Abstract
Hepatitis B virus (HBV) genotypes A and D are prevalent in many parts of the world and show overlapping geographic distributions. We amplified the entire HBV genome from sera of patients with genotypes A and D and generated overlength constructs for transient transfection into Huh7 or HepG2 cells. Genotype D clones were associated with less HBsAg in culture supernatant and even less intracellular HBsAg. They produced less 2.1-kb RNA due to a weaker SPII promoter. Chimeric promoter constructs identified three divergent positions as most critical, and their exchange reversed extracellular HBsAg phenotype. The S protein of genotype D was more efficient at secretion, while its L protein possessed greater inhibitory effect. Swapping the S gene diminished genotypic difference in intracellular S protein but widened the difference in secreted HBsAg. In conclusion, HBV genotypes A and D differ in S protein expression, secretion and modulation by L protein.
Keywords: hepatitis B virus, genotype, small envelope protein, large envelope protein, hepatitis B surface antigen, SPII promoter
1. Introduction
Hepatitis B virus (HBV) causes acute and chronic infection of the liver. Chronic HBV infection is associated with a high risk of developing liver cirrhosis and hepatocellular carcinoma, which results in nearly a million deaths annually (Trepo et al., 2014). HBV is an enveloped DNA virus. Its transcriptional template is the 3.2-kb covalently closed circular (ccc) DNA in the nucleus of infected hepatocytes (Seeger and Mason, 2015). Four genes are arranged on this circular genome including precore/core, polymerase (P), envelope (preS1/preS2/S), and X. The precore/core gene is responsible for expression of core protein as well as hepatitis B e antigen (HBeAg), a serological marker of HBV infection correlating with high viral load. The AUG codons at the 5′ end of preS1, preS2, and S regions can all serve as translation initiation sites, thus generating three co-terminal envelope proteins called large (L; preS1+preS2+S), middle (M; preS2+S), and small (S; with S domain alone) envelope proteins. Expression of these proteins requires two of the three subgenomic HBV RNAs transcribed from the cccDNA: the 2.4-kb RNA for L protein and the 2.1-kb RNA for M and S proteins. Core particles with mature (double stranded DNA) genome can be enveloped and released as 42-nm virions, with the three envelope proteins inserted on the surface. The S protein is the most abundant envelope protein expressed. It is also secreted without core particles as the 22-nm empty “subviral particles”, which can be 10,000- to 1,000,000-fold more abundant than virions in the blood of infected persons (Bruss and Ganem, 1991; Ganem and Prince, 2004). Envelope proteins associated with virions and subviral particles are detected by anti-S antibodies as hepatitis B surface antigen (HBsAg), a marker of on-going HBV infection. HBsAg seroconversion (loss of HBsAg followed by detection of anti-HBs antibody) indicates recovery from HBV infection, and is the ultimate goal of hepatitis B treatment (Lok and McMahon, 2009).
HBV isolates worldwide can be grouped into 10 genotypes with nucleotide sequence divergence of 7.5% or greater (Chu and Lok, 2002; Kramvis, 2014; Norder et al., 2004; Okamoto et al., 1988; Tong and Revill, 2016). Genotypes A and D are prevalent in North America and Europe. We previously found that genotype A rarely circulates as HBeAg-negative mutant (Li et al., 1993), and genotype A-specific 6-nucleotide insertion in the core gene caused production of HBeAg of multiple sizes (Ito et al., 2009). To further compare the biological properties between these two HBV genotypes, we amplified full-length HBV genome from sera of patients with chronic HBV infection and performed transient transfection experiments in human hepatoma cell lines.
2. Material and Methods
2.1. Amplification of full-length HBV genome from serum samples and generation of replication competent dimeric constructs
Serum samples from HBV carriers were collected at the Division of Gastroenterology and Hepatology, University of Michigan Medical Center, with informed consent from the patients. The protocol for sample collection was approved by the Institutional Review Board at the University of Michigan. The subjects were HBeAg positive and had not received antiviral therapy. All the samples had an HBV DNA titer of >106 copies/ml and those harboring the A1762T/G1764A core promoter mutations were excluded. DNA was extracted from 200 μl of serum samples by a QIAamp DNA Blood Minikit (Qiagen), and the full-length HBV genome was amplified by 40 cycles of polymerase chain reaction (PCR) using High Fidelityplus polymerase (Roche) and primers located in the precore region (Parekh et al., 2003; Qin et al., 2011). The PCR product was initially cloned to the HindIII and SacI sites of the pUC18 vector using such sites attached to the sense and antisense primers. The cloned HBV DNA was released from the vector by SapI (or BspQI) digestion, and ligated with a modified pUC18 vector to generate a tandem dimer with junctions in the precore region (SapI dimer). The SapI dimer was then digested with SphI, and the 3.2-kb HBV genome was purified and ligated with the SphI-cut, dephosphorylated pUC18 vector to generate a tandem dimer ending at the SphI site (SphI dimer). The SphI dimer is replication competent (Zong et al., 2016). The full-length sequences of 6 genotype A clones and 7 genotype D clones studied here were deposited to GenBank under the accession numbers KX827290 - KX827302.
2.2. 1.3mer replication construct and subgenomic expression constructs for envelope proteins
Besides SphI dimer, we also used 1.3mer construct to study HBV replication. The 1.3mer construct was generated by inserting nucleotide sequence 980-3221/1-1948 of genotype A and 980-3182/1-1948 of genotype D into the pBluescript SK (−) vector (Chen et al., 2016). A KOD-Plus-Mutagenesis Kit (Toyobo) was used to introduce the T3118C or T3118C/G3150C mutations into the 1.3mer genome of two genotype A clones (5.4 and 6.5), as well as the C3079T, T3102C/C3111G, and C3079T/T3102C/C3111G mutations into two genotype D clones (1.2 and 20.11). A 0.7mer construct was obtained by inserting a 2.3-kb genomic fragment covering nucleotide positions 2721-3221/1-1770 of genotype A, or positions 2715-3182/1-1770 of genotype D, into the SacI and HindIII sites of pBluescript vector. The 0.7mer construct can transcribe both the 2.4-kb and 2.1-kb subgenomic RNAs under the SPI and SPII promoters, respectively, leading to the expression of all the three envelope proteins (Garcia et al., 2009). The R169P mutant of the 0.7mer construct was generated by C660T mutation. To prevent L and M protein expression from the 0.7mer construct, a C17T mutation was used to convert the 6th codon in the preS2 region from CAA to TAA.
A 0.6mer construct capable of transcribing the 2.1-kb RNA through the SPII promoter was generated by inserting a 1.9-kb DNA fragment (2957-3221/1-1623 for genotype A, 2918-3182/1-1623 for genotype D) into the SacI-HindIII sites of pBluescript vector. The CMV-S protein construct was created by inserting a 0.7-kb fragment covering nucleotides 135 - 835 (the S region) into the EcoRI and HindIII sites of the pcDNA3.1zeo (−) vector. To generate the CMV-L protein construct, a 1.2-kb DNA fragment (2854-3221/1-835 for genotype A, 2848-3182/1-835 for genotype D) was inserted to the NheI and HindIII sites of pcDNA3.1zeo (−) vector. For this construct both preS2 and S AUG codons were mutated to ATC to prevent expression of M and S proteins.
2.3. Transient transfection, Western blot and ELISA detection of HBV proteins
The human hepatoma cell line Huh7 was cultured in Dulbecco’s Modified Eagle’s Medium (GIBCO) supplemented with 10% fetal bovine serum (Sigma). Transient transfection was performed on cells seeded in 6-well plates using Mirus reagent. Cells were lysed 3 days later in 80μl of lysis buffer (10mM Hepes pH7.5, 100 mM NaCl, 1 mM EDTA, and 1% NP40), and 1/4th was subjected to Western blot analysis. The blots were incubated at 4°C overnight with a 1:4000 dilution of rabbit polyclonal anti-HBs antibody (Novus), or a 1:2000 dilution of horse polyclonal anti-Ad/Ay antibody (Abcam). After further incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody at 1:10,000 dilution, or goat anti-horse antibody at 1:5,000 dilution, signals were revealed by enhanced chemiluminescence (PerkinElmer) (Garcia et al., 2009; Tsai et al., 2009) and visualized by chemiluminescent imaging system (Tanon). The antibodies were removed by the stripping buffer (CWBio), and the blot was incubated sequentially with mouse anti-actin antibody (Proteintech) (1:3000 dilution) and HRP-conjugated goat anti-mouse antibody (1:10,000 dilution). Unless stated otherwise, rabbit antibody from Novus was used for Western blot analysis of S protein.
HBsAg and HBeAg secreted to culture supernatant were measured by ELISA kits from KHB, Shanghai, China with proper dilution to prevent signal saturation. In some experiments, HBsAg was further analyzed by an ELISA kit from Wantai-bio pharm, Beijing, China. To concentrate secreted HBsAg from culture supernatant for Western blot, 800 μl of culture supernatant was mixed with 250 μl of 36% polyethylene glycol (PEG) 8000 (dissolved in PBS). After rotating at 4°C overnight, the samples were centrifuged at 14,000rpm for 1h and the pellet was resuspended in 20μl of lysis buffer (Chen et al., 2016).
2.4. Southern blot analysis of replicative HBV DNA
Huh7 cells seeded in 6-well plates were harvested at day 5 post transfection with HBV DNA constructs. Core particles were precipitated from half of the cell lysate, followed by nuclease treatment, proteinase K digestion, and DNA extraction. DNA was separated in 1.2% agarose gel, transferred to nylon membrane, followed by hybridization with 32P-labeled full-length HBV DNA probe (Parekh et al., 2003). For unbiased detection of both genotypes A and D, genotype A and D probes were mixed at 1:1 ratio. The blots were washed at 65°C in 2×SSC/0.1% SDS solution (Guarnieri et al., 2006; Parekh et al., 2003; Qin et al., 2011).
2.5. Northern blot analysis of HBV RNAs
Huh7 cells were lysed by TRI reagent (Sigma) at day 2 posttransfection, and RNA was extracted according to the manufacturer’s protocol (http://www.sigmaaldrich.com/china-mainland/zh/technical-documents/protocols/biology/tri-reagent.html). Total RNA (10μg) in1×RNA loading buffer was denatured at 65°C for 10 min, and separated in a 1.5% agarose gel with morpholine propane sulfonic acid and formaldehyde. After transfer to the positively charged nylon membrane (Roche), the blot was hybridized using a DIG-Northern Starter Kit (Roche) according to the manufacturer’s instructions. To generate an HBV riboprobe, a 0.7-kb HBV DNA fragment covering positions 1266-1950 was amplified from clone 5.4 of genotype A and clone 1.2 of genotype D, and inserted to the KpnI-XhoI sites of pcDNA3 vector. A DIG-labeled, negative stranded HBV RNA probe was generated by in vitro transcription of the plasmid linearized at the KpnI site, using SP6 RNA polymerase. The blots were washed at 65°C in 2×SSC/0.1% SDS solution for 30 min. The HBV probe was removed by boiling the blots in 0.1× SSC/2% SDS solution for 30 min, and for the loading control the blots were further hybridized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe generated in a similar manner.
2.6. Luciferase reporter constructs and reporter assay
To compare the transcriptional activity of the SPI and SPII promoters between the two genotypes, the 91-bp SPI promoter (position 2716-2806 for genotype A, 2710-2800 for genotype D) and the 221-bp SPII promoter (position 2960-3180 for genotype A, 2921-3141 for genotype D) were PCR amplified from genotype A and D clones and inserted to the KpnI-XhoI sites of pGL2-BASIC promoter luciferase report plasmid (Qin et al., 2016). Huh7 cells grown in 24-well plates were transfected with 0.6μg of the luciferase plasmid constructs and 0.125μg of RLUC reporter plasmid. Cells were harvested 48 hours later and lysed. Activities of the two types of luciferase were measured from cell lysate two days later using Dual-Luciferase Reporter Assay System (Promega). The promoter activity was calculated as the ratio of firefly luciferase activity over renilla luciferase activity.
2.7. Statistical Analysis
Data were expressed as mean ± standard deviation (SD). Statistical analysis was performed by IBM SPSS Statistics (version 23). One-way ANOVA was chosen for comparing the means, and Duncan’s multiple range test was used to analyze the difference among the multiple groups. P values <0.05 were considered as statistically significant.
3. Results
The objective of this study was to compare the production and secretion of HBsAg between genotype A and D isolates. To account for variability within the same genotype, we amplified and cloned full-length HBV genomes from several different patients with genotype A or D infection. Since the full-length HBV genome cloned to a vector is unable to generate pregenomic (pg) RNA essential for genome replication, these clones were converted to tandem dimers via the unique SphI site. Six genotype A clones derived from three patients and seven genotype D clones from seven patients were characterized. Full-length sequencing revealed all the six genotype A clones to be of the A2 subgenotype, whereas the genotype D clones belonged to D1 (2.2; 21.1), D2 (1.2; 30.4), D3 (3.1; 31.9), and D7 (20.11), respectively (Ozaras et al., 2015; Yousif and Kramvis, 2013).
3.1. Genotype D clones released less HBsAg to culture supernatant but retained even less HBsAg inside transfected hepatoma cell lines
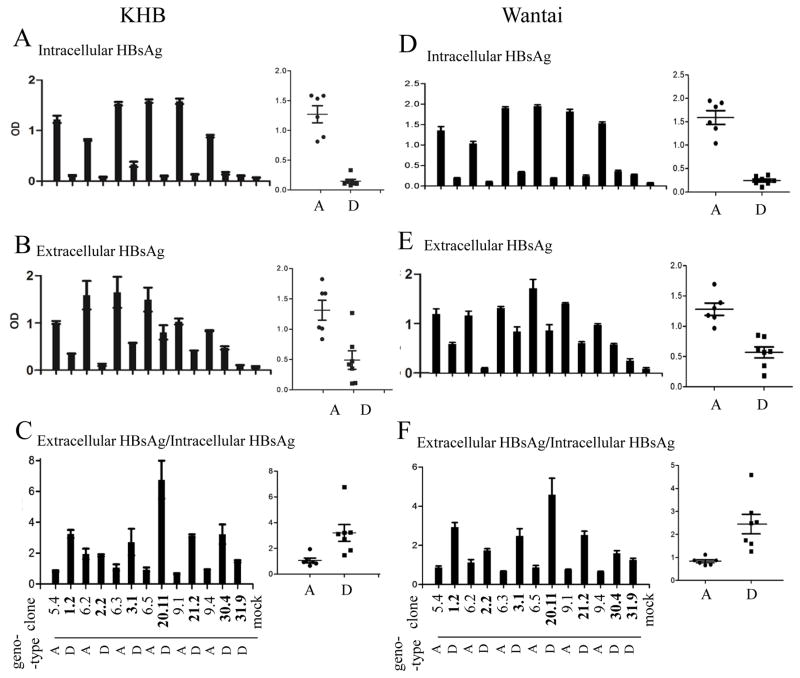
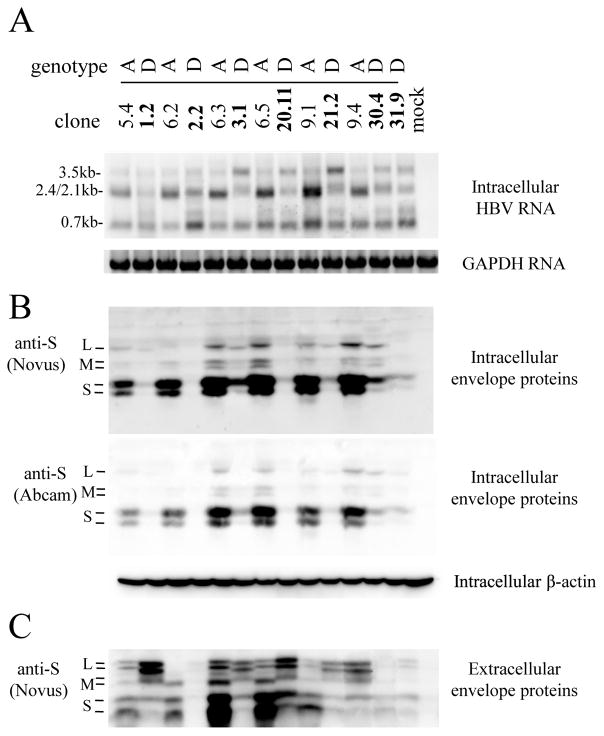
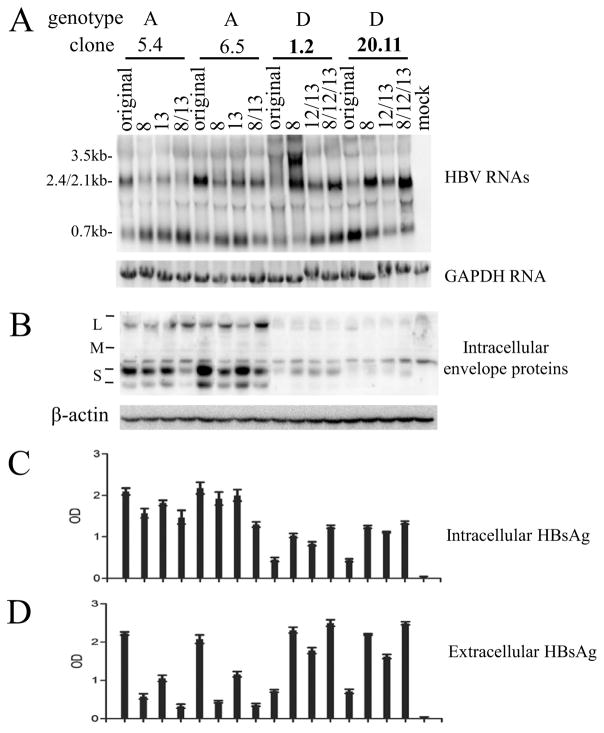
The 13 dimeric HBV DNA constructs of genotype A or D were transiently transfected to Huh7 cells, a human hepatoma cell line. Genotype D clones in general displayed reduced HBsAg titers in culture supernatant than genotype A clones (Fig. 1B and E), and the difference was even greater in cell lysate (Fig. 1A and D). Consequently, genotype D clones displayed higher ratio of extracellular HBsAg/intracellular HBsAg than genotype A clones (Fig. 1C and F). The results from ELISA were confirmed by Western blot analysis (Fig. 2B and C). Less S protein, the major constituent of HBsAg, was detected in lysate of cells transfected with genotype D clones (Fig. 2B). Immuofluorescent staining revealed weaker S protein signals within individual cells transfected with genotype D clones (supplementary Fig. 1), thus ruling out reduced transfection efficiency as the cause for lower HBsAg titer. A potential concern is that the reduced S protein or HBsAg associated with genotype D is an artifact of reduced antibody affinity. We think it highly unlikely for two reasons. First, concordant findings were made through ELISA detection of HBsAg and Western blot analysis of envelope proteins, from both cell lysate and culture supernatant. Second, HBsAg was measured by two ELISA kits from different companies (KHB in Fig. 1A–C and Wantai in Fig. 1D–F), while the Western blot was probed with two different polyclonal antibodies (rabbit anti-S from Novus; horse anti-Ad/Ay from Abcam) (Fig. 2B). Another concern is that the HBsAg phenotype observed was attributed to the particular human hepatoma cell line used. To address this issue, we repeated the transfection experiment in another human hepatoma cell line, HepG2. Similar results were obtained for both intracellular and extracellular HBsAg (supplementary Fig. 2) and viral envelope proteins (supplementary Fig. 3).
Fig 1.
Comparison of intracellular and extracellular HBsAg from Huh7 cells transfected with dimeric constructs of genotype A and genotype D. SphI dimers of 6 genotype A clones and 7 genotype D clones were transiently transfected to Huh7 cells, followed by ELISA detection of intracellular HBsAg (diluted 1:1600 for KHB kit, 1:800 for Wantai kit) and secreted HBsAg (diluted 1:800 for KHB kit, 1:400 for Wantai kit) 5 days posttransfection. Furthermore, the ratio of extracellular HBsAg/intracellular HBsAg was calculated (C and F). The ELISA data were based on 4 independent transfection experiments.
Fig 2.
Comparison of HBV RNA and envelope protein levels in Huh7 cells transfected with dimeric constructs of genotype A and genotype D. SphI dimers of 6 genotype A clones and 7 genotype D clones were transiently transfected to Huh7 cells, followed by Northern blot analysis of HBV RNAs (A) as well as Western blot analysis of intracellular and secreted envelope proteins (B and C). For Northern blot, positions of 3.5-kb, 2.4/2.1-kb and 0.7-kb HBV RNAs are indicated, and the same blot was subsequently hybridized with a GAPDH probe for loading control. For cell lysate, the same blot was sequentially probed with polyclonal anti-S antibody from Novus, Abcam, and anti-actin antibody for loading control. Positions of L, M, and S proteins are indicated. Western blot analysis of viral envelope proteins from culture supernatant was preceded by PEG precipitation.
Southern blot analysis of cell lysate revealed great clonal variability in replicative HBV DNA, with clones 6.5, 9.1 (genotype A) and 31.9 (genotype D) apparently defective in replication (supplementary Fig. 4A). HBeAg titers in culture supernatant also varied greatly, with all genotype A clones except clone 6.2 showing limited HBeAg expression (supplementary Fig. 4B).
3.2. Reduced HBsAg production was attributable to diminished transcription of the 2.1-kb RNA due to a weaker SPII promoter
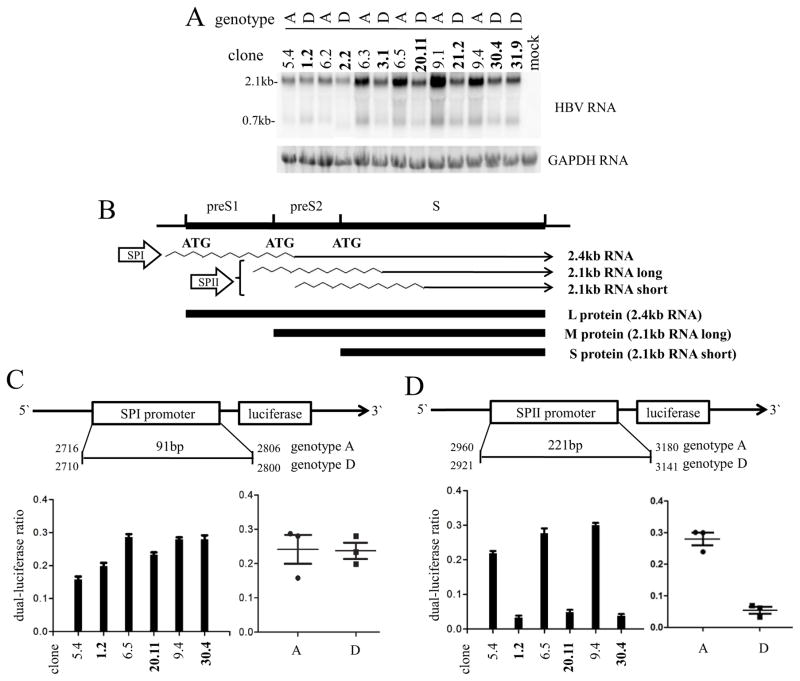
S protein is translated from the 2.1-kb subgenomic RNA. Northern blot analysis revealed reduced levels of the 2.4-kb/2.1-kb RNAs in cells transfected with genotype D clones, although these two RNA species could not be resolved into separate bands (Fig. 2A). To study transcription of the 2.1-kb RNA under its own promoter, a 1.9-kb HBV DNA fragment was inserted to the pBluescript vector to generate a 0.6mer construct capable of transcribing just the 2.1-kb and 0.7-kb RNAs. With such a construct, the 7 genotype D clones in general produced less 2.1-kb RNA than the 6 genotype A clones (Fig. 3A).
Fig 3.
Comparison of 2.1-kb RNA transcription and SPII promoter activities between genotype A and genotype D clones. (A) Northern blot analysis of HBV RNAs from Huh7 cells transiently transfected with 0.6mer constructs of 6 genotype A clones and 7 genotype D clones. Positions of 2.1-kb and 0.7-kb HBV RNAs are indicated. The same blot was hybridized with a GAPDH probe for loading control. (B) Schematic representation of the envelope gene, the SPI promoter driving 2.4-kb RNA, the SPII promoter driving the 2.1-kb RNA, and the three envelope proteins translated from these RNA species. (C and D) Shown on top is the schematic representation of the firefly luciferase reporter construct driven by the SPI or SPII promoter. Due to a 6-bp insertion in genotype A genomes and a 33-bp deletion in genotype D genomes, numbering of SPI and SPII promoters is different for the two genotypes despite identical sizes. Shown at the bottom are results of firefly luciferase/vanilla luciferase ratio from Huh7 cells transiently transfected with such a reporter construct from 3 genotype A clones (plain) and 3 genotype D clones (bold). A total of 4 transfection experiments were performed.
Transcription of the 2.1-kb RNA is driven by the SPII promoter (Fig. 3B). To compare the SPI and SPII promoter activities between the two genotypes, the 221-bp SPII promoter and the 91-bp SPI promoter from three genotype A clones (5.4, 6.5, and 9.4) and three genotype D clones (1.2, 20.11, and 30.4) were respectively inserted to the pGL2-BASIC reporter plasmid. Transient transfection of Huh7 cells confirmed a weaker SPII promoter from the genotype D clones (Fig. 3D), whereas activity of the SPI promoter did not show statistical difference between clones of the two genotypes (Fig. 3C).
3.3. Genotype-specific sequence divergence at three positions contributed to a weaker SPII promoter in genotype D
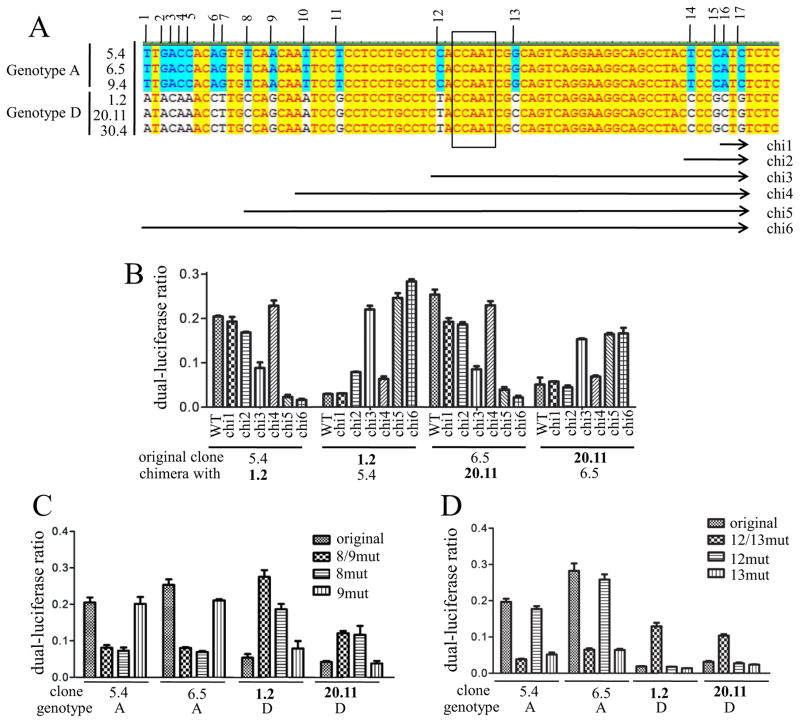
To establish the structural basis for differential SPII activity between genotype A and D clones, a professional online software (http://www.gene-regulation.com/pub/programs.html#pmatch) was used to identify transcription factors that bind selectively to the genotype A or genotype D SPII promoter. Of the 95 transcription factors predicted to bind only to the 221-bp SPII promoter of the three genotype A clones (5.4, 6.5, 9.4), 73 bind to the 86 bp located at the 3′ end (positions 146-221 of the SPII promoter). Among the 51 transcription factors with exclusive affinity for the three clones of genotype D (1.2, 20.11, 30.4), 31 target the 3′ 86 bp. Therefore, the 3′ end of the SPII promoter most likely plays a greater role in differential SPII activity than the 5′ end. Sequence alignment revealed 38 divergent positions in the entire SPII promoter, of which 17 are located at the 3′ end (Fig. 4A). Swapping these 86 bp at the 3′ end of the SPII promoter construct between clone 5.4 of genotype A and clone 1.2 of genotype D, or between clone 6.5 of genotype A and clone 20.11 of genotype D, was sufficient to revert the SPII promoter activities (Fig. 4B; chimera 6).
Fig 4.
Mapping of genotype-specific sequence divergences responsible for different SPII promoter activities. (A) Alignment of the 86 bp located at the 3′ end of the SPII promoter between three genotype A clones (5.4, 6.5, 9.4) and three genotype D clones (1.2, 20.11, 30.4). The 17 divergent positions are numbered, and sequences exchanged in the 6 chimeric constructs (chi1-chi6) are indicated schematically. The CCAAT element is boxed. (B) Luciferase activities of the 6 chimeric SPII reporter constructs relative to the original clones. The original SPII reporter constructs were based on genotype A (clone 5.4 or 6.5) or genotype D (clone 1.2 and 20.11), with various parts of the 86 bp at the 3′ end replaced with that of the other genotype (clone 5.4 with clone 1.2, clone 6.5 with clone 20.11, and vice versa). (C and D) Site-directed mutagenesis to verify the contribution of divergent positions 8/9 or 12/13 in different SPII promoter activities. The reporter constructs were based on genotype A or D, with the specific positions converted to those of the other genotype. Results were averaged from 4 transfection experiments. P <0.05.
To determine which of the 17 divergent positions were responsible for differential SPII promoter activities, progressive sequence exchange between these two pairs of HBV clones was made to cover divergent positions 16/17 (chimera 1), 14–17 (chimera 2), 12–17 (chimera 3), 10–17 (chimera 4), or 8–17 (chimera 5). The promoter activities of the 24 chimeric constructs (6×4), in comparison to the 4 parental constructs, suggested that divergent positions 8/9 and 12/13 contributed to lower SPII promoter activity in genotype D clones (Fig. 4B). Surprisingly, divergent positions 10/11 from genotype D were associated with higher promoter activity, making the profile of genotype D-based chimera 3 and genotype A-based chimera 4 inconsistent with those of the remaining constructs. The role of divergent positions 8/9 and 12/13 in differential promoter activities was further validated by site-directed mutagenesis. Swapping just divergent positions 8 and 9 or 8 was sufficient to reduce the promoter activities of the two genotype A clones or to enhance the promoter activities of the two genotype D clones (Fig. 4C). Mutating just divergent position 13 was sufficient to diminish promoter activity for genotype A clones, while mutating both positions 12 and 13 was needed to increase promoter activity for the two genotype D clones (Fig. 4D).
3.4. Mutating two or three positions in the SPII promoter sufficed to alter S protein and HBsAg production from 1.3mer construct of genotype A and genotype D
To establish the impact of divergent positions in SPII promoter on HBsAg production from the full-length HBV genome, the critical positions identified by the promoter assay were exchanged in the 1.3mer over-length construct. Secreted HBsAg from clones 5.4 and 6.5 of genotype A was diminished by converting the 13th position, and more so by converting the 8th or 8th+13th positions into genotype D sequences (Fig. 5D). Intracellular S protein and HBsAg were also reduced, although the effect was less prominent (Fig. 5B and C). Conversely, replacing the 12th/13th positions and more so replacing the 8th or 8th/12th/13th positions in the two genotype D clones (1.2 and 20.11) with genotype A sequences markedly increased HBsAg in culture supernatant and cell lysate. Northern blot analysis revealed altered level of the 2.4-/2.1-kb RNA in accordance with the changes in HBsAg (Fig 5A). These results confirmed the importance of the 8th divergent position, and to a lesser extent, the 13th or 12th/13th divergent positions, in differential HBsAg production by the two HBV genotypes.
Fig 5.
Contribution of divergent positions 8, 12, and 13 at the 3′ end of SPII promoter on HBsAg production by the 1.3mer HBV DNA construct. Divergent positions 8, 13, or both were converted from two genotype A clones (5.4 and 6.5) to genotype D-specific sequences. Alternatively, divergent positions 8, 12/13, or 8/12/13 from two genotype D clones (1.2 and 20.11) were replaced with the genotype A sequences. The parental clones and the site-directed mutants as 1.3mer construct were transiently transfected to Huh7 cells, followed by Northern blot analysis of HBV RNAs (A), Western blot analysis of intracellular envelope proteins (B), and ELISA for intracellular HBsAg (1:600 dilution) (C) or extracellular HBsAg (1:200 dilution) (D).
3.5. Low intracellular HBsAg titer associated with genotype D was not a consequence of efficient virion formation or release
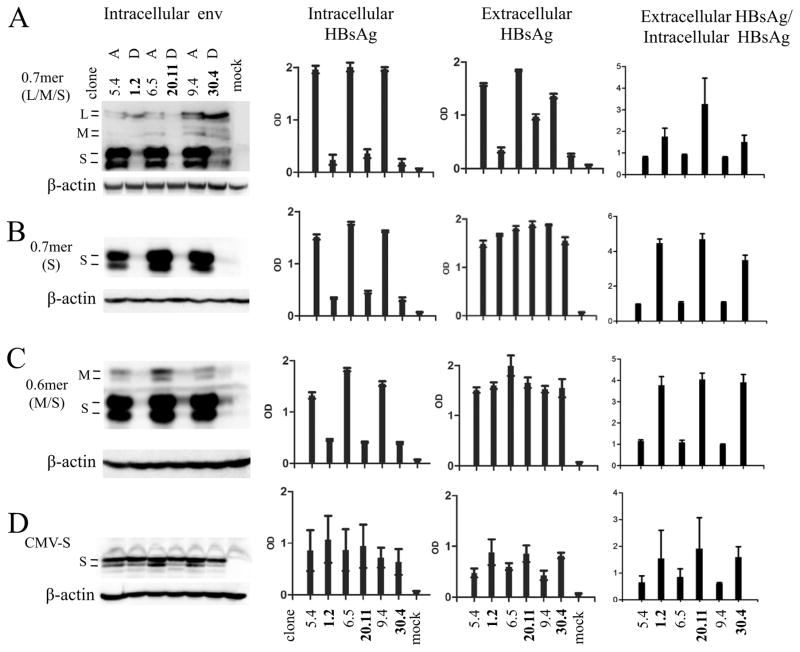
Secretion of subviral particles, which are largely comprised of the S protein, is inhibited by the L protein in a dose-dependent manner (Chisari et al., 1986; Ou and Rutter, 1987; Persing et al., 1986). The level of intracellular S protein might also be affected by the efficiency of virion formation/secretion, which in turn is influenced by the extent of HBV DNA replication. The 13 HBV clones studied here displayed marked variability in replication capacity (supplementary Fig. 4A), but replication capacity did not correlate with intracellular or extracellular HBsAg titers (Fig. 1A, B, D and E). To establish the relative contribution of the L and S proteins to different ratio of extracellular HBsAg/intracellular HBsAg without confounding from viral DNA replication or other viral proteins (core, polymerase, and HBx), a 2.3-kb HBV DNA fragment covering nt 2721-1770 was inserted to pBluescript vector to generate a 0.7mer expression construct for the three envelope proteins. Similar to the SphI dimer, the 0.7mer construct of the three genotype D clones produced lower HBsAg titers than the three genotype A clones in culture supernatant and more so in cell lysate (Fig. 6A). Consequently genotype D clones continued to display a higher ratio of extracellular HBsAg/intracellular HBsAg than genotype A clones. In addition, introduction of an R169P mutation to abolish HBsAg secretion (Ito et al., 2010) did not significantly increase the intracellular S protein level, whether for genotype A or genotype D (Supplementary Figure 5A and B).
Fig 6.
Comparison of intracellular and extracellular levels of HBsAg produced by various forms of subgenomic constructs of genotype A vs. genotype D. Three clones each of genotype A (5.4, 6.5, and 9.4) and genotype D (1.2, 20.11, and 30.4) were examined. (A) 0.7mer construct for L, M, and S proteins. (B) 0.7mer construct unable to express L and M proteins due to a stop codon in the preS2 region. (C) 0.6mer construct for M and S proteins. (D) CMV-S construct for the S protein. These constructs were transiently transfected to Huh7 cells. Shown on the left is Western blot analysis of intracellular envelope proteins. The next two panels show ELISA results of intracellular HBsAg (1:200 dilution for CMV-S construct, and 1:400 dilution for other constructs) and extracellular HBsAg (0.7mer:1:200 dilution; 0.6mer:1:150 dilution; CMV-S:1:100 dilution), respectively. The right panels are calculated ratio of extracellular HBsAg/intracellular HBsAg. ELISA data are based on 6 independent transfection experiments.
3.6. The S protein of genotype D was more efficient at HBsAg secretion, although that was attenuated by stronger inhibitory effect of its L protein
Preventing L and M protein expression from the 0.7mer construct greatly increased HBsAg titers in culture supernatant for genotype D clones, which became comparable to those of genotype A clones (Fig. 6B, third panel). Consequently, the ratio of extracellular HBsAg/intracellular HBsAg was further increased for genotype D (compare the fourth panel of Fig. 6B with that of Fig. 6A). Similar results were observed for the 0.6mer construct (Fig. 6C). The 0.6mer construct can transcribe the 2.1-kb mRNA to express M and S proteins, but not the 2.4-kb RNA for L protein. Taken together, these results suggest that the S protein from genotype D has greater propensity for secretion, but this was offset by the strong inhibitory effect of its L protein. When the S gene was cloned behind the exogenous CMV promoter to ensure comparable level of S protein expression from both genotypes, the intracellular S protein became similar while the HBsAg level in culture supernatant was higher for the three genotype D clones (Fig. 6D, second and third panels). This observation reinforced the greater secretion capacity of the genotype D-derived S protein. At comparable level of intracellular S protein produced from CMV-S construct, the genotype D clones produced a higher ratio of gp27 over p24 (Fig. 6D, first panel), suggesting more efficient N-linked glycosylation.
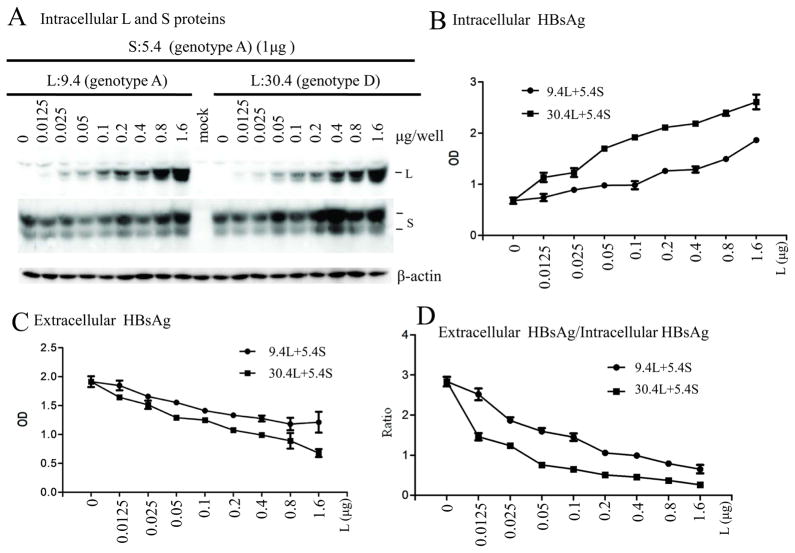
To validate the stronger inhibitory effect of the L protein from genotype D on S protein secretion, CMV promoter-driven expression constructs for the L protein and S protein, respectively, were employed for a co-transfection experiment. Fixed amount of the S protein construct of genotype A (clone 5.4) was cotransfected with increasing amounts of the L protein construct of genotype A (clone 9.4) or genotype D (clone 30.4). Western blot and ELISA indicated that the L protein from genotype D caused more intracellular retention of the S protein and HBsAg thus reducing HBsAg secretion (Fig. 7). We performed similar co-transfection experiment using CMV-S protein construct of clone 20.11 (genotype D), and obtained similar finding (data not shown).
Fig 7.
Comparison of the inhibitory effect of L protein from genotypes A and D on HBsAg secretion. Huh7 cells grown in 6-well plates were transfected with 1μg of CMV-S construct of a genotype A clone (5.4) and 0–1.6μg of CMV-L construct of a genotype A clone (9.4) or a genotype D clone (30.4). (A) Western blot analysis of intracellular envelope proteins. (B and C) ELISA for intracellular HBsAg (1:400 dilution) and extracellular HBsAg (1:150 dilution). (D) Ratio of extracellular HBsAg/intracellular HBsAg. ELISA data were averaged from 6 transfection experiments.
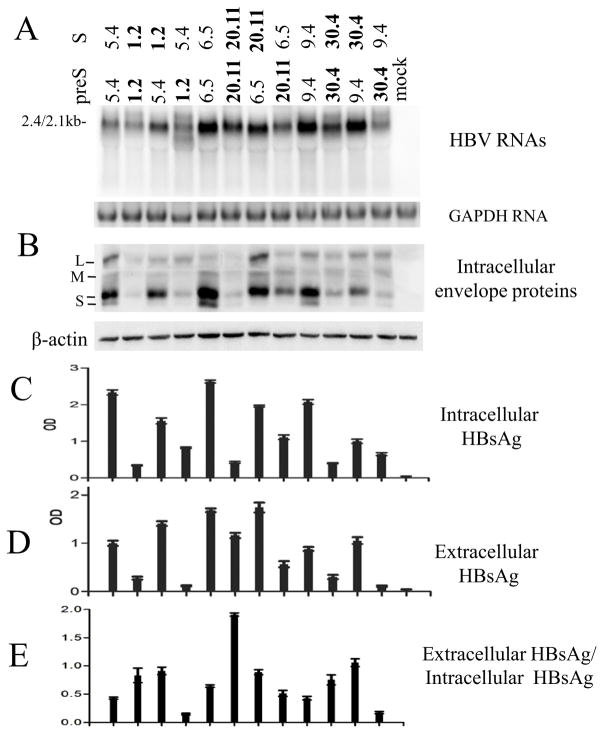
3.7. Swapping the S gene in the 0.7mer construct diminished genotypic difference in intracellular HBsAg but widened the difference in culture supernatant
Contribution of the preS and S regions of the envelope gene from the two HBV genotypes on HBsAg retention vs. secretion was determined by DNA fragment exchange. The S region was reciprocally exchanged between 0.7mer L/M/S expression construct of clones 5.4 and 1.2, 6.5 and 20.11, as well as 9.4 and 30.4. As shown in Fig 8C and D, inserting the S region from genotype D into genotype A clones increased extracellular HBsAg level at the expense of intracellular HBsAg level (lane 3 vs. 1; lane 7 vs. 5; lane 11 vs. 9). Conversely, introducing the S region from genotype A to genotype D clones increased intracellular HBsAg but diminished HBsAg in culture supernatant (lane 4 vs. 2; lane 8 vs. 6; lane 12 vs. 10).
Fig 8.
Impact of exchanging the S region on the ratio of extracellular HBsAg/intracellular HBsAg from 0.7mer expression construct. The S region was exchanged between three pairs of genotype A - genotype D clones: 5.4 (A) and 1.2 (D), 6.5 (A) and 20.11 (D), as well as 9.4 (A) and 30.4 (D). (A) Northern blot analysis of the 2.4-kb/2.1-kb RNAs. (B) Western blot analysis of intracellular envelope proteins. (C and D) ELISA for intracellular HBsAg (1:400 dilution) and extracellular HBsAg (1:200 dilution). (E) Ratio of extracellular HBsAg/intracellular HBsAg. Results were averaged from 6 transfection experiments.
4. Discussion
In the present study, we amplified full-length HBV genome from carriers of genotype A or genotype D. The 6 genotype A clones derived from three patients all belonged to the A2 subgenotype, while the 7 genotype D clones from 7 patients were of the D1, D2, D3, and D7 subgenotypes. Transient transfection experiments in Huh7 cells revealed great variability in levels of intracellular replicative HBV DNA and extracellular HBeAg among the 13 clones, with some clones nearly defective in genome replication or HBeAg expression (supplementary Fig. 4A and B). HBsAg level was also variable, but in general most genotype D clones displayed lower HBsAg titers than subgenotype A2 clones in culture supernatant of transfected Huh7 cells (Fig. 1B and E) and HepG2 cells (supplementary Fig. 2B and E), and even lower HBsAg titers in cell lysate (Fig. 1A and D; supplementary Fig. 2A and D). This was true with both KHB and Wantai ELISA kits. Consequently, genotype D clones displayed a higher ratio of extracellular HBsAg/intracellular HBsAg (Fig. 1C and F; supplementary Fig. 2C and F). The ELISA data were confirmed by Western blot analysis of the S protein, the major constituent of HBsAg, using two different polyclonal anti-S antibodies (Fig. 2B and C; supplementary Fig. 3A and B). Northern blot analysis revealed that dimeric genotype D clones transcribed less 2.4-kb/2.1-kb RNAs (Fig. 2A), while the 0.6mer construct under the SPII promoter transcribed less 2.1-kb RNA (Fig. 3A). Consistent with our findings, Sugiyama et al found reduced HBsAg secretion by the two genotype D clones than the four genotype A clones studied, which correlated with reduced transcript level (Sugiyama et al., 2006). More importantly, a European study of HBeAg-negative patients recruited for interferon therapy found mean pre-treatment HBsAg titer to be much higher for genotype A (14,816±12,633 IU/ml) than genotype D (6413±3653 IU/ml), genotype B (4953±8209 IU/ml), and genotype C (3155±3748 IU/ml) (Brunetto et al., 2013).
Subsequent experiments to pinpoint the underlying mechanisms for differences in intracellular and extracellular HBsAg centered on clones 5.4, 6.5, 9.4 (all of A2 subgenotype), 1.2 (D2), 20.11 (D7), and 30.4 (D2). These six clones originated from six different patients. Luciferase reporter assay confirmed a weaker SPII promoter from the three genotype D clones (Fig. 3D), with its 3′ 86 bp accounting for most if not all the differences in promoter activity (chi6, Fig. 4B). Functional analysis of 6 pairs of chimeric constructs suggested that of the 17 divergent positions within the 86-bp DNA fragment, positions 8, 9, 12, and 13 likely contributed to the differential promoter activities. Site-directed mutagenesis revealed that divergent position 8 but not 9 contributed to different promoter activities between A2 and D (Fig. 4C). On the other hand, mutating just divergent position 13 was sufficient to reduce promoter activity for subgenotype A2 clones, whereas exchanging both positions 12 and 13 was needed to increase promoter activity for genotype D clones (Fig. 4D). Due to a size difference of 39 bp, divergent positions 8, 12, and 13 in the SPII promoter correspond to positions 3118, 3141, and 3150 in a genotype A genome but positions 3079, 3102, and 3111 in a genome of genotype D, respectively. In genotype A, divergent position 8 is the 3rd base of the recognition sequence TGTCAA for transforming growth factor β-induced factor (TGIF). This binding site is absent in genotypes B, C, and D due to the T to C change. Indeed, the T3118C change introduced to the 1.3mer construct of two subgenotype A2 clones reduced secreted HBsAg, whereas the C3079T change in two genotype D clones markedly increased extracellular HBsAg (Fig. 5D). TGIF belongs to a family of evolutionarily conserved homeodomain transcription factor associated with neural development and human cancers, and it is highly expressed in the human liver by Northern blot analysis (Bertolino et al., 1995; Razzaque and Atfi, 2016). However, so far there has been no research on its regulation of viral transcription, let alone HBV.
Divergent positions 12 and 13 flank the CCAAT box (Fig. 4A), which is conserved in all HBV genotypes and required for efficient transcription of the 2.1-kb RNA (Zhou and Yen, 1991). The 13th divergent position is located between the CCAAT box and the transcription start site of the 2.1-kb RNA. Previous studies revealed that CCAAT box binding factor (CBF) regulates the SPII promoter, with transcription diminished by mutations not only within the CCAAT box, but also between the CCAAT box and the transcription start site (Bock et al., 1999; Lu et al., 1995). Other studies found that artificial mutations adjacent to the CCAAT box could alter the helical orientation of the CCAAT box, which was critical for transcription factor binding to the promoter and its transcriptional activity (Wright et al., 1994). Considering that genotypes A, B, and C have identical sequences at divergent positions 12 and 13, these two positions might be responsible for the low HBsAg production associated with genotype D.
Unexpectedly, we discovered that genotype D-specific sequences at divergent positions 10 and 11 conferred higher SPII promoter activity (Fig. 4B). A previous research divided the SPII promoter into seven regions named A to G (Raney, Milich, and McLachlan, 1991), and divergent positions 8/9, 10/11 and 12/13 are located in the E, F and G regions, respectively. The F region has a negative influence on the promoter activity, and this effect is counteracted by factor(s) that bind to region E. Region G cannot prevent the negative influence of transcription factor F but has the capacity to compensate for the effect of the loss of transcription factor E on the transcriptional activity of the surface antigen promoter (Raney, Milich, and McLachlan, 1991). Our results could be explained if the F region in genotype D is no longer functional or less active. Whether swapping the divergent positions 10/11 in the 1.3mer genomes (nucleotide positions 3125 and 3129 in genotype A, 3086 and 3090 in genotype D) further widens the difference in intracellular and secreted HBsAg between the two HBV genotypes warrants further investigation.
Besides the difference in the SPII promoter activities leading to different S protein expression between genotypes A and D, the S and L proteins from these two genotypes also differ in their ability to drive and to block HBsAg secretion, respectively. First, the contrasting pattern of intracellular and extracellular HBsAg between the two HBV genotypes can be reproduced using a 0.7mer expression construct for just L, M, and S proteins, suggesting its separation from genome replication and virion formation. Second, we used 0.7mer construct unable to express L/M proteins, 0.6mer expression construct for M and S proteins, and CMV promoter-driven expression construct for the S protein to demonstrate that the S protein of genotype D was more efficient at HBsAg secretion, which was mitigated to some extent by the stronger inhibitory effect of its L protein (Fig. 6). This conclusion was further corroborated by co-transfecting fixed amount of the S protein expression construct with increasing concentrations of the L protein construct (Fig. 7), and by swapping the S gene between genotypes A and D clones in the 0.7mer expression construct (Fig. 8).
A drawback of the current study is that only HBV clones of the A2 subgenotype were studied, and whether the findings can be extended to the A1 subgenotype circulating in Africa and Southern Asia remains to be determined. Moreover, a comparative study including all the four major HBV genotypes (A, B, C, D) is warranted. According to a previous study (Sugiyama et al., 2006), the two genotype D clones produced the lowest level of HBsAg among the 14 clones of genotypes A–D analyzed. The four genotype A clones released the highest HBsAg level, with the two A2 (called Ae at that time) clones having slightly higher titer than the two A1 clones (called Aa).
Taken together, our results indicate that genotype D clones are less efficient at HBsAg production due to a weaker SPII promoter. The S protein of genotype D is much more efficient at HBsAg secretion than that of subgenotype A2, even though its L protein is a more potent inhibitor of HBsAg release. The cumulative effect is reduced extracellular HBsAg but even less intracellular HBsAg in comparison to subgenotype A2. Based on our in vitro findings, it will be interesting to determine whether liver tissue from patients infected with subgenotype A2 and genotype D differ in HBsAg expression values. Whether differences in S expression and secretion account for differences in rates of spontaneous and antiviral therapy induced HBsAg clearance and recovery from acute HBV infection warrants further investigation.
Supplementary Material
Highlights.
Genotype D clones were associated with less extracellular HBsAg and even less intracellular HBsAg than genotype A clones.
Genotype D produced less 2.1-kb RNA than genotype A due to a weaker SPII promoter.
Swapping three divergent positions in the SPII promoter could reverse the extracellular HBsAg phenotype.
The S protein of genotype D has higher secretion efficiency.
The L protein of genotype D is a more potent inhibitor of HBsAg secretion
Acknowledgments
This study was supported by NIH grants AI103648, AI107618, AI113394, and AI116639, and also by a grant from National Science Foundation of China (81371822).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- Bock CT, Kubicka S, Manns MP, Trautwein C. Two control elements in the hepatitis B virus S-promoter are important for full promoter activity mediated by CCAAT-binding factor. Hepatology. 1999;29:1236–1247. doi: 10.1002/hep.510290426. [DOI] [PubMed] [Google Scholar]
- Brunetto MR, Marcellin P, Cherubini B, Yurdaydin C, Farci P, Hadziyannis SJ, Rothe V, Regep L, Bonino F. Response to peginterferon alfa-2a (40 KD) in HBeAg-negative CHB: on treatment kinetics of HBsAg serum levels vary by HBV genotype. J Hepatol. 2013;59:1153–1159. doi: 10.1016/j.jhep.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci U S A. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jia H, Zhang F, Qin Y, Zong L, Yuan Q, Wang Y, Xia N, Li J, Wen Y, Tong S. Functional characterization of hepatitis B virus core promoter mutants revealed transcriptional interference among co-terminal viral mRNAs. J Gen Virol. 2016;97:1–9. doi: 10.1099/jgv.0.000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari FV, Filippi P, McLachlan A, Milich DR, Riggs M, Lee S, Palmiter RD, Pinkert CA, Brinster RL. Expression of hepatitis B virus large envelope polypeptide inhibits hepatitis B surface antigen secretion in transgenic mice. J Virol. 1986;60:880–887. doi: 10.1128/jvi.60.3.880-887.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CJ, Lok AS. Clinical significance of hepatitis B virus genotypes. Hepatology. 2002;35:1274–1276. doi: 10.1053/jhep.2002.33161. [DOI] [PubMed] [Google Scholar]
- Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- Garcia T, Li J, Sureau C, Ito K, Qin Y, Wands J, Tong S. Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J Virol. 2009;83:11152–11165. doi: 10.1128/JVI.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri M, Kim KH, Bang G, Li J, Zhou Y, Tang X, Wands J, Tong S. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J Virol. 2006;80:587–595. doi: 10.1128/JVI.80.2.587-595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Kim KH, Lok AS, Tong S. Characterization of genotype-specific carboxyl-terminal cleavage sites of hepatitis B virus e antigen precursor and identification of furin as the candidate enzyme. J Virol. 2009;83:3507–3517. doi: 10.1128/JVI.02348-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Qin Y, Guarnieri M, Garcia T, Kwei K, Mizokami M, Zhang J, Li J, Wands JR, Tong S. Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J Virol. 2010;84:12850–12861. doi: 10.1128/JVI.01499-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141–150. doi: 10.1159/000360947. [DOI] [PubMed] [Google Scholar]
- Li JS, Tong SP, Wen YM, Vitvitski L, Zhang Q, Trepo C. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J Virol. 1993;67:5402–5410. doi: 10.1128/jvi.67.9.5402-5410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661–662. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- Lu CC, Chen M, Ou JH, Yen TS. Key role of a CCAAT element in regulating hepatitis B virus surface protein expression. Virology. 1995;206:1155–1158. doi: 10.1006/viro.1995.1042. [DOI] [PubMed] [Google Scholar]
- Norder H, Courouce AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- Ou JH, Rutter WJ. Regulation of secretion of the hepatitis B virus major surface antigen by the preS-1 protein. J Virol. 1987;61:782–786. doi: 10.1128/jvi.61.3.782-786.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaras R, Inanc BI, Yemisen M, Tabak F. Epidemiology of HBV subgenotypes D. Clin Res Hepatol Gastroenterol. 2015;39:28–37. doi: 10.1016/j.clinre.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Parekh S, Zoulim F, Ahn SH, Tsai A, Li J, Kawai S, Khan N, Trepo C, Wands J, Tong S. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J Virol. 2003;77:6601–6612. doi: 10.1128/JVI.77.12.6601-6612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persing DH, Varmus HE, Ganem D. Inhibition of secretion of hepatitis B surface antigen by a related presurface polypeptide. Science. 1986;234:1388–1391. doi: 10.1126/science.3787251. [DOI] [PubMed] [Google Scholar]
- Qin Y, Tang X, Garcia T, Hussain M, Zhang J, Lok A, Wands J, Li J, Tong S. Hepatitis B virus genotype C isolates with wild-type core promoter sequence replicate less efficiently than genotype B isolates but possess higher virion secretion capacity. J Virol. 2011;85:10167–10177. doi: 10.1128/JVI.00819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhou X, Jia H, Chen C, Zhao W, Zhang J, Tong S. Stronger enhancer II/core promoter activities of hepatitis B virus isolates of B2 subgenotype than those of C2 subgenotype. Sci Rep. 2016;6:30374. doi: 10.1038/srep30374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney AK, Milich DR, McLachlan A. Complex regulation of transcription from the hepatitis B virus major surface antigen promoter in human hepatoma cell lines. J Virol. 1991;65:4805–4811. doi: 10.1128/jvi.65.9.4805-4811.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaque MS, Atfi A. TGIF function in oncogenic Wnt signaling. Biochim Biophys Acta. 2016;1865:101–104. doi: 10.1016/j.bbcan.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479–480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, Gish RG, Kramvis A, Shimada T, Izumi N, Kaito M, Miyakawa Y, Mizokami M. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology. 2006;44:915–924. doi: 10.1002/hep.21345. [DOI] [PubMed] [Google Scholar]
- Tong S, Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64:S4–S16. doi: 10.1016/j.jhep.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- Tsai A, Kawai S, Kwei K, Gewaily D, Hutter A, Tong DR, Li J, Wands JR, Tong S. Chimeric constructs between two hepatitis B virus genomes confirm transcriptional impact of core promoter mutations and reveal multiple effects of core gene mutations. Virology. 2009;387:364–372. doi: 10.1016/j.virol.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KL, Vilen BJ, Itoh-Lindstrom Y, Moore TL, Li G, Criscitiello M, Cogswell P, Clarke JB, Ting JP. CCAAT box binding protein NF-Y facilitates in vivo recruitment of upstream DNA binding transcription factors. EMBO J. 1994;13:4042–4053. doi: 10.1002/j.1460-2075.1994.tb06721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousif M, Kramvis A. Genotype D of hepatitis B virus and its subgenotypes: An update. Hepatol Res. 2013;43:355–364. doi: 10.1111/j.1872-034X.2012.01090.x. [DOI] [PubMed] [Google Scholar]
- Zhou DX, Yen TS. The hepatitis B virus S promoter comprises A CCAAT motif and two initiation regions. J Biol Chem. 1991;266:23416–23421. [PubMed] [Google Scholar]
- Zong L, Qin Y, Jia H, Zhou H, Chen C, Qiao K, Zhang J, Wang Y, Li J, Tong S. Two-way molecular ligation for efficient conversion of monomeric hepatitis B virus DNA constructs into tandem dimers. J Virol Methods. 2016;233:46–50. doi: 10.1016/j.jviromet.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.