Abstract
One of our primary goals is to help families who have a child with an 18q deletion anticipate medical issues in order to optimize their child’s medical care. To this end we have narrowed the critical regions for four phenotypic features and determined the penetrance for each of those phenotypes when the critical region for that feature is hemizygous. We completed molecular analysis using oligo-array CGH and clinical assessments on 151 individuals with deletions of 18q and made genotype–phenotype correlations defining or narrowing critical regions. These nested regions, all within 18q22.3 to q23, were for kidney malformations, dysmyelination of the brain, growth hormone stimulation response failure, and aural atresia. The region for dysmyelination and growth hormone stimulation response failure were identical and was narrowed to 1.62 Mb, a region containing five known genes. The region for aural atresia was 2.3 Mb and includes an additional three genes. The region for kidney malformations was 3.21 Mb and includes an additional four genes. Penetrance rates were calculated by comparing the number of individuals hemizygous for a critical region with the phenotype to those without the phenotype. The kidney malformations region was 25% penetrant, the dysmyelination region was 100% penetrant, the growth hormone stimulant response failure region was 90% penetrant with variable expressivity, and the aural atresia region was 78% penetrant. Identification of these critical regions suggest possible candidate genes, while penetrance calculations begin to create a predictive phenotypic description based on genotype.
Keywords: chromosome 18, critical regions, genotype–phenotype correlation, dysmyelination, growth hormone deficiency, aural atresia, kidney malformation
INTRODUCTION
Chromosome abnormalities are common disorders, collectively occurring in one of every 118 live births [Nielson and Wohlert, 1991]. These conditions are major contributors to infant morbidity and mortality. Abnormalities of chromosome 18 are among the most common of the autosomal abnormalities. With the advent of microarray technology, it is now possible with a high degree of precision to rapidly determine exactly which genes are not diploid in an individual with a deletion. However, the role that each of the non-diploid genes plays, if any, in producing the composite phenotype is virtually unknown. One goal of the Chromosome 18 Clinical Research Center is to identify the genes associated with the specific phenotypic features seen in individuals with 18q deletions (18q-) (OMIM 601808). Here, we report the first step in this process: the identification of chromosome regions linked to four of these features.
The phenotype associated with a terminal deletion of 18q has been well described and can include cognitive impairment, hypotonia, short stature, ear canal abnormalities, abnormal genitalia, foot deformities, and delayed myelination (i.e., dysmyelination). Other common features include proximally placed thumbs, kidney malformations, and growth hormone deficiency. Dysmorphic features include relative hypertelorism and a flattened midface. The flattened midface is typically apparent by about age 4 and becomes more noticeable through childhood. The maxilla tends to be underdeveloped, allowing the mandible to over-rotate and giving the individual a prognathic appearance [Cody et al., 1999]. The size of terminal deletions varies between individuals from 30 to 0.5 Mb [Heard et al., 2009].
It has been well established that hemizygosity of different regions of 18q is associated with different phenotypic characteristics. For example, individuals with proximal interstitial deletions of 18q do not have the common features seen in individuals with terminal deletions [Cody et al., 2007] yet both groups have 18q-. A deletion of any portion of 18q is sufficient for a diagnosis of “18q-.”
In order to define the critical regions for phenotypic features, it is necessary to make several assumptions:
The phenotype is not purely the result of a global genomic dysregulation, but instead can be linked to the hemizygosity of specific genomic elements.
Not all genes will produce a phenotype when hemizygous; some are involved in pathways or processes that are haplosufficient.
The missing genetic content of a hemizygous region is necessary, but may not be sufficient, to dictate an abnormal phenotypic outcome.
The first assumption is supported by the work of Amano et al. [2004]. Using DNA microarray analysis, these authors assessed gene expression levels in the brains of Ts1Cje mice, a mouse model for Down syndrome. Although the genes in the trisomic region were universally expressed at 1.5-fold that of the normal littermates, neighboring genes as well as genes in the remainder of the genome, did not appear to be dysregulated. Additionally, determination of mRNA levels of 10 genes in 18q- individuals showed that expression levels of 9 of 10 genes studied correlated with gene copy number [Wang et al., 1999]. One gene showed partial dosage compensation. Taken together, these results imply that a chromosome imbalance does not typically lead to abnormal expression or dysregulation of genes across the genome.
The second assumption is supported by several lines of investigation. It has been estimated that in Drosophila, 90% of mutations are recessive to wild-type [Wilke, 1994]. In other words, one fully functional gene results in haplosufficiency 90% of the time. This implies that only 10% of genes result in haploinsufficiency when present in the hemizygous state. Evidence that humans may be analogous can be found on 17p. The region of 17p that is duplicated in Charcot-Marie-Tooth disease and deleted in hereditary neuropathy with liability to pressure palsy contains 21 genes. However, only one gene (PMP22) is associated with both phenotypes [Inoue et al., 2001]. The other 20 genes in the region do not produce an apparent phenotype when hemizygous or duplicated. Other examples of haplosufficiency have recently been identified. Deletions of gene-containing regions on 2p and 11q have been identified that are not associated with an abnormal phenotype [Li et al., 2002; Barber et al., 2005]. Array-based technology has been used to catalog 2672 loci of large-scale variation in the human genome [Iafrate et al., 2004]. This leads us to hypothesize that most genes on 18q are haplosufficient in a hemizygous state and that relatively few genes will be responsible for the major phenotypic features in a condition like 18q-.
Our third assumption is that hemizygosity for a gene associated with a specific phenotype is not always sufficient to cause that phenotype. This is the well accepted but poorly understood concept of incomplete penetrance [Zlotogora, 2003].
We have performed molecular analysis on 189 individuals with deletions of 18q, 95% of whom are hemizygous for some portion of the most distal 30 Mb of the chromosome. We have performed a comprehensive clinical evaluation on 151 of those individuals who had a net copy number variation involving only 18q [Heard et al., 2009]. Our population’s phenotypic and genotypic variability can be exploited to identify regions of the chromosome associated with specific phenotypes.
Here, we report on the critical regions for two major features of 18q-: kidney malformations and aural atresia. In addition, we narrow our previously reported critical regions for two additional features: growth hormone stimulant response failure and dysmyelination of the brain [Cody et al., 1997; Gay et al., 1997].
MATERIALS AND METHODS
Participant Recruitment
Participants primarily learned of the study through the Chromosome 18 Registry and Research Society, a lay advocacy group for people affected by chromosome 18 abnormalities. The protocol to perform phenotypic and genotypic assessment of individuals with chromosome 18 abnormalities was approved by the Institutional Review Board of the University of Texas Health Science Center at San Antonio, the Research and Development Committee of the Audie L. Murphy VA Hospital, and the advisory committee of the General Clinical Research Center. Informed consent was utilized throughout the study and documented for all participants. Enrollment criteria included a diagnostic karyotype demonstrating a non-mosaic deletion of some portion of 18q including interstitial as well as terminal deletions. Individuals with more complex rearrangements involving other chromosomes were excluded from the analysis, as they may confound results by preventing the identification of a critical region or erroneously identifying a region.
Genotyping
Blood samples were obtained from the affected individual and his/her biological parents for chromosome preparations, DNA isolation, and creation of immortalized cell lines [Cody et al., 1997]. The molecular analysis of DNA from people with 18q deletions was performed using microarray oligonucleotide comparative genomic hybridization (aCGH) using the Agilent Technologies System (Santa Clara, CA) as previously described [Heard et al., 2009]. DNA samples were assessed using a custom array that in the majority of cases contained 32,000 oligonucleotides across chromosome 18 and 12,000 across the remainder of the genome. The average resolution for chromosome 18 copy number changes was 2 kb.
Breakpoints were confirmed using quantitative real-time PCR (QRT-PCR). This technique has been described for detecting both deletions and duplications of the PMP22 gene [Aarskog and Vedeler, 2000]. We used the iCycler iQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA) and designed TaqMan probe/primer sets from the genomic sequence data in the regions of interest.
Quantification of the amount of target sequence in unknown samples is accomplished by measuring the threshold cycle number (Ct value) using the standard curve and a housekeeping gene as an internal control reference. The fractional Ct values at which the amount of amplified target DNA reaches a fixed threshold is directly related to the amount of starting target DNA. A higher starting copy number of the genomic DNA target will result in an earlier and significant increase in fluorescence. DNA for the control samples for the QRT-PCR and the aCGH were purchased from Promega Corp. (Madison, WI) and are pooled samples from 10 individuals.
The 151 participants whose data were analyzed in this study were those who had only an 18q deletion and who have been to our center for evaluation. Individuals who had additional copy number changes were not included. Individuals included in this study would be those included in Table I, rows 1, 3, and 4 of the preceding manuscript [Heard et al., 2009].
TABLE I.
Location of Critical Regions
| Phenotype | Current study
|
Linnankivi et al. [2006]
|
Feenstra et al. [2007]
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Critical region borders
|
Size of region (Mb) | Critical region borders
|
Size of region (Mb) | Critical region borders
|
Size of region (Mb) | ||||
| Proximal | Distal | Proximal | Distal | Proximal | Distal | ||||
| Kidney malformation | 70,079,559 | 73,287,604 | 3.21 | Not assessed | Not assessed | ||||
| Dysmyelination | 71,669,548 | 73,287,604 | 1.62 | 69.4 | 75.0 | 5.6 | 69.9 | 76.0 | 6.1 |
| Growth hormone response failure | 71,669,548 | 73,287,604 | 1.62 | Not assessed | 58.5 | 76.0 | 17.5 | ||
| Aural atresia | 70,983,374 | 73,287,604 | 2.30 | 68.4 | 75.0 | 6.6 | 69.9 | 74.9 | 5.0 |
Phenotyping
Phenotypic assessment was performed by a thorough medical record review and a series of comprehensive assessments performed at The Chromosome 18 Clinical Research Center. The Center was a component of the General Clinical Research Center, Audie Murphy Veterans Hospital, San Antonio, TX and more recently a component of the Children’s Health Advocacy Research and Treatment (CHART) Center of CHRISTUS Santa Rosa Children’s Hospital.
Kidney Malformation
Medical records of the participants were reviewed to determine whether an abdominal sonogram had been performed. The presence or absence of a kidney malformation was determined.
Dysmyelination
Study participants received MRI scans using a 1.9 T Elscint magnet. Scans were analyzed using MRI relaxometry techniques as previously described [Gay et al., 1997; Lancaster et al., 2005]. The diagnosis of dysmyelination was made when the T1 and T2 relaxation times were prolonged relative to age matched controls [Gay et al., 1997].
Growth Hormone Stimulant Response Failure
Study participants were tested for growth hormone deficiency as previously described [Hale et al., 2000]. The criteria for a therapeutic diagnosis of growth hormone deficiency involves additional measures including growth velocity, bone age, and height standard deviation. However, for the purposes of this study we chose to define the phenotype as the failure to produce growth hormone in response to one growth hormone provocative agent (arginine or clonidine) and a height Z-score of less than −1.5.
Aural Atresia
Subjects were evaluated for atretic or stenotic ear canal by a neurotologist at the Clinical Research Center. Congenital aural atresia can be classified in several ways, for the purposes of this study we have followed that of Schuknecht [1989]. Types A and B are stenotic and types C and D are true atresia’s. The presence of one ear with an abnormally narrow or closed external canal was sufficient for the presence of the phenotype.
Determination of Critical Regions
The aCGH data from all participants were converted to custom tracks for the UCSC Genome Browser for ease of analysis [Heard et al., submitted]. Critical regions were determined by selecting all the individuals who had the specific phenotype being analyzed. Then the genotype data for these individuals were compared to determine the common region of chromosome 18q hemizygosity. The largest common hemizygous region defined the critical regions for the respective phenotypes. In each case, two different individuals defined the proximal and distal boundaries of the critical regions.
RESULTS
The data for each of the four phenotypic features (kidney malformations, dysmyelination, growth hormone stimulant response failure, and aural atresia) were analyzed independently. Because the participants have a compound phenotype, any one individual may be in more than one phenotype group. Also, because each of these groups only includes those individuals who were successfully assessed for each phenotype, each phenotype group includes a different number of individuals from our cohort of 151 (e.g., some children were unable to lie still for the MRI).
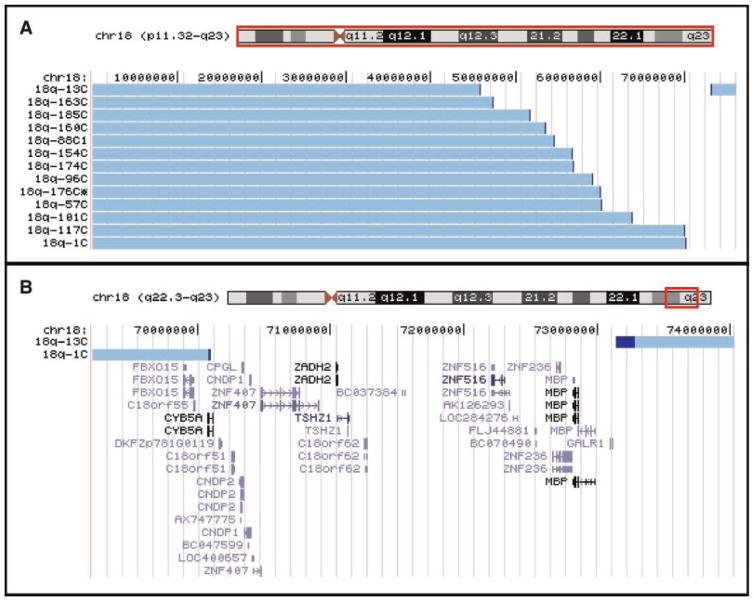
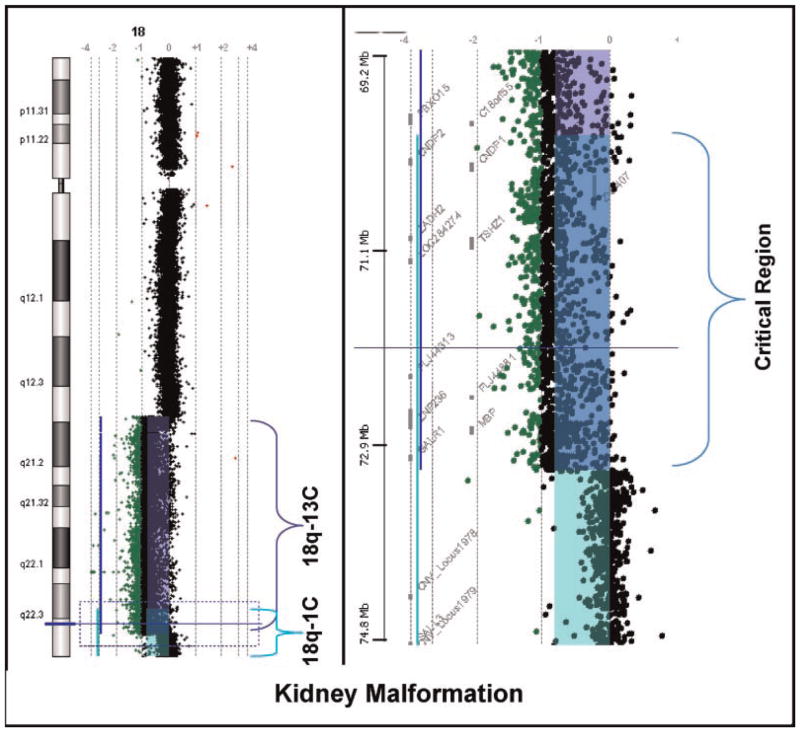
Seventy-two participants had abdominal ultrasound evaluations. Thirteen were found to have kidney malformations (Fig. 1A). The types of malformations included an absent kidney (n = 1), a horseshoe kidney (n = 2), hydronephrosis (n = 3), a polycystic kidney (n = 1) or an aplastic or hypoplastic kidney (n = 6). The largest common region of hemizygosity was defined by individuals 18q-1C and 18q-13C (Fig. 2). This region was 3.21 Mb in size (Table I) and contains 12 known genes (Fig. 1B). Fifty-two of the individuals who had abdominal ultrasounds were hemizygous for all or part of the critical region, yet only 13 have the phenotype for a penetrance of 25% (Table II).
FIG. 1.
Kidney malformation critical region. Panel A illustrates the chromosome content for everyone with an 18q deletion and kidney malformations using the UCSC Genome Browser Custom Tracks feature. The horizontal light blue bars depict the region of chromosome 18 that is present in two copies. The darker blue sections are the breakpoint regions. All individuals who have retained a distal segment of chromosome 18 have interstitial deletions and not translocations to other chromosomes. Panel B depicts the chromosome content data from the two individuals whose deletions define the smallest common region of hemizygosity. This region is shown along with the known genes in the region.
FIG. 2.
Oligonucleotide array comparative genomic hybridization microarray (aCGH) data for the two participants whose deletions define the smallest common hemizygous region for kidney malformations. The colored bars indicate the location of features that are significantly different from 0 on the log2 scale; purple for participant 18q-13C and green for participant 18q-1C. Features exactly on the 0 vertical axis have a 1:1 red green color ratio between the test and reference DNA samples. The region where the purple and green bars overlap (blue) indicates their common region of deletion (i.e., the critical region). The panel on the left shows the whole chromosome 18 view of the data. The panel on the right shows a zoomed in view of the critical region; from position 69.2 to 78.4 Mb. The locations of the genes in that region are shown between the megabase scale and the data points.
TABLE II.
Penetrance Calculations
| Phenotype | Number assessed for each phenotype | Number with phenotype (who define critical region) | Number hemizygous for the critical region without phenotype | Number hemizygous for critical region (both with and without the phenotype) | % Non-penetrance | % Penetrance |
|---|---|---|---|---|---|---|
| Kidney malformation | 72 | 13 | 39 | 52 | 39/52 = 75 | 25 |
| Dysmyelination | 88 | 80 | 0 | 80 | 0/80 = 0 | 100 |
| Growth hormone response failure | 99 | 46 | 9 | 89 | 9/89 = 10 | 90 |
| Aural atresia | 116 | 81 | 23 | 104 | 23/104 = 22 | 78 |
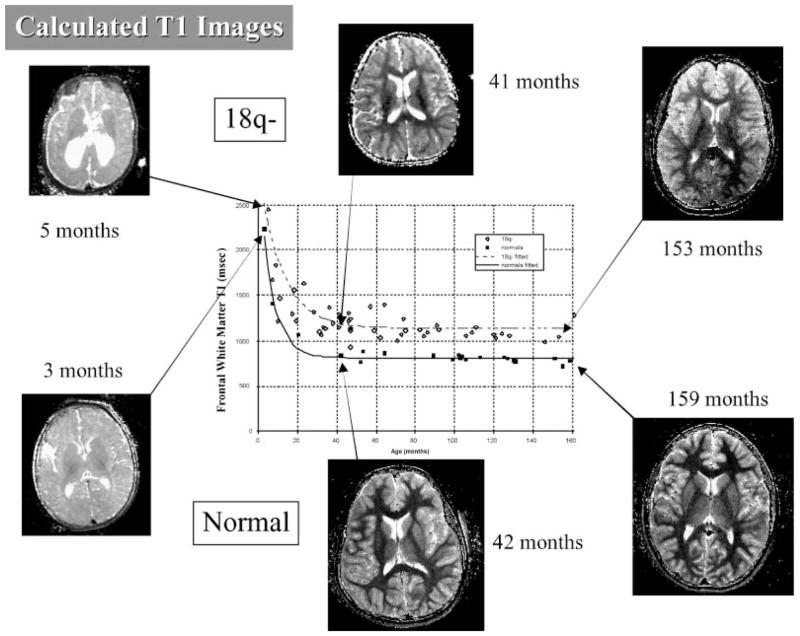
Detailed modeling of myelination revealed later onset and a lower myelination rate, leading to equilibrium myelin levels in children with 18q that are slightly less than 50% of the levels seen in typically developing children [Lancaster et al., 2005]. An illustration of these data is shown in Figure 3. MRI scans and determinations of myelination status were performed on 88 individuals with 18q deletions. The chromosome content of the 81 individuals identified as having dysmyelination of the brain by an MRI scan are shown in supporting information Figure 1A which may be found in the online version of this article. This analysis identified two regions. One individual (18q-140C) has a unique region of hemizygosity and dysmyelination. In our cohort, we have one other participant with a similar deletion who does not have dysmyelination so it is unclear if this is a new critical region associated with a gene involved in myelination or dysmyelination caused by an unrelated defect. The remaining 80 individuals with dysmyelination have a single region of hemizygosity defined by participants 18q-100C and 18q-13C (supporting information Fig. 2 may be found in the online version of this article). This region is 1.62 Mb in size (Table I) and contains five known genes (supporting information Fig. 1B may be found in the online version of this article). There were no individuals in our study who were hemizygous for this entire region and had normal myelination, making this phenotype 100% penetrant with regard to this critical region (Table II).
FIG. 3.
Calculated T1 images from individuals with 18q deletion and from typically developing children. Fitted model curves show higher T1 values in frontal white matter for children with 18q- indicating less myelin during development.
Ninety-nine individuals were evaluated for growth deficiency. Forty-six met the criteria of a height Z-score of less than −1.5 and failure to produce growth hormone in response to one provocative agent. Their chromosome content is shown in supporting information Figure 3 which may be found in the online version of this article. The critical region is defined by participants 18q-13C and 18q-100C; the same two individuals who define the dysmyelination region (supporting information Figs. 1B and 2 may be found in the online version of this article). The nucleotide borders of this region are shown in Table I. Because growth can be influenced by many factors this phenotype had variable expressivity. Therefore we defined the critical region using the data from only the 46 individuals exhibiting the complete phenotype. However, when we calculated the penetrance we defined non-penetrance as those individuals who had no evidence of growth failure; a group of nine. There were a total of 89 individuals who were hemizygous for this critical region yet only 9 had unequivocally normal growth and response to growth hormone stimulants. There was a group of 34 individuals who did not meet the criteria for growth hormone stimulant failure yet who did not have all normal growth parameters either. Therefore 90% of individuals hemizygous for this region will have some evidence of growth hormone deficiency, but will have only a 52% chance (46/89 = 52) of being clearly growth hormone deficient (Table II).
One hundred sixteen individuals were assessed by a neurotologist for the presence of atretic or stenotic ear canals. The vast majority of participants had congenital aural atresia types A or C. Eighty-one individuals had at least one ear canal that was atretic or stenotic (supporting information Fig. 4A may be found in the online version of this article). Individuals 18q-105C and 18q-13C defined the largest common region of hemizygosity (supporting information Fig. 2 may be found in the online version of this article). This region was 2.30 Mb in size (Table I) and contains seven known genes (supporting information Fig. 4B may be found in the online version of this article). There were a total of 104 individuals in our study who were evaluated for the phenotype and were hemizygous for this critical region yet only 81 had the phenotype; making the aural atresia phenotype 78% penetrant (Table II).
FIG. 4.
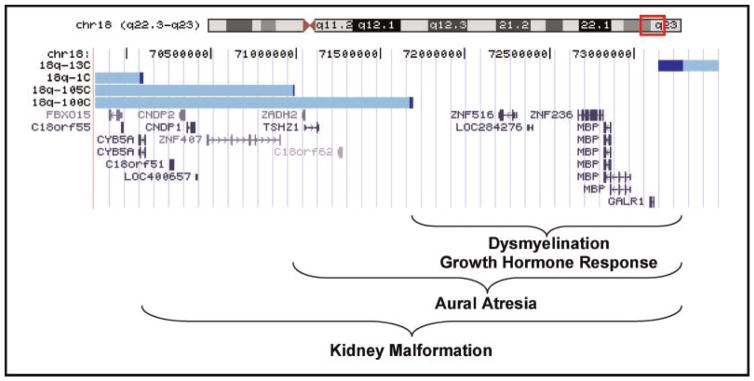
Aligned critical regions. The red box on the chromosome 18 ideogram defines the region displayed below. The aCGH data from the four individuals whose breakpoints define the boundaries of the critical regions have been converted to custom tracks on the UCSC Genome Browser. The light blue bars depict the intact chromosome for each individual with their study identification number to the left. The adjoining dark blue bars depict the breakpoint region for each individual. Below the Custom tracks is the RefSeq Gene track showing the known genes aligned within the region.
DISCUSSION
In this study, we completed molecular and clinical assessments on 151 individuals with deletions of 18q in an effort to draw genotype/phenotype correlations. We have defined critical regions of chromosome 18q that encompass the genes associated with four different phenotypic features seen in individuals with 18q deletions (Fig. 4). We previously reported critical regions for both dysmyelination and growth hormone deficiency [Cody et al., 1997; Gay et al., 1997]. In our 1997 report on dysmyelination [Gay et al., 1997], we identified a critical region by what we now consider to be a flawed strategy. We initially identified the smallest common region of hemizygosity for the individuals with dysmyelination that we now know to be 3.2 Mb (by applying the old data to today’s maps). In that study, we further narrowed that region by identifying an individual who had normal myelination and an interstitial deletion that included only a portion of the smallest common region of hemizygosity. We suggested that normal myelination indicated that the individual was not hemizygous for the causative gene for dysmyelination, thus narrowing the critical region. Including individuals without the phenotype to define the critical region is reasonable, however, only for phenotypes that are 100% penetrant in those hemizygous for the critical region. In retrospect we were lucky because one of the findings in the present study is that dysmyelination is 100% penetrant. However, for phenotypes that are not 100% penetrant, the individual without the phenotype could be either not hemizygous for the causative gene or could be non-penetrant. For this reason we have changed our strategy and only use the data from individuals with a particular phenotype in order to define a critical region. Critical regions are defined as the largest common region of hemizygosity in people with the phenotype. Our revised strategy often makes it more difficult to define small critical regions, because for regions other than those near the telomere, it relies on identifying those very rare individuals with interstitial deletions. However, the resulting critical regions are more likely to prove accurate.
In this study, the critical region for dysmyelination was narrowed by analyzing additional individuals, thereby reducing the size of the region to 1.62 Mb. This region is completely within the dysmyelination critical region identified by Feenstra et al. [2007] which was 6 Mb and the region identified by Linnankivi et al. [2006] which was 5.6 Mb in size (Table II). The more focused region described here contains five known genes. The genes are ZNF516, LOC284276, ZNF236, myelin basic protein (MBP) and the galanin receptor 1 (GALR1). The first three genes are virtually undescribed with regard to their biological function and no animal model orthologous genes have been identified. Based on their nucleotide sequence, ZNF516 and ZNF236 are presumed to be zinc finger transcription factors and as such are predicted to have important roles in the regulation of other genes. The gene expression profiles of ZNF516 show highest expression in bone marrow and blood, while the profile for ZNF236 suggests it is expressed in the superior cervical ganglion and the appendix. There is no expression data available for LOC284276.
In contrast there is extensive information about MBP and GALR1 and their biological significance. The galanin receptor 1 has been implicated in the regulation of feeding, learning and memory, seizures, pain, anxiety and mood disorders [Mitsukawa et al., 2008]. The consequences of hemizygosity of this gene are not clear. The GalR1 homozygous null mutation mouse has normal growth, but low IGF-1 as well as spontaneous tonic–clonic seizures [Jacoby et al., 2002]. A gene product involved so many diverse regulatory functions may turn out to be haplosufficient for some processes and haploinsufficient for others.
The obvious candidate gene for the dysmyelination phenotype is the myelin basic protein gene (MBP); located at 72.9 Mb. Myelin basic protein plays a key role in the compaction of central nervous system myelin. The shiverer mouse is a naturally occurring mutant with a deletion of exons 7–11 of the MBP gene, eliminating wild-type MBP mRNA production [Roach et al., 1985; Mikoshiba et al., 1991]. The homozygous shiverer mice fail to make compact myelin, develop severe tremors and have a shortened life span [Griffiths, 1996]. The heterozygous mice (analogous to 18q-) make MBP at 50% of normal and have delayed myelination but an otherwise normal phenotype [Roch et al., 1987].
Evidence suggests that a deletion of the MBP gene is not a major cause of dysmyelination in humans. In a study of 195 individuals with dysmyelination and without PLP1 mutations, none were shown to have copy number changes in MBP using BAC clone FISH and quantitative PCR [Vaurs-Barriere et al., 2006]. In addition, we have performed a similar analysis of fifty-seven individuals with dysmyelination. No deletions of MBP were identified (unpublished data). This may indicate that hemizygosity for the MBP gene is not associated with dysmyelination or that, if it is, it accounts for only the very rare cases. Alternatively, it is possible that the phenotype resulting from the hemizygosity of the MBP gene does not include developmental delay. Although the children enrolled in these studies had dysmyelination as the only physical finding, they came to the attention of clinicians because of developmental delay which could not be explained as a part of an identifiable syndrome. If the phenotype resulting from isolated hemizygosity of MBP does not include developmental delay, these studies have assessed the wrong population for MBP gene deletions.
The region associated with growth hormone stimulant response failure has been narrowed to the same 1.62 Mb dysmyelination region defined by the same two individuals and thereby includes the same genes. Feenstra et al. [2007] reported a 7.2 Mb region that includes the region we identified. However, they only assessed two individuals for growth hormone response failure. One individual was diagnosed with growth hormone deficiency who had a 17.5 Mb terminal deletion.
The aural atresia region included the dysmyelination region and extends proximally to include three additional genes; ZADH2, TSHZ1, and C18orf62. The biological role of ZADH2, the zinc binding alcohol dehydrogenase domain containing protein 2, is implied based on sequence similarity only. The TSHZ1 gene is teashirt family zinc finger 1 and in mice is involved in the development of the axial skeleton, middle ear and soft palate [Coré et al., 2007]. This is a possible candidate gene for this phenotype, however, in the mouse; the heterozygous null mutants have a normal phenotype. C18orf62 is an uncharacterized protein.
The variable presentation of kidney malformations in our study population is not unique. A similar spectrum of kidney malformations from horseshoe to aplastic kidney to polycystic kidney is seen in other single gene disorders such as PAX2 and EYA1 mutations [Ichikawa et al., 2002]. The critical region associated with kidney malformations overlaps with the other two regions and extends proximally to include six additional genes. These genes are CYB5A, C18orf51, CNDP2, CNDP1, LOC400657, and ZNF407. The genes C18orf51, LOC400657, and ZNF407 produce uncharacterized proteins, although ZNF407 is predicted to be a zinc finger protein based on sequence homology. CYB5A is the gene for cytochrome b-5 isoform 2 which causes recessive methemoglobinemia (OMIM 250790). The genes CNDP2, CNDP1 produce proteins carnosinase 1 and 2. Homozygous mutations in the CNDP1 genes can cause homocarnosinosis [Willi et al., 1997], while carnosinase 2 is essentially uncharacterized.
It is interesting to note that all the phenotypic features discussed in this study have a relatively high penetrance except for the kidney malformations. This is not surprising, since the more individuals there are with a particular phenotypic feature, the more likely there will be two individuals whose deletions define a small critical region. In a sense, critical regions for these highly penetrant phenotypes are the “low-hanging fruit.” Critical regions for phenotypes with lower penetrance, such as cleft palate, will require larger sample sizes. Of course, truly accurate calculations of the penetrance for each phenotype are not possible until the causative gene is identified. This is because there are individuals who do not have the phenotype in question, yet have a deletion for a portion of a critical region. Until the causative gene is determined, it is not known if these individuals are non-penetrant or if their deletion does not include the causative gene.
Although these features share a critical region, we do not hypothesize that a single gene is responsible for all of the features. Until we can prove definitively that the same gene is responsible for more than one phenotype, we will approach the problem as if each phenotype is caused by the hemizygosity of a different gene. We also do not think of this region as the critical region for the “18q-syndrome.” Many individuals have 18q deletions that do not include this region yet have an 18q deletion with clinical consequences. Therefore defining one region versus another region as “the critical region for the syndrome” is merely a matter of semantics and not particularly informative or valuable to those affected or their healthcare providers.
The next step in defining genotype–phenotype correlations is to identify genes responsible for the various phenotypes. Although we continue to assess additional individuals with 18q deletions, it is unlikely that additional genotyping and phenotyping will enable further restriction of the critical regions for these phenotypes. Rather, singling out a candidate gene can be accomplished by identifying individuals with a microdeletion of 18q and a single phenotypic feature of 18q-. We are actively seeking individuals with apparently normal karyotypes or small deletion by aCGH who have a single phenotypic feature of 18q-.
Once the key dosage-sensitive genes are identified, devising treatment options for affected children may be relatively straightforward in comparison to many other types of genetic conditions. For example, in recessive conditions there are no functional copies of the causative gene and in dominant negative conditions the deleterious effects of mutated gene product need to be reversed. In most cases, the affected child with an 18q deletion has one normal gene, and treatment would focus on increasing the expression of this gene. In chromosome abnormalities, modest changes in gene expression may have substantial phenotypic benefit. Of course, because these are genomic diseases, the expression of multiple genes may need to be altered. However, as previously discussed, it is most likely that only a minority of these genes will be responsible for the phenotype and therefore will be in need of upregulation.
Acknowledgments
Grant sponsor: MacDonald Family; Grant sponsor: The Chromosome 18 Registry & Research Society; Grant sponsor: NIH/NICHD; Grant number: R01-HD-045907; Grant sponsor: National Center for Research Resources; Grant number: M01-RR-001346.
We would like to thank the families who have so generously supported this work with their time and enthusiasm. This work was supported by the MacDonald family, The Chromosome 18 Registry & Research Society, NIH/NICHD grant R01-HD-045907, National Center for Research Resources grant M01-RR-001346 for the Frederic C. Bartter General Clinical Research Center.
Footnotes
WEB RESOURCES
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/.
Additional supporting information may be found in the online version of this article.
References
- Aarskog NK, Vedeler CA. Real-time quantitative polymerase chain reaction. A new method that detects both the peripheral myelin protein 22 duplication in Charcot-Marie-Tooth type 1A disease and the peripheral myelin protein 22 deletion in hereditary neuropathy with liability to pressure palsies. Hum Genet. 2000;107:494–498. doi: 10.1007/s004390000399. [DOI] [PubMed] [Google Scholar]
- Amano K, Sago H, Uchikawa C, Suzuki T, Kotliarova SE, Nukina N, Epstein CJ, Yamakawa K. Dosage-dependant over-expression of genes in the trisomic region of Ts1Cje mouse model for Down syndrome. Hum Mol Genet. 2004;13:1333–1340. doi: 10.1093/hmg/ddh154. [DOI] [PubMed] [Google Scholar]
- Barber JCK, Thomas NS, Collinson MN, Dennis NR, Liehr T, Weise A, Belitz B, Pfeiffer L, Kirchhoff M, Krag-Olsen B, Lundsteen C. Segmental haplosufficiency: Transmitted deletions of 2p12 include a pancreatic regeneration gene cluster and have no apparent phenotypic consequences. Eur J Hum Genet. 2005;13:283–291. doi: 10.1038/sj.ejhg.5201267. [DOI] [PubMed] [Google Scholar]
- Cody JD, Hale DE, Brkanac Z, Kaye CI, Leach RJ. Growth hormone insufficiency associated with a deletion of 2 Mb at 18q23. Am J Med Genet. 1997;71:420–425. [PubMed] [Google Scholar]
- Cody JD, Ghidoni PD, DuPont B, Hale DE, Hilsenbeck SG, Stratton RF, Hoffman DS, Muller S, Schaub RL, Leach RJ, Kaye CI. Congenital anomalies and anthropometry of 42 individuals with deletions of 18q. Am J Med Genet. 1999;85:455–462. doi: 10.1002/(sici)1096-8628(19990827)85:5<455::aid-ajmg5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Cody JD, Sebold C, Malik A, Heard P, Carter E, Crandall A, Soileau B, Semrud-Clikeman M, Cody CM, Hardies LJ, Li J, Lancaster J, Fox PT, Stratton RF, Richards K, Perry B, Hale DE. Recurrent interstitial deletions of 18q: A new syndrome involving expressive speech delay. Am J Med Genet Part A. 2007;143A:1181–1190. doi: 10.1002/ajmg.a.31729. [DOI] [PubMed] [Google Scholar]
- Coré N, Caubit X, Metchat A, Boned A, Djabali M, Fasano L. Tshz1 is required for axial skeleton, soft palate and middle ear development in mice. Dev Biol. 2007;308:407–420. doi: 10.1016/j.ydbio.2007.05.038. [DOI] [PubMed] [Google Scholar]
- Feenstra I, Vissers LELM, Orsel M, van Kessel AG, Brunner HG, Veltman JA, van Ravenswaaij-Arts CMA. Genotype-phenotype mapping of chromosome 18q deletions by high-resolution array CGH: An update of the phenotypic map. Am J Med Genet Part A. 2007;143A:1858–1867. doi: 10.1002/ajmg.a.31850. [DOI] [PubMed] [Google Scholar]
- Gay CT, Hardies LJ, Rauch RA, Lancaster JL, Plaetke R, DuPont BR, Cody JD, Herndon RC, Ghidoni PD, Schiff JM, Kaye CI, Leach RJ, Fox PT. Magnetic resonance imaging demonstrates incomplete myelination in the 18q- syndrome: Evidence for myelin basic protein haploin-sufficiency. Neuropsychiatr Genet. 1997;74:422–431. doi: 10.1002/(sici)1096-8628(19970725)74:4<422::aid-ajmg14>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Griffiths IR. Myelin mutants: Model systems for the study of normal and abnormal myelination. Bioessays. 1996;18:789–797. doi: 10.1002/bies.950181005. [DOI] [PubMed] [Google Scholar]
- Hale DE, Cody JD, Baillargeon J, Schaub RL, Danney MM, Leach RJ. The spectrum of growth abnormalities in children with 18q deletions. J Clin Endocrinol Metab. 2000;85:4450–4454. doi: 10.1210/jcem.85.12.7016. [DOI] [PubMed] [Google Scholar]
- Heard PL, Carter EM, Crandall AC, Sebold C, Hale DE, Cody JD. High resolution analysis of 18q- using oligo-microarray comparative genomic hybridization. Am J Med Genet Part A. 2009 doi: 10.1002/ajmg.a.32900. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nature. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Ichikawa I, Kuwayama F, Pope JC, Stephens FD. Paradigm shift from classic anatomic theories to contemporary cell biological views of CA-KUT. Kidney Int. 2002;61:898–899. doi: 10.1046/j.1523-1755.2002.00188.x. [DOI] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B. The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provided insights into the recent evolution of new genes. Genome Res. 2001;11:1018–1033. doi: 10.1101/gr.180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby AS, Hort YJ, Constantinescu G, Shine J, Iismaa TP. Critical role for GALR1 galanin receptor in galanin regulation of neuroendocrine function and seizure activity. Brain Res Mol Brain Res. 2002;107:195–200. doi: 10.1016/s0169-328x(02)00451-5. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Cody JD, Andrews T, Hardies LJ, Hale DE, Fox PT. Myelination in children with partial deletions of chromosome 18q. Am J Neuroradiol. 2005;26:447–454. [PMC free article] [PubMed] [Google Scholar]
- Li L, Moore P, Ngo C, Petrovic V, White SM, Northrup E, Ioannou PA, McKinlay Gardner RJ, Slater HR. Identification of a haplosufficient 3.6 Mb region of human chromosome 11q14.3 → q21. Cytogenet Genome Res. 2002;97:158–162. doi: 10.1159/000066612. [DOI] [PubMed] [Google Scholar]
- Linnankivi T, Tienari P, Somer M, Kähkönen M, Lönnqvist T, Valanne L, Pihko H. 18q deletions: Clinical, molecular, and brain MRI findings of 14 individuals. Am J Med Genet Part A. 2006;140A:331–339. doi: 10.1002/ajmg.a.31072. [DOI] [PubMed] [Google Scholar]
- Mikoshiba K, Okana H, Tamura T, Ikenaka K. Structure and function of the myelin protein genes. Ann Rev Neurosci. 1991;14:201–217. doi: 10.1146/annurev.ne.14.030191.001221. [DOI] [PubMed] [Google Scholar]
- Mitsukawa K, Lu X, Bartfai T. Galanin, galanin receptors and drug targets. Cell Mol Life Sci. 2008;65:1796–1805. doi: 10.1007/s00018-008-8153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson J, Wohlert M. Chromosome abnormalities found among 34910 newborn children: Results from a 13-year incidence study in Arhus, Denmark. Hum Genet. 1991;87:81–83. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- Roach AN, Takahashi D, Pravtcheva F, Ruddle F, Hood LE. Chromosomal mapping of mouse myelin basic protein gene and structure and transcription of the partially deleted gene in shiverer mutant mice. Cell. 1985;42:149–155. doi: 10.1016/s0092-8674(85)80110-0. [DOI] [PubMed] [Google Scholar]
- Roch JM, Cooper BJ, Ramirez M, Matthieu JM. Expression of only one myelin basic protein allele in mouse is compatible with normal myelination. Mol Brain Res. 1987;3:61–68. doi: 10.1016/0169-328x(87)90045-3. [DOI] [PubMed] [Google Scholar]
- Schuknecht HF. Congenital aural atresia. Laryngoscope. 1989;99:908–917. doi: 10.1288/00005537-198909000-00004. [DOI] [PubMed] [Google Scholar]
- Vaurs-Barriere C, Bonnet-Dupeyron MN, Combes P, Gauther-Barichard F, Reveles XT, Schiffmann R, Bertini E, Rodriguez D, Vago P, Armour JA, Saugier-Veber P, Frebourg T, Leach RJ, Boespflug-Tanguy O. Golli-MBP copy number analysis by FISH, QMPSF, and MAPH in 195 patients with hypomyelinating leukodystrophies. Ann Hum Genet. 2006;70:66–77. doi: 10.1111/j.1529-8817.2005.00208.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Cody JD, Leach RJ, O’Connell P. Gene expression patterns in cell lines from patients with 18q- syndrome. Hum Genet. 1999;104:467–475. doi: 10.1007/s004390050989. [DOI] [PubMed] [Google Scholar]
- Wilke AOM. The molecular basis of dominance. J Med Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi SM, Zhang Y, Hill JB, Phelan MC, Michaelis RC, Holden KR. A deletion in the long arm of chromosome 18 in a child with serum carnosinase deficiency. Pediatr Res. 1997;41:210–213. doi: 10.1203/00006450-199702000-00009. [DOI] [PubMed] [Google Scholar]
- Zlotogora J. Penetrance and expressivity in the molecular age. Genet Med. 2003;5:347–352. doi: 10.1097/01.gim.0000086478.87623.69. [DOI] [PubMed] [Google Scholar]