Abstract
Whether disclosure of genetic risk for coronary heart disease (CHD) influences shared decision-making (SDM) regarding use of statins to reduce CHD risk is unknown. We randomized 207 patients, age 45– 65 years, at intermediate CHD risk, and not on statins, to receive the 10-year risk of CHD based on conventional risk factors alone (n=103) or in combination with a genetic risk score (n=104). A genetic counselor disclosed this information followed by a physician visit for SDM regarding statin therapy. A novel decision aid was used in both encounters to disclose the CHD risk estimates and facilitate SDM regarding statin use. Patients reported their decision quality and physician visit satisfaction using validated surveys. There were no statistically significant differences between the two groups in the SDM score, satisfaction with the clinical encounter, perception of the quality of the discussion or of participation in decision-making and physician visit satisfaction scores. Quantitative analyses of a random subset of 80 video-recorded encounters using the OPTION5 scale also showed no significant difference in SDM between the two groups. Disclosure of CHD genetic risk using an electronic health record-linked decision aid did not adversely affect SDM or patients' satisfaction with the clinical encounter.
Trial registration number
NCT01936675; Results.
Introduction
Shared decision-making (SDM) is increasingly being recognized as an important factor in patient-centered care.1 Decision aids have been proven effective in improving knowledge transfer, decisional conflict, and patient involvement in SDM.2 The newly issued guidelines on management of dyslipidemia and prevention of coronary heart disease (CHD) emphasize the need for SDM when considering initiation of statin therapy.3 However, whether incorporating probabilistic genetic risk information for CHD risk estimation influences SDM regarding statin therapy is unknown.
Genome-wide association studies (GWAS) have revealed multiple genetic susceptibility variants that influence risk of common diseases such as CHD.4 Knowledge of increased genetic susceptibility to a disease, such as CHD, for which drug therapy and lifestyle interventions exist, may influence SDM regarding initiation of medications to lower the risk and potentially help motivate patients to adopt healthy lifestyle behaviors. However, interpretation of results of multiplex genetic testing for common diseases can be challenging since patients as well as physicians are often lacking in numeracy5 and poorly equipped to comprehend statistical probabilities related to risk of disease.6 Decision aids that incorporate genetic risk for CHD into the SDM process could help address these potential hurdles.
Incorporating genetic susceptibility variants in CHD risk estimating equations may help refine stratification of CHD risk.7 Little is known about how such information may affect SDM. There is also a larger challenge of transforming genetic data into an edible format to facilitate implementation of genomic information into the everyday practice of medicine.8 There is a need to establish best practices for disclosing genetic risk and on motivating patients to adopt actionable health interventions.6,9,10 In particular, effective and content-based approaches that promote comprehension of genetic information are needed to support informed and SDM.
In this report, we describe the feasibility of using a decision aid linked to the electronic health record (EHR), to visually disclose genetic risk for CHD and enable SDM regarding statin therapy. We modified a previously developed decision aid11 to convey the probabilistic nature of CHD genetic risk estimates and to facilitate SDM. This tool is meant to help patients and physicians visually comprehend CHD genetic risk and the extent of risk reduction with statins. We assessed whether disclosure of genetic risk for CHD influenced SDM and physician visit satisfaction in the Myocardial Infarction GENES (MI-GENES) study. MI-GENES is a randomized controlled trial to assess the effect of disclosing CHD genetic risk on low-density lipo-protein cholesterol (LDL-C levels). MI-GENES demonstrated that disclosure of genetics-informed CHD risk led to lower LDL-C levels than disclosure of CHD risk based on conventional risk factors alone.
Methods
MI-GENES study
The MI-GENES study was approved by the Mayo Clinic IRB and is registered at ClinicalTrials.gov (NCT01936675). MI-GENES was submitted to ClinicalTrials.gov on September 3, 2013. The study was performed in accordance with relevant guidelines and regulations. The MI-GENES study protocol, design, and primary outcome have been previously published.12,13
Study population
Among 29,352 participants of the Mayo Clinic BioBank,14 we identified 2026 individuals who met the following inclusion criteria: 45–65 years old, no prior history of CHD or other atherosclerotic vascular diseases, not on statins, intermediate risk for CHD (conventional risk score, CRS: 5–20%) and residency in Olmsted County, Minnesota. A random subset of 1000 was genotyped for the 28 SNPs associated with CHD independent of blood pressure (BP) and lipid levels to calculate a genetic risk score (GRS) for each individual. Genotyping results were valid for 968 individuals who became the recruitment pool for this study. Enrolment was performed by invitation and was continued with a target of recruiting 110 participants with a high GRS (≥1.1) and 110 participants with average/low GRS (<1.1).12 EMB and HJ recruited participants for the MI-GENES clinical trial.
Study design and randomization
Design of the MI-GENES clinical trial has been published previously.12 At the first study visit, patients completed written informed consent forms and baseline characteristics were obtained. Randomization was performed by means of a computer-generated random sequence with stratification for age, gender, and positive family history for CHD using the Pocock and Simon15 method. Once a study participant completed the first study visit and scheduled the second study visit, he/she was randomized to either receive CRS only or the genetics-informed risk of CHD (CRS*GRS). One of the investigators (HJ) generated the random allocation sequence and study arm assignment using the computer software described earlier.
The second study visit included meeting with a genetic counselor for 30 min during which study participants discussed their risk profile and how their family history and GRS affected their overall CHD risk (for patients randomized to receive CRS*GRS) versus discussion of effects of a positive family history for CHD overall risk (for patient randomized to receive CRS). After meeting with the genetic counselor, each study participant met with a physician in the Cardiovascular Health Clinic to further discuss means to reduce their CHD risk. Disclosure of risk in both arms was facilitated by the use of a modified version of the Statin Choice decision aid (described below). It was emphasized that CHD risk assessment was probabilistic and not deterministic. Encounters with the genetic counselor and physician were video-recorded in 187 patients who consented to the recording.
The primary end point was LDL-C level at 6 months after disclosure of CHD risk in the two arms. The primary end point of MI-GENES was previously reported. Participants randomized to CRS*GRS had lower LDL-C levels at 6-months due to a higher percentage being prescribed statin medications.13 Secondary outcomes include changes in dietary habits, physical activity and psychosocial measures related to CHD risk disclosure. SDM and physician visit satisfaction reported in this manuscript are among the secondary outcomes of this clinical trial. Recruitment started in October 2013 and ended in May 2014.
Genetic risk score and 10-year risk of CHD
Meta-analyses of GWAS have revealed 46 single nucleotide polymorphisms (SNPs) associated with CHD.4 Among these 46 loci, 29 are associated with CHD independent of risk factors such as hypertension, lipid levels, and diabetes.4 We genotyped 28 SNPs in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory (the complete list of genotyped SNPs can be found in online supplementary table S1). Genotypes at susceptibility loci were used to derive a GRS that takes into account the average population risk.7,12 In brief, a GRS of 1.0 indicates a combination of risk and non-risk alleles that does not alter susceptibility relative to the population average. A GRS of 1.2 implies a 20% increased risk while a score of 0.8 implies a 20% reduction in genetic risk for CHD compared to the population average. Since each CHD risk variant has different odds of CHD risk, we reported GRS as a weighted cumulative score rather than the total number of risk alleles.
The 10-year CRS for CHD was estimated based on known cardiovascular risk factors including age, sex, smoking, diabetes, high BP, total and high-density lipoprotein cholesterol (HDL-C) as described by Wilson et al.16 The GRS was integrated into CRS, as previously described,7 to generate an genotype-informed probability of adverse CHD events over the next 10 years (CRS*GRS).
Linkage to the EHR
The generic disease management system (GDMS), developed by the Mayo Clinic in collaboration with VitalHealth software,17 is a web-based guideline reminder system used at the point of care at Mayo Clinic Rochester. GDMS is integrated into the Mayo EHR by means of a web viewer system named ‘Synthesis’ and assists with guideline compliance and improvement of quality metrics.17 In order to incorporate GRS into the 10-year CHD risk estimate, GDMS was modified to deliver a web link to the genomic decision aid tool. When the link is clicked, GDMS transmits pertinent risk factors and the GRS to the online tool via a secure link without any patient identifiers (see online supplementary figure S1). A risk report was also placed in the EHR (see online supplementary figure S2).
Statin Choice decision aid
The Statin Choice decision aid was originally developed to disclose CHD risk and help patients as well as clinicians review the benefits and downsides of taking a statin medication to reduce CHD risk.11,18,19 The tool displays the 10-year probability of CHD based on CRS in addition to the absolute risk reduction with the use of statin drugs, and the associated costs/side effects. The original Statin Choice decision aid has been evaluated previously in three randomized controlled trials. 18–20 It can be freely accessed online at http://statindecisionaid.mayoclinic.org.
In order to implement the GRS into SDM, the Statin Choice decision aid was modified to include a variable for GRS for incorporation into the 10-year CRS (figure 1). A feature was added to the tool enabling the physician as well as the patient to visualize the effect of implementing GRS into CRS (figure 2). The modified tool can be accessed online at http://migenesstudy.mayoclinic.org (password: migenes—use of this decision aid should be limited to research purposes only). Afterwards, the provider can discuss the benefits of starting standard versus high dose statins as well as potential side effects (figure 3). CHD risk was disclosed using scripted language as follows: ‘Out of 100 people like you…’. The benefit of statins was conveyed in a similar manner stressing the absolute risk reduction while minimizing framing by presenting the groups helped and not helped by using statins. The tool was also equipped with a report generating function and a frequently asked questions page that includes additional information about GRS.
Figure 1.
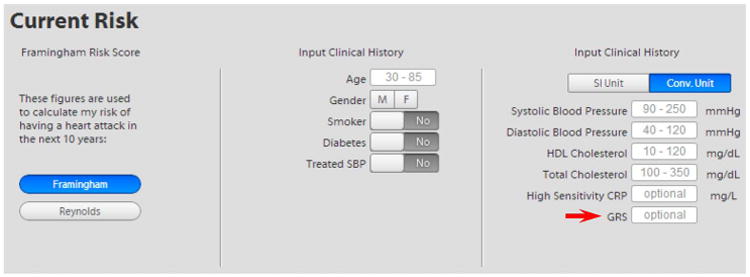
Data entry screen for the decision aid. The risk factor entry screen of the decision aid was modified to implement the GRS as highlighted in the figure. Implementation of GRS into the conventional risk score was embedded into the coding of the decision aid application. GRS, genetic risk score.
Figure 2.
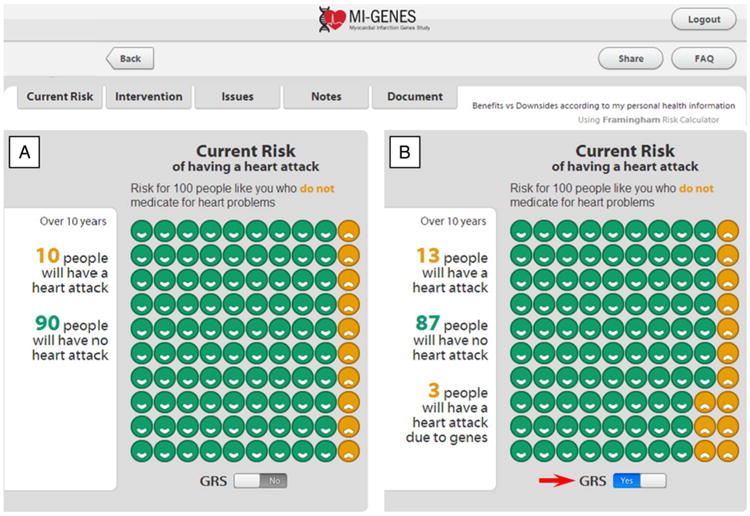
Disclosure of CHD risk. Disclosure of CHD risk estimates based on the CRS (panel A) and after implementing the genetic risk score (CRS*GRS, panel B) by clicking on the GRS button (arrow). In this example, the patient's 10-year CHD risk based on CRS is displayed as 10% (A). With a GRS of 1.3, the overall risk increases to 13% as shown in (B). CHD, coronary heart disease; CRS, conventional risk score; GRS, genetic risk score; MI-GENES, Myocardial Infarction GENES.
Figure 3.
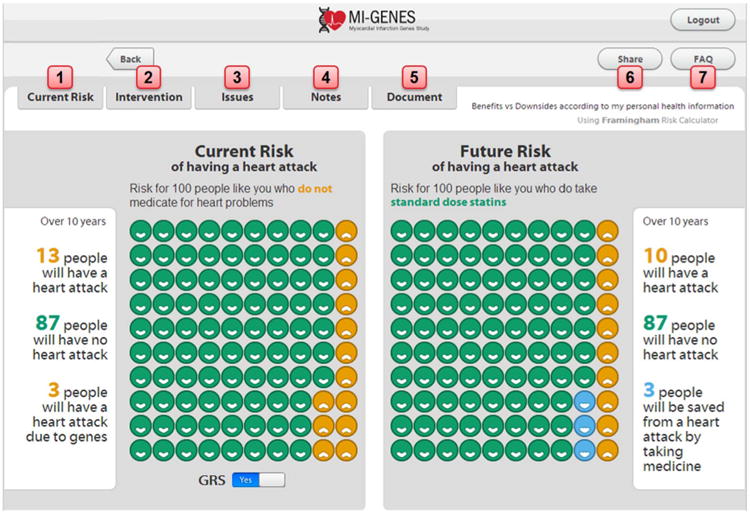
Summary of features of the decision aid. These features include: (1) CHD risk estimates which can be modified to show patients how risk can change according to their risk factors. (2) The healthcare provider can select an intervention such as standard dose versus high dose statins. (3) Statin side effects can be discussed with the patient. (4) There is also a section where the healthcare provider and patient can input notes regarding CHD risk assessment and associated interventions. (5) A complete risk assessment statement can be generated and includes the patient's estimated 10-year CHD risk. This statement can be copied and pasted into an electronic medical note if desired. (6) The displayed risk report can be exported as an email or printed as a PDF document. The exported data include the patient's CHD estimate risk and impact of using statins, without any patient identifiers. (7) A page dedicated to frequently asked questions (including questions regarding the genetic risk score for CHD and how it was calculated). CHD, coronary heart disease; MI-GENES, Myocardial Infarction GENES.
SDM survey measure
SDM was evaluated using the SDM Questionnaire (SDM-Q),21 a validated 11-question survey that assesses patients' perception of the SDM process during clinical encounters.21 The 4-point Likert scale was converted to ‘0’ for ‘disagree’ and ‘strongly disagree’ responses vs ‘1’ for ‘strongly agree’ and ‘agree’ responses. The SDM score ranges from 0 corresponding to least effective SDM, to 11 which corresponds to optimal SDM. If a participant did not answer all of the 11 questions, he/she was excluded from the analyses. If some questions were answered, the missing answers were assigned a score of ‘0’.
SDM quantitative video analysis
For an in-depth evaluation of SDM, we performed quantitative analyses of video recordings of the genetic counselor and physician encounters using the OPTION5 scoring scale.22 A random sample of 40 CRS encounters and 40 age- and sex-matched CRS*GRS encounters was obtained and video recordings were analyzed by one of the authors (TSM). For each participant, the genetic counselor and physician encounters were combined for analysis. Scores were rescaled as percentages ranging from 0 to 100.
Physician visit satisfaction
Physician visit satisfaction was assessed by having study participants complete a 6-statement survey adapted from the Consumer Assessment of Health Plans and Systems (CAHPS).23 This survey has been previously validated and assesses patient satisfaction with their physician encounter in the ambulatory setting. The 3-point Likert scale response to each statement was transformed to ‘0’ for ‘No’ and ‘1’ for ‘Yes, somewhat’ and ‘Yes, definitely’. Physician visit satisfaction score was calculated by summation of participants' responses to these 6 questions yielding a physician visit satisfaction score ranging from 0 to 6 indicating poor to optimal satisfaction. If a participant did not answer all of the 6 questions, he/she was excluded from the analyses. The missing answers were assigned a score of ‘0’.
Statistical methods
Statistical analyses were performed using computer software JMP V.10.0 (SAS Institute, Cary, North Carolina, USA). Continuous variables were expressed as mean (SD), whereas dichotomous variables were expressed as percentages. Analyses were performed with randomization groups as the independent variables and physician visit satisfaction and SDM scores as the dependent variables. For continuous characteristics, Wilcoxon rank sum test or two-sample t-test were used as appropriate to assess differences between the two study arms. Fisher's exact test was used to assess for differences in each question's two-level response.
The study was designed to primarily detect an LDL-C difference of 10 mg with 80% power and 5% Type I error rate. Please refer to the clinical trial design and primary end point reports for additional details.12,13 With 80% power and 5% Type I error, our study was able to detect a difference of 0.69 in the SDM score (6.2% of maximum score 11), 2.9 in the OPTION5 score (2.9% of maximum score 100) and 0.04 in the physician satisfaction score (0.4% of maximum score 9).
Results
Of 216 enrolled participants (mean age 58.9±5.0 years, 46.8% males), 207 were randomized and completed the second study visit including the genetic counselor and physician appointments. Study participants were randomized to receive either CRS based on conventional CHD risk factors (n=103, mean age 59.0±5.2 years, 48.5% male) or CRS*GRS based on conventional risk factors as well as genetic risk (n=104, 59.0±4.9 years, 46.2% male) (figure 4). Baseline characteristics including age, sex, smoking status, and other CHD conventional risk factors were similar between the two groups (table 1). The SDM survey was completed by 206 study participants, and the physician visit satisfaction survey was completed by all study participants.
Figure 4.
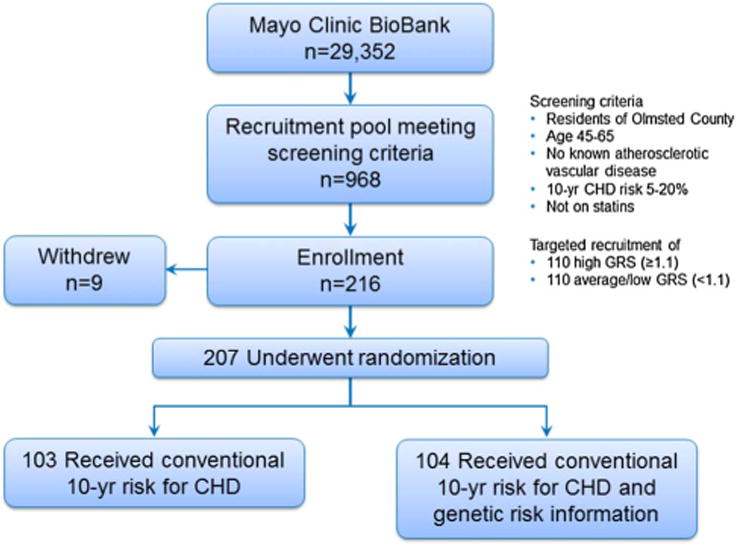
The MI-GENES study flow diagram. Study participants were drawn from the Mayo Clinic BioBank (n=29,352) and were residents of Olmsted County, Minnesota, USA; of 2026 biobank participants who met the eligibility criteria, a random subset of 1000 was genotyped. After quality control checks, genotyping results were valid for 968 individuals who served as the recruitment pool for this study. Targeted recruitment of 216 individuals was completed. Patients who were scheduled for the second study visit were randomized in a 1:1 fashion to receive conventional CHD risk versus conventional and genetic CHD risk. Both arms completed surveys to assess SDM and physician visit satisfaction at the conclusion of visit 2. CHD, coronary heart disease; MI-GENES, Myocardial Infarction GENES; SDM, shared-decision-making.
Table 1. Participant characteristics (n=207).
| CRS, n=103 | CRS*GRS, n=104 | p Value | |
|---|---|---|---|
| Age, years | 59.0±5.2 | 59.0±4.9 | 0.91 |
| Male gender | 50 (48.5%) | 48 (46.2%) | 0.78 |
| Ever smoker | 41 (39.8%) | 32 (30.8%) | 0.19 |
| Family history of CHD | 30 (29.1%) | 25 (24%) | 0.43 |
| BMI, kg/m2 | 30.4±7.0 | 30.3±6.1 | 0.79 |
| SBP, mm Hg | 130.0±14.0 | 131.7±17.6 | 0.81 |
| Waist circumference, cm | 101.6±15.9 | 100.5±14.0 | 0.76 |
| Total cholesterol, mg/dL | 201.4±29.9 | 202.9±27.7 | 0.74 |
| LDL-C, mg/dL | 119.2±23.7 | 120.5±25.7 | 0.80 |
| HDL-C, mg/dL | 55.5±15.8 | 56.4±16.7 | 0.86 |
| Triglycerides, mg/dL | 133.8±69.3 | 132.5±78.4 | 0.47 |
| College education or higher | 68 (66%) | 53 (56.7%) | 0.25 |
BMI, body mass index; CHD, coronary heart disease; CRS, conventional risk score; GRS, genetic risk score; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; SBP, systolic blood pressure.
SDM scores were similar among both groups (SDM score for CRS 10.40±1.76 vs 10.61±1.55 for CRS*GRS, p=0.09), although there was a trend toward higher SDM scores in the CRS*GRS group. Similarly, there were no differences between the two groups when individual questions in the SDM-Q were evaluated (table 2). Specifically, there was no difference in participants' perception of adequate consultation time between CRS and CRS*GRS (p=1.0). Incorporation of GRS into the decision-making process did not affect the participant's perception of sufficient discussion of different treatment options including use of statins versus other medications or lifestyle modifications, selection of the treatment, advantages of each treatment, and feeling of inclusion in the treatment decision (p=NS for all). Analyses of video recordings of 80 encounters using the OPTION5 scale revealed similar SDM scores among the CRS*GRS and CRS groups (72.7±6.0 vs 70.1±7.4, respectively; p=0.13).
Table 2. Shared decision-making*.
| CRS | CRS*GRS | p Value | |
|---|---|---|---|
| 1. In the selection of the treatment method, my thoughts were taken into account just as much as the considerations of my doctor. | 101/103 | 101/103 | 1.00 |
| 2. There was enough time for questions. | 102/103 | 102/103 | 1.00 |
| 3. My doctor and I weighed up the different treatment options thoroughly. | 98/103 | 98/103 | 1.00 |
| 4. I was able to discuss the different treatment options with my doctor in detail. | 98/103 | 97/103 | 1.00 |
| 5. My doctor and I selected a treatment option together. | 93/103 | 99/103 | 0.16 |
| 6. I now know the advantages of the individual treatment options. | 97/103 | 101/103 | 0.28 |
| 7. I now know which treatment option is the best one for me. | 92/103 | 99/103 | 0.11 |
| 8. During the consultation, I felt included in the treatment decision. | 99/103 | 100/103 | 1.00 |
| 9. Through the consultation with the doctor, I felt jointly responsible for my further treatment. | 100/103 | 100/103 | 1.00 |
| 10. My doctor and I discussed the next steps of the treatment plan in detail. | 93/103 | 97/103 | 0.44 |
| 11. My doctor and I reached an agreement as to how we will proceed. | 98/103 | 99/103 | 1.00 |
| Achieved an SDM score of 11 | 83/103 | 92/103 | 0.12 |
| Overall SDM score | 10.40±1.76 | 10.61±1.55 | 0.09 |
| OPTION5 score | 70.1±7.4 | 72.7±6.0 | 0.13 |
Displayed data depict the number of study participants who answered ‘agree’ or ‘strongly agree’ to the overall number of respondents to each question. One study participant in the CRS*GRS group did not complete the SDM survey.
CRS, conventional risk score; GRS, genetic risk score; SDM, shared decision-making.
Overall, physician visit satisfaction scores were similar among both arms (physician visit satisfaction score for CRS 5.99±0.10 vs 5.97±0.17 for CRS*GRS, p=0.32). When individual responses to the physician visit satisfaction survey were assessed, there were no differences between the CRS and CRS*GRS arms (table 3). Participants felt that physicians: (1) explained things in a way easy to understand (100% vs 100%, respectively, p=1.0); (2) listened carefully to them (100% vs 99%, p=1.0); (3) gave easy to understand information (99% vs 100%, p=0.5); (4) knew important information about the patient's medical history (100% vs 100%, p=1.0); (5) showed respect for what the patient had to say (100% vs 99%, p=1.0) and (6) spent enough time with the patient (99% vs 97%, p=0.62).
Table 3. Physician visit satisfaction*.
| CRS | CRS*GRS | p Value | |
|---|---|---|---|
| 1. During your visit, did your physician explain things in a way that was easy to understand? | 103/103 | 104/104 | 1.00 |
| 2. Did your physician listen carefully to you? | 103/103 | 103/104 | 1.00 |
| 3. Did your physician give you easy to understand information about these health questions or concerns? | 102/103 | 104/104 | 0.50 |
| 4. Did your physician seem to know the important information about your medical history? | 103/103 | 104/104 | 1.00 |
| 5. Did this physician show respect for what you had to say? | 103/103 | 103/104 | 1.00 |
| 6. Did this provider spend enough time with you? | 103/103 | 103/104 | 1.00 |
| Achieved a physician visit satisfaction score of 6 | 102/103 | 101/104 | 0.62 |
| Overall physician visit satisfaction score | 5.99±0.10 | 5.97±0.17 | 0.32 |
Displayed data depict the number of study participants who answered ‘yes, definitely’ or ‘yes, somewhat’ to the overall number of respondents.
CRS, conventional risk score; GRS, genetic risk score.
Discussion
With continuing advances in genomics, there will be an increasing need to integrate genomic information in clinical practice. Currently, most patients and physicians are not familiar with interpreting or communicating genetic testing results.24,25 In this report, we demonstrated the feasibility of using a novel decision aid, linked to the EHR, to disclose genetic risk for CHD, thereby allowing SDM at the point of care. Incorporation of CHD genetic risk information in the decision aid enabled physicians and patients to visually discern the effects of genes on CHD risk without the need to discuss the technical details of calculating genetic risk. More importantly, disclosure of CHD genetic risk did not adversely affect patient-perceived SDM or physician visit satisfaction. To the best of our knowledge, our study is the first to describe use of a genomic decision aid to disclose genetic risk information in the clinical setting and facilitate SDM regarding the use of statin therapy to reduce CHD risk.
Recent advances in genomics have the potential to improve the health of the population.26 However, implementing genomic medicine in the busy flow of everyday clinical practice poses several challenges and may only be feasible by leveraging the EHR.27 Although adoption of EHRs is increasing due to federal mandates, the majority of EHR systems are not yet configured to deal with genomic data. To implement genomic medicine, EHRs will need to be modified to receive, store, present complex genomic information for clinical use, incorporate genomic clinical decision support systems to help providers practice individualized medicine and provide links to relevant knowledge resources.
In contrast to single-gene Mendelian disorders, the genetic architecture of ‘complex’ diseases such as CHD includes a large number of susceptibility loci with varying effect sizes. Physicians may not be familiar with using probabilistic genetic risk of common diseases in the clinical setting. Current CHD risk prediction equations such as the Framingham risk score and the atherosclerotic cardiovascular disease risk (ASCVD) score are based on conventional risk factors.3,16 Incorporating genetic risk information for CHD may further help stratify patients' risk for CHD.
EHRs are an important resource for conducting genomic research using novel electronic data mining and phenotyping methods.28–30 However, how to harness the EHR to implement genomic medicine remains an area of active investigation. We integrated a genomic decision aid in the EHR to facilitate SDM regarding CHD risk reduction. The decision aid: (1) allowed implementation of genetic risk information in the clinical setting; (2) transformed genomic results into a user-friendly, readily comprehensible format for patients and physicians; (3) used pictograms for disclosing risk; and (4) provided links to additional resources for patients and providers. Since the decision aid is web-based, it is portable to other EHRs. Moreover, as new genetic susceptibility loci are discovered, GRS can be updated accordingly.
In a systematic review assessing use of genetic testing in the primary care setting,31 the most common barriers to genetic testing among primary care providers included inadequate genetic knowledge, lack of confidence in counseling about risk, in addition to lack of confidence and ability to order and interpret test results. Our approach to implementing CHD genetic risk in the EHR, using a GRS rather than raw genetic test results, simplifies multiplex genetic testing results while simultaneously transforming the data into user-friendly and clinically actionable information. More importantly, the pictographic component of the decision aid helps physicians as well as patients better understand CHD absolute risk ‘out of a 100 similar patients’ and discern the effects of genes on overall risk for CHD when GRS is incorporated into risk estimates.
By taking into account patients' experiences and preferences at the time of decision-making, SDM promotes patient-centered care.1,32 SDM has also been proven to reduce decisional conflict, improve patient knowledge, and even increase patient adherence to medications.11 However, there is concern that disclosing genetic information could impair SDM as most physicians are not comfortable dealing with genomic information.31 On the basis of participant-perceived SDM scores as well as quantitative analysis of video recordings using the OPTION5 scale, we found no difference in SDM between the two study groups. This suggests that disclosure of genetic risk is feasible and does not adversely affect SDM compared to disclosure of conventional CHD risk estimates. Indeed, a recent meta-analysis reported that tools like Statin Choice can be reliably implemented in clinical practice with minimum clinician training.33
There is concern that patients may have limited ability to retain and understand complex genomic information,24 which in turn may lead to uninformed decision-making and ineffective patient–physician interaction. In a study by Kaphingst et al,34 patients were assessed for their understanding and reactions to genetic testing of eight common diseases including CHD. About 80% of participants recalled their genetic test results and they were unlikely to interpret these results as deterministic. In our study, there was no difference in physician visit satisfaction whether patients received conventional risk for CHD (CRS) or CRS*GRS. This further supports the notion that properly disclosed genetic information need not add complexity to patient–physician encounters.
Several limitations need to be recognized. Study participants were recruited from the Mayo Clinic BioBank and may not be fully representative of the general population. Results from our study cannot be generalized unless the same disclosure process and decision aid are used in the risk disclosure process. Owing to the high SDM scores, we were not able to perform an in-depth evaluation of the SDM theoretical framework. Our study used the Framingham risk equation rather than the new ASCVD equation since the MI-GENES study protocol was finalized and recruitment had already been started when the ASCVD risk calculator was published. Our study participants had a higher than average educational and socioeconomic background and may have been more adept in understanding genetic test results. The disclosure process described in this manuscript describes one approach that would need to be compared against other approaches in future studies.
Conclusion
We demonstrate the use of a decision aid embedded in the EHR to disclose CHD genetic risk information in a randomized controlled trial setting. Disclosure of CHD genetic risk did not adversely affect SDM or patients' satisfaction with their physician encounters. Our findings suggest that disclosing probabilistic genetic risk of common diseases is feasible leveraging the EHR and that subsequent SDM can be facilitated by the use of appropriate decision aids.
Supplementary Material
Significance of this study.
What is already known about this subject?
Shared decision-making (SDM) is integral to patient-centered care. Decision aids have been proven to facilitate SDM. Decision aids that use genetic risk information have not been previously studied.
Understanding genetic risk of coronary heart disease (CHD) in the clinical setting can be challenging for patients and providers.
How disclosure of CHD genetic risk influences SDM and physician visit satisfaction is not known.
What are the new findings?
We describe the development of a novel decision aid that implements CHD genetic risk into the SDM process and is readily integrated in the electronic health record.
SDM did not differ between patients randomized to receive a conventional risk score for CHD versus those randomized to receive the genetics-informed risk.
Contrary to beliefs that inclusion of genetic information into the clinical setting may threaten patient–physician interaction and decision-making, we have found that SDM and patient satisfaction with the clinical encounter were not affected by disclosure of genetic risk information.
How might these results change the focus of research or clinical practice?
These findings should reassure providers and patients that inclusion of CHD genetic risk information in the clinical setting is feasible using an appropriately designed decision aid and does not affect the quality of SDM or patient satisfaction with the clinical encounter.
Acknowledgments
Funding: This study was funded as part of the National Human Genome Research Institute-supported eMERGE (Electronic Records and Genomics) Network (U01HG04599 and U01HG006379) and the Mayo Clinic Center for Individualized Medicine. Data management was performed with the use of REDCap, which is funded by the Center for Clinical and Translational Science (UL1TR000135).
Footnotes
Contributors: IJK and VMM contributed to study concept and design. HJ, RAH, TSM, S-AB, TMK, EEA, KS and EMB were involved in acquisition, analysis, and interpretation of data. EEA was involved in statistical analysis. HJ, RAH, TSM, S-AB, EEA, RC, VMM and IJK contributed to drafting of the manuscript. All authors were involved in critical revision of the manuscript for important intellectual content.
Competing interests: None declared.
Ethics approval: This study was approved by the Mayo Clinic institutional review board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Spatz ES, Spertus JA. Shared decision making: a path toward improved patient-centered outcomes. Circ Cardiovasc Qual Outcomes. 2012;5:e75–7. doi: 10.1161/CIRCOUTCOMES.112.969717. [DOI] [PubMed] [Google Scholar]
- 2.Coylewright M, Branda M, Inselman JW, et al. Impact of sociodemographic patient characteristics on the efficacy of decision AIDS: a patient-level meta-analysis of 7 randomized trials. Circ Cardiovasc Qual Outcomes. 2014;7:360–7. doi: 10.1161/HCQ.0000000000000006. [DOI] [PubMed] [Google Scholar]
- 3.Goff DC, Jr, Lloyd-Jones DM, Bennett G, et al. American College of Cardiology/American Heart Association Task Force on Practice Guidelines.2013 ACC/AHA Guideline on the Assessment of Cardiovascular Risk: a Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129(Suppl 2):S49–73. doi: 10.1161/01.cir.0000437741.48606.98. [DOI] [PubMed] [Google Scholar]
- 4.Deloukas P, Kanoni S, Willenborg C, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulos JA. Innumeracy: mathematical illeteracy and its consequences. New York: Hill and Wang; 1988. [Google Scholar]
- 6.McBride CM, Alford SH, Reid RJ, et al. Putting science over supposition in the arena of personalized genomics. Nat Genet. 2008;40:939–42. doi: 10.1038/ng0808-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding K, Bailey KR, Kullo IJ. Genotype-informed estimation of risk of coronary heart disease based on genome-wide association data linked to the electronic medical record. BMC Cardiovasc Disord. 2011;11:66. doi: 10.1186/1471-2261-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gigerenzer G, Edwards A. Simple tools for understanding risks: from innumeracy to insight. BMJ. 2003;327:741–4. doi: 10.1136/bmj.327.7417.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Doherty K, Suthers GK. Risky communication: pitfalls in counseling about risk, and how to avoid them. J Genet Couns. 2007;16:409–17. doi: 10.1007/s10897-006-9077-9. [DOI] [PubMed] [Google Scholar]
- 11.Montori VM, Breslin M, Maleska M, et al. Creating a conversation: insights from the development of a decision aid. PLoS Med. 2007;4:e233. doi: 10.1371/journal.pmed.0040233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kullo IJ, Jouni H, Olson JE, et al. Design of a randomized controlled trial of disclosing genomic risk of coronary heart disease: the Myocardial Infarction Genes (MI-GENES) study. BMC Med Genomics. 2015;8:51. doi: 10.1186/s12920-015-0122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kullo IJ, Jouni H, Austin EE, et al. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES Clinical Trial) Circulation. 2016;133:1181–8. doi: 10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson JE, Ryu E, Johnson KJ, et al. The Mayo Clinic Biobank: a building block for individualized medicine. Mayo Clin Proc. 2013;88:952–62. doi: 10.1016/j.mayocp.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pocock SJ, Simon R. Sequential treatment assignment with balancing for prognostic factors in the controlled clinical trial. Biometrics. 1975;31:103–15. [PubMed] [Google Scholar]
- 16.Wilson PW, D'Agostino RB, Levy D, et al. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–47. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhry R, Peters SG, Wagholikar K, et al. Innovations in the delivery of primary care services using a software solution: the Mayo Clinic's generic disease management system. Int J Pers Cent Med. 2012;2:361–7. [Google Scholar]
- 18.Mann DM, Ponieman D, Montori VM, et al. The Statin Choice decision aid in primary care: a randomized trial. Patient Educ Couns. 2010;80:138–40. doi: 10.1016/j.pec.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Weymiller AJ, Montori VM, Jones LA, et al. Helping patients with type 2 diabetes mellitus make treatment decisions: statin choice randomized trial. Arch Intern Med. 2007;167:1076–82. doi: 10.1001/archinte.167.10.1076. [DOI] [PubMed] [Google Scholar]
- 20.Branda ME, LeBlanc A, Shah ND, et al. Shared decision making for patients with type 2 diabetes: a randomized trial in primary care. BMC Health Serv Res. 2013;13:301. doi: 10.1186/1472-6963-13-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon D, Schorr G, Wirtz M, et al. Development and first validation of the shared decision-making questionnaire (SDM-Q) Patient Educ Couns. 2006;63:319–27. doi: 10.1016/j.pec.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Elwyn G, Tsulukidze M, Edwards A, et al. Using a ‘talk’ model of shared decision making to propose an observation-based measure: observer OPTION 5 Item. Patient Educ Couns. 2013;93:265–71. doi: 10.1016/j.pec.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 23.McInnes DK, Brown JA, Hays RD, et al. Development and evaluation of CAHPS questions to assess the impact of health information technology on patient experiences with ambulatory care. Med Care. 2012;50(Suppl):S11–19. doi: 10.1097/MLR.0b013e3182610a50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttmacher AE, McGuire AL, Ponder B, et al. Personalized genomic information: preparing for the future of genetic medicine. Nat Rev Genet. 2010;11:161–5. doi: 10.1038/nrg2735. [DOI] [PubMed] [Google Scholar]
- 25.Lanie AD, Jayaratne TE, Sheldon JP, et al. Exploring the public understanding of basic genetic concepts. J Genet Couns. 2004;13:305–20. doi: 10.1023/b:jogc.0000035524.66944.6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 27.Marsolo K, Spooner SA. Clinical genomics in the world of the electronic health record. Genet Med. 2013;15:786–91. doi: 10.1038/gim.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kullo IJ, Ding K, Jouni H, et al. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kullo IJ, Ding K, Shameer K, et al. Complement receptor 1 gene variants are associated with erythrocyte sedimentation rate. Am J Hum Genet. 2011;89:131–8. doi: 10.1016/j.ajhg.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kullo IJ, Jarvik GP, Manolio TA, et al. Leveraging the electronic health record to implement genomic medicine. Genet Med. 2013;15:270–1. doi: 10.1038/gim.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikat-Stevens NA, Larson IA, Tarini BA. Primary-care providers' perceived barriers to integration of genetics services: a systematic review of the literature. Genet Med. 2015;17:169–76. doi: 10.1038/gim.2014.101. [DOI] [PubMed] [Google Scholar]
- 32.Ting HH, Brito JP, Montori VM. Shared decision making: science and action. Circ Cardiovasc Qual Outcomes. 2014;7:323–7. doi: 10.1161/CIRCOUTCOMES.113.000288. [DOI] [PubMed] [Google Scholar]
- 33.Wyatt KD, Branda ME, Anderson RT, et al. Peering into the black box: a meta-analysis of how clinicians use decision aids during clinical encounters. Implement Sci. 2014;9:26. doi: 10.1186/1748-5908-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaphingst KA, McBride CM, Wade C, et al. Patients' understanding of and responses to multiplex genetic susceptibility test results. Genet Med. 2012;14:681–7. doi: 10.1038/gim.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


