Abstract
Intermediary metabolism studies have typically concentrated on four major regulatory mechanisms—substrate availability, allosteric enzyme regulation, post-translational enzyme modification, and regulated enzyme synthesis. Although transcriptional control has been a big focus, it is becoming increasingly evident that many post-transcriptional events are deeply embedded within the core regulatory circuits of enzyme synthesis/breakdown that maintain metabolic homeostasis. The prominent post-transcriptional mechanisms affecting intermediary metabolism include alternative pre-mRNA processing, mRNA stability and translation control, and the more recently discovered regulation by noncoding RNAs. In this review, we discuss the latest advances in our understanding of these diverse mechanisms at the cell-, tissue- and organismal-level. We also highlight the dynamics, complexity and non-linear nature of their regulatory roles in metabolic decision making, and deliberate some of the outstanding questions and challenges in this rapidly expanding field.
Keywords: Alternative splicing, microRNA, lncRNA, post-transcriptional gene regulation, metabolic syndrome
Graphical Abstract
1. Introduction
Metabolism is an essential process integral to all forms of life. Broadly speaking, it entails all of the biochemical and physical processes that occur within a living system. This includes the chemical breakdown of food to yield energy, but also the synthesis of important macromolecules, transport of substances and excretion of waste. Central to many of these metabolic processes are three basic classes of molecules: carbohydrates, amino acids, and lipids. Owing to their importance in sustaining life, it was crucial for early organisms to develop mechanisms allowing for their uptake, storage and metabolism. Mammals today have highly dynamic and complex metabolic networks that integrate signaling with a variety of gene regulatory mechanisms. This allows the cell to orchestrate various processes in a graded manner providing continuous and efficient feedback.
Studies over the years have revealed that in addition to allosteric regulation, metabolic pathways are also strongly regulated via post-translational covalent modifications (PTMs) and gene transcription [1, 2]. PTMs are by far one of the fastest means by which cells can respond to external and internal stimuli. This is because PTMs tend to be reversible and they act directly on the effector protein to modulate properties such as protein activity, stability, interaction partners, and localization. Although a multitude of PTMs have been identified, the most extensively studied and relevant modification in regards to metabolic regulation is protein phosphorylation. Cells can also respond to metabolic cues by modulating transcriptional rates of key genes within specific pathways. This form of regulation is fulfilled by transcription factors (TFs), which bind directly to specific DNA sequences near a gene and can either promote or repress the recruitment of RNA polymerase. For instance, Sterol regulatory element binding proteins (SREBPs) and Carbohydrate response element binding protein (ChREBP) are TFs, which play major regulatory functions in lipid and carbohydrate metabolism, respectively [3, 4]. Although transcriptional and post-translational regulations contribute greatly in coordinating metabolic processes, recent studies are now identifying a highly diverse set of control occurring at the post-transcriptional level.
Eukaryotic transcription and translation take place in separate cellular compartments, the nucleus, and cytoplasm, respectively. This physical uncoupling forces newly transcribed mRNA to journey into the cytoplasm before being translated into protein. It is now known that along this journey, an mRNA transcript experiences numerous interactions with various factors and undergoes extensive remodeling [5]. This diverse set of processing events includes capping, splicing, and polyadenylation and as with most biological processes, they are highly regulated, dynamic and dependent upon the state and type of the cell. In general, these mechanisms regulate gene expression by altering the following properties of RNA: its stability, localization, translation efficiency, and nucleotide sequence. Collectively, these are referred to as post-transcriptional gene regulatory mechanisms.
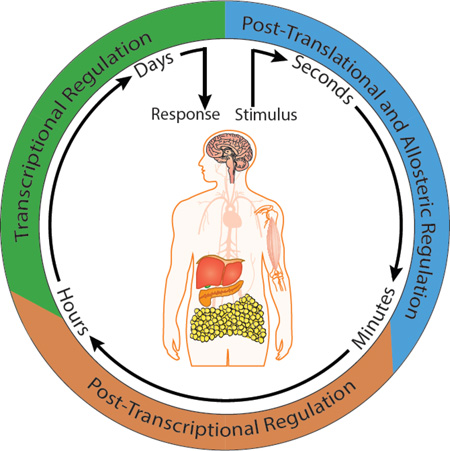
A simple question one might ask is why do living systems utilize post-transcriptional gene regulation to control metabolic pathways if cells can regulate activity using transcriptional and post-translational mechanisms? These overlapping mechanisms may at first seem unnecessarily redundant; however, each of them operates on a different time-scale, providing the organism flexibility to adapt to a wide range of physiological situations. Allosteric changes typically occur within seconds-to-minutes; post-translational modifications and post-transcriptional gene regulatory events function between minutes-to-hours; whereas new transcription and protein synthesis generally require hours-to-days. In theory, every step of gene expression has the potential to be regulated. However, it is unlikely that any given gene will exhibit regulation at all of these steps. Nevertheless, despite the mechanistic diversity of the different forms of regulation, they all have one thing in common. That is, they generally act in response to stimuli and adjust the activity of a biological process to maintain homeostasis. In this article, we review the evidence that these principles also apply to regulation of metabolic pathways by post-transcriptional mechanisms. We further explore the integration of these mechanisms within different metabolic circuits while considering their dynamics, complexity and non-linear nature in affecting physiological outcomes.
2. Non-coding RNAs tune complex networks of metabolic regulation
Mammalian metabolism is a complex process that takes place at multiple levels ranging from the cellular to the organismal. Moreover, metabolic activities vary widely among different types of tissues. However, the metabolic contributions from each tissue act synergistically providing robust homeostasis. Deficiency of a single tissue within the system can cause global physiological imbalance and result in the development of metabolic syndrome. At the cellular level, homeostasis is maintained by the intricate signaling and gene expression networks, which sense and respond to various metabolic cues and nutrients. Currently, much of our understanding regarding metabolic regulation is focused at the layer of gene transcription. Nonetheless, since the completion of the Human Genome Project, it has become clear that a majority of the human transcriptome consists of transcripts, which are not translated into protein [6]. These RNAs are referred to as non-coding RNAs (ncRNAs). Studies elucidating the role of these RNA species revealed that certain classes of ncRNAs could directly modulate gene expression providing an additional layer of gene regulation on top of transcriptional control mechanisms [7, 8]. An increasing number of studies have demonstrated that expression of many ncRNAs is highly dynamic and can be affected by feeding and nutrient availability. Moreover, aberrant expression of ncRNAs is found in several metabolic disorders, which in some cases could lead to pathological states like diabetes and obesity.
2.1. Metabolic Regulation by microRNAs
A class of ncRNAs, known as microRNAs (miRNAs), is now considered to have key regulatory roles in maintaining metabolic homeostasis. An emerging body of evidence is revealing that miRNA profiles can change drastically depending on circulating hormones such as insulin or the metabolic state of the cell. Originally discovered to regulate development of C. elegans, miRNAs are now found to influence almost every biological process from X-chromosome inactivation to cell proliferation and apoptosis. These ubiquitous regulators are transcribed and processed into short (~22 bases) fragments by the Dicer and Drosha ribonucleases before binding to Argonaute (AGO) proteins to form the RNA-induced silencing complex (RISC). miRNAs bind primarily to the 3’ untranslated region (3’UTR) of their target mRNAs, subsequently inhibiting protein expression through translational repression or mRNA degradation as determined by seed sequence complementarity [9, 10]. In general, a single miRNA can target hundreds of genes, and multiple miRNAs can target a single gene. While an individual miRNA may repress the expression of hundreds of proteins, the downregulation of these proteins are typically mild [11, 12]. However, it has been shown that perturbing the in-vivo levels of a single miRNA can induce significant phenotypic consequences. If this is the case, then how are individual miRNAs exerting large-scale effects despite their marginal influence on protein expression? Studies over the past few years have shown that miRNA-based regulation in some cases can involve targeted inhibition of multiple genes within a given pathway or select genes within multiple related pathways. The cumulative effect is thus markedly coherent, modulating the metabolic flux within the cell.
2.1.1. Lipid Metabolism
The first miRNA identified to play a role in regulating metabolism was discovered over a decade ago. Initially identified in Drosophila melanogaster, loss of miR-14 resulted in the abnormal accumulation of lipids within adipocytes [13]. Since then, numerous tissue-specific miRNAs have been discovered which play essential roles in regulating metabolism (Figure 1). For example, miR-33a and miR-33b are two miRNAs expressed in the liver, brain, pancreas, and adipose tissue, and are involved in controlling lipid and cholesterol homeostasis. Interestingly, in humans these two miRNAs are located within the intronic regions of the SREBP-coding genes: SREBF1 encodes miR-33b in intron 17, while SREBF2 encodes miR-33a in intron 16 [14]. SREBPs are master regulators of lipid metabolism; in particular, they modulate the expression of numerous enzymes involved in synthesis and transport of cholesterol and fatty acids [3]. Remarkably, it has been shown that both miR-33a/b are extensively co-expressed with their SREBP host genes allowing them to work together in regulating cellular levels of lipids and cholesterol [15]. Especially, when sterols or fatty acids levels are low, activated SREBPs promote the transcription of lipogenic and cholesterogenic genes. This response is further bolstered by a simultaneous shutdown of pathways involved in fatty acid degradation and cholesterol export by miR-33a/b activity. Specifically, miR-33 inhibits the expression of several genes involved in beta-oxidation of fatty acids including carnitine O-octanoyltransferase (CROT), trifunctional enzyme subunit beta (HADHB), and carnitine palmitoyltransferase 1A (CPT1A) [16, 17]. miR-33a/b also repress the expression of several members of the ATP-binding cassette superfamily, including ABCA1 and ABCG1 which are primary mediators of cholesterol efflux, resulting in increased levels of cellular cholesterol and reduced formation of HDL particles [18]. Additionally, miR-33b also targets and inhibits rate-limiting enzymes involved in gluconeogenesis leading to significant reduction in glucose production [19]. Although this link between glucose and lipid metabolism needs to be studied further, this tuning of glucose metabolism by miR-33b may allow the cell to focus efforts on increasing lipid levels.
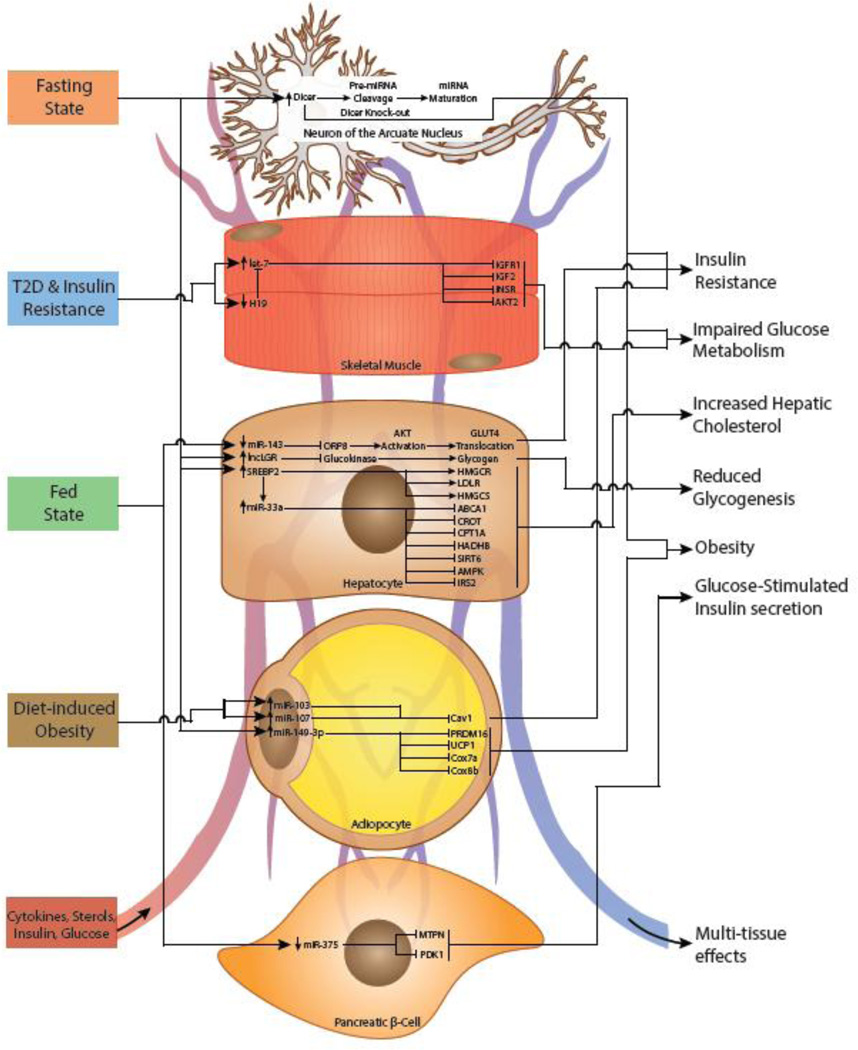
Figure 1. Non-coding RNA regulation of metabolic activity in multiple tissues.
Non-coding RNAs (ncRNAs) are central to metabolic regulation in the brain, skeletal muscle, liver, adipose tissue, and pancreas. Under fasting conditions, neurons of the arcuate nucleus increase DICER expression. Knockout of DICER in POMC-expressing cells impairs glucose metabolism, insulin resistance, and obesity. Fasting also upregulates hepatic lncRNA liver GCK repressor (lncLGR), leading to glucokinase inhibition and reduced glycogenesis. Decreased cholesterol coordinately increases hepatic sterol regulatory element-binding protein 2 (SREBP2) with its intronic miR-33a to increase cholesterol biosynthesis and uptake through (3-Hydroxy-3-Methylglutaryl-CoA Reductase (HMGCR), low density lipoprotein receptor (LDLR), hydroxymethylglutaryl-CoA synthase (HMGCS)), and decrease cholesterol efflux (ATP-binding cassette, sub-family A, member 1 (ABCA1)), β-oxidation (carnitine O-octanoyltransferase (CROT), trifunctional enzyme subunit beta (HADHB), and carnitine palmitoyltransferase 1A (CPT1A)), and SREBP inhibition (Sirtuin 6 (SIRT6), 5' AMP-activated protein kinase (AMPK), insulin receptor substrate 2 (IRS2)). Fasting also increases miR-149-3p, inhibiting genes involved in brown adipocyte thermogenesis (PR domain-containing protein 16 (PRDM16), uncoupling protein 1 (UCP1), cytochrome c oxidase polypeptide 7A1 (COX7A), cytochrome c oxidase polypeptide 8B (COX8B)). Type II Diabetes (T2D) and insulin resistance lead to decreased H19 and increased lethal-7 (let-7) expression in skeletal muscle. Let-7 inhibits insulin-phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)-mammalian target of rapamycin (mTOR) signaling, causing impaired glucose metabolism. Under fed conditions, decreased hepatic miR-143 leads to increased glucose uptake and glucose transporter type 4 (GLUT4) translocation. Overexpression of miR-143 has been linked to insulin resistance. Under fed conditions, increased liver X receptor (LXR) activation upregulates LXR-responsive lncRNA (LeXis) expression, inhibiting SREBP2 and HMGCR, leading to reduced serum cholesterol. Additionally, fed conditions increase liver-specific triglyceride regulator lncRNA (lncLSTR) expression which upregulates Apolipoprotein C2 (APOC2). Upregulated APOC2 leads to increased Lipoprotein Lipase (LPL) activity and subsequent triacylglyceride (TAG) uptake in peripheral tissues. Diet-induced obesity has been shown to upregulate miR-103 and miR-107 in adipocytes, leading to inhibition of insulin receptor stabilization Caveolin 1 (Cav1)) and insulin resistance. Under fed conditions, downregulation of miR-375 in pancreatic β-cells inhibits Myotrophin (MTPN) and 3-phosphoinositide dependent protein kinase-1 (PDK1), and subsequently decreases glucose-stimulated insulin secretion.
While miR-33a/b directly influences cellular concentrations of sterols and lipids, several other miRNAs have been implicated in regulating circulating levels of cholesterol and triglycerides. Recently, Goedeke et al. have shown that miR-148 controls circulating lipoprotein levels through decreased expression of key lipoprotein trafficking proteins, low-density lipoprotein receptor (LDLR) and ABCA1 [20]. Similarly, Wagschal et al. exploited available data from genome-wide association studies (GWAS) of a large patient cohort in which common single-nucleotide polymorphisms (SNPs) were associated with blood lipid levels [21, 22]. Analysis of this GWAS dataset identified 69 miRNAs located in proximity to SNPs linked to abnormal blood lipid levels. These were further narrowed down to four conserved miRNAs—miR-128-1, miR-148a, miR-130b, and miR-301b—that were predicted to strongly target genes involved in lipid metabolism and cardiometabolic disorders. Transfecting HepG2 cells with precursor oligonucleotides of these miRNAs resulted in decreased expression of LDLR and ABCA1. Using lentiviral vectors, in-vivo overexpression of miR-128-1 and miR-148a in diet-induced obese C57BL/J mice resulted in strong reduction of circulating HDL-C levels, as expected with diminished ABCA1 activity [21]. Taken together, these studies provide strong evidence that miRNA can influence blood lipid levels and misregulation of these regulatory factors can predispose individuals to cardiometabolic disorders.
2.1.2. Insulin signaling and Glucose Metabolism
In addition to lipid regulation, there are now many miRNAs shown to regulate multiple aspects of glucose metabolism and insulin signaling (Figure 1). Proper metabolism of glucose is critical for normal physiology and misregulation at any step can cause disease. In humans, blood glucose concentration is maintained by two opposing hormones released by the pancreas known as insulin and glucagon. Under fasting conditions, when blood glucose levels are low, the pancreas secretes glucagon and stimulates the liver to break down glycogen into glucose and releases it into the bloodstream. On the contrary, when glucose levels are elevated following a meal, the pancreas responds by releasing insulin and thus lowers blood glucose concentration. This physiological response is mediated by the pleiotropic effects of insulin on a number of tissues which include: increasing uptake of glucose in adipose and muscle tissue, inhibiting gluconeogenesis in liver, and promoting the synthesis of fat, protein and glycogen.. At the molecular level, binding of insulin to the insulin receptor (INSR) activates a complex network of signaling cascades within the cell. A key event within this cascade is the phosphorylation of serine/threonine-specific protein kinase (AKT) which promotes nutrient uptake and growth [23]. Inadequacy at any step of insulin production and signaling can potentially result in the development of diabetes. Type 2 diabetes (T2D) is one of the most widespread metabolic disorders in the developing world today. Improper management of this disease can lead to overwhelming complications such as kidney failure, cardiovascular disease and blindness. The typical hallmark of T2D is the development of insulin resistance, often resulting from obesity and sedentary lifestyle. Although T2D is a highly prevalent disease, the underlying mechanisms of obesity-associated insulin resistance are still not clear.
To identify misregulated miRNAs, which might contribute to T2D, many studies have taken an unbiased ‘miRNome’ expression profiling approach. Using transgenic or diet-induced animal models of insulin resistance, alterations in transcriptome-wide miRNA levels can be measured simultaneously in a tissue-specific manner. These studies have shown that many miRNAs misregulated in obesity are involved in controlling tissue insulin sensitivity. For example, Kornfeld et al. have shown that expression of miR-802 is upregulated in livers of obese mice as well as obese human subjects [24]. In-vivo loss- and gain-of-function studies revealed that miR-802 impaired hepatic response to insulin and glucose metabolism. While inducible overexpression of miR-802 in lean mice diminished insulin-induced phosphorylation of AKT, suppression of miR-802 in obese mice using locked nucleic acids enhanced this response. These findings were further corroborated by improved glucose tolerance and insulin sensitivity with inhibition of miR-802, and vice versa. Subsequent bioinformatics analysis identified 26 potential target genes, of which hepatocyte nuclear factor 1 beta (HNF1B) was functionally validated to be a bonafide miR-802 target [24]. Although in-vivo knockdown of HNF1B in liver recapitulated the effects of miR-802 overexpression, further investigation is needed to elucidate the detailed mechanisms by which miR-802 asserts its regulatory effects.
Whereas miR-802 is specific to liver, several miRNAs have now been described to maintain insulin sensitivity in other tissues as well. Work by Zhu et al. showed that global overexpression of lethal-7 (let-7) in mice resulted in impaired glucose tolerance [25]. However, it was later reported by Frost et al. that tissue-specific overexpression of let-7 in muscle, liver, neurons, and adipocytes did not cause impaired glucose tolerance [26]. Although let-7 is not altered in obesity-induced insulin resistant mice, treatment of obese mice with let-7 antimiR was sufficient to improve glucose tolerance. This result suggests that let-7 miRNA might be improving insulin sensitivity in peripheral tissues. Importantly, it has been predicted that let-7 can target INSR and Insulin Receptor Substrate 2 (IRS2). Indeed, antimiR treatment in obese mice brought levels of INSR back to normal physiological levels in both muscle and liver and IRS2 in liver [26]. Another study on human patients found miR-1 and miR-133 significantly decreased in skeletal muscles after insulin stimulation [27]. Further investigation into the mechanism of this repression revealed that it was due to decreased transcriptional activity of Myocyte Enhancer Factor 2C (MEF2C). However, in diabetic patients, this insulin-mediated decrease of miR-1 and miR-133 expression was blunted due to decreased transcription. The physiological effect of insulin-mediated downregulation of these two miRNA in skeletal muscle has not been investigated. There are now several known miRNAs, which are misregulated in T2D and have been shown to play a role in modulating insulin sensitivity in tissues. This includes miR-143 and miR-26a in liver, miR-103/107 in both adipose and liver tissue, and miR-16 in skeletal muscle [28–31]. Moreover, future investigations also need to focus on the upstream regulators, which are altering miRNA levels in tissues in disease conditions. This knowledge will be crucial in understanding if altered expression of miRNA is protective or damaging.
2.1.3. MicroRNAs in Pancreas and Brain
Aside from affecting insulin sensitivity within peripheral tissues, miRNAs are also important for regulating the production and secretion of insulin from pancreatic β cells (Figure 1). The importance of miRNA in pancreatic function was revealed from a study in which a pancreas-specific knock out of DICER resulted in developmental defects [32]. Studies are now focusing on identifying specific miRNAs that drive these phenotypes. miR-375 is an important pancreatic miRNA regulating insulin secretion [33]. Overexpression of this miRNA resulted in a reduction of glucose-stimulated insulin secretion. Myotrophin, a protein associated to exocytosis, was identified as a target of miR-375 providing a possible mechanism for miR-375 control in insulin secretion.
The nervous system can often be overlooked when discussing metabolic regulation; however, it plays a crucial role in maintaining whole-body energy balance. Neurons, such as POMC and AgRP, within this region of the brain, have the ability to sense humoral factors and nutrient levels to evaluate energy status. Depending on the energy status, the activity of these neurons will then modulate appetite and energy usage. Exciting new findings suggest that miRNA may contribute to feeding behaviors. It was shown that levels of DICER are upregulated in the hypothalamus under fasting conditions [34]. Furthermore, ablating DICER in a sub-region of the hypothalamus known as the arcuate nucleus caused hyperphagia and obesity that was not due to neuronal cell death [35]. These mice exhibited increased activation of the phosphoinositide 3-kinase (PI3K)-Akt-mammalian target of rapamycin (mTOR) pathway, which previously was shown to result in hyperphagic behavior. Additional computational analysis suggested miR-103 as a likely candidate resulting in over-activation of the pathway. Introduction of miR-103 mimic into the brain reduced the activity of PI3K and diminished the hyperphagic obesity [35]. Multiple studies are now beginning to show that expression of many miRNAs is altered in the hypothalamus depending on the overall energy status [36–38]. Future studies interrogating the specific role of these miRNAs will be crucial to understanding their physiological effects.
2.2. Metabolic Regulation by lncRNAs
Another class of ncRNAs, known as long noncoding RNA (lncRNA), has emerged to regulate gene expression. But, do any lncRNAs contribute to the complex regulatory networks of metabolism? If so, how might these RNAs direct changes in metabolic states and vice versa? Unlike miRNA, lncRNAs closely resemble a typical mRNA transcript; once transcribed, these RNAs are 5’-capped, spliced, and polyadenylated. To date, the GENCODE consortium has annotated almost 15,000 unique lncRNAs. However only a handful of them have been functionally characterized [39]. Elucidating functional roles of lncRNAs has posed a unique challenge within the field due to their lack of conservation across species. This has made it difficult to study relevant human lncRNAs in model organisms. Moreover, lncRNAs exert their regulation on gene expression through a variety of mechanisms such as influencing transcriptional activity, remodeling chromatin, molecular scaffolding or acting as molecular sponges for miRNAs [40].
2.2.1. Cholesterol Metabolism
Within the past few years, several liver-enriched lncRNAs have been identified in controlling cholesterol homeostasis. It is well established that the transcriptional regulators liver X receptor (LXR) and SREBP2 tightly regulate hepatic levels of cholesterol. Under conditions of elevated cholesterol, LXRs protect the cell by upregulating genes involved in cholesterol efflux and converting it to bile acids [41]. However, a recent transcriptome profiling study on mouse hepatocytes revealed a previously uncharacterized lncRNA to be highly induced upon LXR activation [42]. Hence, it was given the name LeXis, which stands for liver-expressed LXR-induced sequence. Expression of LeXis in mouse livers downregulated genes involved in cholesterol biosynthesis, such as Srebf2 and Hmgcr, resulting in reduced serum cholesterol levels (Figure 1). Although the mechanism is not well defined, LeXis mediated transcriptional repression of its target genes was shown to be dependent on interactions with a heterogeneous ribonucleoprotein (hnRNP) associated with lethal yellow protein homolog (RALY)[42]. Analogous to the SREBP2/miR-33a axis, LXR and LeXis act in harmony by simultaneously regulating separate but related pathways to lower cellular cholesterol. Whereas a functional human orthologue of LeXis is yet to be found, a recently discovered lncRNA also involved in cholesterol metabolism called lnc-HC is highly conserved in mice, rats, and humans both by sequence and chromosomal position [43]. Elevated levels of cholesterol stimulate expression of lnc-HC in hepatocytes. However, unlike LeXis, functional characterization of lnc-HC revealed it to behave antagonistically to LXR activity. It was shown that lnc-HC forms a complex with hnRNPA2B1 and reduces the stability of mRNA targets cholesterol 7 alpha-hydroxylase (Cyp7a1) and Abca1 which are involved in bile acid synthesis and cholesterol efflux, respectively [43]. These results are unexpected and suggest that lnc-HC is counteracting the effects of LXR which upregulates the expression of Cyp7a1 and Abca1 under conditions of elevated cholesterol. A possible explanation for the antagonizing effects of lnc-HC and LXR is that it allows the cell to fine-tune cholesterol metabolism to an optimal level. However, further investigation of lnc-HC’s targets and regulatory mechanism is needed to fully understand its functional role.
2.2.2. Lipid Metabolism
In addition to cholesterol, lncRNAs can also regulate aspects of lipid homeostasis. Recently, Li et al. have identified a mouse liver-enriched lncRNA termed liver-specific triglyceride regulator (lncLSTR) which is involved in clearance of circulating plasma triglycerides (TAGs) [44]. They have shown that increased hepatic expression of lncLSTR ultimately results in upregulation of apoC2 in a farnesoid X receptor (FXR) dependent manner. This leads to subsequent elevation of serum apolipoprotein C2 (APOC2), which enhances lipoprotein lipase activity and allows for increased uptake of TAGs into peripheral tissues like the heart, skeletal muscle, and adipose tissue (Figure 1). Interestingly, physiological expression of lncLSTR is generally low during fasting but is highly induced upon feeding [44]. Thus, it is likely that lncLSTR’s physiological role is to move lipids into various tissues accommodating for recently ingested lipids. Although not discussed in this review, a large number of lncRNAs have been identified to control various aspects of adipogenesis [45]. Some are specific to activation thermogenic circuits in brown adipose tissue such as Brown fat lncRNA 1 (Blnc1) [46].
2.2.3. Glucose Metabolism
LncRNAs have also been implicated in glucose metabolism. H19 is a lncRNA that was originally found to control gene expression of the Imprinted Gene Network involved in embryo growth [47]. During normal development, expression of H19 is strongly downregulated in most tissues, except for skeletal muscle and heart [48]. It was recently discovered that H19 contains binding sites for let-7 allowing it to act as a molecular sponge sequestering endogenous let-7 [49]. As mentioned earlier, let-7 is involved in modulating insulin sensitivity of muscle tissue suggesting H19 might play a role in glucose homeostasis. Measuring H19 levels in skeletal muscle of T2D patients and insulin-resistant obese mice revealed a strong decrease in expression [50]. In agreement with the sponging model, let-7 targets, Insr and lipoprotein lipase (Lpl), were also downregulated while levels of let-7 remained unchanged in the obese mice. In non-diabetic mice, it was observed that acute hyperinsulinemia resulted in diminished levels of H19 in muscle causing an increase in let-7 availability [50]. Since Insr is a known target of let-7, increased levels of let-7 results in reduced expression of INSR and diminished muscle insulin sensitivity and therefore decreases uptake of glucose. This regulatory axis is analogous to the lnc-HC in which the effects of the lncRNA are antagonizing the initial response. It is hypothesized that the H19/let-7 axis acts as a safety mechanism preventing muscle to draw too much glucose at later time points post insulin stimulation [50] (Figure 1). Another lncRNA implicated in glucose metabolism is liver glucokinase repressor (lncLGR) which is induced in liver under fasting conditions [51]. During fasting, the physiological response of the liver is to transition from a glucose consuming to a glucose producing state. This is in part facilitated by the downregulation of glucokinase, an enzyme that catalyzes the first step of the glycolysis and glycogen synthesis pathway. Reduced synthesis of glycogen allows the liver to start producing glucose and releasing it into the blood to supply peripheral tissues. Overexpression of lncLGR in mice causes downregulation of glucokinase and reduction of hepatic glycogen stores [51]. It was shown that lncLGR forms a complex with hnRNPL and represses the transcriptional activity of glucokinase.
While these findings establish the unique role of ncRNAs in eliciting rapid responses to metabolic perturbations, recent studies suggest a greater trans-generational impact of ncRNA-mediated regulation on metabolic activity. This new body of evidence has focused on the epigenetic inheritance of acquired phenotypes through sperm RNAs [52]. More specifically, miRNAs and an exciting new class of small ncRNAs known as tRNA fragments (BOX 1) have emerged as critical regulators of trans-generational metabolic reprogramming. Paternal stress and diet may result in epigenetic modifications that influence ncRNA expression, altering the metabolic activity of the offspring [53]. Although ncRNA-based regulation of inheritance is poorly understood, it is clear that ncRNAs are central to metabolic regulation at multiple levels, ranging from the cellular to the organismal, and the short-term to the trans-generational.
BOX ITEM 1: A class of small RNAs involved in metabolism and transgenerational inheritance.
One of the newest classes of RNAs to be discovered is a set of small RNAs (~28 – 34 nt) distinct from miRNAs known as tRNA fragments (tRFs). Development of deep sequencing technologies has catalyzed the discovery and understanding of tRFs [121]. Initially thought to be random byproducts of tRNA degradation or biogenesis, recent studies are now showing that tRFs can exhibit selective expression patterns and elicit potent metabolic effects. These small RNAs are highly abundant, second to miRNAs, and are derived from mature tRNAs. Although tRF biogenesis is not completely understood, the abundance of fragments with precise start and end points suggest that they are most likely generated from cleavage by specific endonucleases [122]. Depending on their origin, tRFs have been further subdivided into three distinct types and are referred to as tRF-5, tRF-3 and tRF-1 [123]. As suggested by their classification, tRF-5 and tRF-3 are derived from the 5’ and 3’ ends of mature tRNA, respectively. tRF-1, however, arise from the 3’ trailer region which is present in tRNA prior to processing (pre-tRNA) [123].
Recently, two independent studies have now provided evidence that tRFs can behave as epigenetic factors and contribute to the intergenerational inheritance of metabolic phenotypes. It has long been understood that parental environments and nutrition can influence the health of the offspring. In fact, controlled studies in mice have shown that glucose and lipid metabolism of the F1 offspring can be affected depending on the diet fed to the father [124]. These observations strongly suggested that nutrition could alter factors within gametes resulting in heritable phenotypes that are independent of genotype. While these factors involved in reprogramming offspring phenotype have remained elusive, work by both Sharma et al. and Chen et al. show that tRFs may be the major contributors [125, 126]. Using a paternal mouse model fed a low-protein diet which results in offspring with impaired cholesterol metabolism; Sharma et al. showed that protein restriction caused an increase in tRF-Gly-CCC abundance within mature sperm [127]. To further assess its biological role, microinjection of synthetic tRF-Gly-CCC into control zygotes resulted in the repression of genes known to be targets of MERVL, a transposable element that regulates genes needed for pre-implantation development [125]. While repression of MERVL targets is seen in offspring of low-protein fed fathers, it is still not clear whether this is responsible for the marked impairment in cholesterol metabolism. While Sharma et al. studied nutrient restriction, Chen et al. utilized a model of nutrient abundance in which male mice were fed a high-fat diet (HFD) [126]. Offspring fathered from these mice are known to exhibit impairment in glucose metabolism with the onset of glucose tolerance and insulin resistance [128]. Similar to Sharma et al., they also found changes in tRF levels in the sperm of HFD mice. Indeed, injection of tRFs isolated from these mice into naïve zygotes was capable of transmitting the metabolic disorder to the F1 generation [126]. Taken together, these studies have shown that tRFs may be major contributors in nutrition-induced transgenerational inheritance.
There are now numerous studies showing that stress conditions such as hypoxia, osmotic stress, and nutrient deficiency can up-regulate the production of tRFs [129–131]. However, mechanistic understandings of how tRFs influence physiological effects on cells are still in the early phases. Some studies suggest tRFs might behave like miRNAs since they were found to be associated with DICER and AGO proteins [132]. However, this notion has been challenged by other studies in which knock down of a tRF resulted in decreased expression of its predicted targets. Whether tRFs follow a particular course of action or exhibit a diverse range of mechanisms is yet to be determined.
3. RNA processing directs the correct expression of key metabolic enzymes
It is becoming increasingly evident that RNA processing plays an important role in metabolic regulation. Nascent transcripts go through a wide range of remodeling and processing steps prior to being translated. This includes capping, splicing, polyadenylation and editing. Although essential for RNA maturation, each step is highly regulated and has the potential to process each transcript differentially, thus, increasing both transcriptome and proteome diversity. Moreover, RNA processing plays a significant role in regulating gene expression as it can directly alter properties of an RNA molecule, such as its sequence, stability, localization, and translation efficiency. Essentially, this allows a cell to control gene expression without perturbing its transcriptional network. Furthermore, changes mediated by RNA processing occur at shorter time-scales as compared to transcriptional regulation. Therefore in terms of metabolic control, these mechanisms allow a cell respond in a graded manner to the changing metabolic environment.
3.1. Alternative Splicing
Splicing of nascent pre-mRNA is a crucial process in gene expression involving the removal of introns and ligation of exons within the nucleus by a large macromolecular complex known as the spliceosome. This process is highly regulated and certain exons can be spliced differentially resulting in different mature transcripts in a process known as alternative splicing (AS). This process allows a single gene to produce multiple variants, thus, expanding the diversity of the eukaryotic proteome. Recent studies estimate that about 95% of human genes are alternatively spliced resulting in greater than 100,000 proteins from less than 20,000 genes [54, 55]. Although AS essentially changes the sequence of short segments within the transcript, the affect on the protein output can be drastic. A recent study profiling the protein-protein interactions of protein isoforms revealed that a majority share less than 50% of their interactions [56]. There are multiple modes of AS with the simplest being exon skipping which involves the inclusion or exclusion of a single exon. However, other types of AS exons also exist including mutually exclusive exons, retained introns, and exons with alternative 5’/3’ splice site. Alternative splicing is regulated by a special class of RNA binding proteins (RBP) known as splicing factors which bind to cis-acting motifs within the transcript [57]. The pattern of splicing for a given exon is dependent on the splicing factor and the context of its binding. For instance, certain cis-acting elements can promote the inclusion of an exon and are known as splicing enhancers, while other elements, which promote exclusion, are known as splicing silencers. Mechanistically, binding of these factors to the RNA regulates the splicing of an exon by either promoting or inhibiting the recruitment of the spliceosome. AS is central to postnatal maturation of tissues and their normal physiological function, however, aberrant splicing has been implicated in several disease pathologies [58, 59].
Recent studies are now revealing that AS can strongly impact major metabolic pathways and control the flux of metabolites within a cell. One way this can be achieved by AS is through the alteration of the activities of key metabolic enzymes. Glycolysis, which converts glucose to pyruvate, is one pathway that is regulated by AS in a tissue-specific manner. Not only is this pathway crucial for cellular respiration, it also provides intermediates for other essential pathways such as gluconeogenesis. Pyruvate kinase (PK), the last enzyme in glycolysis responsible for converting phosphoenolpyruvate (PEP) to pyruvate, exists as multiple isozymes which are expressed in a tissue dependent manner. Two of the isozymes, PKM1 and PKM2, are derived from the alternative splicing of the mutually exclusive exons 9 and 10 in the PKM gene (Figure 2) [60]. While PKM1 is highly expressed in muscle, heart and brain, PKM2 is mainly detected in early fetal tissue and proliferating cells. Cells with increased proliferation naturally require greater amounts of building block precursors to grow. Significant sources of these precursors are derived from the glycolytic intermediates. One simple strategy to increase the amounts of these intermediates is to block glycolysis at the last step, resulting in the accumulation of upstream intermediates. Such is the case with the PKM2 isozyme. In highly proliferative cells the splicing factor SRSF3 promotes the inclusion of exon 10 resulting in the expression of the PKM2 isozyme [61]. Unlike the tetrameric PKM1, PKM2 can also exist as a dimer with a significantly diminished affinity for PEP [62]. This prevents the formation of pyruvate allowing the cell to generate the necessary intermediates for the biosynthesis of nucleotides, phospholipids, and amino acids. PKM2 activity is also highly dependent on amino acid concentration, glycolytic intermediates, and phosphorylation [63]. For this reason, PKM2 isozyme serves an important role in proliferating tissues and fetal development.
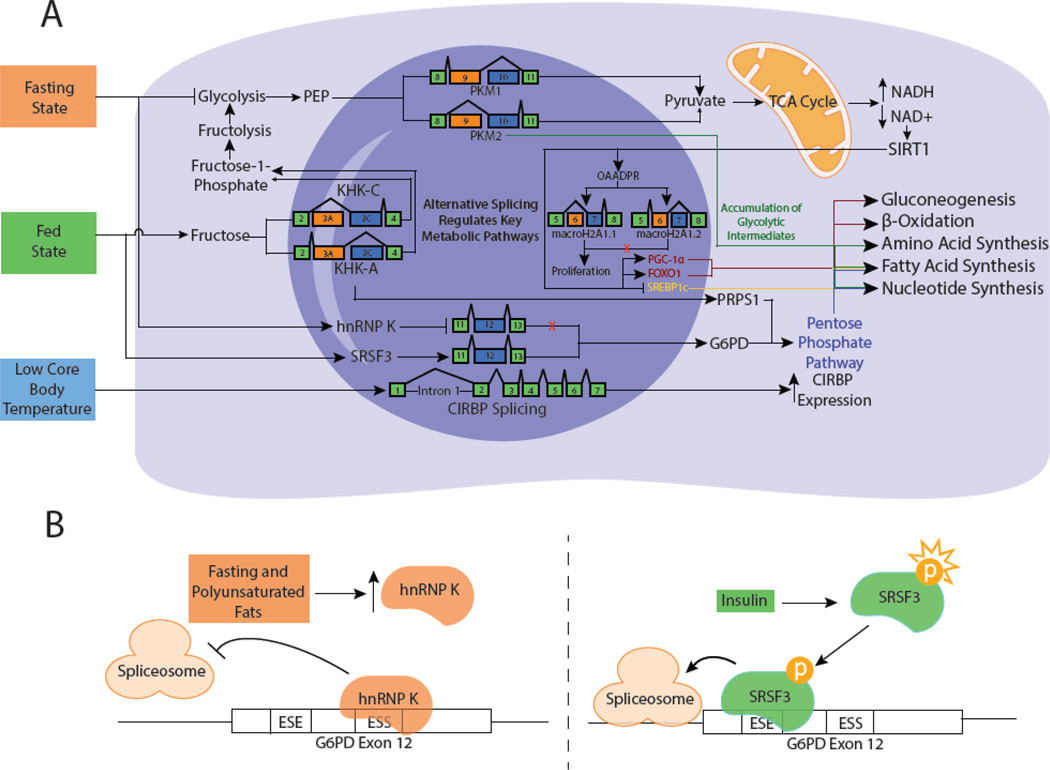
Figure 2. Regulation of cellular metabolism by alternative splicing.
A) Alternative splicing (AS) regulates metabolic activity through the production of functionally distinct protein isoforms. Fasting decreases glycolytic activity, increasing cellular NAD+ levels. Sirtuin-1 (SIRT1) binds NAD+, activating the transcription of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and forkhead box protein O1 (FOXO1) while inhibiting sterol regulatory element-binding protein 1C (SREBP1c). SIRT1 activity increases gluconeogenesis, β-oxidation, and decreases fatty acid synthesis. The NAD+ metabolite, 2-O-acetyl-ADP-ribose (OAADPR) is bound by macroH2A1.1, leading to a transcriptional increase in proliferation-associated genes. The less active pyruvate kinase M2 (PKM2) isoform of pyruvate kinase shunts glycolytic intermediates into amino acid, fatty acid, and nucleotide synthesis. The less active ketohexokinase-A (KHK-A) isoform of ketohexokinase inhibits phosphate depletion and promotes nucleotide synthesis through activation of the pentose phosphate pathway (PPP). The macroH2A1.2 histone isoform is unable to bind NAD+ metabolites, resulting in loss of transcriptional activation. Under fasting conditions, upregulation of heterogeneous nuclear ribonucleoprotein K (hnRNP K) inhibits glucose-6-phosphate dehydrogenase (G6PD) splicing and PPP activation, reducing fatty acid and nucleotide synthesis. B) hnRNP K binds to the exonic splicing suppressor (ESS) in exon 12 of G6PD, inhibiting splicing. Fed states have opposite effects through the activation of Serine/Arginine-Rich Splicing Factor 3 (SRSF3) and increased G6PD expression. Activated SRSF3 binds the exonic splicing enhancer (ESE) in exon 12, promoting splicing. Low core body temperature induces Cold Inducible RNA Binding Protein (CIRBP) expression through a cold-responsive element in intron 1 that regulates splicing efficiency.
AS mechanisms are also involved in regulating key enzymes part of the pentose phosphate pathway (PPP). Branching off from glycolysis, PPP is an essential pathway involved in generating precursors for nucleotide and amino acid synthesis. Glucose-6-phosphate dehydrogenase (G6PD) which converts glucose-6-phosphate to 6-phosphogluconolactone is the rate-limiting enzyme of the PPP. Moreover, G6PD also produces nicotinamide adenine dinucleotide phosphate (NADPH), a reducing equivalent required for fatty acid and nucleotide synthesis. Therefore, it is not surprising that the level of this enzyme is highly dependent on the nutritional status of the organism. Under fed conditions, increased expression of G6PD in the liver promotes lipogenesis by providing necessary reducing equivalents [64]. Surprisingly, regulation of this gene by varying nutritional status is mediated by changing its splicing efficiency and not transcriptional rates. Specifically, reducing the efficiency of intron removal from the G6PD primary transcript can attenuate expression of the enzyme. Regulation of the splicing efficiency of this gene is accomplished by a bifunctional exonic splicing enhancer (ESE) and silencer (ESS) element located within exon 12 of the transcript (Figure 2). Under fasting conditions, upregulation of the splicing factor hnRNP K and binding to ESS element inhibits splicing, subsequently reducing expression of G6PD enzyme. Remarkably, this motif also functions as an ESE site for Serine/Arginine-Rich Splicing Factor 3 (SRSF3). Under nutrient-rich conditions, insulin induces SRSF3 activity through the phosphorylation of SR proteins. Phosphorylated SRSF3 can then bind G6PD pre-mRNA and promote exon inclusion thus increasing G6PD expression [65, 66].
Studies have also outlined the role of AS in regulating cholesterol metabolism. The rate-limiting enzyme in the cholesterol biosynthetic pathway, 3-Hydroxy-3-Methylglutaryl-CoA Reductase (HMGCR), is a target of heterogeneous nuclear ribonucleoproteins A1 (hnRNP A1), a prominent regulatory protein and splicing factor. An HMGCR SNP in the binding area of hnRNP A1 was shown to promote skipping of exon 13, leading to inactivation of HMGCR activity and increased LDL-C uptake [67]. Additionally, sterol loading has been demonstrated to induce the splicing and subsequent inactivation of HMGCR and LDLR by another prominent splicing factor Polypyrimidine Tract Binding Protein 1 (PTBP1) [68]. SRSF3 is also important for lipid and cholesterol metabolism in the liver. Ablation of this splicing factor in mouse liver caused significant damage including misregulation of lipid and cholesterol metabolism. Loss of hepatic SRSF3 resulted in aberrant splicing of many metabolic genes including SREBP cleavage-activating proteins, which are important regulatory genes in cholesterol metabolism. These mice exhibited reduced hepatic and serum cholesterol with elevated levels of triglycerides [69]. Taken together, these studies show that splicing factors serve important roles in maintaining normal metabolism.
Besides regulating metabolic enzymes, AS can be used to tune the properties of various factors in response to external metabolic cues. Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) is a major transcriptional activator that has been shown to be extremely important for skeletal muscle remodeling, mitochondrial biogenesis, and angiogenesis. Variations in skeletal muscle training were found to induce alternative promoter utilization and splicing of PGC-1α mRNA, producing the α1 and α4 isoforms. Endurance training generates the α1 isoform, which leads to increases in oxidative phosphorylation genes and myosin switching. In contrast, the α4 isoform of PGC-1α interacts with separate targets like insulin-like growth factor 1 (IGF-1) and genes in the myostatin pathways to induce skeletal muscle hypertrophy in response to resistance training [70] (Figure 2).
Interestingly, changes in core body temperature (CBT) at the organismal level can also impact splicing efficiency. While many have reported on the effects of heat shock and splicing regulation, a recent study by Gotic et al. has introduced for the first time a cold-induced change in splicing efficiency that directs the expression of the cold-induced RNA binding protein (CIRBP) [71]. A number of studies over the last two decades have determined that CIRBP has a role in proliferation, both activating and inhibiting cancer formation in a cell type-specific manner. During low CBT fluctuations (33°C), CIRBP pre-mRNA is efficiently spliced and expressed. However, high CBT (38°C) conditions decreases splicing efficiency of the transcript and subsequently reducing protein expression. Splicing of CIRBP pre-mRNA is mediated by a temperature-dependent regulatory element within the first intron that is contextually independent, suggesting that a cold-induced response takes advantage of this element to induce splicing changes (Figure 2).
While regulation of gene expression by AS is important for normal physiology, aberrant splicing can contribute to the development of metabolic syndrome [69, 72]. The expression of several genes involved in splicing is downregulated in human obesity within various metabolic tissues, suggesting its importance for normal metabolism [73]. For instance, splicing factor arginine/serine-rich 10 (SFRS10) was shown to be downregulated in the liver and skeletal muscle tissues in obese patients. The key regulator of lipid metabolism, Lipin-1 (LPIN1), is a putative target of SFRS10 and exists in two major splice isoforms, LPIN1A and LPIN1A. Downregulation of SFRS10 favors the LPIN1B isoform, which increases lipogenesis by upregulating the levels of Srebp1c and Fatty acid synthase (FASN). SFRS10 heterozygosity in mice results in increased VLDL secretion and hypertriglyceridemia [73]. Besides obesity, cancer is another pathological condition, which is known to have misregulated AS. An example of an enzyme misregulated in hepatocellular cancer is ketohexokinase (fructokinase). This enzyme is responsible for the initial phosphorylation of fructose to fructose-1-phosphate and thus enables the utilization of fructose by the cell. Interestingly, the splicing of the gene encoding this enzyme is misregulated in hepatocellular carcinoma cells by the splicing regulators hnRNPH1/2. This generates the fructokinase-A isoform instead of fructokinase-C [74]. Interestingly, the activity of fructokinase-A is significantly diminished as compared to fructokinase-C. Importantly, this isoform switch to fructokinase-A also stimulates the pentose-phosphate pathway through phosphorylation of phosphoribosyl pyrophosphate synthetase 1 (PRPS1). This induces de novo nucleic acid synthesis necessary for cancer proliferation and prevents intracellular phosphate depletion (Figure 2).
Ultimately, the regulation of metabolic enzymes directs the fluxes in metabolite concentrations within a cell and its environment. However, it is often found that the levels of metabolites can also feedback and tune gene regulatory network. An example of this can be seen with the ubiquitous coenzyme nicotinamide adenine dinucleotide (NAD+). The levels of NAD+ strongly correlate with cellular energy states. Studies have shown that nuclear NAD+ levels can affect gene expression epigenetically. During periods of energy crisis, increased NAD+ concentrations upregulate the activity of Sirtuin 1, a histone deacetylase, and promote transcriptional repression of loci associated with energy expenditure processes [75][76]. Interestingly, emerging studies have now shown that NAD+ may also be linked to splice isoforms of histones. The H2A family of histone proteins produces a number of histone variants with significant implications in regulating chromatin structure and transcriptional repression. MacroH2As are H2A variants with a unique macro domains, which can bind NAD+ metabolites [77]. Of the two macroH2A genes (macroH2A1/H2AFY), H2AFY is known to be alternatively spliced, producing two distinct isoforms (macroH2A1.1 and macroH2A1.2) that differ in only a single exon within the macro domain. Interestingly, macroH2A1.2 is incapable of binding NAD+ metabolites suggesting that splicing may modify the activity of macroH2A proteins in response to NAD+ metabolites (Figure 2). Although the function of macroH2A1.2 has not been fully explored, it is possible that alternative splicing of macroH2A1 is a homeostatic mechanism used to fine-tune transcriptional regulation in response to cellular metabolites.
Finally, important metabolic hormones can also affect the AS of genes. A recent study by Malakar et al. has shown that alternative splicing of INSR protects pancreatic β cells from lipotoxicity and stress-induced apoptosis. Insulin-mediated activation of the rat sarcoma (Ras)-Mitogen-Activated Protein Kinase 1 (MAPK) signaling pathway led to upregulation of the splicing factor Serine/Arginine-Rich Splicing Factor 1 (SRSF1), favoring the INSR- β isoform [78]. Collectively, these studies point to a splicing mechanism by which cells may respond to changes in nutrient availability.
3.2. RNA Modifications
In addition to AS, chemical base modification of RNA also significantly influences its metabolism and downstream expression. The 5’ untranslated terminal region (UTR) of nascent mRNAs is immediately capped with a 7-methylguanosine (m7g) residue by the RNA-binding enzyme (guanine-N7-)-methyltransferase [79]. The co-transcriptional, chemical modification of RNA bases within the nucleus gives rise to the RNA epitranscriptome, directly impacting the expression of nearly all cellular RNAs. Over 100 distinct chemical modifications have been identified [80], most of which were observed in tRNAs and other non-coding RNAs [81, 82]. Modified nucleosides have also been observed in mRNAs, including the abundant N6-methyladenosine (m6A) and pseudouridine (Ψ) [83, 84]. M6A and Ψ residues are the most prevalent modifications in the mammalian transcriptome [84, 85], affecting their host RNAs by modulating splicing, stability, and translation [86–90]. Additionally, downstream interactions such as RBP and ncRNA binding may be impacted by enriched modification of 3’ UTR nucleosides [91].
Given the tremendous impact that these RNA marks can have, how does their regulation influence metabolic activity within the cell? A seminal study by Zhao et al. illuminated the role of RNA modification in energy homeostasis [86]. Fat mass and obesity associated protein (FTO) was the first mRNA demethylase to be identified in humans and was incorrectly grouped amongst “obesity genes” correlated with obesity and impaired adipose metabolism [92]. In fact, a non-coding region of the FTO gene originally identified in GWAS studies functions as an enhancer for the Iroquois-class homeodomain proteins 3 and 5 (IRX3 and IRX5) promoters. These genes are closely linked with obesity and body mass composition, leading to the misclassification of FTO. As a result, an interest in the FTO gene sparked a number of studies exploring RNA modification and the epitranscriptome. The FTO demethylase targets m6A residues enriched in exonic regions flanking 5’ and 3’ splice sites. These sites often contain overlapping sequences regulated by the conserved splicing factor Serine/Arginine-Rich Splicing Factor 2 (SRSF2). FTO depletion in pre-adipocytes enhanced the levels of m6A, promoted binding of SRSF2, and increased target exon inclusion within the adipogenic regulatory factor RUNX1 Translocation Partner 1 (RUNX1T1). These results suggest a prominent link between the regulation of RNA modification at the cellular level and its impact on adipogenesis in whole tissues.
These modifications may be further regulated by changes in metabolic state. Cellular stress, and to a lesser extent nutrient starvation, increased the level of Ψ in HEK293T cells, suggesting that the extent of Ψ is responsive to metabolic cues [84]. These results point to an interesting mechanism by which cells may respond quickly and efficiently to outside factors at the post-transcriptional level through RNA modification. Although the role of these modifications in human metabolism is still poorly understood, their ability to induce widespread downstream changes in response to changing metabolic conditions suggests an integral role in metabolic regulation.
4. Modulation of metabolic pathways using translational regulation
The final stage of gene expression, which occurs in the cytoplasm, involves the translation of mRNA into protein. Regulation of translation provides a rapid and efficient way for cells to directly alter rates of protein synthesis in response to changing environments and stimuli. Instead of synthesizing, processing and exporting mRNA, considerable time can be saved by tuning the translation efficiency of existing mRNAs. In principle, this improves fitness by conferring additional plasticity and adaptability to the cell. Translational control is widespread and essential to many biological processes such as embryogenesis, neural plasticity and memory [93–95]. Likewise, it is becoming increasingly evident that regulatory mechanisms of translation are vastly integrated into various aspects of metabolic control. Indeed, major nutrient and energy sensing pathways, such as the mTOR pathway, are tightly coupled to translation regulation [96]. Activation of these pathways by growth factors and excess nutrients stimulates protein synthesis to meet the demands of cell and tissue growth. Alternatively, conditions of stress or nutrient deficiency reduce translation efficiency allowing the cell to reallocate resources or facilitate energy conservation (Figure 3). Although translation encompasses a singular event utilizing mRNA to synthesize protein, the mechanisms involved in modulating the rate of this process are numerous and diverse.
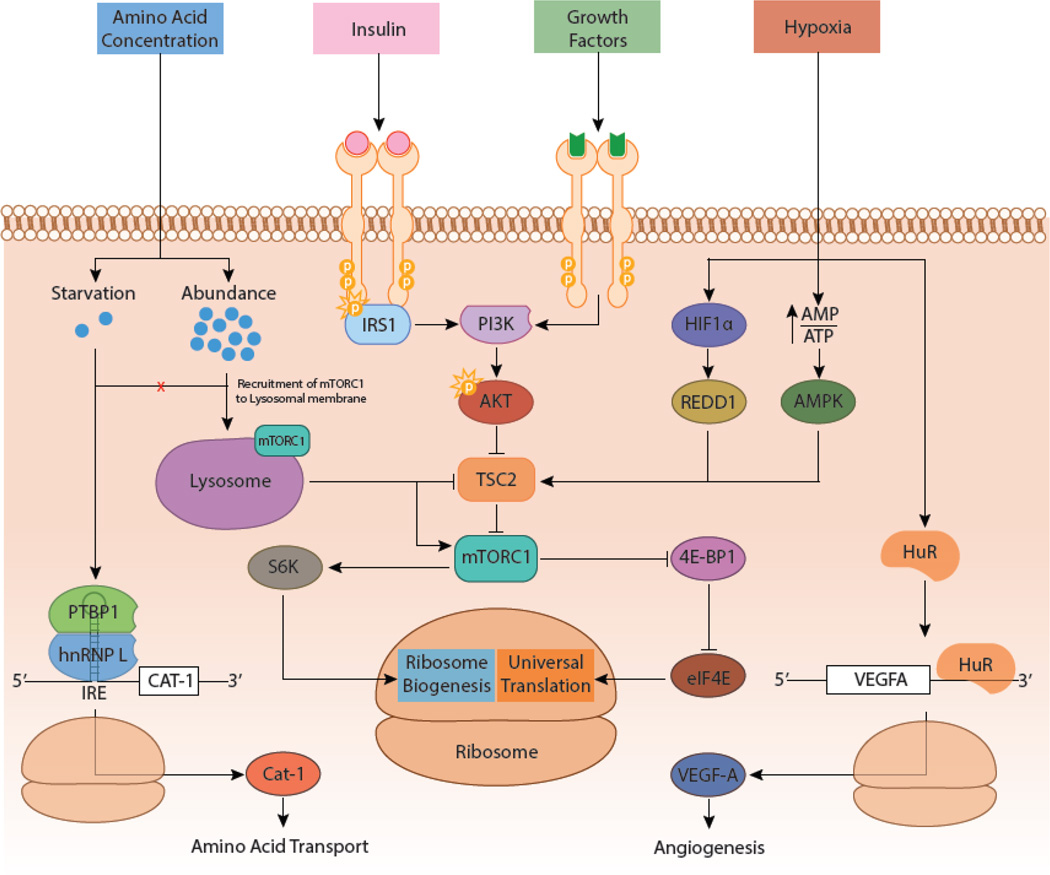
Figure 3. Translational regulation in response to metabolic state.
Changes in metabolic conditions can impact translational regulation. During nutrient abundance, amino acids, insulin, and growth factors activate mammalian target of rapamycin complex 1 (mTORC1), upregulating total translation and ribosome biogenesis. Amino acid starvation is associated with increased polypyrimidine tract binding protein 1 (PTBP1) and heterogeneous nuclear ribonucleoprotein L (hnRNP L) binding to the iron-responsive-element (IRE) in the 5’ un-translated region (UTR) of of cationic amino acid transporter-1 (CAT-1) mRNA. IRE binding increases translation efficiency, CAT-1 expression, and amino acid transport. Hypoxia results in a high AMP:ATP ratio and activates hypoxia-inducible factor 1 (HIF1α), leading to mTORC1 inhibition. Additionally, hypoxia induces human antigen R (HuR) expression, which binds to the 3’-UTR of vascular endothelial growth factor A (VEGFA), increasing translation efficiency. Increased VEGF-A expression increases angiogenesis.
Translation can be divided into four phases: initiation, elongation, termination, and recycling. Of the four phases, elongation requires the highest amount of energy. The process is driven by a set of proteins known as elongation factors which utilize energy stored in guanosine triphosphate to propel the ribosome forward, elongating the growing peptide one amino acid at a time [97]. Therefore, with regards to regulating translation, most mechanisms target initiation, thus allowing the cell to adjust protein synthesis without fruitlessly expending energy. Translation initiation in itself is a multi-step process involving a coordinated assembly of numerous eukaryotic initiation factors (eIFs) onto the mRNA [98]. In theory, interfering or augmenting any factor involved with initiation can potentially cause repression or enhancement of translation, respectively.
It is well known that regulation of translation rates by metabolic signals are mainly imparted by post-translational mechanisms. For instance, mTOR is an important kinase that controls global anabolic activities within the cell arbitrated by the information it receives from multiple nutrient sensing pathways [96]. Activation of mTOR results in the phosphorylation of numerous downstream substrates, including components associated with translation such as ribosomal protein S6 kinase beta-1 (S6K1) and eIF4E binding protein 1 (4E-BP) [99]. Phosphorylation of S6K1 and 4E-BP promotes global translation by enhancing RNA helicase activity of eIF4A and preventing inhibition of 5’ cap-binding factor eIF4E, respectively. In contrast, global translation can be repressed under conditions of stress and nutrient deficiency by the phosphorylation of eIF2α and thus inhibiting the binding of initiator methionine to the 40S ribosome [100]. In general, these post-translational mechanisms alter rates of global protein synthesis as they influence the activity of general ribosomal factors. However, translation of specific transcripts can also be controlled. This involves post-transcriptional mechanisms, which are governed by elements within the 5’ and 3’ UTRs. Eukaryotes have evolved longer UTRs allowing for greater complexity of translation regulation. These UTRs allow specific trans-acting factors to bind cis-regulatory elements within the UTRs and alter rates of protein synthesis by either inhibiting or activating certain aspects of translation. One such mechanism discussed earlier is the repression of translation by miRNA. These miRNA inhibit translation of select transcripts by binding to specific sites within their 3’ UTR. RBPs are the other major type of trans-acting factor that directly interact with mRNA and affects its stability and translation. In addition to canonical RBPs, there is now emerging evidence that many metabolic enzymes can also act as RBPs and illicit translation regulation upon binding to an mRNA (BOX 2). In some cases, a sequence element within the UTR alone can confer regulation of translation through the formation of secondary structures, which may block or promote translation [101].
BOX ITEM 2: Enzymes in intermediary metabolism can bind to mRNAs.
Owing to their direct involvement in energy production and macromolecule biosynthesis, enzymatic expression and activity is highly regulated at multiple levels within the cell. However, studies over the past three decades have revealed that these housekeeping enzymes may also have non-canonical regulatory roles. For instance, certain enzymes have the capacity to bind specific mRNAs, and regulate their expression post-transcriptionally [133][134]. One of the first moonlighting enzymes to be identified, thymidylate synthase (TS), normally serves a critical role in de novo nucleotide biosynthesis by catalyzing the conversion of dUMP to dTMP. However, under conditions of low dUMP concentration, TS can inhibit the translation of its own mRNA, creating a negative feedback loop [135]. In contrast, some RNA-binding enzymes have non-canonical functions distinct from their traditional roles. The iron regulatory protein 1 (IRP1) is identical to cytosolic aconitase (ACO1), an enzyme which interconverts citrate and isocitrate for NADPH production [136]. Under conditions of low intracellular iron, the protein functions as IRP1 and regulates the translation of mRNAs involved in iron homeostasis and utilization by binding to iron response elements (IREs) located within their UTRs [137]. In contrast, under conditions of high intracellular iron, IRP1 is bound to a 4Fe-4S cluster, disrupting its ability to bind IREs [138]. As a result, the protein returns to its canonical function as ACO1. In this way, IRP1 controls iron homeostasis by promoting iron intake and release from iron-storage proteins in response to iron depletion.
The regulatory impact of moonlighting enzymes may extend well beyond local regulatory circuits. The glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH) converts glyceraldehyde-3-phosphate to glyceraldeyhyde-1,3-bisphosphate and utilizes an NAD+ cofactor to generate NADH. Importantly, this housekeeping enzyme also plays a non-canonical role in DNA repair, transcription, and cell death [139]. Moreover, GAPDH functions can also behave as an RBP, with a range of putative targets including mRNAs, tRNAs, and rRNAs [140, 141]. For instance, the RNA-binding ability of GAPDH may contribute to its role in suppressing inflammatory mRNA translation as a component of the gamma interferon inhibitor of translation (GAIT) complex [142]. Many reported moonlighting enzymes, including GAPDH, share a common dinucleotide-binding domain known as the Rossmann fold. Previously thought to bind only confactors, such as FAD, NAD+ and NADP+, these folds also facilitate interactions with RNA [143]. This property provides the enzyme to have bifunctional roles depending on the cellular conditions. For instance, when energy is depleted, increased levels of NAD+ inhibit the RNA-binding function of GAPDH and pushes it towards glycolytic function. A recent study by Chang et al. showed that a metabolic switch from oxidative phosphorylation to aerobic glycolysis disrupted the GAPDH-mediated repression of interferon gamma (IFNγ) mRNA [144]. In conclusion, these findings suggest that many well studied enzymes may in fact have additional functionalities in regulating gene expression which still need to be identified and characterized.
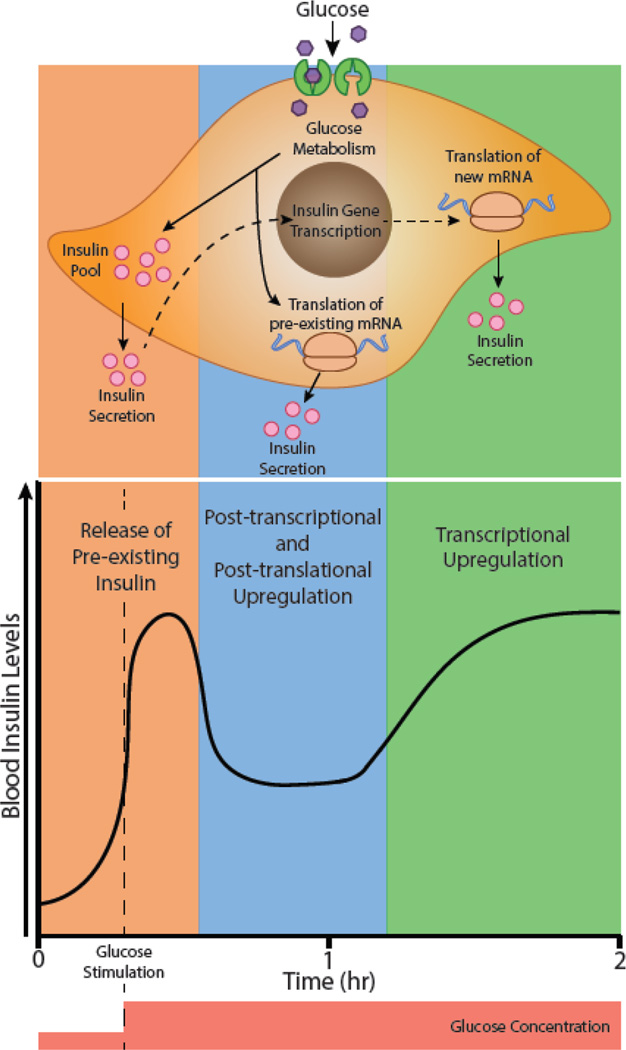
The earliest examples of metabolic signals perturbing rates of translation were from studies examining the mechanism of glucose-stimulated insulin secretion in pancreatic cells. Initial observations showed that treatment of β cells with glucose-induced rapid secretion and synthesis of insulin, taking place within minutes, and preceding synthesis of new mRNA [102, 103]. Although transcription of insulin mRNA eventually follows, early phases of glucose stimulated release of insulin are predominantly governed by increased translation of pre-existing insulin mRNAs [104] (Figure 4). After this observation, the existence of elements was discovered within the 5’ and 3’ UTRs of the insulin mRNA responsible for its increased translation [105, 106]. Glucose stimulation of β cells causes the nucleocytoplasmic translocation of PTBP1 [107]. Within the cytoplasm, PTBP1 increases insulin levels in two ways. First, it binds to sites within the 3’ UTR of insulin mRNA and increases its stability [106]. Second, PTBP1 binds within the 5’ UTR and enhances the translation of insulin mRNA in a non-canonical manner [108]. Typically, translation is cap-dependent which involves the binding of eIF4E to the 5’ cap of mRNA resulting in the subsequent assembly of the ribosome. However, mRNAs containing internal ribosome entry sites (IRESs) can form secondary and tertiary structures that can directly recruit the ribosome independently of the 5’ cap structure. Formation and activity of IRES elements are often aided by additional factors referred to as IRES trans-acting factors (ITAFs) [109]. In the case of insulin mRNA, PTBP1 acts as an ITAF and promotes its cap-independent translation upon stimulation by glucose [108]. This mechanism of translation activation mediated through a trans-factor binding is pervasive and utilized in other metabolic conditions. For instance, a similar mechanism is employed to increase expression of cationic amino acid transporter 1 (CAT-1) during conditions of amino acid starvation (Figure 3). This is achieved through the binding of PTBP1 and heterogeneous nuclear ribonucleoprotein L (hnRNP L) to the 5’ UTR and promotes IRES activity [110]. Since amino acids are essential for cell viability and function, this form of regulation allows the cell to quickly generate transporters for amino acids to increase uptake when availability is low. Regulation of translation by similar mechanisms for other metabolically relevant genes is summarized in Table 1.
Figure 4. Glucose-stimulated insulin response occurs at multiple regulatory levels.
Upon glucose stimulation, β-cells of the pancreas utilize multiple methods of insulin production that operate at different time-scales. In response to increased glucose metabolism, pre-existing insulin molecules within β-cells are released into the blood stream within minutes, causing an initial peak in blood insulin levels shortly after glucose stimulation. Glucose metabolism further induces the stabilization and translation of pre-existing insulin mRNA, allowing β-cells to produce and secrete more insulin [145]. Insulin levels decrease after this initial peak but remain above their resting state due to this increased production at the post-transcriptional and post-translational level. Furthermore, extracellular insulin sensed by β-cells increases the transcriptional activation of the insulin gene [102–104102–104]. After more than an hour of stimulation, this feed-forward signal increases insulin production at the transcriptional level.
Table 1.
List of metabolically relevant genes and their translation regulators.
| mRNA | Stimulus/ Pathway |
Trans-acting factor |
Binding location |
Metabolic Response |
|---|---|---|---|---|
| insulin 1/2 | Elevated glucose |
PTBP1 | 5’ UTR | Increases translation of insulin1/2 upon stimulation of β cells by glucose [108]. |
| 3’ UTR | Increases stability of insulin and cargo genes [106]. | |||
| cat-1 | Amino acid starvation |
hnRNPL PTBP1 |
5’ UTR | Enhances translation of cat-1 under conditions of amino acid starvation allowing for improved uptake [110]. |
| HuR | 3’ UTR | Increases stability of cat-1 mRNA under AA starvation [111]. | ||
| srebp1a | Lipid accumulation |
hnRNPA1 | 5’ UTR | hnRNPA1 behaves as an ITAF and promotes the translation of SREBP-1a. Increased levels of SREBP-1a promote reduction of cellular lipid levels [112]. |
| vegfa | Hypoxia | HuR | 3’ UTR | Enhances translation of VEGFA, a master regulator of angiogenesis, mRNA under hypoxic conditions [113]. |
| abca1 | Cholesterol accumulation |
HuR | 3’ UTR | Increases translation of ABCA1 mRNA and allows for greater efflux of cholesterol and HDL biogenesis [114]. |
| insig1 | Lipid synthesis |
HuD | 3’ UTR | Increases translation of insig1 in pancreatic β cells resulting in the downregulation of triglyceride synthesis [115]. |
|
pten and stat3 |
Insulin signaling |
CPEB1 | 3’ UTR | Represses translation of pten and stat3, which are negative regulators of insulin signaling. Deficiency of CPEB1 in mice induced hepatic insulin resistance [116]. |
5. Conclusions and Outlook
A growing number of studies have now demonstrated that post-transcriptional regulatory mechanisms expand the organism’s capacity to fine-tune gene expression for maintaining energy homeostasis. In particular, these mechanisms seem to serve critical roles in providing appropriate buffering capacity in response to changes such as nutrient availability, exercise, and thermoregulation. They are also programmed to generate rapid responses and sustain physiological requirements before the transcriptional machinery is activated to achieve precise metabolic states. An excellent example of this is the release of insulin in response to glucose stimulation where enhanced mRNA stabilization and translation serve as back up means for sustaining insulin levels after the pre-existing pool of mRNA is depleted but the new transcription of insulin gene has not yet initiated (Figure 4). In this review, we have presented multiple examples that highlight the dynamics and impact of post-transcriptional control of intermediary metabolism, however; many questions still remain to be answered. First, how are different post-transcriptional events integrated with broader signaling and transcriptional networks to produce coherent metabolic outcomes? Second, just as master TFs, are there also master RBPs or ncRNAs that work in a concerted manner to activate particular metabolic pathways? Third, how are overlapping and sometimes competing posttranscriptional mechanisms consolidated to achieve the correct mRNA levels and kinetics under diverse metabolic contexts? Finally, do metabolic intermediates influence the activity/expression of RBPs to support feed-forward or feedback loops and control metabolic flux within certain pathways?
Recently, Gerstberger et al. have determined that approximately 7.5% of the ~20,500 human protein-coding genes encode for RBPs that directly bind to and/or process RNA [117]. Nearly 50% of these RBPs are involved in mRNA metabolism and intriguingly; they are amongst the most abundant RBPs in the cell. The Encyclopedia of DNA Elements (ENCODE) project is employing next-generation sequencing methods to generate detailed information about the extent and impact of individual RBPs on gene expression in model organisms and human cells [118]. Further intersection of these genomics and large-scale metabolomics datasets must probe which RBPs have direct roles in regulating energy homeostasis and beyond. Target-based classification of RBPs along with GWAS should likewise guide new investigations to determine exact relationships between certain human mutations and metabolic diseases. With the advent of CRISPR-based genome editing technologies, we are not only poised to interrogate the metabolic effects of these mutations in cell and animal models[119] but also possess the toolkit to correct them in human patients [120].
Highlights.
Metabolic regulatory mechanisms operate at overlapping yet distinct timescales
Post-transcriptional events fine-tune gene expression and energy homeostasis
Metabolic flux is buffered by adaptive, post-transcriptional responses
Aberrant post-transcriptional regulation contributes to metabolic disease
Acknowledgments
A.K. is supported by grants from the US National Institute of Health (R01HL126845), March of Dimes (5-FY14-112), and the Center for Advanced Study at the University of Illinois. W.A. is supported by the US National Institutes of Health pre-doctoral NRSA fellowship (F30DK108567). We thank people in the Kalsotra lab for helpful discussions and comments on the manuscript. We sincerely apologize to colleagues whose work was not discussed in this review because of space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest:
The authors declare no conflict of interest.
References
- 1.Karve TM, Cheema AK. Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. Journal of amino acids. 2011;2011:207691. doi: 10.4061/2011/207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiological reviews. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 3.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Filhoulaud G, Guilmeau S, Dentin R, Girard J, Postic C. Novel insights into ChREBP regulation and function. Trends in endocrinology and metabolism: TEM. 2013;24:257–268. doi: 10.1016/j.tem.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Hocine S, Singer RH, Grunwald D. RNA processing and export. Cold Spring Harb Perspect Biol. 2010;2:a000752. doi: 10.1101/cshperspect.a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. International Human Genome Sequencing. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 7.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 9.Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews. Molecular cell biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 10.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nature reviews. Genetics. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 11.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 13.Xu PZ, Vernooy SY, Guo M, Hay BA. The Drosophila MicroRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 14.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. The Journal of biological chemistry. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramirez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison JA, Suarez Y, de Cabo R, Gorospe M, Fernandez-Hernando C. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol. 2013;33:2891–2902. doi: 10.1128/MCB.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, Horton JD, Lasuncion MA, Naar AM, Suarez Y, Fernandez-Hernando C. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nature medicine. 2015;21:1280–1289. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagschal A, Najafi-Shoushtari SH, Wang L, Goedeke L, Sinha S, deLemos AS, Black JC, Ramirez CM, Li Y, Tewhey R, Hatoum I, Shah N, Lu Y, Kristo F, Psychogios N, Vrbanac V, Lu YC, Hla T, de Cabo R, Tsang JS, Schadt E, Sabeti PC, Kathiresan S, Cohen DE, Whetstine J, Chung RT, Fernandez-Hernando C, Kaplan LM, Bernards A, Gerszten RE, Naar AM. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nature medicine. 2015;21:1290–1297. doi: 10.1038/nm.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Global Lipids Genetics. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O'Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Doring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR. Discovery and refinement of loci associated with lipid levels. Nature genetics. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 24.Kornfeld JW, Baitzel C, Konner AC, Nicholls HT, Vogt MC, Herrmanns K, Scheja L, Haumaitre C, Wolf AM, Knippschild U, Seibler J, Cereghini S, Heeren J, Stoffel M, Bruning JC. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494:111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 25.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, D. Consortium, M. Investigators. Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granjon A, Gustin MP, Rieusset J, Lefai E, Meugnier E, Guller I, Cerutti C, Paultre C, Disse E, Rabasa-Lhoret R, Laville M, Vidal H, Rome S. The microRNA Signature in Response to Insulin Reveals Its Implication in the Transcriptional Action of Insulin in Human Skeletal Muscle and the Role of a Sterol Regulatory Element-Binding Protein-1c/Myocyte Enhancer Factor 2C Pathway. Diabetes. 2009;58:2555–2564. doi: 10.2337/db09-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu X, Dong B, Tian Y, Lefebvre P, Meng Z, Wang X, Pattou F, Han W, Wang X, Lou F, Jove R, Staels B, Moore DD, Huang W. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. The Journal of clinical investigation. 2015;125:2497–2509. doi: 10.1172/JCI75438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Bottger T, Braun T, Seibler J, Bruning JC. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nature cell biology. 2011;13:434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 30.Lee DE, Brown JL, Rosa ME, Brown LA, Perry RA, Wiggs MP, Nilsson MI, Crouse SF, Fluckey JD, Washington TA, Greene NP. microRNA-16 Is Downregulated During Insulin Resistance and Controls Skeletal Muscle Protein Accretion. J Cell Biochem. 2016;117:1775–1787. doi: 10.1002/jcb.25476. [DOI] [PubMed] [Google Scholar]
- 31.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 32.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 33.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma XS, MacDonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 34.Schneeberger M, Altirriba J, Garcia A, Esteban Y, Castano C, Garcia-Lavandeira M, Alvarez CV, Gomis R, Claret M. Deletion of miRNA processing enzyme Dicer in POMC-expressing cells leads to pituitary dysfunction, neurodegeneration and development of obesity. Mol Metab. 2013;2:74–85. doi: 10.1016/j.molmet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinnikov IA, Hajdukiewicz K, Reymann J, Beneke J, Czajkowski R, Roth LC, Novak M, Roller A, Dorner N, Starkuviene V, Theis FJ, Erfle H, Schutz GN, Grinevich V, Konopka W. Hypothalamic miR-103 Protects from Hyperphagic Obesity in Mice. J Neurosci. 2014;34:10659–10674. doi: 10.1523/JNEUROSCI.4251-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crepin D, Benomar Y, Riffault L, Amine H, Gertler A, Taouis M. The over-expression of miR-200a in the hypothalamus of ob/ob mice is linked to leptin and insulin signaling impairment. Mol Cell Endocrinol. 2014;384:1–11. doi: 10.1016/j.mce.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 37.Benoit C, Ould-Hamouda H, Crepin D, Gertler A, Amar L, Taouis M. Early leptin blockade predisposes fat-fed rats to overweight and modifies hypothalamic microRNAs. J Endocrinol. 2013;218:35–47. doi: 10.1530/JOE-12-0561. [DOI] [PubMed] [Google Scholar]
- 38.Sangiao-Alvarellos S, Pena-Bello L, Manfredi-Lozano M, Tena-Sempere M, Cordido F. Perturbation of Hypothalamic MicroRNA Expression Patterns in Male Rats After Metabolic Distress: Impact of Obesity and Conditions of Negative Energy Balance. Endocrinology. 2014;155:1838–1850. doi: 10.1210/en.2013-1770. [DOI] [PubMed] [Google Scholar]
- 39.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan XA, Ruan YJ, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 41.Hong C, Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov. 2014;13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 42.Sallam T, Jones MC, Gilliland T, Zhang L, Wu XH, Eskin A, Sandhu J, Casero D, Vallim TQD, Hong C, Katz M, Lee R, Whitelegge J, Tontonoz P. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534:124-+. doi: 10.1038/nature17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lan X, Yan JD, Ren J, Zhong B, Li J, Li Y, Liu L, Yi J, Sun QZ, Yang XD, Sun J, Meng LS, Zhu WH, Holmdahl R, Li DM, Lu SM. A Novel Long Noncoding RNA Lnc-HC Binds hnRNPA2B1 to Regulate Expressions of Cyp7a1 and Abca1 in Hepatocytic Cholesterol Metabolism. Hepatology. 2016;64:58–72. doi: 10.1002/hep.28391. [DOI] [PubMed] [Google Scholar]
- 44.Li P, Ruan XB, Yang L, Kiesewetter K, Zhao Y, Luo HT, Chen Y, Gucek M, Zhu J, Cao HM. A Liver-Enriched Long Non-Coding RNA, lncLSTR, Regulates Systemic Lipid Metabolism in Mice. Cell metabolism. 2015;21:455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu S, Chen P, Sun L. Regulatory networks of non-coding RNAs in brown/beige adipogenesis. Bioscience reports. 2015;35 doi: 10.1042/BSR20150155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 49.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu LG, Liu CC, Yi JS, Zhang HF, Min W, Bennett AM, Gregory RI, Ding Y, Huang YQ. The Imprinted H19 LncRNA Antagonizes Let-7 MicroRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Y, Wu FJ, Zhou JC, Yan L, Jurczak MJ, Lee HY, Yang LH, Mueller M, Zhou XB, Dandolo L, Szendroedi J, Roden M, Flannery C, Taylor H, Carmichael GG, Shulman GI, Huang YQ. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic acids research. 2014;42:13799–13811. doi: 10.1093/nar/gku1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan X, Li P, Cangelosi A, Yang L, Cao H. A Long Non-coding RNA, lncLGR, Regulates Hepatic Glucokinase Expression and Glycogen Storage during Fasting. Cell reports. 2016;14:1867–1875. doi: 10.1016/j.celrep.2016.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rando OJ. Intergenerational Transfer of Epigenetic Information in Sperm. Cold Spring Harbor perspectives in medicine. 2016;6 doi: 10.1101/cshperspect.a022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu L, Lu Y, Jiao Y, Liu B, Li S, Li Y, Xing F, Chen D, Liu X, Zhao J, Xiong X, Gu Y, Lu J, Chen X, Li X. Paternal Psychological Stress Reprograms Hepatic Gluconeogenesis in Offspring. Cell metabolism. 2016;23:735–743. doi: 10.1016/j.cmet.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 55.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annual review of biochemistry. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray RR, Spirohn K, Begg BE, Duran-Frigola M, MacWilliams A, Pevzner SJ, Zhong Q, Trigg SA, Tam S, Ghamsari L, Sahni N, Yi S, Rodriguez MD, Balcha D, Tan G, Costanzo M, Andrews B, Boone C, Zhou XJ, Salehi-Ashtiani K, Charloteaux B, Chen AA, Calderwood MA, Aloy P, Roth FP, Hill DE, Iakoucheva LM, Xia Y, Vidal M. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell. 2016;164:805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nature reviews. Genetics. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nature reviews. Genetics. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong G, Mao Q, Xia W, Xu Y, Wang J, Xu L, Jiang F. PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncology letters. 2016;11:1980–1986. doi: 10.3892/ol.2016.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Z, Chatterjee D, Jeon HY, Akerman M, Vander Heiden MG, Cantley LC, Krainer AR. Exon-centric regulation of pyruvate kinase M alternative splicing via mutually exclusive exons. Journal of molecular cell biology. 2012;4:79–87. doi: 10.1093/jmcb/mjr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Critical reviews in oncogenesis. 1992;3:91–115. [PubMed] [Google Scholar]
- 63.Gupta V, Wellen KE, Mazurek S, Bamezai RN. Pyruvate kinase M2: regulatory circuits and potential for therapeutic intervention. Current pharmaceutical design. 2014;20:2595–2606. doi: 10.2174/13816128113199990484. [DOI] [PubMed] [Google Scholar]
- 64.Salati LM, Amir-Ahmady B. Dietary regulation of expression of glucose-6-phosphate dehydrogenase. Annual review of nutrition. 2001;21:121–140. doi: 10.1146/annurev.nutr.21.1.121. [DOI] [PubMed] [Google Scholar]
- 65.Cyphert TJ, Suchanek AL, Griffith BN, Salati LM. Starvation actively inhibits splicing of glucose-6-phosphate dehydrogenase mRNA via a bifunctional ESE/ESS element bound by hnRNP K. Biochimica et biophysica acta. 2013;1829:905–915. doi: 10.1016/j.bbagrm.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walsh CM, Suchanek AL, Cyphert TJ, Kohan AB, Szeszel-Fedorowicz W, Salati LM. Serine arginine splicing factor 3 is involved in enhanced splicing of glucose-6-phosphate dehydrogenase RNA in response to nutrients and hormones in liver. The Journal of biological chemistry. 2013;288:2816–2828. doi: 10.1074/jbc.M112.410803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu CY, Theusch E, Lo K, Mangravite LM, Naidoo D, Kutilova M, Medina MW. HNRNPA1 regulates HMGCR alternative splicing and modulates cellular cholesterol metabolism. Human molecular genetics. 2014;23:319–332. doi: 10.1093/hmg/ddt422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Medina MW, Gao F, Naidoo D, Rudel LL, Temel RE, McDaniel AL, Marshall SM, Krauss RM. Coordinately regulated alternative splicing of genes involved in cholesterol biosynthesis and uptake. PloS one. 2011;6:e19420. doi: 10.1371/journal.pone.0019420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sen S, Talukdar I, Webster NJ. SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Mol Cell Biol. 2009;29:871–880. doi: 10.1128/MCB.01709-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruas JL, White JP, Rao RR, Kleiner S, Brannan KT, Harrison BC, Greene NP, Wu J, Estall JL, Irving BA, Lanza IR, Rasbach KA, Okutsu M, Nair KS, Yan Z, Leinwand LA, Spiegelman BM. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151:1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gotic I, Omidi S, Fleury-Olela F, Molina N, Naef F, Schibler U. Temperature regulates splicing efficiency of the cold-inducible RNA-binding protein gene Cirbp. Genes & development. 2016 doi: 10.1101/gad.287094.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaminska D, Kakela P, Nikkola E, Venesmaa S, Ilves I, Herzig KH, Kolehmainen M, Karhunen L, Kuusisto J, Gylling H, Pajukanta P, Laakso M, Pihlajamaki J. Regulation of alternative splicing in human obesity loci. Obesity. 2016 doi: 10.1002/oby.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pihlajamaki J, Lerin C, Itkonen P, Boes T, Floss T, Schroeder J, Dearie F, Crunkhorn S, Burak F, Jimenez-Chillaron JC, Kuulasmaa T, Miettinen P, Park PJ, Nasser I, Zhao Z, Zhang Z, Xu Y, Wurst W, Ren H, Morris AJ, Stamm S, Goldfine AB, Laakso M, Patti ME. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell metabolism. 2011;14:208–218. doi: 10.1016/j.cmet.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li X, Qian X, Peng LX, Jiang Y, Hawke DH, Zheng Y, Xia Y, Lee JH, Cote G, Wang H, Wang L, Qian CN, Lu Z. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nature cell biology. 2016;18:561–571. doi: 10.1038/ncb3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends in endocrinology and metabolism: TEM. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell metabolism. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Till S, Ladurner AG. Sensing NAD metabolites through macro domains. Frontiers in bioscience. 2009;14:3246–3258. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- 78.Malakar P, Chartarifsky L, Hija A, Leibowitz G, Glaser B, Dor Y, Karni R. Insulin receptor alternative splicing is regulated by insulin signaling and modulates beta cell survival. Scientific reports. 2016;6:31222. doi: 10.1038/srep31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 80.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA Modification Database, RNAMDB: 2011 update. Nucleic acids research. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annual review of genetics. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 82.Karijolich J, Kantartzis A, Yu YT. RNA modifications: a mechanism that modulates gene expression. Methods in molecular biology. 2010;629:1–19. doi: 10.1007/978-1-60761-657-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perry RP, Kelley DE. Existence of Methylated Messenger-Rna in Mouse L Cells. Cell. 1974;1:37–42. [Google Scholar]
- 84.Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nature chemical biology. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 85.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 86.Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, Jin KX, Wang X, Huang CM, Fu Y, Ge XM, Song SH, Jeong HS, Yanagisawa H, Niu Y, Jia GF, Wu W, Tong WM, Okamoto A, He C, Rendtlew Danielsen JM, Wang XJ, Yang YG. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell research. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, Vagbo CB, Kussnierczyk A, Klungland A, Darnell JE, Jr, Darnell RB. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes & development. 2015;29:2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nature reviews. Endocrinology. 2014;10:51–61. doi: 10.1038/nrendo.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuersten S, Goodwin EB. The power of the 3 ' UTR: Translational control and development. Nature Reviews Genetics. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 94.Lasko P. mRNA Localization and Translational Control in Drosophila Oogenesis. Csh Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational Control of Long-Lasting Synaptic Plasticity and Memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Bio. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 97.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 98.Sonenberg N, Hinnebusch AG. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baird TD, Wek RC. Eukaryotic Initiation Factor 2 Phosphorylation and Translational Control in Metabolism. Adv Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Adeli K. Translational control mechanisms in metabolic regulation: critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. Am J Physiol-Endoc M. 2011;301:E1051–E1064. doi: 10.1152/ajpendo.00399.2011. [DOI] [PubMed] [Google Scholar]
- 102.Itoh N, Sei T, Nose K, Okamoto H. Glucose stimulation of the proinsulin synthesis in isolated pancreatic islets without increasing amount of proinsulin mRNA. FEBS letters. 1978;93:343–347. doi: 10.1016/0014-5793(78)81136-3. [DOI] [PubMed] [Google Scholar]
- 103.Welsh M, Scherberg N, Gilmore R, Steiner DF. Translational control of insulin biosynthesis. Evidence for regulation of elongation, initiation and signal-recognition-particle-mediated translational arrest by glucose. The Biochemical journal. 1986;235:459–467. doi: 10.1042/bj2350459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Itoh N, Okamoto H. Translational control of proinsulin synthesis by glucose. Nature. 1980;283:100–102. doi: 10.1038/283100a0. [DOI] [PubMed] [Google Scholar]
- 105.Muralidharan B, Bakthavachalu B, Pathak A, Seshadri V. A minimal element in 5'UTR of insulin mRNA mediates its translational regulation by glucose. FEBS letters. 2007;581:4103–4108. doi: 10.1016/j.febslet.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 106.Tillmar L, Carlsson C, Welsh N. Control of insulin mRNA stability in rat pancreatic islets - Regulatory role of a 3 '-untranslated region pyrimidine-rich sequence. Journal of Biological Chemistry. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- 107.Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkruger A, Verkade P, Solimena M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nature cell biology. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- 108.Knoch KP, Nath-Sain S, Petzold A, Schneider H, Beck M, Wegbrod C, Sonmez A, Munster C, Friedrich A, Roivainen M, Solimena M. PTBP1 is required for glucose-stimulated cap-independent translation of insulin granule proteins and Coxsackieviruses in beta cells. Mol Metab. 2014;3:518–530. doi: 10.1016/j.molmet.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hellen CUT, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & development. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 110.Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang CP, Caprara MG, Venema RC, Komar AA, Snider MD, Hatzoglou M. The hnRNA-Binding Proteins hnRNP L and PTB Are Required for Efficient Translation of the Cat-1 Arginine/Lysine Transporter mRNA during Amino Acid Starvation. Mol Cell Biol. 2009;29:2899–2912. doi: 10.1128/MCB.01774-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yaman I, Fernandez J, Sarkar B, Schneider RJ, Snider MD, Nagy LE, Hatzoglou M. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. Journal of Biological Chemistry. 2002;277:41539–41546. doi: 10.1074/jbc.M204850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Siculella L, Tocci R, Rochira A, Testini M, Gnoni A, Damiano F. Lipid accumulation stimulates the cap-independent translation of SREBP-1a mRNA by promoting hnRNP A1 binding to its 5 '-UTR in a cellular model of hepatic steatosis. Bba-Mol Cell Biol L. 2016;1861:471–481. doi: 10.1016/j.bbalip.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 113.Osera C, Martindale JL, Amadio M, Kim J, Yang XL, Moad CA, Indig FE, Govoni S, Abdelmohsen K, Gorospe M, Pascale A. Induction of VEGFA mRNA translation by CoCl2 mediated by HuR. RNA biology. 2015;12:1121–1130. doi: 10.1080/15476286.2015.1085276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ramirez CM, Lin CS, Abdelmohsen K, Goedeke L, Yoon JH, Madrigal-Matute J, Martin-Ventura JL, Vo DT, Uren PJ, Penalva LO, Gorospe M, Fernandez-Hernando C. RNA binding protein HuR regulates the expression of ABCA1. J Lipid Res. 2014;55:1066–1076. doi: 10.1194/jlr.M044925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim C, Lee H, Kang H, Shin JJ, Tak H, Kim W, Gorospe M, Lee EK. RNA-binding protein HuD reduces triglyceride production in pancreatic beta cells by enhancing the expression of insulin-induced gene 1. Bba-Gene Regul Mech. 2016;1859:675–685. doi: 10.1016/j.bbagrm.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alexandrov IM, Ivshina M, Jung DY, Friedline R, Ko HJ, Xu M, O'Sullivan-Murphy B, Bortell R, Huang YT, Urano F, Kim JK, Richter JD. Cytoplasmic Polyadenylation Element Binding Protein Deficiency Stimulates PTEN and Stat3 mRNA Translation and Induces Hepatic Insulin Resistance. Plos Genet. 2012;8 doi: 10.1371/journal.pgen.1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nature reviews. Genetics. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sundararaman B, Zhan L, Blue SM, Stanton R, Elkins K, Olson S, Wei X, Van Nostrand EL, Pratt GA, Huelga SC, Smalec BM, Wang X, Hong EL, Davidson JM, Lecuyer E, Graveley BR, Yeo GW. Resources for the Comprehensive Discovery of Functional RNA Elements. Mol Cell. 2016;61:903–913. doi: 10.1016/j.molcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, Karumbayaram S, Kumagai-Cresse C, Wang D, Zack JA, Kohn DB, Nakano A, Nelson SF, Miceli MC, Spencer MJ, Pyle AD. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell stem cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, Carninci P, Hayashizaki Y. Hidden layers of human small RNAs. Bmc Genomics. 2008;9 doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pederson T. Regulatory RNAs derived from transfer RNA? Rna. 2010;16:1865–1869. doi: 10.1261/rna.2266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes & development. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rando OJ, Simmons RA. I'm Eating for Two: Parental Dietary Effects on Offspring Metabolism. Cell. 2015;161:93–105. doi: 10.1016/j.cell.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun FY, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen Q, Yan MH, Cao ZH, Li X, Zhang YF, Shi JC, Feng GH, Peng HY, Zhang XD, Zhang Y, Qian JJ, Duan EK, Zhai QW, Zhou Q. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 127.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li RW, Bock C, Li CJ, Gu HC, Zamore PD, Meissner A, Weng ZP, Hofmann HA, Friedman N, Rando OJ. Paternally Induced Transgenerational Environmental Reprogramming of Metabolic Gene Expression in Mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wei YC, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic acids research. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, Guan BJ, Yuan YY, Jankowsky E, Feng ZY, Hu GF, Pusztai-Carey M, Gorla M, Sepuri NBV, Pan T, Hatzoglou M. Angiogenin-Cleaved tRNA Halves Interact with Cytochrome c, Protecting Cells from Apoptosis during Osmotic Stress. Mol Cell Biol. 2014;34:2450–2463. doi: 10.1128/MCB.00136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li Q, Hu B, Hu GW, Chen CY, Niu X, Liu J, Zhou SM, Zhang CQ, Wang Y, Deng ZF. tRNA-Derived Small Non-Coding RNAs in Response to Ischemia Inhibit Angiogenesis. Scientific reports. 2016;6 doi: 10.1038/srep20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. Bmc Biol. 2014;12 doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Castello A, Hentze MW, Preiss T. Metabolic Enzymes Enjoying New Partnerships as RNA-Binding Proteins. Trends in endocrinology and metabolism: TEM. 2015;26:746–757. doi: 10.1016/j.tem.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liao Y, Castello A, Fischer B, Leicht S, Foehr S, Frese CK, Ragan C, Kurscheid S, Pagler E, Yang H, Krijgsveld J, Hentze MW, Preiss T. The Cardiomyocyte RNA-Binding Proteome: Links to Intermediary Metabolism and Heart Disease. Cell reports. 2016;16:1456–1469. doi: 10.1016/j.celrep.2016.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chu E, Koeller DM, Casey JL, Drake JC, Chabner BA, Elwood PC, Zinn S, Allegra CJ. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:8977–8981. doi: 10.1073/pnas.88.20.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rouault TA, Stout CD, Kaptain S, Harford JB, Klausner RD. Structural relationship between an iron-regulated RNA-binding protein (IRE-BP) and aconitase: functional implications. Cell. 1991;64:881–883. doi: 10.1016/0092-8674(91)90312-m. [DOI] [PubMed] [Google Scholar]
- 137.Casey JL, Hentze MW, Koeller DM, Caughman SW, Rouault TA, Klausner RD, Harford JB. Iron-responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science. 1988;240:924–928. doi: 10.1126/science.2452485. [DOI] [PubMed] [Google Scholar]
- 138.Constable A, Quick S, Gray NK, Hentze MW. Modulation of the RNA-binding activity of a regulatory protein by iron in vitro: switching between enzymatic and genetic function? Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4554–4558. doi: 10.1073/pnas.89.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cellular signalling. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ryazanov AG. Glyceraldehyde-3-phosphate dehydrogenase is one of the three major RNA-binding proteins of rabbit reticulocytes. FEBS letters. 1985;192:131–134. doi: 10.1016/0014-5793(85)80058-2. [DOI] [PubMed] [Google Scholar]
- 141.Singh R, Green MR. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- 142.Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL. The GAIT system: a gatekeeper of inflammatory gene expression. Trends in biochemical sciences. 2009;34:324–331. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nagy E, Rigby WF. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AUrich RNA in the NAD(+)-binding region (Rossmann fold) The Journal of biological chemistry. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 144.Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O'Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, Weber JD, Pearce EJ, Jones RG, Pearce EL. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fred RG, Welsh N. The importance of RNA binding proteins in preproinsulin mRNA stability. Mol Cell Endocrinol. 2009;297:28–33. doi: 10.1016/j.mce.2008.06.007. [DOI] [PubMed] [Google Scholar]