Summary
HSP90 acts as a protein-folding buffer that shapes the manifestations of genetic variation in model organisms. Whether HSP90 influences the consequences of mutations in humans, potentially modifying the clinical course of genetic diseases, remains unknown. Mining data for >1,500 disease-causing mutants, we found strong correlation between reduced phenotypic severity and a dominant (HSP90≥HSP70) increase in mutant engagement by HSP90. Examining the cancer predisposition syndrome Fanconi Anemia in depth revealed that mutant FANCA proteins engaged predominantly by HSP70 had severely compromised function. In contrast, the function of less severe mutants was preserved by a dominant increase in HSP90 binding. Reducing HSP90’s buffering capacity with inhibitors or febrile temperatures destabilized HSP90-buffered mutants, exacerbating FA-related chemosensitivities. Strikingly, a compensatory FANCA somatic mutation from an “experiment of nature” in monozygotic twins both prevented anemia and reduced HSP90 binding. These findings provide one plausible mechanism for the variable expressivity and environmental sensitivity of genetic diseases.
Keywords: Gene-Environment Interaction, HSP90-Buffering, HSP70, Fanconi Anemia, Cancer
Graphical abstract
Introduction
HSP90 is a protein-folding chaperone that exerts a powerful influence on the biological consequences of genetic variation in several model organisms and links these consequences to changes in the environment (Jarosz et al., 2010). In general, chaperones (also known as heat shock proteins or HSPs) transiently bind other proteins to protect them from misfolding and aggregation. Indeed, the abundant chaperones HSP90 and HSP70 cooperatively assist a major fraction of the human proteome to fold and function (Taipale et al., 2012; Taipale et al., 2014).
Several unique aspects of HSP90 chaperone function make it particularly effective in connecting phenotypes to the cellular environment, at least in model organisms. Unlike most other chaperones, HSP90 is normally present in large excess over requirements for growth (Borkovich et al., 1989). This creates a protein-folding “buffer” that supports the function of a specialized suite of proteins with critical roles in protein quality control and trafficking, signal transduction, apoptosis, cell cycle regulation, gene silencing, and genome maintenance (Mayer and Bukau, 2005; Taipale et al., 2012; Taipale et al., 2014). While the need for HSP90 increases greatly with stress, its expression is only modestly induced compared to many other chaperones (Borkovich et al., 1989). Thus, under some conditions HSP90 levels may become insufficient to buffer the effects of proteotoxic stresses on the folding and function of mutant protein variants. Moreover, HSP90’s chaperone activity requires a large variety of client-specific cofactors (>30) (Taipale et al., 2014), the levels of which can also become limiting under stress. Finally, HSP90 engages many of its clients in an unusual way that enables this chaperone to act as an environmental sensor. Most proteins interact with chaperones only transiently during their initial folding steps. However, many HSP90 clients require continuous cycles of chaperoning by HSP90 to maintain their functional conformations (Pratt and Toft, 1997). For all these reasons, environmental insults that tax HSP90 function can have immediate and profound effects on cell physiology.
Previous work by us and others in various models has shown that the protein-folding buffer activity of HSP90 permits it to serve as both a potentiator and a capacitor of genetic variation (Cowen and Lindquist, 2005; Jarosz and Lindquist, 2010; Queitsch et al., 2002; Rohner et al., 2013; Rutherford and Lindquist, 1998; Yeyati et al., 2007). As a potentiator, the HSP90 buffer allows variation to have immediate phenotypic manifestations. For example, HSP90 promotes the oncogenic hyperactivity and malignant potential of many kinases by assisting these intrinsically unstable proteins to fold and function (Whitesell and Lindquist, 2005). HSP90 also supports the function of unstable signal transducers (i.e. calcineurin) that enable the evolution of drug resistance in fungi and cancer cell lines (Cowen and Lindquist, 2005; Whitesell et al., 2014). Such potentiated traits are disrupted by clinically relevant environmental stresses (Cowen and Lindquist, 2005; Vincent et al., 2013).
As a capacitor, the buffering effects of HSP90’s folding activities render genetic alterations phenotypically silent, until environmental stresses overwhelm chaperone function and reveal them. For example, low salinity water uncovers preexisting, cryptic genetic variation in an HSP90-dependent way, contributing to the adaptive loss of eyes in cavefish (Rohner et al., 2013). Depending on the specific environment, HSP90-buffered genetic variation can lead to deleterious or beneficial traits, with the potential to shape evolutionary processes (Cowen and Lindquist, 2005; Jarosz et al., 2010; Lindquist, 2009; Rohner et al., 2013; Rutherford, 2003; Whitesell et al., 2014).
In the realm of human genetic variation, very little is known about the potential role(s) of HSP90 or HSP70, the other major cytosolic chaperone, in shaping clinical manifestations. Recently we used quantitative high throughput methods to measure protein::protein interactions (PPI) for >2,300 mutant and cognate wild-type proteins. About 30% of the disease-causing mutants showed increased interaction with chaperones suggesting they are protein-folding mutants (Sahni et al., 2015). Whether the increased chaperone binding of mutant human proteins has any functional consequences or has the potential to modify the clinical course of human genetic diseases has been speculated upon by others and by us, but has yet to be shown.
To address this, we first correlated chaperone interactions from our quantitative dataset with available literature on the clinical and cellular severity of diverse mutations. A clear distinction in severity was associated with HSP90 vs. HSP70 binding across disorders. We then chose to focus on a broad array of mutations in a single disorder, namely Fanconi Anemia (FA). FA is a complex autosomal recessive human cancer-predisposition syndrome. It results from point mutations in any one of over 19 genes known to participate in the so-called FA genome maintenance pathway (Ceccaldi et al., 2016; Joenje and Patel, 2001).
The FA pathway provides a near ideal system for studying the role of HSP90 in buffering human genetic variation because of the large number of causal alleles identified. Many known FA-causing mutations have been functionally characterized in genetically tractable cellular models that recapitulate key features of the disease (Adachi et al., 2002; Joenje and Patel, 2001; Yagasaki et al., 2004). Most notably, cells deficient in FA pathway function are hypersensitive to genotoxic chemotherapeutic agents, such as mitomycin C (MMC), providing an opportunity for quantitative assessment of the trait in vitro in cells obtained from patients. Indeed, this effect is the preferred clinical test for diagnosing FA (Joenje and Patel, 2001).
FA genes are highly polymorphic (Castella et al., 2011) and deleterious mutations in the very same FA gene manifest with great variability across affected individuals (Joenje and Patel, 2001). These variable phenotypes are shaped by genetic and environmental factors that create extremely complex genotype-phenotype relationships. Herein, we present a quantitative analysis of mutant protein::chaperone interactions that reveals a direct and specific role for HSP90 in buffering the effects of human genetic variation and linking its functional consequences to clinically relevant environmental stresses.
Results
Distinct pattern of chaperone interactions for different human disease mutants
HSP90 and HSP70 bind different types of polypeptide conformations, with HSP70 recognizing extended hydrophobic chains and HSP90 recognizing partially folded proteins (Figure 1A). To probe their relative contributions to shaping genotype-phenotype relationships for mutant human proteins, we compared the physical interaction of HSP90 and HSP70 with mutant proteins causally linked to a diverse range of human genetic diseases. Primary data for this comparison were mined from our previous study (Sahni et al., 2015), in which we used a LUMIER (luminescence-based mammalian interactome mapping) (Taipale et al., 2012) (Figure 1B) approach to measure protein::protein interactions (PPIs) across a library of over 2,300 mutant and cognate wild-type proteins.
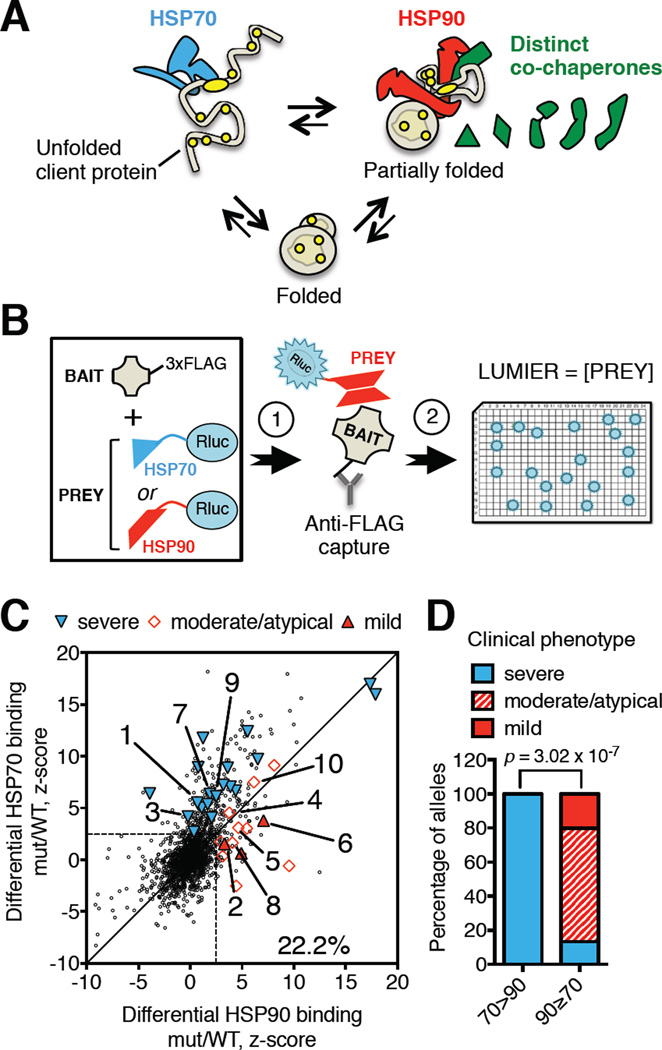
Figure 1. Pattern of increased chaperone engagement reflects mutant severity across diverse human diseases.
(A) Schematic protein-folding pathway modeling HSP70- or HSP90-bound client polypeptide conformations. HSP70 (blue) recognizes an extended hydrophobic (yellow) chain (unfolded client protein). HSP90 (red) recognizes structured polypeptides that are less hydrophobic (partially folded) with assistance from specialized co-chaperones (green). The fully folded, active state (folded) binds neither.
(B) Schematic of LUMIER assays. HEK293T cells stably expressing Renilla luciferase-tagged fusions of the constitutive chaperones HSP90 (HSP90β) or HSP70 (HSPA8) (PREY) are transiently transfected with a library of plasmids encoding FLAG-tagged proteins (BAIT) (arrow 1). Bait proteins are captured by incubation of whole cell lysates on anti-FLAG antibody-coated plates. The relative amount of co-captured chaperone is measured by luciferase assays (arrow 2). Bait protein levels are subsequently measured by FLAG-ELISA to determine expression levels and calculate chaperone interaction scores.
(C) Plot of chaperone interaction scores of 1628 missense mutants relative to the corresponding wild-type protein for HSP90 (x-axis) and HSP70 (y-axis) (dataset from (Sahni et al., 2015)). ∼ 22% of disease-causing mutants exhibit an increased interaction with HSP90 or HSP70 (area outside of the dashed lines). Literature curated clinical phenotypes of comparable mutants are grouped into severe, moderate or mild phenotypic classes. Severe: 1: SOD1-G41S; 3: GGCX-T591K; 7: AKR1D1-L106F, 9: GNAS-I103T. Moderate: 4: GGCX-W157R; 5: AAAS-S263P, 8: AKR1D1-P198L; 10: GNAS-A366S. Mild: 2: SOD1-G37R; 6: AAAS-L430F.
(D) Correlation between reported clinical phenotype and the pattern of chaperone engagement of mutant proteins (HSP70-preferring: 70>90, compared to HSP90-preferring: 90≥70). Reported p value was determined by Fisher’s exact 2×3 extension test.
We parsed these previous data for differential association with the HSP90 or HSP70 chaperones. As a positive control, we used a FLAG-tagged version of the classical HSP90 client kinase v-SRC. This oncogenic mutant associates to a much greater extent with HSP90 than HSP70 (Figure S1A). The shift in equilibrium chaperone engagement seen for v-SRC reflects impairment of its normal folding cycle caused by the mutation driving its hyperactivity (Figure S1A). Analysis of the entire dataset revealed that some mutations appeared to stall folding at an intermediate stage characterized by increased binding to both chaperones, while others impaired folding at earlier (HSP70-engaging) or later (HSP90-engaging) stages in the classical conformation-maturation process (Figures 1C and S1B). Different mutations, even in the same protein, caused different protein-folding problems, which were reflected as divergent changes in relative chaperone engagement (Figures S1B–C).
Pattern of increased chaperone engagement reflects severity of protein-folding mutations
To determine whether increased engagement of HSP90 vs. HSP70 correlated with the severity of human genetic disorders, we examined the clinical phenotypes reported for the specific mutant proteins that had increased chaperone binding. Data were found for 32 mutations across 20 genes of diverse biological functions and a broad spectrum of diseases. These include the various diseases listed in Table S1A. With one exception (the membrane-bound retinol dehydrogenase RDH12), the proteins underlying these disorders normally localize to the cytoplasm or the nucleus. Key to our analysis, the severity of clinical presentation varies extensively within this group of mutations.
Strikingly, the clinical severity of specific mutations correlated with their relative chaperone-binding patterns. Mutants that preferentially bound HSP70 were associated with more severe disease than mutants that preferentially bound HSP90 or mutants with comparably increased binding to both chaperones (Figures 1C and S1C). “HSP70-engaged” (70>90) protein-folding mutants are thus associated with more severe phenotypes than “HSP90-engaged” mutants which show increases in HSP90 binding equal or greater than that of HSP70 (90≥70, Figure 1D; Table S1B). Five proteins exhibited pairs of mutations with divergent clinical severities. In each case, altered levels of chaperone engagement correlated with the severity of the phenotype (mutants 1 – 10; Figures 1C–D; Table S1A). For superoxide dismutase (SOD1) mutants linked to ALS, the HSP70-engaged SOD1 mutant G41S is typically associated with far more severe disease than the HSP90-engaged G37R mutant (mutants 1 vs. 2; Figures 1C and S1C–D; Table S1A). The HSP90-engaged G37R mutant also exhibited a broad spectrum of disease severity across the 27 patients reported (Figure S1D) (Wang et al., 2008), suggesting likely phenotypic modification by here-to-fore elusive genetic and environmental factors.
In many genetic diseases modifier loci and environmental factors alter the expressivity of a causative mutation thereby confounding simple genotype to phenotype relationships. To disentangle these relationships, we focused on 13 diseases driven by mutant proteins that have well-defined, disease-relevant biochemical functions measurable in vitro (Table S1A). For example, single amino-acid substitutions in the Vitamin-K-dependent carboxylase GGCX result in a bleeding diathesis of variable severity. In this disorder, clinical severity correlates well with the extent of the specific mutant’s reduction in biochemical carboxylase activity (mutants 3 vs. 4; Table S1A). In 11 of the 13 disorders examined, inactive/severe mutants preferentially bound HSP70, while mutants retaining some physiological activity had similar increases in binding to both HSP70 and HSP90 (Tables S1A–B).
FANCA variants differentially associate with HSP90 and HSP70
To further examine the relationship between differential chaperone binding and functional severity in a single, genetically tractable system, we focused on FANCA, the most frequently compromised protein in FA patients (Figure 2A) (Joenje and Patel, 2001). We transiently expressed a library of 90 FLAG-tagged FANCA variants in human cells. The library consisted of wild-type FANCA, 38 disease-causing mutants and 51 polymorphic variants found in healthy individuals (Table S2A) and included only missense substitutions, which often have more subtle effects on protein function than truncations or deletions. Wild-type FANCA protein stably interacts with HSP90 and HSP70 in living cells (Oda et al., 2007) (Figure S2A). However, our quantitative assessment showed that these interactions were relatively weak compared to those of other well-established clients and co-chaperones of HSP90 and HSP70 (Figure S1A).
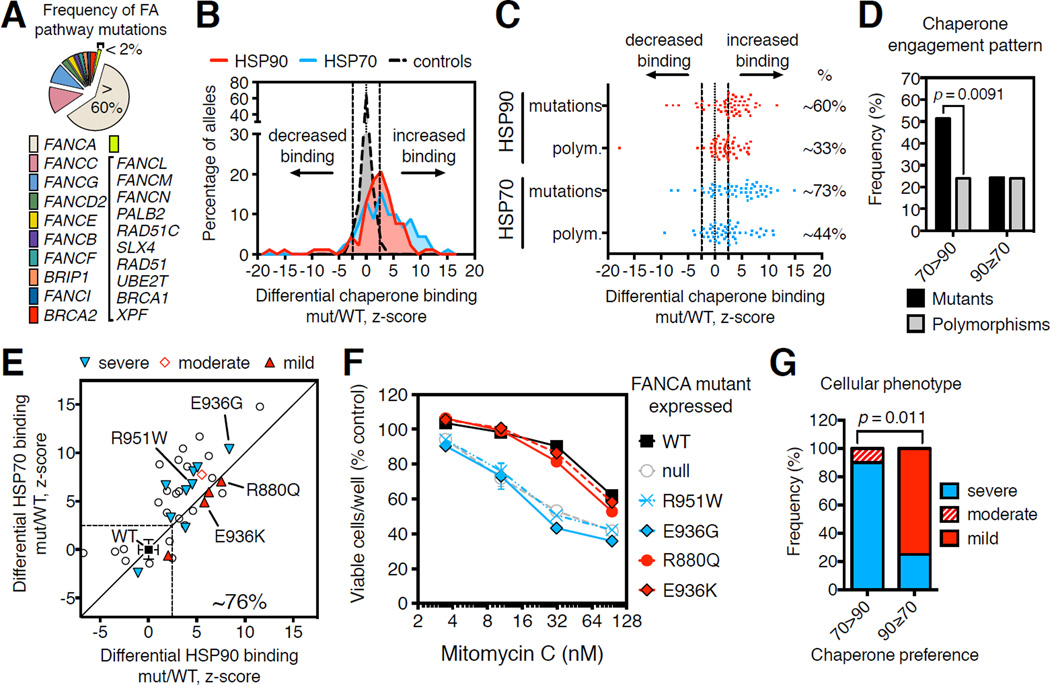
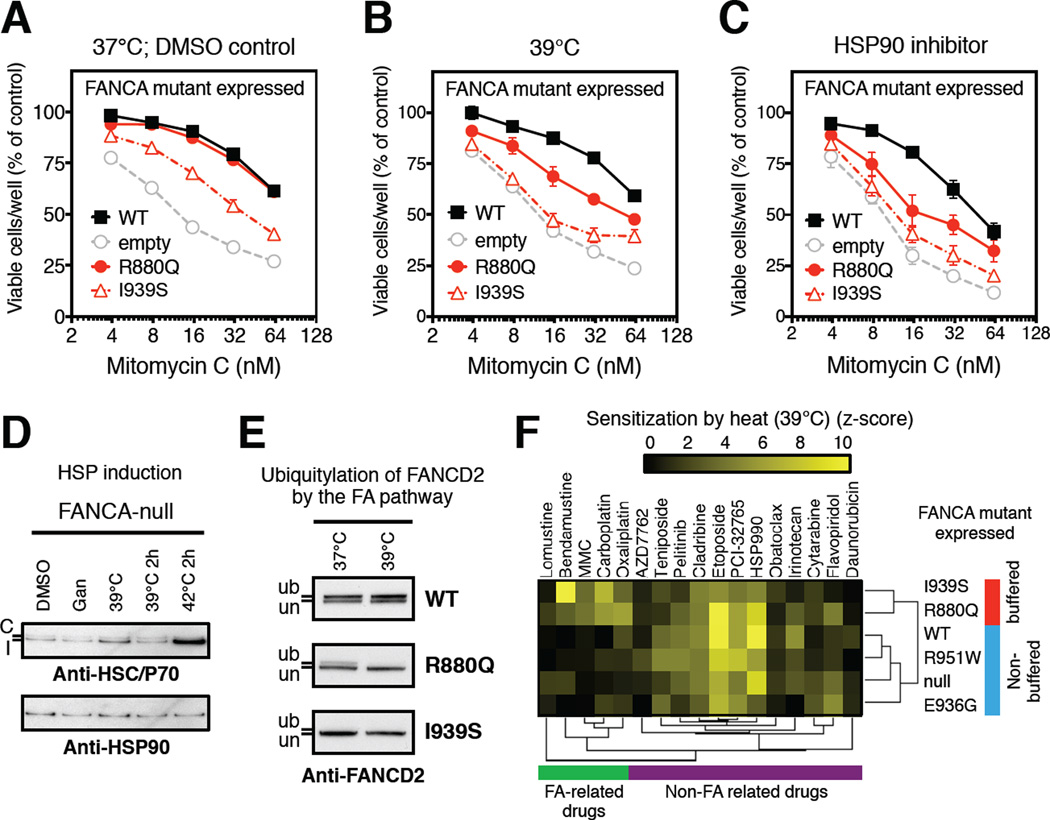
Figure 2. Pattern of increased chaperone engagement reflects functional severity of FA-causing mutants.
(A) Estimated frequency of inactivating mutations in FA genes across FA patients.
(B) Distribution of LUMIER interaction scores for 90 FANCA variants normalized by the respective HSP90 (red area) or HSP70 (blue area) interaction scores of wild-type FANCA (mut/WT).
(C) Scatter plot of differential (mut/WT) LUMIER interaction scores for 90 FANCA variants. HSP90 (red dots) and HSP70 (blue dots) interactions are parsed by their association with disease. Fractions of variants with significantly increased chaperone interaction compared to wild-type FANCA (%) are shown on the right for each category: p = 0.0095, HSP90; p = 0.0063, HSP70; determined by Fisher’s exact one-sided test.
(D) Frequency of engagement patterns of HSP70 (70>90) or HSP90 (90≥70) chaperones with FANCA missense variants, grouped into FA-causing mutants (mutants) and polymorphisms of unknown significance (polymorphisms). Reported p value determined by Fisher’s exact one-sided test.
(E) Scatter plot of differential LUMIER interaction z-scores for 38 FA-causing FANCA mutants versus HSP90 (x-axis) and HSP70 (y-axis) binding. 29 of 38 mutants (∼76%) exhibit increased binding to either chaperone (P1194L falls out of the indicated z-score range). Functionally characterized mutants are reported as “severe”, “moderate” or “mild” based on their functional severity.
(F) Relative viability after exposure to mitomycin C (MMC) of GM6914 FANCA-null cells transduced with retroviruses encoding the indicated FANCA variants.
(G) Correlation between pattern of chaperone engagement and observed cellular phenotypic severity of FANCA mutants. Reported p value determined by Fisher’s exact 2×3 extension test.
A large fraction of FANCA variants displayed significant increases in HSP90 or HSP70 binding compared to the wild-type protein (Figure 2B). These interactions were highly reproducible across independent experiments (Figures S2B–D; Table S2B), and were not observed with the Renilla luciferase tag alone (Figure S2E). We validated these differences in chaperone interactions with 15 representative FANCA mutants by coimmunoprecipitation experiments (Figure S2F). We further noted that increased binding to HSP90 correlated with a greater reduction in steady-state expression of FANCA mutants upon HSP90 inhibition (Figure S2G). In addition, mutations causing changes in chaperone binding did not cluster spatially within the protein, suggesting that no single domain in FANCA is enriched in protein-folding mutations (Figure S2H).
Both HSP90 and HSP70 bound clinically recognized mutants much more frequently than they bound polymorphisms (Figure 2C). Thus, most clinically important FANCA mutations appear to introduce a protein-folding defect. Notably, many FANCA variants bound to HSP70 much more than they bound to HSP90, while some bound similarly to both chaperones (Figure S2I). These differences in chaperone binding did not correlate with the sensitivity of FANCA mutant protein levels to HSP90 inhibition (Figure S2G). However, preferential binding to HSP70 was greatly enriched among known disease-causing FA mutations as compared to polymorphisms (51.35% vs. 24.32%; Figure 2D). In contrast, equal or greater binding to HSP90 was evenly distributed across FA-causing mutations and polymorphisms (Figure 2D), reinforcing our finding that HSP90 engagement is a feature of less severely compromised variants.
Pattern of chaperone engagement reflects functional severity across FANCA variants
In contrast to their clinical severity, the functional severity of FANCA variants can be precisely assigned experimentally by comparing biochemical assays in exactly the same cellular backgrounds. The functional severity of FANCA variants is classically assessed in patient-derived cell lines as a reduced ability to support ubiquitylation of the FA pathway component FANCD2 and reduced cell survival when exposed to genotoxic stress. On this basis, three phenotypic classes of FANCA variant have previously been described (referred to herein as severe, moderate and mild) (Adachi et al., 2002; Yagasaki et al., 2004).
Five of seven mutants previously characterized as severe preferentially bound HSP70 (Figure S2J). One mutant characterized as moderate to severe (I939S) also bound HSP70 preferentially (Figure S2J; Tables S2B–C). To standardize our own assays, we engineered patient-derived GM6914 cells (which express no functional FANCA protein) to express a severe (T724P) or moderate (I939S) FANCA mutant. Both cell lines behaved as previously reported (Xie et al., 2015; Yagasaki et al., 2004) (Figures S2K–L). Of two FANCA clinical mutants previously characterized as mild, one (D598N) showed no increase in chaperone binding, while the second (F868V) showed a comparable increase in HSP90 and HSP70 binding (Figure S2J; Table S2C). Two common, benign FANCA polymorphisms (ClinVar: rs75501942, I311T and rs17232973, S674L) (Table S2A) were predominantly engaged by HSP90 (Table S2B). Thus, increased binding of one chaperone in relation to the other appears to better define the stage at which folding is stalled and reflect the functional severity of FANCA variants as opposed to the increased binding of either chaperone considered in isolation (Figure S2J).
Can the pattern of chaperone engagement predict the functional severity of previously uncharacterized FANCA mutants? We engineered GM6914 cell lines to stably express mutants from the various experimentally defined chaperone-engagement categories (Figure 2E). Although HSP70-engaged mutants R951W and E936G were expressed at levels similar to wild-type FANCA, they were functionally null in both MMC survival and FANCD2 ubiquitylation assays (Figures 2F and S2L). The same mutants conferred hypersensitivity to other genotoxic agents related to FA, such as carboplatin and oxaliplatin (Figure S2M). In contrast, the R880Q and E936K mutants, which showed increased binding to both HSP90 and HSP70, did not increase sensitivity to MMC and other FA-related chemotherapeutics (Figures 2F and S2M), and only moderately reduced FANCD2 ubiquitylation in the presence of hydroxyurea (Figure S2L).
Notably, two FANCA mutants with divergent cellular phenotypes carried different amino acids at the same codon. The HSP70-preferring E936G mutant was much more severe than the E936K mutant, which showed similar increases in binding to both chaperones, even though E936K was expressed at lower levels (Figures 2F and S2L). Predicting the functional consequences of such same-codon substitutions is particularly challenging for sequence/structure-based algorithms (Table S2C). Our findings with these previously uncharacterized mutants confirm what we observed studying FANCA mutants characterized phenotypically by others; namely chaperone-binding pattern reflects functional severity. Mutants that preferentially bind HSP70 cause significantly more severe phenotypes than those showing an equal or greater increase in HSP90 binding (Figure 2G).
HSP90 buffers FANCA mutants thereby preserving FA pathway function
To investigate the role of HSP90 in buffering FANCA mutants, we first defined the HSP90-dependence of the wild-type protein. FANCA function in the FA pathway was previously shown to be sensitive to first-generation inhibitors of HSP90 (Oda et al., 2007). We titrated second-generation HSP90 inhibitors to identify the lowest concentration of each that could impair wild-type FANCA function without affecting cell proliferation or survival. These concentrations impaired the MMC tolerance of FANCA wild-type cells but had little effect on isogenic FANCA-null cells (Figures 3A, S3A–C). Notably, low-level HSP90 inhibition did not appreciably increase the expression of HSP70 or HSP90 (Figure S3D).
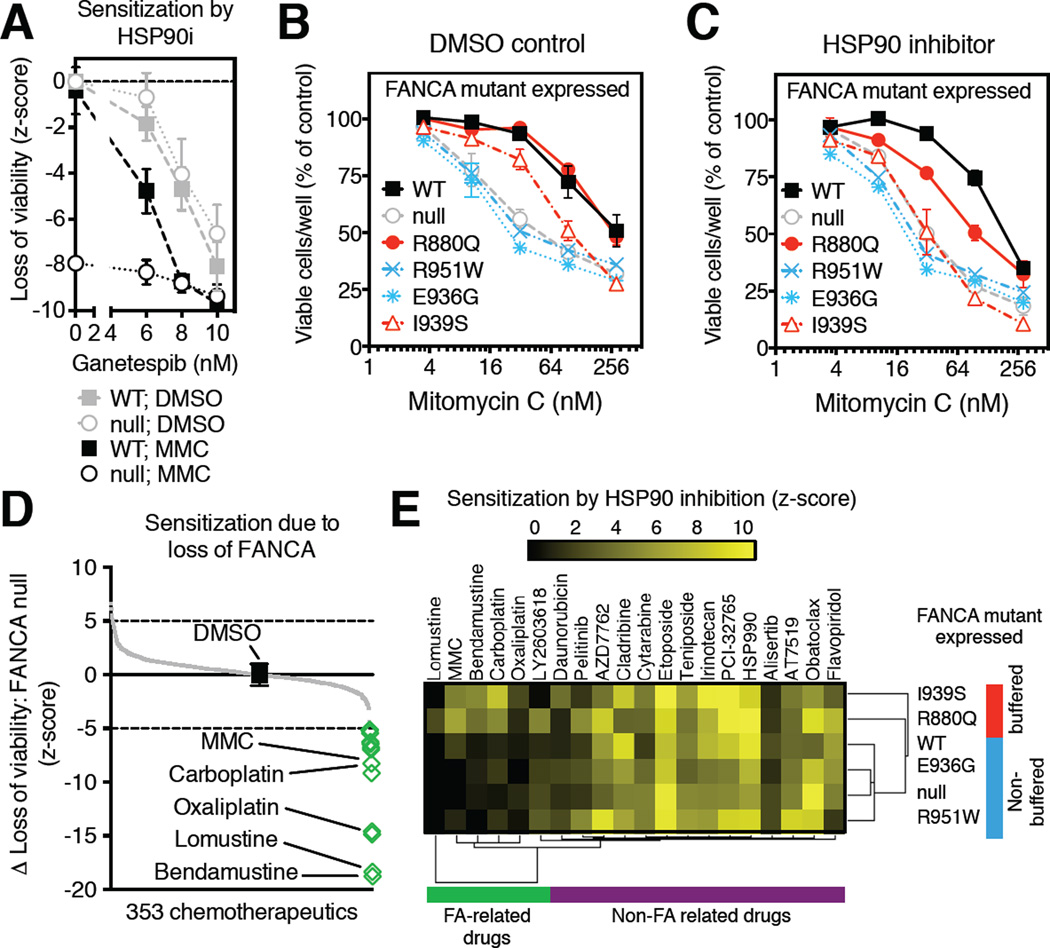
Figure 3. Non-toxic, low-level HSP90 inhibition induces mutant-specific drug sensitivities.
(A) Loss of viability (z-score) resulting from HSP90 inhibition (HSP90i; ganetespib, 5 nM) in FANCA-null GM6914 cells transduced with retroviruses encoding FANCA wild-type (squares; WT) or empty vector control (circles; null) in the presence (black symbols) of MMC (31.6nM) or DMSO control (grey symbols). Data from 3 independent experiments are presented as mean ± SEM.
(B–C) Relative viability of GM6914 FANCA-null cells transduced with retroviruses encoding the indicated FANCA mutants, wild-type control (WT; black squares) or empty vector control (empty) in the presence of mitomycin C (MMC) under normal growth conditions (B) or low-level HSP90 inhibition (ganetespib, 5 nM) (C). Data presented as mean ± SEM from 2 independent experiments. Error-bars not visible are smaller in size than the symbol.
(D) Screen for chemotherapeutics particularly toxic to FA. Highlighted drug treatment regimes (green diamonds) are significantly more toxic to FANCA-null GM6914 cells than to isogenic cells expressing wild-type FANCA. Black square indicates DMSO alone.
(E) Mutant-specific chemo-sensitization by low-level HSP90 inhibition. Low-level HSP90 inhibition (ganetespib, 5 nM) sensitizes FANCA-null (GM6914) cells expressing the indicated FANCA mutants to diverse chemotherapeutic agents. Hierarchical clustering of significant relative sensitization effects of HSP90 inhibition (z-score >2) reveals two FANCA mutant clusters (HSP90-buffered mutants: red; non-buffered mutants: blue), on the basis of their differential sensitization to FA-related drugs (green cluster). HSP90 inhibition also induced sensitization to non-FA related drugs (purple cluster), but these effects were not mutant specific.
We next measured the sensitivity to HSP90 inhibition of various FANCA mutants expressed in FANCA-null cells. HSP90 inhibitor concentrations that had no effect on wild-type function markedly exacerbated the MMC sensitivity of cells expressing the R880Q mutant that exhibits increased HSP90 binding (Figures 3B–C). We observed this same mutant-specific sensitization effect in replicate mutant cell lines (Figure S3E) and using alternative HSP90 inhibitor chemotypes (Figures S3F–G). Mutant-specific sensitization to MMC with low-level HSP90 inhibition was accompanied by increased cell death (Figure S3H). It also abrogated residual genotoxic stress-induced FANCD2 ubiquitylation in R880Q cells (Figure S3I). We conclude that a single amino-acid change in FANCA can increase the dependence of the entire FA pathway on HSP90.
The sensitization of specific FANCA mutants to MMC by low-level HSP90 inhibition extended to other genotoxic stresses. We screened FANCA-null cells against 353 mechanistically diverse, clinically relevant anticancer drugs. As anticipated, the cells were hypersensitive to the DNA-damaging agents carboplatin and oxaliplatin in addition to MMC (Figure 3D). Bendamustine and lomustine, classical alkylating agents not previously known to be FA-relevant, were also identified in this screen (Table S3A).
We next tested select FANCA mutants against the hits from this screen. Low-level HSP90 inhibition hyper-sensitized cells expressing the HSP90-buffered R880Q to the same compounds that FANCA-null cells were hypersensitive to (Figures 3E and S3J). Low-level HSP90 inhibition had no effect on the sensitivity of cells expressing FANCA mutants with complete loss of FANCA function, R951W or E936G, (Figure 3B–C and 3E). For I939S, the HSP70-preferring FANCA mutant that retains partial function and has increased HSP90-binding, low-level HSP90 inhibition eliminated residual function. This was manifest both by sensitivity to FA-related drugs and loss of FANCD2 ubiquitylation (Figures 3B–C, 3E and S3I–J).
Low-level HSP90 inhibition also sensitized FANCA mutant cells lines to drugs that were not related to FA (e.g. topoisomerase poisons, cell cycle inhibitors, kinase inhibitors and anti-metabolites; Figures 3E and S3K; Table S3B). However the sensitivities of FANCA mutants to these drugs were all affected equally. Presumably, then, these sensitivities are due to the impairment of diverse HSP90-supported pathways that are unrelated to FANCA. Taken together, our results establish that HSP90 selectively buffers the effects of particular genetic variants of FANCA thereby preserving FA pathway function.
Environmental stress compromises HSP90’s ability to buffer FANCA mutants
To determine whether environmental stresses that compromise protein homeostasis alter the ability of human HSP90 to buffer the effects of genetic variation, we chose heat treatments as an example of a common and physiologically relevant stress. To investigate the effect of increased temperature on the function of FANCA and its mutants, we exposed cells to cytotoxic agents at 37°C and allowed them to recover at 39°C or 40°C. These temperature increases are well within the range experienced by patients with fever. They had no effect on the proliferation or survival of any cell lines on their own (Figures S4A and S4B), and they did not appreciably increase expression of HSP70 or HSP90 (compared to severe heat stresses at 42°C) in FANCA-null or wild-type cells (Figures 4D and Figure S3D). This emphasizes the mild nature of these treatments. However, 40°C clearly imposed a greater demand on H SP90 function because growth became sensitive to low-level HSP90 inhibition (Figure S4A).
Figure 4. Exposure to febrile-range temperatures phenocopies low-level HSP90 inhibition.
(A–C) MMC sensitivity of FANCA-null GM6914 cell-lines expressing the indicated FANCA mutants recovering under normal conditions (DMSO control) (A), at a febrile-range temperature (39°C) (B), or in the presence of low-level HSP90 inhibition (ganetespib, 5 nM) (C). Data presented as mean ± SEM from 4 independent experiments; these experiments are distinct from those in Figure 3B–C.
(D) Effect of low-level HSP90 inhibition (Gan: ganetespib, 5 nM O/N) vs. exposure to febrile-range temperatures on HSP70 or HSP90 levels in FANCA-null (GM6914) cell. Cells underwent transient (39°C 2h or 42°C 2h; including 11-hour recovery at 37°C to allow for HSP induction; heat-shock protein) or prolonged overnight (39°C) exposure. Both constitutive (C, upper band, Anti-HSC70) and inducible (I, lower band, Anti-HSP70) forms of HSP70 are indicated, compared to HSP90, as determined by Western blotting with specific antibodies.
(E) Effects of low-level HSP90 inhibition (Gan: ganetespib, 5 nM vs. 39°C) on FANCD2 ubiquitylation in cells exposed to hydroxyurea (1 mM; 24 h). Samples from FANCA wild-type GM6914 cells were analyzed by Western blotting with antibodies against the indicated proteins. un indicates the unmodified FANCD2 protein, and ub the monoubiquitinated species.
(F) Mutant-specific chemo-sensitization by temperature increase to febrile-range (39°C). Hierarchical clustering of significant relative sensitization by febrile-range temperatures (z-score >2) of FANCA-null (GM6914) cells expressing the indicated FANCA mutants reveals two FANCA mutant clusters (HSP90-buffered mutants: red; non-buffered mutants: blue) on the basis of their differential sensitization to FA-related drugs (green cluster). Febrile-range temperature also sensitized cells to non-FA related drugs (purple cluster), but these effects were not mutant-specific. See also Figure S4
FA pathway function in wild-type cells, as measured by MMC sensitivity, was unaffected by 39°C and was only mildly impaired by 40°C (Figure S4C). The HSP70-preferring mutant proteins, R951W, E936G and T724P, which were highly sensitive to MMC at 37°C (see above) remained so at elevated temperatures (Figure S4B). In contrast, increased temperatures selectively sensitized cells expressing the HSP90-buffered R880Q and I939S FANCA mutants to MMC (Figures 4A–B and S4D). In addition, elevated temperatures eliminated residual FANCD2 ubiquitylation in the presence of a standard genotoxic stress in R880Q and I939S but not wild-type cells, also phenocopying low-level HSP90 inhibition (Figure 4E).
The selective sensitivity of the buffered mutants to elevated temperatures held true for other FA-relevant agents such as carboplatin, bendamustine and lomustine (Figures S4E–J). Indeed, the effect of increased temperature was comparable in magnitude to that of pharmacological HSP90 inhibitors (Figures 4C and S4K–L). Elevated temperatures also sensitized cells to the same non-FA related drugs that HSP90 inhibitors sensitized cells to (Figures 4F and Figure S3K). But these effects were not specific to FANCA mutations. We conclude that HSP90 specifically supports the function of HSP90-buffered FANCA mutants under basal conditions, and that the increased HSP90-dependence of these mutants renders the entire FA pathway more sensitive to a physiologically relevant environmental stress.
An anemia-preventing compensatory mutation in twins eliminates HSP90-dependence and environmental sensitivity
Somatic mutations in pathway components frequently arise spontaneously in the bone marrow of FA patients. The nature of some of these mutations is compensatory or protective. Such mutations progressively rescue bone marrow failure, as the hematopoietic compartment slowly becomes a genetic mosaic through Darwinian selection of the hematopoietic stem cells that have acquired the protective mutations. In most of these cases the compensatory mechanism involves frame restoration or back mutation of the FA-causing mutation to the wild-type residue (Joenje and Patel, 2001). However, in rare cases compensatory somatic mutations at separate sites can restore the function of mutant FA proteins while retaining the disease-causing missense substitutions. We took advantage of such a case involving monozygotic twins to examine the role of HSP90 in buffering naturally occurring genetic variation.
Both of the twins displayed atypical clinical courses due to the acquisition of a compensatory mutation (Mankad et al., 2006). At birth, each twin had different, moderately severe congenital anomalies (skeletal, skin and urinary tract abnormalities) characteristic of FA (Poole et al., 1992). Both carried a FANCA null mutant and the HSP90-buffered R880Q mutant (patients FA-A19 and FA-A25) (Mankad et al., 2006). The R880Q mutant has been confirmed to cause FA in several unrelated cases (Castella et al., 2011) (see also FA Mutation Database). One twin had no blood phenotype and the other had a blood defect that became corrected later in life (Poole et al., 1992). Correction of the twins’ blood defects is attributed to the early acquisition of compensatory mutation E966K in one twin’s bone marrow that was transferred to the other twin via intra-uterine circulation later in the pregnancy (Mankad et al., 2006).
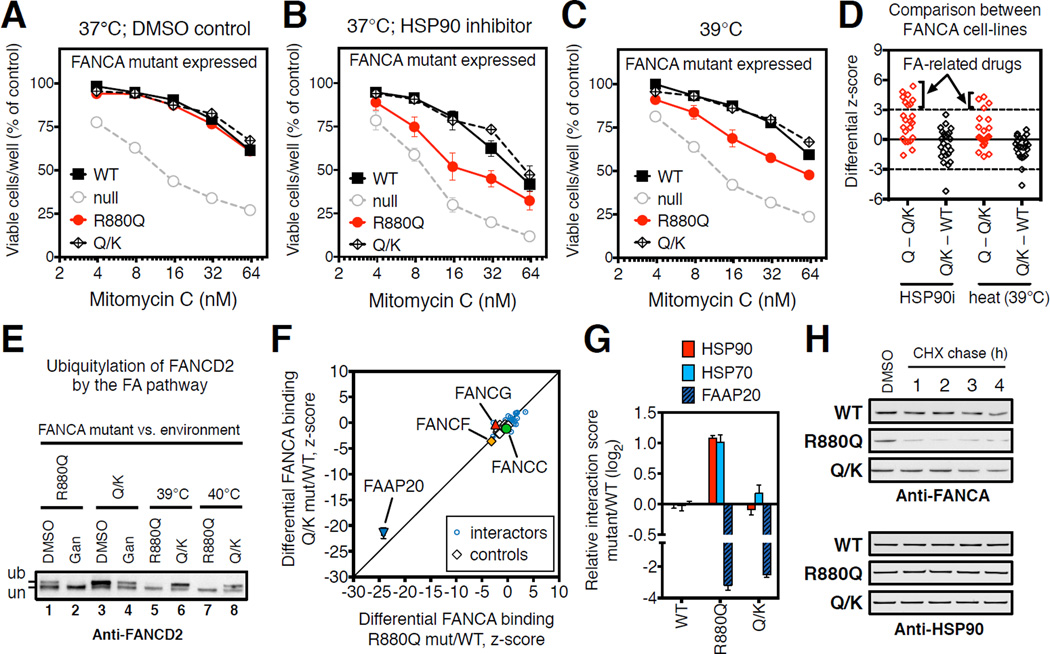
To determine the effect of the compensatory mutation on the HSP90-dependence of mutant FANCA, we compared FANCA-null cells stably expressing the R880Q or the R880Q/E966K double mutant (Q/K). The double mutant was expressed similarly to the single R880Q mutant and wild-type proteins (Figure S5A). Under basal conditions, the double mutant, the R880Q single mutant and wild-type cells had the same MMC tolerance (Figures 5A and S5B). However, double mutant cells retained normal MMC tolerance even in the presence of low-level HSP90 inhibition, which strongly sensitized cells carrying the R880Q single mutant (Figures 5B and S5C). The double mutant also retained MMC tolerance at the same febrile-range temperatures that impaired tolerance in the R880Q single mutant (Figure 5C).
Figure 5. Compensatory mutation prevents progression of anemia by restoring normal HSP90-dependence of FANCA protein.
(A–C) Sensitivity to MMC of FANCA-null GM6914 cells expressing the indicated FANCA mutants. FANCA-Q/K encodes both the R880Q FA-causing mutation and the E966K compensatory mutation. Sensitization by pharmacological low-level HSP90 inhibition (ganetespib, 5 nM) (B) is compared to sensitization by febrile-range temperature (39°C) (C). Data presented as mean ± SEM from 4 independent experiments and obtained from the same experiment as Figures 4A–C; data for WT, R880Q and null were repeated here for comparison.
(D) Differential chemosensitization of the indicated FANCA mutant cell-line pairs (R880Q vs. R880Q/E966K, Q – Q/K; R880Q/E966K vs. wild-type, Q/K – WT) by low-level HSP90 inhibition (HSP90i: ganetespib, 5 nM) and heat (39°C). Each point represents a different drug from Figure 3E. Dotted lines indicate significance thresholds (differential z-score >3 and <-3) in both biological replicates. Means are plotted. Arrows indicate mutant-specific sensitivities that are significantly different between single (Q, R880Q) and double (Q/K, R880Q/E966K) mutant cell-lines.
(E) FANCD2 ubiquitylation in response to DNA replication stress (hydroxyurea, 1mM; 24h) is impaired in FANCA-R880Q but not FANCA-Q/K cells by low-level HSP90 inhibition (ganetespib, 5 nM; Gan) and increased temperatures (39°C or 40°C) as compared to DMSO controls (DMSO). un indicates the unmodified FANCD2 protein, and ub the monoubiquitinated species.
(F) Differential LUMIER interaction scores of FANCA-R880Q mutant (x-axis) and FANCA-Q/K mutant (y-axis) as compared to wild-type FANCA. Specific FANCA partner proteins are indicated.
(G) Differential LUMIER interaction scores for the indicated FANCA mutants relative to wild-type for binding to FAAP20, HSP90 and HSP70. Data presented as mean ± SEM from 4 independent experiments.
(H) Cycloheximide (CHX) chase of the indicated FANCA mutant-expressing cells in the presence of hydoxyurea (1 mM). Samples were collected at the indicated time-points after CHX addition (20 µg/ml) and analyzed by Western for FANCA and HSP90. See also Figure S5 and Table S4
These results suggest that the compensatory E966K mutation makes the entire pathway less HSP90-dependent and less environmentally sensitive. Indeed, this same compensatory mutation imparted functional robustness to the FA pathway alleviating all chemosensitizing effects of low-level HSP90 inhibition and heat (Figure 5D). In addition, the compensatory mutant fully restored FANCD2 ubiquitylation in the presence of DNA replication stress (compare lanes 1 and 3; Figures 5E and S5A). FANCD2 ubiquitylation in cells with the double mutant remained robust even upon reducing HSP90 buffer capacity by low-level HSP90 inhibition or heat (compare lanes 2 to 4, 5 to 6 and 7 to 8; Figure 5E). Hence, a single coding substitution in the 1,455 amino-acid-long FANCA protein contributes to disease by increasing the HSP90-dependence and environmental sensitivity of the FA pathway.
HSP90 directly links genotype to phenotype in Fanconi Anemia
To investigate HSP90’s mechanistic connection to FA in the context of these twins, we examined the scaffolding function of FANCA in a more comprehensive way than has previously been undertaken. FA has no catalytic activity of its own, but is an essential coordinator of the FA core complex (11–14 reported members). We constructed an expression library of 383 human FLAG-tagged proteins involved in some aspect of genome maintenance, processes related to or parallel to the FA pathway (Table S4A).
First, we measured the ability of these genome maintenance factors to stably associate with wild-type FANCA. In addition to the established, direct FANCA partners, FANCG and FAAP20 (Ceccaldi et al., 2016), we identified 27 interactions that could be detected with confidence, many of which are novel (Figure S5D; Table S4B). Next, we compared the interactome of wild-type FANCA to that of the single and double mutants. The R880Q mutant bound to FAAP20 much more weakly (∼8-fold) and bound FANCF slightly more weakly than did the wild-type protein (Figures 5F–G and S5E). Interactions with the remaining 27 genome maintenance factors were not affected (Figures 5F and S5E). Increased binding of the R880Q mutant to the general chaperone HSP90 may thus be a consequence of the reduced ability of the mutant to bind FANCA-specific proteins with chaperone-like function such as FAAP20. The double mutant exhibited a similar reduction in its association with FAAP20 (Figure 5G) and FANCF proteins. Interactions with the other 27 genome maintenance factors were also unchanged by the compensatory mutation. Furthermore, in assaying the entire library of 383 maintenance factors the double mutant did not gain any new interactions (Figure S5F). Thus, changes in the normal FANCA interactome do not explain the phenotypic consequences of these mutations.
In contrast to the absence of effects on the broad interactome of FANCA, the compensatory mutant markedly decreased the HSP90 binding of the R880Q mutant as well as its HSP70 binding (Figures 5G and S5G). Consistent with normalization of its HSP90 binding, the double mutant was also much more resistant to proteasomal degradation upon low-level HSP90 inhibition than the R880Q single mutant protein (Figures 5H and S5H). Further supporting the concept of HSP90-mediated buffering, we identified another compensatory mutation (E966A) (Gross et al., 2002) which reduced HSP90 binding to an unrelated FANCA mutant (R951Q), but did not restore its binding to FAAP20 (Figures S5I–J). Interestingly, both this compensatory mutant and the compensatory E996K mutant, which we found to reduce the HSP90-dependence of disease-causing human FANCA mutants, are actually the wild-type sequences in other organisms. These compensatory mutations also reduced the low level of chaperone binding to FANCA detectable even in the absence of a disease-causing mutation, but they had no measurable effect on the HSP90-dependence of the FA-pathway in their own right (Figures S5I–K).
Overall, we conclude that genetic variations affecting the folding of proteins shape their HSP90-dependence and the environmental sensitivity of the FA pathway. These effects can directly contribute to the evolving clinical course of this disease.
Discussion
This work demonstrates that HSP90 can modify the consequences of genetic variation in man, something long hypothesized but never shown. It also provides insights into the mechanisms by which HSP90 buffering can alter the course of human diseases. HSP90 buffering appears to be broadly protective, mitigating the deleterious effects of missense mutations. In our survey of over 600 Mendelian diseases and >2,300 normal and mutant proteins, 11% showed increased interaction with HSP90 to an extend equal or greater than HSP70, which is characteristic of buffering. For FANCA alleles, HSP90 rescues FA pathway function by directly binding and stabilizing the encoded mutant proteins. The beneficial ramifications of HSP90 buffering, however, come at the cost of linking genetic variation to common, mildly stressful changes in the cellular environment. The environmentally sensitive ability of HSP90 to shape the manifestations of genetic variation therefore provides a plausible mechanism to explain, at least in part, the variable expressivity of many Mendelian disorders.
A long-standing question has been whether HSP90’s ability to buffer genetic variation is special compared, for example, to the other abundant cytosolic chaperone HSP70. It is well accepted that the biochemical functions of the two proteins are distinct. During forward folding of the proteome, HSP70 binds to extended hydrophobic stretches of amino acids before they become buried within the core of folded domains (Mayer and Bukau, 2005). In contrast, HSP90 preferentially binds to partially structured and metastable domains present in multi-domain late-stage folding intermediates (Pratt and Toft, 1997; Taipale et al., 2012) (Figure 1A). Our quantitative, high throughput analysis of protein::protein interactions indicates that HSP90, but not HSP70, can buffer human genetic variation by binding to the encoded proteins. In fact, HSP70 prefers to bind inactive mutant proteins. Such HSP70-engaged, functionally severe mutants presumably expose hydrophobic stretches typical of early-stage folding. Mutants with less severe protein-folding defects populate later-stage, metastable conformations typically recognized by HSP90. Consequently, increased binding to HSP90 to a degree equal or greater than HSP70 discriminates between HSP90-buffered environmentally sensitive mutations and non-buffered functionally inactive mutant proteins (HSP70 binding > HSP90) (Figure S1B). Thus, increased binding of a mutant protein to HSP90 cannot always rescue its function. Adding consideration of HSP70 engagement as a refinement greatly improves the ability to distinguish buffered from non-buffered alleles. Although not a focus of this work, it is important to recognize that these findings in human cells do not preclude a potential role for HSP70 in buffering, especially in prokaryotes where the role of HSP90 is much less evident.
Focusing on FA as a paradigmatic genetic disorder, we show that HSP90 buffering can alter the mapping of specific genotypes onto disease phenotypes. Many mutations in FANCA yield simple loss-of-function alleles and thereby cause highly penetrant clinical symptoms. Some mutations, however, have a more modest effect because HSP90’s protein-folding activities help maintain the function of the proteins they encode. Reducing the availability of free functional HSP90, either by specific chemical inhibitors or by exposure to protein homeostatic stresses, reveals the detrimental effects of buffered FANCA mutations on FA pathway function. These effects increase sensitivity to febrile range temperatures and a diverse yet highly specific set of drugs. The ability of HSP90 to render genetic variation conditional on the environment may provide one explanation for why the same FA-causing missense alleles manifest with enormous clinical variability even within kindreds of genetically similar patients.
A particularly compelling example is provided by a serendipitous “experiment of nature” involving the compensatory mutation we studied in twins with FA. Compared to wild-type FANCA, the germline FANCA (R880Q) mutant in these twins is much more dependent on HSP90 folding and much more sensitive to mild clinically relevant increases in temperature. The compensatory mutation (E966K) arising in the bone marrow of these patients, which prevented the progression of anemia (Mankad et al., 2006), also corrects the protein-folding defect of the original single mutant. Likewise it reduces HSP90-dependence to wild-type levels and eliminates the environmental sensitivity of the entire FA pathway. This somatic mutation occurred early in the hematopoietic compartment of one fetus and was transferred to the other via twin-twin transfusion at a later point in gestation (Mankad et al., 2006). However, the somatic tissues of these twins did not acquire the compensatory mutation, and thus expressed only the environmentally sensitive R880Q single mutant form of FANCA protein.
Notably, the twins had relatively mild clinical presentations, but their individual phenotypes were highly discordant. In fact, the clinical criteria for FA diagnosis were satisfied only when the FA features of both twins were considered in aggregate. This unusual qualitative discordance led to the suggestion that the twins were exposed to FA-modifying environmental factors early in life (Poole et al., 1992). In light of our findings now, the increased environmental sensitivity of the R880Q mutant could have exacerbated the clinical severity of this mutant in response to proteotoxic stresses that reduce free functional HSP90. Elevated temperatures and many other environmental stressors are known to influence HSP90’s buffering activity (Jarosz et al., 2010). Thus, although unknowable stochastic factors likely also made a contribution (Biesecker and Spinner, 2013; Burga et al., 2011), we suggest that proteotoxic, possibly preventable environmental stressors contributed to the discordant phenotypes by perturbing HSP90 buffering.
In addition to their developmental anomalies, patients with FA and other DNA repair defects are greatly predisposed to developing cancers due to genomic instability (Hoeijmakers, 2001). By buffering deleterious mutations within DNA repair pathways HSP90 could provide a powerful link between genomic instability and the severity of proteotoxic stresses within tumors. That is, when a tumor cell is not well adapted to its environment and protein homeostasis becomes compromised, novel genetic alterations would appear at higher rates than when it is well adapted. Such mechanisms could also facilitate the rapid emergence of drug resistance within tumors (Whitesell et al., 2014). Thus, HSP90 buffering may contribute to cancer initiation and progression through its role as a capacitor for both germline and somatic mutations within DNA repair genes.
Does the literature provide any additional evidence for environmentally sensitive HSP90-buffered mutations shaping the trajectory of other human diseases? Indeed, some of the missense mutants we identified as HSP90-engaging in our high-throughput assay have been reported to manifest variable and intriguingly fever- or temperature-dependent phenotypes at both the cellular and clinical levels (Tables S1A–B). We suggest that by buffering these kinds of human genetic variation, HSP90 enables gene-environment interactions capable of shaping disease trajectories. Exploiting chaperone interactions as indicators of the disease susceptibility conferred by sequence variations within an individual’s genome could increase the precision of patient diagnosis, risk stratification and management.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Please contact the Lead Contact Luke Whitesell, whitesell@wi.mit.edu, with any request regarding reagents used in this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture and Stable Cell Lines
HEK293T and GM6914 derivative cell lines were propagated in DMEM supplemented with 10% fetal bovine serum and penicillin/streptomycin. GM6914 FANCA-null cells (SV40-transformed human skin fibroblasts from a 12 year-old patient with severe pancytopenia) were engineered by transduction with retroviruses (pMMP vector backbone) encoding various forms of the FANCA polypeptide, as previously described (Adachi et al., 2002). Sensitivity to various drugs was subsequently measured in duplicate by celltiter-glo (Promega) and propidium iodide staining. Drugs were stored in DMSO accordingly to the manufacturer’s specifications. Reporter cell lines stably expressing Renilla luciferase (Rluc)-tagged HSP90 (HSP90β), Rluc-HSP70 (HSPA8), or Rluc alone (negative control) were previously described (Taipale et al., 2012). The stable polyclonal HEK293T reporter cell lines expressing Rluc-FANCA or mutant Rluc-FANCA mutants R880Q or Q/K (R880Q/E966K) were created using lentiviral infection. All cell-lines have been authenticated using PCR-sequencing and Western blotting.
METHOD DETAILS
Selection of FANCA variants
50 FANCA disease-causing missense mutations and an equal number of common FANCA variants and polymorphisms of unknown clinical significance were collected from multiple sources a detailed description of which can be found in Table S2A (related to Figure 2). Additional FANCA mutations were derived from the literature as indicated. Full-length human FANCA cDNA sequence corresponds to NCBI reference transcript NM_000135.
High-Throughput Mutagenesis
To generate the FANCA variant library, we applied a previously described high-throughput site-directed mutagenesis pipeline (Sahni et al., 2015). We performed a 3-step PCR experiment, involving two primary PCRs and one fusion PCR to obtain a desired mutated ORF. All amplification reactions were performed with with KOD HotStart Polymerase (Novagen). For the primary PCRs, two universal primers, Tag1 (5’-GGCAGACGTGCCTCACTACTCGCGTTTGGAATCACTACAGGG-3’) and Tag2 (5’-CTGAGCTTGACGCATTGCTAGGAGACTTGACCAAACCTCTGGCG-3’), and two mutation-specific internal forward (MutF) and reverse (MutR) primers (∼40bp) were employed. The two ORF fragments flanking the mutation of a FA gene were amplified using the primer pair Tag1 and MutR, and the primer pair Tag2 and MutF, respectively. For the fusion PCR, the two primary PCR fragments were fused together using the primer pair Tag1 (5’-GGCAGACGTGCCTCACTACT-3’) and Tag2 (5’-CTGAGCTTGACGCATTGCTA-3’) to generate the mutated allele. The final product was a full-length FANCA ORF harboring the desired mutation. To create mutation Entry clones, all mutant ORFs were cloned into the Gateway donor vector, pDONR223, by Gateway recombination cloning. ENTRY vector encoding the full-length FANCA was obtained from Harvard Plasmid ID Database. Inserts were validated using restriction digestion (BsrGI, NEB) and sequencing.
Confirmation of Mutations By Sequencing
For sequence confirmation mutant ORFs from single colonies were PCR amplified with KOD HotStart Polymerase (Novagen) using M13G-FOR (5’-CCCAGTCACGACGTTGTAAAACG-3’) and M13G-REV (5’-GTGTCTCAAAATCTCTGATGTTAC-3’) as primers. To confirm mutations, the resulting PCR products were processed for Illumina next-generation sequencing. Reads from the next-generation sequencing runs were assembled and aligned to the reference ORF sequences. Clones with full-length coverage that contained only the desired mutation were selected and consolidated.
Selection of Genome Maintenance Factors
A list of 1,269 human genes with a known function in genome maintenance was generated as defined by KEGG pathway, Gene Ontology ID (EMBL-EBI Quick GO), Uniprot annotation, PPI analysis (BioGrid) and literature curation. This list of genome maintenance factors was refined by excluding genes encoding for specialized meiotic factors, mitochondrial components, and proteins with a primary function in apoptosis, metabolism, stress signaling, protein homeostasis, cell-cycle regulation, chromatin remodeling or transcription. 792 ORF clones were collected from the human ORFeome (Yang et al., 2011) (related to Figure 5).
Expression Library Cloning
All inserts were cloned using Gateway recombination into a pcDNA3.1-based mammalian expression vector containing an N- (for the case of FANCA variants) or C-terminal 3 × FLAG tag (Sahni et al., 2015; Taipale et al., 2012). Inserts were verified with restriction digestion and clones that could not be validated by restriction digestion or Sanger sequencing were excluded from further analysis. Some of these ORFs sequence-verified full-length clones were available and were obtained from Harvard Plasmid ID Database and Addgene. We generated two libraries of FLAG-tagged expression plasmids that passed all quality controls. The FANCA-Variant library contains 90 FLAG-tagged plasmids (Table S2A, related to Figure 2). The Genome-Maintenance-Factor library contains 544 FLAG-tagged plasmids encoding isoforms of 383 distinct genome maintenance proteins, and 44 unrelated control proteins (Table S4A, related to Figure 5). Inserts were validated using restriction digestion (BsrGI, NEB) and Sanger sequencing.
LUMIER Assay
PPI scores were determined using a quantitative LUMIER (luminescence-based mammalian interactome mapping) assay (Barrios-Rodiles et al., 2005; Taipale et al., 2012) that reports binary PPI scores. Stable HEK293T cell lines were expressing Renilla luciferease (Rluc)-tagged prey proteins (in this work, chaperones, FANCA wild-type, FANCA R880Q, or FANCA Q/K) were generated by lentiviral infection. These stable cell lines were transfected with libraries of plasmids encoding for FLAG-tagged bait proteins (FANCA wild-type vs. variants, and genome maintenance factors) in an arrayed fashion. Following cell lysis and capture of FLAG-tagged proteins, luminescence signals were measured and levels of bait proteins were assessed.
LUMIER assays were performed essentially as previously described (Taipale et al., 2012). 3×FLAG-tagged constructs were transfected in 96-well format in duplicates into HEK293T-derived reporter cell lines using polyethylenimine (Polysciences 24765). To avoid spatial gradients in transfection efficiency, plates were incubated at room temperature for 30 minutes before transfer to 37°C (5% CO2). Two days after transfection, cells were washed with 1×PBS using an automated plate washer (Biotek ELx406) and lysed in lysis buffer: 50mM HEPES [pH 7.9]/150mM NaCl/10mM MgCl2/20mM Na2MoO4s to stabilize HSP90::client interactions/0.7% TritonX-100/5% glycerol, protease and phosphatase inhibitors. We included in the lysis buffer Benzonase (1 U) to reduce the viscosity of the samples by degrading nucleic acids (∼20 minutes at 23°C). This treatment facilitated the release chromatin-bound proteins, thereby increasing specific LUMIER signal for PPIs involving certain genome maintenance proteins. Subsequently, lysates were transferred with an automated liquid handler (Tecan) into 384-well plates that had been coated with monoclonal anti-FLAG M2 antibody (Sigma-Aldrich, F1804) and blocked with 3% BSA/5% sucrose/0.5% Tween 20. Plates were incubated at 4°C for 3 hours, after which plates were washed with lysis buffer using an automated plate washer. Luminescence was measured with a plate reader (Perkin-Elmer Envision) using Renilla luciferase kits (Promega).
To determine FLAG bait protein levels, after luminescence measurement, HRP-conjugated anti-FLAG antibody in ELISA buffer (1×PBS, 2% goat serum, 5% Tween 20) was added to wells. Plates were incubated for 90 minutes at room temperature and were subsequently washed in 1×PBS/0.05% Tween 20 with an automated plate washer. ELISA signal was detected using a chemiluminescent substrate (Thermo Scientific, 37069). Changing the position expression constructs occupied within the plate had no effect on protein expression levels or LUMIER signal. All FLAG-tagged protein baits were tested for unspecific binding to Renilla luciferase in cells stably expressing the luciferase (Rluc-only) and proteins that exhibited greater than background signal were excluded from further analysis.
Estimation of Functional Impact of Mutation
The functional impact of a mutation on a human protein was determined as previously (Sahni et al., 2015). Results are presented in Tables S1B and S2C, respectively for previously characterized disease mutants and FANCA variants. Using a qualitative measurement, we classified each mutation as probably damaging, possibly damaging, or benign, based on the false positive rate (FPR) thresholds: Mutations with their PolyPhen-2 scores associated with estimated FPRs at or below 10% are predicted to be probably damaging, between 10% and 20% as possibly damaging, above 20% as benign. Otherwise, the prediction would return an NA result. These predictions were similar to those obtained by SuSPect, which employs sequence conservation and multiple network-level features to calculate a probability score for the functional severity of missense mutations. SuSPect results are reported on a 0–100 scale with the highest number characterizing mutations predicted to be most damaging.
Co-immunoprecipitation and Western Blotting
1.2×106 or 107 HEK293T or GM6914-derivative cells, respectively, were lysed in the same buffer as for LUMIER (300 µl and 1 ml, respectively) and were subjected to co-immunoprecipitation (co-IP) as previously described (Taipale et al., 2012). Total cell extracts were incubated on ice for 20 minutes to digest nucleic acids and spun at 14,000 x g for 10 minutes to precipitate insoluble material. A small aliquot of the soluble extracts were collected as input control and the remaining extracts were subjected to immunoprecipitation. HEK293T cellular extracts containing different FLAG-tagged FANCA variants were incubated with anti-FLAG beads at 4°C for 3 ho urs. For extracts from GM6914-derivative cells, non-tagged FANCA variants were immunoprecipitated with a specific anti-FANCA antibody (2 µg, Bethyl) with subsequent capture of the immune complexes on protein G magnetic beads (50 µl, Thermo Fischer). Beads were then washed in lysis buffer to remove non-specific interactions, boiled at 65°C for 10 minutes in 8 M urea-based loading buffer, loaded on Bis-Tris 4–12% PAGE, and analyzed by Western blotting on PVDF membranes. For Figures 4D–E, 5E, 5H, S2L, S3D, S3I, S5A and S5H cells were cultured in 6-well plates and collected by trypsinization (Accumax), pelleted and directly lysed by boiling in 8 M urea-based loading buffer in order to preserve ubiquitylated FANCD2 species.
Chemosensitivity Assays
GM6914-derivative cells expressing different FANCA variants were seeded in 384-well plates at density of 500 per well (20 µl media volume) and were incubated for 24 hours at 37°C 5% CO2 in a humidified chamber. The next day, DMSO or Ganetespib (5 nM) was first added to each well using an Evoware liquid handling robot (Tecan). Immediately after, additional compounds were administered using nano-dispensers, as follows. For the anti-cancer compound library, drugs were arrayed in four doses in 10-fold dilutions and 200 nl each of four concentrations (starting stock concentration 10 mM) were transferred to each well using an automatic nanoliter 384-well dispensing system in duplicates. For all other viability experiments HSP90i and compounds were administered either simultaneously using the Evoware system or sequentially using a digital dispenser D300 (Hewlett-Packard). Results were comparable irrespective of the order of treatments. Cells were incubated at the indicated temperature in a humidified chamber for 3 days. Relative cell viability was calculated as the mean luminescence of treated samples normalized by the signal for untreated controls. Sensitization z-scores were calculated based on 64–144 DMSO-treated controls in each 384-well plate; z-scores >3 were considered significant. Relative fold-increase in positive staining with propidium iodide compared to untreated controls was measured in duplicate in 96-well plates using a flow cytometer in duplicates (Guava Technologies). Pearson coefficient of determination threshold for high correlation between biological duplicates was set to R2 > 0.92. Plotted values represent mean ± SEM from multiple independent experiments; error-bars that are not visible are smaller than the symbol representing the mean.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are expressed as the mean ± the standard error of the mean (SEM), unless otherwise stated in the figure legend.
LUMIER Data Normalization and Scoring
Luminescence z-scores were calculated for each separate well and significant protein::protein interactions (z-score >2.5) were averaged on an individual protein basis from 2 or 4 biological replicate values from each individual experiment. Correlation between replicate values was consistently greater than 0.96 (R2). To avoid subsequent normalization artifacts due to low expression of bait proteins, only luminescence values from wells with significant ELISA signal (z-score of log2 values >2.5) were considered, yielding at least 2 biological replicate interaction values for most protein variants. HSP90 and HSP70 binding z-scores were corrected for prey-dependent differences in background luminescence to make them comparable (Figure 1C) (Sahni et al., 2015). To determine absolute chaperone preference, HSP70 binding z-scores were normalized by HSP90-binding z-scores (HSP70 – HSP90) for mutant and corresponding wild-type proteins, respectively (Figure S1C).
For FANCA variants (related to Figure 2), interaction scores for each bait::prey pair were defined as the log2 value of luminescence over ELISA ratio. Interaction scores for each FANCA variant were normalized to 8–12 FANCA wild-type control wells per plate per experiment to calculate differential interaction scores from multiple independent experiments against HSP90 or HSP70 binding (Table S2B). Differential HSP90 and HSP70 interaction scores for FANCA wild-type, similarly to log-transformed luminescence signals, fit normal distributions, and thus a z-score value for each differential interaction score was calculated. Differential chaperone binding between mutant and corresponding wild-type protein, reported as differential z-scores (mut/WT) for FANCA variants, also reports on absolute chaperone preference, because interactions scores between wild-type FANCA protein and HSP90 or HSP70 and their standard deviations are comparable. Here we focus on relative increases in chaperone engagement caused by protein-folding missense mutations. Relative reduction in chaperone binding could not be examined, due to smaller effect sizes and increased signal variability. A z-score cutoff equal to 2 was employed to identify variants causing greater engagement for HSP70 over HSP90 (70>90).
To compare the protein::protein interactomes of different FANCA protein variants (Figures 5F, S5D–F), instead of calculating interaction scores that are susceptible to artifacts introduced by low protein expression, luminescence z-score values were adjusted against differential expression levels (z-scores) of bait (anti-FLAG ELISA) and prey (10% input luminescence). This approach also improves comparison between preys by avoiding errors due to differences in ELISA efficiency across different bait proteins, which do not apply when FANCA variants are used as baits. Adjusted luminescence z-scores were highly reproducible across independent replicate experiments (R2 > 0.92).
Comparing Chaperone Engagement to Functional Severity
To compare chaperone binding to clinical phenotype we employed a publically available dataset generated in our previous work (Sahni et al., 2015). A list of 3,324 mutant and cognate wild-type proteins assayed for relative binding (mut/WT z-scores) to HSP90 (HSP90β) and HSP70 (HSPA8) was refined as follows. Data were filtered for protein expression using an ELISA threshold z-score >2.5. Among the remaining 1,628 FA-unrelated mutants (and 714 cognate wild-type proteins) ∼30% exhibit increased binding to key components of the protein homeostasis machinery, suggesting they are protein-folding mutants (related to Figure 1). Mutants exhibiting no change in binding to the protein homeostasis machinery may still be functionally compromised, yet by different mechanisms than protein-folding defects the examined protein homeostasis components recognize (Sahni et al., 2015).
Here we investigate how the two major cytosolic chaperones, HSP90 and HSP70, “see” mutant proteins. Our analysis does not take into account the binding of mutants to ER-resident chaperones, cofactors and partner proteins. Among 360 mutants (∼22%) in our dataset of primarily cytosolic proteins with increased binding to HSP90 or HSP70 (z-score >2.5), we found 32 mutants (in 20 different proteins) for which clinical severity had been previously reported (OMIM, http://omim.org/). Mutants were assigned to “severe”, “moderate/atypical”, or “mild” clinical severity classes, to most accurately represent previous assignments. References (PubMed ID) on the clinical and molecular data employed for this analysis are provided in Table S1A. Pairs of mutations with disparate clinical phenotypes and differential chaperone engagement were identified within 5 genes (labeled 1–10, Figure 1C). Statistical significance was determined by two-sided Fisher’s exact 2×3 extension test.
Comparing our original threshold delta z-score 2.5 (which indicates a probability of 0.00621 of being false positive) to increasingly stringent thresholds (delta Z = 3, p = 0.00135; delta Z = 4, p = 3.2 ×10−3; delta Z = 5, p = 3.0019 × 10−5) we find that variants with increased chaperone binding can be found in the polymorphism group at each threshold. Even for the most stringent threshold, ∼11% of variants show increased binding to HSP90 and ∼15% to HSP70. This suggests that some of the polymorphisms we examined here may introduce protein-folding defects and may thus be deleterious. Also, the fraction of variants with increased chaperone binding is significantly enriched in the disease group as compared to the polymorphism group at all thresholds (Figure 2D; HSP90: z=3, p= 0.0039; z=4, p = 0.0142; z=5, p=0.05; and HSP70: z=3, p= 0.0004; z=4, p<0.0001; z=5, p=0.0001). Similarly, the same pattern of chaperone engagement for mutants and polymorphisms is retained at all thresholds (z=3 p= 0.0122, z=4 p= 0.0007, z=5 p=0.001). The functional analyses for HSP70-engaging vs. HSP90-engaging mutants (Figures 1D and 2G) are also robust to moving thresholds. We kept differential z-score 2.5 as our default threshold because this value is in best agreement with our validation experiments involving co-immunoprecipitation of endogenous chaperones.
For determining differential chaperone engagement across FANCA missense mutants and polymorphisms, variants with available interaction scores for both HSP90 and HSP70 from at least two independent experiments were included (mutants N = 37, polymorphisms N = 46). Statistical significance was determined by one-sided Fisher’s exact test. Mutants were grouped on the basis of their functional severity as determined by the ability to support FA pathway function when expressed in GM6914 FANCA-null cells. This metric of mutant functional severity was compared to their differential chaperone engagement. Statistical significance was determined by two-sided Fisher’s exact 2×3 extension test. Conclusions are robust to changing the chaperone-binding significance and differential chaperone engagement cutoffs.
ADDITIONAL RESOURCES
Plasmids encoding genome maintenance factors and chaperones were obtained from the human ORFeome collection (Yang et al., 2011), Harvard PlasmID Database and Addgene.
Constructs encoding FLAG-tagged, disease-associated, mutant and variant, non-FA, proteins were previously described by (Sahni et al., 2015).
Supplementary Material
(A) LUMIER interaction scores of the indicated FLAG-tagged bait proteins against HSP90 and HSP70. HSP90 client, v-SRC; HSP90 co-chaperone, CDC37; HSP70 client, p53 (TP53); HSP70 co-chaperone, BAG1, were used as controls. Interactions not significantly over background are indicated as ND (not detected). Data presented as the mean ± SD from 4-12 independent biological replicates.
(B) Protein-folding mutations vary in their severity as defined by the extent of protein destabilization they cause. Missense mutations shift the folding equilibrium towards a partially folded state thus increasing the cellular fraction of chaperone-bound molecules relatively to the wild-type protein (right of the WT protein on the chaperone-binding chart). Mild protein-folding mutants (Mut. 1) shift the folding equilibrium only slightly and as a result increase the abundance of partially folded chaperone-bound states. These include late-folding, HSP90-engaging and early-folding, HSP70-engaging conformations, as well as their intermediate states (HSP90::Hop::HSP70 complexes indicated by asterisks; Hop, shown in green, is an HSP90 co-chaperone required for the transfer of protein clients from HSP70 to HSP90). Severe protein-folding mutants (Mut. 2) more frequently occupy conformations that expose hydrophobic residues that are normally present in the core of the folded domain. Such conformations are the preferred substrate for HSP70. Thus Mut. 2 proteins remain at an equilibrium point distant from the functionally active conformations. Other mutations reduce chaperone binding by stabilizing the protein fold (left of the WT protein in the chaperone-binding chart).
(C) Absolute HSP70 vs. HSP90 binding preference for >1,500 disease-associated mutants as compared to their cognate wild-type proteins. Mutants with well-documented clinical phenotypes are grouped into three classes based on clinical severity (severe: filled blue triangles; moderate/atypical: open red diamonds; mild: filled red triangles). Two SOD1 mutants with distinct chaperone preferences and clinical severities are indicated.
(D) Clinical phenotype of the indicated SOD1 mutations as summarized by Wang et al., 2008. Number of patients from each severity class is indicated in parentheses. Symbols as indicated in (C).
(A) Luminescence z-scores of wild-type FANCA and negative control (PCNA protein, critical for DNA replication and repair) proteins for binding to HSP90 and HSP70. Data presented as the mean ± SEM from 12 independent biological replicates. Significance cutoff (z-score > 2) indicated by dotted line.
(B) Correlation between biological replicates of normalized LUMIER interaction scores for 61 FANCA variants relative to FANCA wild-type for binding to HSP90 (red triangles) and HSP70 (blue triangles). Correlations determined by Pearson coefficient of determination, R2.
(C-D) Differential interaction scores (log2) of HSP90 (C) and HSP70 (D) with FANCA variants exhibiting increased chaperone binding relative to wild-type FANCA protein.
Data presented as the mean ± SEM from multiple (3-5), independent LUMIER experiments.
(E) Distribution of luminescence signals (z-score) of 61 FANCA variants assayed by LUMIER in cells stably expressing Renilla luciferase alone.
(F) Validation of HSP90 binding to FANCA variant proteins by immunoprecipitation with anti-FLAG and Western blotting for endogenous HSP90. Mutants with increased HSP90 interactions are indicated in red/bold.
(G) Comparison between the relative binding of 61 FANCA variants to HSP90 and the effect of HSP90 inhibition (HSP90i: ganetespib, 100 nM, 20 h) on mutant FANCA protein steady-state expression levels relative to wild-type FANCA protein. FANCA variants with disparate chaperone engagement patterns are indicated (90≥70: HSP90-preferring; 70>90: HSP70-preferring; none: no increase in chaperone binding). Data presented as the mean of 2 independent biological replicates. Correlations by Pearson coefficient of determination, r.
(H) A linear representation of the FANCA protein shows the positions of 90 variant amino acids (x-axis) and their effect on chaperone binding relative to FANCA wild-type (mut/WT, mutant vs. wild-type; z-score). Differential interaction scores (z-scores) for HSP90 (red triangles) and HSP70 (blue triangles) (y-axis) were determined by LUMIER. (I) Differential HSP90 (x-axis) and HSP70 (y-axis) interaction scores for 90 FANCA variants categorized based on chaperone engagement pattern (70>90: preferential binding to HSP70 over HSP90; 90≥70: increased binding to HSP90 equal or greater than that of HSP70; none: no increase in chaperone binding).
(J) Differential HSP90 (x-axis) and HSP70 (y-axis) interaction scores for 11 previously characterized FANCA mutants belonging to the indicated phenotypic classes.
(K) Relative cell viability of FANCA null GM6914 cell-lines expressing the indicated FANCA mutants, wild-type FANCA protein or empty vector control (null). Cells were seeded at a low density and were incubated at 37°C for 24 hours. Cells were subsequently exposed to increasing concentrations of MMC and immediately incubated at 37°C for 3 days to allow recovery. Viability was then measured. Data presented as the mean ± SEM from 2 independent experiments. Error bars smaller than the symbol are not shown.
(L) FANCA mutant expression levels and FANCD2 ubiquitylation in the indicated GM6914-derived FANCA mutant cell-lines in response to DNA replication stress induced by hydroxyurea treatment (1 mM, 24h). Samples were analyzed by Western blot using the indicated antibodies. un indicates the unmodified FANCD2 protein; ub indicates the monoubiquitinated species. HSP90 serves as a loading control.
(M) Sensitivity (z-score) to various chemotherapeutic agents of FANCA null GM6914 cell-lines expressing the indicated FANCA mutants relative to wild-type. R951W, E936G and I939S cell lines, but not R880Q or wild-type, are hypersensitive to FA-related drugs.
(A-C) Relative effect of pharmacological HSP90 inhibition by (A) ganetespib, (B) onalespib (AT13387, 10-40 nM) or (C) PU-H71 (30-50 nM) on the viability of FANCA wild-type and FANCA-null cells under normal conditions or with increasing concentrations of MMC. Left panels in (A-C) indicate the effects of the indicated inhibitor on cell growth; middle panels the effects on MMC tolerance in FANCA-wild-type cells; right panels the effects on MMC tolerance in FANCA-null cells Data presented as the mean ± SEM from 2 independent experiments.
(D) Effects of low-level HSP90 inhibition (by ganetespib, 2-5 nM; PU-H71, 40-70 nM) or febrile-range temperature (39°C) on HSP70 and HSP90 protein levels in FANCA wild-type cells. High-level HSP90 inhibition (ganetespib, 25 nM; PU-H71, 300 nM) as well as proteotoxic proteasomal inhibition (MG132, 2.5 μM) induced the expression of HSP70. Constitutive (upper band: C) and inducible forms (lower band: I) of HSP70 are indicated. HSP90 levels are shown for comparison.
(E-G) Relative viability of MMC-treated, GM6914-derived R880Q FANCA mutant cells exposed to DMSO control or to low-level HSP90 inhibition with ganetespib, 5 nM (E), or low-level HSP90 inhibition by onalespib (AT13387) (F) or PU-H71 (G). Data presented as the mean ± SEM from 3 independent experiments for panel (E) and from 2 independent experiments for panels (F-G). Panels (F) and (G) are directly comparable to (B) and (C), respectively.
(H) Increased cell death of FANCA WT, R880Q, or null cells as determined by propidium iodide staining in the presence of mitomycin C (MMC) and DMSO control or ganetespib at 4 nM and 5 nM. Values presented as the mean ± SEM from 2 independent biological replicates.
(I) Effects of low-level HSP90 inhibition (Gan: ganetespib, 5 nM vs. PU-H71, 70 nM) on FANCD2 ubiquitylation in the presence of hydroxyurea (1 mM; 24 h). Samples from GM6914 cells expressing WT, R880Q or I939S FANCA proteins were analyzed by Western blotting with antibodies against the indicated proteins. un indicates the unmodified FANCD2 protein, and ub the indicates the monoubiquitinated species.
(J) Sensitivity of the indicated FANCA mutant-expressing cell lines to carboplatin in the presence of DMSO or low-level HSP90 inhibition (ganetespib, 5 nM). Data presented as the mean ± SEM from 3 independent experiments.
(K) Screen of 353 chemotherapeutics for drug sensitivities induced by HSP90 inhibition (HSP90i: ganetespib, 5 nM). Chemotherapeutics highlighted by purple triangles are significantly more toxic to FANCA wild-type cells upon HSP90i, as compared to the remaining drugs (grey squares). DMSO control is indicated by the black square. As an internal control for the assay itself, low-level HSP90i strongly sensitized the cells to other HSP90 inhibitors (17-AAG, 17-DMAG) as expected.
(A) Relative cell viability of the indicated FANCA mutant cell lines exposed to mild HSP90 inhibition (ganetespib, 5 nM; 3 days) or DMSO control at different temperatures. Data presented as the mean ± SD from 4 independent biological replicates.
(B) Relative cell viability of FANCA-null GM6914 cells expressing the indicated FANCA mutants after treatment with MMC followed by recovery at 39°C vs. 40°C. Cells were allowed to recover from exposure to increasing concentrations of MMC at 39°C or 40°C for 3 days. Data presented as the mean ± SEM from 2 independent experiments.
(C) Relative viability of FANCA wild-type cells after exposure to increasing concentrations of MMC and subsequent recovery (3 days) at the indicated temperatures. Data presented as the mean ± SD from 4 independent experiments.
(D-L) Relative cell viability of FANCA-null GM6914 cells expressing the indicated the FANCA mutants after exposure to the indicated chemotherapeutics at different temperatures, as described in (C). Effect of 40°C on MMC (D) and carboplatin tolerance (E). Bendamustine tolerance at normal temperature (37°C) (F), compared to 39°C (G) and 40°C (H). Recovery from lomustine at 37°C (I) or at 40°C (J). Recovery from exposure to bendamustine (K) or lomustine (L) at 37°C in the presence of low-level HSP90 inhibition (ganetespib, 5 nM). Data presented as the mean ± SEM from 3 independent experiments.
(A) FANCD2 ubiquitylation in response to hydroxyurea (0.2 mM for 24 h at 37°C) in null, WT, R880Q and Q/K FANCA mutant-expressing cell-lines. Western blotting against the indicated proteins; HSP90 is used as loading control. un indicates unmodified FANCD2 protein, and ub the monoubiquitinated species.
(B) Sensitivity (z-score) of FANCA-R880Q/E966K (Q/K) cells to various chemotherapeutics as compared to FANCA-null (red triangles) and FANCA wild-type cells (black spheres). Each dot indicates a different chemotherapeutic agent. MMC is indicated. Correlation between chemotolerance of Q/K cells and wild-type cells by Pearson coefficient of determination, R2.
(C) Effect of onalespib (AT13387) exposure on MMC tolerance of FANCA-Q/K-expressing cells (directly comparable to Figures S3B and S3F). Data presented as the mean ± SEM from 2 independent experiments.
(D) Correlation between biological replicate LUMIER interaction z-scores of FANCA protein partners (blue circles) among 385 human proteins involved in genome maintenance. Established FANCA partners FANCG, FAAP20, FANCC and FANCF are indicated. Correlations by Pearson coefficient of determination, R2.
(E) LUMIER z-scores of the indicated Renilla luciferase-tagged FANCA mutants interacting with the indicated FLAG-tagged FA pathway components. Data presented as the mean ± SD from 3 (or 9 for the case of FAAP20) biological replicates.
(F) LUMIER interaction z-scores for wild-type and Q/K mutant FANCA proteins against 385 human proteins involved in genome maintenance. Established FANCA partners FANCG, FAAP20, FANCC and FANCF are indicated. Error-bars indicate SD among biological replicates.
(G) Validation of HSP90 binding to the indicated FLAG-tagged FANCA variant proteins by immunoprecipitation with anti-FLAG and blotting for endogenous HSP90.
(H) FANCA depletion upon exposure to low-level HSP90 inhibition is proteasome-dependent. R880Q and Q/K cells were exposed to cycloheximide (CHX, 20 ug/ml, 2 hours) and hydroxyurea (1 mM) in the presence of DMSO (control), 5 nM ganetespib (Gan), or 5 nM ganetespib plus 20 uM proteasome inhibitor MG132 (Gan + MG132). Samples were analyzed by Western blotting for the indicated proteins, un indicates the unmodified FANCD2 protein, and ub the monoubiquitinated species.
(I) Differential LUMIER interaction scores for the indicated FANCA mutants relative to wild-type FANCA. Data presented as mean ± SEM from 3 independent experiments.
(J) HSP90 and FAAP20 binding to the indicated FLAG-tagged FANCA variant proteins by immunoprecipitation with anti-FLAG and blotting for endogenous HSP90 in stable cell lines expressing FAAP20 in fusion with Renilla luciferase (Rluc).
(K) Effect of ganetespib (5nM; dotted lines) on the MMC tolerance of FANCA-null cells engineered to stably express the FANCA-E966K mutant (red lines) as compared to cells expressing wild-type FANCA (black lines). Ganetespib-treated cells are indicated by the dotted lines, and DMSO-treated controls by the continuous lines. Data presented as the mean ± SEM from 6 independent experiments.
(A) Clinical data for indicated missense mutants and literature. (B) Chaperone engagement and predictions of functional severity for the same mutants.
(A) FANCA missense variants and variant sources. (B) Chaperone engagement of FANCA variants. (C) Phenotypic characterization of FANCA variants and literature.
(A) Chemosensitivities related to FA only. (B) Chemosensitivities related to HSP90 inhibition alone (HSP90i) or common to FA and HSP90 (both FA and HSP90i)
(A) List of Genome Maintenance Factors screened for the ability to bind FANCA. Each positive interaction represents a FANCA partner protein and is indicated as “HIT”. (B) Binding z-scores of FANCA partners to indicated FANCA mutant proteins.
Acknowledgments
We thank L. Clayton for reviewing and editing the manuscript. We are grateful to D. Hill for providing the human ORFeome and for constructive feedback. We thank J. Peng, G.L. Moldovan, members of the DFCI Department of Radiation Oncology, and J. Cheah and C. Soule at The David H. Koch Institute’s High Throughput Screening Facility for providing expertise and reagents. We also thank M. Taipale, D. Pincus, S. Lourido, M. Mendillo, C.A. McLellan and C. Kayatekin for constructive comments on the manuscript.
This work was supported by the Fanconi Anemia Research Fund (to S.L.), the Department of Defense (CDMRP BCRP W81XWH-14-1-0157 to S.L.), the Harold and Leila Y. Mathers Foundation, and the NIH (R37HL052725, RO1DK43889 and PO1HL048546 to A.D.D.; P50HG004233 and R01HG001715 to M.V.; RC4HG006066 to M.V., L.W. and S.L.). N.S. is supported by Cancer Prevention and Research Institute of Texas (CPRIT) New Investigator Award RR160021. S.L. was an investigator of the Howard Hughes Medical Institute. G.I.K. was supported by EMBO and HFSP Long-Term Fellowships.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
G.I.K., L.W., S.L. conceived the project. G.I.K. designed and performed the experiments with assistance from M.F. in performing the LUMIER assays, L.W. in performing chemosensitivity assays. N.S. and S.Y., under the direction of M.V., constructed the FANCA variant library and performed computational analyses. J.X., supervised by A.D.D., established FANCA mutant cell lines. A.D.D. and M.V. provided advice and constructive feedback. G.I.K., L.W., and S.L. wrote the manuscript, with significant contributions from all other co-authors.
References
- Adachi D, Oda T, Yagasaki H, Nakasato K, Taniguchi T, D’Andrea AD, Asano S, Yamashita T. Heterogeneous activation of the Fanconi anemia pathway by patient-derived FANCA mutants. Human molecular genetics. 2002;11:3125–3134. doi: 10.1093/hmg/11.25.3125. [DOI] [PubMed] [Google Scholar]
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- Biesecker LG, Spinner NB. A genomic view of mosaicism and human disease. Nature reviews Genetics. 2013;14:307–320. doi: 10.1038/nrg3424. [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Molecular and cellular biology. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480:250–253. doi: 10.1038/nature10665. [DOI] [PubMed] [Google Scholar]
- Castella M, Pujol R, Callen E, Trujillo JP, Casado JA, Gille H, Lach FP, Auerbach AD, Schindler D, Benitez J, et al. Origin, functional role, and clinical impact of Fanconi anemia FANCA mutations. Blood. 2011;117:3759–3769. doi: 10.1182/blood-2010-08-299917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nature reviews Molecular cell biology. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- Gross M, Hanenberg H, Lobitz S, Friedl R, Herterich S, Dietrich R, Gruhn B, Schindler D, Hoehn H. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet Genome Res. 2002;98:126–135. doi: 10.1159/000069805. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Taipale M, Lindquist S. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annual review of genetics. 2010;44:189–216. doi: 10.1146/annurev.genet.40.110405.090412. [DOI] [PubMed] [Google Scholar]
- Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nature reviews Genetics. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- Lindquist S. Protein folding sculpting evolutionary change. Cold Spring Harbor symposia on quantitative biology. 2009;74:103–108. doi: 10.1101/sqb.2009.74.043. [DOI] [PubMed] [Google Scholar]
- Mankad A, Taniguchi T, Cox B, Akkari Y, Rathbun RK, Lucas L, Bagby G, Olson S, D’Andrea A, Grompe M. Natural gene therapy in monozygotic twins with Fanconi anemia. Blood. 2006;107:3084–3090. doi: 10.1182/blood-2005-07-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cellular and molecular life sciences : CMLS. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Hayano T, Miyaso H, Takahashi N, Yamashita T. Hsp90 regulates the Fanconi anemia DNA damage response pathway. Blood. 2007;109:5016–5026. doi: 10.1182/blood-2006-08-038638. [DOI] [PubMed] [Google Scholar]
- Poole SR, Smith AC, Hays T, McGavran L, Auerbach AD. Monozygotic twin girls with congenital malformations resembling fanconi anemia. American journal of medical genetics. 1992;42:780–784. doi: 10.1002/ajmg.1320420606. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocrine reviews. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, Borowsky RL, Lindquist S, Tabin CJ. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL. Between genotype and phenotype: protein chaperones and evolvability. Nature reviews Genetics. 2003;4:263–274. doi: 10.1038/nrg1041. [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, Peng J, Weile J, Karras GI, Wang Y, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161:647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Tucker G, Peng J, Krykbaeva I, Lin ZY, Larsen B, Choi H, Berger B, Gingras AC, Lindquist S. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS biology. 2013;11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Johnson JL, Agar NY, Agar JN. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS biology. 2008;6:e170. doi: 10.1371/journal.pbio.0060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature reviews Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Santagata S, Mendillo ML, Lin NU, Proia DA, Lindquist S. HSP90 empowers evolution of resistance to hormonal therapy in human breast cancer models. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1421323111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Kim H, Moreau LA, Puhalla S, Garber J, Al Abo M, Takeda S, D’Andrea AD. RNF4-mediated polyubiquitination regulates the Fanconi anemia/BRCA pathway. The Journal of clinical investigation. 2015;125:1523–1532. doi: 10.1172/JCI79325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagasaki H, Hamanoue S, Oda T, Nakahata T, Asano S, Yamashita T. Identification and characterization of novel mutations of the major Fanconi anemia gene FANCA in the Japanese population. Human mutation. 2004;24:481–490. doi: 10.1002/humu.20099. [DOI] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, et al. A public genome-scale lentiviral expression library of human ORFs. Nature methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. Hsp90 selectively modulates phenotype in vertebrate development. PLoS genetics. 2007;3:e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) LUMIER interaction scores of the indicated FLAG-tagged bait proteins against HSP90 and HSP70. HSP90 client, v-SRC; HSP90 co-chaperone, CDC37; HSP70 client, p53 (TP53); HSP70 co-chaperone, BAG1, were used as controls. Interactions not significantly over background are indicated as ND (not detected). Data presented as the mean ± SD from 4-12 independent biological replicates.
(B) Protein-folding mutations vary in their severity as defined by the extent of protein destabilization they cause. Missense mutations shift the folding equilibrium towards a partially folded state thus increasing the cellular fraction of chaperone-bound molecules relatively to the wild-type protein (right of the WT protein on the chaperone-binding chart). Mild protein-folding mutants (Mut. 1) shift the folding equilibrium only slightly and as a result increase the abundance of partially folded chaperone-bound states. These include late-folding, HSP90-engaging and early-folding, HSP70-engaging conformations, as well as their intermediate states (HSP90::Hop::HSP70 complexes indicated by asterisks; Hop, shown in green, is an HSP90 co-chaperone required for the transfer of protein clients from HSP70 to HSP90). Severe protein-folding mutants (Mut. 2) more frequently occupy conformations that expose hydrophobic residues that are normally present in the core of the folded domain. Such conformations are the preferred substrate for HSP70. Thus Mut. 2 proteins remain at an equilibrium point distant from the functionally active conformations. Other mutations reduce chaperone binding by stabilizing the protein fold (left of the WT protein in the chaperone-binding chart).
(C) Absolute HSP70 vs. HSP90 binding preference for >1,500 disease-associated mutants as compared to their cognate wild-type proteins. Mutants with well-documented clinical phenotypes are grouped into three classes based on clinical severity (severe: filled blue triangles; moderate/atypical: open red diamonds; mild: filled red triangles). Two SOD1 mutants with distinct chaperone preferences and clinical severities are indicated.
(D) Clinical phenotype of the indicated SOD1 mutations as summarized by Wang et al., 2008. Number of patients from each severity class is indicated in parentheses. Symbols as indicated in (C).
(A) Luminescence z-scores of wild-type FANCA and negative control (PCNA protein, critical for DNA replication and repair) proteins for binding to HSP90 and HSP70. Data presented as the mean ± SEM from 12 independent biological replicates. Significance cutoff (z-score > 2) indicated by dotted line.
(B) Correlation between biological replicates of normalized LUMIER interaction scores for 61 FANCA variants relative to FANCA wild-type for binding to HSP90 (red triangles) and HSP70 (blue triangles). Correlations determined by Pearson coefficient of determination, R2.
(C-D) Differential interaction scores (log2) of HSP90 (C) and HSP70 (D) with FANCA variants exhibiting increased chaperone binding relative to wild-type FANCA protein.
Data presented as the mean ± SEM from multiple (3-5), independent LUMIER experiments.
(E) Distribution of luminescence signals (z-score) of 61 FANCA variants assayed by LUMIER in cells stably expressing Renilla luciferase alone.
(F) Validation of HSP90 binding to FANCA variant proteins by immunoprecipitation with anti-FLAG and Western blotting for endogenous HSP90. Mutants with increased HSP90 interactions are indicated in red/bold.
(G) Comparison between the relative binding of 61 FANCA variants to HSP90 and the effect of HSP90 inhibition (HSP90i: ganetespib, 100 nM, 20 h) on mutant FANCA protein steady-state expression levels relative to wild-type FANCA protein. FANCA variants with disparate chaperone engagement patterns are indicated (90≥70: HSP90-preferring; 70>90: HSP70-preferring; none: no increase in chaperone binding). Data presented as the mean of 2 independent biological replicates. Correlations by Pearson coefficient of determination, r.
(H) A linear representation of the FANCA protein shows the positions of 90 variant amino acids (x-axis) and their effect on chaperone binding relative to FANCA wild-type (mut/WT, mutant vs. wild-type; z-score). Differential interaction scores (z-scores) for HSP90 (red triangles) and HSP70 (blue triangles) (y-axis) were determined by LUMIER. (I) Differential HSP90 (x-axis) and HSP70 (y-axis) interaction scores for 90 FANCA variants categorized based on chaperone engagement pattern (70>90: preferential binding to HSP70 over HSP90; 90≥70: increased binding to HSP90 equal or greater than that of HSP70; none: no increase in chaperone binding).
(J) Differential HSP90 (x-axis) and HSP70 (y-axis) interaction scores for 11 previously characterized FANCA mutants belonging to the indicated phenotypic classes.
(K) Relative cell viability of FANCA null GM6914 cell-lines expressing the indicated FANCA mutants, wild-type FANCA protein or empty vector control (null). Cells were seeded at a low density and were incubated at 37°C for 24 hours. Cells were subsequently exposed to increasing concentrations of MMC and immediately incubated at 37°C for 3 days to allow recovery. Viability was then measured. Data presented as the mean ± SEM from 2 independent experiments. Error bars smaller than the symbol are not shown.
(L) FANCA mutant expression levels and FANCD2 ubiquitylation in the indicated GM6914-derived FANCA mutant cell-lines in response to DNA replication stress induced by hydroxyurea treatment (1 mM, 24h). Samples were analyzed by Western blot using the indicated antibodies. un indicates the unmodified FANCD2 protein; ub indicates the monoubiquitinated species. HSP90 serves as a loading control.
(M) Sensitivity (z-score) to various chemotherapeutic agents of FANCA null GM6914 cell-lines expressing the indicated FANCA mutants relative to wild-type. R951W, E936G and I939S cell lines, but not R880Q or wild-type, are hypersensitive to FA-related drugs.
(A-C) Relative effect of pharmacological HSP90 inhibition by (A) ganetespib, (B) onalespib (AT13387, 10-40 nM) or (C) PU-H71 (30-50 nM) on the viability of FANCA wild-type and FANCA-null cells under normal conditions or with increasing concentrations of MMC. Left panels in (A-C) indicate the effects of the indicated inhibitor on cell growth; middle panels the effects on MMC tolerance in FANCA-wild-type cells; right panels the effects on MMC tolerance in FANCA-null cells Data presented as the mean ± SEM from 2 independent experiments.
(D) Effects of low-level HSP90 inhibition (by ganetespib, 2-5 nM; PU-H71, 40-70 nM) or febrile-range temperature (39°C) on HSP70 and HSP90 protein levels in FANCA wild-type cells. High-level HSP90 inhibition (ganetespib, 25 nM; PU-H71, 300 nM) as well as proteotoxic proteasomal inhibition (MG132, 2.5 μM) induced the expression of HSP70. Constitutive (upper band: C) and inducible forms (lower band: I) of HSP70 are indicated. HSP90 levels are shown for comparison.
(E-G) Relative viability of MMC-treated, GM6914-derived R880Q FANCA mutant cells exposed to DMSO control or to low-level HSP90 inhibition with ganetespib, 5 nM (E), or low-level HSP90 inhibition by onalespib (AT13387) (F) or PU-H71 (G). Data presented as the mean ± SEM from 3 independent experiments for panel (E) and from 2 independent experiments for panels (F-G). Panels (F) and (G) are directly comparable to (B) and (C), respectively.
(H) Increased cell death of FANCA WT, R880Q, or null cells as determined by propidium iodide staining in the presence of mitomycin C (MMC) and DMSO control or ganetespib at 4 nM and 5 nM. Values presented as the mean ± SEM from 2 independent biological replicates.
(I) Effects of low-level HSP90 inhibition (Gan: ganetespib, 5 nM vs. PU-H71, 70 nM) on FANCD2 ubiquitylation in the presence of hydroxyurea (1 mM; 24 h). Samples from GM6914 cells expressing WT, R880Q or I939S FANCA proteins were analyzed by Western blotting with antibodies against the indicated proteins. un indicates the unmodified FANCD2 protein, and ub the indicates the monoubiquitinated species.
(J) Sensitivity of the indicated FANCA mutant-expressing cell lines to carboplatin in the presence of DMSO or low-level HSP90 inhibition (ganetespib, 5 nM). Data presented as the mean ± SEM from 3 independent experiments.
(K) Screen of 353 chemotherapeutics for drug sensitivities induced by HSP90 inhibition (HSP90i: ganetespib, 5 nM). Chemotherapeutics highlighted by purple triangles are significantly more toxic to FANCA wild-type cells upon HSP90i, as compared to the remaining drugs (grey squares). DMSO control is indicated by the black square. As an internal control for the assay itself, low-level HSP90i strongly sensitized the cells to other HSP90 inhibitors (17-AAG, 17-DMAG) as expected.
(A) Relative cell viability of the indicated FANCA mutant cell lines exposed to mild HSP90 inhibition (ganetespib, 5 nM; 3 days) or DMSO control at different temperatures. Data presented as the mean ± SD from 4 independent biological replicates.
(B) Relative cell viability of FANCA-null GM6914 cells expressing the indicated FANCA mutants after treatment with MMC followed by recovery at 39°C vs. 40°C. Cells were allowed to recover from exposure to increasing concentrations of MMC at 39°C or 40°C for 3 days. Data presented as the mean ± SEM from 2 independent experiments.
(C) Relative viability of FANCA wild-type cells after exposure to increasing concentrations of MMC and subsequent recovery (3 days) at the indicated temperatures. Data presented as the mean ± SD from 4 independent experiments.
(D-L) Relative cell viability of FANCA-null GM6914 cells expressing the indicated the FANCA mutants after exposure to the indicated chemotherapeutics at different temperatures, as described in (C). Effect of 40°C on MMC (D) and carboplatin tolerance (E). Bendamustine tolerance at normal temperature (37°C) (F), compared to 39°C (G) and 40°C (H). Recovery from lomustine at 37°C (I) or at 40°C (J). Recovery from exposure to bendamustine (K) or lomustine (L) at 37°C in the presence of low-level HSP90 inhibition (ganetespib, 5 nM). Data presented as the mean ± SEM from 3 independent experiments.
(A) FANCD2 ubiquitylation in response to hydroxyurea (0.2 mM for 24 h at 37°C) in null, WT, R880Q and Q/K FANCA mutant-expressing cell-lines. Western blotting against the indicated proteins; HSP90 is used as loading control. un indicates unmodified FANCD2 protein, and ub the monoubiquitinated species.
(B) Sensitivity (z-score) of FANCA-R880Q/E966K (Q/K) cells to various chemotherapeutics as compared to FANCA-null (red triangles) and FANCA wild-type cells (black spheres). Each dot indicates a different chemotherapeutic agent. MMC is indicated. Correlation between chemotolerance of Q/K cells and wild-type cells by Pearson coefficient of determination, R2.
(C) Effect of onalespib (AT13387) exposure on MMC tolerance of FANCA-Q/K-expressing cells (directly comparable to Figures S3B and S3F). Data presented as the mean ± SEM from 2 independent experiments.
(D) Correlation between biological replicate LUMIER interaction z-scores of FANCA protein partners (blue circles) among 385 human proteins involved in genome maintenance. Established FANCA partners FANCG, FAAP20, FANCC and FANCF are indicated. Correlations by Pearson coefficient of determination, R2.
(E) LUMIER z-scores of the indicated Renilla luciferase-tagged FANCA mutants interacting with the indicated FLAG-tagged FA pathway components. Data presented as the mean ± SD from 3 (or 9 for the case of FAAP20) biological replicates.
(F) LUMIER interaction z-scores for wild-type and Q/K mutant FANCA proteins against 385 human proteins involved in genome maintenance. Established FANCA partners FANCG, FAAP20, FANCC and FANCF are indicated. Error-bars indicate SD among biological replicates.
(G) Validation of HSP90 binding to the indicated FLAG-tagged FANCA variant proteins by immunoprecipitation with anti-FLAG and blotting for endogenous HSP90.
(H) FANCA depletion upon exposure to low-level HSP90 inhibition is proteasome-dependent. R880Q and Q/K cells were exposed to cycloheximide (CHX, 20 ug/ml, 2 hours) and hydroxyurea (1 mM) in the presence of DMSO (control), 5 nM ganetespib (Gan), or 5 nM ganetespib plus 20 uM proteasome inhibitor MG132 (Gan + MG132). Samples were analyzed by Western blotting for the indicated proteins, un indicates the unmodified FANCD2 protein, and ub the monoubiquitinated species.
(I) Differential LUMIER interaction scores for the indicated FANCA mutants relative to wild-type FANCA. Data presented as mean ± SEM from 3 independent experiments.
(J) HSP90 and FAAP20 binding to the indicated FLAG-tagged FANCA variant proteins by immunoprecipitation with anti-FLAG and blotting for endogenous HSP90 in stable cell lines expressing FAAP20 in fusion with Renilla luciferase (Rluc).
(K) Effect of ganetespib (5nM; dotted lines) on the MMC tolerance of FANCA-null cells engineered to stably express the FANCA-E966K mutant (red lines) as compared to cells expressing wild-type FANCA (black lines). Ganetespib-treated cells are indicated by the dotted lines, and DMSO-treated controls by the continuous lines. Data presented as the mean ± SEM from 6 independent experiments.
(A) Clinical data for indicated missense mutants and literature. (B) Chaperone engagement and predictions of functional severity for the same mutants.
(A) FANCA missense variants and variant sources. (B) Chaperone engagement of FANCA variants. (C) Phenotypic characterization of FANCA variants and literature.
(A) Chemosensitivities related to FA only. (B) Chemosensitivities related to HSP90 inhibition alone (HSP90i) or common to FA and HSP90 (both FA and HSP90i)
(A) List of Genome Maintenance Factors screened for the ability to bind FANCA. Each positive interaction represents a FANCA partner protein and is indicated as “HIT”. (B) Binding z-scores of FANCA partners to indicated FANCA mutant proteins.