SUMMARY
Liver fibrosis, a form of scarring, gradually develops in chronic liver diseases when hepatocyte regeneration cannot compensate for hepatocyte death. At earlier stages, collagen produced by activated myofibroblasts (MFs) functions to maintain tissue integrity, but upon repeated injury, collagen accumulation suppresses hepatocyte regeneration, ultimately leading to liver failure. As a strategy to generate new hepatocytes and limit collagen deposition in the chronically injured liver, we developed in vivo reprogramming of MFs into hepatocytes using adeno-associated virus (AAV) vectors expressing hepatic transcription factors. We first identified the AAV6 subtype as effective in transducing MFs in mouse models of chronic liver disease. We then use lineage-tracing approaches to show that hepatocytes reprogrammed from MFs replicate primary hepatocyte function, and that liver fibrosis in AAV treated animals is reduced. Because AAV vectors are already used for liver-directed human gene therapy, our strategy has potential for clinical translation into a therapy for liver fibrosis.
INTRODUCTION
Liver fibrosis is a complication of chronic liver diseases associated with repeated hepatocyte death like viral hepatitis B and C, alcoholic liver disease and fatty liver disease (Bataller and Brenner, 2005; Friedman, 2008; Iredale, 2007; Schuppan and Afdhal, 2008). Liver fibrosis leads to liver cirrhosis, a common cause of death worldwide (Hoyert and Xu, 2012; Lim and Kim, 2008). In the United States alone, liver cirrhosis affects more than 600,000 patients (Scaglione et al., 2015). The only available cure is liver transplantation. Liver cell therapy is ineffective because of impaired engraftment (Hughes et al., 2012). The shortage of donor livers means only 6,000 patients benefit from this therapy and more than 35,000 patients die in the United States every year (Kim et al., 2014; Yoon et al.).
The biology of liver fibrosis has been studied extensively (Bataller and Brenner, 2005; Friedman, 2008; Iredale, 2007). Liver fibrosis is a repair mechanism that maintains the integrity of the injured liver through deposition of extracellular matrices such as collagen. Persistent liver injury causes collagen accumulation, which suppresses hepatocyte function and restricts intrahepatic blood flow, leading to liver failure, portal hypertension and liver cancer (Liu et al., 2012; Zhang and Friedman, 2012). Most of the collagen is produced by myofibroblasts (MFs), a mesenchymal liver cell type generated in large numbers by hepatic stellate cells (HSCs) or portal fibroblasts when they become activated in response to liver injury (Iwaisako et al., 2014; Mederacke et al., 2013). Because MFs are essential for liver fibrosis development and progression, we reasoned that in vivo reprogramming of MFs into hepatocytes would be effective as a therapy for liver fibrosis by replenishing the hepatocyte mass and limiting collagen production. For this, we built on previous studies of in vitro reprogramming of mouse fibroblasts into induced hepatocytes (iHeps) by lentiviral or retroviral expression of the hepatic transcription factor (TF) genes Foxa3, Gata4 and Hnf1a (Huang et al., 2011) or Foxa1/Foxa2/Foxa3 and Hnf4a (Sekiya and Suzuki, 2011), respectively.
A prerequisite for clinical translation of in vivo reprogramming strategies is that delivery of TFs to the targeted cells is both efficient and safe (Addis and Epstein, 2013; Heinrich et al., 2015). We therefore explored the use of adeno-associated virus (AAV) vectors, which have emerged as a safe and effective tool for gene delivery—including in clinical trials of liver-directed gene therapy of hemophilia B (Nathwani et al., 2014; Nathwani et al., 2011)—because they do not integrate into the genome or exhibit the strong immunogenicity that has hampered adenoviral vectors (Crystal, 2014).
RESULTS
In Vivo Gene Delivery to MFs with an AAV Vector
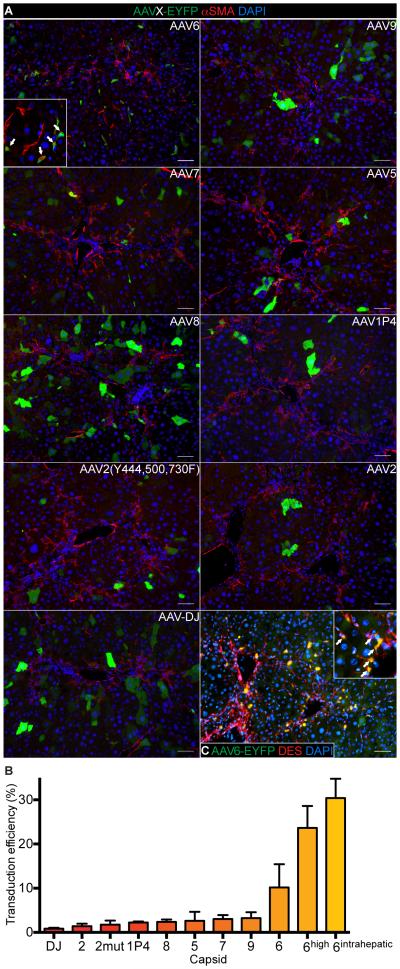
Because AAV capsids with MF tropism were unknown, we performed a screen in the carbon tetrachloride (CCl4) mouse model of liver fibrosis (Liedtke et al., 2013). We focused on AAV capsids effective in transducing fibroblasts or other mesenchymal cell types, including the naturally occurring AAV2, AAV5, AAV6, AAV7, AAV8 and AAV9 capsids (Blankinship et al., 2004; Di Pasquale et al., 2003; Gao et al., 2004; Grimm and Kay, 2003; Rutledge et al., 1998) and the engineered capsids AAV1P4 (seven amino acid re-targeting peptide displayed in an exposed capsid loop) (Borner et al., 2013), AAV2(Y444,500,730F) (mutation of three exposed tyrosines to phenylalanines) (Li et al., 2010) and AAV-DJ (chimera of AAV2/8/9) (Grimm et al., 2008). After treating wildtype mice with 12 doses of CCl4 we intravenously injected them with 1 × 1011 viral genomes of an AAV-EYFP vector pseudotyped with one of the nine capsids and analyzed the mouse organs after seven additional doses of CCl4. Only AAV6 had a relevant MF tropism, transducing 10.2 ± 5.3% of α-smooth muscle actin (αSMA)-positive MFs (Figure 1A,B). Increasing the vector dose to 4 × 1011 viral genomes or intrahepatic injection increased the MF transduction efficiency to 23.6 ± 5.0% or 30.4 ± 4.36 %, respectively (Figure 1B,C). At a dose of 4 × 1011 viral genomes, AAV6 also transduced 7.9 ± 7.15% of Kupffer cells and 1.2 ± 0.53% of hepatocytes, but did not transduce cholangiocytes or endothelial cells (Figure S1A,B). Interestingly, the same AAV6 dose transduced only 2.3 ± 1.99% of desmin (DES)-positive HSCs in mice without liver injury (Figure S1C). As previously reported (Zincarelli et al., 2008), intravenously injected AAV6 showed organ tropism for liver, skeletal muscle, heart and spleen (Figure S1D).
Figure 1. In Vivo Screen For AAV Capsids Effective in Transducing MFs.
(A) EYFP and αSMA co-immunofluorescence (IF) of livers of recipients of AAV-EYFP vectors pseudotyped with indicated capsids. Arrows in inset point at transduced MFs. Size bars, 75 μm.
(B) Quantification of MFs transduced in vivo. Results are means ± SD for biological replicates (n = 3 for 6, 9, 7, 5, 8 and 1P4, n = 2 for AAV2(Y444,500,730F) (2mut), 2, DJ, 6high and 6intrahepatic). The transduction efficiency of 6intrahepatic was quantified in the injected liver lobe.
(C) EYFP and desmin (DES) co-IF of liver after intrahepatic injection of AAV6-EYFP. Arrows in inset point at transduced MFs. Size bar, 75 μm.
Hepatic Reprogramming of MFs
We used the AAV6 capsid to generate six AAV vectors expressing the TF genes Foxa1, Foxa2, Foxa3, Gata4, Hnf1a or Hnf4a from the CMV promoter (combination referred to as AAV6-6TFs). To test the efficacy of AAV6-6TFs, we transduced MFs generated by in vitro activation of primary mouse HSCs, which produced clones of cells that lost most of their original identity and acquired hepatocyte gene expression and function (Figure S1E-J).
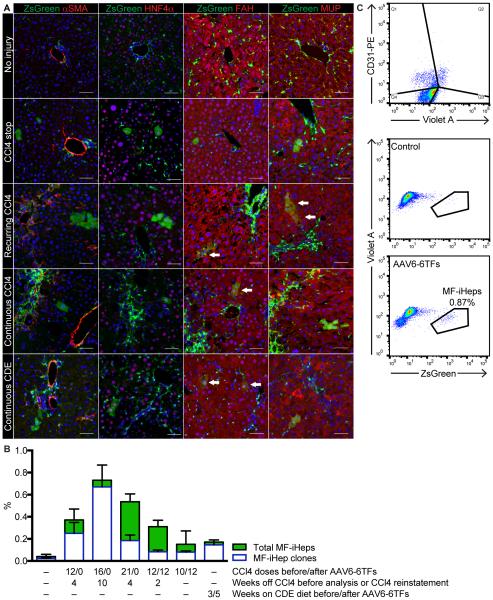
To establish in vivo efficacy of AAV6-6TFs, we used a mouse model of MF fate tracing in which a constitutive reporter (R26R-ZsGreen) is activated by Cre recombinase expressed from the lecithin retinol acyltransferase (Lrat) promoter (Mederacke et al., 2013). We treated these mice with 12 doses of CCl4, intravenously injected them with 4 × 1011 viral genomes of AAV6-6TFs and analyzed their livers four weeks later (CCl4 stop protocol). We found cells expressing the MF fate-tracing marker ZsGreen and the hepatocyte markers HNF4α, fumarylacetoacetate hydrolase (FAH) and major urinary protein (MUP) (Figure 2A,B), suggesting they were iHeps derived from MFs (MF-iHeps). To confirm this finding, we determined the specificity and efficiency of MF labeling in Lrat-Cre;R26R-ZsGreen mice treated with 12 doses of CCl4. As previously reported (Mederacke et al., 2013), we found that most (92.8%) MFs and few (0.021%) hepatocytes were labeled in these mice (Figure S2A). We also excluded cell fusion as the mechanism of MF-iHep formation (Figure S2B).
Figure 2. In Vivo Hepatic Reprogramming of MFs.
(A) ZsGreen fluorescence and αSMA, HNF4α, FAH or MUP IF of livers of mice treated with the indicated protocols consisting of intravenous injection of AAV6-6TFs and no, CCl4-induced or CDE diet-induced liver injury. Nuclei are stained blue with DAPI. Size bars, 50 μm. Arrows point at examples of MF-iHeps.
(B) Hepatic reprogramming efficiency and liver repopulation assessed by quantification of MF-iHep clones and total MF-iHeps, respectively. Results are means ± SEM for biological replicates (n = 4, 2, 2, 2, 3, 3, 4 from no to CDE-induced liver injury).
(C) Quantification by flow cytometry of MF-iHeps in hepatocytes isolated from mice treated with 16 doses of CCl4 and intravenously injected with AAV6-6TFs or vector diluent (control). Potential contamination with vitamin A (Violet A)-positive MFs or abundant CD31-positive endothelial cells was excluded.
Because AAV6 also transduced Kupffer cells (Figure S1A,B), we tested AAV6-6TFs in LysM-Cre;R26R-ZsGreen Kupffer cell fate-tracing mice (Clausen et al., 1999) (Figure S2A), which showed rare (< 0.01%) fate-traced hepatocytes after treatment with the CCl4 stop protocol (Figure S2C). Moreover, AAV6-6TFs induced very few cells expressing hepatocyte markers in skeletal muscle, heart and spleen; none of these cells showed hepatocyte morphology (Figure S2D).
Next, we determined the efficiency of hepatic reprogramming of MFs by AAV6-6TFs under different conditions of fibrotic liver injury. In accord with our finding of low transduction of HSCs, we found only baseline levels of labeled hepatocytes in mice not treated with CCl4 (Figure 2A,B). We found the most MF-iHeps—0.87% of all hepatocytes in the liver—in mice treated with 16 doses of CCl4 (Figure 2B,C). MF-iHeps were mostly single cells in these mice but clonally expanded in mice treated with 21 doses of CCl4, leading to a similar overall number of MF-iHeps despite a lower number of MF-iHep clones (Figure 2B). Findings were similar in mice in which we induced liver injury with 12 doses of CCl4 and reinstated it after a two-week break with additional 12 doses (recurring CCl4 protocol), and in mice continuously treated with CCl4—10 doses before and 12 doses after intravenous injection of AAV6-6TFs (continuous CCl4 protocol) (Figure 2A,B). We validated these results in mice with liver fibrosis due to steatohepatitis induced by continuous feeding of a choline-deficient ethionine-supplemented (CDE) diet (Knight et al., 2007; Liedtke et al., 2013) (Figure 2A,B). These findings suggest that severe liver fibrosis and ongoing liver injury reduce the efficiency of hepatic reprogramming of MFs, most likely because collagen-engulfed MFs are less accessible to AAV vectors and because nonintegrating AAV vectors are lost from proliferating MFs (Li et al., 2011; Nakai et al., 2001). However, these findings also show that MF-iHep proliferation can compensate for reduced MF-iHep formation.
Normal Proliferation of MF-iHeps
Prompted by our finding of clonal expansion of MF-iHeps, we determined whether MF-iHeps proliferate normally. For this, we compared clone size of MF-iHeps and primary hepatocytes between Lrat-Cre;R26R-ZsGreen mice treated with the recurring CCl4 protocol and R26R-ZsGreen mice receiving the same CCl4 treatment but intravenous injection of 2 × 1010 viral genomes of hepatocyte-targeted AAV8-Ttr-Cre vector to label hepatocytes at low frequency (Tarlow et al., 2014a). We found a similar clone size distribution in MF-iHeps and primary hepatocytes (Figure S3A).
Because nonintegrating AAV vectors are lost from proliferating cells (Li et al., 2011; Nakai et al., 2001), these results also suggested that MF-iHeps are stably reprogrammed. We confirmed this hypothesis by showing that AAV6-6TFs were present in newly formed MF-iHeps and absent in clonally expanded MF-iHeps isolated by laser-capture microdissection (LCM) (Figure S3B-D).
Normal Function of MF-iHeps
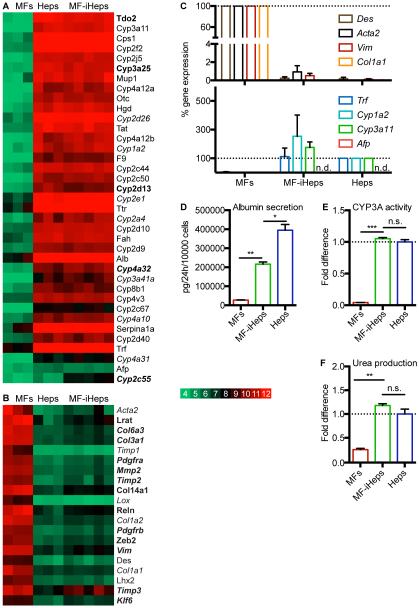
To determine whether MF-iHeps acquire full hepatocyte differentiation, we profiled their global gene expression with microarrays. For this, we isolated MF-iHeps by LCM from Lrat-Cre;R26R-ZsGreen mice four to 10 weeks after treatment with 16 doses of CCl4 and intravenous injection of AAV6-6TFs. As controls we used primary hepatocytes from the same mice and MFs from CCl4-treated littermates, isolated by LCM and FACS, respectively. We found that MF-iHeps closely resembled primary hepatocytes (Figures S3E). Few of the 908 significantly differentially expressed (P ≤ 0.025) genes were associated with hepatocyte differentiation, as illustrated by mostly normal expression of cytochrome P450 (CYP) genes specific for immature or mature hepatocytes (Peng et al., 2012) (Table S1 and Figure 3A,C). However, MF-iHeps expressed some genes associated with MF or HSC differentiation (Figure 3B,C), suggesting that they retained some original identity. Importantly, this MF/HSC memory was minimal and did not translate into impaired hepatocyte function, as evidenced by normal albumin secretion, CYP3A activity and urea production in MF-iHeps isolated from Lrat-Cre;R26R-ZsGreen mice by FACS (Figure 3D-F).
Figure 3. Characterization of MF-iHep Differentiation and Function.
(A-C) Microarray and qRT-PCR analysis of MFs, primary hepatocytes (Heps) and MF-iHeps (n = 3, 3 and 5 biological replicates). Color key includes log2 microarray values.
(A) Heatmap of genes reflecting hepatocyte differentiation in microarray analysis (brown cells in column A of Table S1). All CYP genes specific for immature or mature primary hepatocytes, i.e., enriched in neonatal or adult mouse liver (Peng et al., 2012), and expressed > 15-fold higher in Heps than in MFs were included. In addition, examples of intermediate CYP genes that peak in the adolescent mouse liver and decrease moderately (Cyp1a2) or markedly (Cyp2e1 and Cyp2c55) in the adult mouse liver were included (Peng et al., 2012). Immature or adolescent CYP genes are in italics. Other genes were derived from the literature (Huang et al., 2011; Tarlow et al., 2014b). Significantly differentially expressed (P ≤ 0.025) genes are bold.
(B) Heatmap of genes reflecting MF/HSC differentiation in microarray analysis (gray cells in column A in Table S1). All collagen genes expressed > 10-fold higher in MFs than in Heps were included. Other genes were derived from the literature; genes expressed higher in MFs than in HSCs are in italics (Duarte et al., 2015; Hayes et al., 2014; Henderson et al., 2013; Iwaisako et al., 2014; Lua et al., 2016; Mederacke et al., 2013; Schuppan et al., 2001). Significantly differentially expressed (P ≤ 0.025) genes are bold.
(C) qRT-PCR analysis of hepatocyte and MF/HSC genes in the microarray samples. Results are means ± SEM for biological replicates (n = 3 for MFs, n = 5 for MF-iHeps and n = 3 for Heps). n.d., not detected.
(D-F) Analysis of albumin secretion (D), CYP3A activity (E) and urea production (F) in FACS-isolated MFs, MF-iHeps and primary hepatocytes (Heps). Results are means ± SEM for biological replicates (n = 2). Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant.
Antifibrotic Effect and Further Development of In Vivo Hepatic Reprogramming of MFs
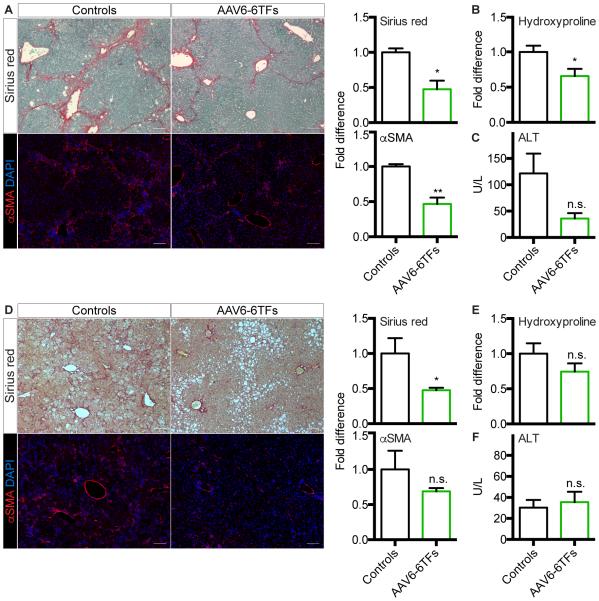
To test the efficacy of our strategy in a model of chronic fibrotic liver injury, the most challenging clinical scenario, we used the continuous CCl4 protocol (Figure 2B). We found reduced liver fibrosis in AAV6-6TF recipients as assessed by Sirius red staining, αSMA IF and hydroxyproline measurement (Figure 4A,B). In addition, serum levels of the hepatocyte injury marker alanine aminotransferase (ALT) were reduced in these mice (Figure 4C). Findings were similar in mice continuously fed a CDE diet (Figures 2B and 4D-F).
Figure 4. Therapeutic Efficacy of In Vivo Hepatic Reprogramming of MFs.
(A-C) Therapeutic efficacy of AAV6-6TFs in mice treated with the continuous CCl4 protocol.
(A) Sirius red staining and αSMA IF with quantification. Size bars, 100 μm. Results are means ± SEM for biological replicates (n = 3). Student’s t test; *P < 0.05, **P < 0.01.
(B) Analysis of whole liver collagen content by hydroxyproline assay. Results are means ± SEM for biological replicates (n = 3). Student’s t test; *P < 0.05.
(C) Serum levels of ALT. Results are means ± SEM for biological replicates (n = 3). Student’s t test; n.s., not significant.
(D-F) Therapeutic efficacy of AAV6-6TFs in mice fed a CDE diet.
(D) Sirius red staining and αSMA IF with quantification. Size bars, 100 μm. Results are means ± SEM for biological replicates (n = 3 for controls and n = 4 for AAV6-6TFs). Student’s t test; *P < 0.05; n.s., not significant.
(E) Analysis of whole liver collagen content by hydroxyproline assay. Results are means ± SEM for biological replicates (n = 3 for controls and n = 4 for AAV6-6TFs). Student’s t test; n.s., not significant.
(F) Serum levels of ALT. Results are means ± SEM for biological replicates (n = 3 for controls and n = 4 for AAV6-6TFs). Student’s t test; n.s., not significant.
Next, we defined the contribution of MF-iHeps to reduced liver fibrosis and injury. Because the number of Kupffer cell-derived hepatocytes induced by AAV6-6TFs was negligible (Figure S2C), we investigated the contribution of increased expression of hepatic TFs in hepatocytes (Nishikawa et al., 2015). For this, we generated hepatocyte-targeted AAV8 vectors expressing the six TFs (AAV8-6TFs) and intravenously injected them—at a dose at which each of the six vectors transduced 1.2% of hepatocytes (Figure S1B and Table S2)—into mice treated with the continuous CCl4 protocol (Figure 2B). We found that liver fibrosis and injury were not significantly reduced in these mice (Figure S4A-C), although whole liver gene expression of fibrolytic MMP9 and MMP12 was increased (Ramachandran et al., 2012) (Figure S4D). We also used AAV8-6TFs to exclude unspecific activation of MF fate tracing in hepatocytes of Lrat-Cre;R26R-ZsGreen mice (Figure S4E). These results show that the therapeutic efficacy of our strategy is mainly due to hepatic reprogramming of MFs.
To identify the essential TFs, we determined which of the six AAV6-TF vectors are enriched in newly formed MF-iHeps in vivo (Figure S4F). Based on this analysis and in vitro and in vivo validation, we arrived at FOXA3, GATA4 and HNF1α as the most effective minimal TF combination (Figure S4G-M). These findings accord with a previous study that used lentiviral vectors for hepatic reprogramming of fibroblasts in vitro, except that AAV6-mediated in vitro or in vivo hepatic reprogramming of MFs did not require p19 inactivation (Huang et al., 2011).
Finally, we established efficient transduction and hepatic reprogramming of primary human MFs with AAV6 vectors in vitro (Figure S4N-Q).
DISCUSSION
Our study establishes the feasibility of in vivo reprogramming of MFs into fully functional hepatocytes using AAV vectors, a gene delivery tool that proved to be safe and effective in clinical trials of liver-directed gene therapy (Nathwani et al., 2014; Nathwani et al., 2011). To develop this strategy into a therapy for patients with liver fibrosis, the number of MFs that express the essential TFs needs to be increased. To maximize the efficacy of the wildtype AAV6 capsid, the essential TFs could be delivered with one vector, which would be feasible if a short promoter, self-cleaving 2A peptides and a synthetic polyA signal are used (Ostedgaard et al., 2005). In addition, the MF transduction efficiency could be improved, which—as illustrated by our findings made after intrahepatic vector injection—could be achieved by detargeting AAV6 from extrahepatic cells. For this, technologies like AAV capsid DNA shuffling or peptide display could be used (Grimm et al., 2008; Lisowski et al., 2014). As a safety measure, residual extrahepatic TF expression could be prevented by tagging with target sequences of tissue-specific microRNAs (Xie et al., 2011). Further aided by the ability of MF-iHeps to expand in response to liver injury, our strategy of repurposing MFs has potential as a therapy for liver fibrosis that addresses both insufficient hepatocyte function and collagen accumulation.
EXPERIMENTAL PROCEDURES
Mice
All procedures involving mice were approved by the Institutional Animal Care and Use Committee at the University of California San Francisco. All mice were housed under barrier conditions. Wildtype mice and mice heterozygous for Lrat-Cre (Mederacke et al., 2013) or LysM-Cre (Clausen et al., 1999) and heterozygous for R26R-ZsGreen (Madisen et al., 2010) were used. All mice were C57BL/6 and 5-16 weeks old. Littermates and male and female mice were equally distributed between experimental and control groups. Mice were not randomized and investigators were not blinded.
Liver Fibrosis Models
Mice received intraperitoneal injections of 0.5 μL/g body weight carbon tetrachloride (CCl4) diluted 1:4 in corn oil (both Sigma-Aldrich) every 2 days. Livers were analyzed 2 or 3 days after the last CCl4 dose. Alternatively, mice were fed a diet deficient in choline (MP Biomedicals) supplemented with 0.15% (w/v) ethionine (Sigma-Aldrich) in drinking water.
AAV Vectors
AAV vectors were produced in HEK293T cells (Agilent Technologies) and purified using iodixanol (Sigma-Aldrich) density gradients (Zolotukhin et al., 1999). To reduce iodixanol thickness for intravenous injection, AAV vectors were diluted in phosphate-buffered saline (PBS)/5% sorbitol at a ratio of 5:1. AAV vectors were also produced by Virovek in insect cells using the baculovirus expression system (Chen, 2008). AAV vectors from Virovek were purified using cesium chloride gradients and diluted in PBS containing 0.001% pluronic F-68 (Sigma-Aldrich) for intravenous injection. Mice were intravenously injected via the tail vein with 4 × 1011 viral genomes of each vector unless otherwise specified. Vector injections were performed slowly and the volume was limited to 300 μL for tail vein injection and 50 μl for intrahepatic injection.
Calculation of Hepatic Reprogramming Efficiency and Liver Repopulation
The percentage of MF-iHep clones (single cells or nodules) of all hepatocytes in the liver is the hepatic reprogramming efficiency. The total number of hepatocytes per 10x field in a liver section was determined by FAH immunofluorescence and DAPI staining. After correcting for hepatocyte-free areas in a field, e.g., blood vessels and mesenchyme, MF-iHep clones were quantified in > 2,000 hepatocytes per mouse in randomly taken 10x images of ≥ 2 liver sections from ≥ 2 liver lobes. MF-iHep clones were identified by co-expression of ZsGreen and FAH. The percentage of all MF-iHeps of all hepatocytes in the liver is the extent of liver repopulation.
Statistical Analyses
Data are expressed as means ± standard error of the mean (SEM) or standard deviation (SD). Statistical differences between experimental and control groups were determined by Student’s t test (unpaired, two-tailed) unless otherwise specified. A P value of less than 0.05 was considered significant. Prism 6.0 (Graphpad) was used for statistical analyses.
Supplementary Material
ACKNOWLEDGEMENTS
H.W. was supported by NIH grants R21 AA022158 and P30 DK26743 (UCSF Liver Center). M.R. was supported by the Deutsche Forschungsgemeinschaft postdoctoral fellowship RE 3749/1-1. R.E.S. was supported by the Wallonie-Bruxelles International World postdoctoral fellowship and the Fonds Spécial de Recherche de la Fédération Wallonie-Bruxelles. A.A.G. was supported by the California Institute for Regenerative Medicine training program in stem cell research at UCSF (TG2-01153). The authors thank Donghui Wang in the UCSF Preclinical Therapeutics Core, Vinh Nguyen and Ninnia Lescano in the Flow Cytometry Core and Chris Her in the Cell Biology Core of the UCSF Liver Center for technical support, Alexander Williams in the Gladstone Bioinformatics Core for bioinformatic support, Pamela Derish in the UCSF Department of Surgery for manuscript editing and Kinji Asahina from the University of Southern California for advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
H.W. conceived the project. H.W., M.R., R.E.S., Y.M., L.D. and A.A.G. designed the experiments. H.W. and L.D. designed the AAV vector constructs. L.D., E.K., E.W., Y.M. and D.G. produced the AAV vectors. R.E.S., Y.M., M.R. and J.G.B. performed and analyzed the AAV capsid screen. M.R. and R.E.S. performed and analyzed the experiments related to in vivo hepatic reprogramming and therapeutic efficacy. Y.M., L.D. and A.A.G. performed and analyzed other in vivo experiments. M.R., R.E.S., L.D., A.A.G., B.Y.H. and S.J.N. performed and analyzed the in vitro experiments. C.U.C. und R.R.S. provided techniques and reagents. H.W., M.R., R.E.S., Y.M., L.D. and A.A.G. wrote the manuscript. H.W. and D.G. edited the manuscript. All authors read and approved the final manuscript.
REFERENCES
- Addis RC, Epstein JA. Induced regeneration--the progress and promise of direct reprogramming for heart repair. Nat Med. 2013;19:829–836. doi: 10.1038/nm.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankinship MJ, Gregorevic P, Allen JM, Harper SQ, Harper H, Halbert CL, Miller AD, Chamberlain JS. Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Mol Ther. 2004;10:671–678. doi: 10.1016/j.ymthe.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Borner K, Niopek D, Cotugno G, Kaldenbach M, Pankert T, Willemsen J, Zhang X, Schurmann N, Mockenhaupt S, Serva A, et al. Robust RNAi enhancement via human Argonaute-2 overexpression from plasmids, viral vectors and cell lines. Nucleic Acids Res. 2013;41:e199. doi: 10.1093/nar/gkt836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Intron splicing-mediated expression of AAV Rep and Cap genes and production of AAV vectors in insect cells. Mol Ther. 2008;16:924–930. doi: 10.1038/mt.2008.35. [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Crystal RG. Adenovirus: the first effective in vivo gene delivery vector. Hum Gene Ther. 2014;25:3–11. doi: 10.1089/hum.2013.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pasquale G, Davidson BL, Stein CS, Martins I, Scudiero D, Monks A, Chiorini JA. Identification of PDGFR as a receptor for AAV-5 transduction. Nat Med. 2003;9:1306–1312. doi: 10.1038/nm929. [DOI] [PubMed] [Google Scholar]
- Duarte S, Baber J, Fujii T, Coito AJ. Matrix metalloproteinases in liver injury, repair and fibrosis. Matrix Biol. 2015;44-46:147–156. doi: 10.1016/j.matbio.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Kay MA. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr Gene Ther. 2003;3:281–304. doi: 10.2174/1566523034578285. [DOI] [PubMed] [Google Scholar]
- Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, Kay MA. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BJ, Riehle KJ, Shimizu-Albergine M, Bauer RL, Hudkins KL, Johansson F, Yeh MM, Mahoney WM, Jr., Yeung RS, Campbell JS. Activation of platelet-derived growth factor receptor alpha contributes to liver fibrosis. PLoS One. 2014;9:e92925. doi: 10.1371/journal.pone.0092925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C, Spagnoli FM, Berninger B. In vivo reprogramming for tissue repair. Nat Cell Biol. 2015;17:204–211. doi: 10.1038/ncb3108. [DOI] [PubMed] [Google Scholar]
- Henderson NC, Arnold TD, Katamura Y, Giacomini MM, Rodriguez JD, McCarty JH, Pellicoro A, Raschperger E, Betsholtz C, Ruminski PG, et al. Targeting of alphav integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med. 2013;19:1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyert DL, Xu J. Deaths: preliminary data for 2011. Natl Vital Stat Rep. 2012;61:1–51. [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Hughes RD, Mitry RR, Dhawan A. Current status of hepatocyte transplantation. Transplantation. 2012;93:342–347. doi: 10.1097/TP.0b013e31823b72d6. [DOI] [PubMed] [Google Scholar]
- Iredale JP. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest. 2007;117:539–548. doi: 10.1172/JCI30542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaisako K, Jiang C, Zhang M, Cong M, Moore-Morris TJ, Park TJ, Liu X, Xu J, Wang P, Paik YH, et al. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A. 2014;111:E3297–3305. doi: 10.1073/pnas.1400062111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WR, Smith JM, Skeans MA, Schladt DP, Schnitzler MA, Edwards EB, Harper AM, Wainright JL, Snyder JJ, Israni AK, et al. OPTN/SRTR 2012 Annual Data Report: liver. Am J Transplant. 2014;14(Suppl 1):69–96. doi: 10.1111/ajt.12581. [DOI] [PubMed] [Google Scholar]
- Knight B, Akhurst B, Matthews VB, Ruddell RG, Ramm GA, Abraham LJ, Olynyk JK, Yeoh GC. Attenuated liver progenitor (oval) cell and fibrogenic responses to the choline deficient, ethionine supplemented diet in the BALB/c inbred strain of mice. J Hepatol. 2007;46:134–141. doi: 10.1016/j.jhep.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jayandharan GR, Li B, Ling C, Ma W, Srivastava A, Zhong L. High-efficiency transduction of fibroblasts and mesenchymal stem cells by tyrosine-mutant AAV2 vectors for their potential use in cellular therapy. Hum Gene Ther. 2010;21:1527–1543. doi: 10.1089/hum.2010.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke C, Luedde T, Sauerbruch T, Scholten D, Streetz K, Tacke F, Tolba R, Trautwein C, Trebicka J, Weiskirchen R. Experimental liver fibrosis research: update on animal models, legal issues and translational aspects. Fibrogenesis Tissue Repair. 2013;6:19. doi: 10.1186/1755-1536-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YS, Kim WR. The global impact of hepatic fibrosis and end-stage liver disease. Clin Liver Dis. 2008;12:733–746. doi: 10.1016/j.cld.2008.07.007. vii. [DOI] [PubMed] [Google Scholar]
- Lisowski L, Dane AP, Chu K, Zhang Y, Cunningham SC, Wilson EM, Nygaard S, Grompe M, Alexander IE, Kay MA. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yannam GR, Nishikawa T, Yamamoto T, Basma H, Ito R, Nagaya M, Dutta-Moscato J, Stolz DB, Duan F, et al. The microenvironment in hepatocyte regeneration and function in rats with advanced cirrhosis. Hepatology. 2012;55:1529–1539. doi: 10.1002/hep.24815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lua I, Li Y, Zagory JA, Wang KS, French SW, Sevigny J, Asahina K. Characterization of hepatic stellate cells, portal fibroblasts, and mesothelial cells in normal and fibrotic livers. J Hepatol. 2016;64:1137–1146. doi: 10.1016/j.jhep.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai H, Yant SR, Storm TA, Fuess S, Meuse L, Kay MA. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stableliver transduction in vivo. J Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, Della Peruta M, Lheriteau E, Patel N, Raj D, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Bell A, Brooks JM, Setoyama K, Melis M, Han B, Fukumitsu K, Handa K, Tian J, Kaestner KH, et al. Resetting the transcription factor network reverses terminal chronic hepatic failure. J Clin Invest. 2015;125:1533–1544. doi: 10.1172/JCI73137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostedgaard LS, Rokhlina T, Karp PH, Lashmit P, Afione S, Schmidt M, Zabner J, Stinski MF, Chiorini JA, Welsh MJ. A shortened adeno-associated virus expression cassette for CFTR gene transfer to cystic fibrosis airway epithelia. Proc Natl Acad Sci U S A. 2005;102:2952–2957. doi: 10.1073/pnas.0409845102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong XB. RNA sequencing reveals dynamic changes of mRNA abundance of cytochromes P450 and their alternative transcripts during mouse liver development. Drug Metab Dispos. 2012;40:1198–1209. doi: 10.1124/dmd.112.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Pellicoro A, Vernon MA, Boulter L, Aucott RL, Ali A, Hartland SN, Snowdon VK, Cappon A, Gordon-Walker TT, et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci U S A. 2012;109:E3186–3195. doi: 10.1073/pnas.1119964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge EA, Halbert CL, Russell DW. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione S, Kliethermes S, Cao G, Shoham D, Durazo R, Luke A, Volk ML. The Epidemiology of Cirrhosis in the United States: A Population-based Study. J Clin Gastroenterol. 2015;49:690–696. doi: 10.1097/MCG.0000000000000208. [DOI] [PubMed] [Google Scholar]
- Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppan D, Ruehl M, Somasundaram R, Hahn EG. Matrix as a modulator of hepatic fibrogenesis. Semin Liver Dis. 2001;21:351–372. doi: 10.1055/s-2001-17556. [DOI] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Tarlow BD, Finegold MJ, Grompe M. Clonal tracing of Sox9(+) liver progenitors in mouse oval cell injury. Hepatology. 2014a;60:278–289. doi: 10.1002/hep.27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow BD, Pelz C, Naugler WE, Wakefield L, Wilson EM, Finegold MJ, Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014b;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Xie Q, Zhang H, Ameres SL, Hung JH, Su Q, He R, Mu X, Seher Ahmed S, Park S, et al. MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol Ther. 2011;19:526–535. doi: 10.1038/mt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Chen CM, Yi H. Liver cirrhosis mortality in the United States: National, state, and regional trends, 2000–2011. NIAAA Surveillance Report #100. 2014 Dec; ( http://pubs.niaaa.nih.gov/publications/Surveillance100/Cirr11.htm). [Google Scholar]
- Zhang DY, Friedman SL. Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology. 2012;56:769–775. doi: 10.1002/hep.25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Zolotukhin S, Byrne BJ, Mason E, Zolotukhin I, Potter M, Chesnut K, Summerford C, Samulski RJ, Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.