subcloned into RP1B as previously described (Peti and Page, 2007). PTP1B variants— PTP1BΔ7 (PTP1B 1-284, residues 1-284), Y152AY153A, Y176G, T177G, T178A, V184G, P185G, L192A and N193A (single and double variants in both PTP1B and PTP1BΔ7)— were generated using the Quikchange (Agilent) site directed mutagenesis kit. For protein expression, plasmid DNAs were transformed into E. coli BL21 (DE3) RIL cells (Agilent). Cells were grown in Luria Broth in the presence of selective antibiotics at 37°C to an OD600 of ~0.6, and expression was induced by the addition of 1 mM IPTG. Induction proceeded for ~20 h at 18°C prior to harvesting by centrifugation at 6,000 × g. Cell pellets were stored at - 80°C until purification. For NMR measurements, expression of uniformly 2H,15N- or 2H,15N,13C-labeled PTP1B was facilitated by growing cells in D2O based M9 minimal media containing 1 g/L 15NH4Cl and/or 4 g/L [2H,13C]-D-glucose (CIL or Isotec) as the sole nitrogen and carbon sources, respectively. Multiple rounds (25%, 50%, 75%, 90% and 100%) of D2O adaptation (Peti and Page, 2016) was necessary for high-yield expression.
Cell pellets were lysed in Lysis Buffer (25 mM Tris pH 8.0, 500 mM NaCl, 5 mM imidazole, 0.1% Triton X-100) containing EDTA-free protease inhibitor cocktail (Roche) using high-pressure homogenization (Avestin). The lysate was clarified by centrifugation at 50,000 × g and filtered through a 0.22 µm filter before loading onto a His-trap HP column (GE Healthcare). Bound proteins were washed with Buffer A (50 mM Tris pH 8.0, 500 mM NaCl, 5 mM imidazole) and eluted with increasing amounts of Buffer B (50 mM Tris pH 8.0, 500 mM NaCl, 500 mM imidazole) using a 5-500 mM imidazole gradient. Peak fractions were pooled and dialyzed overnight at 4°C in Dialysis Bu ffer (50 mM Tris pH 8.0, 500 mM NaCl) with 5:1 volume ratio of TEV protease overnight. The next day, a ‘subtraction’ his-tag purification was performed to remove TEV and the cleaved His-tag. Cleaved PTP1B was further purified using Size Exclusion Chromatography (SEC, Superdex 75 26/60) in either Assay Buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP), protein Crystallization Buffer (20 mM Tris pH 7.5, 50 mM NaCl, 0.5 mM TCEP) or NMR Buffer (50 mM HEPES pH 6.8, 150 mM NaCl, 0.5 mM TCEP for PTP1B and variants; 50 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP for PTP1BΔ7). Purified protein was either used immediately or flash frozen in liquid nitrogen for storage. Typical PTP1B protein yields were ~46 mg/L in Luria Broth, ~34 mg/L in 2H,15N M9 medium and ~19 mg/L in 2H,15N,13C M9 medium.
pNPP activity assay
The activities of freshly prepared (0.15 µM) PTP1B/PTP1BΔ7 and variants were measured in assay buffer containing varying concentrations of p-nitrophenyl phosphate (pNPP; 0 to 6000 µM) either with or without TCS401 (100 or 300 µM). Enzymes were incubated with the substrate at 30°C for 30 minutes. The reaction was stopped using 0.5 M NaOH and the absorbance was measured at 405 nm using a plate reader (BioTek). TCS401 stock solution (25 mM) was prepared in 100% DMSO, resulting in 0.4% (v/v) and 1.2% (v/v) of DMSO in the reactions. Measured absorbance from blanks that contained substrate and inhibitors but no protein was subtracted from all measurements. The rate of dephosphorylation of pNPP was analyzed using the molar extinction coefficient for pNPP of 18000 M−1cm−1 and an optical path length of 0.3 cm (96 well plates). Km and Vmax were determined by fitting to the Michaelis-Menten equation, y = Vmax*x/(Km + x); kcat was extracted using y = Et*kcat*x/(Km + x). The catalytic efficiency was obtained as kcat /Km. The inhibition constant Ki, was obtained using a Lineweaver-Burk plot, Km,obs = Km*[1+[I]/Ki], where y = Vmax*x/(Kmobs + x) (Table S1,S4). Sigma Plot 12.5 and GraphPad Prism 6 were used for data analysis including the statistical analysis using unpaired (between PTP1B/PTP1BΔ7 and all variants) or paired t-test (presence and absence of inhibitor). The experiments were carried out in duplicate and repeated 3 to 6 times.
Affinity measurement of PTP1B to TCS401 or PTP1B allosteric inhibitor
PTP1B/PTP1BΔ7 and all variants (2.5 µM) in assay buffer were incubated with either TCS401 or the PTP1B allosteric inhibitor (3-(3,5-Dibromo-4-hydroxy-benzoyl)-2-ethylbenzofuran-6-sulfonicacid-(4-(thiazol-2-ylsulfamyl)-phenyl)-amide) at varying concentrations (0-140 µM) or with DMSO (0 – 1%) as control. Stock solutions of the PTP1B allosteric inhibitor (13 mM in 100% DMSO) and TCS401 (25 mM in 100% DMSO) were prepared immediately prior to each experiment. Affinity measurements were made using a FluoroMax-4 spectrofluorometer (Horiba) with the following parameters: excitation = 280 nm, slit width 5 nm, emission = 300 – 550 nm, slit width 5 nm. KD was obtained from LR= ((X+Rtot+KD)-√((X+Rtot+KD)2-4*X*Rtot))/2, where, L=(X – LR) and Y= BKG + MF*L + FR*MF*LR); Rtot is total receptor concentration, BKG is background fluorescence w/o ligand, MF is molar fluorescence of the free ligand, KD is dissociation constant, FR is fluorescence ratio, MF of bound ligand=MF*FR (unitless ratio), so FR > 1 means binding increases fluorescence and FR < 1 means binding quenches fluorescence. GraphPad Prism 6 was used to perform the binding analysis. Sigma Plot 12.5 was used for the statistical analysis using the unpaired t-test between PTP1B/PTP1BΔ7 and all variants. All affinity measurements were repeated in triplicate.
Circular Dichroism (CD)
Proteins were dialyzed overnight into CD Buffer (10 mM Na-phosphate pH 6.8, 150 mM NaCl). CD spectra were measured using 2.5 µM protein at 25°C, with wavelength scan 260- 190 nm. The melting temperature (TM) was recorded at 220 nm and temperature scan from 20 to 90°C (Table S2). TM values were calculated using a two-state transition model for a monomer from a folded to unfolded state (the heat capacities of the folded and unfolded state were either assumed to be equal or corrected for linear changes in ellipticity as a function of temperature). The CD measurements were performed on a Jasco J-815 CD spectropolarimeter.
Protein Crystallization and Data Collection
Crystallization trials of PTP1B/PTP1BΔ7 and all variants, including PTP1BYAYA, PTP1BT178A, PTP1BL192A, PTP1BN193A and PTP1BP185G were performed at 10 mg/mL using vapor diffusion sitting drop experiments. For TCS401-bound structures, 3.3 mg/mL purified PTP1B was incubated with a 10 molar excess of TCS401 (25 mM stock in 100% DMSO) for an hour on ice, and then concentrated to 10 mg/mL for crystallization. Apo PTP1B and TCS401-bound PTP1B crystals were grown using a fine screen with pH ranging from 7.4 – 8.0 (0.1 M Tris/HEPES) and PEG8000 concentrations ranging from 13% - 24% (w/v). The MgCl2 concentration was 0.2 M in all conditions. All crystals were grown at 4°C. Crystals were cryoprotected by a quick soak (few seconds) in mother liquor supplemented with 30% glycerol and immediately flash frozen in liquid nitrogen. X-ray data were collected using either a home source (Rigaku FR-E+ Superbright, 1.54 Å wavelength; Saturn944 CCD detector) or the GM/CA@APS beamline 23-ID-B (0.98 Å wavelength; MAR 300 CCD detector. The data was processed using either HKL2000, HKL3000R or XDS (Kabsch, 2010).
X-ray Structure Determination
The crystal structure (PTP1B 1-301) was solved by molecular replacement (search model PDBID: 1SUG) using Phaser as implemented in PHENIX (Adams et al., 2010). For TCS401-bound structures, clear density of the ligand was observed after PHENIX.AutoBuild and PHENIX.Refine. PHENIX.LigandFit was used to fit TCS401 into the electron density. There was a strong anomalous signal in the anomalous maps at the position of the sulfur atom in TCS401 (when data was recorded on Cu source). All structures were subjected to iterative rounds of refinement in PHENIX and manually built using Coot (Emsley et al., 2010). All data collection and refinement statistics are reported in Table 1.
Table 1.
| Data collection and refinement statistics (1/4). | ||||
|---|---|---|---|---|
| PTP1B | PTP1B:TCS401 | PTP1BΔ7 | PTP1BΔ7:TCS401 | |
| Data collection | ||||
| X-ray beam line | home source | home source | home source | home source |
| Wavelength (Å) | 1.54 | 1.54 | 1.54 | 1.54 |
| Space group | P 32 2 1 | P 31 2 1 | P 32 2 1 | P 32 2 1 |
| Cell dimensions | ||||
| a, b, c (Å) | 88.5, 88.5, 73.0 | 88.6, 88.6, 103.9 | 88.4, 88.4, 72.5 | 88.0, 88.0, 73.0 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50.00–1.90 (1.93–1.90) | 50.00–2.01 (2.04–2.01) | 50.00–1.99 (2.02–1.99) | 50.00–1.84 (1.87–1.84) |
| Rsym Or Rmerge | 0.06 (0.37) | 0.10 (0.59) | 0.59 (0.22) | 0.06 (0.22) |
| I/σI | 53.73 (4.29) | 25.32 (2.67) | 63.37 (9.74) | 40.65 (3.28) |
| Completeness (%) | 98.9 (89.8) | 99.9 (99.1) | 99.8 (97.5) | 97.0 (77.2) |
| Redundancy | 19.0 (7.6) | 10.4 (6.2) | 20.5 (11.6) | 9.0 (2.7) |
| # molecule in asymmetrical unit | 1 | 1 | 1 | 1 |
| Refinement | ||||
| Resolution (Å) | 38.34–1.90 (1.97–1.90) | 35.98–2.01 (2.07–2.01) | 28.93–1.99 (2.08–1.99) | 28.82–1.84 (1.91–1.84) |
| No. reflections | 26070 | 31782 | 22687 | 27862 |
| Rwork/Rfree | 0.18 (0.19)/0.21 (0.26) | 0.18 (0.23)/0.21 (0.26) | 0.16 (0.17)/0.21 (0.27) | 0.17 (0.20)/0.19 (0.25) |
| No. atoms | ||||
| Protein | 2256 | 2420 | 2267 | 2285 |
| Ligand/ion | 27 | 19 | 35 | 74 |
| TCS401 | 0 | 18 | 0 | 18 |
| Water | 213 | 171 | 213 | 200 |
| Total | 2496 | 2628 | 2515 | 2559 |
| B-factors | ||||
| Macro Molecule | 29.51 | 29.27 | 26.60 | 24.20 |
| Ligand/ion | 40.72 | 23.84 | 43.13 | 41.41 |
| TCS401 | NA | 35.99 | NA | 18.71 |
| Water | 35.07 | 32.31 | 31.05 | 30.70 |
| All atoms | 29.99 | 29.47 | 26.98 | 24.71 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.004 | 0.014 | 0.011 | 0.007 |
| Bond angles (°) | 0.650 | 1.255 | 1.047 | 0.864 |
| Ramachandran | ||||
| Outliers (%) | 0.36 | 0.34 | 0.36 | 0.35 |
| Allowed (%) | 2.86 | 2.03 | 2.51 | 2.12 |
| Favored (%) | 96.79 | 97.64 | 97.13 | 97.53 |
| Rotamer Outliers (%) | 0.41 | 0.37 | 0.40 | 0.40 |
| Clashscore | 2.22 | 3.50 | 2.42 | 3.43 |
| PDB accession no. | 5K9V | 5K9W | 5KA0 | 5KA1 |
| Data collection and refinement statistics (2/4). | |||
|---|---|---|---|
| PTP1BL192A | PTP1BL192A:TCS401 | PTP1BN193A:TCS401 | |
| Data collection | |||
| X-ray beam line | home source | home source | home source |
| Wavelength (Å) | 1.54 | 1.54 | 1.54 |
| Space group | P 31 2 1 | P 31 2 1 | P 21 |
| Cell dimensions | |||
| a, b, c (Å) | 88.9, 88.9, 105.8 | 88.7, 88.7, 106.2 | 47.6, 73.7, 88.1 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 104.4, 90.0 |
| Resolution (Å) | 50.00–1.97 (2.00–1.97) | 50.00–2.07 (2.11–2.07) | 50.00–1.90 (1.93–1.90) |
| Rsym Or Rmerge | 0.06 (0.45) | 0.14 (0.55) | 0.09 (0.27) |
| I/σI | 62.01 (4.26) | 20.58 (2.63) | 15.54 (2.51) |
| Completeness (%) | 99.1 (89.7) | 97.8 (93.8) | 89.1 (44.7) |
| Redundancy | 20.2 (11.4) | 10.9 (8.8) | 3.5 (2.4) |
| # molecule in asymmetrical unit | |||
| Refinement | 1 | I | Z |
| Resolution (Å) | 36.18–1.97 (2.03–1.97) | 44.36–2.07 (2.14–2.07) | 28.80–1.90 (1.94–1.90) |
| No. reflections | 33453 | 29179 | 41746 |
| Rwork/Rfree | 0.18 (0.22)/0.22 (0.28) | 0.19 (0.22)/0.23 (0.29) | 0.18 (0.23)/0.21 (0.32) |
| No. atoms | |||
| Protein | 2328 | 2430 | 4589 |
| Ligand/ion | 15 | 36 | 61 |
| TCS401 | 0 | 18 | 36 |
| Water | 201 | 189 | 322 |
| Total | 2544 | 2655 | 4972 |
| B-factors | |||
| Macro Molecule | 33.90 | 30.84 | 22.25 |
| Ligand/ion | 55.04 | 40.59 | 30.94 |
| TCS401 | NA | 28.70 | 18.48 |
| Water | 41.07 | 32.86 | 25.44 |
| All atoms | 34.57 | 30.98 | 22.46 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.018 | 0.007 | 0.006 |
| Bond angles (°) | 1.369 | 0.841 | 0.863 |
| Ramachandran | |||
| Outliers (%) | 0.35 | 0.67 | 0.36 |
| Allowed (%) | 2.42 | 2.35 | 2.14 |
| Favored (%) | 97.23 | 96.98 | 97.50 |
| Rotamer Outliers (%) | 0.39 | 0.37 | 1.19 |
| Clashscore | 2.56 | 1.23 | 1.52 |
| PDB accession no. | 5KA8 | 5KA9 | 5KAD |
| Data collection and refinement statistics (3/4). | ||||
|---|---|---|---|---|
| PTP1BYAYA | PTP1BYAYA:TCS401 | PTP1BT178A | PTP1BT178A:TCS401 | |
| Data collection | ||||
| X-ray beam line | home source | home source | APS | home source |
| Wavelength (Å) | 1.54 | 1.54 | 1.03 | 1.54 |
| Space group | P 31 2 1 | P 31 2 1 | C 2 | P 31 2 1 |
| Cell dimensions | ||||
| a, b, c (Å) | 87.3, 87.3, 98.6 | 88.6, 88.6, 104.2 | 117.2 46.5 59.9 | 88.6, 88.6, 104.1 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0 90.3 90.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50.00–2.07 (2.11–2.07) | 50.00–2.14 (2.18–2.14) | 50.00–2.17 (2.21–2.17) | 50.00–2.06 (2.10–2.06) |
| Rsym Or Rmerge | 0.11 (0.73) | 0.13 (0.89) | 0.12 (0.50) | 0.072 (0.390) |
| I/σI | 26.76 (2.30) | 30.40 (3.40) | 11.90 (2.11) | 35.73 (4.89) |
| Completeness (%) | 99.8 (98.8) | 100.0 (100.0) | 91.3 (87.5) | 98.2 (94.6) |
| Redundancy | 10.7 (8.6) | 21.5 (18.0) | 2.4 (2.6) | 10.8 (8.9) |
| # molecule in asymmetrical unit | 1 | 1 | 1 | 1 |
| Refinement | ||||
| Resolution (Å) | 39.93–2.07 (2.15–2.07) | 44.29–2.14 (2.23–2.14) | 35.10–2.19 (2.35–2.19) | 44.29–2.06 (2.14–2.06) |
| No. reflections | 26729 | 26422 | 15407 | 28952 |
| Rwork/Rfree | 0.19 (0.21)/0.24 (0.25) | 0.19 (0.22)/0.23 (0.25) | 0.21 (0.27)/0.24 (0.31) | 0.18 (0.20)/0.21 (0.24) |
| No. atoms | ||||
| Protein | 2233 | 2367 | 2215 | 2402 |
| Ligand/ion | 26 | 26 | 0 | 19 |
| TCS401 | 0 | 18 | 0 | 18 |
| Water | 149 | 122 | 80 | 165 |
| Total | 2408 | 2533 | 2295 | 2604 |
| B-factors | ||||
| Macro Molecule | 35.07 | 38.94 | 32.44 | 29.00 |
| Ligand/ion | 39.83 | 54.92 | NA | 34.73 |
| TCS401 | NA | 30.87 | NA | 24.17 |
| Water | 38.31 | 42.08 | 30.94 | 31.74 |
| All atoms | 35.27 | 39.13 | 32.39 | 29.18 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.002 | 0.004 | 0.002 | 0.010 |
| Bond angles (°) | 0.511 | 0.717 | 0.440 | 1.083 |
| Ramachandran | ||||
| Outliers (%) | 0.36 | 0.34 | 0.36 | 0.34 |
| Allowed (%) | 2.16 | 2.73 | 2.16 | 2.02 |
| Favored (%) | 97.48 | 96.93 | 97.48 | 97.64 |
| Rotamer Outliers (%) | 0.41 | 0.00 | 1.70 | 0.00 |
| Clashscore | 2.68 | 2.71 | 1.85 | 1.04 |
| PDB accession no. | 5KA2 | 5KA3 | 5KA4 | 5KA7 |
| Data collection and refinement statistics (4/4). | |||
|---|---|---|---|
| PTP1BP185G | PTP1BΔ7P185G | PTP1BΔ7P185G:TCS401 | |
| Data collection | |||
| X-ray beam line | home source | home source | home source |
| Wavelength (Å) | 1.54 | 1.54 | 1.54 |
| Space group | P 32 2 1 | P 32 2 1 | P 32 2 1 |
| Cell dimensions | |||
| a, b, c (Å) | 88.4, 88.4, 72.3 | 88.3, 88.3, 72.4 | 88.2, 88.2, 71.9 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50.00–1.90 (1.93–1.90) | 50.00–1.97 (2.00–1.97) | 50.00–1.97 (2.00–1.97) |
| Rsym Or Rmerge | 0.04 (0.16) | 0.06 (0.22) | 0.05 (0.22) |
| I/σI | 67.64 (10.89) | 42.57 (6.26) | 45.27 (5.75) |
| Completeness (%) | 87.3 (40.7) | 99.3 (92.6) | 99.6 (93.8) |
| Redundancy | 10.8 (8.4) | 10.1 (5.4) | 10.1 (5.4) |
| # molecule in asymmetrical unit | 1 | 1 | 1 |
| Refinement | |||
| Resolution (Å) | 38.28–1.90 (1.99–1.90) | 38.26–1.97 (2.06–1.97) | 38.19–1.97 (2.06–1.97) |
| No. reflections | 22716 | 23301 | 23144 |
| Rwork/Rfree | 0.17 (0.20)/0.21 (0.29) | 0.17 (0.19)/0.21 (0.27) | 0.18 (0.20)/0.21 (0.25) |
| No. atoms | |||
| Protein | 2234 | 2286 | 2283 |
| Ligand/ion | 27 | 28 | 25 |
| TCS401 | 0 | 0 | 18 |
| Water | 202 | 206 | 161 |
| Total | 2463 | 2520 | 2487 |
| B-factors | |||
| Macro Molecule | 32.83 | 25.59 | 29.24 |
| Ligand/ion | 46.38 | 37.26 | 37.94 |
| TCS401 | NA | NA | 34.83 |
| Water | 31.83 | 28.57 | 29.62 |
| All atoms | 32.13 | 25.83 | 29.26 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.006 | 0.010 | 0.010 |
| Bond angles (°) | 0.789 | 1.055 | 1.042 |
| Ramachandran | |||
| Outliers (%) | 0.72 | 0.36 | 0.36 |
| Allowed (%) | 2.15 | 2.14 | 2.14 |
| Favored (%) | 97.13 | 97.51 | 97.50 |
| Rotamer Outliers (%) | 2.07 | 0.79 | 2.39 |
| Clashscore | 4.95 | 4.13 | 4.98 |
| PDB accession no. | 5KAC | 5KAA | 5KAB |
All data was collected from single crystal. Values in parentheses are for highest resolution shell. NA= Not Applicable.
NMR Spectroscopy
NMR data were collected on Bruker Avance II 500- and Avance IIIHD 850 MHz spectrometers equipped with TCI HCN Z-gradient cryoprobes at 298 K. NMR measurements of PTP1B were recorded using either 2H,15N- or 2H,15N,13C-labeled protein at a final concentration of 0.2 mM in NMR buffer and 90% H2O/10% D2O. The sequence-specific backbone assignments of PTP1B and variants, as well as inhibitor-bound PTP1B were achieved using 3D triple resonance experiments including 2D [1H,15N] TROSY, 3D TROSYHNCA, 3D TROSY-HN(CO)CA, 3D TROSY-HN(CO)CACB and 3D TROSY-HNCACB. All NMR data were processed using NMRPipe (Delaglio et al., 1995) and analyzed using CcpNMR (Vranken et al., 2005).
NMR Analysis of inhibitor binding
Inhibitors were titrated into 0.2 mM 2H,15N-PTP1B at molar ratios of 0:1, 0.5:1, 1:1, 2:1 and 3:1 inhibitor:PTP1B and 2D [1H,15N] TROSY spectra were recorded for each titration point. Inhibitors (TCS401 and the allosteric inhibitor, 3-(3,5-Dibromo-4-hydroxy-benzoyl)-2-ethylbenzofuran- 6-sulfonicacid-(4-(thiazol-2-ylsulfamyl)-phenyl)-amide) were solubilized in d6-DMSO (25 mM). No chemical shift differences were identified in the PTP1B 2D [1H,15N] TROSY spectrum upon the addition of d6-DMSO. Chemical shift perturbations (Δδ) between apo PTP1B and inhibitor-bound PTP1B spectra were calculated using:
Relaxation measurements and analysis
Relaxation measurements were performed at 500 & 850 MHz 1H Larmor frequency using 2H,15N-labeled ligand-free and inhibitor-bound (at saturating 3:1 molar ratios) PTP1B at a final concentration of 0.2 mM in 50 mM HEPES pH 6.8, 150 mM NaCl, 0.5 mM TCEP and 90% H2O/10% D2O. 850 MHz 1H Larmor frequency data was recorded for all PTP1B variants. 15N longitudinal (R1) and transverse (R2) relaxation rates measurements were acquired using sensitivity-enhanced experiments. TROSY versions of T1 and T2 experiments were acquired with a recycle delay of 2.5 seconds between experiments, and the following relaxation delays for T1: 100, 600, 1000, 1600, 1800, 2000, 2400, and 3000 ms; and T2: 3.6, 7.2, 14.4, 21.6, 25.2, 28.8, 32.4, 36.0, 54.0, and 64.8 ms. T1, T2 values were calculated using CcpNMR by fitting the intensity of peaks to exponential decay function, and errors were determined via relaxation curve fitting. ~150-190 residues were analyzed for each sample, excluding peaks that overlapped or could not be accurately fitted.
Molecular Dynamics Simulations
Both unbiased and biased MD simulations we performed. In the former, dynamics is governed by a physics-based molecular mechanics force field, whereas the biased simulations also integrate information from experimental data. In particular, we used the NMR chemical shifts as a “system-specific force field correction” (Tiberti et al., 2015), so that the resulting conformational ensemble simultaneously integrates information from the most accurate force fields (Lindorff-Larsen et al., 2012) and the experimental chemical shifts (Camilloni et al., 2012) following the maximum entropy principle (Boomsma et al., 2014). All simulations were performed using the Amber ff99sb*-ILDN force field for the protein (Lindorff-Larsen et al., 2012) and TIP3P model for water. The system was neutralized by the addition of Na+ or Cl− ions at a salinity of 150 mM. Water molecules were added with an isotropic buffer distance of 1.2 nm in a cubic box. VdW and electrostatic short-range interactions were cut off at 1.0 nm and shifted from 0.8 nm to 1.0 nm. Long-term electrostatics were treated using PME with a 0.12 nm grid spacing. Velocity rescaling was used to maintain the simulation temperature. We used an integration time step of 2 fs, and bonds to hydrogen were constrained using P-LINCS. Production MD runs were then performed in the NVT ensemble after reaching the correct density using the NpT ensemble with a Berendsen barostat at 1 bar.
The chemical shift restrained simulations were performed using GROMACS 5.0.1 (Hess et al., 2008) in combination with PLUMED2. Linear restraints were employed and stepwise increased as a means of equilibrating the system with the restraints until a SLOPE value of 24 was reached. Replica averaged restraints were introduced using the ENSEMBLE flag in PLUMED2 (Tribello et al., 2014). Unbiased simulations where performed using ACEMD (Harvey et al., 2009) with a 4 fs time step following a default protocol.
QUANTIFICAITON AND STATISTICAL ANALYISIS
For the affinity measurements of PTP1B and variants with inhibitors, Sigma Plot 12.5 was used for the statistical analysis using the unpaired t-test between PTP1B/PTP1BΔ7 and all variants. All affinity measurements were repeated in triplicate.
DATA AND SOFTWARE AVAILABILITY
All NMR chemical shifts have been deposited in the BioMagResBank (BMRB 2681)4, 26832, 26835, 26836, 26837, 26838). Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB 5K9V, 5K9W, 5KA0, 5KA1, 5KA2, 5KA3, 5KA4, 5KA7, 5KA8, 5KA9, 5KAA, 5KAB, 5KAC, 5KAD).
Supplementary Material
Acknowledgments
This work was supported by the American Diabetes Association Pathway to Stop Diabetes Grant 1-14-ACN-31 to WP and R01GM098482 to RP. WP, MBAK and KLL were supported by grants from the Lundbeck and Novo Nordisk Foundations. The authors thank Drs. Nicholas Tonks (CSHL) and Michael Clarkson (Brown) for input. Crystallographic data were collected at the APS on beamline 23ID-B. GM/CA@APS has been funded in whole or in part with funds from NCI (ACB-12002) and NIGMS (AGM-12006). This research used resources of the APS, a DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory (DE-AC02-06CH11357). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information includes 5 figures and 5 tables.
Author Contributions. WP, RP and KLL designed research; MSC, YL, LESFM, MBAK, CRC, XW, WP carried out and analyzed experiments; KLL, RP and WP wrote the paper; all authors discussed the data and manuscript.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto M, Selvaratnam R, McNicholl ET, Verma G, Taylor SS, Melacini G. Signaling through dynamic linkers as revealed by PKA. Proc Natl Acad Sci U S A. 2013;110:14231–14236. doi: 10.1073/pnas.1312644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21:7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay D, Kusari A, Kenner KA, Liu F, Chernoff J, Gustafson TA, Kusari J. Protein-tyrosine phosphatase 1B complexes with the insulin receptor in vivo and is tyrosine-phosphorylated in the presence of insulin. J Biol Chem. 1997;272:1639–1645. doi: 10.1074/jbc.272.3.1639. [DOI] [PubMed] [Google Scholar]
- Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- Biedermannova L, K ER, Berka K, Hobza P, Vondrasek J. Another role of proline: stabilization interactions in proteins and protein complexes concerning proline and tryptophane. Phys Chem Chem Phys. 2008;10:6350–6359. doi: 10.1039/b805087b. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- Boomsma W, Ferkinghoff-Borg J, Lindorff-Larsen K. Combining experiments and simulations using the maximum entropy principle. PLoS Comput Biol. 2014;10:e1003406. doi: 10.1371/journal.pcbi.1003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandao TA, Johnson SJ, Hengge AC. The molecular details of WPD-loop movement differ in the protein-tyrosine phosphatases YopH and PTP1B. Arch Biochem Biophys. 2012;525:53–59. doi: 10.1016/j.abb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterwick JA, Loria JP, Astrof NS, Kroenke CD, Cole R, Rance M, Palmer AG., 3rd Multiple time scale backbone dynamics of homologous thermophilic and mesophilic ribonuclease HI enzymes. J Mol Biol. 2004;339:855–871. doi: 10.1016/j.jmb.2004.03.055. [DOI] [PubMed] [Google Scholar]
- Camilloni C, Robustelli P, De Simone A, Cavalli A, Vendruscolo M. Characterization of the conformational equilibrium between the two major substates of RNase A using NMR chemical shifts. J Am Chem Soc. 2012;134:3968–3971. doi: 10.1021/ja210951z. [DOI] [PubMed] [Google Scholar]
- Dadke S, Kusari J, Chernoff J. Down-regulation of insulin signaling by protein-tyrosine phosphatase 1B is mediated by an N-terminal binding region. J Biol Chem. 2000;275:23642–23647. doi: 10.1074/jbc.M001063200. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhammer M, Uetani N, Miranda-Saavedra D, Tremblay ML. PTP1B: a simple enzyme for a complex world. Crit Rev Biochem Mol Biol. 2013;48:430–445. doi: 10.3109/10409238.2013.819830. [DOI] [PubMed] [Google Scholar]
- Harvey MJ, Giupponi G, Fabritiis GD. ACEMD: Accelerating Biomolecular Dynamics in the Microsecond Time Scale. J Chem Theory Comput. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Iversen LF, Andersen HS, Branner S, Mortensen SB, Peters GH, Norris K, Olsen OH, Jeppesen CB, Lundt BF, Ripka W, et al. Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B. J Biol Chem. 2000;275:10300–10307. doi: 10.1074/jbc.275.14.10300. [DOI] [PubMed] [Google Scholar]
- Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller JM, Lu M, Bracken C. An effective method for the discrimination of motional anisotropy and chemical exchange. J Am Chem Soc. 2002;124:1852–1853. doi: 10.1021/ja017461k. [DOI] [PubMed] [Google Scholar]
- Krishnan N, Koveal D, Miller DH, Xue B, Akshinthala SD, Kragelj J, Jensen MR, Gauss CM, Page R, Blackledge M, et al. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat Chem Biol. 2014;10:558–566. doi: 10.1038/nchembio.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Depetris RS, Barford D, Chernoff J, Hubbard SR. Crystal structure of a complex between protein tyrosine phosphatase 1B and the insulin receptor tyrosine kinase. Structure. 2005;13:1643–1651. doi: 10.1016/j.str.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang J, Lu S, Huang W, Geng L, Shen Q, Zhang J. The mechanism of allosteric inhibition of protein tyrosine phosphatase 1B. PLoS One. 2014;9:e97668. doi: 10.1371/journal.pone.0097668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Maragakis P, Piana S, Eastwood MP, Dror RO, Shaw DE. Systematic validation of protein force fields against experimental data. PLoS One. 2012;7:e32131. doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisi GP, Loria JP. Using NMR spectroscopy to elucidate the role of molecular motions in enzyme function. Prog Nucl Magn Reson Spectrosc. 2016;92–93:1–17. doi: 10.1016/j.pnmrs.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel AM, Akke M, Palmer AG., 3rd Dynamics of ribonuclease H: temperature dependence of motions on multiple time scales. Biochemistry. 1996;35:16009–16023. doi: 10.1021/bi962089k. [DOI] [PubMed] [Google Scholar]
- Masterson LR, Cheng C, Yu T, Tonelli M, Kornev A, Taylor SS, Veglia G. Dynamics connect substrate recognition to catalysis in protein kinase. A. Nat Chem Biol. 2010;6:821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Olmez EO, Alakent B. Alpha7 helix plays an important role in the conformational stability of PTP1B. J Biomol Struct Dyn. 2011;28:675–693. doi: 10.1080/07391102.2011.10508599. [DOI] [PubMed] [Google Scholar]
- Pannifer AD, Flint AJ, Tonks NK, Barford D. Visualization of the cysteinyl-phosphate intermediate of a protein-tyrosine phosphatase by x-ray crystallography. J Biol Chem. 1998;273:10454–10462. doi: 10.1074/jbc.273.17.10454. [DOI] [PubMed] [Google Scholar]
- Peti W, Page R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr Purif. 2007;51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Peti W, Page R. NMR Spectroscopy to Study MAP Kinase Binding to MAP Kinase Phosphatases. Methods Mol Biol. 2016;1447:181–196. doi: 10.1007/978-1-4939-3746-2_11. [DOI] [PubMed] [Google Scholar]
- Piana S, Klepeis JL, Shaw DE. Assessing the accuracy of physical models used in protein-folding simulations: quantitative evidence from long molecular dynamics simulations. Curr Opin Struct Biol. 2014;24:98–105. doi: 10.1016/j.sbi.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Shinde RN, Sobhia ME. Binding and discerning interactions of PTP1B allosteric inhibitors: novel insights from molecular dynamics simulations. J Mol Graph Model. 2013;45:98–110. doi: 10.1016/j.jmgm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Tiberti M, Papaleo E, Bengtsen T, Boomsma W, Lindorff-Larsen K. ENCORE: Software for Quantitative Ensemble Comparison. PLoS Comput Biol. 2015;11:e1004415. doi: 10.1371/journal.pcbi.1004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks NK, Diltz CD, Fischer EH. Purification of the major protein-tyrosine-phosphatases of human placenta. J Biol Chem. 1988;263:6722–6730. [PubMed] [Google Scholar]
- Tribello GA, Bonomi M, Branduardi D, Camilloni C, Bussi G. PLUMED 2: New feathers for an old bird. Comput Phys Commun. 2014;185:604–613. [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Whittier SK, Hengge AC, Loria JP. Conformational motions regulate phosphoryl transfer in related protein tyrosine phosphatases. Science. 2013;341:899–903. doi: 10.1126/science.1241735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W, Fahr BJ, Zhong M, Taylor L, Randal M, et al. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat Struct Mol Biol. 2004;11:730–737. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- Xiang J, Jung JY, Sampson NS. Entropy effects on protein hinges: the reaction catalyzed by triosephosphate isomerase. Biochemistry. 2004;43:11436–11445. doi: 10.1021/bi049208d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.