Abstract
Purpose of Review
Plasma lipids, namely cholesterol and triglyceride, and lipoproteins, such as low-density lipoprotein (LDL) and high-density lipoprotein, serve numerous physiological roles. Perturbed levels of these traits underlie monogenic dyslipidemias, a diverse group of multisystem disorders. We are on the verge of having a relatively complete picture of the human dyslipidemias and their components.
Recent Findings
Recent advances in genetics of plasma lipids and lipoproteins include the following: (1) expanding the range of genes causing monogenic dyslipidemias, particularly elevated LDL cholesterol; (2) appreciating the role of polygenic effects in such traits as familial hypercholesterolemia and combined hyperlipidemia; (3) accumulating a list of common variants that determine plasma lipids and lipoproteins; (4) applying exome sequencing to identify collections of rare variants determining plasma lipids and lipoproteins that via Mendelian randomization have also implicated gene products such as NPC1L1, APOC3, LDLR, APOA5, and ANGPTL4 as causal for atherosclerotic cardiovascular disease; and (5) using naturally occurring genetic variation to identify new drug targets, including inhibitors of apolipoprotein (apo) C-III, apo(a), ANGPTL3, and ANGPTL4.
Summary
Here, we compile this disparate range of data linking human genetic variation to plasma lipids and lipoproteins, providing a “one stop shop” for the interested reader.
Keywords: Dyslipidemia, DNA variants, Monogenic, Polygenic, Atherosclerosis
Introduction
Plasma lipids, namely cholesterol and triglyceride (TG), are carried within complex lipoprotein particles, such as low-density lipoprotein (LDL) and high-density lipoprotein (HDL) [1]. Lipids and lipoproteins serve numerous physiological roles. Although the terms “lipids” and “lipoproteins” are often used interchangeably, especially by clinicians for convenience, they are different biochemical entities [1]. Plasma lipid levels represent the integrated lipid component of various lipoprotein species. For instance, plasma TG is the sum of TG carried within chylomicrons and very-low-density lipoprotein (VLDL) particles and their metabolic remnants, while plasma total cholesterol is the sum of cholesterol carried within these particles and also within LDL and HDL. In contrast, lipoproteins are discrete molecular entities that are likewise subject to manifold genetic and environmental influences and show complex metabolic interrelationships with one another [1]. Thus, observations that genetic determinants of lipids and lipoproteins often overlap between phenotypes were predictable a priori based on understanding the metabolism of these complex traits. Here, we attempt to respect the biochemical distinction between “lipid” and “lipoprotein.”
Recent technological advances have helped identify numerous genetic variants, ranging from ultra-rare to common, which have significant effects, ranging from large to small, on inter-individual differences in plasma lipid and lipoprotein levels. Over the past 3 years, the range of causative genes and mutations underlying rare familial dyslipidemia syndromes has expanded, while new insights have emerged from genotyping and next-generation sequencing (NGS) studies in unrelated individuals. Here we summarize recent findings of (1) rare genetic variants underlying monogenic dyslipidemias in clinically ascertained patients; (2) common variants contributing to a polygenic component of clinical dyslipidemias; (3) common and rare variants contributing to variations of plasma lipids and lipoproteins in epidemiologic samples; (4) common and rare variants from (3) that have been implicated as causative for atherosclerosis; and (5) targets for drug development to treat dyslipidemia and possibly to prevent atherosclerosis. We also summarize various lines of evidence supporting the biological and clinical significance of genetic variants and loci underlying perturbed lipid and lipoprotein metabolism.
Expanding the Genetic Etiologies in Monogenic Dyslipidemias: Focus on Familial Hypercholesterolemia
Monogenic dyslipidemias are classified according to the primary lipid or lipoprotein disturbance: elevated or depressed concentrations of LDL cholesterol (LDL-C) or HDL cholesterol (HDL-C), or elevated TG [1]. Currently, 27 monogenic dyslipidemias are defined by extreme deviations of plasma lipid or lipoprotein values typically with discrete clinical signs and symptoms caused by numerous rare mutations affecting a total of 25 genes (Table 1) [2]. Over the past 3 years, no new monogenic dyslipidemias have been added to this list, although some new genes for known dyslipidemias have been identified. For instance, recent high throughput NGS of 213 selected family members from 41 kindreds with suspected Mendelian inheritance of extreme levels of LDL-C and no previously detected mutation, only revealed mutations in known causative genes [3]. However, other NGS efforts have identified a few rare large-effect variants in new genes underlying some of these disorders, particularly familial hypercholesterolemia (FH) [4].
Table 1.
Monogenic dyslipidemias and dyslipoproteinemias
| Phenotype | Disorder | Alternative name | Gene symbol | Chr |
|---|---|---|---|---|
| High LDL-C | Familial hypercholesterolemia | Hyperlipoproteinemia type 2A | LDLR | 19p13.3 |
| Familial defective apolipoprotein B | Autosomal dominant hypercholesterolemia type 2 (binding-defective apo B) | APOB | 2p24-p23 | |
| Autosomal dominant hypercholesterolemia | Autosomal dominant hypercholesterolemia type 3 (PCSK9 gain-of-function) | PCSK9 | 1p32.3 | |
| Autosomal dominant hypercholesterolemia | Autosomal dominant hypercholesterolemia type 4 | STAP1 | 4q13.2 | |
| Autosomal dominant hypercholesterolemia | Autosomal dominant hypercholesterolemia type 5 | APOE | 19q13 | |
| Autosomal recessive hypercholesterolemia | LDLRAP1 (ARH) | 1p36-p35 | ||
| Cholesterol ester storage disease | Includes Wolman disease | LIPA | 10q21.31 | |
| Sitosterolemia | Phytosterolemia | ABCG5/ABCG8 | 2p21 | |
| Low LDL-C | Abetalipoproteinemia | Bassen-Kornzweig syndrome | MTTP | 4q24 |
| Hypobetalipoproteinemia | APOB | 2p24-p23 | ||
| PCSK9 deficiency with low LDL-C | Hypobetalipoproteinemia (PCSK9 loss-of-function) | PCSK9 | 1p32.3 | |
| Familial combined hypolipidemia | ANGPTL3 deficiency | ANGPTL3 | 1p31.1-p22.3 | |
| Chylomicron retention disease | Anderson disease | SAR1B | 5p31.1 | |
| High HDL-C | Cholesteryl ester transfer protein deficiency | Hyperalphalipoproteinemia | CETP | 16q21 |
| Hepatic lipase deficiency | LIPC | 15q21-q23 | ||
| Scavenger receptor B1 deficiency | SCARB1 | 12q23.31 | ||
| Endothelial lipase deficiency | LIPG | 18q21.1 | ||
| Low HDL-C | Tangier disease | ABCA1 | 9q31 | |
| Apolipoprotein A-I deficiency | APOA1 | 11q23 | ||
| Familial LCAT deficiency (complete or partial) | Includes Fish-eye disease | LCAT | 16q22 | |
| High TG | Lipoprotein lipase deficiency | Familial chylomicronemia | LPL | 8p22 |
| Apolipoprotein C-II deficiency | Familial chylomicronemia | APOC2 | 19q13 | |
| Apolipoprotein A-V deficiency | Severe hypertriglyceridemia | APOA5 | 11q23 | |
| Lipase maturation factor deficiency | Severe hypertriglyceridemia | LMF1 | 16p13.3 | |
| Glycosylphosphatidylinositol anchored HDL binding protein 1 | Severe hypertriglyceridemia | GPIHBP1 | 8q23 | |
| Glycerol-3-phosphate dehydrogenase-1 | Infantile hypertriglyceridemia | GPD1 | 12q13.12 | |
| Dysbetalipoproteinemia | Hyperlipoproteinemia type 3 | APOE | 19q13 |
Chr chromosome, LDL-C low-density lipoprotein cholesterol, HDL-C high-density lipoprotein cholesterol, TG triglyceride
Previous estimates that heterozygous FH (HeFH) occurs in 1 in 500 individuals underestimated the actual prevalence of 1 in ~217 individuals as determined from carrier status of pathogenic variants in the LDL receptor gene (LDLR) [5•]. However, FH mutation carriers show a relatively wide range of LDL-C levels; while a cut-point of 4.4 mmol/L (169.9 mg/dL) was proposed to discriminate between carriers and non-carriers, there was considerable overlap of carrier status at lower levels of LDL-C [5•]. A specific association with mutation status was superior at higher levels, i.e., LDL-C >7 mmol/L (270.3 mg/dL) [5•]. Others have suggested a diagnostic cut-point for HeFH of total cholesterol >8.6 mmol/L (332.1 mg/dL) [6]. Higher numbers of mutation carriers are captured at a lower LDL-C cut-point—i.e., higher sensitivity—at the expense of lower specificity of association with true causative mutations. The goals of implementing particular cut-points in clinical practice need to be carefully considered. The revised population prevalence may affect prevention strategies for atherosclerotic cardiovascular disease (CVD) risk in HeFH patients and their families [7].
Currently, nine genes underlie FH or FH-like phenotypes (Table 1). In addition to canonical causative genes, namely LDLR, APOB, and PSCK9 for co-dominant forms of FH, and LDLRAP1 (alias ARH) for purely recessive FH [8], NGS revealed that the APOE p.Leu167del variant causes a dominant presentation of FH [9, 10], while recessive mutations in ABCG5 (and likely ABCG8) [11] and LIPA can cause an FH-like phenotype [12]. Exome sequencing in uncharacterized FH families also showed that ultra-rare mutations in STAP1 encoding signal transducing adaptor family member 1, likely causes autosomal dominant FH [13]. The heterozygous APOE p.Leu167del mutation was previously associated with combined hyperlipidemia, splenomegaly and sea-blue histiocytosis [14]; its association now with simple hypercholesterolemia suggests that secondary genetic or environmental factors modulate phenotypic expression. Similarly, ABCG5 and LIPA cause the discrete syndromes sitosterolemia (or phytosterolemia) and lysosomal acid lipase deficiency (also known as Wolman disease in its more severe form), respectively [15]. So among genes that have been implicated in FH, the primarily causative genes, in order of importance and prevalence, remain LDLR, APOB, and PCSK9, while causative mutations in the rest are quite rare [2].
Exome sequencing has also identified potential new FH loci. For instance, whole exome sequencing (WES) of 554 individuals with extreme LDL-C levels found a high burden of rare or low-frequency variants in PNPLA5 encoding a phospholipase-domain-containing protein implicated in fatty liver disease [16]. In particular, 3.1 % of individuals with LDL-C >98th percentile carried a rare missense variant in PNPLA5 compared to 1.2 % of non-extreme individuals and only 0.5 % of individuals with LDL-C <2nd percentile; each PNPLA5 variant allele increased LDL-C by ~1 mmol/L (38.6 mg/dL). The association was replicated in an independent sample, although neither vertical transmission in families nor mechanistic impairment was demonstrated [16]. Exome sequencing in other unrelated FH subjects identified other possible new genes, namely CH25H and INSIG2, which have yet to be validated [17].
The role of polygenic determinants in FH was recently clarified [18]. Up to 40 % of HeFH individuals diagnosed clinically have no monogenic mutation identified through sequencing, but instead have accumulated a burden of small-effect, LDL-C-raising single-nucleotide polymorphism (SNP) alleles that cumulatively raise LDL-C levels into the HeFH range [19]. Polygenic effects explain the high LDL-C levels in many but not all patients with clinically diagnosed FH who lack a monogenic mutation [18]. Thus, polygenic determinants should be included when screening for molecular causes of FH [2]. The contribution of polygenic factors to familial combined hyperlipidemia is much greater than in FH [20].
Common and Low-Frequency Variants Associated with Lipid and Lipoprotein Traits
The Global Lipids Genetics Consortium (GLGC) genome-wide association studies (GWASs) identified common genetic variants governing plasma lipids and lipoproteins in essentially normolipidemic populations [21, 22]. The 157 loci identified by GLGC explain 10–20 % of the total variation in total, LDL-C, HDL-C, and TG, and a higher proportion of variation attributable to genetic factors [22]. Evaluating the polygenic determinants of plasma lipids typically starts with the lead SNP genotypes identified in these landmark publications [23].
Many of the genetic loci identified by GWAS—perhaps one-third—harbor genes whose products were already implicated in plasma lipoprotein metabolism [23]. Other associated loci have generated hypotheses to evaluate new pathways and mechanisms, as exemplified by molecular and biochemical studies of such GWAS specified genes as SORT1 [24], TRIB1 [25], and GCKR [26]. Another new mechanistic lead was the association of lower total cholesterol and LDL-C levels with a SNP at the haptoglobin locus (HP) [21], which marks haplotypes with exonic deletions that likely affect expression of haptoglobin and possibly interaction with apolipoprotein (apo) E-containing lipoproteins [27]. Each new locus identified from GLGC could lead to comparable lines of investigation.
The microarrays used in the first GLGC studies surveyed primarily common variants, defined as those with minor allele frequencies >5 % in the general population. A few low-frequency variants, defined as those with minor allele frequencies between 0.5 and 5 %, were represented on earlier microarrays. However, newer platforms, such as the “exome array,” allowed for a more systematic evaluation of low-frequency variants, some of which had larger phenotypic effects [28]. For instance, using the exome array to screen >200,000 low-frequency and rare coding sequence variants across the genome in 56,538 individuals of varied ancestries identified four low-frequency (frequencies between 0.1 and 2 %) variants, namely ANGPTL8 rs145464906, PAFAH1B2 rs186808413, COL18A1 rs114139997, and PCSK7 rs142953140, with relatively large effects on HDL-C and TG, although none of these was associated with atherosclerotic CVD [28].
Another recent insight is that frequencies of genetic variants differ across human populations: a variant considered as low frequency or ultra-rare in certain populations can be common in others. For instance, the LDLR p.G116S missense variant is absent from virtually all populations except Inuit from the circumpolar north [29]. This variant is absent from microarrays and was identified by candidate gene sequencing of Greenland Inuit with high LDL-C levels. Genotyping p.G116S in 3324 Inuit from Alaska, Canada, and Greenland showed an allele frequency of ~10 % [29]. Each allele raised LDL-C by 0.54 mmol/L (20.9 mg/dL) and carriers had a 3.0-fold increased risk of hypercholesterolemia [29]. In vitro, p.G116S showed 60 % reduced ligand-binding compared with wild-type receptor, indicating causality for elevated LDL-C [29]. These findings suggest that such terminology as “common,” “low frequency,” and “rare” is context-dependent, often reflecting ascertainment bias and the sampling strategy that led to the initial detection of the variant.
Rare Variants Associated with Lipid and Lipoprotein Traits
In contrast to common and low-frequency variants, rare variants are defined as having a population prevalence of <1 % and sometimes much lower [30, 31]. Their low frequency might result from a higher probability of biochemical dysfunction, reflecting effects of recent population explosion or of negative (purifying) selection [32]. However, in the lipid field, WES has revealed numerous examples of rare—even personal—variants with no obvious functional consequences upon health or survival [33]. Rare variants present logistic barriers to investigation of genetic determinants of plasma lipids and lipoproteins. The first issue is technical: rare variants need to be detected through high throughput sequencing technology, since they are generally not represented on microarrays. The second issue is statistical: any individual rare variant may not be statistically associated with the trait of interest, even in large samples. Solutions include (1) increasing the sample size to tens or hundreds of thousands of subjects, with its attendant burden on resources; and (2) bundling together likely causative rare variants within a particular gene or rationally grouped genes according to mechanisms or pathways, and testing the association of the bundle of variants with the trait of interest.
An early rare variant association study demonstrated an 8-fold increased frequency of a bundle of heterozygous rare nonsynonymous sequence variants in ABCA1, LCAT, and APOA1 genes in individuals with low HDL-C compared to those with high HDL-C levels [34]. Another study showed a 2-fold increased frequency of a bundle of rare variants in LPL, APOA5, GCKR, and APOB genes in individuals with severe hypertriglyceridemia compared to normal controls [35]. A wrinkle complicating this approach was seen when PCSK9 was sequenced in patients with extremes of LDL-C [36]. While several variants were found at each extreme, it soon became clear that uncommon and rare loss-of-function variants (premature truncations) were cumulatively more prevalent in individuals with low LDL-C, while rare missense mutations shown to result in a gain-of-function were enriched in individuals with high LDL-C. Testing the association of bundled uncommon and rare loss-of-function variants in PCSK9 with atherosclerosis end-points is a now classic example of the Mendelian randomization (MR) approach [37], which identified PSCK9 as a drug target to reduce LDL-C and CVD risk.
Association of Lipid and Lipoprotein Variants with Atherosclerotic Cardiovascular Disease
Many genetic determinants of plasma lipoproteins are also significant determinants of atherosclerotic CVD. A recent synthesis of coronary heart disease (CHD) GWAS results indicated 58 significantly associated loci [38], of which about one-quarter overlapped with GWAS loci for lipids, outnumbering the contributions of loci associated with blood pressure or diabetes [30]. Because lipid-associated GWAS loci were determinants of either LDL-C, TG, or lipoprotein(a) (Lp[a]), the complexity of the biology seemed reducible to the common presence of apo B in these particles; genetic determinants of apo B-containing lipoproteins seemed to be the unifying element underlying these observations [30]. However, discordance between levels of apo B and LDL-C or TG is well-known [39]; furthermore, apo B and Lp(a) levels are uncorrelated. Until a GWAS of apo B (or non-HDL-C) concentration is performed, invoking apo B as the unifying intermediate phenotype for genetic determinants of CHD seems premature.
Genetic Evidence for a Causal role for LDL-C in Coronary Heart Disease Susceptibility
The causal relationship between LDL-C and CHD has been supported by early observations in families and cohorts with FH due to rare variants in LDLR [40]. More recently, an evaluation of 164 heterozygous carriers of 16 different rare gain-of-function variants in PCSK9 [41] showed a high prevalence of early onset CHD, with 33 % of carriers expressing symptoms and hard end-points at mean age of 49 years. But other types of genetic evidence have superseded observational studies in families and cohorts as arbiters of causality.
For instance, MR experiments using common or low-frequency variants of candidate genes have both supported and refuted causative roles for various lipids and lipoproteins in atherosclerosis. Theoretical underpinnings of the MR approach, and its strengths and limitations are discussed elsewhere [42, 43]. In contrast to other criteria for causality, such as Koch’s postulates [44] or Bradford Hill’s criteria [45], Mendel likely never envisioned that laws of inheritance would provide a litmus test for a causal relationship between a genetically determined intermediate trait and a temporally remote disease outcome. Nonetheless, MR now routinely arbitrates causal relationships between markers and disease. Numerous examples of MR experiments examine either common or rare variants, individually or bundled together, affecting lipid and lipoprotein metabolic pathways. In the case of rare variants, probable loss-of-function or “inactivating” variants (i.e., truncation, nonsense or splicing) are preferred when testing concurrently for association with lipids and atherosclerosis end-points, since functional compromise is more easily inferred without the need for mechanistic confirmation. The preferred MR study design concentrates on favorable outcomes, such as protection from CHD; positive results are more easily translated since a drug or biological intervention can pharmacologically mimic the genetic deficiency linked to the favorable outcome.
As mentioned, the MR design proved that protection from CHD was associated with uncommon loss-of-function variants in PCSK9 [37, 46]. This motivated development of PCSK9 inhibitors, which recently became available for prescription to reduce LDL-C, proving that MR can identify causal relationships and drug targets [47]. Interestingly, extending the MR approach in Copenhagen cohorts showed no causal relationship between low LDL-C and either cancer or gallstones [48, 49]. Another MR experiment assembled 13 common genetic determinants of LDL-C into a polygenic score and found that an ~0.8 mmol/L (30.9 mg/dL) genetically determined increase in LDL-C was associated with a 2.1-fold increased risk of CHD [50]. MR evaluation of 10,464 CHD events occurring in 108,376 individuals from 14 studies showed that common polymorphisms in NPC1L1 or HMGCR both reduced LDL-C and CHD risk [51•]. Furthermore, WES showed that heterozygotes for very rare NPC1L1 inactivating mutations had LDL-C that was 0.31 mmol/L (12.0 mg/dL) lower than non-carriers and a 53 % reduced CHD risk [52]. These observations supported the contemporaneous publication of randomized clinical trial results showing that ezetimibe, which targets the NPC1L1 transporter, reduced both LDL-C and CHD risk [53]. A study of common NPC1L1 variants genotyped in 67,385 individuals not only replicated reductions in LDL-C and CHD risk, but also documented increased risk of gallstone disease [54].
A final illustration of how MR can illuminate causal pathways was seen in a meta-analysis of genotypes of HMGCR, which encodes the target of statin drugs. Statins slightly increase the risk of type 2 diabetes [55]. To address a possible causal relationship, HMGCR genotyping in 223,463 individuals from 43 studies showed associations with reduced LDL-C and increased body weight, waist circumference, plasma insulin and glucose [55]. This supported a causal relationship between altered HMG coenzyme A reductase activity and worsened glycaemia and increased diabetes risk, perhaps mediated by slightly increased body weight.
Genetic Evidence for a Diminished Causal Role for HDL-C in Coronary Heart Disease Susceptibility
MR data for a causal relationship between CHD and HDL-C are much weaker than for LDL-C. The Copenhagen group first showed that lower HDL-C in carriers of rare loss-of-function variants in ABCA1 were not associated with increased CHD risk [56]. They showed similar neutral results for low-frequency variants in LIPC encoding hepatic lipase [57] and LCAT encoding lecithin cholesterol acyl transferase [57]. A meta-analysis using several independent approaches showed that a polygenic score that raised HDL-C by one standard deviation had no effect on CHD risk, while a comparable polygenic score for LDL-C was strongly associated [50]. The same study reported borderline association of variants in CETP encoding cholesteryl ester transfer protein that raised HDL-C and reduced CHD risk [50]. Furthermore, the Copenhagen group showed that common variants associated with reduced CETP activity were associated with increased HDL-C and reduced LDL-C, together with reduced risk of CHD, other CVD end-points and increased longevity [58], which sustains hope for the strategy of CETP inhibition. However, the termination of three drug development programs for CETP inhibitors supports the idea that simply increasing HDL-C quantity is not associated with CHD risk, although results of the Randomized Evaluation of the Effects of Anacetrapib Through Lipid-Modification (REVEAL) trial with the CETP inhibitor anacetrapib are pending [59]. In observational studies in families with monogenic extreme HDL-C deviations, where association with CVD was also inconsistent [60]. The complexities of HDL-C levels were further highlighted following the discovery of the loss-of-function variant, p.P367L, in SCARB1 encoding scavenger receptor BI [61]. This variant not only was identified in individuals with significantly elevated levels of HDL-C, but carriers of this poorly functioning variant also demonstrated a significantly increased risk for CHD [61]. Perhaps genetic determinants of HDL function, such as efflux capacity, are more mechanistically related to atherosclerosis susceptibility than determinants of simple HDL-C quantity [62, 63•].
Genetic Evidence for a Causal Role for TG in Coronary Heart Disease Susceptibility
The ying-yang relationship between concentrations of TG and HDL-C and underlying mechanisms have been appreciated for decades [64•]. Before the MR era, elevated TG levels in this joint phenotype were considered to be an innocent bystander, with depressed HDL-C levels thought to underlie disease susceptibility. Concurrent with diminished recent enthusiasm for HDL-C, MR studies suggest that high TG plays a proximal causal role in atherosclerosis, although disentangling the association with low HDL-C is challenging. GLGC version 1.0 indicated that common TG-raising variants were associated with atherosclerosis risk; however almost all of these had joint effects, particularly depressed HDL-C [21]. Furthermore, common APOA5 variants associated with higher TG and lower HDL-C were also associated with increased CHD risk [65]. Also, the common LPL p.S447X gain-of-function variant (also known as p.S474X) has long been associated with reduced TG, increased HDL-C, and reduced CHD risk in small cohorts [66], while the relatively common LPL p.D9N loss-of-function variant (also known as p.D36N) has been associated with increased TG, reduced HDL-C, and increased CHD risk [67]. Associations of these two LPL variants with the high TG/low HDL-C atherogenic dyslipidemia complex and with CHD risk were recently confirmed in a large case-control sample [68•]. Using statistical models, genetic determinants with predominantly TG-related effects were correlated with increased CHD risk, while genetic determinants with predominantly HDL-C-related effects were not [69]. These associations might be related in part to the cholesterol content of TG-rich lipoprotein remnant particles [70].
Exome sequencing showed that rare heterozygous loss-of-function APOC3 mutations are primarily associated with reduced plasma TG levels: mutation carriers had significantly reduced CHD risk, again supporting the idea that TG might contribute directly to atherosclerosis [71, 72]. However, these rare variants were almost always associated with reduced LDL-C and increased HDL-C [71]. In addition, carriers of heterozygous rare loss-of-function mutations in APOA5 that increased plasma TG levels had a 2-fold increased risk of early CHD [73], but these variants were associated with increased LDL-C and decreased HDL-C. Furthermore, inactivating variants in ANGPTL4 [74] were associated with reduced TG and reduced CHD risk [68•, 75•]. This clarified earlier inconsistent observations of ANGPTL4 E40K variant association with CHD in small cohorts [76, 77].
While recent genetic studies support longstanding prior biological knowledge of the importance of the joint high TG/low HDL-C phenotype as an integrated read-out of a complex network of underlying processes [70, 71], some questions remain. For instance, why are patients with severely elevated TG and depressed HDL-C due to familial chylomicronemia from homozygous LPL variants not at increased risk of atherosclerosis [78, 79]? Also, why are common or rare variants in some other LPL-associated genes, namely APOC2, LMF1, and GPIHBP1 (Table 1), which all cause severe hypertriglyceridemia and have well established roles in lipolysis [70, 71], not associated with increased CHD risk in GWAS or WES studies?
Renaissance of Lipoprotein(a)
The relationship between Lp(a) and atherosclerosis has been appreciated for decades [80]. Recent meta-analyses in case-control studies using SNPs at the LPA locus on chromosome 6q25-26 confirm association with CHD [81]. GWAS also shows that LPA genotype is significantly associated with CHD [82•]. The range of associated phenotypes has recently been broadened to include calcific aortic stenosis [83, 84] and heart failure [85]. This convergence of genetic data suggesting that isolated reduction of Lp(a) can protect against CHD and other adverse phenotypes has prompted development of antisense therapy targeting Lp(a), which in a recent phase 1 study showed reductions of up to 78 % with no apparent adverse effects [86]. This agent should help advance evaluation of Lp(a)’s role in pathogenesis and of possible benefits of targeted reduction. It is also interesting that PCSK9 monoclonal antibodies reduce plasma Lp(a) levels by ~30 % [87], which may explain part of their clinical benefits [88].
Translation to Other Therapies
PCSK9 inhibitors, whose development could be traced to human genetic observations, are now in widespread clinical use and appear to have beneficial effects on outcomes [88, 89], as well as potential adverse effects [90] and economic implications [91]. Analogous advances include development of an anti-sense inhibitor of APOC3 (volanesorsen) as a treatment for familial chylomicronemia due to homozygous mutations in LPL and other genetically undefined forms of severe hypertriglyceridemia [64•, 65]. While volanesorsen may prove to be useful clinically for severe hypertriglyceridemia with its attendant risk of life-threatening pancreatitis, suggestions that it might also be used to prevent CHD need to be carefully considered. The specific targeting of APOC3 has pleiotropic effects on the lipid profile, including reductions of apo C-III-containing subfractions of LDL, HDL, and Lp(a) [92]. Also, ANGPTL3 and ANGPTL4 are active drug targets as they modulate LPL activity, and because of the apparently favorable phenotypes observed in carriers of inactivating or loss-of-function variants [68•, 75•, 93].
As mentioned above, some patients with mutations in LIPA that otherwise cause cholesterol ester storage disease or Wolman syndrome can present clinically with a phenotype that resembles recessive FH (Table 1) [12]. Patients with lysosomal acid lipase deficiency (LALD) classically express some combination of hepatomegaly, elevated transaminases, and dyslipidemia (usually elevated LDL-C) [94]. Liver biopsy shows hepatosteatosis that can progress to fibrosis, cirrhosis, and liver failure [94]. A recent multicentre randomized, double-blind, placebo-controlled study in LALD patients showed that enzyme-replacement therapy with sebelipase alfa was associated with improved plasma lipids and transaminases and reduced hepatic fat, together with only mild adverse effects [95]. The long-term benefits, risks, and costs of this interesting new treatment remain to be determined.
Integrated Overview of Genetic Determinants of Plasma Lipids and Lipoproteins
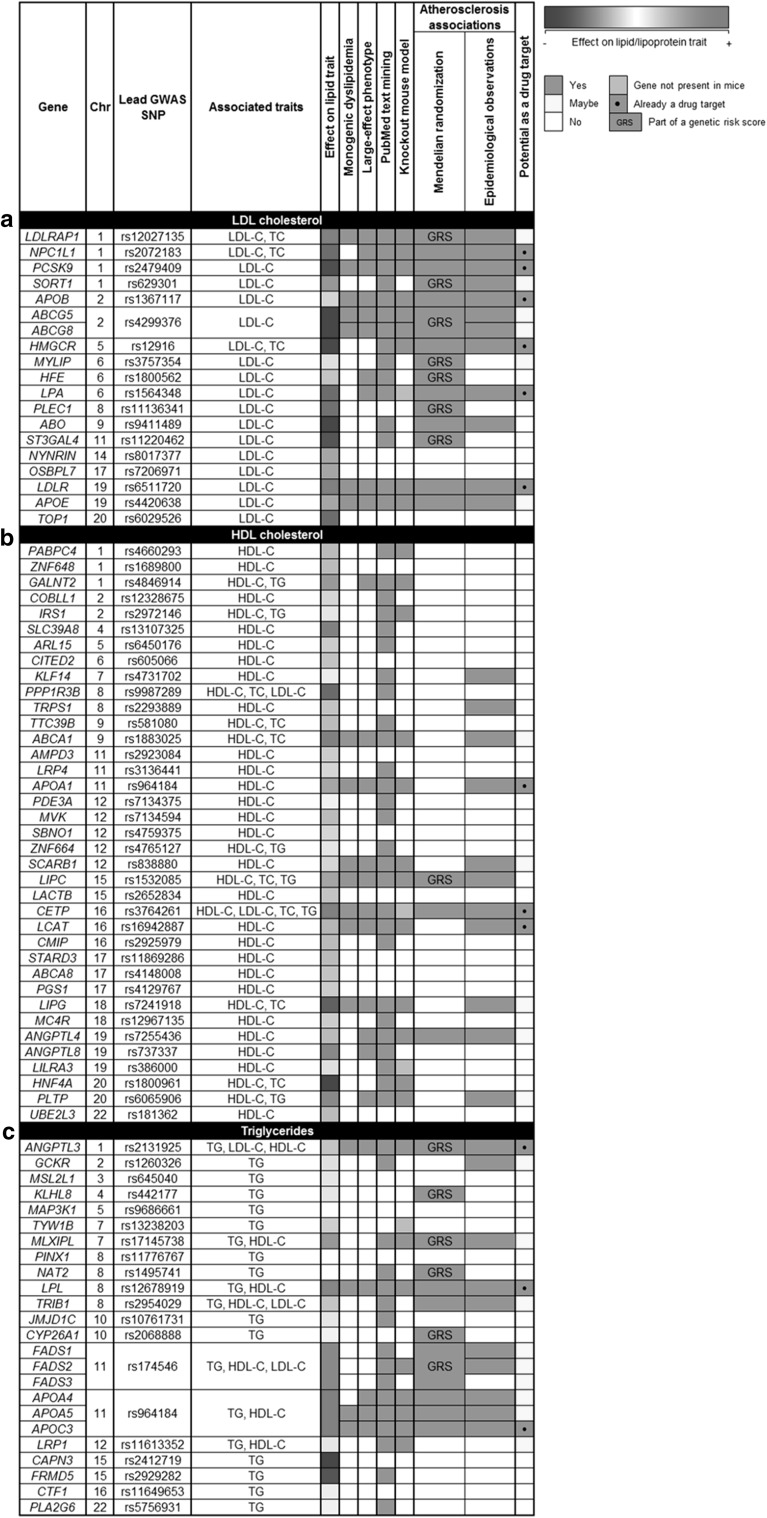
A PubMed search on March 5, 2016 using the terms “human genetics” and “lipoproteins” yielded 17,660 hits. Has this investment in resources and effort been worth it? In a first attempt to summarize and integrate some of the accumulated knowledge of the genetics of lipid and lipoprotein traits, we have compiled results from various types of studies for LDL-C (Fig. 1a), HDL-C (Fig. 1b), and TG (Fig. 1c). Because GWAS have been the largest single source of trait-associated variants, the rows of each figure are populated by GWAS loci identified from the GLGC meta-analyses [22], sorted by chromosomal location. The lead SNP at each locus is shown, although some loci had more than one lead SNP or significant association signal. Because most loci had joint associations with other traits, these are also shown. The relative effect size and direction of association of the lead SNP with the trait are indicated. Association of each locus with a monogenic disorder or syndrome, or with an extreme biochemical deviation in the absence of other systemic involvement, are indicated. The extent to which the non-genetic literature (i.e., biochemical, cell biological, pathological, epidemiological) supported the candidacy of the gene locus was gaged and noted. Of particular importance was the recapitulation of the representative phenotype in knock-out mouse models. Because of the importance of lipids and lipoproteins in atherosclerosis risk, evidence of association with these end-points from MR and observational studies in affected kindreds and cohorts of variant carriers is highlighted. Finally, rapid prioritization of loci that have led to drug development projects or could represent promising targets for such development are shown.
Fig. 1.
Summary of current information for genes associated with: a LDL cholesterol (LDL-C), b HDL cholesterol (HDL-C), c Triglyceride (TG)
Conclusions and Future Directions
The scope of genetic determinants of inter-individual phenotypic variation both in dyslipidemia syndromes and the general population as revealed by GWAS and NGS is staggering. While a few more rare monogenic dyslipidemias may still be identified and characterized, by and large the knowledge of these disorders is complete. Furthermore, we have a good start on understanding genomic variants contributing to lipid and lipoprotein levels, although the proportion of variation unexplained remains large. While rare variants underlying lipid phenotypes are proving to be informative, they only explain a small amount of variation; factors such as gene-gene, gene-environment, epigenetic modifications, and perhaps new aspects of biology may fill in our knowledge gaps. Human genetics has also inspired development of new interventions to improve the quality and quantity of life for patients with dyslipidemia, but also more generally for those at risk of the devastating end-points of atherosclerotic CVD.
Acknowledgments
RAH is supported by the Jacob J. Wolfe Distinguished Medical Research Chair, the Martha G. Blackburn Chair in Cardiovascular Research, and operating grants from the Canadian Institutes of Health Research (Foundation Grant), the Heart and Stroke Foundation of Ontario (T-000353) and Genome Canada through Genome Quebec (award 4530).
Compliance with Ethical Standards
Disclosure
Jacqueline S. Dron declares that she has no conflict of interest. Robert A. Hegele is a consultant and speaker’s bureau member for Aegerion, Amgen, Sanofi, Pfizer, and Valeant.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical collection on Cardiovascular Genetics.
An erratum to this article is available at http://dx.doi.org/10.1007/s40142-017-0117-6.
References
Recently published papers of particular interest have been highlighted as: • Of importance
- 1.Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10(2):109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 2.Hegele RA, Ban MR, Cao H, McIntyre AD, Robinson JF, Wang J. Targeted next-generation sequencing in monogenic dyslipidemias. Curr Opin Lipidol. 2015;26(2):103–113. doi: 10.1097/MOL.0000000000000163. [DOI] [PubMed] [Google Scholar]
- 3.Stitziel NO, Peloso GM, Abifadel M, Cefalu AB, Fouchier S, Motazacker MM, et al. Exome sequencing in suspected monogenic dyslipidemias. Circ Cardiovasc Genet. 2015;8(2):343–350. doi: 10.1161/CIRCGENETICS.114.000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gidding SS, Ann Champagne M, de Ferranti SD, Defesche J, Ito MK, Knowles JW, et al. The agenda for familial hypercholesterolemia: a scientific statement from the American Heart Association. Circulation. 2015;132(22):2167–2192. doi: 10.1161/CIR.0000000000000297. [DOI] [PubMed] [Google Scholar]
- 5.• Benn M, Watts GF, Tybjaerg-Hansen A, Nordestgaard BG. Mutations causative of familial hypercholesterolaemia: screening of 98,098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur Heart J. 2016;37(17):1384–94. This study is the first to report on the frequency of FH-causing mutations in a general population. [DOI] [PubMed]
- 6.Futema M, Kumari M, Boustred C, Kivimaki M, Humphries SE. Would raising the total cholesterol diagnostic cut-off from 7.5 mmol/L to 9.3 mmol/L improve detection rate of patients with monogenic familial hypercholesterolaemia? Atherosclerosis. 2015;239(2):295–298. doi: 10.1016/j.atherosclerosis.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vallejo-Vaz AJ, Kondapally Seshasai SR, Cole D, Hovingh GK, Kastelein JJ, Mata P, et al. Familial hypercholesterolaemia: a global call to arms. Atherosclerosis. 2015;243(1):257–259. doi: 10.1016/j.atherosclerosis.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 8.Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 2013;34(45):3478–3490. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Awan Z, Choi HY, Stitziel N, Ruel I, Bamimore MA, Husa R, et al. APOE p.Leu167del mutation in familial hypercholesterolemia. Atherosclerosis. 2013;231(2):218–222. doi: 10.1016/j.atherosclerosis.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Marduel M, Ouguerram K, Serre V, Bonnefont-Rousselot D, Marques-Pinheiro A, Erik Berge K, et al. Description of a large family with autosomal dominant hypercholesterolemia associated with the APOE p.Leu167del mutation. Hum Mutat. 2013;34(1):83–87. doi: 10.1002/humu.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rios J, Stein E, Shendure J, Hobbs HH, Cohen JC. Identification by whole-genome resequencing of gene defect responsible for severe hypercholesterolemia. Hum Mol Genet. 2010;19(22):4313–4318. doi: 10.1093/hmg/ddq352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stitziel NO, Fouchier SW, Sjouke B, Peloso GM, Moscoso AM, Auer PL, et al. Exome sequencing and directed clinical phenotyping diagnose cholesterol ester storage disease presenting as autosomal recessive hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2013;33(12):2909–2914. doi: 10.1161/ATVBAHA.113.302426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier SW, Dallinga-Thie GM, Meijers JC, Zelcer N, Kastelein JJ, Defesche JC, et al. Mutations in STAP1 are associated with autosomal dominant hypercholesterolemia. Circ Res. 2014;115(6):552–555. doi: 10.1161/CIRCRESAHA.115.304660. [DOI] [PubMed] [Google Scholar]
- 14.Rahalkar AR, Wang J, Sirrs S, Dimmick J, Holmes D, Urquhart N, et al. An unusual case of severe hypertriglyceridemia and splenomegaly. Clin Chem. 2008;54(3):606–610. doi: 10.1373/clinchem.2007.097139. [DOI] [PubMed] [Google Scholar]
- 15.Rahalkar AR, Hegele RA. Monogenic pediatric dyslipidemias: classification, genetics and clinical spectrum. Mol Genet Metab. 2008;93(3):282–294. doi: 10.1016/j.ymgme.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Lange LA, Hu Y, Zhang H, Xue C, Schmidt EM, Tang ZZ, et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet. 2014;94(2):233–245. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Futema M, Plagnol V, Li K, Whittall RA, Neil HA, Seed M, et al. Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J Med Genet. 2014;51(8):537–544. doi: 10.1136/jmedgenet-2014-102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Futema M, Shah S, Cooper JA, Li K, Whittall RA, Sharifi M, et al. Refinement of variant selection for the LDL cholesterol genetic risk score in the diagnosis of the polygenic form of clinical familial hypercholesterolemia and replication in samples from 6 countries. Clin Chem. 2015;61(1):231–238. doi: 10.1373/clinchem.2014.231365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talmud PJ, Shah S, Whittall R, Futema M, Howard P, Cooper JA, et al. Use of low-density lipoprotein cholesterol gene score to distinguish patients with polygenic and monogenic familial hypercholesterolaemia: a case-control study. Lancet. 2013;381(9874):1293–1301. doi: 10.1016/S0140-6736(12)62127-8. [DOI] [PubMed] [Google Scholar]
- 20.Brahm AJ, Hegele RA. Combined hyperlipidemia: familial but not (usually) monogenic. Curr Opin Lipidol. 2015;22:247–253. doi: 10.1097/MOL.0000000000000270. [DOI] [PubMed] [Google Scholar]
- 21.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen CT, Dube JB, Loyzer MN, MacDonald A, Carter DE, McIntyre AD, et al. LipidSeq: a next-generation clinical resequencing panel for monogenic dyslipidemias. J Lipid Res. 2014;55(4):765–772. doi: 10.1194/jlr.D045963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strong A, Ding Q, Edmondson AC, Millar JS, Sachs KV, Li X, et al. Hepatic sortilin regulates both apolipoprotein B secretion and LDL catabolism. J Clin Invest. 2012;122(8):2807–2816. doi: 10.1172/JCI63563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer RC, Sasaki M, Cohen DM, Cui J, Smith MA, Yenilmez BO, et al. Tribbles-1 regulates hepatic lipogenesis through posttranscriptional regulation of C/EBPalpha. J Clin Invest. 2015;125(10):3809–3818. doi: 10.1172/JCI77095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees MG, Raimondo A, Wang J, Ban MR, Davis MI, Barrett A, et al. Inheritance of rare functional GCKR variants and their contribution to triglyceride levels in families. Hum Mol Genet. 2014;23(20):5570–5578. doi: 10.1093/hmg/ddu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boettger LM, Salem RM, Handsaker RE, Peloso GM, Kathiresan S, Hirschhorn JN, et al. Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat Genet. 2016;48(4):359–366. doi: 10.1038/ng.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94(2):223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dube JB, Wang J, Cao H, McIntyre AD, Johansen CT, Hopkins SE, et al. Common low-density lipoprotein receptor p.G116S variant has a large effect on plasma low-density lipoprotein cholesterol in circumpolar inuit populations. Circ Cardiovasc Genet. 2015;8(1):100–108. doi: 10.1161/CIRCGENETICS.114.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musunuru K, Kathiresan S. Surprises from genetic analyses of lipid risk factors for atherosclerosis. Circ Res. 2016;118(4):579–585. doi: 10.1161/CIRCRESAHA.115.306398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeff JM, Peloso GM, Do R. What can we learn about lipoprotein metabolism and coronary heart disease from studying rare variants? Curr Opin Lipidol. 2016;27(2):99–104. doi: 10.1097/MOL.0000000000000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohmueller KE. The impact of population demography and selection on the genetic architecture of complex traits. PLoS Genet. 2014;10(5):e1004379. doi: 10.1371/journal.pgen.1004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farhan SM, Hegele RA. Exome sequencing: new insights into lipoprotein disorders. Curr Cardiol Rep. 2014;16(7):507. doi: 10.1007/s11886-014-0507-2. [DOI] [PubMed] [Google Scholar]
- 34.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 35.Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42(8):684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 37.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 38.McPherson R, Tybjaerg-Hansen A. Genetics of coronary artery disease. Circ Res. 2016;118(4):564–578. doi: 10.1161/CIRCRESAHA.115.306566. [DOI] [PubMed] [Google Scholar]
- 39.Wilkins JT, Li RC, Sniderman A, Chan C, Lloyd-Jones DM. Discordance between apolipoprotein B and LDL-cholesterol in young adults predicts coronary artery calcification: the CARDIA study. J Am Coll Cardiol. 2016;67(2):193–201. doi: 10.1016/j.jacc.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Versmissen J, Oosterveer DM, Yazdanpanah M, Defesche JC, Basart DC, Liem AH, et al. Efficacy of statins in familial hypercholesterolaemia: a long term cohort study. BMJ. 2008;337:a2423. doi: 10.1136/bmj.a2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hopkins PN, Defesche J, Fouchier SW, Bruckert E, Luc G, Cariou B, et al. Characterization of autosomal dominant hypercholesterolemia caused by PCSK9 gain of function mutations and its specific treatment with alirocumab, a PCSK9 monoclonal antibody. Circ Cardiovasc Genet. 2015;8(6):823–831. doi: 10.1161/CIRCGENETICS.115.001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jansen H, Samani NJ, Schunkert H. Mendelian randomization studies in coronary artery disease. Eur Heart J. 2014;35(29):1917–1924. doi: 10.1093/eurheartj/ehu208. [DOI] [PubMed] [Google Scholar]
- 43.Rosenson RS, Koenig W. Mendelian randomization analyses for selection of therapeutic targets for cardiovascular disease prevention: a note of circumspection. Cardiovasc Drugs Ther. 2016;30(1):65–74. doi: 10.1007/s10557-016-6642-9. [DOI] [PubMed] [Google Scholar]
- 44.Brown MS, Goldstein JL. Koch’s postulates for cholesterol. Cell. 1992;71(2):187–188. doi: 10.1016/0092-8674(92)90346-E. [DOI] [PubMed] [Google Scholar]
- 45.Calvert GD. A review of observational studies on the relationship between cholesterol and coronary heart disease. Aust N Z J Med. 1994;24(1):89–91. doi: 10.1111/j.1445-5994.1994.tb04442.x. [DOI] [PubMed] [Google Scholar]
- 46.Benn M, Nordestgaard BG, Grande P, Schnohr P, Tybjaerg-Hansen A. PCSK9 R46L, low-density lipoprotein cholesterol levels, and risk of ischemic heart disease: 3 independent studies and meta-analyses. J Am Coll Cardiol. 2010;55(25):2833–2842. doi: 10.1016/j.jacc.2010.02.044. [DOI] [PubMed] [Google Scholar]
- 47.Bergeron N, Phan BA, Ding Y, Fong A, Krauss RM. Proprotein convertase subtilisin/kexin type 9 inhibition: a new therapeutic mechanism for reducing cardiovascular disease risk. Circulation. 2015;132(17):1648–1666. doi: 10.1161/CIRCULATIONAHA.115.016080. [DOI] [PubMed] [Google Scholar]
- 48.Benn M, Tybjaerg-Hansen A, Stender S, Frikke-Schmidt R, Nordestgaard BG. Low-density lipoprotein cholesterol and the risk of cancer: a mendelian randomization study. J Natl Cancer Inst. 2011;103(6):508–519. doi: 10.1093/jnci/djr008. [DOI] [PubMed] [Google Scholar]
- 49.Stender S, Frikke-Schmidt R, Benn M, Nordestgaard BG, Tybjaerg-Hansen A. Low-density lipoprotein cholesterol and risk of gallstone disease: a Mendelian randomization study and meta-analyses. J Hepatol. 2013;58(1):126–133. doi: 10.1016/j.jhep.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 50.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380(9841):572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.• Ference BA, Majeed F, Penumetcha R, Flack JM, Brook RD. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both: a 2 × 2 factorial Mendelian randomization study. J Am Coll Cardiol. 2015;65(15):1552–61. This study found that CHD risk in response to LDL-C levels is more dependent on the actual measurable concentration of LDL-C, rather than the mechanism that lowers it. [DOI] [PMC free article] [PubMed]
- 52.Stitziel NO, Won HH, Morrison AC, Peloso GM, Do R, Lange LA, et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371(22):2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cannon CP, Blazing MA, Giugliano RP, McCagg A, White JA, Theroux P, et al. Ezetimibe added to statin therapy after acute coronary syndromes. N Engl J Med. 2015;372(25):2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 54.Lauridsen BK, Stender S, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Genetic variation in the cholesterol transporter NPC1L1, ischaemic vascular disease, and gallstone disease. Eur Heart J. 2015;36(25):1601–1608. doi: 10.1093/eurheartj/ehv108. [DOI] [PubMed] [Google Scholar]
- 55.Preiss D, Sattar N. Statins and the risk of new-onset diabetes: a review of recent evidence. Curr Opin Lipidol. 2011;22(6):460–466. doi: 10.1097/MOL.0b013e32834b4994. [DOI] [PubMed] [Google Scholar]
- 56.Frikke-Schmidt R, Nordestgaard BG, Stene MC, Sethi AA, Remaley AT, Schnohr P, et al. Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA. 2008;299(21):2524–2532. doi: 10.1001/jama.299.21.2524. [DOI] [PubMed] [Google Scholar]
- 57.Haase CL, Tybjaerg-Hansen A, Qayyum AA, Schou J, Nordestgaard BG, Frikke-Schmidt R. LCAT, HDL cholesterol and ischemic cardiovascular disease: a Mendelian randomization study of HDL cholesterol in 54,500 individuals. J Clin Endocrinol Metab. 2012;97(2):E248–E256. doi: 10.1210/jc.2011-1846. [DOI] [PubMed] [Google Scholar]
- 58.Johannsen TH, Frikke-Schmidt R, Schou J, Nordestgaard BG, Tybjaerg-Hansen A. Genetic inhibition of CETP, ischemic vascular disease and mortality, and possible adverse effects. J Am Coll Cardiol. 2012;60(20):2041–2048. doi: 10.1016/j.jacc.2012.07.045. [DOI] [PubMed] [Google Scholar]
- 59.Barter PJ, Rye KA. Cholesteryl ester transfer protein inhibition is not yet dead-pro. Arterioscler Thromb Vasc Biol. 2016;36(3):439–441. doi: 10.1161/ATVBAHA.115.306879. [DOI] [PubMed] [Google Scholar]
- 60.Hovingh GK, Rader DJ, Hegele RA. HDL re-examined. Curr Opin Lipidol. 2015;26(2):127–132. doi: 10.1097/MOL.0000000000000161. [DOI] [PubMed] [Google Scholar]
- 61.Zanoni P, Khetarpal SA, Larach DB, Hancock-Cerutti WF, Millar JS, Cuchel M, et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 2016;351(6278):1166–1171. doi: 10.1126/science.aad3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agarwala AP, Rodrigues A, Risman M, McCoy M, Trindade K, Qu L, et al. High-density lipoprotein (HDL) phospholipid content and cholesterol efflux capacity are reduced in patients with very high HDL cholesterol and coronary disease. Arterioscler Thromb Vasc Biol. 2015;35(6):1515–1519. doi: 10.1161/ATVBAHA.115.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.• Saleheen D, Scott R, Javad S, Zhao W, Rodrigues A, Picataggi A, et al. Association of HDL cholesterol efflux capacity with incident coronary heart disease events: a prospective case-control study. Lancet Diabetes Endocrinol. 2015;3(7):507–13. An important demonstration of the concept that dynamic HDL function rather than simple HDL concentration is the analyte that plays a more proximal causative role in atherogenesis. [DOI] [PMC free article] [PubMed]
- 64.• Nordestgaard BG. Triglyceride-rich lipoproteins and atherosclerotic cardiovascular disease: new insights from epidemiology, genetics, and biology. Circ Res. 2016;118(4):547–63. An outstanding review that integrates multifaceted knowledge of TG-rich lipoproteins and theor role in atherosclerosis. [DOI] [PubMed]
- 65.Sarwar N, Sandhu MS, Ricketts SL, Butterworth AS, Di Angelantonio E, Boekholdt SM, et al. Triglyceride-mediated pathways and coronary disease: collaborative analysis of 101 studies. Lancet. 2010;375(9726):1634–1639. doi: 10.1016/S0140-6736(10)60545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clee SM, Loubser O, Collins J, Kastelein JJ, Hayden MR. The LPL S447X cSNP is associated with decreased blood pressure and plasma triglycerides, and reduced risk of coronary artery disease. Clin Genet. 2001;60(4):293–300. doi: 10.1034/j.1399-0004.2001.600407.x. [DOI] [PubMed] [Google Scholar]
- 67.Mailly F, Fisher RM, Nicaud V, Luong LA, Evans AE, Marques-Vidal P, et al. Association between the LPL-D9N mutation in the lipoprotein lipase gene and plasma lipid traits in myocardial infarction survivors from the ECTIM Study. Atherosclerosis. 1996;122(1):21–28. doi: 10.1016/0021-9150(95)05736-6. [DOI] [PubMed] [Google Scholar]
- 68.• Investigators MIGaCEC. Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N Engl J Med. 2016;374(12):1134. A large study using the exome array and sequencing showing that the uncommon ANGPTL4 E40 K variant was associated with reduced plasma triglycerides, raised HDL-C and reduced atherosclerosis risk; this was also seen with very rare inactivating variants in ANGPTL4. Concurrent analyses showed similar observations for LPL variants, confirming results from many earlier studies of LPL.
- 69.Do R, Willer CJ, Schmidt EM, Sengupta S, Gao C, Peloso GM, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45(11):1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varbo A, Benn M, Tybjaerg-Hansen A, Jorgensen AB, Frikke-Schmidt R, Nordestgaard BG. Remnant cholesterol as a causal risk factor for ischemic heart disease. J Am Coll Cardiol. 2013;61(4):427–436. doi: 10.1016/j.jacc.2012.08.1026. [DOI] [PubMed] [Google Scholar]
- 71.Crosby J, Peloso GM, Auer PL, Crosslin DR, Stitziel NO, Lange LA, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371(1):22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jorgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371(1):32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 73.Do R, Stitziel NO, Won HH, Jorgensen AB, Duga S, Angelica Merlini P, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518(7537):102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39(4):513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.• Dewey FE, Gusarova V, O’Dushlaine C, Gottesman O, Trejos J, Hunt C, et al. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N Engl J Med. 2016;374(12):1123–33. A large study showing that the uncommom E40K variant of ANGPTL4 is associated with reduced plasma triglycerides, raised HDL-C and reduced atherosclerosis risk. Inhibition of ANGPTL4 with a monoclonal antibody in primates resulted in an improved plasma lipid profile, but abdominal lymphomatosis, raising concerns about ANGPTL4 as a target. [DOI] [PMC free article] [PubMed]
- 76.Folsom AR, Peacock JM, Demerath E, Boerwinkle E. Variation in ANGPTL4 and risk of coronary heart disease: the Atherosclerosis Risk in Communities Study. Metabolism. 2008;57(11):1591–1596. doi: 10.1016/j.metabol.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Talmud PJ, Smart M, Presswood E, Cooper JA, Nicaud V, Drenos F, et al. ANGPTL4 E40K and T266M: effects on plasma triglyceride and HDL levels, postprandial responses, and CHD risk. Arterioscler Thromb Vasc Biol. 2008;28(12):2319–2325. doi: 10.1161/ATVBAHA.108.176917. [DOI] [PubMed] [Google Scholar]
- 78.Brunzell J, Deeb S. Familial lipoprotein lipase deficiency, apo C-II deficiency, and hepatic lipase deficiency. In: Scriver C, Beaudet A, Sly W, Valle D, editors. The metabolic and molecular bases of inherited disease. New York: McGraw-Hill; 2001. pp. 2789–2816. [Google Scholar]
- 79.Ebara T, Endo Y, Yoshiike S, Tsuji M, Taguchi S, Murase T, et al. A 60-y-old chylomicronemia patient homozygous for missense mutation (G188E) in the lipoprotein lipase gene showed no accelerated atherosclerosis. Clin Chim Acta. 2007;386(1–2):100–104. doi: 10.1016/j.cca.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 80.Dube JB, Boffa MB, Hegele RA, Koschinsky ML. Lipoprotein(a): more interesting than ever after 50 years. Curr Opin Lipidol. 2012;23(2):133–140. doi: 10.1097/MOL.0b013e32835111d8. [DOI] [PubMed] [Google Scholar]
- 81.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361(26):2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 82.• Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121–30. State of the art of loci associated with coronary artery disease risk. [DOI] [PMC free article] [PubMed]
- 83.Thanassoulis G, Campbell CY, Owens DS, Smith JG, Smith AV, Peloso GM, et al. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med. 2013;368(6):503–512. doi: 10.1056/NEJMoa1109034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arsenault BJ, Boekholdt SM, Dube MP, Rheaume E, Wareham NJ, Khaw KT, et al. Lipoprotein(a) levels, genotype, and incident aortic valve stenosis: a prospective Mendelian randomization study and replication in a case-control cohort. Circ Cardiovasc Genet. 2014;7(3):304–310. doi: 10.1161/CIRCGENETICS.113.000400. [DOI] [PubMed] [Google Scholar]
- 85.Kamstrup PR, Nordestgaard BG. Elevated lipoprotein(a) levels, LPA risk genotypes, and increased risk of heart failure in the general population. JACC Heart Fail. 2016;4(1):78–87. doi: 10.1016/j.jchf.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 86.Tsimikas S, Viney NJ, Hughes SG, Singleton W, Graham MJ, Baker BF, et al. Antisense therapy targeting apolipoprotein(a): a randomised, double-blind, placebo-controlled phase 1 study. Lancet. 2015;386(10002):1472–1483. doi: 10.1016/S0140-6736(15)61252-1. [DOI] [PubMed] [Google Scholar]
- 87.Gaudet D, Kereiakes DJ, McKenney JM, Roth EM, Hanotin C, Gipe D, et al. Effect of alirocumab, a monoclonal proprotein convertase subtilisin/kexin 9 antibody, on lipoprotein(a) concentrations (a pooled analysis of 150 mg every two weeks dosing from phase 2 trials) Am J Cardiol. 2014;114(5):711–715. doi: 10.1016/j.amjcard.2014.05.060. [DOI] [PubMed] [Google Scholar]
- 88.Navarese EP, Kolodziejczak M, Schulze V, Gurbel PA, Tantry U, Lin Y, et al. Effects of proprotein convertase subtilisin/kexin type 9 antibodies in adults with hypercholesterolemia: a systematic review and meta-analysis. Ann Intern Med. 2015;163(1):40–51. doi: 10.7326/M14-2957. [DOI] [PubMed] [Google Scholar]
- 89.Lipinski MJ, Benedetto U, Escarcega RO, Biondi-Zoccai G, Lhermusier T, Baker NC, et al. The impact of proprotein convertase subtilisin-kexin type 9 serine protease inhibitors on lipid levels and outcomes in patients with primary hypercholesterolaemia: a network meta-analysis. Eur Heart J. 2016;37(6):536–545. doi: 10.1093/eurheartj/ehv563. [DOI] [PubMed] [Google Scholar]
- 90.Kolodziejczak M, Navarese EP. Role of PCSK9 antibodies in cardiovascular disease: critical considerations of mortality and neurocognitive findings from the current literature. Atherosclerosis. 2016;15(247):189–192. doi: 10.1016/j.atherosclerosis.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 91.Tice JA, Kazi DS, Pearson SD. Proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors for treatment of high cholesterol levels: effectiveness and value. JAMA Intern Med. 2016;176(1):107–108. doi: 10.1001/jamainternmed.2015.7248. [DOI] [PubMed] [Google Scholar]
- 92.Yang X, Lee SR, Choi YS, Alexander VJ, Digenio A, Yang Q, et al. Reduction in lipoprotein-associated apoC-III levels following volanesorsen therapy: Phase 2 randomized trial results. J Lipid Res. 2016;57(4):706–713. doi: 10.1194/jlr.M066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363(23):2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reiner Z, Guardamagna O, Nair D, Soran H, Hovingh K, Bertolini S, et al. Lysosomal acid lipase deficiency—an under-recognized cause of dyslipidaemia and liver dysfunction. Atherosclerosis. 2014;235(1):21–30. doi: 10.1016/j.atherosclerosis.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 95.Burton BK, Balwani M, Feillet F, Baric I, Burrow TA, Camarena Grande C, et al. A phase 3 trial of sebelipase alfa in lysosomal acid lipase deficiency. N Engl J Med. 2015;373(11):1010–1020. doi: 10.1056/NEJMoa1501365. [DOI] [PubMed] [Google Scholar]