SUMMARY
Environmental cues provoke rapid transitions in gene expression to support growth and cellular plasticity through incompletely understood mechanisms. Lin28 RNA-binding proteins have evolutionarily conserved roles in post-transcriptional coordination of pro-growth gene expression, but signaling pathways allowing trophic stimuli to induce Lin28 have remained uncharacterized. We find that Lin28a protein exhibits rapid basal turnover in neurons and that mitogen-activated protein kinase (MAPK)-dependent phosphorylation of the RNA-silencing factor HIV TAR-RNA-binding protein (TRBP) promotes binding and stabilization of Lin28a, but not Lin28b, with an accompanying reduction in Lin28-regulated miRNAs, downstream of brain-derived neurotrophic factor (BDNF). Binding of Lin28a to TRBP in vitro is also enhanced by phospho-mimic TRBP. Further, phospho-TRBP recapitulates BDNF-induced neuronal dendritic spine growth in a Lin28a-dependent manner. Finally, we demonstrate MAPK-dependent TRBP and Lin28a induction, with physiological function in growth and survival, downstream of diverse growth factors in multiple primary cell types, supporting a broad role for this pathway in trophic responses.
In Brief
Amen et al. show that Lin28a is rapidly stabilized by BDNF through co-association with the miRNA-processing factor TRBP, promoting the pro-growth effects of BDNF on dendritic spines. This MAPK-responsive regulatory mechanism for Lin28a is broadly generalizable across trophic stimuli and primary cell types.
INTRODUCTION
The cellular complement of proteins instructs cell identity, structure, and function. Mechanisms to rapidly tailor cellular protein composition in reaction to external trophic cues facilitate effective physiological responses supporting survival, growth, and plasticity. The heterochronic Lin28 RNA-binding proteins, first discovered in C. elegans, have evolutionarily conserved roles as determinants of transitions in biological growth (Buganim et al., 2012; Shyh-Chang and Daley, 2013a; Viswanathan et al., 2009; Zhu et al., 2010), but understanding their regulation by environmental stimuli has remained a challenge. Lin28 proteins coordinate selective protein synthesis from suites of genes that control cellular and organismal growth, in part through a mutually antagonistic relationship with the Let-7 family of micro-RNAs (miRNAs). Lin28 exerts widespread effects on mammalian growth and development, including governing body size, reproductive maturation, metabolism, tissue regeneration, and cellular reprogramming (Buganim et al., 2012; Shyh-Chang et al., 2013b; Zhu et al., 2010). Elevated Lin28 expression is a common oncogenic feature associated with advanced malignancy in humans (Mao et al., 2013; Urbach et al., 2014; Viswanathan et al., 2009) and is implicated in highly prevalent and age-related diseases such as insulin insensitivity and type II diabetes (Frost and Olson, 2011; Zhu et al., 2010, 2011).
Despite understanding the Lin28/Let-7 pathway as a key regulator of growth-related genes, mechanisms by which stimuli might target Lin28 for biological responses are lacking. Lin28 levels are high in progenitor cells and gradually decline during development to negligible basal levels in differentiated cells, leading to assumptions of protracted Lin28 regulatory mechanisms. However, a prior observation from our laboratory (Huang et al., 2012) demonstrated that brain-derived neurotrophic factor (BDNF)-mediated induction of both Dicer and Lin28a produced dual regulation of microRNA biogenesis, which created a bi-directional change in the neuronal microRNA profile and generated post-transcriptional specificity in gene expression. The finding of rapid Lin28a upregulation by BDNF in fully differentiated neurons presented an opportunity to uncover cellular signaling pathways controlling Lin28. In this study, we demonstrate that Lin28a undergoes rapid transcription-independent induction by protein stabilization in complex with a previously unknown binding partner, TRBP. BDNF induces mitogen-activated protein kinase (MAPK)-dependent TRBP phosphorylation, which both protects TRBP from proteasomal degradation and enhances TRBP binding to Lin28a. Lin28b, a paralog of Lin28a, does not associate with TRBP and is not induced by TRBP phosphorylation, exposing a mechanism for distinct stimulus-responsive regulation of Lin28a and Lin28b. These studies identify TRBP phosphorylation downstream of activated MAPK, and concomitant Lin28a stabilization, as a central mediator in neuronal pro-growth responses to BDNF. Further, our data show that this rapid Lin28a induction pathway is broadly employed downstream of multiple growth factors in diverse primary cells, highlighting an unappreciated post-transcriptional induction mechanism for the pluripotency-associated Lin28/Let-7 pathway in rapid trophic responses.
RESULTS
Rapid Post-transcriptional Induction of Lin28a, but Not Lin28b
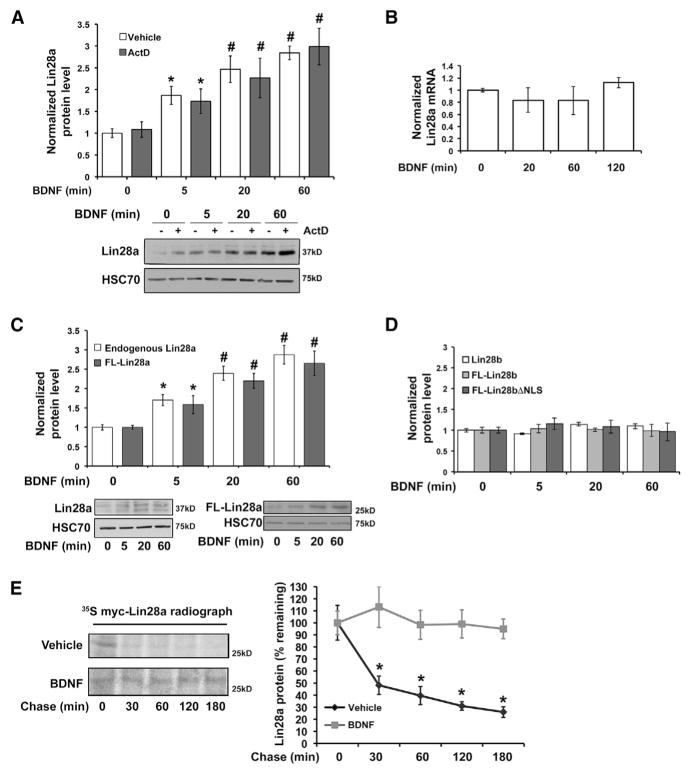
To address the regulatory mechanisms supporting stimulus-dependent Lin28a induction, we first evaluated the possibilities of altered Lin28a transcription, translation, and protein stability. Primary murine hippocampal neurons stimulated with BDNF in the presence or absence of the transcription inhibitor Actino-mycin-D displayed no significant difference in the rate or amplitude of rapid Lin28a protein induction (Figure 1A). Lin28a protein is post-transcriptionally induced 2- to 2.5-fold by BDNF over this time course, a magnitude sufficient to mediate the physiological effects of BDNF on neuronal growth and protein synthesis (Huang et al., 2012; Kelleher et al., 2004; Kanhema et al., 2006). A band near the predicted molecular weight of Lin28a protein (25 kD) typically predominates in undifferentiated cells (e.g., embryonic stem cells and cell lines), whereas, in differentiated primary cells, we detect endogenous Lin28a protein as a predominant band or bands near 37 kD, as reported previously (Huang et al., 2012; Moss and Tang, 2003; Nowak et al., 2014; Seggerson et al., 2002) and validated by knockdown (KD) (Figure S1A, available online). This banding pattern is observed using multiple antibodies raised against distinct Lin28a epitopes (Figure S1A) and has been attributed to post-translational modification and alternative splicing of Lin28a (Seggerson et al., 2002).
Figure 1. BDNF Post-translationally Induces Lin28a, but Not Paralog Lin28b, through Protein Stabilization.
(A) Quantification (top) and representative immunoblot (bottom) of Lin28a protein levels in lysates from hippocampal neurons exposed to BDNF with or without transcription blockade (Actinomycin-D) for the indicated times, normalized to GAPDH and plotted relative to 0-min BDNF without Actino-mycin-D (set as 1.0). #p < 0.05, ANOVA; *p < 0.05, t test.
(B) Quantification of Lin28a mRNA level by individual TaqMan quantitative real-time PCR reaction normalized to β-tubulin III mRNA in hippocampal neurons subjected to BDNF for the indicated times.
(C) Quantification (top) and representative immunoblots (bottom) of endogenous Lin28a and expressed FL-Lin28a in lysates from hippocampal neurons subjected to BDNF stimulation with Actinomycin-D for the indicated times, normalized to HSC70. #p < 0.05, ANOVA; *p < 0.05, t test. FL-Lin28a migrates as a single band near 25 kD, consistent with the higher apparent molecular weight of endogenous Lin28a in neurons resulting from alternative splicing, post-transcriptional modification, or both.
(D) Protein levels of endogenous Lin28b and expressed FL-Lin28b or FL-Lin28bΔNLS, normalized to GAPDH, quantified from immunoblot of lysates from hippocampal neurons subjected to BDNF stimulation with Actinomycin-D.
(B–D) Data plotted relative to 0-min BDNF (set as 1.0).
(E) Immunoprecipitated [35S]Cys/Met-labeled myc-Lin28a protein was assessed following a chase time course in vehicle (growth medium) or BDNF (added in chase). Left: representative radiograph. Right: quantification of percent remaining 35S-labeled myc-Lin28a protein at specified chase time points.
n = 3–8 independent replicates for all panels. *p < 0.05 by t test for all experiments unless otherwise noted. Quantified data represent mean ± SEM. See also Figure S1.
Concordant with the Actinomycin-D results, quantitative real-time PCR demonstrated that Lin28a mRNA levels were not significantly altered in a 2-hr time course following BDNF stimulation without transcription inhibition (Figure 1B). These results indicate that post-transcriptional mechanisms likely mediate rapid upregulation of Lin28a protein. The UTRs of mRNAs can regulate mRNA stability, localization, and translation efficiency. To examine a potential role for the Lin28a mRNA UTRs in conferring rapid Lin28a induction by BDNF, we next compared the induction of endogenous Lin28a with that of a FLAG-tagged Lin28a (FL-Lin28a) construct lacking the Lin28a UTRs. Rapid induction kinetics of endogenous Lin28a and FL-Lin28a were indistinguishable from 5 to 60 min following neuronal BDNF stimulation (Figure 1C).
Most vertebrates possess two Lin28 paralogs, Lin28a and Lin28b, both of which can reduce mature Let-7 miRNA levels by inhibiting distinct steps in Let-7 miRNA biogenesis (Piskounova et al., 2011) to influence mRNA translation. In contrast to Lin28a, protein levels of endogenous Lin28b or a FLAG-tagged Lin28b (FL-Lin28b) lacking the Lin28b UTRs did not undergo BDNF-dependent induction over a similar time course (Figures 1D and S1B). Although both Lin28a and Lin28b can shuttle between the nucleus and cytoplasm, Lin28a is localized predominantly to the cytoplasm, whereas Lin28b is believed to be primarily nuclear. We tested the possibility that nuclear localization of Lin28b prevents its induction by BDNF using a Lin28b construct in which mutation of both nuclear localization sequences (FL-Lin28bΔNLS, a gift from R. Gregory) results in predominantly cytoplasmic localization (Piskounova et al., 2011). No BDNF-responsive induction was observed in protein levels of FL-Lin28bΔNLS in hippocampal neurons (Figures 1D and S1B).
The lack of a requirement for the Lin28a UTRs in BDNF-dependent induction suggested direct regulation at the level of the Lin28a protein. To test this, we examined the effects of BDNF on Lin28a protein stability by radiolabel ([35S]Cys/Met) pulse-chase analysis. Hippocampal neurons were incubated with BDNF only during the chase period to exclude effects on radio-label incorporation because of altered translation. The median half-life of mammalian proteins has been estimated to be 46 hr (Schwanhausser et al., 2011). We observed that Lin28a protein undergoes fast turnover under basal conditions, with the Lin28a signal reduced to 48.2% ± 7.6% in 30 min (Figure 1E). Lin28a immunoprecipitated from HEK293T cells under basal conditions demonstrates K48-linked ubiquitination (Figure S1C), consistent with basal Lin28a turnover through the ubiquitin-proteasome system. In contrast, no significant loss of Lin28a was observed over 3 hr in the presence of BDNF (Figure 1E). BDNF and excitatory neuronal activity are generally reported to increase ubiquitin-proteasome-mediated turnover of cellular proteins (Bingol and Schuman, 2006; Ehlers, 2003; Jia et al., 2008; Lin et al., 2011; but see Santos et al., 2015). However, taken together, these results indicate that rapid induction of Lin28a protein levels by BDNF occurs by enhanced Lin28a protein stability.
MAPK-Mediated TRBP Phosphorylation Promotes Induction of Lin28a, but Not Lin28b
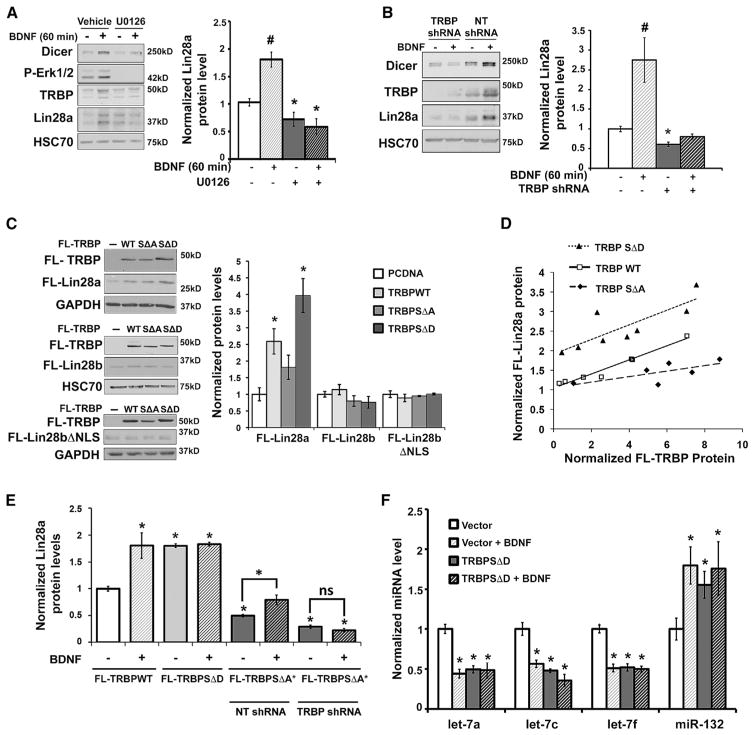
We noted that the time course of Lin28a protein induction paralleled our previous observations of rapid BDNF-induced activation of extracellular signal-regulated protein kinase (Erk) and phosphorylation of TRBP (Huang et al., 2012), which is a cofactor and binding partner of the miRNA-processing enzyme Dicer. TRBP is reported to stabilize Dicer protein through co-association, and Erk-dependent phosphorylation of TRBP can increase both TRBP and Dicer levels (Chendrimada et al., 2005; Paroo et al., 2009; Haase et al., 2005; Melo et al., 2009; but see Kim et al., 2014). To examine whether Erk activity also participates in rapid induction of Lin28a protein, we tested the effects of the MEK/Erk inhibitor U0126 on BDNF-dependent induction of Lin28a in neurons. Interestingly, Erk inhibition eliminated Lin28a induction by BDNF and also caused a modest reduction in basal Lin28a protein levels (Figure 2A), consistent with known low-level basal BDNF signaling in neurons (Jia et al., 2008; McAllister et al., 1997). As anticipated, U0126 also prevented BDNF-dependent elevation of TRBP and Dicer protein levels (Figure 2A). These results suggest that TRBP phosphorylation might underlie a requirement for Erk activity in post-translational Lin28a induction by BDNF. To directly test a requirement for TRBP in Lin28a induction, we used lentivirus-mediated KD of TRBP in hippocampal neurons, which prevented BDNF-mediated induction of both Lin28a and Dicer proteins. TRBP deficiency also produced a modest but significant reduction in the low basal levels of Lin28a (Figure 2B). In contrast, Dicer KD did not alter basal Lin28a protein levels in hippocampal neurons (Figure S2A). We conclude that both MEK/Erk activity and TRBP are required for BDNF-mediated Lin28a induction.
Figure 2. TRBP Phosphorylation by BDNF Regulates Lin28a Induction.
(A) Representative immunoblot (left) and quantified Lin28a protein levels (right) normalized to HSC70 from immunoblot of lysates from hippocampal neurons pre-treated with vehicle (DMSO) or the MEK/Erk inhibitor U0126 (30 min) prior to BDNF. Erk1/2 inhibition prevents Lin28a induction by BDNF (#p < 0.01, ANOVA) and decreases basal Lin28a protein levels (*p < 0.05, t test).
(B) Representative immunoblot (left) and quantified Lin28a protein levels (right), normalized to GAPDH, from immunoblot of lysates from hippocampal neurons treated with non-target shRNA (NT shRNA, control) or TRBP shRNA in the presence or absence of BDNF. Loss of TRBP precludes Lin28a induction by BDNF compared with the control condition (p < 0.01, ANOVA) and causes a small but significant decrease in basal Lin28a protein levels (*p < 0.01, t test).
(C) Representative immunoblots (left) and quantified FL-Lin28 levels (right) in lysates from HEK293T cells expressing FL-Lin28a, FL-Lin28b, and FL- Lin28bΔNLS in the presence of FL-TRBPWT, SΔA, SΔD, or PCDNA3.1 (control), normalized to GAPDH and plotted relative to the PCDNA3.1 condition (set as 1.0). *p < 0.05 by t test.
(D) Scatterplot of FL-Lin28a protein levels relative to titrated FL-TRBPWT, SΔA, or SΔD constructs, quantified from FLAG immunoblots of HEK293T cell lysates and normalized to GAPDH. FL-Lin28a protein levels were positively correlated with increasing expression of all TRBP constructs, but the phosphorylation status of TRBP had a significant effect (F-test(2,17) = 47.98, p < 0.0001, analysis of covariance [ANCOVA]). TRBPSΔD caused the greatest increase in FL-Lin28a (Mean [M] = 2.65) compared with TRBPWT (M = 1.73) and TRBPSΔA (M = 1.20).
(E) Hippocampal neurons co-expressing lentiviral FL-Lin28a and either FL-TRBPWT, SΔD, or shRNA-resistant SΔA (SΔA*) were treated with vehicle or BDNF (60 min). Under the FL-TRBPSΔA* condition, neurons were treated with either NT shRNA (control) or shRNA targeting endogenous TRBP (TRBP shRNA). The graph shows quantification of FL-Lin28a protein levels normalized to HSC70, plotted relative to the effect of FL-TRBPWT + vehicle, set as 1.0 (*p < 0.05, ANOVA). Immunoblots are shown in Figures S2B and S2C.
(A–E) n = 3–16 independent replicates each.
(F) Quantification of miRNA levels following vehicle or BDNF (60 min) treatment in neurons expressing control virus or TRBPSΔD. miRNA levels normalized to U6 snRNA are plotted relative to each vehicle-stimulated vector control, set as 1.0 (*p < 0.05, ANOVA). n = 7–24 independent replicates.
Quantified data represent mean ± SEM. See also Figure S2.
We next investigated the mechanism by which TRBP might upregulate Lin28a protein and the role of TRBP phosphorylation in this process. Mass spectrometry analysis has shown TRBP phosphorylation at four serine residues (142,152,283,286) (Paroo et al., 2009), two of which are potential consensus Erk phosphorylation sites. Expression of either FLAG-tagged wild-type (TRBPWT) or phosphomimic (TRBPSΔD), but not phosphomutant, TRBP (TRBPSΔA) (a gift from Z. Paroo; Paroo et al., 2009) in HEK293T cells significantly elevated Lin28a protein expression from a construct lacking the Lin28a UTRs (FL-Lin28a), showing that these effects require only the Lin28a protein coding region (Figure 2C). In contrast, the levels of FL-Lin28b protein were not altered by expression of TRBP constructs even when Lin28b was localized to the cytoplasm through mutation of its nuclear localization sequences (FL-Lin28bΔNLS) (Figure 2C). Dose titration of the TRBP constructs revealed that, at equivalent protein levels, phospho-mimic TRBP produced significantly greater elevation of Lin28a protein than either wild-type or phospho mutant TRBP (Figure 2D).
If BDNF signaled through phosphorylation of TRBP to rapidly induce Lin28a, then we anticipated that phospho-mimic TRBP might similarly induce Lin28a, whereas phospho-mutant TRBP might inhibit induction. To test this prediction, we subjected neurons to lentivirus-mediated expression of equivalent levels of FL-TRBPWT, SΔD, and SΔA under control of a neuron-specific synapsin promoter. In neurons expressing wild-type TRBP, BDNF was still able to upregulate FL-Lin28a protein levels; however, phospho-mimic TRBP expression elevated basal Lin28a levels and occluded further induction by BDNF (Figures 2E and S2B). BDNF-dependent induction of Lin28a could still be observed in neurons expressing phospho-mutant TRBP in the presence of endogenous TRBP (non-target short hairpin RNA [shRNA]), but this was eliminated by concomitant KD of endogenous TRBP (TRBP shRNA) (Figures 2E, S2C, and S2D). We conclude that TRBP deficient in serine sites for Erk phosphorylation does not support BDNF induction of Lin28a. Our laboratory demonstrated previously that BDNF achieves translational specificity in part by increasing levels of many mature miRNAs through elevated Dicer and TRBP while simultaneously decreasing biogenesis of Lin28-targeted miRNAs through Lin28a induction (Huang et al., 2012). Neuronal expression of phospho-mimic TRBP mimicked this downstream effect of elevated Lin28a by lowering the levels of Lin28-targeted miRNAs (e.g., Let-7 family members) (Figure 2F) and occluding further reduction by BDNF. As expected, a Dicer-dependent but non-Lin28-targeted control miRNA (miR-132) underwent BDNF-mediated induction that was mimicked and occluded by neuronal expression of phospho-mimic TRBP (Figure 2F). These results indicate that TRBP phosphorylation is necessary and sufficient to functionally upregulate Lin28a and link BDNF-induced TRBP phosphorylation to rapid post-transcriptional changes in the Let-7 family miRNAs that govern synthesis of many pro-growth proteins.
TRBP Phosphorylation Reduces Its Polyubiquitination
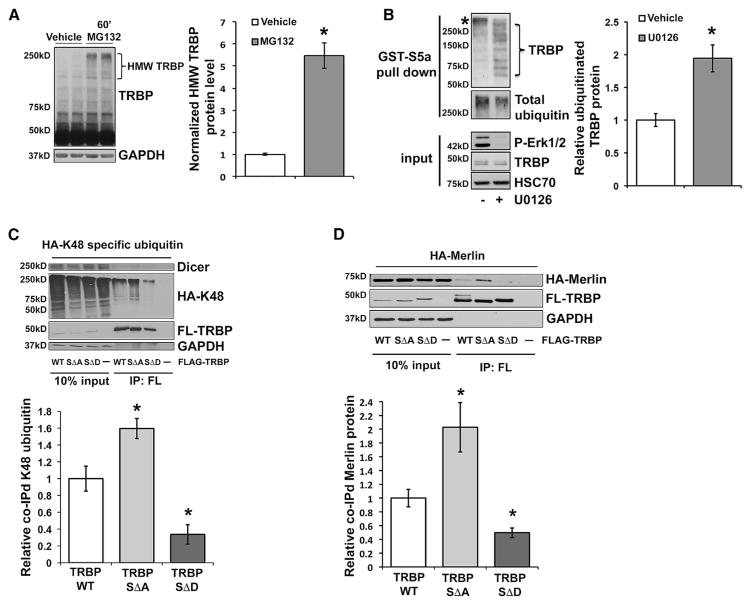
Our results implicated Erk-mediated TRBP phosphorylation and the concomitant increase in total TRBP protein (Figures 2A–2E) as a critical signaling event producing rapid Lin28a induction. Previous reports using cell lines indicated that TRBP was subject to cell density-dependent turnover by the 26S proteasome during a prolonged 15-hr time course (Lee et al., 2006) and that phosphorylation might enhance TRBP stability (Paroo et al., 2009). To examine the possibility that BDNF-induced TRBP phosphorylation might alter its proteasomal regulation, we first evaluated whether TRBP underwent rapid proteasome-dependent turnover in primary cells. Brief inhibition of 26S proteasomal activity (MG132, 60 min) in hippocampal neurons resulted in the accumulation of high-molecular-weight (HMW) forms of TRBP, suggesting basal ubiquitin-mediated turnover (Figure 3A). To investigate the mechanism for regulation of TRBP stability by phosphorylation, we first assessed whether MEK/Erk inhibition (U0126) altered ubiquitinated TRBP levels in neurons responding to BDNF. We isolated K48-linked polyubiquitin-conjugated proteins from neuronal lysates by pull-down with a glutathione S-transferase (GST)-S5a ubiquitin interaction motif and detected ubiquitinated TRBP species that were elevated by MEK/Erk inhibition and ran in a laddering pattern, consistent with protein polyubiquitination (Figure 3B). S5a is an integral component of the 26S proteasome that interacts selectively with K48-linked ubiquitin chains (associated with proteasomal degradation) (Zhang et al., 2009). We next evaluated wild-type, phospho-mimic, and phospho-mutant forms of TRBP for K48-linked ubiquitination. Stringent immunoprecipitation (IP) of FL-TRBP, FL-TRBPSΔD, or FL-TRBPSΔA from HEK293T cells co-expressing an hemagglutinin (HA)-tagged ubiquitin mutant exclusive for K48-linkages (Lim et al., 2005) showed a ladder of anti-HA immunoreactivity with wild-type and phospho-mutant TRPB, consistent with polyubiquitination. Quantification normalized to the amount of immunoprecipitated TRBP showed that K48-linked ubiquitination of phospho-mutant TRBP was significantly elevated relative to wild-type TRBP. In contrast, K48-linked ubiquitination of phospho-mimic TRBP was reduced by nearly 5-fold relative to phospho-mutant TRBP and was also significantly reduced relative to wild-type TRBP (Figure 3C). IP stringency was validated by loss of TRBP association with a Dicer-containing complex (Figure 3C). These results are consistent with phosphorylation of TRBP regulating the process of TRBP ubiquitination and proteasomal degradation. To understand how this regulation might occur, we considered the known binding interactions of TRBP.
Figure 3. TRBP Phosphorylation Reduces Merlin Binding and TRBP Polyubiquitination.
(A) Representative immunoblot (left) and HMW TRBP protein levels, quantified from immunoblot of lysates from hippocampal neurons treated with vehicle (DMSO) or MG132 (60 min), normalized to GAPDH and plotted relative to vehicle alone (set as 1.0).
(B) Representative immunoblot (left) and quantification of TRBP protein bound to GST-S5a (ubiquitin binding motif) (right) using lysates from hippocampal neurons pre-treated with either U0126 or vehicle control (30 min), followed by BDNF (60 min). Ubiquitinated TRBP protein was normalized to total ubiquitin pull-down and plotted relative to vehicle (set as 1.0). *non-TRBP specific band reactive with secondary antibody.
(C) Lysates from HEK293T cells co-expressing FL-TRBPWT, SΔA, SΔD, or PCDNA3.1 alone (control) with HA-K48 ubiquitin underwent stringent IP with anti-FLAG antibody, followed by immunoblot with anti-FLAG and anti-HA antibodies. Shown are representative immunoblots (left) and quantification (right) of co-associated HA-K48 ubiquitin normalized to the amount of immunoprecipitated FL-TRBP for each construct, plotted relative to the FL-TRBPWT condition (set as 1.0).
(D) Lysates from HEK293T cells co-expressing FL-TRBPWT, SΔA, SΔD, or PCDNA3.1 alone (control) with HA-Merlin were immunoprecipitated with anti-FLAG antibody, followed by immunoblot with anti-FLAG and anti-HA antibodies. Shown are representative immunoblots (left) and quantification (right) of co-associated HA-Merlin protein normalized first to the amount of input HA-Merlin protein and next to the immunoprecipitated FL-TRBP for each construct, plotted relative to the FL-TRBPWT condition (set as 1.0).
*p < 0.05 by t test for all experiments. Quantified data represent mean ± SEM. n = 3–8 independent replicates each. See also Figure S3.
The third double-stranded RNA binding domain of TRBP does not bind RNA but is, instead, thought to mediate interactions with protein binding partners such as Dicer (Haase et al., 2005) and a tumor suppressor protein, Merlin (Lee et al., 2004). Merlin binding to TRBP has been shown to facilitate ubiquitination and proteasomal degradation of TRBP (Lee et al., 2006). As anticipated, we observed that Merlin KD in HEK293T cells resulted in the detection of significantly less ubiquitinated TRBP compared with cells expressing a non-target shRNA control (Figure S3A). Accordingly, we asked whether Merlin played a role in BDNF-mediated regulation of TRBP phosphorylation and abundance. Although treatment of neurons with BDNF did not alter the total levels of Merlin protein (Figure S3B), pull-down assays showed that HA-tagged Merlin (Zhao et al., 2007) expressed in HEK293T cells exhibited a co-association with FL-TRBPWT that was robustly enhanced with FL-TRBPSΔA and greatly reduced with FL-TRBPSΔD (Figure 3D). This result suggests that phosphorylation of TRBP inhibits TRBP binding to Merlin, a mechanism that could underlie the reduced ubiquitination and enhanced stability of phosphorylated TRBP and, in turn, mediate enhanced Lin28a stability.
TRBP Is a Lin28a Binding Partner
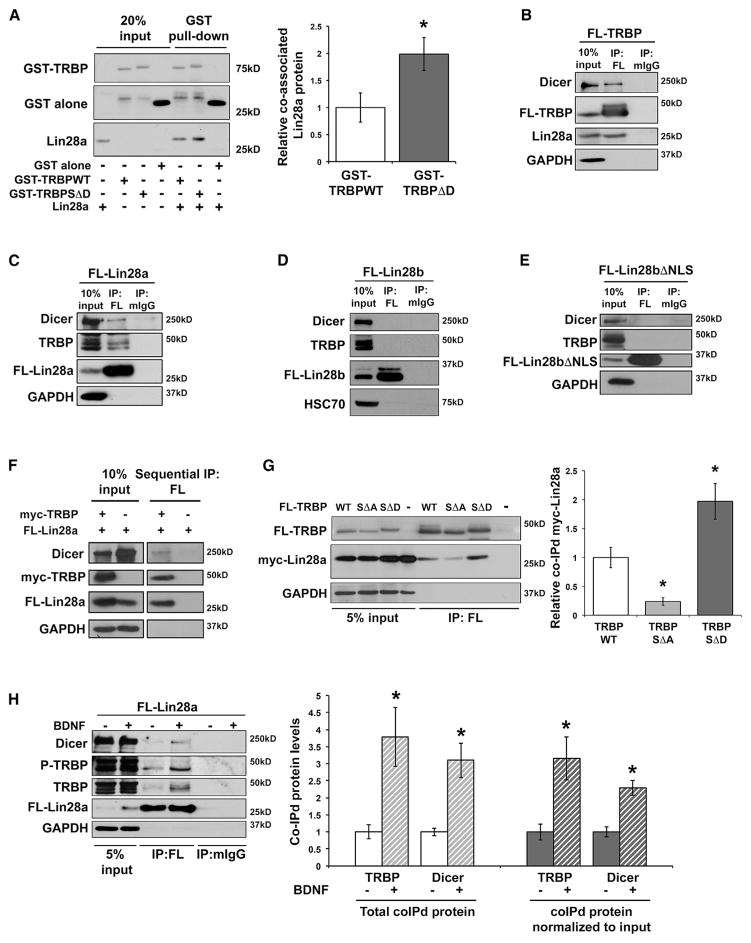
Although precise roles of TRBP in RNA processing are incompletely delineated, TRBP has been shown to regulate miRNA biogenesis and to assist in generating the small RNA-induced silencing complex (RISC) by stabilizing associations with Dicer-containing complexes (Chendrimada et al., 2005; Daniels et al., 2009; Paroo et al., 2009; Fukunaga et al., 2012; Wilson et al., 2015). To further elucidate the role of TRBP, and in particular phospho-TRBP, in Lin28a induction, we asked whether TRBP might also stabilize Lin28a through direct co-association using recombinant TRBP and Lin28a proteins expressed and purified from bacteria. Purified Lin28a protein bound to a pull-down of purified GST-TRBP protein but not to GST alone (Figures 4A and S4A), demonstrating direct protein-protein interaction between Lin28a and TRBP. Lin28a protein showed significantly increased binding to phospho-mimic TRBP protein (GST-TRBPSΔD) compared with wild-type TRBP protein (Figure 4A). This result is consistent with our intracellular expression titration (Figure 2D) and indicates that TRBP phosphorylation may stabilize Lin28a both through increased stability of TRBP itself and also by increasing binding affinity for Lin28a. We next tested whether endogenous Lin28a associates with TRBP-containing protein complexes in HEK293T cells, which, as we found, express Lin28a at a molecular weight of ~25 kD (Figure S4B), although a previous study suggested that HEK293T cells may lack Lin28a (Heo et al., 2012). Endogenous Lin28a and Dicer both co-immunoprecipitated with FL-TRBP and not with control immunoglobulin G (IgG) in HEK293T cells, and the reverse association of endogenous TRBP and Dicer with immunoprecipitated FL-Lin28a was also observed (Figures 4B and 4C). Because TRBP expression did not induce the Lin28a paralog, Lin28b (Figure 2C), we tested whether this might be due to an inability of Lin28b to associate with complexes containing TRBP and Dicer. IP of FL-Lin28b or cytoplasmic Lin28b (FL-Lin28bΔNLS) did not demonstrate association with endogenous Dicer or TRBP (Figures 4D and 4E) and provided additional corroboration of the specificity of the FL-Lin28a IP. Specific co-association of FL-TRBP with myc-tagged Lin28a (myc-Lin28a) and endogenous Dicer, but not with myc-Lin28bΔNLS, was also observed (Figure S4C). These results highlight a critical node of differential regulation by TRBP between the mammalian Lin28 paralogs.
Figure 4. TRBP and Lin28a Co-association Is Enhanced by TRBP Phosphorylation.
(A) Representative immunoblot (left) and densitometric quantification (right) of purified Lin28a protein co-associated with purified GST-TRBP, normalized to TRBP pull-down and plotted relative to GST-TRBPWT (set as 1.0). A lower band in the GST-TRBP conditions that migrates similarly to GST alone reflects partial cleavage of the GST tag through a protease site between GST and TRBP. Uncropped blots are shown in Figure S4A.
(B–E) Lysates from HEK293T cells expressing (B) FL-TRBPWT, (C) FL-Lin28a, (D) FL-Lin28b, and (E) FL-Lin28bΔNLS were immunoprecipitated with anti-FLAG or control rabbit IgG (rIgG) or mouse IgG (mIgG) antibodies. Representative immunoblots (A–D) show immunoprecipitated proteins and co-immunoprecipitated endogenous proteins.
(F) Sequential IP of HEK293T cells lysates involving initial IP for myc-TRBP followed by subsequent IP for FL-Lin28a. A flowchart and experimental details are shown in Figure S4D.
(G) Representative immunoblot (left) and quantification (right) of lysates from HEK293T cells co-expressing FL-TRBPWT, SΔA, SΔD, or PCDNA3.1 alone (control) with myc-Lin28a and immunoprecipitated with anti-FLAG antibody. Myc-Lin28a was overexpressed to minimize differential the effects of the FL-TRBP constructs on total Lin28a levels. Co-immunoprecipitated myc-Lin28a was normalized first to myc-Lin28a input levels and then to immunoprecipitated FL-TRBP for each construct and plotted relative to the FL-TRBPWT condition (set as 1.0).
(H) Lysates from hippocampal neurons expressing FL-Lin28a and treated with either vehicle (growth medium) or BDNF (90 min) were immunoprecipitated with anti-FLAG or control mIgG antibody. Left: representative immunoblot. Right: co-immunoprecipitated TRBP and Dicer proteins quantified as both total co-immunoprecipitated protein (left) and input-normalized co-immunoprecipitated protein (right), plotted relative to vehicle alone, set as 1.0.
*p < 0.05 by t test for all experiments. Quantified data represent mean ± SEM. n = 3–5 independent replicates for each panel. See also Figure S4.
We next used a sequential co-immunoprecipitation (coIP) strategy to distinguish whether Lin28a can exist in a single complex with both TRBP and Dicer or whether the observed coIP might reflect separate complexes of Lin28a with either TRBP or Dicer. Lysates from HEK293T cells co-expressing myc-TRBP and FL-Lin28a were subjected to initial IP for the myc epitope, followed by elution and secondary IP of eluates for the FLAG epitope (Figures 4F and S4D). Immunoblot of eluates from secondary FLAG IP showed that Lin28a and Dicer were both associated with the TRBP-containing complex, indicating that these components can reside in a single complex (Figure 4F). Just as previous RISC characterizations show that RNase treatment reduces (but does not eliminate) TRBP association with Dicer (Haase et al., 2005; Daniels et al., 2009), we observed a striking reduction in the co-association of cellular Lin28a with TRBP and Dicer by RNaseA (Figure S4E). Intracellular associations between RNA-binding proteins are often enhanced by cooperative complexes with RNA and other proteins, which can facilitate interaction at low intracellular concentrations. The high conservation of RNA-binding domains between Lin28a and Lin28b and the failure of Lin28b to co-associate with TRBP (Figures 4D, 4E, and S4C) suggest that RNA-binding is not the sole mechanism of intracellular Lin28a and TRBP interaction. Taken together, our results are consistent with direct interactions of TRBP and Lin28a (observed with recombinant, purified proteins in vitro) that are facilitated at intracellular protein concentrations by the presence of RNA.
BDNF-Mediated TRBP Phosphorylation Promotes Lin28a Co-association
Our results indicated that phospho-mimic, compared with phospho-mutant, TRBP significantly elevated total Lin28a levels (Figures 2C and 2D) and also exhibited an increased apparent binding affinity for Lin28a (Figure 4A). Based on these observations, we suspected that phosphorylation of TRBP might convey signal-induced Lin28a regulation and that intracellular Lin28a might also preferentially co-associate with phospho-mimic TRBP. IP of lysates from HEK293T cells co-expressing myc-Lin28a with FL-tagged wild-type, phospho-mimic, or phospho-mutant TRBP revealed robust association of Lin28a with phospho-mimic TRBP, which was enhanced relative to wild-type TRBP, and faint association of Lin28a with phospho-mutant TRBP (Figure 4G). Overexpression of Lin28a was employed in these experiments to minimize possible artifacts resulting from differential induction of total Lin28a levels among the TRBP constructs. FL-TRBP constructs were transfected to achieve approximately equal expression, and quantitation of co-associated Lin28a was normalized to both Lin28a input and immuno-precipitated FL-TRBP level for each construct.
We next tested whether Lin28a might undergo enhanced association with phosphorylated TRBP as the physiological mechanism for Lin28a induction in neurons responding to BDNF. Depletion IP with lentivirus-mediated expression of FL-Lin28a under control of the neuron-specific synapsin promoter revealed that BDNF significantly enhanced the association of FL-Lin28a with endogenous total TRBP, phospho-TRBP, and Dicer and produced the expected elevation in total levels of all three proteins (Figure 4H; depleted lysates shown in Figure S4F). These effects of BDNF were blocked by MEK/Erk inhibition using U0126 (Figure S4G). The increased co-association of Lin28a with TRBP and Dicer was partially attributable to elevation in the total levels of all three proteins, but co-association remained significantly increased by BDNF even when co-immunoprecipitated proteins were normalized to their respective input levels (Figure 4H). These findings suggest that BDNF can induce the Lin28a-TRBP-Dicer complex both by increasing total levels of TRBP and by enhancing the ability of Lin28a to associate with phosphorylated TRBP. Phospho-TRBP was detected using a phospho-specific TRBP polyclonal antibody developed against an amino-terminal perfect Erk phosphorylation consensus site located adjacent to a putative Erk docking site in TRBP (Figure S4H); we observed loss of antibody signal in lysates from Erk-inhibited cells (Figure S4I) and increased antibody signal in lysates from BDNF-stimulated neurons (Figure S4J). Although our data indicate that phospho-TRBP can function as a hub for parallel regulation of both Lin28a and Dicer, Dicer might also be independently regulated, potentially through additional known Dicer partner proteins (Wilson et al., 2015; Rybak-Wolf et al., 2014; Pépin et al., 2012; Lee et al., 2013). Collectively, these data establish a context in which a stimulus, BDNF, employs TRBP phosphorylation as a central molecular mechanism coordinating induction of Lin28a and Dicer, which, together, produce gene target specification in protein synthesis (Huang et al., 2012; Ruiz et al., 2014).
Phospho-TRBP Requires Lin28a to Regulate Dendritic Spine Growth
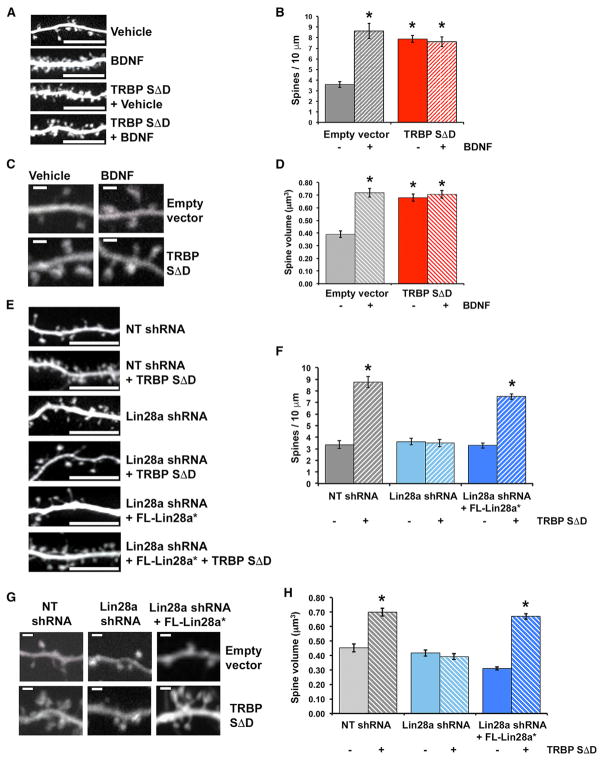
BDNF-regulated protein synthesis plays a prominent role in the synaptic plasticity of terminally differentiated excitatory neurons, in part by promoting growth of dendritic spines, which are the primary sites of excitatory synaptic contacts (Alonso et al., 2004; Je et al., 2009; Tanaka et al., 2008). We used this biological readout to test whether a pro-growth response to BDNF requires phospho-TRBP-mediated induction of Lin28a. We initially assessed the effect of phospho-TRBP on spine growth by comparing the density and volumes of spines in vehicle-or BDNF-stimulated hippocampal pyramidal neurons (days in vitro [DIV] 17–19) expressing either phospho-mimic TRBP or empty vector control. Hippocampal pyramidal neurons expressing empty vector exhibited robust BDNF-induced elevations in both dendritic spine density (increased by 140.6%; Figures 5A and 5B) and spine head volume (increased by 78.2%; Figures 5C and 5D), consistent with previous observations of enhanced excitatory synaptic function by BDNF (Tanaka et al., 2008; Lauterborn et al., 2007). In contrast, neurons expressing phospho-mimic TRBP showed increases in dendritic spine density and spine head volume that mimicked BDNF-mediated elevations and occluded further induction by BDNF (Figures 5A–5D).
Figure 5. TRBP Phosphorylation Downstream of BDNF Regulates Neuronal Dendritic Spine Growth through Lin28a.
(A–D) Hippocampal neurons expressing either empty vector or TRBPSΔD were treated with vehicle (growth medium) or BDNF (12 hr).
(E–H) Hippocampal neurons expressing either empty vector (control) or TRBPSΔD and either NT shRNA (control), Lin28a shRNA, or Lin28a shRNA plus an shRNA-resistant FL-Lin28a construct (FL-Lin28a*).
(A and E) Representative confocal projections of hippocampal pyramidal dendrites. Scale bar, 10 μm.
(B and F) Quantification of dendritic spine density.
(C and G) Representative confocal images of spines. Scale bar, 1.0 μm.
(D and H) Quantification of average spine volume.
*p < 0.001, ANOVA. n = 10–18 dendritic segments and 81–321 individual spines from 4–11 neurons for each condition. Quantified data represent mean ± SEM See also Figure S5.
We hypothesized that phospho-TRBP might support dendritic spine growth by producing a pro-growth program of protein synthesis through elevated Lin28a, which could relieve Let-7 miRNA-mediated repression of pro-growth mRNAs (Huang et al., 2012). Accordingly, we next tested the role of Lin28a in the promotion of spine growth by phospho-mimic TRBP. KD of Lin28a through RNAi, but not expression of a control hairpin (non-target shRNA), prevented the induction of elevated spine density and spine head volume by phospho-TRBP (Figures 5E–5H). The effects of phospho-TRBP on spine density and volume were rescued by expression of an shRNA-resistant Lin28a construct (FL-Lin28a*) in the presence of Lin28a shRNA (Figures 5E–5H and S5A), supporting specificity of the requirement for Lin28a. Neurons expressing control non-target shRNA displayed increased spine growth in response to phospho-TRBP, similar to wild-type neurons that were not expressing shRNA (compare Figures 5F and 5H with Figures 5B and 5D). These experiments place Lin28a as an essential downstream mediator of phospho-TRBP in a physiological growth response, which is consistent with our data showing BDNF-induction of a Lin28a-stabilizing complex through TRBP phosphorylation. We conclude that TRBP phosphorylation and subsequent Lin28a protein stabilization provide an essential function in neurotrophin-induced structural plasticity of terminally differentiated neurons.
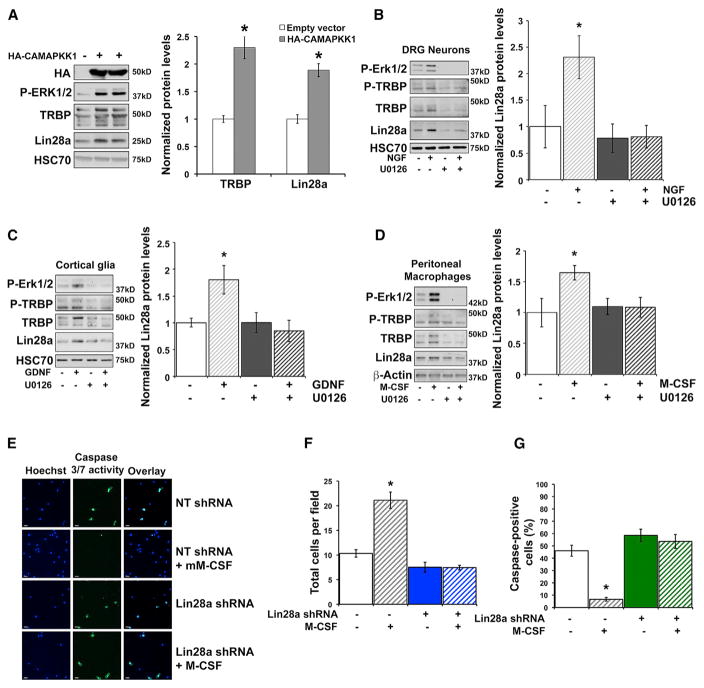
MAPK-Dependent Lin28a Induction Is a Shared Feature in Trophic Responses
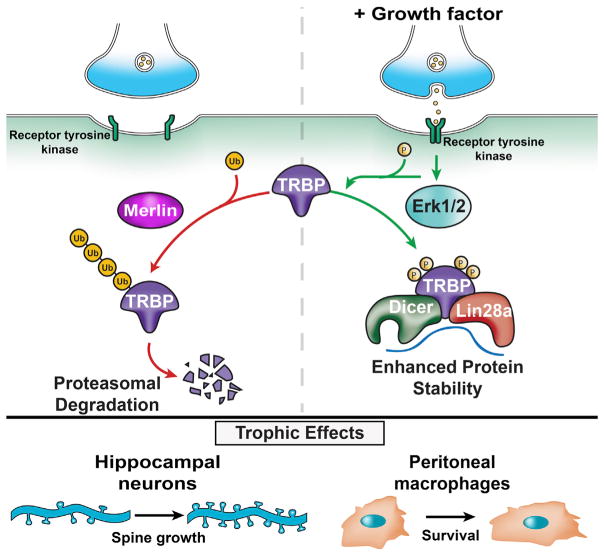
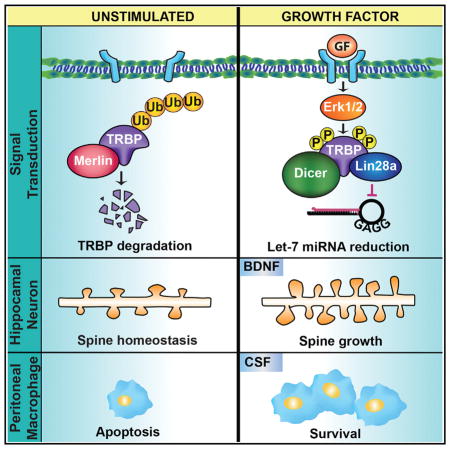
Our data collectively implicated Erk-mediated TRBP phosphorylation and the concomitant increase in total TRBP protein as a critical signaling event capable of producing rapid Lin28a induction to generate a pro-growth program. The observation that phospho-TRBP could induce Lin28a protein not only in neurons but also in an HEK293T cell line (Figures 2C–2E) suggested broader use of this regulatory pathway in trophic responses. Because MAPK pathway activation is a common feature of growth factor signaling cascades, we tested whether this was sufficient to induce Lin28a. Expression of a constitutively active MEK construct (HA-CAMAPKK1) (Mansour et al., 1994) in HEK293T cells induced both TRBP and Lin28a proteins (Figure 6A), consistent with the potential for MAPK pathway activation to control TRBP and Lin28a protein regulation more widely. We next tested whether MAPK-mediated TRBP phosphorylation might serve as a gateway to dynamic receptor-mediated regulation of Lin28a in a variety of cell types. Stimulation of primary murine cultures of dorsal root ganglion (DRG) neurons with nerve growth factor (NGF), cortical glia with glial-derived neurotrophic factor (GDNF), and peritoneal macrophages with murine macrophage colony stimulating factor (M-CSF) all resulted in TRBP and Lin28a protein induction (Figures 6B–6D and S6A–6C). As in hippocampal neurons, Lin28a protein in these primary cells migrates as predominant bands near 37 kD (Figure S1A), as reported previously in differentiated cells (Huang et al., 2012; Moss and Tang, 2003; Nowak et al., 2014; Seggerson et al., 2002). In each cell type, TRBP and Lin28a induction by growth factor was blocked by treatment with the MEK/Erk inhibitor U0126 (Figures 6B–6D and S6A–S6C). These results are consistent with TRBP phosphorylation, downstream of MAPK activation, serving as a regulatory hub for Lin28a stabilization and subsequent trophic effects in diverse primary cells. We further tested this premise by assessing whether Lin28a KD might prevent physiological effects of trophic signaling not only on neuronal dendritic spines but also in an unrelated cell type, murine peritoneal macrophages. Although M-CSF can promote proliferation of less differentiated macrophages, we and others observed minimal effects of M-CSF on the proliferation of differentiated peritoneal macrophages (Figure S6D; Wang et al., 2013). Instead, M-CSF trophic support can promote peritoneal macrophage survival, particularly in the context of environmental stress such as serum deprivation (Jin et al., 1995). We exposed primary peritoneal macrophages to serum deprivation for 48 hr and examined rescue of survival by concurrent incubation with M-CSF. In peritoneal macrophages expressing a control non-target shRNA, M-CSF significantly rescued survival, as measured by cell number, and significantly reduced the percentage of cells undergoing apoptosis, as measured by caspase activity (Figures 6E–6G). In contrast, KD of Lin28a through RNAi (Lin28a shRNA) prevented M-CSF-mediated effects on survival and reduced caspase activity in peritoneal macrophages responding to serum deprivation (Figures 6E–6G). These data indicate that Lin28a expression is necessary for M-CSF trophic signaling to support peritoneal macrophage survival. Collectively, our findings lead us to propose a model in which MAPK/Erk-mediated phosphorylation of TRBP reduces its association with Merlin, leading to decreased proteasomal degradation of TRBP and enhanced levels of a phospho-TRBP and Lin28a protein complex (Figure 7), which can also include Dicer. Activation of this pathway downstream of multiple receptor tyrosine kinases is implicated in diverse physiological contexts, including growth of neuronal dendritic spines and enhanced survival of peritoneal macrophages. We conclude that TRBP phosphorylation is a previously unrecognized mechanism for rapid post-translational induction of Lin28a protein and downstream control of miRNA biogenesis that has biological relevance in acute trophic factor responses.
Figure 6. MAPK Pathway Activation by Diverse Growth Factors Induces TRBP and Lin28a Proteins in Multiple Cell Types.
(A) Representative immunoblot (left) and quantification (right) of immunoblots from HEK293T cell lysates expressing empty vector (control) or constitutively active MEK (HA-CAMAPKK1), normalized to HSC70 and plotted relative to empty vector (set as 1.0).
(B–D) Primary mouse (B) DRGs, (C) cortical glia, and (D) peritoneal macrophages were stimulated with (B) NGF, (C) GDNF, or (D) M-CSF for 90 min following a 30-min pretreatment with either DMSO (control) or the pharmacological MEK/Erk inhibitor U0126. Left: representative immunoblots. Right: densitometric quantification of immunoblots, normalized to (B and C) HSC70 or (D) β-Actin, plotted relative to no growth factor vehicle control (set as 1.0). Uncropped immunoblots of Lin28a protein are shown in Figures S6A–S6C.
(A–D) *p < 0.05, t test.
(E–G) Survival assay in peritoneal macrophages expressing either NT shRNA (control) or shRNA targeting Lin28a.
(E) Macrophages were cultured (48 hr) in serum-starvation medium with or without M-CSF and then analyzed for cell number with Hoechst 33342 nuclear stain (blue) and caspase-3/7 activity (green).
(F) Average number of cells per field for each condition.
(G) Average percent caspase-3/7-positive cells per field for each condition. Quantified data represent mean ± SEM.
(F and G) *p < 0.01, ANOVA.
n = 16–17 microscope fields from 5 independent dishes. See also Figure S6.
Figure 7. Growth Factor-Mediated TRBP Phosphorylation Rapidly Stabilizes Lin28a Protein.
Shown is a graphical model. Left: at baseline, TRBP undergoes polyubiquitination and degradation facilitated by Merlin protein. Lin28a protein levels are negligible as it undergoes rapid turnover. Right: growth factor binding to cognate receptors activates Erk1/2 signaling and subsequent TRBP phosphorylation, which stabilizes TRBP and enhances the TRBP/Lin28a protein complex, elevating Lin28a protein levels. Downstream inhibition of Let-7 miRNAs facilitates protein synthesis from pro-growth mRNAs. Bottom: The rapid Erk1/2-dependent induction pathway exerts Lin28a-dependent trophic effects on neuronal spines and peritoneal macrophage survival.
DISCUSSION
The conserved importance of Lin28 proteins in processes of growth and development has led to their widespread study in diverse organisms. A post-translational control mechanism enabling elevation of Lin28a may have escaped previous recognition because the understanding that Lin28a may undergo rapid stimulus-dependent induction is relatively recent (Huang et al., 2012; Ruiz et al., 2014; Shyh-Chang and Daley, 2013a). Genetic studies in C. elegans, where Lin28 was first discovered, have reported the developmental decline of Lin28 levels as a result of miRNA-mediated repression (Morita and Han, 2006; Shyh-Chang and Daley, 2013a), and changes in transcription have been associated with altered mammalian Lin28 levels during differentiation and in tumors (Iliopoulos et al., 2009; Marson et al., 2008; Shyh-Chang and Daley, 2013a). In addition to development, Lin28 levels are also altered in a variety of biological contexts, including onco-genesis, injury, and cellular reprogramming, to generate pluripotent stem cells (Rehfeld et al., 2015; Viswanathan et al., 2009; Ramachandran et al., 2010; Buganim et al., 2012). The potentially broader significance of phospho-TRBP as a Lin28a regulatory linchpin is underscored by our finding that upregulation of phospho-ERK and TRBP downstream of diverse growth factor receptors in multiple cell types is also accompanied by Lin28a protein induction. Further, we find this pathway to be required in biological responses to trophic factors in neurons and a non-neuronal cell type, peritoneal macrophages.
The vertebrate paralogs Lin28a and Lin28b are thought to have arisen through gene duplication, and their distinct and overlapping roles remain incompletely understood. Lin28a and Lin28b proteins share similar, although not identical, expression patterns and a high degree of sequence identity (66% in mice, 73% in humans) (Balzer et al., 2010). Both Lin28 paralogs can lower mature Let-7 miRNA levels to de-repress growth-related transcripts, but the extent of redundancy in their net effects on gene expression and biological responses is not yet clear. Our finding of differential stabilization of Lin28a, but not Lin28b, through protein association with phospho-TRBP reveals a regulatory difference that may have physiological relevance in growth and cellular self-renewal and is also intriguing in light of literature reporting discrete oncogenic roles of Lin28a and Lin28b in different tumor settings (Thornton and Gregory, 2012).
Post-transcriptional regulation of gene expression is currently understood to substantially, if not predominantly, control cellular protein abundance, but the knowledge of pathways operating at this level to coordinate growth responses is relatively limited. Here we define a post-translational MAPK/Erk- and TRBP-dependent Lin28a induction mechanism that provides dynamic receptor-mediated regulation of Lin28a capable of producing rapid transitions in growth, development, and pluripotency. Previous work indicating that mitogenic signaling in tumor cell lines also depends on TRBP phosphorylation is consistent with this pathway as a determinant of Lin28a function in broader biological contexts (Paroo et al., 2009). Our data also highlight the unexpected co-regulation of core factors in small RNA biogenesis as a molecular event underlying pro-growth protein synthesis. Improved understanding of Lin28 regulatory mechanisms may provide insight to dysregulated growth control by the Lin28/Let-7 pathway and opportunities for therapeutic manipulation in human disease and stem cell biology.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Lin28a (A177) | Cell Signaling Technology | Cat#3978; RRID: AB_2297060 |
| Rabbit polyclonal anti-Lin28a | LifeSpan Biosciences | Cat#LS-C165782 now LS-B11566 |
| Rabbit polyclonal anti-Lin28a | Novus | Cat#NBP1-49537; RRID: AB_10011591 |
| Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat#F3165; RRID: AB_259529 |
| Rabbit polyclonal anti-FLAG | Sigma-Aldrich | Cat#F7425; RRID: AB_439687 |
| Rabbit polyclonal anti-HA | Thermo Fisher Scientific | Cat#71-5500; RRID: AB_2533988 |
| Mouse monoclonal anti-GAPDH (6C5) | Millipore | Cat#CB1001; RRID: AB_2107426 |
| Rabbit polyclonal anti-TRBP | Abcam | Cat#ab72110; RRID: AB_1271291 |
| Rabbit polyclonal anti-TRBP | Proteintech Group | Cat#15753-1-AP; RRID: AB_2200485 |
| Mouse monoclonal anti-TRBP (46D1) | Thermo Fisher Scientific | Cat#LF-MA0209; RRID: AB_2200483 |
| Rabbit polyclonal anti-Dicer | Sigma-Aldrich | Cat#SAB4200087; RRID: AB_10603336 |
| Rabbit polyclonal anti-Dicer | Santa Cruz Biotechnology | Cat#sc-30226; RRID: AB_639122 |
| Rabbit polyclonal anti-Lin28b | Cell Signaling Technology | Cat#5422; RRID: AB_10697489 |
| Mouse monoclonal anti-c-Myc | Sigma-Aldrich | Cat#M4439; RRID: AB_439694 |
| Rabbit polyclonal anti-c-Myc | Sigma-Aldrich | Cat#C3956; RRID: AB_439680 |
| Mouse monoclonal anti-HSC70 | Santa Cruz Biotechnology | Cat#sc-7298; RRID: AB_627761 |
| Mouse monoclonal anti-NF2/Merlin | Abcam | Cat#ab88957; RRID: AB_2042307 |
| Rabbit polyclonal anti-β-Actin | Cell Signaling Technology | Cat#4967; RRID: AB_330288 |
| Rabbit polyclonal anti-Phospho-p44/42 MAP kinase (phosphorylated Erk1/2) | Cell Signaling Technology | Cat#9101; RRID: AB_331646 |
| Mouse monoclonal anti-BrdU | Sigma-Aldrich | Cat#B8434; RRID: AB_476811 |
| Normal Mouse IgG antibody | Santa Cruz Biotechnology | Cat#sc-2025; RRID: AB_737182 |
| Normal Rabbit IgG antibody | Santa Cruz Biotechnology | Cat#sc-2027; RRID: AB_737197 |
| Rabbit polyclonal anti-Phospho-TRBP | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Brain-Derived Neurotrophic Factor (hBA-238; human) | Santa Cruz Biotechnology | Cat#sc-4554 |
| Nerve Growth Factor (2.5S; mouse) | Millipore | Cat#01-125 |
| Glial-Derived Neurotrophic Factor (mouse) | Sigma-Aldrich | Cat#SRP3200 |
| Macrophage Colony Stimulating Factor (mouse) | Cell Signaling Technology | Cat#5228 |
| U0126 | Cell Signaling Technology | Cat#9903 |
| Z-Leu-Leu-Leu-al (MG132) | Sigma-Aldrich | Cat#C2211 |
| Factor Xa protease | New England Biolabs | Cat#P8010 |
| RNasin Ribonuclease Inhibitor | Promega | Cat#N2111 |
| PR-619 | LifeSensors | Cat#SI9619 |
| 35S-methionine/cysteine EasyTag Mix | PerkinElmer | Cat#NEG772014MC |
| EDTA-free Protease Inhibitor Cocktail | Roche | Cat#11836170001 |
| N-ethylmaleimide (NEM) | Sigma-Aldrich | Cat#04260 |
| GST-S5a (UIM) | UBPBio | Cat#I1620 |
| 5-bromo-2′-deoxyuridine (BrdU) | Sigma-Aldrich | Cat#B5002 |
| NucBlue Fixed Cell ReadyProbes Reagent (DAPI) | Thermo Fisher Scientific | Cat#R37606 |
| NucBlue Live ReadyProbes Reagent (Hoechst 33342) | Thermo Fisher Scientific | Cat#R37605 |
| TaqMan mRNA qPCR probe: Lin28a | Applied Biosystems | Assay ID: Mm00524077_m1 |
| TaqMan mRNA qPCR probe: β-Tubulin | Applied Biosystems | Assay ID: Mm00727586_s1 |
| TaqMan mRNA qPCR probe: GAPDH | Applied Biosystems | Assay ID: Mm99999915_g1 |
| TaqMan MicroRNA Assay: Let7a | Applied Biosystems | Assay ID: 000377 |
| TaqMan MicroRNA Assay: Let7c | Applied Biosystems | Assay ID: 000379 |
| TaqMan MicroRNA Assay: Let7f | Applied Biosystems | Assay ID: 000382 |
| TaqMan MicroRNA Assay: miR132 | Applied Biosystems | Assay ID: 000457 |
| TaqMan MicroRNA Assay: sno234 | Applied Biosystems | Assay ID: 001234 |
| TaqMan MicroRNA Assay: U6 snRNA | Applied Biosystems | Assay ID: 002282 |
| FLAG peptide | Sigma-Aldrich | Cat#F3290 |
| 3X FLAG peptide | Sigma-Aldrich | Cat#F4799 |
| Critical Commercial Assays | ||
| CellEvent Caspase-3/7 Green Detection Reagent | Thermo Fisher Scientific | Cat#C10423 |
| TaqMan High-Capacity RNA to cDNA Kit | Applied Biosystems | Cat#4387406 |
| Experimental Models: Cell Lines | ||
| Human: HEK293T/17 cell line | ATCC | CRL-11268; RRID: CVCL_1926 |
| Experimental Models: Organisms/Strains | ||
| E. Coli: BL21-Gold (E3) Competent Cells | Agilent Technologies | Cat#230132 |
| Mus musculus: ICR (CD-1) | ENVIGO (formerly Harlan laboratories) | N/A |
| Recombinant DNA | ||
| Plasmid: 3XFLAG-TRBPWT | (Paroo et al., 2009) | N/A |
| Plasmid: 3XFLAG-TRBPSΔD | (Paroo et al., 2009) | N/A |
| Plasmid: 3XFLAG-TRBPSΔA | (Paroo et al., 2009) | N/A |
| Plasmid: pMCL-HA-MAPKK1-R4F[(Δ31-51)S218E/S222D | (Mansour et al., 1994) | Addgene Plasmid #40810 |
| Plasmid: pRK5-HA-Ubiquitin-K48 | (Lim et al., 2005) | Addgene Plasmid #17605 |
| Plasmid: FL-Lin28bΔNLS | (Piskounova et al., 2011) | N/A |
| Plasmid: HA-Merlin | (Zhao et al., 2007) | Addgene plasmid #32836 |
| Plasmid: FLAG-Lin28a | (Xu et al., 2009) | N/A |
| Plasmid: GST-TRBPWT | This paper | N/A |
| Plasmid: GST-TRBPSΔD | This paper | N/A |
| Plasmid: MBP-Lin28a | This paper | N/A |
| Plasmid: myc-Lin28a | This paper | N/A |
| Plasmid: shRNA-resistant TRBPSΔA* | This paper | N/A |
| Plasmid: shRNA-resistant FLAG-Lin28a* | This paper | N/A |
| Control shRNA: pLKO.1 non-target shRNA | Sigma | SHC002 |
| Mouse TRBP shRNA: pLKO.1 TRBP shRNA | Thermo Fisher Scientific | TRCN0000102563 |
| Human/Mouse Lin28a shRNA#1: pLKO.1 Lin28a shRNA | Thermo Fisher Scientific | TRCN0000102576 |
| Human/Mouse Lin28a shRNA#2: pLKO.1 Lin28a shRNA | Thermo Fisher Scientific | TRCN0000102577 |
| Human Lin28a shRNA#3: pLKO.1 Lin28a shRNA | Thermo Fisher Scientific | TRCN0000021802 |
| Human Merlin shRNA: pLKO.1 Merlin shRNA | Thermo Fisher Scientific | TRCN0000039975 |
| Mouse Dicer shRNA: pLKO.1 Dicer shRNA | Thermo Fisher Scientific | TRCN0000071320 |
| Software and Algorithms | ||
| Imaris 7.6.5 | Bitplane, Inc. | N/A |
| Stratagene Mx3000P | Agilent Technologies | N/A |
| Other | ||
| Glutathione Sepharose 4 Fast Flow | GE Healthcare | Cat#17-5132-01 |
| Amylose Resin | New England Biolabs | Cat#E80215 |
| Methionine- and cysteine-free DMEM | Mediatech, Inc. | Cat#17-204-CI |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the corresponding author, Dr. Mollie K Meffert, at Johns Hopkins University (mmeffer1@jhmi.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Animal procedures conformed to care guidelines approved by the Johns Hopkins University Institutional Animal Care and Use Committee. Male and female wild-type ICR (CD-1) mice from ENVIGO (formerly Harlan laboratories) were used for all experiments involving primary cell culture.
Cell lines
HEK293T cells were obtained from ATCC (CRL-11268) and were cultured in DMEM supplemented with 10% FBS and 1% glutamine at 37°C, 4% CO2.
Microbe strains
BL21 (E3) chemically comptetent cells were obtained from Agilent Technologies (230132). Cells were stored at −80°C and grown in LB medium at 37°C. BL21 cells were used for purification of recombinant proteins.
METHOD DETAILS
Primary cell culture and stimulation
Stimulation
BDNF (Santa Cruz sc-4554), NGF (Millipore 01-125), and GDNF (Sigma SRP3200) were applied to a final concentration of 100 ng/ml; M-CSF (Cell Signaling 5228) to a final concentration of 50ng/ml; U0126 (Cell Signaling, 9903) to a final concentration of 30 μM (hippocampal neurons) or 10 μM (DRGs, glia, and macrophages); MG132 (Sigma C2211) to a final concentration of 10 μM.
Hippocampal neurons
Dissociated hippocampal cultures were prepared from postnatal day 0 (P0) mice as previously described (Meffert et al., 2003), and were maintained in Neurobasal A medium (GIBCO 10888) with 2% B27 Supplement (GIBCO 17504-44) and 0.5% glutamine. Cells were cultured at 37°C in 4% CO2. On DIV 14–19, hippocampal cultures were changed into Serum Reduced Media (NBA + 0.5% B27 + glutamine) for 2 hr prior to stimulation, followed by pre-incubation with Actinomycin-D (Life Technologies A7592) at a final concentration of 0.5 μg/ml for 10 min if required.
Dorsal Root Ganglion
Acutely dissociated DRG neurons from 4-week old mice were cultured as previously described (Li et al., 2014). After trituration and centrifugation, cells were resuspended in DH10, plated in 24-well plates with poly-L-lysine, and cultured at 37°C, 4% CO2 for 24 hr with nerve growth factor (25 ng/ml) and glial cell-derived neurotrophic factor (50 ng/ml). After 24 hr, cells were changed into DMEM + 10% FBS + 1% glutamine. 48 hr after plating (DIV 2), cells were changed into serum starvation media (1% FBS) for 2 hr prior to stimulation.
Cortical glia
Dissociated cortical cultures were prepared from postnatal day 0 (P0) mice as previously described (Meffert et al., 2003), plated onto 10 cm tissue culture dishes, and maintained in DMEM + 10% FBS + 1% glutamine at 37°C, 4% CO2. Dishes were rinsed 24 hr after plating and passaged 48 hr after plating. Surviving cells are a mixed glial population. Cells were used within 4 passages and were changed into serum starvation media (1% FBS) for 2 hr prior to stimulation.
Peritoneal Macrophages
Mice (12–20 weeks old) were euthanized and peritoneal cells were harvested in RPMI media with 10% FBS and 1% glutamine as previously described (Link et al., 2010). Cells were plated on 24-well plates at 6.0 × 105 cells per well and rinsed 1 hr later with fresh media. Adherent cells are > 99% macrophages. 24 hr after plating (DIV 1), cells were changed into serum starvation media (1% FBS) for 2 hr prior to stimulation.
Plasmids
The following plasmids were obtained as generous gifts: Wild-type (WT) TRBP, phospho mutant (SΔA) TRBP, and phospho-mimic (SΔD) TRBP (from Dr. Zain Paroo (Paroo et al., 2009)); FLAG-Lin28a (from Dr. Yinqun Huang (Xu et al., 2009)); HA-Ubiquitin K48 (from Dr. Ted Dawson (Lim et al., 2005)); FL-Lin28bΔNLS mutant (from Dr. Richard Gregory (Piskounova et al., 2011)); HA-CAMAPKK (pMCL-HA-MAPKK1-R4F[(Δ31-51)S218E/S222D] from Dr. Natalie Ahn, Addgene plasmid # 40810 (Mansour et al., 1994)); HA-Merlin (from Dr. Kunliang Guan, Addgene plasmid # 32836 (Zhao et al., 2007)). GST-TRBPWT and GST-TRBPSΔD were subcloned using the pGEX-6P-1 vector. MBP-Lin28a was subcloned using the pMAL-c5X vector. Myc-Lin28a was subcloned using myc-pcDNA3.1. Fl-Lin28a, FL-Lin28b, FL-Lin28bΔNLS, FL-TRBPWT, FL-TRBPSΔA, and FL-TRBPSΔD were all subcloned into the synapsin promoter driven lentiviral vector FSW. Quikchange site-directed mutagenesis of a single base pair was used to generate a silent mutation for shRNA-resistant TRBPSΔA (FL-TRBPSΔA*). The sequence for FSW-TRBPSΔA* following quikchange was GGCAATGAGGTGG AGCCCGATGATGACCACTTC. The shRNA-resistant Lin28a construct (FL-Lin28a*) was generated by introducing three silent mutations into the cDNA sequence that is targeted by Lin28a shRNA. Mutagenesis was performed using the Quikchange method. The sequence for FL-Lin28a* following quickchange was CTGCCACCCCAGCCCAAAAAATGTCA CTTCTGCCAGAGC.
Transduction
Lentiviral-mediated delivery of FL-Lin28a, myc-Lin28a, FL-Lin28b, FL-Li28bΔNLS, FL-TRBPWT, FL-TRBPSΔD, FL-TRBPSΔA, or FL-TRBPSΔA* was used to achieve expression of tagged proteins in hippocampal cultures for biochemical analysis at a multiplicity of infection (MOI) of 5–10, 2 – 4.5 days before harvest, depending on required expression level. Lentiviral stocks were prepared and lentiviral KD carried out as previously described (Meffert et al., 2003). KD for biochemical analysis used lentiviral-mediated delivery of non-target shRNA (Sigma SHC002), shRNA targeting TRBP (Thermo Scientific TRCN0000102563), or shRNA targeting Lin28a (Thermo Scientific TRCN0000102576 (Lin28a shRNA#1) and TRCN0000102577 (Lin28a shRNA#2)) to infect hippocampal cultures at an MOI of 5–10, 4–5 days before harvest. 24 hr prior to imaging experiments, neurons were transiently transfected with PCDNA3.1 or TRBPSΔD, non-target shRNA or Lin28a shRNA#1, and PCDNA3.1 or FL-Lin28a* using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). HEK293T cells were infected with lentivirus expressing non-target shRNA, Lin28a shRNA#2, Lin28a shRNA#3 (TRCN0000021802), or shRNA targeting Merlin (TRCN0000039975) 96 hr prior to harvest for biochemical analysis of KD. Expression of all tagged constructs in HEK293T cells was achieved by calcium phosphate transfection.
Immunoblotting
Primary cultures of mouse hippocampal neurons, DRG neurons, cortical glia, peritoneal macrophages, or cultured HEK293T cells were washed 2 times in cold PBS with MgCl2 (0.9 mM) and harvested on ice with lysis buffer (50 mM HEPES, 150 mM NaCl, 10% Glycerol, 1 mM EDTA, 1% Triton X-100, 0.2% SDS) plus freshly added protease inhibitor cocktail (Roche 11836170001), phosphatase inhibitors (sodium orthovanadate 0.2 mM, sodium pyrophosphate 1 mM), and NEM (20 mM Sigma 04260). Protein concentration was determined by Bicinchoninic acid (BCA) assay and equal protein amounts resolved on SDS-PAGE gels and electrotransferred to PVDF membrane. Membrane was blocked with 5% BSA in Tris-buffered saline tween 20 (TBST 0.1%) for 2–4 hr and probed with primary antibodies: Lin28a (Cell Signaling A177, Lifespan LS-C165782 now LS-B11566, or Novus NBP-149537), FLAG (M2 Sigma F3165 or Sigma F7425), HA (Thermo Fisher Scientific 71-5500), GAPDH (Millipore 6C5), TRBP (Abcam ab72110, Pierce LF-MA0209, or Proteintech 15753-1-AP), Dicer (Sigma SAB4200087 or Santa Cruz sc-30226), Phospho-Erk1/2 (Cell Signaling 9101), Lin28b (Cell Signaling 5422), c-Myc (Sigma M4439 or Sigma C3956), HSC70 (Santa Cruz sc-7298), NF2/Merlin (abcam ab88957), β-Actin (Cell Signaling 4967). All immunoblots were scanned and quantified without image adjustment. For representative image figures, image levels were uniformly and minimally adjusted for visual clarity in some instances.
Purification and pull-down of recombinant bacterial proteins
GST-TRBPWT, GST-TRBPSΔD, and GST alone in the PGEX-6P-1 vector were expressed in BL21 bacteria (Agilent Technologies 230132) and purified as described in Pedersen et al. (Pedersen et al., 2015). Bacteria were grown in LB-amp until OD600 = 0.75 after which 1 mM IPTG was applied for 3 hr at 30°C to induce protein expression. Cultures were centrifuged at 4,000 × g for 20 min at 4°C and lysed in phosphate-buffered saline (PBS) pH = 7.8 with lysozyme (0.5 mg/ml) for 30 min, followed by sonication 5 times with 20 s pulses at 20% amplitude. 1% Triton-X was added, and lysates were centrifuged at 4000 × g for 30 min at 4°C. RnaseA was added to lysates to a final concentration of 20 μg/ml. Lysates were applied to 500 μL bed volume of glutathione Sepharose beads (GE Health-care 17-5132-01) and rotated at 4°C overnight. Beads were then added to a poly-prep column and washed, and GST-fusion proteins were eluted with glutathione elution buffer (50 mM Tris HCL, pH = 7.5, 10 mM glutathione). MBP-Lin28a in the pMAL-c5X vector was expressed in BL21 bacteria and purified as described, but rotated with amylose resin (NEB E80215). Lin28a was cleaved from the amylose-bound MBP tag with factor Xa protease (NEB P8010).
Purified protein concentrations were determined by Bradford Protein Assay, and equivalent amounts of GST-TRBPWT, GST-TRBPSΔD, and GST alone were re-bound to glutathione Sepharose beads by rotation at 4°C for 2 hr. Equal amounts of purified Lin28a protein taken from the same protein aliquot were added to each Sepharose-bound GST construct and rotated at 4°C for 2 hr. Beads were washed in cold PBS, pH = 7.8, followed by boiling in sample buffer at 95°C for 10 min to elute. Eluents were subjected to SDS-PAGE electrophoresis and immunoblotting.
Immunopurification
The following antibody and peptide reagents were used for immunopurification (IP): FLAG antibody (M2 Sigma F3165 or Sigma F7425), 3X FLAG peptide (Sigma F4799), FLAG peptide (Sigma F3290), c-Myc (Sigma M4439), c-Myc peptide (Sigma M2435), Control Mouse IgG (Santa Cruz sc-2025), Control Rabbit IgG (Santa Cruz sc-2027).
Co-Immunoprecipitation
coIP lysis buffer
100 mM KCl, 4 mM MgCl2, 10 mM HEPES (pH 7.3), 50 μM ZnCl, 0.5% NP-40, protease inhibitor cocktail (Roche), phosphatase inhibitor (sodium orthovanadate 0.2 mM, sodium pyrophosphate 1 mM), 20 mM NEM.
coIP wash buffer
150 mM NaCl, 1 mM MgCl2, 50 mM HEPES (pH = 7.8), 50 μM ZnCl, 0.05% NP-40, protease inhibitor cocktail (Roche), phosphatase inhibitor (sodium orthovanadate 0.2 mM, sodium pyrophosphate 1 mM), 50 mM NEM.
Binding partners of TRBP and Lin28a proteins were isolated through IP of FL-TRBP or FL-Lin28a. Protein G-Sepharose beads were coated with anti-FLAG antibody or control isotype-specific antibody in coIP wash buffer overnight after blocking with coIP wash buffer plus 5% BSA for 1 hr. Antibody amount was estimated as 1 μg antibody per 100 μg protein lysate in order to deplete the IPd protein. HEK293T cells: 1–2 days prior to transfection, cells were plated into either 24 well plates or 10cm dishes. Cells were trans-fected with appropriate constructs at ~70% confluency for 48 hr. Media was changed 8 hr post-transfection. Primary hippocampal cultures: Neurons (DIV 12–13) were infected with lentivirus expressing FL-Lin28a under the synapsin promoter for 86 hr. Cultures were then incubated in serum-reduced medium (0.5% B27 supplement) for 2 hr, followed by pre-treatment with vehicle (DMSO) or U0126 for 20 min (some cases), and bath application of vehicle (neuronal growth media) or BDNF (100 ng/ml) for 60–90 min (all cases). All cell types: Cell lysates were harvested in coIP lysis buffer with freshly added 1 mM DTT and RNasin (Promega N2111, 0.5 units/ml). Lysates were centrifuged (13,000xg, 15 min) and the supernatants pre-cleared by 30 min incubation with recombinant protein G-Sepharose beads pre-washed in coIP wash buffer. Input samples (5%–10% of IPd protein) were saved for immunoblot analysis. For IP, equal amounts of lysate protein (0.5–2mg, determined by Bradford protein assay) were incubated with antibody-coated beads and tumbled for 3–4 hr at 4°C, followed by 3 washes in coIP wash buffer and 1 wash in coIP wash buffer without protease and phosphatase inhibitors. Elutions from the washed beads were performed using a FLAG peptide competitive inhibitor (Sigma, F4799 or F3290) diluted in coIP wash buffer without protease and phosphatase inhibitors, and samples were subjected to SDS-PAGE electrophoresis and immunoblotting.
Sequential IP
HEK293T cells were transfected with either myc-TRBPWT and FL-Lin28a or FL-Lin28a alone (control) for 48 hr. Protein G-Sepharose beads were bound to anti-FLAG or anti-myc antibodies, and cells from both conditions were lysed and subjected to IP as previously described in “Co-Immunoprecipitation” using anti-myc conjugated G-Sepharose beads. Elutions from the washed beads were performed using a myc peptide competitive inhibitor (Sigma M2435). Eluates then underwent a second IP using anti-FLAG conjugated beads. Following the second IP, washed beads were incubated in sample buffer at 95°C for 10 min, eluates resolved by SDS-PAGE electrophoresis, and immunoblotted.
Co-Immunoprecipitation with RNase
HEK293T cells were transfected with FL-Lin28a for 48 hr, then lysed and subjected to IP as described in Co-Immunoprecipitation. During the IP tumbling step, lysates were incubated with either RNAsin (Promega, 0.5 units/ml) to maximize RNA stability, or RNaseA (200 μg/ml) to inhibit RNA.
FL-TRBP limiting immunoprecipitation
Protein G-Sepharose beads were coated with anti-FLAG antibody in intentionally limiting amounts, estimated at about 1 μg antibody per 400 μg protein lysate. Cells were then lysed and subjected to IP as previously described in Co-Immunoprecipitation. Limiting antibody allowed for equal pull-down of each FL-TRBP mutant construct - which express at highly different levels - in order to allow visual appreciation and more accurate quantification of TRBP-associated proteins.
Stringent immunoprecipitation (for ubiquitination assessment)
Stringent lysis buffer
100mM KCl, 10 mM HEPES (pH = 7.3), 4 mM MgCl2, 50 μM ZnCl2, 1% Triton-X, 0.25% SDS, 50 μM PR-619, protease inhibitor cocktail (Roche), phosphatase inhibitor (sodium orthovanadate 0.2 mM, sodium pyrophosphate 1 mM), 20 mM NEM.
Stringent wash buffer
1M NaCl, 1 mM MgCl2, 50 mM HEPES (pH = 7.8), 50 μM ZnCl2, 20% Glycerol, 0.2% SDS, protease inhibitor cocktail (Roche), phosphatase inhibitor (sodium orthovanadate 0.2 mM, sodium pyrophosphate 1 mM), 20 mM NEM.
FLAG antibody coating of protein G-Sepharose beads was carried out as previously described in Co-Immunoprecipitation. HEK293T cells were transfected with HA-K48 specific ubiquitin and either PCDNA3.1, FL-TRBPWT, FL-TRBPSΔA, or FL-TRBPSΔD constructs for TRBP ubiquitination assessment, or HA-K48 specific ubiquitin and either PCDNA3.1 or FL-Lin28a for Lin28a ubiquitination assessment. Cells underwent bath application of the deubiquitinase inhibitor PR-619 (10 μM; LifeSensors SI9619) for 1 hr prior to harvest. Cell lysates were harvested in stringent IP lysis buffer with freshly added 1mM DTT, and underwent centrifugation, pre-clearing, and BCA assay. Equal amounts of protein (1.5–2mg) were incubated with antibody-coated beads and tumbled for 4 hr at 4°C, followed by washing with stringent wash buffer. The washed beads were incubated in sample buffer at 95°C for 10 min and subjected to SDS-PAGE electrophoresis and immunoblotting.
35S pulse chase
Cultured hippocampal neurons were infected at DIV 12–14 with lentiviral myc-Lin28a for 72 hr. Following viral-mediated expression, neurons were pre-incubated in media containing reduced serum as previously described, followed by 2 washes and 10 min incubation with methionine- and cysteine-free DMEM (Mediatech, Inc. 17-204-CI). 35S labeling was performed in the same methionine- and cysteine-free DMEM with the addition of 35S-methionine/cysteine (35S Met/Cys EasyTag Mix, Perkin Elmer NEG772014MC) to a final concentration of 100 μCi/ml for 3 hr. Following the 3 hr labeling period, 35S-methionine/cysteine containing media was washed out and replaced with normal serum-reduced media (NBA + 0.5% B27 + glutamine), after which neurons were subjected to either vehicle (neuronal growth media) or BDNF bath application, as previously described, for a designated period lasting 30–180 min. Cells were washed and lysed with stringent IP lysis buffer. As described in Co-Immunoprecipitation and Stringent Immunoprecipitation, Lysates were processed for IP and stringent IP was performed using anti-myc coated protein G-Sepharose beads. The washed IP beads were incubated in sample buffer at 95°C for 10 min and subjected to SDS-PAGE electrophoresis. Radioactive signal of IPd myc-Lin28a was quantified using a Typhoon phosphoimager (Molecular Devices). Myc-Lin28a protein expression and IP was verified via immunoblot.
Ubiquitin-interacting motif (S5a) immunoprecipitation
Cultured hippocampal neurons were pre-treated with either MEK/Erk inhibitor U0126 or a vehicle control for 30 min, followed by BDNF for 1 hr to stimulate neuronal activity, and finally MG132 for an additional 2 hr to accumulate polyubiquitinated proteins. HEK293T cells were infected with either a non-target shRNA control or shRNA targeting Merlin for 96 hr, and incubated with MG132 for 2 hr prior to harvest. Both cell types were lysed in Ub-IP lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10% glycerol, 0.05% SDS) plus freshly added protease inhibitor cocktail (Roche 11836170001), phosphatase inhibitors (sodium orthovanadate 0.2 mM, sodium pyrophosphate 1 mM), and NEM (20 mM; Sigma 04260). The lysates were centrifuged (13,000xg, 15 min), and protein concentration determined by BCA assay. 5% aliquots were removed for input samples. The lysates were then tumbled overnight at 4°C with 10 μg of a GST-S5a fusion protein (UBPBio, #I1620; S5a is an integral component of the 26S proteasome that recognizes K48-linked polyubiquitinylated proteins), pre-bound to glutathione Sepharose. Beads were washed 4 times in cold Ub-IP lysis buffer without SDS and supplemented with NEM (20 mM). Elutions were performed by boiling in sample buffer at 95°C, and protein ubiquitination was assessed via immunoblot.
Macrophage proliferation and survival assays
Peritoneal macrophages were harvested as described in Primary Cell Culture, plated at 1.0 × 105 cells per 10mm glass coverslip, and washed 1 hr after plating with fresh media. For proliferation assay, macrophages were incubated for 8 hr in serum starvation (0% FBS) media 24 hr after plating, followed by incubation with 10 μM 5-bromo-2′-deoxyuridine (BrdU; Sigma, B5002) for 12 hr, in the presence or absence of 50 ng/ml M-CSF. Cells were then fixed in 4% PFA/4% sucrose/PBS for 40 min, permeabilized for 40 min in 0.2% Triton X-100/PBS, and incubated in 2 M HCL for 10 min at 37°C, followed by incubation in 0.1 M borate buffer for 5 min, twice, to neutralize acid. Macrophages were incubated with primary anti-BrdU antibody (Sigma B8434) overnight at 4°C, followed by Alexa 488 secondary antibody for 1 hr at room temperature. During the last wash following secondary antibody incubation, DAPI nuclear stain (Thermo Scientific R37606) was added for 10 min.
For survival assay, 24 hr after plating (DIV 1), peritoneal macrophages were infected with lentivirus expressing either non-target shRNA (Sigma SHC002) or shRNA targeting Lin28a (Thermo Scientific TRCN0000102576, Lin28a shRNA#1). After 8 hr of infection, macrophages were changed into serum starvation media (0% FBS) with or without M-CSF (50 ng/ml) for 48 hr. Fresh media with or without M-CSF was replaced once a day. After 48 hr, macrophages were incubated with Hoechst 33342 live nuclear stain for 20 min at RT (Thermo Scientific R37605), followed by incubation with Caspase-3/7 activity detection reagent (Thermo Scientific C10423; fluorogenic caspase 3/7 substrate) for 45 min at 37°C.
Imaging and quantification
For live cell imaging, confocal images of hippocampal pyramidal neurons (determined by morphology) were acquired using a 40x (whole cell) or a 100x (dendrite) objective. Confocal images of peritoneal macrophages were acquired using a 25x objective. All images were taken using a Yokogawa spinning disk system (Cell Observer, Carl Zeiss) at 37°C. Laser power and exposure time were adjusted to minimize photobleaching and avoid saturation. For pyramidal neurons, all experiments were from a minimum of 3 independent cultures, no more than 4 neurons per dish, and no more than 3 dendritic segments per neuron. Only tertiary dendritic branches were used for dendritic spine analysis to control for distance from the soma. Z stacks containing the entire process of interest were analyzed using Imaris (7.6.5; Bitplane). For peritoneal macrophages, all experiments were from 3–4 independent cultures, no more than 2 dishes per condition per culture, no more than 4 fields per dish. Blue (Hoechst) and green (caspase active) cells in each field were manually counted by a researcher blinded to experimental condition. For representative images, image levels were uniformly and minimally adjusted in photoshop to maximize visual clarity. Within each color channel, levels were adjusted equally both across the entire image, and between experimental conditions.
Dendritic spine analysis
Neuronal cultures were randomly assigned to treatment conditions and the experimenter was blinded during data acquisition and analysis. 3D reconstructions of hippocampal neurons were analyzed with Filament Tracer (Imaris 7.6.5 Bitplane, Inc.). Tertiary dendritic segments were traced and volume rendered using automatic thresholds and dendritic protrusions 0.4–2.0 μm in length with or without a spine head were assigned as spines. Tertiary dendritic branch segments were chosen for spine density analysis. For each dendritic segment a manual spine count was conducted in Imaris Surpass mode. Dendritic spine density was calculated by counting the number of spines per 10 microns of dendritic length. The dendrite length was measured using Filament Tracer and each spine was marked using Spots (Imaris 7.6.5 Bitplane, Inc.). Dendritic protrusion length was measured using Measurement Points. Individual dendritic spine length was manually marked from its point of insertion in the dendritic shaft to the distal tip of the spine. Spine volume was measured by manually tracing 3D images of dendritic spines using Filament Tracer’s AutoDepth feature. An accurate 3-D model of dendritic spines was generated using the diameter function, which calculates the correct diameter and represents the complete morphology of the selected spine based on the 3-D image. Imaris MeasurementPro provided spine volume measurements.
RNA analysis
Total RNA from primary cultures of mouse hippocampal neurons was isolated in Tri-Reagent (Molecular Research Center, Inc.) according to the manufacturer’s protocol. Cultures were homogenized in Tri-Reagent directly. RNA pellets were air-dried and resuspended in nuclease-free water. RNA concentration and quality were assayed by spectrophotometric measurements at optical density (OD) 260/280/230 nm. Reverse transcription of Lin28a mRNA was completed using 2 μg total RNA with the TaqMan High Capacity RNA to cDNA Kit (Applied Biosystems 4387406). The following reverse transcription program was used: 1) 37°C incubation for 60 min, 2) 95°C inactivation for 5 min. Products of RT reaction were then assayed via qPCR using TaqMan mRNA qPCR probes: Lin28a (Applied Biosystems Mm00524077_m1), β-tubulin 3 (Applied Biosystems Mm00727586_s1), and GAPDH (Applied Bio-systems Mm99999915_g1). The qPCR program used was the following: 1) 95°C denature for 15 s, 2) 60°C reanneal and extension for 60 s, 3) repeat of steps 1 and 2 for 40 cycles. For individual microRNA abundance assays (Applied Biosystems), 20 ng of total isolated RNA was prepared for reverse transcription with stem-looped primers specific for individual mature miRNAs in a final volume of 15 μL according to manufacturer’s protocol: 1) 4°C for 5 min, 16°C for 30 min, 2) 42°C for 30 min, 3) 85°C for 5 min, and subjected to TaqMan MicroRNA Assays (Applied Biosystems) for Let-7a (000377), Let-7c (000379), Let-7f (000382), and miR-132 (000457). The abundance of U6 snRNA (002282) and sno234 (001234) in each sample was used as an internal control to normalize all miRNA species. RT-qPCR was performed using a Stratagene Mx3000P machine and software. Quantification was carried out using the standard-curve method and no preamplification. RQ was calculated as 2−ΔCtBDNF/2−ΔCtmock where ΔCt = (cycle threshold for miRNA of interest) − (cycle threshold for reference control).
QUANTIFICATION AND STATISTICAL ANALYSIS
Two-tailed unpaired Student’s t tests were used with α = 0.05. Where specified, statistical analysis included two-way ANOVA for independent samples with a Bonferroni-Holm post hoc test, α = 0.05. Statistical significance between scatterplot conditions (Figure 2d) was determined using one-way ANCOVA for independent samples. α = 0.05. All quantified data represent mean ± SEM, as referenced in figure legends. Values of ‘n’ are detailed in figure legends. For protein and RNA analysis, ‘n’ refers to the number of independent sample replicates. For imaging experiments, ‘n’ refers to either the number of dendritic segments (neuronal dendritic spine density), the number of dendritic spines (neuronal dendritic spine volume), or the number of imaged fields of view (peritoneal macrophages). Conditions in all experiments were assigned informally at random.
Supplementary Material
Highlights.
BDNF rapidly stabilizes Lin28a, but not Lin28b, by MAPK-dependent TRBP phosphorylation
Lin28a, but not Lin28b, associates with TRBP in the miRNA-generating complex
Phospho-TRBP requires Lin28a to mimic neurotrophin-induced dendritic spine growth
MAPK-dependent Lin28a induction by trophic cues is conserved in diverse primary cells
Acknowledgments
We thank Z. Paroo and R. Gregory for generously sharing reagents. We thank X. Dong, Z. Li, and M. Caterina for assistance with cell culture and J. Pomerantz for technical advice. We thank R. Green, A. Kolodkin, and J. Pomerantz for valuable input on the manuscript and scientific suggestions. This work was supported by the Braude Foundation, NIH MH098016 and MH109341 (to M.K.M.), NIH F31 MH0103902 (to A.M.A.), NIH F31 MH098634 (to C.R.R.-G.), and MH084020 (to the Neuroscience Multiphoton Facility).
Footnotes
Supplemental Information includes six figures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.12.025.
AUTHOR CONTRIBUTIONS
A.M.A., C.R.R.-G., and M.K.M. conceived and designed the experiments. A.M.A., C.R.R.-G., J.S., M.S., and D.L.P. performed all experiments and data analyses in the laboratory of M.K.M. M.K.M., A.M.A., and C.R.R.-G. wrote the manuscript. M.K.M conceptualized and supervised the project.
References
- Alonso M, Medina JH, Pozzo-Miller L. ERK1/2 activation is necessary for BDNF to increase dendritic spine density in hippocampal CA1 pyramidal neurons. Learn Mem. 2004;11:172–178. doi: 10.1101/lm.67804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SM, Melendez-Peña CE, Scarborough RJ, Daher A, Christensen HS, El Far M, Purcell DF, Lainé S, Gatignol A. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol Biol. 2009;10:38. doi: 10.1186/1471-2199-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Huang YW, Ruiz CR, Eyler EC, Lin K, Meffert MK. Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell. 2012;148:933–946. doi: 10.1016/j.cell.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je HS, Lu Y, Yang F, Nagappan G, Zhou J, Jiang Z, Nakazawa K, Lu B. Chemically inducible inactivation of protein synthesis in genetically targeted neurons. J Neurosci. 2009;29:6761–6766. doi: 10.1523/JNEUROSCI.1280-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JM, Chen Q, Zhou Y, Miao S, Zheng J, Zhang C, Xiong ZQ. Brain-derived neurotrophic factor-tropomyosin-related kinase B signaling contributes to activity-dependent changes in synaptic proteins. J Biol Chem. 2008;283:21242–21250. doi: 10.1074/jbc.M800282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DI, Jameson SB, Reddy MA, Schenkman D, Ostrowski MC. Alterations in differentiation and behavior of monocytic phagocytes in transgenic mice that express dominant suppressors of ras signaling. Mol Cell Biol. 1995;15:693–703. doi: 10.1128/mcb.15.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhema T, Dagestad G, Panja D, Tiron A, Messaoudi E, Håvik B, Ying SW, Nairn AC, Sonenberg N, Bramham CR. Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. J Neurochem. 2006;99:1328–1337. doi: 10.1111/j.1471-4159.2006.04158.x. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Kim Y, Yeo J, Lee JH, Cho J, Seo D, Kim JS, Kim VN. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep. 2014;9:1061–1074. doi: 10.1016/j.celrep.2014.09.039. [DOI] [PubMed] [Google Scholar]
- Lauterborn JC, Rex CS, Kramár E, Chen LY, Pandyarajan V, Lynch G, Gall CM. Brain-derived neurotrophic factor rescues synaptic plasticity in a mouse model of fragile X syndrome. J Neurosci. 2007;27:10685–10694. doi: 10.1523/JNEUROSCI.2624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kim H, Ryu CH, Kim JY, Choi BH, Lim Y, Huh PW, Kim YH, Lee KH, Jun TY, et al. Merlin, a tumor suppressor, interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity. J Biol Chem. 2004;279:30265–30273. doi: 10.1074/jbc.M312083200. [DOI] [PubMed] [Google Scholar]
- Lee JY, Moon HJ, Lee WK, Chun HJ, Han CW, Jeon YW, Lim Y, Kim YH, Yao TP, Lee KH, et al. Merlin facilitates ubiquitination and degradation of transactivation-responsive RNA-binding protein. Oncogene. 2006;25:1143–1152. doi: 10.1038/sj.onc.1209150. [DOI] [PubMed] [Google Scholar]
- Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013;41:6568–6576. doi: 10.1093/nar/gkt361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, He SQ, Xu Q, Yang F, Tiwari V, Liu Q, Tang Z, Han L, Chu YX, Wang Y, et al. Activation of MrgC receptor inhibits N-type calcium channels in small-diameter primary sensory neurons in mice. Pain. 2014;155:1613–1621. doi: 10.1016/j.pain.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MY, Lin YM, Kao TC, Chuang HH, Chen RH. PDZ-RhoGEF ubiquitination by Cullin3-KLHL20 controls neurotrophin-induced neurite outgrowth. J Cell Biol. 2011;193:985–994. doi: 10.1083/jcb.201103015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link TM, Park U, Vonakis BM, Raben DM, Soloski MJ, Caterina MJ. TRPV2 has a pivotal role in macrophage particle binding and phagocytosis. Nat Immunol. 2010;11:232–239. doi: 10.1038/ni.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande Woude GF, Ahn NG. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- Mao XG, Hütt-Cabezas M, Orr BA, Weingart M, Taylor I, Rajan AK, Odia Y, Kahlert U, Maciaczyk J, Nikkhah G, et al. LIN28A facilitates the transformation of human neural stem cells and promotes glioblastoma tumorigenesis through a pro-invasive genetic program. Oncotarget. 2013;4:1050–1064. doi: 10.18632/oncotarget.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. Retraction. Nat Genet. 2009;48:221. doi: 10.1038/ng0216-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 2006;25:5794–5804. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Nowak JS, Choudhury NR, de Lima Alves F, Rappsilber J, Michlewski G. Lin28a regulates neuronal differentiation and controls miR-9 production. Nat Commun. 2014;5:3687. doi: 10.1038/ncomms4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen SM, Chan W, Jattani RP, Mackie S, Pomerantz JL. Negative regulation of CARD11 signaling and lymphoma cell survival by the E3 ubiquitin ligase RNF181. Mol Cell Biol. 2015;36:794–808. doi: 10.1128/MCB.00876-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pépin G, Perron MP, Provost P. Regulation of human Dicer by the resident ER membrane protein CLIMP-63. Nucleic Acids Res. 2012;40:11603–11617. doi: 10.1093/nar/gks903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeld F, Rohde AM, Nguyen DT, Wulczyn FG. Lin28 and let-7: ancient milestones on the road from pluripotency to neurogenesis. Cell Tissue Res. 2015;359:145–160. doi: 10.1007/s00441-014-1872-2. [DOI] [PubMed] [Google Scholar]
- Ruiz CR, Shi J, Meffert MK. Transcript specificity in BDNF-regulated protein synthesis. Neuropharmacology. 2014;76:657–663. doi: 10.1016/j.neuropharm.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Jens M, Murakawa Y, Herzog M, Landthaler M, Rajewsky N. A variety of dicer substrates in human and C. elegans. Cell. 2014;159:1153–1167. doi: 10.1016/j.cell.2014.10.040. [DOI] [PubMed] [Google Scholar]
- Santos AR, Mele M, Vaz SH, Kellermayer B, Grimaldi M, Colino-Oliveira M, Rombo DM, Comprido D, Sebastião AM, Duarte CB. Differential role of the proteasome in the early and late phases of BDNF-induced facilitation of LTP. J Neurosci. 2015;35:3319–3329. doi: 10.1523/JNEUROSCI.4521-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013a;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013b;155:778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–482. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Yermalovich A, Zhang J, Spina CS, Zhu H, Perez-Atayde AR, Shukrun R, Charlton J, Sebire N, Mifsud W, et al. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev. 2014;28:971–982. doi: 10.1101/gad.237149.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yu X, Cao Q, Wang Y, Zheng G, Tan TK, Zhao H, Zhao Y, Wang Y, Harris DCh. Characterization of murine macrophages from bone marrow, spleen and peritoneum. BMC Immunol. 2013;14:6. doi: 10.1186/1471-2172-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell. 2015;57:397–407. doi: 10.1016/j.molcel.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, Haririnia A, Storaska AJ, Cole JL, Fushman D, Walters KJ. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009;35:280–290. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.