Abstract
A majority of human papillomavirus (HPV) infections are asymptomatic and self-resolving in the absence of medical interventions. Various innate and adaptive immune responses, as well as physical barriers, have been implicated in controlling early HPV infections. However, if HPV overcomes these host immune defenses and establishes persistence in basal keratinocytes, it becomes very difficult for the host to eliminate the infection. The HPV oncoproteins E5, E6, and E7 are important in regulating host immune responses. These oncoproteins dysregulate gene expression, protein-protein interactions, posttranslational modifications, and cellular trafficking of critical host immune modulators. In addition to the HPV oncoproteins, sequence variation and dinucleotide depletion in papillomavirus genomes has been suggested as an alternative strategy for evasion of host immune defenses. Since anti-HPV host immune responses are also considered to be important for antitumor immunity, immune dysregulation by HPV during virus persistence may contribute to immune suppression essential for HPV-associated cancer progression. Here, we discuss cellular pathways dysregulated by HPV that allow the virus to evade various host immune defenses.
1. Introduction
Papillomaviruses (PV) are small DNA viruses that have coevolved for millions of years with various host species, including mammals, reptiles, and birds (Van Doorslaer, 2013). As of now, over 300 PV genotypes have been discovered. Human papillomavirus (HPV) infects mucosal and/or cutaneous skin and causes benign or malignant tumors. Defined as group 1 or 2 carcinogens by the International Agency for Research on Cancer (IARC), ~25 high-risk HPV genotypes are causally associated with multiple human cancers including cervical cancer (CxCa) and oral squamous cell carcinomas (OSCC) (Forman et al., 2012). HPV-associated cancer is a major global health burden causing nearly half a million deaths worldwide every year. Recent studies have shown that HPV-positive OSCC incidence is increasing at an epidemic rate (Chaturvedi et al., 2011; Stein et al., 2014), suggesting that HPV-positive OSCC will likely comprise the majority of all head and neck cancers (HNC) by 2020 (Auluck et al., 2010; Chaturvedi et al., 2011). This rapid rise in HPV-positive OSCC cases increases the need to improve standard-of-care therapies for this particular subtype of HNC.
While expression of the HPV oncoproteins E6 and E7 quickly inactivates several tumor suppressors, the development of invasive cancer requires many years of viral persistence and disease progression in immunocompetent individuals. Our recent global gene expression analysis of human cervical tissues in different disease stages (normal, early and late precancerous lesions, and cancer) revealed dynamic gene expression changes in a series of cellular pathways, including the cell cycle, translation, mitochondrial energy metabolism, and estrogen signaling (den Boon et al., 2015). Aside from cell cycle- and proliferation-related genes, which are significantly upregulated immediately with high-risk HPV E6 and E7 expression, most host gene expression changes influenced by HPV persistence are slow progressing and accumulating throughout cancer progression. Many of the genes altered slowly and continuously are involved in immune responses and inflammation, such as cytokines and chemokines (den Boon et al., 2015). Our study has further revealed that restoration of the chemokine CXCL14 in mouse OSCC cells, which is downregulated by HPV E7, significantly suppresses HPV-positive OSCC growth in vivo by enhancing NK and T cell infiltration into tumor-draining lymph nodes (Cicchini et al., 2016). These results suggest that HPV-mediated immune dysregulation during virus persistence is important to prevent the elimination of HPV-infected cells during cancer progression. Thus, furthering our understanding of virus-directed immune dysregulation would be critical to develop preventive and therapeutic tools for treating virus-associated cancer as well as eliminating virus-infected cells.
2. Host defense mechanisms against HPV
While the majority of the human population acquires HPV infections, only about 10% to 15% of infected individuals establish life-long persistent infection, and only a subset of which has the potential to progress to invasive cancer (Schiffman 2007). This suggests that, for a majority of HPV-infected individuals, host defense mechanisms are largely effective at eliminating initial HPV infection.
2.1. Physical barriers
The unique lifecycle and strict tropism of HPV generates significant physical barriers for virus entry into basal keratinocytes, the native host cells of HPV. To initiate infection, HPV first needs to translocate across skin and mucous membranes, a process that is facilitated by tissue damage. The mucous membrane poses a major physical barrier to virus infection due to the secretion of a viscous protective fluid and antimicrobial peptides found therein. Once HPV reaches the extracellular matrix, extracellular proteases trigger conformational changes in the virus capsid that facilitates virus internalization. Following uptake of virus particles by macropinocytosis, viruses travel along the endocytic pathway to acidic late endosomes/lysosomes, and retrograde traffics through the Golgi to reach the nucleus (DiGiuseppe et al., 2016; Lipovsky et al., 2013; Pyeon et al., 2009; Schelhaas et al., 2012). During intracellular trafficking, a vast majority of virus particles are degraded and eliminated by host autophagy (Griffin et al., 2013). The nuclear envelope poses another physical barrier to HPV DNA entry into the nucleus. Nuclear envelope breakdown during prometaphase is required for the successful establishment of HPV infection (Pyeon et al., 2009; Schelhaas et al., 2012). Beyond these physical barriers, human α-defensins, particularly α-defensin 5, were found as potent antagonists of HPV infection through inhibition of furin cleavage of the HPV minor capsid protein L2 at the cell surface (Buck et al., 2006; Wiens and Smith, 2015).
2.2. Innate immunity
Once HPV enters a host cell, HPV DNA can be recognized by innate pathogen sensors. Absent in melanoma 2 (AIM2), interferon-γ (IFN-γ) inducible protein 16 (IFI16), and cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS) are cytosolic DNA sensors, while IFI16 also detects foreign DNA in the nucleus (Hornung et al., 2009; Kerur et al., 2011; Li et al., 2013; Unterholzner et al., 2010). Triggering of the AIM2 inflammasome by viral DNA leads to the maturation of caspase-1 and interleukin-1β (IL-1β), which are commonly found activated in HPV16-infected lesions and keratinocytes (Reinholz et al., 2013). IFI16 restricts HPV genome replication and gene transcription by enhancing heterochromatin association with the early and late promoters (Lo Cigno et al., 2015). Further, there is a significant correlation in women between clearance of initial HPV infection and higher expression of nucleic acid-sensing toll-like receptors (TLR3, TLR7, TLR8, and TLR9) as well as TLR2 (Daud et al., 2011). One of the downstream effects of pathogen recognition by pattern recognition receptors (PRRs) is the production of type I interferons (IFN-α and -β). IFN-β treatment hinders HPV entry and promotes clearance of latent HPV episomes in persistently infected cells (Chang et al., 2002; Herdman et al., 2006; Warren et al., 2014). HPV genomes in HPV-positive cervical lesions and IFN-β treated cervical keratinocytes are edited by several IFN-inducible cytidine deaminase APOBEC3 family members (Vartanian et al., 2008; Wang et al., 2014). We and other groups have demonstrated that one of these family members, APOBEC3A, significantly restricts HPV infection (Ahasan et al., 2015; Warren et al., 2015b).
Innate immune cells are also involved in early host responses against HPV infection. HPV infection recruits dendritic (DC), Langerhans (LC), natural killer (NK), and natural killer T (NKT) cells to the HPV-infected microenvironment (Amador-Molina et al., 2013). In addition, plasmacytoid dendritic cells (pDC) have been shown to respond to the presence of HPV16 virus-like particles (VLPs) in CxCa tissue and secrete various cytokines, including IFN-α, IL-6, tumor necrosis factor α (TNF-α), and IL-8 following activation of MyD88-dependent signaling (Bontkes et al., 2005; Lenz et al., 2005; Yang et al., 2004). Additionally, increased HPV infection and HPV-associated cancer incidence has been observed in individuals with various functional NK cell deficiencies (Orange, 2013). Taken together, these findings suggest that the early inflammatory response might be critical for initiating a robust host defense against HPV infection.
2.3. Adaptive immunity
The HPV lifecycle is strictly intraepithelial and virions are produced only from the fully differentiated upper layer of skin. Thus, there is no virus-induced cytolysis or viremia, which limits the exposure of HPV to systemic immune responses. Nevertheless, results from many studies have agreed that host T cell responses are required to eliminate HPV-infected cells. The regression rate of cervical precancerous lesions strongly correlates with the presence of intraepithelial granzyme B-positive cytotoxic T cells (Woo et al., 2008). A recent study has shown that LCs isolated from women with persistent HPV16 infection can present HPV antigens and activate HPV16-specific CD8+ T cells (Da Silva et al., 2015). Additionally, using the recently established mouse papillomavirus (Mus musculus papillomavirus 1 or MmuPV1) model, Handisurya et al. revealed that productive MmuPV1 infection and papilloma formation require both CD4+ and CD8+ T cell functions; while CD4- or CD8-knockout C57BL/6 mice were resistant to productive MmuPV infection, depletion of CD4+ and CD8+ T cells from immunocompetent mice led to infection and papilloma formation (Handisurya et al., 2014). In addition, UVB irradiation-induced systemic immune suppression causes mice to become highly susceptible to MmuPV1 infection, which ultimately results in the development of squamous cell carcinoma (Uberoi et al., 2016). Taken together, these findings exemplify the pivotal roles of T cell-mediated immune responses in host clearance of HPV infection. The immunogenicity and efficacy of current HPV vaccines further confirmed that HPV infection can also be prevented by antibody-driven immunological memory (Stanley, 2012). However, antibody titers from natural HPV infection are usually too low to show a protective effect, suggesting that HPV efficiently evades the host antibody response during natural infection (Viscidi et al., 2004).
3. Immune evasion by altering host gene expression
In order to evade host immune defenses and establish persistence, HPV modulates host gene expression by deregulating host DNA methylation, histone modification, and transcription factors.
3.1. Deregulation of DNA methylation by HPV
Epigenetic regulation by DNA methylation provides distinct gene expression patterns in different developmental stages, organ tissue types, and disease states (Hochedlinger and Plath, 2009; Kim et al., 2009; Robertson, 2005). DNA methylation also functions as a host defense mechanism. By methylating CpG residues of foreign DNA in eukaryotic cells, pathogen activity can be shut down due to alterations in pathogen transcriptional profiles (Shalginskikh et al., 2013; Waterhouse et al., 2001). HPV DNA is also frequently methylated and viral gene transcription is repressed following DNA methylation (Badal et al., 2003; Kalantari et al., 2004). Previous studies have shown that several pathogens hijack the host DNA methylation machinery to manipulate host transcription to their benefit (Hino et al., 2009; Liu et al., 2013; Silmon de Monerri and Kim, 2014). Interestingly, the HPV oncoprotein E7 interacts with the DNA methyltransferase DNMT1, and stimulates its methyltransferase activity (Burgers et al., 2007) (Fig. 1A). This may partially explain the global changes in the host methylome that we have observed in HPV-positive keratinocytes (Cicchini et al., unpublished results). We have recently shown that expression of the chemokine CXCL14 is downregulated during HPV-associated cancer progression by promoter hypermethylation, in an E7-dependent manner (Cicchini et al., 2016). Since restoration of CXCL14 expression in HPV-positive HNC cells significantly suppresses tumor growth in vivo, HPV E7-mediated CXCL14 promoter methylation may represent an immune evasion strategy during HPV persistence. Our and other studies have shown that CXCL14 induces direct chemotaxis of various immune cells including DCs, LCs, NK, and T cells (Cicchini et al., 2016; Schaerli et al., 2005; Shurin et al., 2005). Additionally, using transgenic mouse models, it has been demonstrated that HPV16 E7 expression in the epidermis creates a strong immunosuppressive area where LC and CD8+ T cell functions are significantly suppressed (Abd Warif et al., 2015; Doan et al., 1999; Tindle et al., 2001). This immune suppression appears to be mediated by an influx of HPV16 E7-induced regulatory T (Treg) cells and tolerization of cytotoxic T cells (Doan et al., 1999; Narayan et al., 2009). In addition to HPV E7, HPV E6 is also involved in deregulation of host DNA methylation. High-risk HPV E6 represses IFN-κ, which is constitutively expressed in keratinocytes and serves to reinforce IFN-stimulated gene expression (Fig. 1A). E6-mediated repression of IFN-κ can be reversed by treatment with a DNA methyltransferase inhibitor (Reiser et al., 2011; Rincon-Orozco et al., 2009). These findings suggest that HPV manipulates host DNA methylation to evade host immune defenses.
Fig. 1.
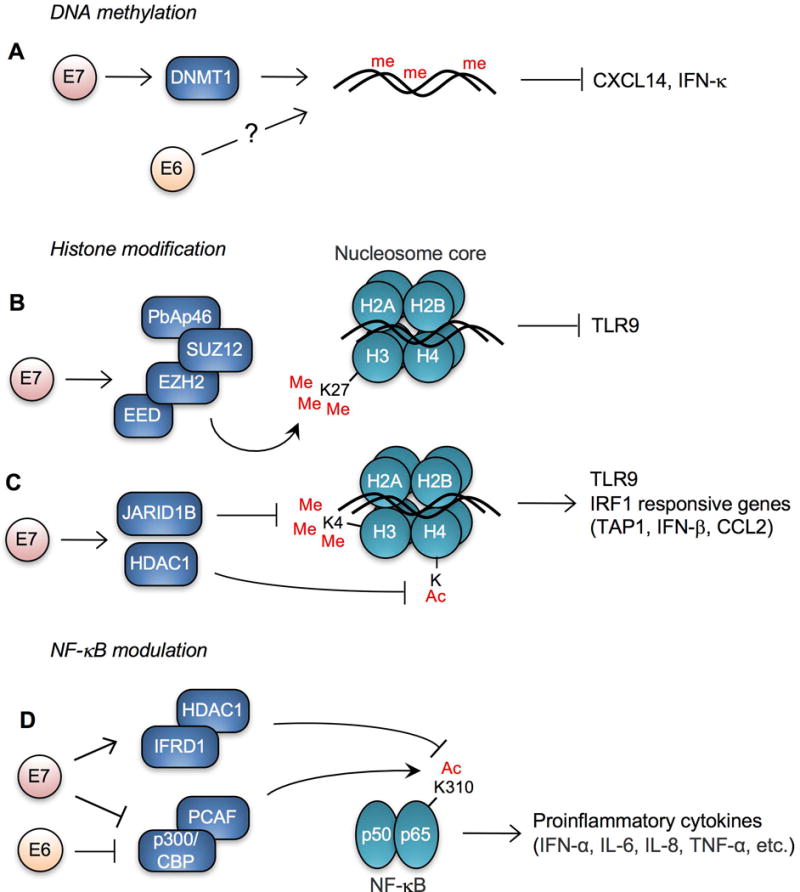
HPV alters transcription of key immune modulators. (A) Gene transcription of IFN-κ and CXCL14 is repressed by high-risk HPV E6 and E7, respectively, through DNA methylation. HPV16 E7 interacts with DNMT1 and enhances its activity, but the ability of E6 to influence host DNA methylation is unknown. High-risk HPV E7 represses transcription of TLR9 and/or IRF1 responsive genes by (B) inducing K27 methylation and (C) reducing K4 methylation and acetylation of histone molecules. (D) NF-κB signaling is inhibited by high-risk HPV E6 and E7 through inhibiting K310 acetylation of p65. me, 5-methylcytidine; Me, methyl lysine; Ac, acetyl lysine; E2BS, E2 binding site.
3.2. Deregulation of histone modification by HPV
Histone modification is another mechanism of epigenetic reprogramming altered by HPV. Cellular gene expression is tightly regulated by chromosome remodeling through histone methylation, phosphorylation, acetylation, ubiquitination, and sumoylation (Kouzarides, 2007). HPV16 E7 binds to E2F6 and interferes with the formation of E2F6-containing polycomb transcriptional repressor complexes (McLaughlin-Drubin et al., 2008). HPV-mediated modulation of host histone methylation has been well described (McLaughlin-Drubin et al., 2011; McLaughlin-Drubin et al., 2008). Interestingly, HPV38 E7 recruits polycomb protein enhancer of zeste homolog 2 (EZH2) to the TLR9 promoter region, resulting in histone 3 trimethylation at lysine 27 (H3K27me3) and repression of TLR9 transcription (Pacini et al., 2015) (Fig. 1B). Another study has shown that HPV16 E7 recruits histone deacetylase HDAC1 and histone demethylase JARID1B (also known as KDM5B) to the regulatory region upstream of the TLR9 transcriptional start site and reduces H4 acetylation and H3K4me3, leading to downregulation of TLR9 expression (Hasan et al., 2013) (Fig. 1C). Since TLR9 recognizes viral DNA and induces innate immune responses, TLR9 repression may represent an important host immune response pathway antagonized by HPV. Additionally, both high-risk and low-risk HPV E7 proteins inhibit the transcriptional activity of IRF1 by recruiting HDAC1 to promoters that contain IRF1 response elements (Park et al., 2000). Consequently, IRF1 responsive genes, including the ATP-binding cassette (ABC) peptide transporter associated with antigen processing 1 (TAP1), IFN-β, and CCL2, are significantly downregulated in HPV16 E7 expressing cells and mice (Georgopoulos et al., 2000; Um et al., 2002). Consistently, treatment of HPV-positive tumor cells with an HDAC inhibitor increased the surface expression of the major histocompatibility complex class I (MHC-I) molecules, enhancing the susceptibility of tumor cells to E7-specific CD8+ T cells (Lee et al., 2013).
3.3. Modulation of NF-κB by HPV
HPV deregulates the activity of transcription factors to repress proinflammatory cytokine production. Nuclear factor-κB (NF-κB) is one of the most important transcription factors involved in immune signaling (Ghosh et al., 1998; Lee and Kim, 2007). Accordingly, NF-κB is frequently targeted by many different viruses, including HPV (Brady and Bowie, 2014; Correia et al., 2013; Qu and Lemon, 2010; Vandermark et al., 2012). High-risk HPVs decrease K310 acetylation of NF-κB in keratinocytes by enhancing IFN related developmental regulator 1 (IFRD1) expression, resulting in downregulation of proinflammatory cytokines (Tummers et al., 2015) (Fig. 1D). E7 alone is capable of inhibiting imiquimod-induced NF-κB transactivation of IFN-α, IL-6, and TNF-α (Richards et al., 2015). Impairing the acetylation at K310 in the p65 subunit of NF-κB requires the CR1 and CR3 domains of E7 and leads to reduced nuclear translocation of NF-κB (Richards et al., 2015). An earlier study has shown that HPV16 E6 and E7 also bind to p300/CBP-associated factor (PCAF) and inhibits NF-κB activation, resulting in IL-8 downregulation (Huang and McCance, 2002) (Fig. 1D).
3.4. Transcriptional regulation by HPV through undefined pathways
While tremendous progress has been made in understanding the molecular mechanisms by which HPV deregulates host gene expression, many pathways still remain undefined. Previous studies have shown that E6 and E7 downregulates the expression of proinflammatory cytokines and chemokines, IL-8, IL-18, CCL2, and CCL20 (Cho et al., 2001; Guess and McCance, 2005; Huang and McCance, 2002; Kleine-Lowinski et al., 2003). In addition to repression of proinflammatory gene expression, the HPV oncoproteins E6 and E7 also upregulate expression of immunosuppressive genes in host cells. Previous studies have shown that HPV16 E7 expression in vivo is sufficient to trigger local immune suppression (Dunn et al., 1997; Matsumoto et al., 2004). Mittal et al., have revealed that indoleamine 2,3-dioxygenase 1 (IDO1) expression in dermal DCs is highly upregulated by HPV16 E7 expression in mouse skin (Mittal et al., 2013). IDO1 is an IFN-γ-inducible immune checkpoint molecule that promotes the development of functional regulatory T (Treg) cells and acts as a suppressor of antitumor immunity (Bonanno et al., 2012; Munn and Mellor, 2013; Schmidt and Schultze, 2014; van Baren and Van den Eynde, 2015). Drugs targeting the IDO1 pathways are already in clinical trials to reverse cancer-induced immune suppression (Platten et al., 2014). Treg expansion and DC tolerance are also driven by secretion of receptor activator of NF-κB ligand (RANKL) during cervical cancer progression (Demoulin et al., 2015). RANKL is a member of the TNF superfamily that regulates DC maturation and survival and is frequently upregulated in several cancers (Chino et al., 2009; Gonzalez-Suarez and Sanz-Moreno, 2016). Interestingly, while IL-18 is downregulated by HPV16 E6 (Cho et al., 2001), expression of its natural antagonist, IL-18 binding protein (IL-18BP), is significantly increased in primary human keratinocytes expressing either high- or low-risk HPV E7 (Richards et al., 2014). IL-18BP prevents the deleterious effects of excessive IL-18 secretion and reduces activation of primary Th1 CD4+ T cells (Novick et al., 1999; Richards et al., 2014). A proximal gamma interferon activation site (GAS) element in the IL-18BP promoter is responsible for IL-18BP transactivation by E7, indicating that IFN-γ signaling might be critical for increased expression of immunosuppressive genes, including IL-18BP and IDO1. A recent study has shown that transgenic mice expressing HPV16 E7 have increased epidermal secretion of the chemokines CCL2 and CCL5, which recruit mast cells expressing CCR2 and CCR5, the receptors of CCL2 and CCL5, respectively (Bergot et al., 2014). Mast cells in HPV16 E7-expressing transgenic skin generate an immunosuppressive environment resulting in significantly reduced graft rejection (Bergot et al., 2014). Contrastingly, a previous study has shown that retroviral transduction of HPV16 E6 and E7 almost completely shut down CCL2 expression in primary cervical epithelial cells (Kleine-Lowinski et al., 2003). These conflicting results may be caused by differential effects from expression of both E6 and E7, and E7 alone, or from an in vivo mouse model vs. cultured human cells. Interestingly, another study showed that CCL2 expression is downregulated by HPV16 E6 and E7 in primary human keratinocytes, but upregulated by HPV16 E6 and E7 in transformed HaCaT keratinocytes (De Andrea et al., 2007). These results indicate that HPV regulation of CCL2 expression might be complex and different between normal and cancer cells. To better understand the physiological relevance of HPV-induced immune evasion by chemokine deregulation, it is critical to determine the molecular pathways by which HPV modulates expression of these genes.
The HPV E2 protein is a transcription factor, containing DNA binding and transactivation domains (McBride, 2013). Normally, it binds to E2 binding motifs within the viral upstream regulatory region (URR) to regulate early and late gene expression (McBride et al., 1991). However, in addition to viral gene regulation, HPV E2 also regulates the transcription of numerous host genes (Bellanger et al., 2011). A recent genome-wide gene expression analysis revealed that the high-risk HPV E2 protein downregulates Stimulator of Interferon Genes (STING) and IFN-κ expression in primary human keratinocytes (Sunthamala et al., 2014). Downregulation of STING and IFN-κ expression is also observed in HPV-positive patient tissue samples (Sunthamala et al., 2014). STING is an endoplasmic reticulum (ER) adaptor protein induced by cytosolic DNA. STING binds with TANK binding kinase (TBK1) and IFN regulatory factor 3 (IRF3), resulting in phosphorylation of IRF3 and thereby promoting type I IFN production (Tanaka and Chen, 2012). However, it is still unclear if downregulation of STING and IFN-κ expression is directly caused by E2 or indirectly by interactions of E2 with other cellular factors that mediate transcription repression.
4. Immune evasion by dysregulating protein functions
4.1. Protein-protein interaction/sequestration
The HPV oncoproteins E6 and E7 lack enzymatic activity. Instead, they act as adaptor proteins that interact with multiple host proteins, recruit them to desired locations, and hijack their activities to enhance virus replication and persistence, and inadvertently induce malignancy (Roman and Munger, 2013; Vande Pol and Klingelhutz, 2013). HPV E6 and E7 frequently inhibit host protein functions by direct binding. The HPV18 E6 protein directly interacts with non-receptor tyrosine-protein kinase 2 (TYK2), resulting in a reduction of IFN-α-induced phosphorylation in TYK2 and Signal Transducer and Activator of Transcription 2 (STAT2) proteins (Li et al., 1999) (Fig. 2A). TYK2 is a member of the JAK family and is important for signal transduction following receptor binding by various cytokines, including IL-6, IL-12, and type I IFNs. The IFN regulatory transcription factor IRF3 is selectively bound and inhibited by HPV16 E6 protein, while HPV6, HPV11, and HPV18 E6 proteins bind poorly to IRF3 (Ronco et al., 1998). IRF3 plays a central role in inducing innate immune response against viral infections (Haller et al., 2006). The interaction of HPV16 E6 to IRF3 does not lead to ubiquitination or degradation of IRF3, but significantly represses its activity on target gene transcription and IFN-β production. This suggests that HPV16 E6 binding is sufficient for inhibition of IRF3 functions. Cytosolic DNA can be detected by cGAS, which initiates innate immune signaling through the adapter proteins STING and IRF3 (Cai et al., 2014; Sun et al., 2013). Interestingly, HPV18 E7 antagonizes the activation of the cGAS-STING pathway and limits the production of type I IFNs by binding with STING through its LXCXE motif (Lau et al., 2015) (Fig. 2B). Additionally, HPV16 E7 protein interacts with IRF1, probably at its N-terminal DNA-binding domain, resulting in decreased expression of IRF1 responsive genes, including TAP1, IFN-β, and CCL2 (Park et al., 2000; Perea et al., 2000; Um et al., 2002) (Fig. 2C). IFN signaling is further repressed by HPV16 E7 as it also binds to IRF9, another IFN regulatory factor (Antonsson et al., 2006) (Fig. 2A). IFN signaling lies at the core of antiviral innate immune responses and appears to be heavily targeted by the HPV oncoproteins E6 and E7. Our previous studies have shown that IFN-β treatment significantly inhibits HPV infection in primary and immortalized keratinocytes (Warren et al., 2014; Warren et al., 2015b). These findings indicate that evasion of IFN signaling mediated through HPV E6 and E7 interactions with host proteins might be critical for HPV persistence.
Fig. 2.
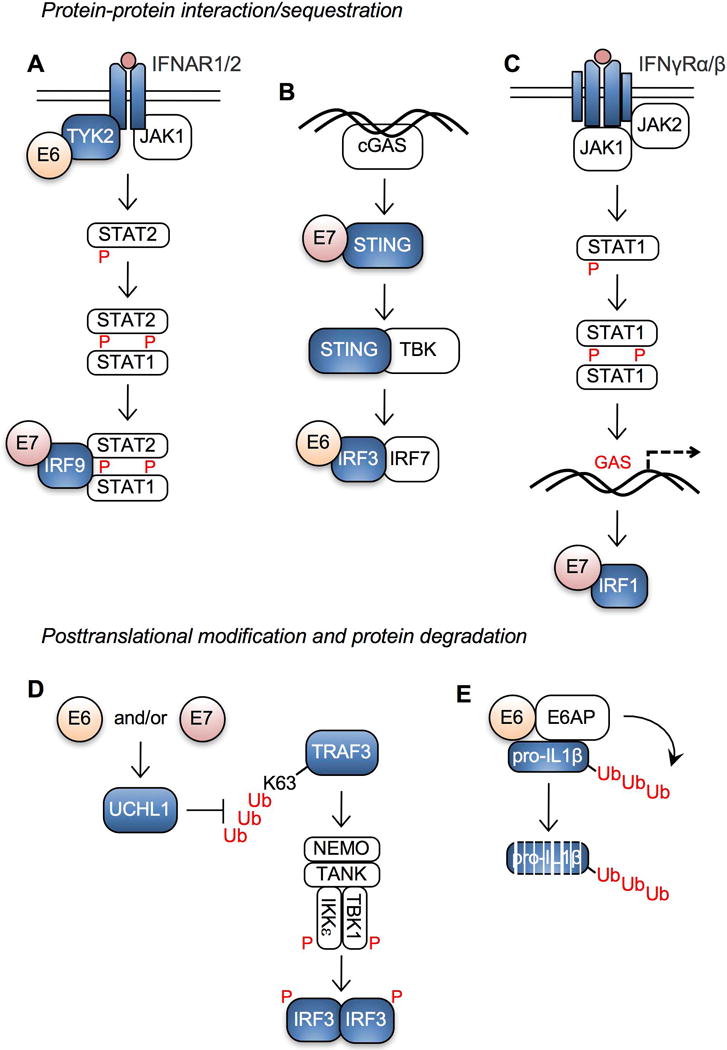
HPV E6 and E7 interacts with core components of the interferon pathway. (A) HPV inhibits type I IFN receptor signaling by HPV E6 and E7 interactions with TYK2 and IRF9, respectively. (B) HPV inhibits signaling by the DNA sensor cGAS via high-risk HPV E6 and E7 interactions with IRF3 and STING, respectively. (C) HPV inhibits type II IFN receptor signaling by high-risk E7 interaction with IRF1. (D) The HPV oncoproteins E6 and/or E7 suppress IRF3 activation by inhibiting K63 ubiquitination of TRAF3 through downregulation of UCHL1. (E) Interaction between high-risk HPV E6 and E6AP facilitates poly-ubiquitination and degradation of pro-IL-1β. P, phosphorylation; GAS, gamma interferon activation site; Ub, ubiquitination.
4.2. Posttranslational modification and protein degradation
The mechanisms of host protein degradation by the HPV oncoproteins E6 and E7 are well established (Howley, 2006). However, posttranslational modification of immunoregulatory proteins is relatively understudied. Karim et al. have shown that high-risk HPVs upregulate the cellular protein ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) in keratinocytes (Karim et al., 2013). UCHL1 inhibits K63-linked ubiquitination of tumor necrosis factor receptor-associated factor 3 (TRAF3) resulting in decreased TRAF3-TBK1 complex formation and IRF3 phosphorylation (Fig. 2D). UCHL1 also mediates degradation of the essential modulator of NF-κB, NEMO, and suppresses p65 phosphorylation and NF-κB signaling. HPV16 E6 also induces degradation of pro-IL-1β in a proteasome-dependent manner (Niebler et al., 2013) (Fig. 2E). E6-mediated pro-IL-1β degradation is independent from caspase-1 activation, autophagy, or lysosomal degradation, but requires E6AP expression and poly-ubiquitination of pro-IL-1β. Since the proinflammatory cytokine IL-1β plays important roles in antiviral defense (Gram et al., 2012; Poeck and Ruland, 2012), these results suggest that high-risk HPV suppresses the proinflammatory response by inducing proteasome-mediated degradation of intracellular cytokines. Interestingly, a number of studies have shown that polymorphism in the IL-1β gene cluster is tightly linked to CxCa and HNC risk (Wu et al., 2014; Xu et al., 2013). Specifically, women with IL-1β T-allele containing genotypes show increased risk of CxCa (Dutta et al., 2015). Our unpublished global gene expression data shows that IL-1β expression is significantly downregulated in HPV-positive normal keratinocytes but upregulated in CxCa tissues (Cicchini et al., unpublished results). These results imply that IL-1β may play dual roles in inhibiting HPV infection by its antiviral function in early stages of disease, but facilitating cancer progression through chronic inflammation in late stages.
5. Immune evasion by altered cytoplasmic trafficking of host proteins
5.1. Intracellular sequestration of MHC molecules by HPV
Antigen presentation of viral epitopes by MHC-I molecules is critical to elicit cell-mediated immune responses that eliminate virus-infected cells. In order to escape host recognition, many viruses have various strategies to suppress surface expression of MHC-I molecules (Blagoveshchenskaya et al., 2002; Hewitt, 2003; Jackson et al., 2011), and HPV is no exception. HPV and bovine papillomavirus (BPV) E5s are small transmembrane proteins that bind to several host transmembrane proteins, including MHC-I. E5 binding to MHC-I leads to its downregulation at the host cell surface (Ashrafi et al., 2006; Ashrafi et al., 2005; Ashrafi et al., 2002; DiMaio and Petti, 2013). Upon binding of antigenic peptides delivered by TAP1 in the ER, MHC-I molecules, composed of a heavy α chain and a soluble subunit β2-microglobulin (β2m), traffic through the secretory pathway and imbed within the cell membrane (Donaldson and Williams, 2009). Several quality control mechanisms are involved in this process for accurate antigen presentation. HPV16 E5 physically interacts with the heavy chain of the MHC-I molecule through the hydrophobic region of E5 protein and retains MHC-I in the Golgi and ER (Ashrafi et al., 2006; Ashrafi et al., 2005) (Fig. 3). While HPV16 E5 does not affect expression of the heavy chain and TAP1 proteins, enhanced expression of the MHC I heavy chain by IFN-β treatment restores surface expression of MHC-I molecules. Interestingly, HPV16 E5 specifically downregulates HLA-A and -B expression, but not HLA-C and HLA-E (Ashrafi et al., 2005) (Fig. 3). Our unpublished data showed that HLA-C and HLA-E are the two most transcriptionally downregulated MHC-I molecules in HPV-positive keratinocytes, and this downregulation is dependent on HPV16 E7 expression (Cicchini et al., unpublished results). Further, HPV E5 downregulation of MHC-I molecules at the cell surface correlates with poor CD8+ T cell responses in E5 expressing cells (Campo et al., 2010). An additional study has described a correlation between surface expression of MHC-I molecules and decreased TAP1 expression, suggesting that TAP1 downregulation by E7 may also interfere with MHC-I trafficking (Li et al., 2010). HPV16 E5 also downregulates the surface expression of MHC-II molecules by preventing degradation of the invariant chain that blocks peptide loading (Zhang et al., 2003). Both high-risk HPV16 and low-risk HPV6 E5s also downregulate surface expression of the non-classical MHC molecule CD1d (Miura et al., 2010) (Fig. 3). HPV E5 directly interacts with calnexin in the ER and redirects CD1d to the cytosolic proteolytic pathway (Miura et al., 2010). CD1d expression on the cell surface activates NKT cell responses to various viral, bacterial, and fungal infections (Brigl and Brenner, 2004). Thus, HPV may evade host immune responses by downregulating the surface expression of CD1d. Further investigation is necessary to determine if CD1d presents HPV-related antigens to induce protective antiviral activity.
Fig. 3.
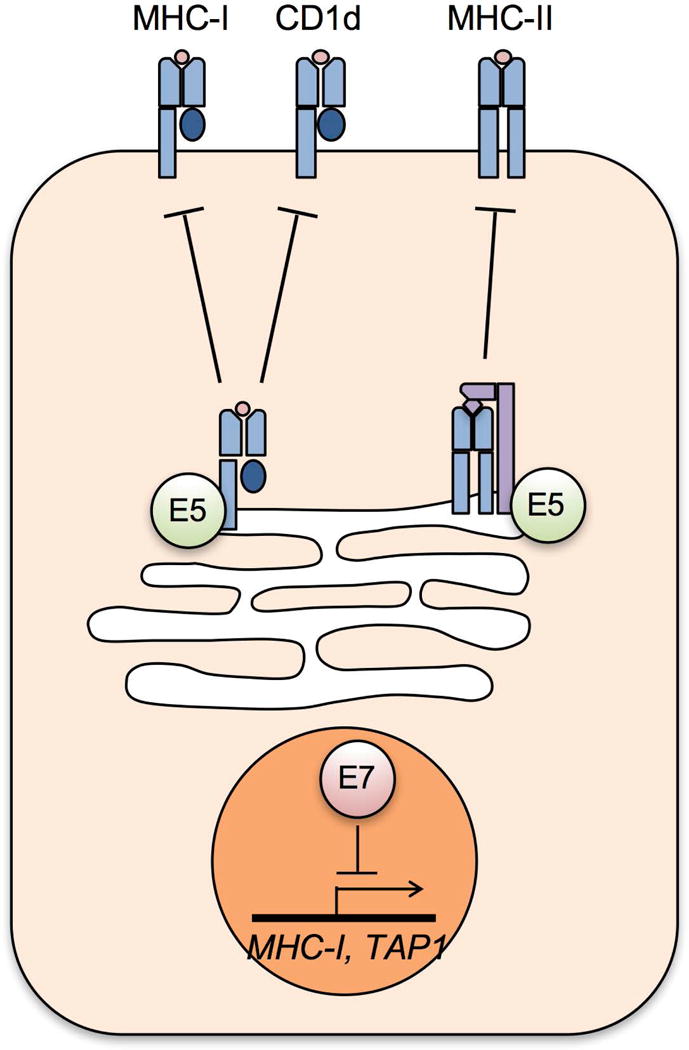
HPV E5 and E7 downregulate surface expression of MHC molecules. HPV E5 binds to MHC-I, CD1d, and the invariant chain of MHC-II in the Golgi and ER, preventing their trafficking to the cell surface. High-risk HPV E7 represses transcription of MHC-I genes.
5.2. Virus trafficking hindered by HPV in antigen presenting cells
The HPV minor structural protein L2 disrupts normal endocytic trafficking of virus particles in DCs and LCs (Fahey et al., 2009). LCs, which are skin resident DCs, uptake L1-only virus-like particles (VLPs) and efficiently induce expression of proinflammatory cytokines and costimulatory molecules leading to the activation of cytotoxic T cell responses (Fausch et al., 2003; Lenz et al., 2001; Rudolf et al., 2001). In contrast, HPV VLPs containing both L1 and L2 proteins did not activate LCs to present viral antigens (Fausch et al., 2002). Interestingly, HPV16 L2 facilitates internalization of VLPs through interaction with the annexin A2 (ANXA2) heterotetramer (Fahey et al., 2009; Woodham et al., 2014). ANXA2 is associated with S100A10 as a heterotetramer and critical for HPV16 VLP internalization (Dziduszko and Ozbun, 2013; Woodham et al., 2012). Thus, it is likely that HPV16 L2 blocks LC maturation and viral antigen presentation by modulating intracellular trafficking of viral particles. Previously, we have shown that overexpression of the interferon-inducible transmembrane protein 1 (IFITM1) delays viral capsid protein degradation and significantly enhances HPV16 VLP entry into primary keratinocytes (Warren et al., 2014). IFITMs, induced by IFN treatment, inhibit a series of RNA viruses by redirecting virus particles to low-pH late endosomes and lysosomes during intracellular trafficking (Brass et al., 2009; Perreira et al., 2013). These results imply that HPV not only bypasses IFITM restriction but may utilize certain IFN-inducible proteins to facilitate virus infection.
6. Roles of host defense in papillomavirus genome evolution
It is well known that RNA viruses and retroviruses evolve rapidly throughout the course of an infection. Rapid mutation allows these viruses to efficiently evade host immune recognition and develop drug resistance. Viral genome mutation rates determine whether host immune responses clear viral infections, subsequently resulting in dramatic changes in viral fitness and pathogenesis (Pfeiffer and Kirkegaard, 2005; Vignuzzi et al., 2006). Accordingly, virus evolution is driven by strong selective pressures elicited by various host immune defenses (Woo and Reifman, 2012). Compared to the mutation rates of RNA viruses (10−4 to 10−6 mutations/bp/generation), double-stranded DNA viruses have substantially lower mutation rates (10−6 to 10−8 mutations/bp/generation). The mutation rate of DNA viruses is approximately a log fold higher than the mutation rates of the human genome (Lauring et al., 2013; Sanjuan et al., 2010). Nevertheless, throughout the course of virus-host coevolution for millions of years, DNA viruses have established large and diverse families that persistently infect countless host species. We and others have identified evolutionary clues within papillomavirus genomes that suggest an important role for host immunity in virus-host coevolution.
6.1. Depletion of CpG dinucleotides in HPV genomes
CpG dinucleotides in viral genomes are frequently targeted for DNA methylation, which negatively affects viral gene expression and occasionally triggers viral genome integration and excision (Karlin et al., 1994). Like other viruses, CpG motifs in HPV genomes can be methylated, and the viral CpG methylation levels in CxCa tissues are negatively correlated with CxCa progression (Badal et al., 2004; Badal et al., 2003). HPV gene transcription is repressed by hypermethylation of the viral upstream regulatory region (URR) (Rosl et al., 1993). An earlier study has shown that in vitro methylation of BPV DNA reduces the transformation rate of mouse fibroblasts (Christy and Scangos, 1986). In contrast, other studies have revealed that DNA methylation in the HPV L1 and L2 regions is correlated with HPV integration and cervical malignancies (Kalantari et al., 2004; Kalantari et al., 2010; Mirabello et al., 2012). These results suggest that HPV gene expression and pathogenesis are closely regulated by viral DNA methylation. Additionally, DNA containing unmethylated CpG dinucleotides is recognized by TLR9 and induces TLR9-mediated antiviral signaling through NF-κB, resulting in type I IFN production (Kumagai et al., 2008). Currently, there is no evidence of direct activation of TLR9 signaling by recognition of unmethylated HPV DNA. However, since TLR9 expression is significantly downregulated in cells expressing the HPV oncoprotein E7 as described above, it is likely that HPV infection and viral gene expression may be influenced by TLR9 responses. Similar to many other small DNA viruses, such as adenovirus and parvovirus, papillomavirus genomes have significantly lower contents of CpG dinucleotides than expected by random chance (Upadhyay et al., 2013; Upadhyay and Vivekanandan, 2015; Warren et al., 2015a). In a recent publication, we calculated the ratio of observed vs. expected (O/E) counts of all dinucleotides in the genomes of all 274 PVs deposited in the Papillomavirus Episteme (PaVE) database (Warren et al., 2015a). We revealed that CpG dinucleotides are the most underrepresented dinucleotide in the genomes of all PV types, except two bird PVs. These results suggest that the loss of CpG dinucleotides in PV genomes may be tied to evolutionary pressures elicited by host DNA methylation and/or TLR9 recognition (Fig. 4A).
Fig. 4.
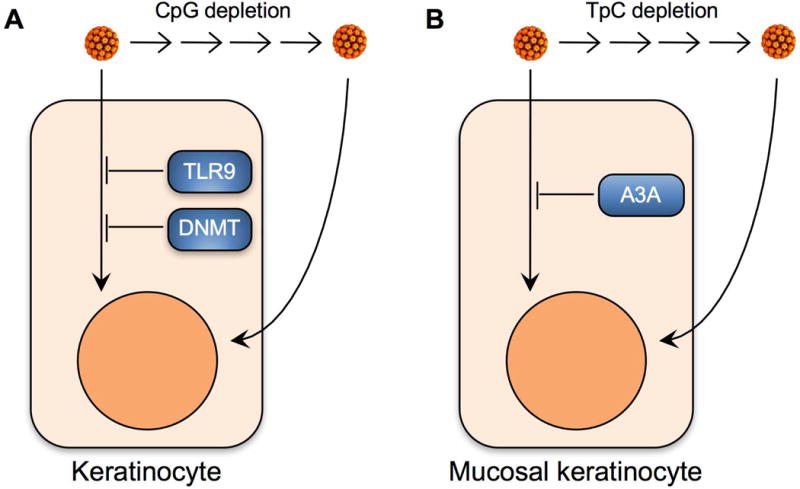
Bias in dinucleotide frequencies suggest that host restriction may drive papillomavirus genome evolution. (A) Unmethylated papillomavirus genomes can be recognized by TLR9 and/or targeted for methylation-induced gene silencing. During coevolution with their hosts, papillomaviruses have been selected and enriched for variants that contain reduced CpG motifs. CpG dinucleotide depletion may reflect an important immune evasion strategy. (B) Mucosal skin expresses high levels of nearly all A3 isoforms, including the HPV restriction factor A3A. The Alphapapillomaviruses are a genus of papillomaviruses that preferentially infect mucosal skin. TpC depletion amongst the Alphapapillomaviruses may allow for evasion of A3A at mucosal sites. DNMT, DNA methyltransferases.
6.2. Depletion of TpC dinucleotides in HPV genomes
The TpC dinucleotide motif is preferably targeted by several APOBEC3 (A3) cytidine deaminase family members that convert cytidine (C) to uridine (U) in single stranded DNA and RNA (Chelico et al., 2006; Yu et al., 2004). The human A3 family consists of seven members: A3A, A3B, A3C, A3D, A3F, A3G, and A3H (Jarmuz et al., 2002; LaRue et al., 2009). Of these seven A3 family members, A3A, A3B, and A3H are expressed in skin keratinocytes (Vartanian et al., 2008). A3s were first discovered as important host restriction factors that block the replication of several retroviruses, including human immunodeficiency virus (HIV) (Hultquist et al., 2011; Sheehy et al., 2002; Zheng et al., 2004). Several studies have shown that A3A can eliminate transfected foreign DNA (Stenglein et al., 2010) and restrict several DNA viruses (Chen et al., 2006; Narvaiza et al., 2009). Editing of episomal HPV genomes by A3A, A3B, and A3H in cervical lesions has been demonstrated (Vartanian et al., 2008; Wang et al., 2014). Our recent study has revealed that A3A expression restricts HPV entry in a cytidine deaminase dependent manner (Warren et al., 2015b). Despite its ability to restrict HPV infection, A3A expression is upregulated in HPV-infected keratinocytes as well as HPV-positive CxCa (Warren et al., 2015b). Interestingly, TpC dinucleotides, the preferred dinucleotide target site of many A3s, are dramatically underrepresented in papillomavirus genomes. TpC depletion was most dramatic amongst the Alphapapillomaviruses which preferably infect mucosal skin such as the cervicovaginal and oral regions (Warren et al., 2015a). Surprisingly, mucosal skin expresses significantly higher basal levels of all A3 isoforms (except A3B) when compared to cutaneous skin. These findings imply that TpC dinucleotide depletion in Alphapapillomaviruses may allow for persistent infection of mucosotropic PV types to avoid A3-mediated antiviral editing (Warren et al., 2015a; Warren and Pyeon, 2015). Additionally, unlike the CpG depletion universally found in the genomes of most papillomavirus types, the degree of TpC dinucleotide depletion varies among PV types infecting different animal species. Taken together, these findings suggest that TpC dinucleotide content variation in PV genomes may limit editing by A3 family members during viral infection and persistence (Fig. 4B).
6.3. Epitope changes in HPV variants
Host adaptive immune responses efficiently neutralize virus particles, eliminate virus-infected cells, and generate immune memory to cope with future infections. Viruses, particularly those with smaller genome capacities, have antigenically altered immune epitopes to evade recognition by adaptive immune cells. On the other hand, viruses with larger genomes encode viral immunomodulatory genes (van de Weijer et al., 2015; Vossen et al., 2002). The rapid accumulation of mutations within the immunodominant epitopes of HIV-1 and hepatitis C virus (HCV) allows for immune escape during the course of an infection, and poses a major challenge to the development of effective vaccines (Goulder et al., 1997; Henn et al., 2012; Timm et al., 2004). Previous studies have described naturally occurring HPV variants in the neutralizing epitopes of L1 and L2 proteins that significantly reduce antigenicity of HPV (Seitz et al., 2013; Yang et al., 2005). For instance, several HPV16 L1 variants isolated from premalignant and malignant cervical tissues show impaired viral capsid assembly, which influences differential B cell class switching and leads to the production of non-neutralizing antibodies (Yang et al., 2005). Although mutations disrupting capsid assembly would diminish viral fitness, they may contribute to virus persistence by evading host immune surveillance (Yang et al., 2005). Additionally, natural epitope variants of the HPV minor capsid protein L2 are also found in high-grade squamous intraepithelial lesions (Seitz et al., 2013). These L2 variants were not efficiently neutralized by anti-L2 antibodies from human sera. Recent studies have also discovered HPV E6 and E7 variants that affect host immune responses (Chagas et al., 2013; Chopjitt et al., 2015). In addition to HPV structural proteins, the HPV oncoproteins E6 and E7 are also potently immunogenic. Several T cell epitopes have been described within E6 and E7, and therapeutic vaccines based on E6 and E7 antigens show promising immune responses (Oosterhuis et al., 2012; Ressing et al., 1995; Welters et al., 2008). A novel E6 variant with an amino acid change in a T cell epitope alters its ability to be bound and presented by MHC-II (Chagas et al., 2013). Additionally, the HPV16 E6 Asian variant upregulates miR-21 expression and suppresses IRF1, 3, and 7 (Chopjitt et al., 2015). These results suggest that variation among HPV genomes may allow for differential evasion of host immune defenses.
7. Conclusion
For optimal control and clearance of HPV infections, both the innate and adaptive arms of the immune response appear to be critical. As described, HPV modulates a series of cellular pathways to evade host immune responses during persistent infection. Some of these mechanisms may lead to virus-mediated immune suppression that creates an immunosuppressive microenvironment in mucosal epithelia, which is vital for cancer progression. Recent clinical trials of novel immunotherapies have promise as cancer therapeutics by reversing tumor-induced immune suppression; however, a majority of treated patients generally do not respond to immune checkpoint inhibitors (Gildener-Leapman et al., 2013; Hamid et al., 2013; Wolchok et al., 2013). Thus, it is very likely that there are additional mechanisms by which tumor cells evade antitumor immune responses. As described above, HPV manipulates various molecular and cellular pathways in host cells to evade host immune surveillance and antiviral immune responses. HPV-mediated immune suppression during virus persistence might also contribute to tumor cell evasion of antitumor immune responses. Thus, an in-depth understanding of the mechanisms of HPV-associated immune evasion potentially lead to the development of novel immunotherapeutic tools that effectively restore antiviral and antitumor immune responses.
Highlights.
HPV dysregulates various molecular and cellular pathways to evade host defenses.
HPV alters transcription of key immune modulators.
HPV E6 and E7 interacts with core proteins of the interferon pathway.
HPV E5 and E7 downregulate surface expression of MHC molecules.
Papillomavirus genome evolution may be driven by virus evasion of host restriction.
Acknowledgments
This work was supported by the National Institutes of Health (grant numbers R01 AI091968 and R01 DE026125), Mary Kay Foundation, Dallas, TX (grant number 041-15), The Colorado Clinical and Translational Sciences Institute, and Cancer League of Colorado (grant number 163354-DP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abd Warif NM, Stoitzner P, Leggatt GR, Mattarollo SR, Frazer IH, Hibma MH. Langerhans cell homeostasis and activation is altered in hyperplastic human papillomavirus type 16 E7 expressing epidermis. PLoS One. 2015;10:e0127155. doi: 10.1371/journal.pone.0127155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahasan MM, Wakae K, Wang Z, Kitamura K, Liu G, Koura M, Imayasu M, Sakamoto N, Hanaoka K, Nakamura M, Kyo S, Kondo S, Fujiwara H, Yoshizaki T, Mori S, Kukimoto I, Muramatsu M. APOBEC3A and 3C decrease human papillomavirus 16 pseudovirion infectivity. Biochem Biophys Res Commun. 2015;457:295–299. doi: 10.1016/j.bbrc.2014.12.103. [DOI] [PubMed] [Google Scholar]
- Amador-Molina A, Hernandez-Valencia JF, Lamoyi E, Contreras-Paredes A, Lizano M. Role of innate immunity against human papillomavirus (HPV) infections and effect of adjuvants in promoting specific immune response. Viruses. 2013;5:2624–2642. doi: 10.3390/v5112624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A, Payne E, Hengst K, McMillan NA. The human papillomavirus type 16 E7 protein binds human interferon regulatory factor-9 via a novel PEST domain required for transformation. J Interferon Cytokine Res. 2006;26:455–461. doi: 10.1089/jir.2006.26.455. [DOI] [PubMed] [Google Scholar]
- Ashrafi GH, Haghshenas M, Marchetti B, Campo MS. E5 protein of human papillomavirus 16 downregulates HLA class I and interacts with the heavy chain via its first hydrophobic domain. Int J Cancer. 2006;119:2105–2112. doi: 10.1002/ijc.22089. [DOI] [PubMed] [Google Scholar]
- Ashrafi GH, Haghshenas MR, Marchetti B, O’Brien PM, Campo MS. E5 protein of human papillomavirus type 16 selectively downregulates surface HLA class I. Int J Cancer. 2005;113:276–283. doi: 10.1002/ijc.20558. [DOI] [PubMed] [Google Scholar]
- Ashrafi GH, Tsirimonaki E, Marchetti B, O’Brien PM, Sibbet GJ, Andrew L, Campo MS. Down-regulation of MHC class I by bovine papillomavirus E5 oncoproteins. Oncogene. 2002;21:248–259. doi: 10.1038/sj.onc.1205008. [DOI] [PubMed] [Google Scholar]
- Auluck A, Hislop G, Bajdik C, Poh C, Zhang L, Rosin M. Trends in oropharyngeal and oral cavity cancer incidence of Human Papillomavirus (HPV)-related and HPV-unrelated sites in a multicultural population: The British Columbia experience. Cancer. 2010;116:2635–2644. doi: 10.1002/cncr.25087. [DOI] [PubMed] [Google Scholar]
- Badal S, Badal V, Calleja-Macias IE, Kalantari M, Chuang LS, Li BF, Bernard HU. The human papillomavirus-18 genome is efficiently targeted by cellular DNA methylation. Virology. 2004;324:483–492. doi: 10.1016/j.virol.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Badal V, Chuang LS, Tan EH, Badal S, Villa LL, Wheeler CM, Li BF, Bernard HU. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression. J Virol. 2003;77:6227–6234. doi: 10.1128/JVI.77.11.6227-6234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger S, Tan CL, Xue YZ, Teissier S, Thierry F. Tumor suppressor or oncogene? A critical role of the human papillomavirus (HPV) E2 protein in cervical cancer progression. Am J Cancer Res. 2011;1:373–389. [PMC free article] [PubMed] [Google Scholar]
- Bergot AS, Ford N, Leggatt GR, Wells JW, Frazer IH, Grimbaldeston MA. HPV16-E7 expression in squamous epithelium creates a local immune suppressive environment via CCL2- and CCL5- mediated recruitment of mast cells. PLoS Pathog. 2014;10:e1004466. doi: 10.1371/journal.ppat.1004466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell. 2002;111:853–866. doi: 10.1016/s0092-8674(02)01162-5. [DOI] [PubMed] [Google Scholar]
- Bonanno G, Mariotti A, Procoli A, Folgiero V, Natale D, De Rosa L, Majolino I, Novarese L, Rocci A, Gambella M, Ciciarello M, Scambia G, Palumbo A, Locatelli F, De Cristofaro R, Rutella S. Indoleamine 2,3-dioxygenase 1 (IDO1) activity correlates with immune system abnormalities in multiple myeloma. J Transl Med. 2012;10:247. doi: 10.1186/1479-5876-10-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontkes HJ, Ruizendaal JJ, Kramer D, Meijer CJ, Hooijberg E. Plasmacytoid dendritic cells are present in cervical carcinoma and become activated by human papillomavirus type 16 virus-like particles. Gynecol Oncol. 2005;96:897–901. doi: 10.1016/j.ygyno.2004.10.040. [DOI] [PubMed] [Google Scholar]
- Brady G, Bowie AG. Innate immune activation of NFkappaB and its antagonism by poxviruses. Cytokine Growth Factor Rev. 2014;25:611–620. doi: 10.1016/j.cytogfr.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- Buck CB, Day PM, Thompson CD, Lubkowski J, Lu W, Lowy DR, Schiller JT. Human alpha-defensins block papillomavirus infection. Proc Natl Acad Sci U S A. 2006;103:1516–1521. doi: 10.1073/pnas.0508033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers Wa, Blanchon L, Pradhan S, de Launoit Y, Kouzarides T, Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Campo MS, Graham SV, Cortese MS, Ashrafi GH, Araibi EH, Dornan ES, Miners K, Nunes C, Man S. HPV-16 E5 down-regulates expression of surface HLA class I and reduces recognition by CD8 T cells. Virology. 2010;407:137–142. doi: 10.1016/j.virol.2010.07.044. [DOI] [PubMed] [Google Scholar]
- Chagas BS, Batista MV, Crovella S, Gurgel AP, Silva Neto Jda C, Serra IG, Amaral CM, Balbino VQ, Muniz MT, Freitas AC. Novel E6 and E7 oncogenes variants of human papillomavirus type 31 in Brazilian women with abnormal cervical cytology. Infect Genet Evol. 2013;16:13–18. doi: 10.1016/j.meegid.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Chang YE, Pena L, Sen GC, Park JK, Laimins LA. Long-term effect of interferon on keratinocytes that maintain human papillomavirus type 31. J Virol. 2002;76:8864–8874. doi: 10.1128/JVI.76.17.8864-8874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi AK, Engels Ea, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. Journal of Clinical Oncology. 2011;9:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ –> 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chino T, Draves KE, Clark EA. Regulation of dendritic cell survival and cytokine production by osteoprotegerin. J Leukoc Biol. 2009;86:933–940. doi: 10.1189/jlb.0708419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Kang JW, Cho M, Cho CW, Lee S, Choe YK, Kim Y, Choi I, Park SN, Kim S, Dinarello CA, Yoon DY. Down modulation of IL-18 expression by human papillomavirus type 16 E6 oncogene via binding to IL-18. FEBS Lett. 2001;501:139–145. doi: 10.1016/s0014-5793(01)02652-7. [DOI] [PubMed] [Google Scholar]
- Chopjitt P, Pientong C, Bumrungthai S, Kongyingyoes B, Ekalaksananan T. Activities of E6 Protein of Human Papillomavirus 16 Asian Variant on miR-21 Up-regulation and Expression of Human Immune Response Genes. Asian Pac J Cancer Prev. 2015;16:3961–3968. doi: 10.7314/apjcp.2015.16.9.3961. [DOI] [PubMed] [Google Scholar]
- Christy BA, Scangos GA. In vitro methylation of bovine papillomavirus alters its ability to transform mouse cells. Mol Cell Biol. 1986;6:2910–2915. doi: 10.1128/mcb.6.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchini L, Westrich JA, Xu T, Vermeer DW, Berger JN, Clambey ET, Lee D, Song JI, Lambert PF, Greer RO, Lee JH, Pyeon D. Suppression of Antitumor Immune Responses by Human Papillomavirus through Epigenetic Downregulation of CXCL14. MBio. 2016;7 doi: 10.1128/mBio.00270-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia S, Ventura S, Parkhouse RM. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013;173:87–100. doi: 10.1016/j.virusres.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Da Silva DM, Woodham AW, Skeate JG, Rijkee LK, Taylor JR, Brand HE, Muderspach LI, Roman LD, Yessaian AA, Pham HQ, Matsuo K, Lin YG, McKee GM, Salazar AM, Kast WM. Langerhans cells from women with cervical precancerous lesions become functionally responsive against human papillomavirus after activation with stabilized Poly-I:C. Clin Immunol. 2015;161:197–208. doi: 10.1016/j.clim.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daud II, Scott ME, Ma Y, Shiboski S, Farhat S, Moscicki AB. Association between toll-like receptor expression and human papillomavirus type 16 persistence. Int J Cancer. 2011;128:879–886. doi: 10.1002/ijc.25400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Andrea M, Mondini M, Azzimonti B, Dell’Oste V, Germano S, Gaudino G, Musso T, Landolfo S, Gariglio M. Alpha- and betapapillomavirus E6/E7 genes differentially modulate pro-inflammatory gene expression. Virus Res. 2007;124:220–225. doi: 10.1016/j.virusres.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Demoulin SA, Somja J, Duray A, Guenin S, Roncarati P, Delvenne PO, Herfs MF, Hubert PM. Cervical (pre)neoplastic microenvironment promotes the emergence of tolerogenic dendritic cells via RANKL secretion. Oncoimmunology. 2015;4:e1008334. doi: 10.1080/2162402X.2015.1008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon JA, Pyeon D, Wang SS, Horswill M, Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R, Schott M, Chung L, He Q, Lambert P, Walker J, Newton MA, Wentzensen N, Ahlquist P. Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal estrogen receptor signaling. Proc Natl Acad Sci U S A. 2015;112:E3255–3264. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiuseppe S, Bienkowska-Haba M, Sapp M. Human papillomavirus entry: hiding in a bubble. J Virol. 2016 doi: 10.1128/JVI.01065-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D, Petti LM. The E5 proteins. Virology. 2013;445:99–114. doi: 10.1016/j.virol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Herd K, Street M, Bryson G, Fernando G, Lambert P, Tindle R. Human papillomavirus type 16 E7 oncoprotein expressed in peripheral epithelium tolerizes E7-directed cytotoxic T-lymphocyte precursors restricted through human (and mouse) major histocompatibility complex class I alleles. J Virol. 1999;73:6166–6170. doi: 10.1128/jvi.73.7.6166-6170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Williams DB. Intracellular assembly and trafficking of MHC class I molecules. Traffic. 2009;10:1745–1752. doi: 10.1111/j.1600-0854.2009.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn LA, Evander M, Tindle RW, Bulloch AL, de Kluyver RL, Fernando GJ, Lambert PF, Frazer IH. Presentation of the HPV16E7 protein by skin grafts is insufficient to allow graft rejection in an E7-primed animal. Virology. 1997;235:94–103. doi: 10.1006/viro.1997.8650. [DOI] [PubMed] [Google Scholar]
- Dutta S, Chakraborty C, Mandal RK, Basu P, Biswas J, Roychoudhury S, Panda CK. Persistent HPV16/18 infection in Indian women with the A-allele (rs6457617) of HLA-DQB1 and T-allele (rs16944) of IL-1beta -511 is associated with development of cervical carcinoma. Cancer Immunol Immunother. 2015;64:843–851. doi: 10.1007/s00262-015-1693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziduszko A, Ozbun MA. Annexin A2 and S100A10 regulate human papillomavirus type 16 entry and intracellular trafficking in human keratinocytes. J Virol. 2013;87:7502–7515. doi: 10.1128/JVI.00519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey LM, Raff AB, Da Silva DM, Kast WM. A major role for the minor capsid protein of human papillomavirus type 16 in immune escape. J Immunol. 2009;183:6151–6156. doi: 10.4049/jimmunol.0902145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausch SC, Da Silva DM, Kast WM. Differential uptake and cross-presentation of human papillomavirus virus-like particles by dendritic cells and Langerhans cells. Cancer Res. 2003;63:3478–3482. [PubMed] [Google Scholar]
- Fausch SC, Da Silva DM, Rudolf MP, Kast WM. Human papillomavirus virus-like particles do not activate Langerhans cells: a possible immune escape mechanism used by human papillomaviruses. J Immunol. 2002;169:3242–3249. doi: 10.4049/jimmunol.169.6.3242. [DOI] [PubMed] [Google Scholar]
- Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, Vignat J, Ferlay J, Bray F, Plummer M, Franceschi S. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30(Suppl 5):F12–23. doi: 10.1016/j.vaccine.2012.07.055. [DOI] [PubMed] [Google Scholar]
- Georgopoulos NT, Proffitt JL, Blair GE. Transcriptional regulation of the major histocompatibility complex (MHC) class I heavy chain, TAP1 and LMP2 genes by the human papillomavirus (HPV) type 6b, 16 and 18 E7 oncoproteins. Oncogene. 2000;19:4930–4935. doi: 10.1038/sj.onc.1203860. [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Gildener-Leapman N, Ferris RL, Bauman JE. Promising systemic immunotherapies in head and neck squamous cell carcinoma. Oral Oncol. 2013;49:1089–1096. doi: 10.1016/j.oraloncology.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Sanz-Moreno A. RANK as a therapeutic target in cancer. FEBS J. 2016;283:2018–2033. doi: 10.1111/febs.13645. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- Gram AM, Frenkel J, Ressing ME. Inflammasomes and viruses: cellular defence versus viral offence. J Gen Virol. 2012;93:2063–2075. doi: 10.1099/vir.0.042978-0. [DOI] [PubMed] [Google Scholar]
- Griffin LM, Cicchini L, Pyeon D. Human papillomavirus infection is inhibited by host autophagy in primary human keratinocytes. Virology. 2013;437:12–19. doi: 10.1016/j.virol.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guess JC, McCance DJ. Decreased migration of Langerhans precursor-like cells in response to human keratinocytes expressing human papillomavirus type 16 E6/E7 is related to reduced macrophage inflammatory protein-3alpha production. J Virol. 2005;79:14852–14862. doi: 10.1128/JVI.79.23.14852-14862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, Dronca R, Gangadhar TC, Patnaik A, Zarour H, Joshua AM, Gergich K, Elassaiss-Schaap J, Algazi A, Mateus C, Boasberg P, Tumeh PC, Chmielowski B, Ebbinghaus SW, Li XN, Kang SP, Ribas A. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. The New England journal of medicine. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handisurya A, Day PM, Thompson CD, Bonelli M, Lowy DR, Schiller JT. Strain-specific properties and T cells regulate the susceptibility to papilloma induction by Mus musculus papillomavirus 1. PLoS Pathog. 2014;10:e1004314. doi: 10.1371/journal.ppat.1004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan UA, Zannetti C, Parroche P, Goutagny N, Malfroy M, Roblot G, Carreira C, Hussain I, Muller M, Taylor-Papadimitriou J, Picard D, Sylla BS, Trinchieri G, Medzhitov R, Tommasino M. The human papillomavirus type 16 E7 oncoprotein induces a transcriptional repressor complex on the Toll-like receptor 9 promoter. J Exp Med. 2013;210:1369–1387. doi: 10.1084/jem.20122394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, Zody MC, Erlich RL, Green LM, Berical A, Wang Y, Casali M, Streeck H, Bloom AK, Dudek T, Tully D, Newman R, Axten KL, Gladden AD, Battis L, Kemper M, Zeng Q, Shea TP, Gujja S, Zedlack C, Gasser O, Brander C, Hess C, Gunthard HF, Brumme ZL, Brumme CJ, Bazner S, Rychert J, Tinsley JP, Mayer KH, Rosenberg E, Pereyra F, Levin JZ, Young SK, Jessen H, Altfeld M, Birren BW, Walker BD, Allen TM. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdman MT, Pett MR, Roberts I, Alazawi WO, Teschendorff AE, Zhang XY, Stanley MA, Coleman N. Interferon-beta treatment of cervical keratinocytes naturally infected with human papillomavirus 16 episomes promotes rapid reduction in episome numbers and emergence of latent integrants. Carcinogenesis. 2006;27:2341–2353. doi: 10.1093/carcin/bgl172. [DOI] [PubMed] [Google Scholar]
- Hewitt EW. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology. 2003;110:163–169. doi: 10.1046/j.1365-2567.2003.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, Morikawa T, Nakaya T, Sakatani T, Takada K, Fukayama M. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766–2774. doi: 10.1158/0008-5472.CAN-08-3070. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley PM. Warts, cancer and ubiquitylation: lessons from the papillomaviruses. Trans Am Clin Climatol Assoc. 2006;117:113–126. discussion 126–117. [PMC free article] [PubMed] [Google Scholar]
- Huang SM, McCance DJ. Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J Virol. 2002;76:8710–8721. doi: 10.1128/JVI.76.17.8710-8721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SE, Mason GM, Wills MR. Human cytomegalovirus immunity and immune evasion. Virus Res. 2011;157:151–160. doi: 10.1016/j.virusres.2010.10.031. [DOI] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Kalantari M, Calleja-Macias IE, Tewari D, Hagmar B, Lie K, Barrera-Saldana HA, Wiley DJ, Bernard HU. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. J Virol. 2004;78:12762–12772. doi: 10.1128/JVI.78.23.12762-12772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari M, Chase DM, Tewari KS, Bernard HU. Recombination of human papillomavirus-16 and host DNA in exfoliated cervical cells: a pilot study of L1 gene methylation and chromosomal integration as biomarkers of carcinogenic progression. J Med Virol. 2010;82:311–320. doi: 10.1002/jmv.21676. [DOI] [PubMed] [Google Scholar]
- Karim R, Tummers B, Meyers C, Biryukov JL, Alam S, Backendorf C, Jha V, Offringa R, van Ommen GJ, Melief CJ, Guardavaccaro D, Boer JM, van der Burg SH. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte’s innate immune response. PLoS Pathog. 2013;9:e1003384. doi: 10.1371/journal.ppat.1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Doerfler W, Cardon LR. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J Virol. 1994;68:2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Friso S, Choi SW. DNA methylation, an epigenetic mechanism connecting folate to healthy embryonic development and aging. J Nutr Biochem. 2009;20:917–926. doi: 10.1016/j.jnutbio.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Lowinski K, Rheinwald JG, Fichorova RN, Anderson DJ, Basile J, Munger K, Daly CM, Rosl F, Rollins BJ. Selective suppression of monocyte chemoattractant protein-1 expression by human papillomavirus E6 and E7 oncoproteins in human cervical epithelial and epidermal cells. Int J Cancer. 2003;107:407–415. doi: 10.1002/ijc.11411. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kumagai Y, Takeuchi O, Akira S. TLR9 as a key receptor for the recognition of DNA. Adv Drug Deliv Rev. 2008;60:795–804. doi: 10.1016/j.addr.2007.12.004. [DOI] [PubMed] [Google Scholar]
- LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, Malik HS, Malim MH, Munk C, O’Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- Lauring AS, Frydman J, Andino R. The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol. 2013;11:327–336. doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- Lee SY, Huang Z, Kang TH, Soong RS, Knoff J, Axenfeld E, Wang C, Alvarez RD, Chen CS, Hung CF, Wu TC. Histone deacetylase inhibitor AR-42 enhances E7-specific CD8(+) T cell-mediated antitumor immunity induced by therapeutic HPV DNA vaccination. J Mol Med (Berl) 2013;91:1221–1231. doi: 10.1007/s00109-013-1054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz P, Day PM, Pang YY, Frye SA, Jensen PN, Lowy DR, Schiller JT. Papillomavirus-like particles induce acute activation of dendritic cells. J Immunol. 2001;166:5346–5355. doi: 10.4049/jimmunol.166.9.5346. [DOI] [PubMed] [Google Scholar]
- Lenz P, Lowy DR, Schiller JT. Papillomavirus virus-like particles induce cytokines characteristic of innate immune responses in plasmacytoid dendritic cells. Eur J Immunol. 2005;35:1548–1556. doi: 10.1002/eji.200425547. [DOI] [PubMed] [Google Scholar]
- Li S, Labrecque S, Gauzzi MC, Cuddihy AR, Wong AH, Pellegrini S, Matlashewski GJ, Koromilas AE. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene. 1999;18:5727–5737. doi: 10.1038/sj.onc.1202960. [DOI] [PubMed] [Google Scholar]
- Li W, Deng XM, Wang CX, Zhang X, Zheng GX, Zhang J, Feng JB. Down-regulation of HLA class I antigen in human papillomavirus type 16 E7 expressing HaCaT cells: correlate with TAP-1 expression. Int J Gynecol Cancer. 2010;20:227–232. doi: 10.1111/IGC.0b013e3181cceec5. [DOI] [PubMed] [Google Scholar]
- Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, Li P. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ, DiMaio D. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A. 2013;110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang Y, Wang X, Sun Z, Li L, Tao Q, Luo B. Epigenetic silencing of WNT5A in Epstein-Barr virus-associated gastric carcinoma. Arch Virol. 2013;158:123–132. doi: 10.1007/s00705-012-1481-x. [DOI] [PubMed] [Google Scholar]
- Lo Cigno I, De Andrea M, Borgogna C, Albertini S, Landini MM, Peretti A, Johnson KE, Chandran B, Landolfo S, Gariglio M. The Nuclear DNA Sensor IFI16 Acts as a Restriction Factor for Human Papillomavirus Replication through Epigenetic Modifications of the Viral Promoters. J Virol. 2015;89:7506–7520. doi: 10.1128/JVI.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Leggatt GR, Zhong J, Liu X, de Kluyver RL, Peters T, Fernando GJ, Liem A, Lambert PF, Frazer IH. Impaired antigen presentation and effectiveness of combined active/passive immunotherapy for epithelial tumors. J Natl Cancer Inst. 2004;96:1611–1619. doi: 10.1093/jnci/djh301. [DOI] [PubMed] [Google Scholar]
- McBride AA. The papillomavirus E2 proteins. Virology. 2013;445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AA, Romanczuk H, Howley PM. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Crum CP, Munger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci U S A. 2011;108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Huh KW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J Virol. 2008;82:8695–8705. doi: 10.1128/JVI.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Sun C, Ghosh A, Rodriguez AC, Schiffman M, Wentzensen N, Hildesheim A, Herrero R, Wacholder S, Lorincz A, Burk RD. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a Costa Rican population. J Natl Cancer Inst. 2012;104:556–565. doi: 10.1093/jnci/djs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal D, Kassianos AJ, Tran LS, Bergot AS, Gosmann C, Hofmann J, Blumenthal A, Leggatt GR, Frazer IH. Indoleamine 2,3-dioxygenase activity contributes to local immune suppression in the skin expressing human papillomavirus oncoprotein e7. J Invest Dermatol. 2013;133:2686–2694. doi: 10.1038/jid.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Kawana K, Schust DJ, Fujii T, Yokoyama T, Iwasawa Y, Nagamatsu T, Adachi K, Tomio A, Tomio K, Kojima S, Yasugi T, Kozuma S, Taketani Y. CD1d, a sentinel molecule bridging innate and adaptive immunity, is downregulated by the human papillomavirus (HPV) E5 protein: a possible mechanism for immune evasion by HPV. J Virol. 2010;84:11614–11623. doi: 10.1128/JVI.01053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013;34:137–143. doi: 10.1016/j.it.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan S, Choyce A, Linedale R, Saunders NA, Dahler A, Chan E, Fernando GJ, Frazer IH, Leggatt GR. Epithelial expression of human papillomavirus type 16 E7 protein results in peripheral CD8 T-cell suppression mediated by CD4+CD25+ T cells. Eur J Immunol. 2009;39:481–490. doi: 10.1002/eji.200838527. [DOI] [PubMed] [Google Scholar]
- Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009;5:e1000439. doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebler M, Qian X, Hofler D, Kogosov V, Kaewprag J, Kaufmann AM, Ly R, Bohmer G, Zawatzky R, Rosl F, Rincon-Orozco B. Post-translational control of IL-1beta via the human papillomavirus type 16 E6 oncoprotein: a novel mechanism of innate immune escape mediated by the E3-ubiquitin ligase E6-AP and p53. PLoS Pathog. 2013;9:e1003536. doi: 10.1371/journal.ppat.1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick D, Kim SH, Fantuzzi G, Reznikov LL, Dinarello CA, Rubinstein M. Interleukin-18 binding protein: a novel modulator of the Th1 cytokine response. Immunity. 1999;10:127–136. doi: 10.1016/s1074-7613(00)80013-8. [DOI] [PubMed] [Google Scholar]
- Oosterhuis K, Aleyd E, Vrijland K, Schumacher TN, Haanen JB. Rational design of DNA vaccines for the induction of human papillomavirus type 16 E6- and E7-specific cytotoxic T-cell responses. Hum Gene Ther. 2012;23:1301–1312. doi: 10.1089/hum.2012.101. [DOI] [PubMed] [Google Scholar]
- Orange JS. Natural killer cell deficiency. J Allergy Clin Immunol. 2013;132:515–525. doi: 10.1016/j.jaci.2013.07.020. quiz 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacini L, Savini C, Ghittoni R, Saidj D, Lamartine J, Hasan UA, Accardi R, Tommasino M. Downregulation of Toll-Like Receptor 9 Expression by Beta Human Papillomavirus 38 and Implications for Cell Cycle Control. J Virol. 2015;89:11396–11405. doi: 10.1128/JVI.02151-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Kim EJ, Kwon HJ, Hwang ES, Namkoong SE, Um SJ. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J Biol Chem. 2000;275:6764–6769. doi: 10.1074/jbc.275.10.6764. [DOI] [PubMed] [Google Scholar]
- Perea SE, Massimi P, Banks L. Human papillomavirus type 16 E7 impairs the activation of the interferon regulatory factor-1. Int J Mol Med. 2000;5:661–666. doi: 10.3892/ijmm.5.6.661. [DOI] [PubMed] [Google Scholar]
- Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. 2013;425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten M, von Knebel Doeberitz N, Oezen I, Wick W, Ochs K. Cancer Immunotherapy by Targeting IDO1/TDO and Their Downstream Effectors. Front Immunol. 2014;5:673. doi: 10.3389/fimmu.2014.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Ruland J. From virus to inflammation: mechanisms of RIG-I-induced IL-1beta production. Eur J Cell Biol. 2012;91:59–64. doi: 10.1016/j.ejcb.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009;5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, Lemon SM. Hepatitis A and hepatitis C viruses: divergent infection outcomes marked by similarities in induction and evasion of interferon responses. Semin Liver Dis. 2010;30:319–332. doi: 10.1055/s-0030-1267534. [DOI] [PubMed] [Google Scholar]
- Reinholz M, Kawakami Y, Salzer S, Kreuter A, Dombrowski Y, Koglin S, Kresse S, Ruzicka T, Schauber J. HPV16 activates the AIM2 inflammasome in keratinocytes. Arch Dermatol Res. 2013;305:723–732. doi: 10.1007/s00403-013-1375-0. [DOI] [PubMed] [Google Scholar]
- Reiser J, Hurst J, Voges M, Krauss P, Munch P, Iftner T, Stubenrauch F. High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J Virol. 2011;85:11372–11380. doi: 10.1128/JVI.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressing ME, Sette A, Brandt RM, Ruppert J, Wentworth PA, Hartman M, Oseroff C, Grey HM, Melief CJ, Kast WM. Human CTL epitopes encoded by human papillomavirus type 16 E6 and E7 identified through in vivo and in vitro immunogenicity studies of HLA-A*0201-binding peptides. J Immunol. 1995;154:5934–5943. [PubMed] [Google Scholar]
- Richards KH, Doble R, Wasson CW, Haider M, Blair GE, Wittmann M, Macdonald A. Human papillomavirus E7 oncoprotein increases production of the anti-inflammatory interleukin-18 binding protein in keratinocytes. J Virol. 2014;88:4173–4179. doi: 10.1128/JVI.02546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KH, Wasson CW, Watherston O, Doble R, Blair GE, Wittmann M, Macdonald A. The human papillomavirus (HPV) E7 protein antagonises an Imiquimod-induced inflammatory pathway in primary human keratinocytes. Sci Rep. 2015;5:12922. doi: 10.1038/srep12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon-Orozco B, Halec G, Rosenberger S, Muschik D, Nindl I, Bachmann A, Ritter TM, Dondog B, Ly R, Bosch FX, Zawatzky R, Rosl F. Epigenetic silencing of interferon-kappa in human papillomavirus type 16-positive cells. Cancer Res. 2009;69:8718–8725. doi: 10.1158/0008-5472.CAN-09-0550. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosl F, Arab A, Klevenz B, zur Hausen H. The effect of DNA methylation on gene regulation of human papillomaviruses. J Gen Virol. 1993;74(Pt 5):791–801. doi: 10.1099/0022-1317-74-5-791. [DOI] [PubMed] [Google Scholar]
- Rudolf MP, Fausch SC, Da Silva DM, Kast WM. Human dendritic cells are activated by chimeric human papillomavirus type-16 virus-like particles and induce epitope-specific human T cell responses in vitro. J Immunol. 2001;166:5917–5924. doi: 10.4049/jimmunol.166.10.5917. [DOI] [PubMed] [Google Scholar]
- Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli P, Willimann K, Ebert LM, Walz A, Moser B. Cutaneous CXCL14 targets blood precursors to epidermal niches for Langerhans cell differentiation. Immunity. 2005;23:331–342. doi: 10.1016/j.immuni.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Schelhaas M, Shah B, Holzer M, Blattmann P, Kuhling L, Day PM, Schiller JT, Helenius A. Entry of human papillomavirus type 16 by actin-dependent, clathrin-and lipid raft-independent endocytosis. PLoS Pathog. 2012;8:e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SV, Schultze JL. New Insights into IDO Biology in Bacterial and Viral Infections. Front Immunol. 2014;5:384. doi: 10.3389/fimmu.2014.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Schmitt M, Bohmer G, Kopp-Schneider A, Muller M. Natural variants in the major neutralizing epitope of human papillomavirus minor capsid protein L2. Int J Cancer. 2013;132:E139–148. doi: 10.1002/ijc.27831. [DOI] [PubMed] [Google Scholar]
- Shalginskikh N, Poleshko A, Skalka AM, Katz RA. Retroviral DNA methylation and epigenetic repression are mediated by the antiviral host protein Daxx. J Virol. 2013;87:2137–2150. doi: 10.1128/JVI.02026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Shurin GV, Ferris RL, Tourkova IL, Perez L, Lokshin A, Balkir L, Collins B, Chatta GS, Shurin MR. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by dendritic cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. Journal of immunology (Baltimore, Md: 1950) 2005;174:5490–5498. doi: 10.4049/jimmunol.174.9.5490. [DOI] [PubMed] [Google Scholar]
- Silmon de Monerri NC, Kim K. Pathogens hijack the epigenome: a new twist on host-pathogen interactions. Am J Pathol. 2014;184:897–911. doi: 10.1016/j.ajpath.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley MA. Epithelial cell responses to infection with human papillomavirus. Clin Microbiol Rev. 2012;25:215–222. doi: 10.1128/CMR.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein AP, Saha S, Yu M, Kimple RJ, Lambert PF. Prevalence of human papillomavirus in oropharyngeal squamous cell carcinoma in the United States across time. Chemical Research in Toxicology. 2014;27:462–469. doi: 10.1021/tx500034c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunthamala N, Thierry F, Teissier S, Pientong C, Kongyingyoes B, Tangsiriwatthana T, Sangkomkamhang U, Ekalaksananan T. E2 proteins of high risk human papillomaviruses down-modulate STING and IFN-kappa transcription in keratinocytes. PLoS One. 2014;9:e91473. doi: 10.1371/journal.pone.0091473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm J, Lauer GM, Kavanagh DG, Sheridan I, Kim AY, Lucas M, Pillay T, Ouchi K, Reyor LL, Schulze zur Wiesch J, Gandhi RT, Chung RT, Bhardwaj N, Klenerman P, Walker BD, Allen TM. CD8 epitope escape and reversion in acute HCV infection. J Exp Med. 2004;200:1593–1604. doi: 10.1084/jem.20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindle RW, Herd K, Doan T, Bryson G, Leggatt GR, Lambert P, Frazer IH, Street M. Nonspecific down-regulation of CD8+ T-cell responses in mice expressing human papillomavirus type 16 E7 oncoprotein from the keratin-14 promoter. J Virol. 2001;75:5985–5997. doi: 10.1128/JVI.75.13.5985-5997.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummers B, Goedemans R, Pelascini LP, Jordanova ES, van Esch EM, Meyers C, Melief CJ, Boer JM, van der Burg SH. The interferon-related developmental regulator 1 is used by human papillomavirus to suppress NFkappaB activation. Nat Commun. 2015;6:6537. doi: 10.1038/ncomms7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberoi A, Yoshida S, Frazer IH, Pitot HC, Lambert PF. Role of Ultraviolet Radiation in Papillomavirus-Induced Disease. PLoS Pathog. 2016;12:e1005664. doi: 10.1371/journal.ppat.1005664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SJ, Rhyu JW, Kim EJ, Jeon KC, Hwang ES, Park JS. Abrogation of IRF-1 response by high-risk HPV E7 protein in vivo. Cancer Lett. 2002;179:205–212. doi: 10.1016/s0304-3835(01)00871-0. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay M, Samal J, Kandpal M, Vasaikar S, Biswas B, Gomes J, Vivekanandan P. CpG dinucleotide frequencies reveal the role of host methylation capabilities in parvovirus evolution. J Virol. 2013;87:13816–13824. doi: 10.1128/JVI.02515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay M, Vivekanandan P. Depletion of CpG Dinucleotides in Papillomaviruses and Polyomaviruses: A Role for Divergent Evolutionary Pressures. PLoS One. 2015;10:e0142368. doi: 10.1371/journal.pone.0142368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baren N, Van den Eynde BJ. Tryptophan-degrading enzymes in tumoral immune resistance. Front Immunol. 2015;6:34. doi: 10.3389/fimmu.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Weijer ML, Luteijn RD, Wiertz EJ. Viral immune evasion: Lessons in MHC class I antigen presentation. Semin Immunol. 2015;27:125–137. doi: 10.1016/j.smim.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer K. Evolution of the papillomaviridae. Virology. 2013;445:11–20. doi: 10.1016/j.virol.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445:115–137. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermark ER, Deluca KA, Gardner CR, Marker DF, Schreiner CN, Strickland DA, Wilton KM, Mondal S, Woodworth CD. Human papillomavirus type 16 E6 and E 7 proteins alter NF-kB in cultured cervical epithelial cells and inhibition of NF-kB promotes cell growth and immortalization. Virology. 2012;425:53–60. doi: 10.1016/j.virol.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscidi RP, Schiffman M, Hildesheim A, Herrero R, Castle PE, Bratti MC, Rodriguez AC, Sherman ME, Wang S, Clayman B, Burk RD. Seroreactivity to human papillomavirus (HPV) types 16, 18, or 31 and risk of subsequent HPV infection: results from a population-based study in Costa Rica. Cancer Epidemiol Biomarkers Prev. 2004;13:324–327. doi: 10.1158/1055-9965.epi-03-0166. [DOI] [PubMed] [Google Scholar]
- Vossen MT, Westerhout EM, Soderberg-Naucler C, Wiertz EJ. Viral immune evasion: a masterpiece of evolution. Immunogenetics. 2002;54:527–542. doi: 10.1007/s00251-002-0493-1. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wakae K, Kitamura K, Aoyama S, Liu G, Koura M, Monjurul AM, Kukimoto I, Muramatsu M. APOBEC3 deaminases induce hypermutation in human papillomavirus 16 DNA upon beta interferon stimulation. J Virol. 2014;88:1308–1317. doi: 10.1128/JVI.03091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C, Van Doorslaer C, Pandey A, Espinosa J, Pyeon D. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus evolution. 2015a doi: 10.1093/ve/vev015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Griffin LM, Little AS, Huang IC, Farzan M, Pyeon D. The antiviral restriction factors IFITM1, 2 and 3 do not inhibit infection of human papillomavirus, cytomegalovirus and adenovirus. PLoS One. 2014;9:e96579. doi: 10.1371/journal.pone.0096579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Pyeon D. APOBEC3 in papillomavirus restriction, evolution and cancer progression. Oncotarget. 2015;6:39385–39386. doi: 10.18632/oncotarget.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Xu T, Guo K, Griffin LM, Westrich Ja, Lee D, Lambert PF, Santiago ML, Pyeon D. APOBEC3A Functions as a Restriction Factor of Human Papillomavirus. Journal of Virology. 2015b;89:688–702. doi: 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- Welters MJ, Kenter GG, Piersma SJ, Vloon AP, Lowik MJ, Berends-van der Meer DM, Drijfhout JW, Valentijn AR, Wafelman AR, Oostendorp J, Fleuren GJ, Offringa R, Melief CJ, van der Burg SH. Induction of tumor-specific CD4+ and CD8+ T-cell immunity in cervical cancer patients by a human papillomavirus type 16 E6 and E7 long peptides vaccine. Clin Cancer Res. 2008;14:178–187. doi: 10.1158/1078-0432.CCR-07-1880. [DOI] [PubMed] [Google Scholar]
- Wiens ME, Smith JG. Alpha-defensin HD5 inhibits furin cleavage of human papillomavirus 16 L2 to block infection. J Virol. 2015;89:2866–2874. doi: 10.1128/JVI.02901-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M. Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HJ, Reifman J. A quantitative quasispecies theory-based model of virus escape mutation under immune selection. Proc Natl Acad Sci U S A. 2012;109:12980–12985. doi: 10.1073/pnas.1117201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YL, Sterling J, Damay I, Coleman N, Crawford R, van der Burg SH, Stanley M. Characterising the local immune responses in cervical intraepithelial neoplasia: a cross-sectional and longitudinal analysis. BJOG. 2008;115:1616–1621. doi: 10.1111/j.1471-0528.2008.01936.x. discussion 1621–1612. [DOI] [PubMed] [Google Scholar]
- Woodham AW, Da Silva DM, Skeate JG, Raff AB, Ambroso MR, Brand HE, Isas JM, Langen R, Kast WM. The S100A10 subunit of the annexin A2 heterotetramer facilitates L2-mediated human papillomavirus infection. PLoS One. 2012;7:e43519. doi: 10.1371/journal.pone.0043519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodham AW, Raff AB, Raff LM, Da Silva DM, Yan L, Skeate JG, Wong MK, Lin YG, Kast WM. Inhibition of Langerhans cell maturation by human papillomavirus type 16: a novel role for the annexin A2 heterotetramer in immune suppression. J Immunol. 2014;192:4748–4757. doi: 10.4049/jimmunol.1303190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Hu G, Chen J, Xie G. Interleukin 1beta and interleukin 1 receptor antagonist gene polymorphisms and cervical cancer: a meta-analysis. Int J Gynecol Cancer. 2014;24:984–990. doi: 10.1097/IGC.0000000000000165. [DOI] [PubMed] [Google Scholar]
- Xu J, Yin Z, Cao S, Gao W, Liu L, Yin Y, Liu P, Shu Y. Systematic review and meta-analysis on the association between IL-1B polymorphisms and cancer risk. PLoS One. 2013;8:e63654. doi: 10.1371/journal.pone.0063654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Murillo FM, Cui H, Blosser R, Uematsu S, Takeda K, Akira S, Viscidi RP, Roden RB. Papillomavirus-like particles stimulate murine bone marrow-derived dendritic cells to produce alpha interferon and Th1 immune responses via MyD88. J Virol. 2004;78:11152–11160. doi: 10.1128/JVI.78.20.11152-11160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Wheeler CM, Chen X, Uematsu S, Takeda K, Akira S, Pastrana DV, Viscidi RP, Roden RB. Papillomavirus capsid mutation to escape dendritic cell-dependent innate immunity in cervical cancer. J Virol. 2005;79:6741–6750. doi: 10.1128/JVI.79.11.6741-6750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- Zhang B, Li P, Wang E, Brahmi Z, Dunn KW, Blum JS, Roman A. The E5 protein of human papillomavirus type 16 perturbs MHC class II antigen maturation in human foreskin keratinocytes treated with interferon-gamma. Virology. 2003;310:100–108. doi: 10.1016/s0042-6822(03)00103-x. [DOI] [PubMed] [Google Scholar]
- Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


