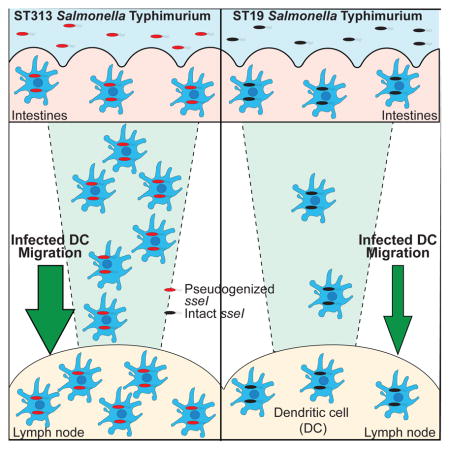
Abstract
Genome degradation correlates with host adaptation and systemic disease in Salmonella. Most lineages of the S. enterica subspecies Typhimurium cause gastroenteritis in humans, however, the recently emerged ST313 lineage II pathovar commonly causes systemic bacteremia in sub-Saharan Africa. ST313 lineage II displays genome degradation compared to gastroenteritis-associated lineages, yet the mechanisms and causal genetic differences mediating these infection phenotypes are largely unknown. We find that the ST313 isolate D23580 hyperdisseminates from the gut to systemic sites such as the mesenteric lymph nodes (MLN) via CD11b+ migratory dendritic cells (DCs). This hyperdissemination was facilitated by the loss of sseI, which encodes an effector that inhibits DC migration in gastroenteritis-associated isolates. Expressing functional sseI in D23580 reduced the number of infected migratory DCs and bacteria in the MLN. Our study reveals a mechanism linking pseudogenization of effectors with the evolution of niche adaptation in a bacterial pathogen.
Keywords: Nontyphoidal Salmonella, pathogenesis, evolution, niche adaptation, iNTS, NTS, pseudogene, SrfH, T3SS effectors, genome degradation
Graphical Abstract
Introduction
Salmonella enterica serovars are important human pathogens, with more than 2300 serovars that are able to cause disease (Porwollik et al., 2004). Salmonella enterica serovars are categorized into typhoidal or nontyphoidal Salmonella (NTS) by the human disease they cause (Feasey et al., 2012). The typhoidal serovars, Salmonella Typhi (S. Typhi) and Salmonella Paratyphi (S. Paratyphi), are human-restricted and cause the systemic disease typhoid fever (Feasey et al., 2012). In contrast, NTS encompass the vast majority of Salmonella enterica serovars and typically cause self-limiting gastroenteritis in humans (Feasey et al., 2012). Invasive NTS infections usually only occur in immunocompromised patients (Feasey et al., 2012). Salmonella serovars with a broad host range, such as Salmonella Typhimurium (S. Typhimurium), rarely cause systemic infections.
Interestingly, bloodstream Salmonella infections in sub-Saharan Africa are mainly nontyphoidal (Feasey et al., 2012; Kingsley et al., 2009). Indeed, NTS infections are among the most common causes of bloodstream infections for children and HIV-infected adults in sub-Saharan Africa (Feasey et al., 2012). The NTS isolates causing these infections in sub-Saharan Africa are collectively referred to as African NTS. For children, risk factors for invasive African NTS infections include HIV or malaria coinfection and malnutrion (Feasey et al., 2015; Gordon, 2011). The relatively immunocompromised sub-Saharan population likely contributes to the high levels of invasive NTS infections. The African NTS are multidrug resistant, which limits therapeutic options and increases the mortality rate (Kariuki et al., 2015). While fever is present in almost all African NTS cases, gastrointestinal (GI) symptoms are found in less than half (Feasey et al., 2012; Porwollik et al., 2004). The lack of GI symptoms in many patients suggests that the disease caused by these African NTS is distinct from that caused by NTS in other parts of the world (Feasey et al., 2012; Gordon, 2011).
S. Typhimurium is responsible for approximately 75% of the invasive NTS infections in Africa (Feasey et al., 2012; Gordon et al., 2008). Phylogenetic analysis of these African S. Typhimurium isolates found that they are primarily sequence type 313 (ST313, Feasey et al., 2012; Kingsley et al., 2009). In contrast, most laboratory strains and gastroenteritis outbreak isolates are sequence type 19 (ST19, Feasey et al., 2012; Kingsley et al., 2009). The ST313 isolates cluster away from other S. Typhimurium to form two closely related lineages (Feasey et al., 2012; Kingsley et al., 2009). Chloramphenicol resistance in ST313 lineage II isolates drove their almost complete replacement of ST313 lineage I isolates in sub-Saharan Africa by 2006 (Feasey et al., 2012; Okoro et al., 2012).
Whole genome analysis of both ST313 S. Typhimurium lineages revealed that major changes in the ST313 lineage include distinct prophage and plasmid repertoires and the insertion of a Tn21-like mobile element containing multiple antibiotic cassettes into the virulence plasmid (Kingsley et al., 2009). Additionally, there was evidence of genome degradation, including pseudogene formation and chromosomal deletions, when compared with ST19 S. Typhimurium genome sequences (Kariuki et al., 2015; Kingsley et al., 2009; Okoro et al., 2015). Increased genome degradation is seen in host-adapted, systemic disease-causing serovars such as S. Typhi and Paratyphi compared to gastrointestinal generalist serovars such as S. Typhimurium (Nuccio and Bäumler, 2014). In addition, S. Typhimurium DT2, which has become host-adapted to pigeons, also has greater genome degradation compared to ST19 S. Typhimurium (Kingsley et al., 2013). Many of the pseudogenes in ST313 S. Typhimurium are also pseudogenes in the human-restricted serovars S. Typhi and Paratyphi (Kingsley et al., 2009; Okoro et al., 2015). These findings suggest that the ST313 S. Typhimurium could be evolving towards causing systemic disease (Kingsley et al., 2009). We and most other groups found that ST313 S. Typhimurium occupy a phenotypically intermediate state between ST19 S. Typhimurium and systemic disease-causing S. Typhi for both host cell invasion and inflammasome activation, suggesting that ST313 S. Typhimurium are becoming phenotypically closer to systemic disease-causing serovars (Carden et al., 2015; Okoro et al., 2015). However, little is known about potential differences in the ability to cause systemic infections between ST313 and ST19 S. Typhimurium. In this work we dissect how genomic differences in the ST313 S. Typhimurium lineage shape host-pathogen interactions and the ability to cause systemic disease in a mouse model.
Results
ST313 isolate D23580 has higher bacterial burden in systemic organs
To determine if ST313 and ST19 isolates differed in their ability to cause systemic disease, we infected mice with ST313 isolate D23580 (ST313 D23580), a lineage II isolate cultured from the blood of a Malawian patient (Kingsley et al., 2009), or ST19 isolate SL1344 (ST19 SL1344), a streptomycin-resistant derivative of an isolate cultured from a septicemic cow (Hoiseth and Stocker, 1981). We focused on ST313 lineage II isolates, as by 2006, lineage II isolates had almost completely replaced ST313 lineage I isolates in sub-Saharan Africa (Okoro et al., 2012).
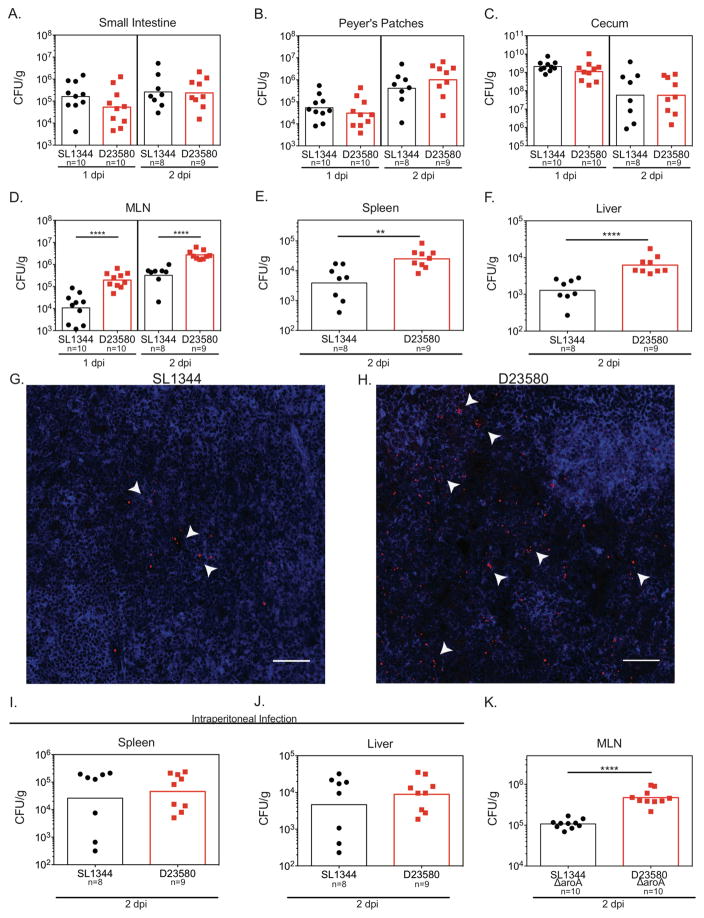
Streptomycin-pretreated C57BL/6 mice were orally infected with either ST313 isolate D23580 or ST19 isolate SL1344. The levels of ST313 D23580 and ST19 SL1344 were similar in the small intestine, Peyer’s patches and cecum at 1 and 2 days post infection (dpi, Figure 1A–C, Figure S1). At 2 dpi, there were no differences in the histopathology in the ceca between mice infected with either isolate (Figure S1). In contrast, the levels of ST313 D23580 were significantly higher than the levels of ST19 SL1344 in the MLN at 1 and 2 dpi (Figure 1D, G–H, Figure S1), and in the spleen and liver at 2 dpi (Figure 1E–F, Table S1). Similar results were seen in mice infected with a ST19 isolate that is commonly associated with gastroenteritis outbreaks, DT104 (ATCC 700408, Figure S1D–I). Collectively, our results indicate that the levels of ST313 and ST19 isolates are similar in the gut. In contrast, the levels of the ST313 isolate were significantly higher than the levels of the ST19 isolates in systemic tissues. Intriguingly, our data correlate with the propensity of ST313 isolates to cause disseminated disease in humans and may reflect differences in the relative abilities of ST313 and ST19 isolates to cause systemic infections.
Figure 1. ST313 S. Typhimurium isolate D23580 hyperdisseminates to systemic sites.
(A–I) Streptomycin-pretreated C57BL/6 mice were orally infected with each Salmonella isolate. Mice were sacrificed at 1 (A–D) or 2 (A–I) days post infection (dpi) as indicated, and Salmonella burden was enumerated by plating. The colony-forming units per gram (CFU/g) for the gut tissues (A–C) and systemic sites (D–F) are presented. CFU/organ for the MLN and Peyer’s patches are presented in Figure S1. (G–H) Representative images of immunostaining of S. Typhimurium (red) and phalloidin (blue) in MLN infected for 2 days with either ST19 SL1344 (G) or ST313 D23580 (H). Arrows indicate select clusters of Salmonella and the scale bars represent 50 μM. Experiments were repeated twice with 2–3 mice per group. (I–J) Streptomycin-pretreated C57BL/6 mice were infected intraperitoneally and sacrificed 2 dpi. Salmonella burden was enumerated by plating in the spleen (I) and liver (J). (K) Streptomycin-pretreated C57BL/6 mice were orally infected with SL1344 ΔaroA or D23580 ΔaroA. Salmonella CFU in the MLN was enumerated 2 dpi. (A–F, I–K) Data is the combination of two independent experiments with 4–5 mice per group. Statistical significance was determined using Mann-Whitney test, bars represent the geometric mean, and dots individual mice.
*P≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.0001. See also Figure S1 and Table S1.
ST313 D23580 hyperdisseminates to systemic sites
Our results indicate that ST313 D23580 is either more efficient at disseminating from the gut to systemic sites or has enhanced survival and/or replication in systemic niches compared to ST19 isolates. To distinguish between these possibilities, we bypassed the gut by injecting mice intraperitoneally (IP) with either ST313 or ST19 isolates. After IP injection, the amount of ST313 and ST19 S. Typhimurium isolates in the spleen and liver were not significantly different (Figure 1I–J). These results are consistent with ST313 D23580 hyperdisseminating from the gut to systemic sites after oral infection.
The higher levels of ST313 D23580 in the MLN after oral infection could be due to either more efficient dissemination or an increase in intracellular replication during and/or after dissemination to the MLN. To test this, we measured the ability of the NTS isolates to disseminate to the MLN in the absence of intracellular replication. To this end, we used aroA mutants of ST313 D23580 and ST19 SL1344 that are attenuated for replication in host cells (Hoiseth and Stocker, 1981). If the higher levels of ST313 bacteria in the MLN were due primarily to increased replication at this site, the levels of ST19 and ST313 ΔaroA mutant strains in the MLN should be similar. However, we found significantly higher levels of ST313 D23580 ΔaroA in the MLN compared to ST19 SL1344 ΔaroA at 2 dpi (Figure 1K). As with the wild type strains, the levels of aroA mutants of ST313 D23580 and ST19 SL1344 were similar in the gut (Figure S1J–L). This suggests that differences in intracellular replication do not drive the higher bacterial load in the MLN. Alternatively, differences in intracellular survival between ST19 SL1344 and ST313 D23580 could play a role in the higher bacterial load in the MLN. Identifying the cell types infected in the MLN will allow us to test whether there are differences in the ability of ST313 D23580 and ST19 SL1344 to survive within those cell types. Since our intraperitoneal infection data show that there are not survival differences in the spleen and liver, it is likely that this is also true in the MLN. Taken together, our results suggest that ST313 isolate D23580 likely hyperdisseminates from the gut to systemic sites, resulting in higher bacterial loads at those sites.
Higher levels of Migratory DCs are infected by ST313 D23580 in the MLN
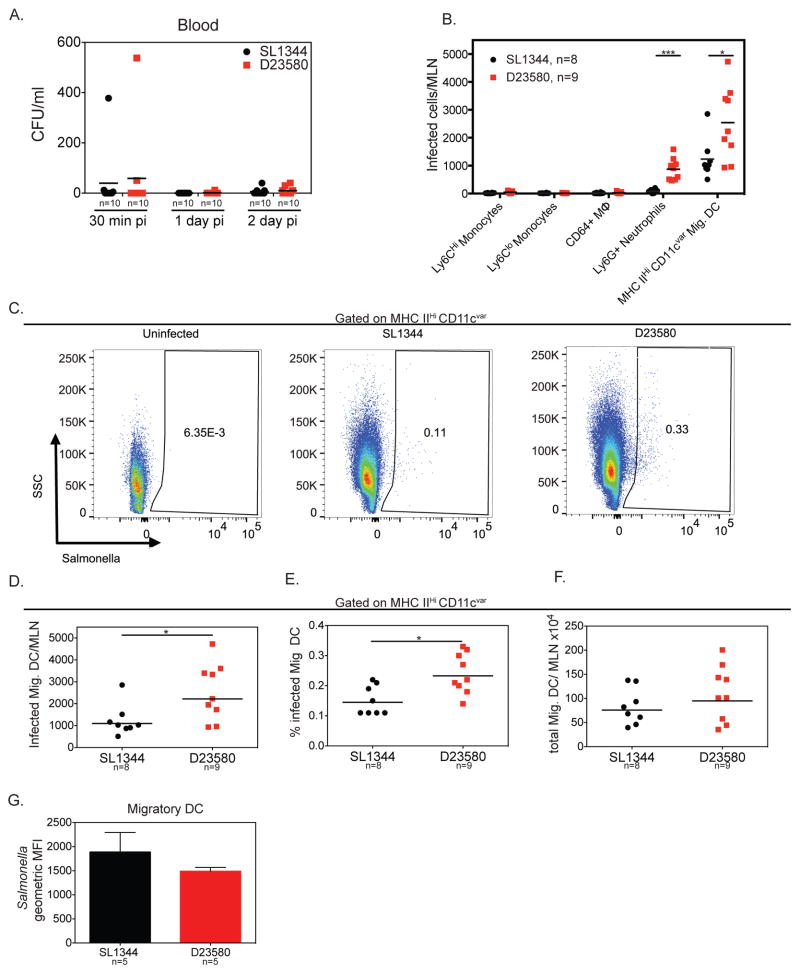
After crossing the gut epithelial barrier, S. Typhimurium ST19 can disseminate to systemic sites via the lymphatics or the bloodstream (Watson and Holden, 2010). Salmonella has been reported to disseminate within CD18+ phagocytes in the bloodstream and within dendritic cells (DCs) in the lymph (Bogunovic et al., 2009; Vazquez-Torres et al., 1999). Dissemination via either route could be important for the hyperdissemination of ST313 D23580. To test whether ST313 D23580 hyperdisseminated via the bloodstream, we measured the levels of Salmonella in the bloodstream of mice orally infected with either ST313 or ST19 isolates. There were no differences in the amount of bacteria in the blood 30 minutes, 1 or 2 dpi between ST19 and ST313 isolates (Figure 2A). Although both isolates can disseminate to distant tissues via the bloodstream, the lack of difference between ST19 and ST313 isolates in the blood suggests that the hyperdissemination of ST313 isolates is not through this route.
Figure 2. Higher levels of Migratory DC are infected by ST313 S. Typhimurium isolate in the MLN.
(A) Streptomycin-pretreated C57BL/6 mice were orally infected and mice were sacrificed at 30 minutes, 1 and 2 dpi. CFU/ml in the blood was enumerated by plating. (B–F) Mice were infected as in A and sacrificed 2 dpi. Single cell suspensions of MLN were stained for cell surface markers and intracellular Salmonella and analyzed by flow cytometry. Salmonella was detected using the anti-Salmonella antibody CSA-1-FITC. (B) The total number of Salmonella-infected cells per MLN for innate immune cell types are presented. (C) Representative pseudocolor dot plots showing the percentage of Salmonella-infected migratory DC are given for uninfected, ST19 SL1344-infected and ST313 D23580-infected mice. (D) Total number of Salmonella-infected migDCs per MLN are shown for ST19 SL1344- and ST313 D23580-infected mice. (E) Percentage of Salmonella-infected migDC was calculated as the percent of total migratory DC that were infected. (F) Total number of migDC per MLN from SL1344- and D23580-infected mice are presented. (A–F) Data presented are the combination of 2 independent experiments with 4–5 mice per group. The geometric mean for each group is shown. Statistical significance was determined by the Mann-Whitney test. (G) Geometric mean fluorescence intensity (MFI) for infected migDCs from a representative experiment with 5 mice per group is presented. Experiment was repeated at least two times. The mean and standard deviation are shown. Statistical significance was determined by unpaired t-test with Welch’s correction. * P≤0.05, ** P≤0.01, *** P≤0.001. See also Figures S1–4.
To determine whether there are differences in the ability of ST313 and ST19 isolates to disseminate within innate immune cells via the lymphatics from the gut to the MLN, we identified Salmonella-infected cells in the MLN by flow cytometry. At 2 dpi, we observed Salmonella inside several different myeloid-derived cell types, such as inflammatory monocytes (Ly6CHiCD11b+Ly6G−), patrolling monocytes (Ly6CloCD11b+Ly6G−), macrophages (CD64+MerTK+), neutrophils (Ly6G+CD11bHiLy6Cint) and migratory DCs (MHC IIHiCD11cvariable, Figure 2B). Although Salmonella was detected in a number of myeloid-derived cells, migratory DCs (migDCs) were the cell type with the highest number of infected cells for both ST313 and ST19 isolates (Figure 2B, Figure S2).
We next compared the levels of infection between ST313 and ST19 isolates in these cell types. ST313 D23580 infected a higher number of neutrophils and migDCs (Figure 2B, Figure S3). The higher amount of infected neutrophils is likely due to the larger bacterial load in ST313 isolate infected-mice, as neutrophil influx is correlated with bacterial load (Cheminay et al., 2003). This notion is supported by an increased number of recruited neutrophils in the MLN in ST313 D23580 infected mice (Figure S3).
Unlike neutrophils, migDCs are likely candidates to carry Salmonella from the gut. MigDCs traffic from the gut and enter the MLN through the afferent lymphatics and can be distinguished from tissue-resident DCs by their high expression of MHC II and variable expression of the integrin CD11c, which is downregulated during migration and activation (Merad et al., 2013). ST313 D23580 infected a higher total number of migDCs per MLN and a larger percentage of migDCs compared to the ST19 SL1344 (Figure 2B–E). In contrast, there was no difference in the total number of migDCs recruited to the MLN by ST313 or ST19 isolates (Figure 2F).
Since we identified migDCs as the major reservoir of intracellular Salmonella at this time point, we next tested whether the higher levels of D23580 in the MLN could be attributed to enhanced intracellular replication and/or survival in migDCs. There was no difference in the geometric mean fluorescence intensity (MFI) of anti-Salmonella staining between migDCs infected with ST313 D23580 and ST19 SL1344 (Figure 2G), indicating that migDCs infected with either isolate have similar intracellular levels of bacteria. This finding supports our observation that differences in intracellular replication do not drive higher bacterial loads in the MLN (Figure 1K). Next we tested if ST313 D23580 and ST19 SL1344 differed in their ability to survive within DCs. To test this, we measured the levels of gentamicin-protected bacteria over time for each isolate in bone-marrow-derived DCs. We found no difference in intracellular survival of ST313 D23580 and ST19 SL1344 (Figure S4A). Taken together, this suggests that hyperdissemination from the gut via infected migDCs is responsible for the higher bacterial load in the MLN.
Similar levels of infected DC are present in gut tissues from mice infected with ST313 or ST19 S. Typhimurium isolates
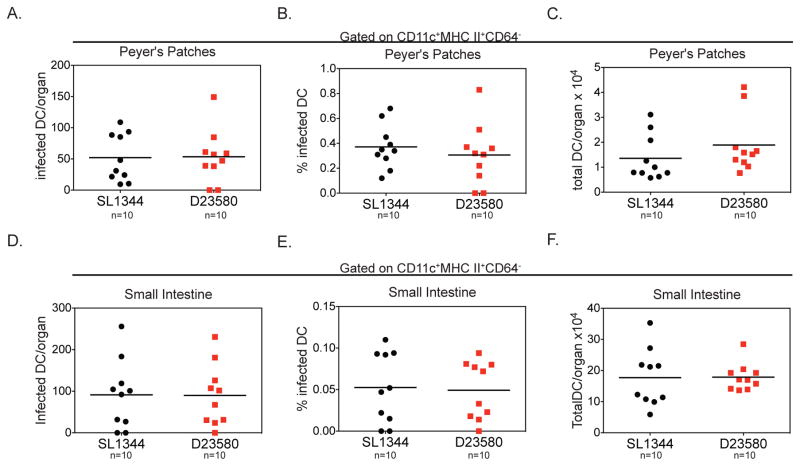
A possible explanation for the higher amount of ST313 D23580-infected migDCs in the MLN is that this isolate infects more DCs in the gut, which then traffic to the MLN. Although we did not observe a difference in the colonization of gut tissues, the vast majority of bacteria are extracellular in gut tissues, which could obscure potential differences in the small fraction of Salmonella that are within DCs (Barthel et al., 2003; Monack et al., 2000). To measure the amount of infected DCs in the gut, mice were infected orally with either ST313 or ST19 isolates and the small intestine and Peyer’s patches were analyzed using flow cytometry to detect S. Typhimurium within DCs (CD45+CD11c+MHC II+CD64−). The total number and percentage of infected DCs in the Peyer’s patches and small intestine were similar for both isolates (Figure 3). Additionally, we found that there was no difference in percent invasion of ST19 SL1344 and ST313 D23580 into bone marrow-derived DCs in vitro (Figure S4B). Since we saw no difference in the infection rate of DC both in vivo and in vitro, our data indicate that the higher amount of infected migDCs in the MLN is not driven by differences in the amount of infected DCs in gut tissues.
Figure 3. Similar levels of infected DC are present in gut tissues from mice infected with ST19 or ST313 S. Typhimurium isolates.
(A–I) Streptomycin-pretreated C57BL/6 mice were orally infected and mice were sacrificed 2 dpi. Single cell suspensions of the Peyer’s patches and small intestine were stained for cell surface markers and intracellular Salmonella and analyzed by flow cytometry. The total number of infected DCs per organ (A, D), percent of DCs infected (B, E,) and total number of DCs per organ (C, F) are presented for Peyer’s patches (A–C) and small intestine (D–F). Data are combined from 2 independent experiments with 5 mice per group. (A–F) The mean for each group is shown and statistical significance was determined by the Mann-Whitney test. * P≤0.05. See also Figure S1 and S4.
Differences in inflammasome activation do not control hyperdissemination to the MLN by ST313 D23580
Another possible explanation for the higher amounts of D23580-infected migDCs is that ST313 D23580 induces less cell death in migDCs, allowing more infected migDCs to reach the MLN. We previously showed that in bone marrow-derived macrophages, ST313 D23580 causes less inflammasome-mediated cell death compared to ST19 SL1344 (Carden et al., 2015). Since Salmonella is also able to cause inflammasome-mediated cell death in DCs (van der Velden et al., 2003), we reasoned that lower levels of inflammasome-mediated DC death triggered by ST313 D23580 could contribute to hyperdissemination to the MLN. To test this hypothesis, we orally infected mice that are deficient for caspase-1 and caspase-11 (Casp1/11−/−), which are required for all forms of inflammasome activation (Broz et al., 2012), with ST313 and ST19 S. Typhimurium. As reported previously Casp1/11−/− mice had a greater Salmonella burden compared to wild type (WT) mice (Broz et al., 2012). There were no differences in bacterial burden between ST19 and ST313 isolates in the gut (Figure S4). However, the levels of ST313 D23580 bacteria were significantly higher than the ST19 isolate SL1344 bacterial levels in the MLN of Casp1/11−/− mice (Figure S4). Consistent with our results in WT mice, ST313 D23580 also infected a higher total number and percentage of migDCs in Casp1/11−/− mice (Figure S4). Taken together, these results indicate that differences in inflammasome activation do not explain the hyperdissemination of ST313 D23580 bacteria to the MLN. By eliminating differences in bacterial uptake in the gut and inflammasome-mediated death of infected DCs as drivers of hyperdissemination, our results suggest that ST313 D23580 is better at co-opting migDC migration to spread from the gut to the MLN.
ST313 D23580 hyperdissemination phenotype correlates with higher levels of infected CD11b+ migratory DC
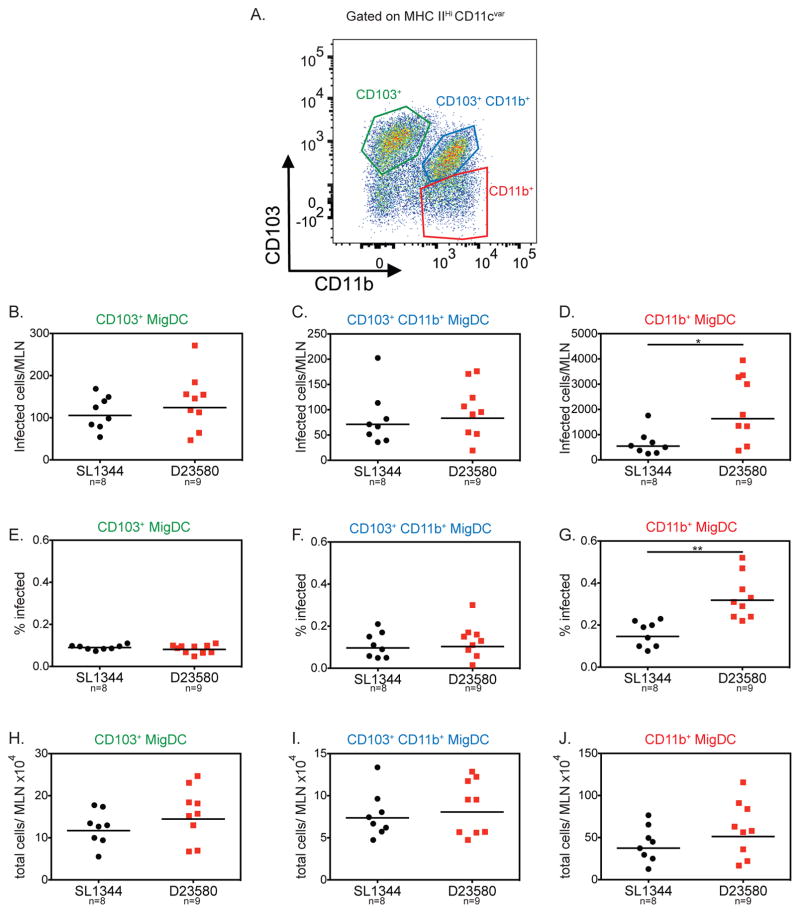
MigDCs in the MLN are categorized into three main subsets defined by their expression of the integrins CD103 and CD11b: CD103+CD11b− (CD103+), CD103+CD11b+, and CD103−CD11b+ (CD11b+, Figure 4A). These subsets have distinct biological functions. The CD103+ and CD103+CD11b+ migDC subsets are important for oral tolerance, while the CD11b+ subset is associated with inducing IFN-γ-producing CD4+ T cells (Coombes et al., 2007) We examined the relative contribution of each of these migDC subsets to the increased dissemination of ST313 bacteria from the gut to the MLN.
Figure 4. ST313 S. Typhimurium infects higher numbers of CD11b+ migratory DCs.
Streptomycin-pretreated C57BL/6 mice were orally infected and mice were sacrificed 2 dpi. Single cell suspensions of the MLN were stained for cell surface markers and intracellular Salmonella. A representative flow cytometry plot of the migratory dendritic cell subsets of the MLN is presented in (A). The total number of infected cells per MLN (B–D), percent of each subset infected (E–G) and total number of cells recruited to the MLN (H–J) are shown for the three migDC subsets. (B–J) Data presented are the combination of 2 independent experiments with 4–5 mice per group. The geometric mean for each group is shown and statistical significance was determined by the Mann-Whitney test. * P≤0.05, ** P≤0.01.
As reported previously, S. Typhimurium infected all three migDC subsets (Bogunovic et al., 2009). At 2 dpi, the CD11b+ migDC subset had the highest number of infected cells compared to the other two subsets (Figure 4). There were no significant differences in the levels of infected CD103+ and CD103+CD11b+ in the MLN of mice infected with either S. Typhimurium isolate (Figure 4). Strikingly, we found significantly higher numbers of CD11b+ migDCs in the MLN infected with ST313 D23580 compared to ST19 SL1344 (Figure 4D). This was also reflected in a higher percentage of CD11b+ migDCs infected with ST313 bacteria compared to ST19 (Figure 4G). Importantly, there were no differences in the total number of CD11b+ migDCs recruited to the MLN between mice infected with either isolate (Figure 4J). The higher amount of ST313 isolate infected CD11b+ migDC suggests that ST313 isolates could use this migDC subset in particular to hyperdisseminate from the gut to the MLN.
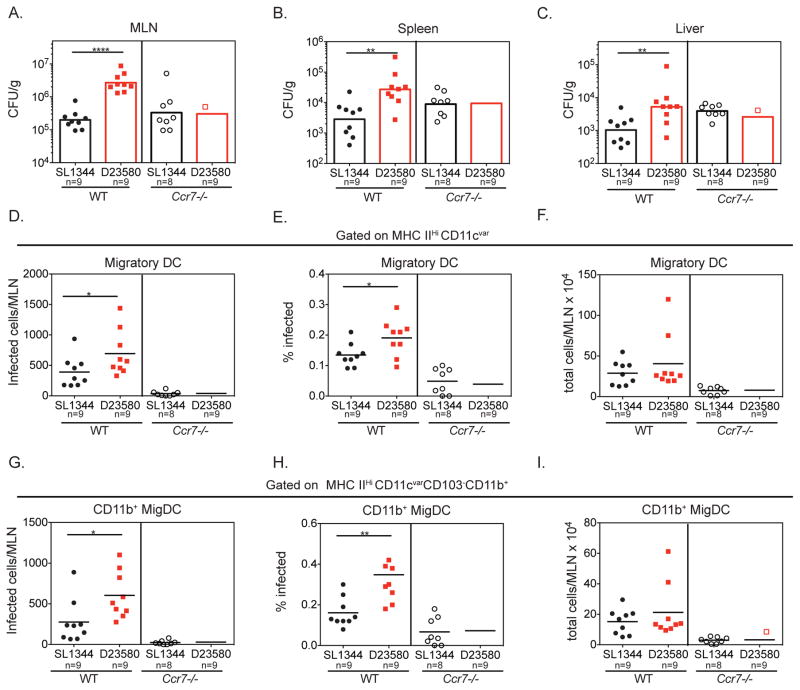
Hyperdissemination of ST313 isolate D23580 from the gut requires CCR7
Ccr7−/− mice are defective for DC and T cell migration to the draining lymph nodes (Förster et al., 1999). To determine whether DC migration mediates the hyperdissemination of ST313 isolate D23580, we orally infected Ccr7−/− mice with ST313 or ST19 S. Typhimurium and measured bacterial loads 2 dpi. In contrast to WT mice, the levels of ST313 D23580 or ST19 SL1344 in the MLN, spleen and liver were the same (Figure 5A–C, Figure S5). Although both ST19 and ST313 isolates disseminated to systemic sites in Ccr7−/− mice, the lack of difference between ST19 and ST313 isolates in this background suggests that CCR7-dependent DC migration is necessary for the hyperdissemination of ST313 D23580.
Figure 5. CCR7-dependent cell migration is necessary for hyperdissemination by ST313 S. Typhimurium.
(A–I) Streptomycin-pretreated WT C57BL/6 or Ccr7−/− mice were orally infected with each Salmonella isolate. Mice were sacrificed at 2 dpi and Salmonella burden was enumerated by plating. The CFU/g for systemic sites (A–C) are presented. CFU/organ for the MLN is presented in Figure S5. (D–I) Single cell suspensions of the MLN were stained for cell surface markers and intracellular Salmonella. The total number of infected cells per MLN (D, G), percent infected (E, H) and total number of cells recruited to the MLN (F, I) are shown for all migDC (D–F) and the CD11b+ migDC subset (G–I), respectively. (A–I) Data are combined from 2 independent experiments with 4–5 mice per group. The geometric mean (A–C) or mean (D–I) for each group is shown and statistical significance was determined by the Mann-Whitney test. * P≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.0001. See also Figure S5.
To further elucidate the role of CCR7-dependent DC migration in hyperdissemination, we analyzed the amount of ST313 D32580- and ST19 SL134 -infected migDCs in the MLN of Ccr7−/− mice. As expected, fewer migDCs were recruited to the MLN of infected Ccr7−/− mice compared to WT mice (Figure 5F). In addition, at 2 dpi, very low numbers of infected migratory DCs were present in the MLN of Ccr7−/− mice and there were no differences between ST19 and ST313 isolates (Figure 5D). This was also reflected in the percentage of infected migDCs in the MLN (Figure 5E). We also analyzed the levels of S. Typhimurium-infected CD11b+ migDCs in the MLN of infected WT and Ccr7−/− mice. In contrast to infections of WT mice, there was no difference in the levels of infected CD11b+ migDC in the MLN of Ccr7−/− mice infected with either S. Typhimurium isolate (Figure 5G–H). Thus, CCR7-dependent migration of migDCs, in particular CD11b+ migDCs, is necessary for hyperdissemination of the ST313 D23580 from the gut to the MLN.
Pseudogenization of the secreted effector SseI is required for ST313 S. Typhimurium to hyperdisseminate via infected migratory DC
We reasoned that lineage-specific manipulation of CCR7-dependent DC migration could account for differences in dissemination of ST313 D23580 and ST19 SL1344 from the gut to the MLN. In our previous studies, we showed that ST19 S. Typhimurium inhibits DC chemotaxis of infected cells by a mechanism that requires the Salmonella Pathogenicity Island-2 (SPI-2) type 3 secretion system (McLaughlin et al., 2014). We identified seven SPI-2 effectors (SseI, SifA, SseF, PipB2, SpiC, SspH2, and SlrP) that inhibit DC migration toward CCL19, a chemokine recognized by CCR7 (McLaughlin et al., 2014; 2009). The genomes of ST313 lineage II isolates contain an insertion of an IS200 element in sseI, rendering this gene a pseudogene (Kingsley et al., 2009; Okoro et al., 2015). Since SseI inhibits the migration of infected DCs, we hypothesized that the pseudogenization of sseI in ST313 D23580 permits DCs infected by this isolate to migrate to the MLN in higher numbers than ST19 SL1344-infected DCs and thus contributes to hyperdissemination from the gut.
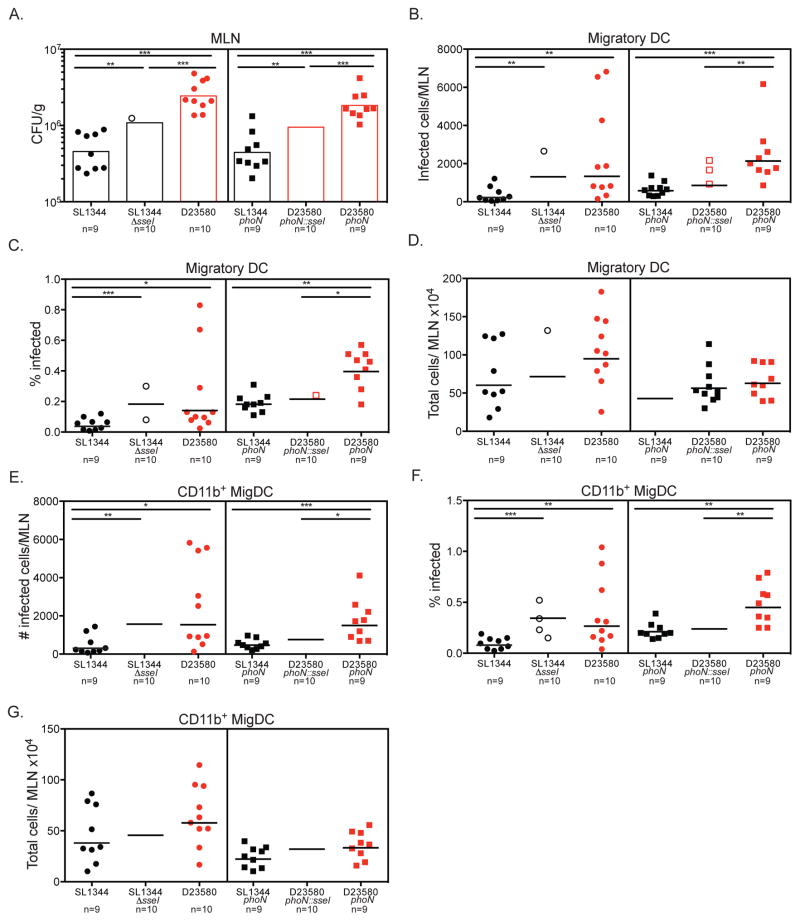
To test the role of sseI in modulating dissemination of S. Typhimurium from the gut to the MLN, we compared the levels of Salmonella in MLN of mice orally infected with either WT ST19 SL1344 or an isogenic ΔsseI mutant 2 days p.i. There were significantly higher levels of the SseI-deficient bacteria compared to WT ST19 SL1344 (Figure 6A, MLN CFU/organ presented in Figure S6). Although the levels of ST313 D23580 bacteria were still higher than the ST19 SL1344 ΔsseI levels in the MLN (Figure 6A), these results indicate that the presence of an intact copy of sseI reduces migration of Salmonella from the gut to MLN. To test this notion further, we complemented sseI in ST313 D23580, designated D23580 phoN::sseI. To control for any potential effects of insertion into the phoN locus, we compared our complemented D23580 phoN::sseI strain to phoN mutants of ST19 SL1344 and ST313 D23580. The level of D23580 phoN::sseI bacteria was significantly lower than the level of D23580 phoN bacteria in the MLN at 2 dpi. The levels of D23580 phoN::sseI bacteria in the MLN were slightly higher than SL1344 phoN, which also contains a functional sseI (Figure 6A, Figure S6). Our results indicate that pseudogenization of sseI contributes to the hyperdissemination of ST313 D23580 to the MLN.
Figure 6. Pseudogenization of SseI in ST313 S. Typhimurium contributes to hyperdissemination in CD11b+ DCs.
(A–G) Streptomycin-pretreated C57BL/6 mice were orally infected with each respective Salmonella strain. To genetically dissect the effect of pseudogenization of sseI, a clean deletion of sseI was made in ST19 background and D23580 was complemented with a functional copy of SseI. Mice were sacrificed 2 dpi, and Salmonella burden was enumerated by plating. CFU/g of each strain in the MLN is shown in A. The CFU/organ in the MLN is presented in Figure S6. (B–G) Single cell suspensions of the MLN were stained for cell surface markers and intracellular Salmonella. The total amount of infected cells per MLN (B, E), percent infected (C, F) and total number of cells recruited to the MLN (D, G) are shown for all migDC (B–D) and the CD11b+ migDC subset (E–G), respectively. (A–G) Data presented are the combination of 2 independent experiments with 4–5 mice per group. The geometric mean for each group is shown and statistical significance was determined by the Mann-Whitney test.
*P≤0.05, ** P≤0.01, *** P≤0.001. See also Figure S6.
To determine whether pseudogenization of sseI is increased the Salmonella burden in the MLN by influencing DC migration, we examined the levels of migDCs infected with S. Typhimurium strains that differed by the presence or absence of SseI at 2 days after oral infection of mice. The number of infected migDCs and CD11b+ migDCs in the MLN of mice infected with ST19 SL1344 ΔsseI was similar to ST313 D23580, which contains pseudogenized sseI (Figure 6B, E). Importantly, the levels of infected migDCs and CD11b+ migDCs in the MLN of mice infected with the S. Typhimurium strains that lack a functional sseI (SL1344 ΔsseI, D23580 and D23580 phoN) were significantly higher than the levels present in mice infected with S. Typhimurium strains that contain a functional sseI (SL1344, SL1344 phoN and D23580 phoN::sseI, Figure 6B, E). Similar results were seen when the percentage of migDCs and CD11b+ migDCs were measured (Figure 6C, F). Taken together, our data indicate that the pseudogenization of the single effector gene, sseI, in the S. Typhimurium ST313 lineage II isolate D23580 facilitates higher levels of infected DC migration from the gut to the MLN, leading to increased bacterial burdens in this tissue.
Discussion
While much is known about the pathogenesis of well-characterized gastroenteritis-associated ST19 S. Typhimurium, few studies examine the pathogenesis of invasive ST313 S. Typhimurium. ST313 S. Typhimurium have emerged to cause invasive disease in sub-Saharan Africa, where the population is relatively immunocompromised. Both genomic changes in ST313 lineage and comorbidities found in the sub-Saharan population are likely important for the systemic disease caused by ST313 isolates. Previous work largely focused on how common comorbidities in the sub-Saharan population, such as HIV or malaria coinfection, contribute to the increased severity of S. Typhimurium infections (Lokken et al., 2014; Mooney et al., 2014; 2015; Raffatellu et al., 2008; Roux et al., 2010). These studies nicely laid the foundation for how differences in the host population exacerbate Salmonella infections. On the pathogen side, little is known about how differences in the ST313 lineage might contribute towards causing systemic disease in humans.
We found that mice infected with the S. Typhimurium ST313 isolate D23580 had higher pathogen levels in the systemic sites, but not in gastrointestinal tissues when compared to S. Typhimurium ST19 isolate SL1344 (Figure 1). This trend of ST313 S. Typhimurium having higher Salmonella burden in systemic tissues compared to ST19 S. Typhimurium has been seen by other groups when infecting chickens and mice (Table S1, Okoro et al., 2015; Parsons et al., 2013; Singletary et al., 2016; Yang et al., 2015). However, the mechanisms mediating higher levels of ST313 S. Typhimurium at systemic sites were unclear. Importantly, we established that increased bacterial colonization at systemic sites is predominantly driven by hyperdissemination from the gut. Our finding that S. Typhimurium ST313 D23580 is better at disseminating from the gut to systemic tissues in the mouse model parallels how ST313 isolates are associated with causing systemic disease in humans.
In this study we identified components of both the host and pathogen that are important for the higher load of ST313 S. Typhimurium to the MLN. Notably, we found that ST313 S. Typhimurium requires CCR7-dependent migration of DCs to hyperdisseminate to the MLN (Figures 2, 5). This suggests that ST313 D23580 bacteria are better at hijacking migDCs to serve as Trojan horses and help them disseminate to the MLN (Figure 2). Intriguingly, both ST313 and ST19 isolates infected all three migDC subsets (CD103+, CD103+CD11b+, CD11b+), but only the CD11b+ subset mediated the enhanced spread from the gut to the MLN by ST313 D23580 (Figure 4). CD11b+ migDCs are proinflammatory, and are the least migratory migDC subset in steady state (Cerovic et al., 2013; Coombes et al., 2007). The low amount of migration during homeostasis likely explains the varied results of studies examining the effect of CCR7 on CD11b+ migDC migration (Bogunovic et al., 2009; Cerovic et al., 2013; Diehl et al., 2013; Scott et al., 2014). Recent work that detected CD11b+ migDCs in the afferent lymph and increased migration of CD11b+ migDC subset after inflammation have helped clarify the migratory capacity of this subset (Cerovic et al., 2013; Diehl et al., 2013). In our study, we see significantly lower levels of CD11b+ migDC in the MLN of infected Ccr7−/− mice compared to infected WT mice, emphasizing that this subset is able to migrate better during an infection. It seems paradoxical that the proinflammatory CD11b+ migDC subset would interact with ST313 and ST19 isolates differently. However, this difference could be mediated by different innate immune responses toward ST313 compared to ST19 isolates. For example, the CD11b+ migDC subset is the only migratory DC subset that expresses significant amounts of TLR5, which recognizes flagellin (Atif et al., 2013). ST313 isolates express less flagellin (Carden et al., 2015), and modulating the amount of flagellin recognition by TLR5 in CD11b+ migDC may allow ST313 isolates to disseminate better within these cells. Supporting this notion, others have shown that introducing the FliC-repressor TviA into ST19 S. Typhimurium led to increased dissemination of Salmonella to the spleen (Atif et al., 2014). Elucidating why ST313 S. Typhimurium preferentially uses the CD11b+ migDC subset to hyperdisseminate will be important for future studies.
Differences in the manipulation of DC migration by ST313 and ST19 isolates likely drive the hyperdissemination to the MLN by ST313 D23580. S. Typhimurium can use several effectors to inhibit DC migration (McLaughlin et al., 2014). The gene encoding one of these effectors, sseI, is a pseudogene in ST313 lineage II S. Typhimurium, like D23580. We tested if the pseudogenization of sseI in ST313 lineage II was important for the hyperdissemination to the MLN via infected DCs by genetically manipulating ST313 and ST19 strains. We demonstrated that the presence of an intact sseI gene correlates with a decrease in the Salmonella burden and amount of infected CD11b+ migDCs in the MLN (Figure 6). Conversely, S. Typhimurium strains that lack an intact sseI disseminated from the gut to the MLN in higher numbers and had higher levels of infected CD11b+ migDC in the MLN (Figure 6). This is consistent with pseudogenization of sseI in ST313 lineage II isolates leading to hyperdissemination to the MLN by allowing DC infected by ST313 isolates to migrate better than DC infected by ST19 isolates. The presence or absence of SseI did not alter the Salmonella burden in the spleen and liver (data not shown), suggesting that other differences in the ST313 lineage besides the pseudogenization of sseI are important for hyperdissemination to the spleen and liver. Since SseI inhibits DC migration, it is possible that the effect of SseI is solely seen in MLN because it is the only tissue that Salmonella directly disseminates to via DCs.
The role of pseudogenization of sseI in hyperdissemination by ST313 S. Typhimurium is particularly intriguing, as gene degradation is enriched in genes encoding secreted effectors in host-adapted serovars of Salmonella enterica (Klemm and Dougan, 2016; Klemm et al., 2016; McClelland et al., 2004; Nuccio and Bäumler, 2014). While the pseudogenization of many genes encoding secreted effectors in host-adapted, systemic disease-causing Salmonella enterica serovars seems counterintuitive, the pseudogenization of effectors is thought to benefit these serovars by either decreasing virulence to promote host survival and pathogen transmission or by getting rid of effectors that are dispensable or deleterious for systemic infections of their adapted host (Klemm and Dougan, 2016). For example, since host-adapted Salmonella enterica serovars cause systemic disease, genes encoding effectors that promote a gastrointestinal lifestyle by inducing gut inflammation are often pseudogenes (Klemm et al., 2016; McClelland et al., 2004; Nuccio and Bäumler, 2014).
Strikingly, we found that the pseudogenization of sseI increased the ability of S. Typhimurium to disseminate from the gut to the draining lymph nodes. Loss of sseI might synergize with infections of immunocompromised hosts, like those often found in sub-Saharan Africa. In immunocompetent hosts, more rapid dissemination likely leads to faster induction of T cell responses and thus enhanced pathogen clearance. As many immunocompromised hosts are not able to mount effective T cell responses, the rapid dissemination induced by sseI pseudogenization is likely not deleterious for Salmonella in these hosts (Nuccio and Bäumler, 2014). This notion is supported by the finding that sseI is a pseudogene in S. Enteritidis isolates that have evolved to cause systemic infection within immunocompromised hosts (Klemm et al., 2016). Loss of sseI might also be important for adaptation to systemic disease in immunocompetent human hosts, as sseI is either absent or a pseudogene in the human-restricted serovars of S. Typhi and Paratyphi. SseI is immunogenic and CD4+ T cell responses to SseI are potent and sustained in a mouse model (Kurtz et al., 2014). Pseudogenization of sseI may allow Salmonella to evade the potent CD4+ T cell response to SseI epitopes. In immunocompetent hosts, evasion of immune recognition of SseI could counterbalance the increased induction of T cell responses due to hyperdissemination when sseI is a pseudogene. Thus, pseudogenization of sseI is likely important for adaptation to cause systemic disease in both immunocompetent and immunocompromised human hosts. Moreover, loss of sseI reveals that pseudogenization of effectors can contribute to Salmonella niche adaptation and expansion by increasing the ability of Salmonella to cause systemic disease. Studying ST313 S. Typhimurium provides insights into how Salmonella evolves to expand and adapt to systemic niches.
Experimental Procedures
Bacterial strains and plasmids
The bacterial isolates and plasmids used in this study are described in Table S2, and culture conditions are described in the Supplemental Experimental Procedures. Construction of Salmonella mutants and plasmids are described in Supplemental Experimental Procedures. Primers used in this study are listed in Table S3.
Animal Experiments
All animal experiments were approved by the Stanford University Administrative Panel on Laboratory Animal Care and overseen by the Institutional Animal Care and Use Committee under Protocol ID 12826.
C57BL/6, B6.129P2(C)-Ccr7tm1Rfor/J (Ccr7−/−), and B6N.129S2-Casp1tm1Flv/J (Casp1/11−/−) mice were purchased from Jackson labs. Ccr7−/− mice were bred and maintained at Stanford. Male and female mice were used at 7–12 weeks of age.
S. Typhimurium infections were performed as previously described (Barthel et al., 2003). Mice were given a single dose of 20 mg of streptomycin (Sigma) orogastrically, and 20 hours later were inoculated orally or intraperitoneally with Salmonella as described in the Supplemental Experimental Procedures. Mice were euthanized at the indicated time points post infection and organs were collected for colony-forming unit (CFU) enumeration and/or analysis by flow cytometry.
Preparation of single cell suspensions
Full details of preparations of single cell suspensions for all organs are in the Supplemental Experimental Procedures. Briefly, MLN were digested in in Ca2+, Mg2+ Hank’s Balanced Salt Solution (HBSS, Gibco) containing 25 μg/ml Liberase TL (Sigma) and 50 μg/ml DNase I (Life Technologies) for 22–25 minutes at 37° C. Peyer’s patches were digested in RPMI (Gibco) with 5% Fetal bovine serum (FBS, Gibco) 12.5 μg/ml Liberase TL and 50 μg/ml DNase I for 15 minutes at 37° C.
Single cell suspensions of the small intestine lamina propria were made as described previously (Geem et al., 2012). In brief, 0.5 cm pieces of small intestines were incubated in Ca2+, Mg2+ free HBSS (Gibco) with 5% FBS, and 2 mM EDTA (Promega) at 37° C for 20 minutes with shaking at 200 rpm. Then, tissues were minced and incubated in Ca2+, Mg2+ free HBSS with 5% FBS, 1.5 mg/ml Collagenase Type VIII (Sigma), and 250 μg/ml Dnase I at 37° C for 12 minutes with shaking at 200 rpm. The amount of live cells were quantified by counting Trypan Blue− cells using a hemocytometer.
Flow cytometry
Antibodies used for flow cytometry can be found in Table S4. Full details on the staining protocol and gating strategies can be found in the Supplemental Experimental Procedures and Figure S2. Single cell suspensions were blocked with 10 μg/ml TruStain fcX (BioLegend) and then stained for viability and surface antigens. Cells were fixed and permeabilized using BD Cytofix/Cytoperm kit and then stained for intracellular Salmonella. Multiparameter analysis of single cell suspensions was performed using BD Biosciencs Fortessa (Becton Dickinson) and analyzed using FlowJo (TreeStar).
Fluorescent Immunohistochemistry
Full details on the protocols for fluorescent immunohistochemistry and the antibodies used for staining can be found in Supplemental Experimental Procedures and Table S4, respectively. Frozen sections of the MLN were blocked with normal goat serum, and stained overnight in primary antibody at 4° C. Sections were then stained with secondary antibody and phalloidin Alexa Fluor 488 (Life technologies) for 2 hours at room temperature. Images were acquired on a Zeiss LSM 880 confocal microscope with the ZEN 2010 software (Zeiss) and processed using FIJI (Schindelin et al., 2012). Scale bars are set at 50 μm.
Statistical Analysis
Data are presented as either geometric mean or mean as specified. For bar graphs, the mean and standard deviation are shown. Statistical analysis was performed using the Mann-Whitney U test or unpaired Student’s t test with Welch’s correction in GraphPad Prism. P-values ≤0.05 were considered significant. * P≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.0001
Supplementary Material
Acknowledgments
SEC was supported by a Stanford Graduate Fellowship and the NSF Graduate Research Fellowship. GTW was supported by T32AI060555. This study was supported by AI116059 (DM) from NIAID, R01AI098078 (RMT) and Paul Allen Discovery Center Funds (DM).
Footnotes
Author Contributions:
Designed experiments: SEC, JI, RT and DM. Performed experiments: SEC, GW, KL, TP, AJ, DB and JH. Analyzed data: SEC, JI and DM. SEC and DM wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atif SM, Uematsu S, Akira S, McSorley SJ. CD103−CD11b+ dendritic cells regulate the sensitivity of CD4 T-cell responses to bacterial flagellin. Mucosal Immunol. 2013;7:68–77. doi: 10.1038/mi.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atif SM, Winter SE, Winter MG, McSorley SJ, Baumler AJ. Salmonella enterica serovar Typhi impairs CD4 T cell responses by reducing antigen availability. Infect Immun. 2014;82:2247–2254. doi: 10.1128/IAI.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect and Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, Stanley ER, Nussenzweig M, Lira SA, Randolph GJ, Merad M. Origin of the Lamina Propria Dendritic Cell Network. Immun. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carden S, Okoro C, Dougan G, Monack D. Non-typhoidal Salmonella Typhimurium ST313 isolates that cause bacteremia in humans stimulate less inflammasome activation than ST19 isolates associated with gastroenteritis. Pathog Dis. 2015;73(4):ftu023. doi: 10.1093/femspd/ftu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerovic V, Houston SA, Scott CL, Aumeunier A, Yrlid U, Mowat AM, Milling SWF. Intestinal CD103(−) dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol. 2013;6:104–113. doi: 10.1038/mi.2012.53. [DOI] [PubMed] [Google Scholar]
- Cheminay C, Chakravortty D, Hensel M. Role of Neutrophils in Murine Salmonellosis. Infect Immun. 2003;72:468–477. doi: 10.1128/IAI.72.1.468-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KRR, Arancibia-Cárcamo CV, Hall J, Sun C-M, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nat Med. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey NA, Everett D, Faragher EB, Roca-Feltrer A, Kang’ombe A, Denis B, Kerac M, Molyneux E, Molyneux M, Jahn A, Gordon MA, Heyderman RS. Modelling the Contributions of Malaria, HIV, Malnutrition and Rainfall to the Decline in Paediatric Invasive Non-typhoidal Salmonella Disease in Malawi. PLoS Negl Trop Dis. 2015;9:e0003979. doi: 10.1371/journal.pntd.0003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster R, Schubel A, Breitfeld D, Kremmer E, Renner-Müller I, Wolf E, Lipp M. CCR7 Coordinates the Primary Immune Response by Establishing Functional Microenvironments in Secondary Lymphoid Organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- Geem D, Medina-Contreras O, Kim W, Huang CS, Denning TL. Isolation and characterization of dendritic cells and macrophages from the mouse intestine. JoVE. 2012;63:e4040. doi: 10.3791/4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MA. Invasive nontyphoidal Salmonella disease- epidemiology, pathogenesis and diagnosis. Curr Opin, Infect Dis. 2011;24:484–489. doi: 10.1097/QCO.0b013e32834a9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MA, Graham SM, Walsh AL, Wilson L, Phiri A, Molyneux E, Zijlstra EE, Heyderman RS, Hart CA, Molyneux ME. Epidemics of Invasive Salmonella enterica Serovar Enteritidis and S. enterica Serovar Typhimurium Infection Associated with Multidrug Resistance among Adults and Children in Malawi. Clin Infect Dis. 2008;46:963–969. doi: 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BAD. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Kariuki S, Gordon MA, Feasey N, Parry CM. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine. 2015;33:C21–C29. doi: 10.1016/j.vaccine.2015.03.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Kay S, Connor T, Barquist L, Sait L, Holt KE, Sivaraman K, Wileman T, Goulding D, Clare S, Hale C, Seshasayee A, Harris S, Thomson NR, Gardner P, Rabsch W, Wigley P, Humphrey T, Parkhill J, Dougan G. Genome and transcriptome adaptation accompanying emergence of the definitive type 2 host-restricted Salmonella enterica serovar Typhimurium pathovar. mBio. 2013;4:e00565–13. doi: 10.1128/mBio.00565-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, Harris D, Clarke L, Whitehead S, Sangal V, Marsh K, Achtman M, Molyneux ME, Cormican M, Parkhill J, MacLennan CA, Heyderman RS, Dougan G. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm E, Dougan G. Advances in Understanding Bacterial Pathogenesis Gained from Whole-Genome Sequencing and Phylogenetics. Cell Host Microbe. 2016;19:599–610. doi: 10.1016/j.chom.2016.04.015. [DOI] [PubMed] [Google Scholar]
- Klemm EJ, Gkrania-Klotsas E, Hadfield J, Forbester JL, Harris SR, Hale C, Heath JN, Wileman T, Clare S, Kane L, Goulding D, Otto TD, Kay S, Doffinger R, Cooke FJ, Carmichael A, Lever AML, Parkhill J, MacLennan CA, Kumararatne D, Dougan G, Kingsley RA. Emergence of host-adapted Salmonella Enteritidis through rapid evolution in an immunocompromised host. Nat Microbiol. 2016;1:15023. doi: 10.1038/nmicrobiol.2015.23. [DOI] [PubMed] [Google Scholar]
- Kurtz JR, Petersen HE, Frederick DR, Morici LA, McLachlan JB. Vaccination with a single CD4 T cell peptide epitope from a Salmonella type III-secreted effector protein provides protection against lethal infection. Infect Immun. 2014;82:2424–2433. doi: 10.1128/IAI.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokken KL, Mooney JP, Butler BP, Xavier MN, Chau JY, Schaltenberg N, Begum RH, Müller W, Luckhart S, Tsolis RM. Malaria Parasite Infection Compromises Control of Concurrent Systemic Non-typhoidal Salmonella Infection via IL-10-Mediated Alteration of Myeloid Cell Function. PLoS Pathog. 2014;10:e1004049. doi: 10.1371/journal.ppat.1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M, Harkins CR, Wang C, Nguyen C, Berghoff A, Elliott G, Kohlberg S, Strong C, Du F, Carter J, Kremizki C, Layman D, Leonard S, Sun H, Fulton L, Nash W, Miner T, Minx P, Delehaunty K, Fronick C, Magrini V, Nhan M, Warren W, Florea L, Spieth J, Wilson RK. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- McLaughlin LM, Govoni GR, Gerke C, Gopinath S, Peng K, Laidlaw G, Chien Y-H, Jeong H-W, Li Z, Brown MD, Sacks DB, Monack D. The Salmonella SPI2 Effector SseI Mediates Long-Term Systemic Infection by Modulating Host Cell Migration. PLoS Pathog. 2009;5:e1000671. doi: 10.1371/journal.ppat.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin LM, Xu H, Carden SE, Fisher S, Reyes M, Heilshorn SC, Monack DM. A microfluidic-based genetic screen to identify microbial virulence factors that inhibit dendritic cell migration. Integr Biol. 2014;6:438–449. doi: 10.1039/c3ib40177d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack DM, Hersh D, Ghori N, Bouley D, Zychlinsky A, Falkow S. Salmonella Exploits Caspase-1 to Colonize Peyer’s Patches in a Murine Typhoid Model. J Exp Med. 2000;192:249–258. doi: 10.1084/jem.192.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney JP, Butler BP, Lokken KL, Xavier MN, Chau JY, Schaltenberg N, Dandekar S, George MD, Santos RL, Luckhart S, Tsolis RM. The mucosal inflammatory response to non-typhoidal Salmonella in the intestine is blunted by IL-10 during concurrent malaria parasite infection. Mucosal Immunol. 2014;7:1302–1311. doi: 10.1038/mi.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney JP, Lokken KL, Byndloss MX, George MD, Velazquez EM, Faber F, Butler BP, Walker GT, Ali MM, Potts R, Tiffany C, Ahmer BMM, Luckhart S, Tsolis RM. Inflammation-associated alterations to the intestinal microbiota reduce colonization resistance against non-typhoidal Salmonella during concurrent malaria parasite infection. Sci Rep. 2015;5:14603. doi: 10.1038/srep14603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio SP, Bäumler AJ. Comparative Analysis of Salmonella Genomes Identifies a Metabolic Network for Escalating Growth in the Inflamed Gut. mBio. 2014;5:e00929–14. doi: 10.1128/mBio.00929-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoro CK, Barquist L, Connor TR, Harris SR, Clare S, Stevens MP, Arends MJ, Hale C, Kane L, Pickard DJ, Hill J, Harcourt K, Parkhill J, Dougan G, Kingsley RA. Signatures of Adaptation in Human Invasive Salmonella Typhimurium ST313 Populations from Sub-Saharan Africa. PLoS Negl Trop Dis. 2015;9:e0003611. doi: 10.1371/journal.pntd.0003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, Kariuki S, Msefula CL, Gordon MA, de Pinna E, Wain J, Heyderman RS, Obaro S, Alonso PL, Mandomando I, MacLennan CA, Tapia MD, Levine MM, Tennant SM, Parkhill J, Dougan G. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44:1215–1221. doi: 10.1038/ng.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons BN, Humphrey S, Salisbury AM, Mikoleit J, Hinton JCD, Gordon MA, Wigley P. Invasive Non-Typhoidal Salmonella Typhimurium ST313 Are Not Host-Restricted and Have an Invasive Phenotype in Experimentally Infected Chickens. PLoS Negl Trop Dis. 2013;7:e2487. doi: 10.1371/journal.pntd.0002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porwollik S, Boyd EF, Choy C, Cheng P, Florea L, Proctor E, McClelland M. Characterization of Salmonella enterica Subspecies I Genovars by Use of Microarrays. J Bacteriol. 2004;186:5883–5898. doi: 10.1128/JB.186.17.5883-5898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixão TA, Gordon MA, Kolls JK, Dandekar S, Baumler AJ. Simian immunodeficiency virus–induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux CM, Butler BP, Chau JY, Paixao TA, Cheung KW, Santos RL, Luckhart S, Tsolis RM. Both Hemolytic Anemia and Malaria Parasite-Specific Factors Increase Susceptibility to Nontyphoidal Salmonella enterica Serovar Typhimurium Infection in Mice. Infect Immun. 2010;78:1520–1527. doi: 10.1128/IAI.00887-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Meth. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, Bain CC, Wright PB, Sichien D, Kotarsky K, Persson EK, Luda K, Guilliams M, Lambrecht BN, Agace WW, Milling SW, Mowat AM. CCR2+CD103− intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal Immunol. 2014;8:327–339. doi: 10.1038/mi.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singletary LA, Karlinsey JE, Libby SJ, Mooney JP, Lokken KL, Tsolis RM, Byndloss MX, Hirao LA, Gaulke CA, Crawford RW, Dandekar S, Kingsley RA, Msefula CL, Heyderman RS, Fang FC. Loss of Multicellular Behavior in Epidemic African Nontyphoidal Salmonella enterica Serovar Typhimurium ST313 Strain D23580. mBio. 2016;7:e02265–15. doi: 10.1128/mBio.02265-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden AWM, Velasquez M, Starnbach MN. Salmonella Rapidly Kill Dendritic Cells via a Caspase-1- Dependent Mechanism. J Immunol. 2003;171:6742–6749. doi: 10.4049/jimmunol.171.12.6742. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres A, Jones-Carson J, Bäumler AJ, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks WT, Fang FC. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- Watson KG, Holden DW. Dynamics of growth and dissemination of Salmonella in vivo. Cell Microbiol. 2010;12:1389–1397. doi: 10.1111/j.1462-5822.2010.01511.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Barrila J, Roland KL, Kilbourne J, Ott CM, Forsyth RJ, Nickerson CA. Characterization of the Invasive, Multidrug Resistant Non-typhoidal Salmonella Strain D23580 in a Murine Model of Infection. PLoS Negl Trop Dis. 2015;9:e0003839. doi: 10.1371/journal.pntd.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.