Abstract
Geographical gradients in selection can shape different genetic architectures in natural populations, reflecting potential genetic constraints for adaptive evolution under climate change. Investigation of natural pH/pCO2 variation in upwelling regions reveals different spatio-temporal patterns of natural selection, generating genetic and phenotypic clines in populations, and potentially leading to local adaptation, relevant to understanding effects of ocean acidification (OA). Strong directional selection, associated with intense and continuous upwellings, may have depleted genetic variation in populations within these upwelling regions, favouring increased tolerances to low pH but with an associated cost in other traits. In contrast, diversifying or weak directional selection in populations with seasonal upwellings or outside major upwelling regions may have resulted in higher genetic variances and the lack of genetic correlations among traits. Testing this hypothesis in geographical regions with similar environmental conditions to those predicted under climate change will build insights into how selection may act in the future and how populations may respond to stressors such as OA.
Keywords: ocean acidification, adaptive evolution, genetic correlations, trade-offs, phenotypic evolution
1. Introduction
Ocean acidification (OA; i.e. increased pCO2 and reduced pH and saturation state of CaCO3-Ω-in the oceans [1]) is expected to be an important driver of phenotypic change [2] and an increasingly important agent of natural selection for many marine organisms [3]. The manifold selective pressures associated with OA are likely to induce microevolutionary changes in natural populations [4], and therefore genetic variation will be essential for population persistence in a changing ocean [5]. Because most populations possess some genetic variation for most characters, the potential for evolutionary change is ubiquitous and has been documented in a range of marine species [6]. However, standing genetic variation, as well as the strength and direction of selection, can vary among populations across the geographical range of species [7]. Thus, natural populations may differ in their adaptive potential, depending on their particular environmental contexts and genetic backgrounds [5]. Variation in the strength, as opposed to direction, of selection among populations is the dominant feature of geographical gradients in natural selection [8]. Harsher environments are generally associated with strong selection regimes that deplete genetic variance and favour resistant genotypes [7]. In contrast, more favourable environments can generate weaker selection and populations may exhibit higher genetic variance [7,9]. Importantly, both genetic variation and selection determine the magnitude of evolutionary responses: when selection and genetic variation are well aligned, evolution will be rapid; when they are not well aligned, evolution will be slow [10]. In this context, genetic correlations among traits under selection become important. Similar to genetic variances, genetic covariances are also shaped in part by local environmental conditions [11], and hence, selection may impact their sign and strength [12]. Understanding how geographical gradients in past selection have shaped the genetic architecture of marine populations along heterogeneous environments (e.g. geographical mosaics of pH/pCO2) can help inform how selection may change in the future and thus how populations may respond to OA.
2. Spatial variation in selection and phenotypic evolution under ocean acidification
Quantitative genetics provides a powerful framework to understand evolutionary dynamics under climate change [4,11], allowing comparison of the strength, direction and form of selection among populations [8]. In this framework, the evolutionary response to selection of a multivariate phenotype ( ) is expected to be biased away from the direction of selection (β) by the magnitude and orientation of the main axis of the genetic (co)variance matrix (G) (i.e. gmax or the line of least resistance, figure 1a,b; more details in the electronic supplementary material). Because both observed and predicted climate change are not uniform across the species range [13], we can expect differences among populations in
) is expected to be biased away from the direction of selection (β) by the magnitude and orientation of the main axis of the genetic (co)variance matrix (G) (i.e. gmax or the line of least resistance, figure 1a,b; more details in the electronic supplementary material). Because both observed and predicted climate change are not uniform across the species range [13], we can expect differences among populations in  generated not only by spatial variation in β, but also by spatial variation in G (figure 1b) [8,10]. From this perspective, populations can evolve towards the same fitness peak with different trajectories and rates (figure 1b), or towards alternate fitness peaks as a result of divergent selection (figure 1c) [8,10]. In both cases, genetic associations among traits (correlations and covariances) have the potential to either facilitate or constrain
generated not only by spatial variation in β, but also by spatial variation in G (figure 1b) [8,10]. From this perspective, populations can evolve towards the same fitness peak with different trajectories and rates (figure 1b), or towards alternate fitness peaks as a result of divergent selection (figure 1c) [8,10]. In both cases, genetic associations among traits (correlations and covariances) have the potential to either facilitate or constrain  and thereby play a causal role in the phenotypic evolution of natural populations [9,12]. Genetic constraints can arise from both positive and negative genetic associations among traits (figure 1b). For example, positive genetic correlations can act as genetic constraints by slowing
and thereby play a causal role in the phenotypic evolution of natural populations [9,12]. Genetic constraints can arise from both positive and negative genetic associations among traits (figure 1b). For example, positive genetic correlations can act as genetic constraints by slowing  if interacting traits are selected in opposite directions (i.e. small values of one trait and large values of the other), whereas negative genetic correlations can induce similar constraints if selection favours high values of both traits [9,11].
if interacting traits are selected in opposite directions (i.e. small values of one trait and large values of the other), whereas negative genetic correlations can induce similar constraints if selection favours high values of both traits [9,11].
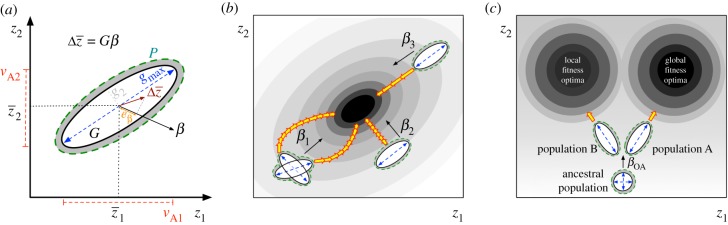
Figure 1.
(a) Phenotypic (P) and genetic (G) (co)variance matrices of two hypothetical traits (z), depicting population means ( ), additive genetic variances (vA), the line of least resistance (gmax), the selection gradient (β, black arrow) and the amount of the predicted response to selection (
), additive genetic variances (vA), the line of least resistance (gmax), the selection gradient (β, black arrow) and the amount of the predicted response to selection ( ) that occurs exactly in the direction of actual selection (eβ). (b) A hypothetical adaptive landscape (grey gradient) with the adaptive optima (peak) in the black oval. The four G matrices exhibit different (co)variance patterns and directions of the two major axes of variation (gmax and g2), so
) that occurs exactly in the direction of actual selection (eβ). (b) A hypothetical adaptive landscape (grey gradient) with the adaptive optima (peak) in the black oval. The four G matrices exhibit different (co)variance patterns and directions of the two major axes of variation (gmax and g2), so  is predicted to: (i) deviate from selection (strong constraint, β1 for two populations with same
is predicted to: (i) deviate from selection (strong constraint, β1 for two populations with same  and G but different sign in covariance); (ii) show little response, because selection acts orthogonal to gmax (moderate constraint, β2) and (iii) show fast change as selection is aligned to gmax (unconstrained, β3). Yellow arrows represent the direction and speed of phenotypic change. (c) Adaptive divergence between two populations with different orientation of G. Evolutionary trajectories are biased by gmax, leading population A (positive covariance) and population B (negative covariance) towards different fitness optima (modified from [10]).
and G but different sign in covariance); (ii) show little response, because selection acts orthogonal to gmax (moderate constraint, β2) and (iii) show fast change as selection is aligned to gmax (unconstrained, β3). Yellow arrows represent the direction and speed of phenotypic change. (c) Adaptive divergence between two populations with different orientation of G. Evolutionary trajectories are biased by gmax, leading population A (positive covariance) and population B (negative covariance) towards different fitness optima (modified from [10]).
In the context of OA, empirical studies have shown that functional and genetic associations among traits can produce correlated responses to selection under elevated pCO2 [14,15]. In the short-term (i.e. within generations), those correlated responses that result from phenotypic plasticity (e.g. changes in energy allocation) are likely to be more important than evolutionary responses, buffering populations against selection and reducing  with only transient physiological costs [14]. However, in the long-term (i.e. among generations), correlated responses from genetic associations (caused by pleiotropy or linkage disequilibrium) are more likely to influence the phenotypic evolution of natural populations [11,12]. Despite their importance, only few examples of correlated genetic responses and evolutionary constraints (trade-offs) to OA have been described, mostly with phytoplankton species [16]. This is perhaps because genetic associations among traits underlying correlated responses are difficult to identify and measure in natural populations [12]. Further research is needed in this area, considering additional model systems and their biogeographic contexts.
with only transient physiological costs [14]. However, in the long-term (i.e. among generations), correlated responses from genetic associations (caused by pleiotropy or linkage disequilibrium) are more likely to influence the phenotypic evolution of natural populations [11,12]. Despite their importance, only few examples of correlated genetic responses and evolutionary constraints (trade-offs) to OA have been described, mostly with phytoplankton species [16]. This is perhaps because genetic associations among traits underlying correlated responses are difficult to identify and measure in natural populations [12]. Further research is needed in this area, considering additional model systems and their biogeographic contexts.
3. Upwelling systems and adaptive evolution
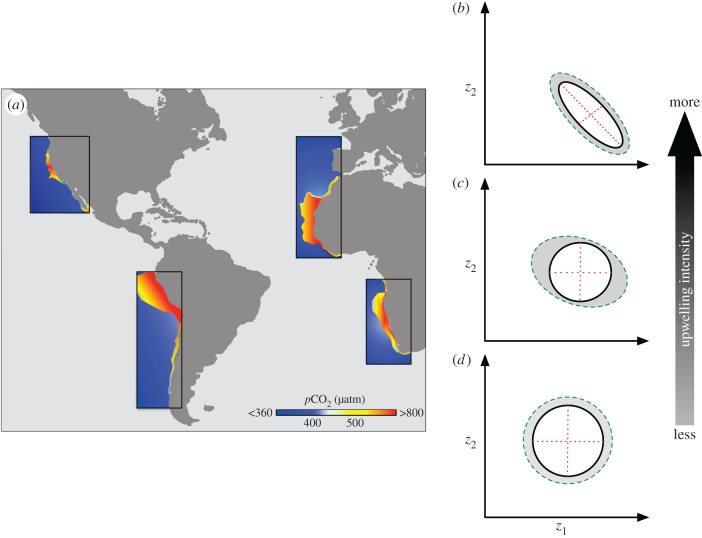
To understand and predict  to OA, it is instructive to consider how organisms cope with natural variations in CO2 and how past selection has shaped the genetic architecture of natural populations in their current environments. In this sense, present-day coastal upwelling systems (e.g. EBUS, figure 2a) represent natural laboratories to explore adaptive evolution in response to low pH and its interaction with other environmental stressors [6,17]. These systems occur over large spatial scales (figure 2a) characterized by strong alongshore variation in temperature, nutrients, pH, oxygen and chlorophyll [18]. The spatio-temporal dynamic of these regions (i.e. subregions with stronger and more continuous upwellings through the year versus subregions with weaker seasonal upwellings) can create complex geographical mosaics [17], offering the possibility to explore selection gradients and local adaptation by comparing sets of populations with differentials in performance and fitness (‘hot and cool spots’; see [17]) that have evolved under similar or different environmental conditions [19]. From an evolutionary perspective, the strength, direction and form of selection are likely to differ along upwelling systems, revealing geographical gradients in selection that may explain the spatial differences in tolerances to low pH and its variability, among populations of some marine species [20]. Historical intense and continuous upwellings may have produced directional selection for resistance to low pH [3], reducing the genetic variation available to selection [21] and creating strong negative genetic correlations among traits (figure 2b). However, not all past selection pressures in upwelling regions have been directional, and genetic correlations that were built in the past may have decayed rapidly over time if selection was relaxed [11]. This could be the case for populations in seasonal upwellings in which both diversifying and fluctuating selection may have been primary drivers of microevolutionary change, acting upon the maintenance of alternatively adaptive phenotypes (higher genetic diversity) [6] or favouring highly plastic phenotypes with broad tolerance to pH variation [22] (figure 2c). In contrast, in more stable pH environments outside of upwelling regions, the absence or weak selection for low-pH and variable-pH resistance may have allowed populations to maintain significant genetic variation to persist in these areas without inducing genetic correlations (figure 2d). This spatio-temporal variation in selection along upwelling systems may have resulted in genotype × environmental interactions in fitness, leading to local adaptation and potential trade-offs (i.e. specialist phenotypes optimally adapted in one habitat and poorly adapted to other habitats) as a result of spatially divergent selection [19]. Although gene flow can impede adaptive divergence [8], local adaptation may still occur if phenotype-specific mortality occurs after dispersal [23].
to OA, it is instructive to consider how organisms cope with natural variations in CO2 and how past selection has shaped the genetic architecture of natural populations in their current environments. In this sense, present-day coastal upwelling systems (e.g. EBUS, figure 2a) represent natural laboratories to explore adaptive evolution in response to low pH and its interaction with other environmental stressors [6,17]. These systems occur over large spatial scales (figure 2a) characterized by strong alongshore variation in temperature, nutrients, pH, oxygen and chlorophyll [18]. The spatio-temporal dynamic of these regions (i.e. subregions with stronger and more continuous upwellings through the year versus subregions with weaker seasonal upwellings) can create complex geographical mosaics [17], offering the possibility to explore selection gradients and local adaptation by comparing sets of populations with differentials in performance and fitness (‘hot and cool spots’; see [17]) that have evolved under similar or different environmental conditions [19]. From an evolutionary perspective, the strength, direction and form of selection are likely to differ along upwelling systems, revealing geographical gradients in selection that may explain the spatial differences in tolerances to low pH and its variability, among populations of some marine species [20]. Historical intense and continuous upwellings may have produced directional selection for resistance to low pH [3], reducing the genetic variation available to selection [21] and creating strong negative genetic correlations among traits (figure 2b). However, not all past selection pressures in upwelling regions have been directional, and genetic correlations that were built in the past may have decayed rapidly over time if selection was relaxed [11]. This could be the case for populations in seasonal upwellings in which both diversifying and fluctuating selection may have been primary drivers of microevolutionary change, acting upon the maintenance of alternatively adaptive phenotypes (higher genetic diversity) [6] or favouring highly plastic phenotypes with broad tolerance to pH variation [22] (figure 2c). In contrast, in more stable pH environments outside of upwelling regions, the absence or weak selection for low-pH and variable-pH resistance may have allowed populations to maintain significant genetic variation to persist in these areas without inducing genetic correlations (figure 2d). This spatio-temporal variation in selection along upwelling systems may have resulted in genotype × environmental interactions in fitness, leading to local adaptation and potential trade-offs (i.e. specialist phenotypes optimally adapted in one habitat and poorly adapted to other habitats) as a result of spatially divergent selection [19]. Although gene flow can impede adaptive divergence [8], local adaptation may still occur if phenotype-specific mortality occurs after dispersal [23].
Figure 2.
(a) pCO2 patterns along eastern boundary upwelling systems (EBUS). (b–d) Variation in G for hypothetical traits (z) along an upwelling cline characterized by differences in the intensity and variability of pH/pCO2.
Considering our hypothetic cline of genetic architectures and selection regimes, we expect that natural populations along upwelling systems would be able to cope with projected OA using several different strategies (i.e. low-pH-resistant phenotypes, alternative adaptive phenotypes and plastic phenotypes). However, there are some caveats in these expectations as coastal upwelling regions are vulnerable to amplifying the global trends of OA [18], perhaps exceeding the evolutionary potential of some populations. Changes in the magnitude of selection pressures associated with OA can strongly affect those populations with low genetic variation and genetic associations among fitness-related traits (figure 2b), because they are already at their evolutionary limit. On the other hand, the exacerbated effects of OA may also affect populations in which past fluctuating selection has favoured highly plastic phenotypes that are not resistant to chronic, persistent low-pH waters. If these populations have enough genetic variation, selection will change the magnitude and orientation of their genetic architectures in response to OA. More accurate predictions about potentials and constraints for adaptive evolution to climate change will require the use of complementary approaches (e.g. reciprocal transplant and multi-generational experiments) involving geographical patterns of co-occurrence and interactions among multiple environmental stressors [17] in order to understand how selection is acting on natural populations along broad spatial scales.
Supplementary Material
Authors' contributions
J.D.G.-E. wrote the manuscript with inputs from co-authors. All authors agree to be held accountable for the content and approve the final version of the manuscript.
Competing interests
We have no competing interests.
Funding
J.D.G.-E. was supported by a CSIRO-OCE post-doctoral fellowship.
References
- 1.Caldeira K, Wickett M. 2003. Anthropogenic carbon and ocean pH. Nature 425, 2003 ( 10.1038/425365a) [DOI] [PubMed] [Google Scholar]
- 2.Reusch TBH. 2014. Climate change in the oceans: evolutionary versus phenotypically plastic responses of marine animals and plants. Evol. Appl. 7, 104–122. ( 10.1111/eva.12109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pespeni M, et al. 2013. Evolutionary change during experimental ocean acidification. Proc. Natl Acad. Sci. USA 110, 6937–6942. ( 10.1073/pnas.1220673110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sunday J, Calosi P, Dupont S, Munday P, Stillman J, Reusch T. 2013. Evolution in an acidifying ocean. Trends Ecol. Evol. 29, 117–125. ( 10.1016/j.tree.2013.11.001) [DOI] [PubMed] [Google Scholar]
- 5.Lande R, Shannon S. 1996. The role of genetic variation in adaptation and population persistence in a changing environment. Evolution (NY) 50, 434–437. ( 10.2307/2410812) [DOI] [PubMed] [Google Scholar]
- 6.Pespeni M, Chan F, Menge B, Palumbi S. 2013. Signs of adaptation to local pH conditions across an environmental mosaic in the California current ecosystem. Integr. Comp. Biol. 53, 857–870. ( 10.1093/icb/ict094) [DOI] [PubMed] [Google Scholar]
- 7.Wilson A, Pemberton JM, Pilkington JG, Coltman DW, Mifsud DV, Clutton-Brock TH, Kruuk LEB. 2006. Environmental coupling of selection and heritability limits evolution. PLoS Biol. 4, 1270–1275. ( 10.1371/journal.pbio.0040216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siepielski A, Gotanda KM, Morrissey MB, Diamond SE, DiBattista JD, Carlson SM. 2013. The spatial patterns of directional phenotypic selection. Ecol. Lett. 16, 1382–1392. ( 10.1111/ele.12174) [DOI] [PubMed] [Google Scholar]
- 9.Etterson J, Shaw R. 2001. Constraint to adaptive evolution in response to global warming. Science 294, 151–154. ( 10.1126/science.1063656) [DOI] [PubMed] [Google Scholar]
- 10.Teplitsky C, Robinson M, Merilä J. 2014. Evolutionary potential and constraints in wild populations. In Quantitative genetics in the wild (eds Charmantier A, Garant D, Kruuk L), pp. 190–208. Oxford, UK: Oxford University Press. [Google Scholar]
- 11.Walsh B, Blows MW. 2009. Abundant genetic variation+strong selection=multivariate genetic constraints: a geometric view of adaptation. Annu. Rev. Ecol. Evol. Syst. 40, 41–59. ( 10.1146/annurev.ecolsys.110308.120232) [DOI] [Google Scholar]
- 12.Sgrò C, Hoffmann A. 2004. Genetic correlations, tradeoffs and environmental variation. Heredity Edinb. 93, 241–248. ( 10.1038/sj.hdy.6800532) [DOI] [PubMed] [Google Scholar]
- 13.Etterson JR. 2004. Evolutionary potential of Chamaecrista fasciculata in relation to climate change. I. Clinal patterns of selection along an environmental gradient in the Great Plains. Evolution (NY) 58, 1446–1456. ( 10.1111/j.0014-3820.2004.tb01726.x) [DOI] [PubMed] [Google Scholar]
- 14.Dupont S, Dorey N, Stumpp M, Melzner F, Thorndyke M. 2013. Long-term and trans-life-cycle effects of exposure to ocean acidification in the green sea urchin Strongylocentrotus droebachiensis. Mar. Biol. 160, 1835–1843. ( 10.1007/s00227-012-1921-x) [DOI] [Google Scholar]
- 15.Lohbeck K, Riebesell U, Reusch T. 2012. Adaptive evolution of a key phytoplankton species to ocean acidification. Nat. Geosci. 5, 346–351. ( 10.1038/ngeo1441) [DOI] [Google Scholar]
- 16.Schlüter L, Lohbeck KT, Gutowska MA, Gröger JP, Riebesell U, Reusch TBH. 2014. Adaptation of a globally important coccolithophore to ocean warming and acidification. Nat. Clim. Change 4, 1024–1030. ( 10.1038/nclimate2379) [DOI] [Google Scholar]
- 17.Kroeker K, et al. 2016. Interacting environmental mosaics drive geographic variation in mussel performance and predation vulnerability. Ecol. Lett. 19, 771–779. ( 10.1111/ele.12613) [DOI] [PubMed] [Google Scholar]
- 18.Feely R, Sabine C, Hernandez-Ayon J, Ianson D, Hales B. 2008. Evidence for upwelling of corrosive ‘acidified’ water onto the continental shelf. Science 320, 1490–1492. ( 10.1126/science.1155676) [DOI] [PubMed] [Google Scholar]
- 19.Kawecki T, Ebert D. 2004. Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241. ( 10.1111/j.1461-0248.2004.00684.x) [DOI] [Google Scholar]
- 20.Padilla-Gamiño J, Gaitan-Espitia J, Kelly M, Hofmann G. 2016. Physiological plasticity and local adaptation to elevated pCO2 in calcareous algae: an ontogenetic and geographic approach. Evol. Appl. 9, 1043–1053. ( 10.1111/eva.12411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd M, Makukhov A, Pespeni M. 2016. Loss of genetic diversity as a consequence of selection in response to high pCO2. Evol. Appl. 9, 1124–1132. ( 10.1111/eva.12404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Futuyma D, Moreno G. 1988. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19, 207–233. ( 10.1146/annurev.es.19.110188.001231) [DOI] [Google Scholar]
- 23.Marshall D, Monro K, Bode M, Keough M, Swearer S. 2010. Phenotype–environment mismatches reduce connectivity in the sea. Ecol. Lett. 13, 128–140. ( 10.1111/j.1461-0248.2009.01408.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.