Abstract
Quantifying the genetic diversity in natural populations is crucial to address ecological and evolutionary questions. Despite recent advances in whole-genome sequencing, microsatellite markers have remained one of the most powerful tools for a myriad of population genetic approaches. Here, we used the 454 sequencing technique to develop microsatellite loci in the fire coral Millepora platyphylla, an important reef-builder of Indo-Pacific reefs. We tested the cross-species amplification of these loci in five other species of the genus Millepora and analysed its success in correlation with the genetic distances between species using mitochondrial 16S sequences. We succeeded in discovering fifteen microsatellite loci in our target species M. platyphylla, among which twelve were polymorphic with 2–13 alleles and a mean observed heterozygosity of 0.411. Cross-species amplification in the five other Millepora species revealed a high probability of amplification success (71%) and polymorphism (59%) of the loci. Our results show no evidence of decreased heterozygosity with increasing genetic distance. However, only one locus enabled measures of genetic diversity in the Caribbean species M. complanata due to high proportions of null alleles for most of the microsatellites. This result indicates that our novel markers may only be useful for the Indo-Pacific species of Millepora. Measures of genetic diversity revealed significant linkage disequilibrium, moderate levels of observed heterozygosity (0.323–0.496) and heterozygote deficiencies for the Indo-Pacific species. The accessibility to new polymorphic microsatellite markers for hydrozoan Millepora species creates new opportunities for future research on processes driving the complexity of their colonisation success on many Indo-Pacific reefs.
Keywords: Genetic diversity, Genetic distance, Cross-species transferability, Microsatellites, Millepora
Introduction
Coral reefs are increasingly threatened by chronic and acute stressors (Bellwood et al., 2004) and are expected to be highly vulnerable to future climate change due to rapidly increasing sea surface temperatures and ocean acidification (Hoegh-Guldberg et al., 2007; Pandolfi et al., 2011; Kuffner et al., 2015). These anthropogenic disturbances can further change the biodiversity in coral reefs and may hamper their capacity to deliver important sources of ecosystem services to millions of people (Wilkinson, 2008; Cardinale et al., 2012). The capacity of reef organisms to survive and adapt to such environmental changes will partially depend on their levels of genetic diversity, which is key for a species’ ability to persist in changing environments (Frankham, 2005; Barrett & Schluter, 2008; Hoffmann & Sgrò, 2011). Many studies have focused on elucidating the underlying mechanisms of the origin and maintenance of genetic variation in populations of scleractinian corals as they provide much of the habitat framework and structural complexity of reefs (e.g., Baums, Miller & Hellberg, 2005; Baums, 2008; Davies, Treml & Matz, 2015).
For long-live sessile organisms, such as reef-building corals, patterns of genetic diversity at both local and global scales are highly governed by the dispersal of sexual larvae (Baird, Guest & Willis, 2009; Harrison, 2011). Molecular studies have uncovered a wide range of dispersal patterns in scleractinian corals, from populations primarily sustained by self-recruitment (Gilmour, Smith & Brinkman, 2009; Mokhtar-Jamaï et al., 2013) through ecologically significant gene flow and connectivity among their populations (Van der Ven et al., 2016; Lukoschek, Riginos & Van Oppen, 2016). Furthermore, the degree of genetic variation in partially clonal reef organisms is heavily influenced by the relative contribution from sexual and asexual reproduction for local population maintenance (e.g., Baums, Miller & Hellberg, 2006; Pinzón et al., 2012; Adjeroud et al., 2014). While our understanding of population genetics in scleractinian corals has improved considerably over the last decade, such information remains unavailable for Millepora hydrocorals (‘fire corals’).
Millepora hydrocorals are an important component of reef communities worldwide where they, similar to scleractinian corals, significantly contribute to reef accretion (Nagelkerken & Nagelkerken, 2004; Lewis, 2006). Although fire corals compete with other reef-building taxa (Wahle, 1980; Dubé, Boissin & Planes, 2016), they also favour coral survival during Acanthaster outbreaks, highlighting their key ecological role in reef resilience (Kayal & Kayal, 2016). Despite their major importance for reef conservation, fire corals have been relatively understudied and not much is known with respect to their genetic diversity, population structure or life history (e.g., reproductive strategies). Few studies have shown that Millepora species can colonise a wide range of reef environments via both sexual propagules (Lewis, 2006; Bourmaud et al., 2013) and asexual reproduction through fragmentation (Edmunds, 1999; Lewis, 2006). While they are sessile and have limited tolerance to environmental changes (Lewis, 2006), species of Milleporidae have a wide distribution range, i.e., circumtropical (Boschma, 1956). Fire corals are also known for their extensive morphological variability, which has caused problems in resolving their systematics (Boschma, 1948). There is currently no agreement regarding the number of Millepora species and no phylogenetic study investigating the genetic relationships among them (but see Ruiz-Ramos, Weil & Schizas, 2014 for a species complex in the Caribbean). Although microsatellite loci have been identified in Millepora alcicornis (Ruiz-Ramos & Baums, 2014), there was a lack of highly variable genetic markers for this genus until very recently (but see Heckenhauer et al., 2014). The development of new molecular markers for Millepora species will increase our knowledge on the genetic diversity of a conspicuous reef-building organism across its geographic range. These microsatellite markers will enable further studies on the biological, ecological and evolutionary processes underlying the population persistence of Millepora hydrocorals.
Microsatellites, also known as simple sequence repeats (SSRs) or short tandem repeats (STRs), have emerged as one of the most powerful genetic markers in population and evolutionary genetics (Selkoe & Toonen, 2006). Improvements in next generation sequencing techniques have provided new opportunities for microsatellite isolation in non-model organisms (i.e., with no genetic information available) (Zhang et al., 2011), with a good representativeness of loci across the genome (Martin et al., 2010). Because microsatellites are codominant (Estoup et al., 1993), highly polymorphic (Schlötterer, 2000) and transferable among closely related species (Cheng et al., 2012), they are commonly used for a remarkable array of applications, such as inferring genetic diversity (Silva & Gardner, 2015; Nakajima et al., 2016) and population structure patterns (Noreen, Harrison & Van Oppen, 2009; Boissin et al., 2016), evaluating reproductive strategies (Baums et al., 2014; Ardehed et al., 2015) and parentage screening (Mourier & Planes, 2013; Warner, Willis & Van Oppen, 2016). Cross-species transferability has been successful in many species (Barbará et al., 2007; Reid, Hoareau & Bloomer, 2012; Maduna et al., 2014; Pirog et al., 2016) allowing for genetic studies in closely related species. However, the few studies that have investigated the efficiency of cross-species transferability of microsatellite loci have demonstrated a negative correlation between the genetic distance and the amplification success (Carreras-Carbonell, Macpherson & Pascual, 2008; Hendrix et al., 2010; Moodley et al., 2015). This constraint can hamper accurate comparisons of genetic diversity among more distantly related species.
Here, we used 454 GS-FLX sequencing technology to develop an additional set of de novo microsatellite markers for Millepora platyphylla to first assess its genetic diversity on Moorea’s reefs in French Polynesia. Secondly, we tested these new microsatellite loci for cross-species amplification in five other Millepora species: the branching Millepora intricata, Millepora dichotoma and Millepora tenera, the plate-like species Millepora complanata and the encrusting Millepora exaesa (Boschma, 1948). Lastly, genetic distances based on the 16S mitochondrial gene were estimated among these species and M. platyphylla to identify the transferability success of these newly developed microsatellites across the Milleporidae.
Materials & Methods
Preparation of genomic DNA for 454 sequencing
The calcification processes (Stanley, 2006) and metabolic pathways (Trench, 1979) of calcareous hydrozoans are supported by a symbiotic association with protozoan dinoflagellate algae of the genus Symbiodinium. To design species-specific markers, genomic DNA of Symbiodinium was removed from the animal tissue using a succession of extraction techniques. Candidate microsatellite repeats were isolated from a pool of 14 partially bleached fragments of M. platyphylla collected in situ on Moorea’s reefs to minimise the quantity of Symbiodinium in their tissues. Further mechanic (centrifugation) and genetic (positive and negative controls in PCR) techniques were applied to ensure microsatellites belonged to the animal only (see below). Fragments were homogenised in 1,000 µL of CHAOS buffer (4 M guanidine thiocyanate; 0.5% N-lauroyl sarcosine; 25 nM Tris–HCl pH 8; 0.1 M 2-mercaptoethanol) modified from Fukami et al. (2004). Samples were incubated at 60 °C for 2 h while rotating and then centrifuged at 1,500 rpms for 30 s to precipitate symbiont algae expelled from host cells. A total of 20 µL of the aqueous phase was examined under microscope to confirm the absence of Symbiodinium. Further potential contamination was tested by running microsatellites on pure cultures of zooxanthellae DNA (see below). 350 µL of CHAOS solution containing animal tissues was transferred to a new vial and 350 µL PEB (protein extraction buffer) was added (100 mM Tris pH 8; 10 mM ethylenediaminetetraacetic acid (EDTA); 0.1% Sodium dodecyl sulfate (SDS)). DNA was purified with phenol/chloroform (24:1) and precipitated with isopropanol as described by Mieog et al. (2009). Samples from the 14 colonies were pooled together to increase detection of polymorphism. A total of 1 µg of genomic DNA was sent to GenoScreen platform (Lille, France) for the development of the microsatellite library using 454 GS-FLX Titanium reagents as described in Malausa et al. (2011). Briefly, total DNA was mechanically fragmented and enriched for TG, TC, AAC, AAG, AGG, ACG, ACAT and ACTC repeat motifs. Enriched fragments were subsequently amplified and PCR products were purified and quantified. GS-FLX libraries were then carried out following manufacturer’s protocols and sequenced on a GS-FLX PTP. The Quality Detection Device (QDD) pipeline (Meglécz et al., 2010) was used to analyse the 454 sequences and to design primers for amplification of the detected microsatellite motifs. Primer pairs were then selected depending on the motif (di-, tri-, tetranucleotide), the number of repeats (≥5) and the product size (≥100 bp) and tested on agarose gels for amplification.
Microsatellite discovery and primer testing
A panel of 16 M. platyphylla colonies was used to optimise PCR amplification and identify polymorphic loci. Small fragments of tissue-covered skeleton (<2cm3) were incubated at 55 °C for 1 h in 450 µL of digest buffer with proteinase K (QIAGEN, Hilden, Germany). Genomic DNA was extracted using a QIAxtractor automated genomic DNA extraction instrument, according to manufacturer’s instructions. PCRs were performed in a final volume of 10 µL including 5 µL Type-it Multiplex PCR Master Mix (1×) (QIAGEN, Hilden, Germany), 3 µL RNase-free water, 1 µL primers (2 µM for both forward and reverse primers diluted in TE buffer) and 1 µL of template (10–50 ng/µL). The PCR program included an initial denaturing step of 5 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 90 s at optimal temperature (57–60 °C) depending on the microsatellite locus (see Table 1), and 30 s at 72 °C, followed by a final 30 min elongation step at 60 °C. The PCR products were electrophoresed on 2% agarose gels. For loci with high-quality and consistent amplification, the PCR was repeated on DNA template isolated from Symbiodinium strains (clade A to F identified based on 23S chloroplast rDNA, Table S1) to identify coral specific loci and to exclude putative Symbiodinium specific loci. Symbiodinium strains were provided by the BURR laboratory at Buffalo, US (BURR; http://www.nsm.buffalo.edu/Bio/burr/). For the loci that are specific to Millepora, the forward primer was fluorescently labelled with the G5 dye set including 6-FAM, VIC, NED and PET (Applied Biosystems, Foster City, CA). Amplified fragments were visualised on an Applied Biosystems 3730 Sequencer using a GeneScan 500 LIZ ladder.
Table 1. Characterisation of de novo microsatellite loci and genetic diversity in the target species Millepora platyphylla collected in Moorea, French Polynesia.
| Locus name | Primer Sequence 5′–3′ | Dye | MP | Motif | TA (°C) | GenBank accession | N | Size (bp) | Null | LD | Na | Ho | He | FIS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mill07 | F: TAGTACATCGGGCATGAGCA | 6-FAM | 3 | (CA)16 | 57 | KX670763 | 50 | 92–144 | – | – | 13 | 0.760 | 0.855 | 0.121∗ |
| R: GTACTCTACGGCGTGTGCGT | ||||||||||||||
| Mill27 | R: CTTTCGTTTCCGATCATTCC | VIC | 3 | (TG)10 | 57 | KX670764 | 50 | 136–148 | – | 0.044 | 5 | 0.600 | 0.636 | 0.067 |
| R: TGCCAGAACTAAGTTATCACAGC | ||||||||||||||
| Mill30 | F: AGTTGGCTCTGAGTGCGAGT | NED | 2 | (TG)11 | 57 | KX670765 | 50 | 203–211 | – | 0.025 | 4 | 0.680 | 0.648 | −0.039 |
| R: CCTCGGTTTATGGCTGAGAT | ||||||||||||||
| Mill47 | F: AAGCGTGTAATGCACTCAAAGA | NED | 2 | (GA)8 | 57 | KX670766 | 50 | 118–162 | 0.101 | 0.057 | 10 | 0.600 | 0.766 | 0.227∗∗ |
| R: AACAGAAGTCGAACTGAGTCAAAA | ||||||||||||||
| Mill52 | F: CCCTGAGGCATCGAAATATAA | 6-FAM | 1 | (AC)9 | 60 | KX670767 | 50 | 94–98 | – | – | 2 | 0.420 | 0.412 | −0.010 |
| R: TGCAATTGATGGTATTTGCATT | ||||||||||||||
| Mill56 | F: TCTGCAGATTTTGCATCTCG | PET | 1 | (AGA)6 | 60 | KX670768 | 50 | 194 | – | – | 1 | 0 | 0 | – |
| R: TAGCAACAATGCTTCGCTGA | ||||||||||||||
| Mill61 | F: AAATGAACTCGCCCAAAAGA | PET | 4 | (CAA)7 | 57 | KX670769 | 50 | 163–166 | – | 0.048 | 2 | 0.480 | 0.467 | 0.044 |
| R: ACACTGTCGATTGTGTTCCAA | ||||||||||||||
| Mill67 | F: TTGCGAGTTTACTTACCAGGC | VIC | 1 | (TAGA)6 | 60 | KX670770 | 50 | 259–359 | 0.144 | 0.039 | 11 | 0.420 | 0.588 | 0.294∗∗ |
| R: TGAAGCAAATGACAAGAGCAA | ||||||||||||||
| Mill86 | F: GCGCGAAAATAAATTAAGGAA | NED | 4 | (GTT)5 | 57 | KX670771 | 50 | 106 | – | – | 1 | 0 | 0 | – |
| R: TCCAATCTGAATTCCACCCT | ||||||||||||||
| Mill91 | F: CACTTTCGCCATTGTTGCTA | PET | 4 | (CAA)6 | 57 | KX670772 | 50 | 116 | – | – | 1 | 0 | 0 | – |
| R: AACGGAATTCGAATCATTGC | ||||||||||||||
| Mill93 | F: TGAAATTTTCCAGTGACATCAAA | 6-FAM | 2 | (TGT)7 | 57 | KX670773 | 50 | 91–100 | – | 0.055 | 3 | 0.260 | 0.339 | 0.243 |
| R: GCTAATTATGAAATAGCAACTCCTAAA | ||||||||||||||
| Mill94 | F: GCATAAAGAATAAAGCAGAGGCA | 6-FAM | 3 | (GAA)7 | 57 | KX670774 | 50 | 131–140 | – | 0.016 | 2 | 0.480 | 0.461 | −0.032 |
| R: CAATTGTGGGGAAGTTCGTT | ||||||||||||||
| Mill95 | F: TCCATAGCTTCTGCCTCCTC | 6-FAM | 1 | (TTG)7 | 60 | KX670775 | 50 | 123–138 | – | 0.022 | 3 | 0.320 | 0.304 | −0.042 |
| R: GCTGATGATGCTGTCGAAGA | ||||||||||||||
| Mill101 | F: AGTCCTTCAATTGGTGGGTG | PET | 2 | (CAA)6 | 57 | KX670776 | 50 | 132–135 | – | – | 2 | 0.640 | 0.493 | −0.289 |
| R: GAGATGATGACTGAGCAGCAG | ||||||||||||||
| Mill103 | F: TTAAAGCCAGAGACAGAGAGACA | VIC | 3 | (AG)7 | 57 | KX670777 | 50 | 94–100 | – | 0.017 | 4 | 0.700 | 0.621 | −0.117 |
| R: ATCAACAGTTTCCCCTGTGC |
Notes.
- MP
- multiplex panel in which each locus was included
- TA
- primer temperature annealing
- N
- number of individuals with reliable amplification
- Null
- proportion of null alleles
- LD
- proportion of allele comparisons showing significant linkage disequilibrium (P < 0.05)
- Na
- number of alleles
- Ho
- observed heterozygosity
- He
- expected heterozygosity
- FIS
- inbreeding coefficient
Significant values of FIS are indicated by bold values with ∗P < 0.05 and ∗∗P < 0.001.
Sampling, genotyping and cross-species amplification
The optimised loci were genotyped in our target species in addition to five other Millepora species to test for their transferability. For the characterisation of newly developed microsatellites, small fragments (<2 cm3) from 50 M. platyphylla colonies were collected on the reefs of Moorea in French Polynesia (CITES - FR1298700028-E). For cross-species amplification transferability tests, samples were collected from various locations in the Indo-Pacific and the Caribbean for five other species of fire corals: 11 M. intricata in Papua New Guinea, 30 M. dichotoma in Europa Island (Mozambique Canal), 30 M. tenera and 14 M. exaesa both in Reunion Island and 30 M. complanata in Curaçao (Table S2). DNA from the 165 Millepora colonies was extracted as described above and optimised loci were combined in four multiplex panels according to their allele size range and primer annealing temperature (see MP in Table 1). PCRs (10 µL) were performed with 2 µM of labelled forward primer and reverse primer with the same amplification conditions described above. PCR products were sent to GenoScreen (Lille, France) for fragment analysis and were visualised using an Applied Biosystems 3730 Sequencer. An internal size ladder (GeneScan 500 LIZ, Applied Biosystems) was used for accurate sizing and alleles were scored and checked manually using GENEMAPPER v.4.0 (Applied Biosystems, Foster City, CA). Samples that were ambiguous in scoring were re-amplified and re-scored. All peak profiles that were faint or ambiguous (i.e., multiple peaks) were considered as missing data.
Phylogenetic analyses
Additionally, a 461 bp portion of the mitochondrial 16S gene was amplified for 30 specimens (five colonies per species) and used to estimate the genetic distances among the six Millepora species. The PCR amplifications were performed using the primers 16S-SHA and 16S-SHB (Cunningham & Buss, 1993) in 20 µL reactions containing: 1.5 mM of MgCl2, 0.2 mM of each dNTP, 1× final concentration of buffer, 0.5 µM of each primer, 0.25 unit of Red Hot Taq polymerase, 2 µL of DNA template (80–100 ng/µL) and sterilised water up to 20 µL. The cycling parameters were as follows: an initial denaturation step of 5 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 50 °C, 1.5 min at 72 °C and a final elongation step of 5 min at 72 °C. Sequencing of the PCR products was performed by GenoScreen (Lille, France).
Data analyses
Control for the presence of null alleles, scoring errors and large allele dropout were performed with MICROCHECKER v.3.7 (Van Oosterhout et al., 2004). To assess the discriminative power of the microsatellite markers, the genotype probability (GP) was estimated for each locus and for a combination of all loci using GENALEX v.6.5 (Peakall & Smouse, 2006). Repeated multilocus genotypes (MLGs) were also identified in GENALEX and were considered as clone mates at GP < 0.001. The probability of identity, P(ID), was computed to evaluate the power of our microsatellites to accurately distinguish closely related genotypes from those produced by asexual reproduction (Waits, Luikart & Taberlet, 2001). Population genetic analyses were then performed after the removal of all clonal replicates.
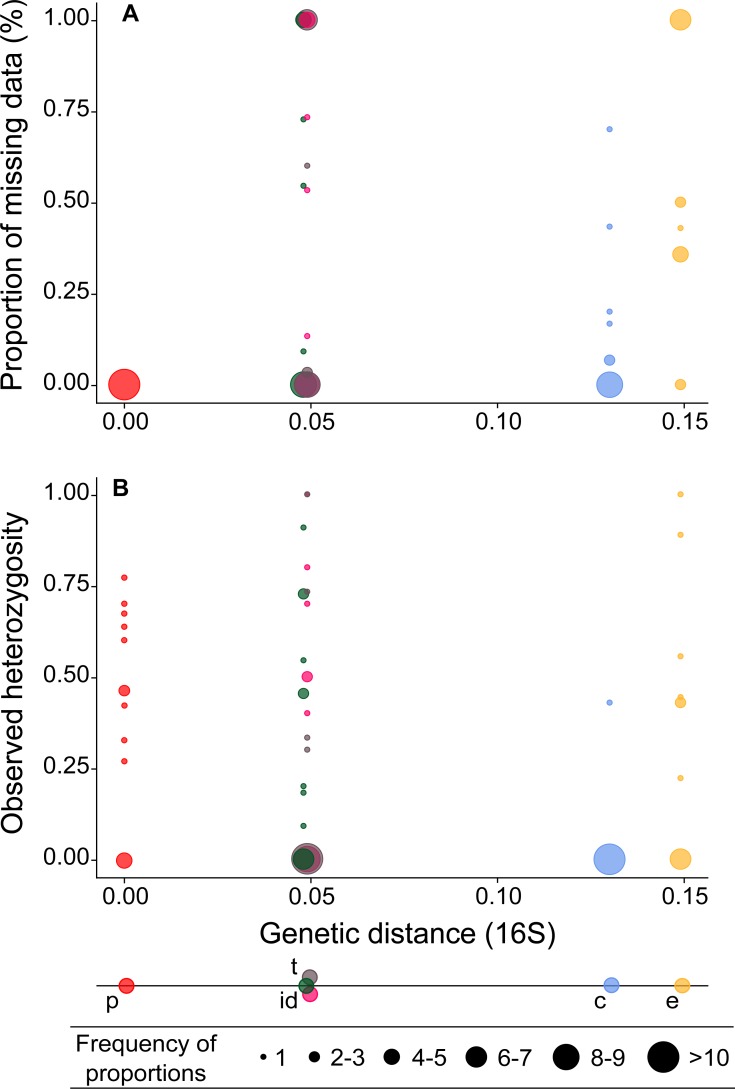
Indices of genetic diversity were estimated for each species in all locations using GENALEX, including Na, the total number of alleles per locus, observed (Ho) and expected (He) heterozygosity (Weir & Cockerham, 1984). The inbreeding coefficient FIS and linkage disequilibrium were estimated using GENETIX v.4.02 (Belkhir et al., 1996), applying a permutation procedure (1,000 permutations) to assess statistical significance. GENETIX was also used to estimate genetic distance among populations of M. platyphylla and the other Millepora species with the microsatellite dataset using the θ estimator of FST (Weir & Cockerham, 1984) based on a permutation procedure (1,000 permutations). Genetic p-distances among species at the mtDNA 16S gene were calculated in Mega v.6 (Tamura et al., 2013). In addition to the p-distance, we also computed other genetic distances (i.e., Kimura-2-Parameters, Tamura & Nei and Maximum composite Likelihood, all available in the software Mega v.6) and found similar species rank among all measures tested. We also examined the cross-species amplification success of the new microsatellite markers by plotting the genetic diversity (Ho) and the proportions of missing data (non amplified loci after 3× repeat PCR, and this at different annealing temperatures) in each species against genetic distance (16S) and relationships were tested using Pearson’s correlation coefficient.
Results
Development of de novo microsatellites in Millepora platyphylla
Sequencing of the microsatellite-enriched library from 14 partially bleached fragments of M. platyphylla yielded 78,784 reads. The Quality Detection Device (QDD) for bioinformatic filtering resulted in a final set of 5,976 sequences containing microsatellite motifs. For the characterisation of new microsatellites, 127 primer pairs (out of the 186 resulting from the QDD filtering, 68.3%) were tested in 16 individuals of M. platyphylla collected on Moorea’s reefs. Fifteen loci showed clear amplification profiles and no Symbiodinium specific locus was identified, proving the efficiency of the DNA extraction technique. For the 50 M. platyphylla colonies collected on Moorea’s reefs, twelve loci were polymorphic (from 2 to 13 alleles) and three additional monomorphic loci were retained for further cross-species transferability tests (Table 1). Contig sequences containing the microsatellites identified in this study are available in GenBank (KX670763 –KX670777, Table 1).
Significant linkage disequilibrium was identified and distributed among all microsatellite loci in M. platyphylla. 9.1% of the pairwise locus combinations showed a significant probability of linkage disequilibrium at P < 0.05 (Table 2). The presence of null alleles was detected at Mill47 and Mill67 with frequencies of null alleles at both loci estimated at 10.1% and 14.4%, respectively. These two loci were removed from our dataset for further genetic analyses, although there was no evidence of scoring error or large allele dropout for any locus. Given the low P(ID) value estimated (1.3E−6), our panel of microsatellites had a high power to distinguish colonies that were clonal replicates. For the ten polymorphic loci showing no evidence of null alleles, the mean number of alleles (Na) per locus was 3.462 and the observed heterozygosity (Ho) was 0.411 (Table 2). Only three loci out of fifteen showed significant deficiency in heterozygotes compared to HWE and only one of them showed no evidence of null alleles (Mill07, FIS: 0.121, Table 1).
Table 2. Summary of genetic distances (GD) based on the 16S gene between the target species and other Millepora species together with indices indicating microsatellite transferability and genetic diversity.
| Species | Locality | N | MLG | Clonal MLG | P(ID) | GD | Amp (%) | Pol (%) | Null (%) | LD (%) | Na | Ho |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. platyphylla | Moorea | 50 | 50 | – | 1.3E−6 | – | 100 | 80.0 | 13.3 | 9.1 | 3.462 | 0.411 |
| M. intricata | Papua | 11 | 10 | 1 | 1.1E−6 | 0.048 | 73.3 | 60.0 | – | 12.1 | 3.909 | 0.405 |
| M. dichotoma | Europa | 30 | 24 | 4 | 4.1E−7 | 0.049 | 86.7 | 60.0 | 7.7 | 10.3 | 3.417 | 0.323 |
| M. tenera | Reunion | 30 | 24 | 6 | 3.1E−7 | 0.049 | 80.0 | 73.3 | 58.3 | 23.0 | 4.833 | 0.439 |
| M. complanata | Curaçao | 30 | 30 | – | 1.3E−6 | 0.130 | 53.3 | 46.7 | 75.0 | 10.2 | 4.000 | 0.250 |
| M. exaesa | Reunion | 14 | 14 | – | 3.9E−6 | 0.149 | 60.0 | 53.3 | 11.1 | 17.6 | 3.625 | 0.496 |
Notes.
- N
- sample size
- MLG
- number of multilocus genotypes
- Clonal MLG
- number of multilocus genotypes with clonal replicates
- P(ID)
- Probability of Identity
- Amp
- percentage of loci amplified
- Pol
- percentage of polymorphic loci
- Null
- percentage of loci showing evidence of null alleles
- LD
- percentage of allele comparisons showing significant linkage disequilibrium (P < 0.05)
- Na
- mean number of alleles
- Ho
- mean observed heterozygosity
Na and Ho were estimated based on loci showing no evidence of null alleles and clonal replicates were removed from our dataset for these measures of genetic diversity.
Cross-species amplification in Milleporidae
Assessment of the mtDNA genetic distances (GD) within the Millepora genus revealed that branching species, i.e., M. intricata, M. dichotoma and M. tenera, were more closely related (0.048–0.049) to our target species, with haplotypes shared between M. dichotoma and M. tenera (Table 3 and see Appendix S1 for the haplotype network). The plate-like M. complanata (0.130) and encrusting M. exaesa (0.149) were more genetically distant from M. platyphylla. The mean amplification success for cross-species amplification was 70.7% (∼11 loci out of 15) and the mean polymorphism was 58.7% (∼9 loci out of 15). Cross-species amplification decreased significantly with mtDNA genetic distance (r = − 0.931, P = 0.007), with a reduced amplification success in the most divergent species, i.e., M. complanata (53.3%) and M. exaesa (60.0%), and higher for M. intricata (73.3%), M. tenera (80.0%) and M. dichotoma (86.7%) (Table 2). Cross-species amplification also revealed a significant decrease of polymorphism with increasing mtDNA distance (r = − 0.857, P = 0.029), with lower percentages of polymorphic loci for non-target species (≤73.3%, Table 2). No relationship was found between the percentage of loci showing evidence of null alleles and genetic distance (r = 0.331, P = 0.521). The highest percentage was recorded for M. complanata (75.0%), while lowest for M. intricata (0%). The proportion of missing data per locus increased significantly with increasing level of genetic distance (Fig. 1A, r = 0.214, P = 0.044).
Table 3. Nuclear (FST) and mitochondrial (p-distance) genetic distances among Millepora species.
Values above the diagonal are the FST calculated on the microsatellite dataset, values with P < 0.001 are in bold and the remaining values are NS. Values below the diagonal are genetic distances (p-distance) based on the mitochondrial 16S gene.
| M. platyphylla | M. intricata | M. dichotoma | M. tenera | M. exaesa | |
|---|---|---|---|---|---|
| M. platyphylla | 0.343 | 0.373 | 0.339 | 0.167 | |
| M. intricata | 0.048 | 0.031 | 0.065 | 0.181 | |
| M. dichotoma | 0.049 | 0.051 | 0.062 | 0.221 | |
| M. tenera | 0.049 | 0.051 | 0.000 | 0.293 | |
| M. exaesa | 0.149 | 0.143 | 0.150 | 0.150 |
Figure 1. Proportion of missing data (A) and observed heterozygosity (B) per microsatellite locus (circles) in five Millepora species plotted against genetic distances (16S gene) from the target species.
Target species Millepora platyphylla (p, red) and non-target species; Millepora intricata (i, green), Millepora dichotoma (d, pink), Millepora tenera (t, purple), Millepora complanata (c, blue) and Millepora exaesa (e, yellow).
Clonal replicates were found in the three branching species: 1 clonal MLG in M. intricata, 4 in M. dichotoma and 6 in M. tenera (Table 2). The mean observed heterozygosity per locus was highly variable in all species, although more limited in M. tenera and M. complanata due to high proportions of null alleles in both species (Fig. 1B and Table S3). No significant correlation was found between the genetic diversity and mtDNA genetic distance (r = − 0.175, P = 0.101). The mean observed heterozygosity was slightly reduced for M. complanata (0.250) compared to other species (0.323 for M. dichotoma ⩽ Ho ⩽ 0.496 for M. exaesa) (Table 2). However, Ho estimate in M. complanata was based on only one microsatellite locus (Mill 103, Fig. 1B). For the four other non-target species, 2 loci out of 15 showed significant deficiencies in heterozygotes compared to HWE in M. dichotoma (Mill07 and Mill67, FIS: 1.000) and another one in M. intricata (Mill101, FIS: 0.500) (Table S3).
The transferability of microsatellites in the Milleporidae also revealed strong genetic differentiation among some species (Table 3 and see Appendix S2 for the Bayesian clustering analysis). No significant genetic differentiation was observed for the closely related branching species (i.e., M. intricata, M. dichotoma and M. tenera). For all comparisons involving our target species M. platyphylla, the lowest value of FST(≤0.167) was recorded for the most divergent species M. exaesa. No relationship (r = 0.150, P = 0.679) was found between the nuclear (FST from microsatellite data) and mitochondrial (p-distance from 16S) genetic distances.
Discussion
Microsatellites’ development and transferability in Milleporidae
To date, there is no study assessing the genetic diversity and population structure of fire coral species. This gap is mostly due to the lack of highly variable genetic markers in the genus until very recently, whereas microsatellite loci have been identified in the Caribbean species Millepora alcicornis (Ruiz-Ramos & Baums, 2014). Heckenhauer et al. (2014) have succeeded in developing eleven microsatellite markers for M. dichotoma from the Great Barrier Reef and showed that their transferability was successful between geographic regions (Red Sea) and the species M. platyphylla. Their study has shown that eight of the eleven microsatellite markers (72.7%) were transferable to M. platyphylla which is less to what we had in the present study (i.e., 86.7% between M. dichotoma and M. platyphylla). Six of their loci had only 2 alleles for M. platyphylla, which is not informative enough depending on the analyses performed (e.g., parentage analyses). Furthermore, most of the microsatellite markers developed by Heckenhauer et al. (2014) were characterised by significant deficiencies in heterozygotes, whereas only two of our loci showed such HWE disequilibrium in both of these species. Depending on the target species, a combination of markers from the two studies would thus seem a good strategy for population genetic approaches in Millepora hydrocorals.
Our cross-amplification tests show a higher cross-taxon transferability success (73.3–86.7%) for a genetic distance below 5% from our target species (i.e., M. intricata, M. dichotoma and M. tenera) and a reduced transferability above this level (≤60% for M. complanata and M. exaesa). Overall, our results show a high probability to amplify a microsatellite locus within the genus Millepora, where 71% of the loci were successfully amplified in the five non-target species. This value is slightly lower to what was demonstrated for the Caribbean Montastraea species complex (scleractinian corals), i.e., ∼80% of amplification success in two other species within the same location (Davies et al., 2013). Our lower value, while still very high, is not surprising as we tested cross-amplification between six species of the genus Millepora (i.e., no species complex as for Montastraea spp), which were also collected throughout their entire geographic range. The non-amplification of some microsatellite loci in the non-target species is most likely due to specific mutations in the primer binding sites in M. platyphylla, i.e., null alleles (Paetkau & Strobeck, 1995). These loci, specific to Moorea’s population, may result from local evolutionary processes at this location, such as bottlenecks, expansions, life history traits, inbreeding and outbreeding (Keller & Waller, 2002; Leffler et al., 2012; Romiguier et al., 2014). Our cross-amplified loci show a high probability to be polymorphic in non-target species (58.7%), which is much higher to what is generally found in other taxa, such as fishes (∼25–30% in Barbará et al., 2007; Reid, Hoareau & Bloomer, 2012) and birds (∼20–50% in Dawson et al., 2010). Many other studies using cross-amplification have shown a significant decrease in the transferability success and polymorphism with evolutionary distance from the target species (Jan et al., 2012; Maduna et al., 2014; Moodley et al., 2015).
Usefulness of cross-species amplification in Indo-Pacific Milleporidae
The level of genetic diversity is key for the persistence of a species in changing environments and represents a fundamental aspect of biodiversity (Romiguier et al., 2014). Quantifying the genetic diversity in natural populations and species is critical to address ecological and evolutionary questions (Nair, 2014), which requires the development of suitable molecular resources. In this study, our cross-species amplification approach for the development of new microsatellites shows no significant evidence of lower genetic diversity nor greater proportion of null alleles with increasing genetic distance from our target species, which is in contradiction with previous studies (Carreras-Carbonell, Macpherson & Pascual, 2008; Hendrix et al., 2010; Moodley et al., 2015). Our results also show that most of our microsatellite markers are not useful to estimate the genetic diversity in the Caribbean species M. complanata due to the high proportion of null alleles. Hence, this study reveals that the transferability of our microsatellites ensures comparable estimations of the genetic diversity among closely related Millepora species, although restricted to the Indo-Pacific region. Further investigations with other Caribbean species, such as M. alcicornis, are needed to test their transferability in this geographic region.
In this study, we also found that genetic distance from interspecific microsatellite data were not congruent with mtDNA distance among the studied species. It is not surprising as such highly variable markers would suffer from homoplasy as one look into higher taxon relationships, while microsatellites are well-known to be mostly useful for intra-specific studies (Selkoe & Toonen, 2006). Nonetheless, assessment of the population structure among closely related Indo-Pacific species revealed a clear genetic differentiation between the branching species and the plate-like M. platyphylla. Our panel of new microsatellite loci is therefore useful for species delineation and can help resolve the century-old species problem in Milleporids (Boschma, 1948).
Patterns of genetic diversity and population structure in Milleporidae
The first evaluation of genetic diversity among species of Millepora across its geographic range in tropical reefs reveals moderate levels of heterozygosity and allelic richness. The lowest genetic diversity was found for the Caribbean species, M. complanata, likely resulting from the low proportion of polymorphic loci (46.7%) and the high proportion of loci showing evidence of null alleles (75.0%). Nonetheless, levels of genetic diversity estimated in this study are similar to what was described for many tropical scleractinian species (Baums, 2008; Shearer, Porto & Zubilaga, 2009) and to what is expected in populations of partially clonal organisms. In this study, linkage disequilibrium, relatively high levels of allelic and genetic diversity, and heterozygote deficiencies were estimated for the six studied hydrocoral species, as previously described in some scleractinian corals (Baums, 2008). Overall, these new microsatellites are suitable to infer genetic diversity and to evaluate reproductive strategies in the partially clonal fire corals.
Conclusions
This study highlights the utility of cross-species amplification of microsatellites in assessing population genetics of the Millepora genus in the Indo-Pacific region. Surprisingly, this approach does not result in lowering genetic diversity (Ho) in non-target species, thus ensuring an unbiased estimation of genetic diversity among fire coral species. The development of microsatellites can be complex and difficult in many taxa, such as birds (Primmer et al., 1997) and plants (Lagercrantz, Ellegren & Adersson, 1993), due to biological constraints that can affect the abundance and motif repeats of microsatellites in the genome (Tóth, Gáspári & Jurka, 2000). A recent study has demonstrated high microsatellite coverage in several species of cnidarians, including Millepora alcicornis (Ruiz-Ramos & Baums, 2014), indicating that there is no biological constraint for the development of microsatellite markers in this phylum. The availability of numerous microsatellite markers for reef-building Millepora species creates new opportunities for future research into the processes driving the complexity of their colonisation success on many Indo-Pacific reefs.
Supplemental Information
See http://www.nsm.buffalo.edu/Bio/burr/ for more details on the Symbiodinium strains.
TA, primer temperature annealing; Sp, Species; N, sample size; Namp, number of individuals with reliable amplification; Null, proportion of null alleles; LD, proportion of allele comparisons showing significant linkage disequilibrium (P < 0.05); Na, number of alleles; Ho, observed heterozygosity; He, expected heterozygosity; FIS, inbreeding coefficient. Significant values of FIS are indicated by bold values with * P < 0.05, ** P < 0.01 and *** P < 0.001. Clonal replicates were removed from our dataset for the measures of genetic diversity.
Each pie represents one 16S haplotype (with its area proportional to the number of individuals in which it was detected). The lengths of the grey lines connecting the 16S haplotypes are proportional to the number of mutations separating them with the number of mutations shown in red on each line. This haplotype network was reconstructed using the median joining algorithm (Bandelt, Forster & Rohl, 1999) in Network v5.0.0.0 (www.fluxus-engineering.com).
Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16:37–48.
Assignment analyses based on Bayesian clustering analysis using STRUCTURE (Pritchard, Stephens & Donnelly, 2000) for five of the six studied species: (1) M. platyphylla, (2) M. exaesa, (3) M. intricata, (4) M. dichotoma and (5) M. tenera. The x-axis shows species identification and y-axis shows the cluster membership ( K = 2). Initial STRUCTURE runs were used to determine the most likely number of clusters (K). Runs were performed with the default setting, a burn-in period of 50000, 50000 MCMC repeats and 10 iterations per K. The results were uploaded to STRUCTURE HARVESTER (Earl & vonHoldt, 2011) and the most likely K was retained for a second run in STRUCTURE with a burn-in period of 500000, 500000 MCMC repeats, 10 iterations and uniform prior setting.
Earl DA, vonHoldt BM. 2011. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361. Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959.
— indicates no amplification.
Acknowledgments
We are grateful to Alexandre Mercière, Maggy Nugues, Nicky Gravier-Bonnet, Chloé Bourmaud, Julia Leung, Suzie Mills and Ricardo Beldade who contributed to sample collection, Nathalie Tolou for technical advice and Jeanine Almany for English improvements on the manuscript. We are also grateful to Mary Alice Coffroth and her laboratory for providing us cultured Symbiodinium strains. We also thank Iliana Baums and one anonymous reviewer for their thoughtful comments on the manuscript.
Funding Statement
Caroline E. Dubé is supported by a graduate scholarship from the Fonds Québécois de Recherche sur la Nature et les Technologies. Emilie Boissin is supported by European Marie Curie Postdoctoral fellowships MC-CIG-618480. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Caroline E. Dubé conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Serge Planes conceived and designed the experiments, contributed reagents/materials/analysis tools, reviewed drafts of the paper.
Yuxiang Zhou performed the experiments.
Véronique Berteaux-Lecellier contributed reagents/materials/analysis tools, reviewed drafts of the paper.
Emilie Boissin conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, reviewed drafts of the paper.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
CITES - FR1298700028-E; field experiments were approved by the Presidency of French Polynesia (#0085).
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
Genbank
BankIt1941788 Seq_Mill1 KX670763
BankIt1941788 Seq_Mill2 KX670764
BankIt1941788 Seq_Mill3 KX670765
BankIt1941788 Seq_Mill4 KX670766
BankIt1941788 Seq_Mill5 KX670767
BankIt1941788 Seq_Mill6 KX670768
BankIt1941788 Seq_Mill7 KX670769
BankIt1941788 Seq_Mill8 KX670770
BankIt1941788 Seq_Mill9 KX670771
BankIt1941788 Seq_Mill10 KX670772
BankIt1941788 Seq_Mill11 KX670773
BankIt1941788 Seq_Mill12 KX670774
BankIt1941788 Seq_Mill13 KX670775
BankIt1941788 Seq_Mill14 KX670776
BankIt1941788 Seq_Mill15 KX670777.
Data Availability
The following information was supplied regarding data availability:
The raw data has been supplied as Data S1.
References
- Adjeroud et al. (2014).Adjeroud M, Guérécheau A, Vidal-Dupiol J, Flot J-F, Arnaud-Haond S, Bonhomme F. Genetic diversity, clonality and connectivity in the scleractinian coral Pocillopora damicornis: a multi-scale analysis in an insular, fragmented reef system. Marine Biology. 2014;161:531–541. doi: 10.1007/s00227-013-2355-9. [DOI] [Google Scholar]
- Ardehed et al. (2015).Ardehed A, Johansson D, Schagerström E, Kautsky L, Johannesson K, Pereyra RT. Complex spatial clonal structure in the macroalgae Fucus radicans with both sexual and asexual recruitment. Ecology and Evolution. 2015;5:4233–4245. doi: 10.1002/ece3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, Guest & Willis (2009).Baird AH, Guest JR, Willis BL. Systematic and biogeographical patterns in the reproductive biology of scleractinian coral. Annual Review in Ecology, Evolution and Systematics. 2009;40:551–571. doi: 10.1146/annurev.ecolsys.110308.120220. [DOI] [Google Scholar]
- Barbará et al. (2007).Barbará T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C. Cross-species transfer of nuclear microsatellite markers: potential and limitations. Molecular Ecology. 2007;16:3759–3767. doi: 10.1111/j.1365-294X.2007.03439.x. [DOI] [PubMed] [Google Scholar]
- Barrett & Schluter (2008).Barrett RD, Schluter D. Adaptation from standing genetic variation. Trends in Ecology & Evolution. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Baums (2008).Baums IB. A restoration genetics guide for coral reef conservation. Molecular Ecology. 2008;17:2796–2811. doi: 10.1111/j.1365-294X.2008.03787.x. [DOI] [PubMed] [Google Scholar]
- Baums et al. (2014).Baums IB, Devlin-Durante M, Laing BAA, Feingold JS, Smith TB, Bruckner A, Monteiro J. Marginal coral populations: the densest known aggregation of Pocillopora in the Galapagos Archipelago is of asexual origin. Frontiers in Marine Science. 2014;1:1–11. [Google Scholar]
- Baums, Miller & Hellberg (2005).Baums IB, Miller MW, Hellberg ME. Regionally isolated populations of an imperiled Caribbean coral, Acropora palmata. Molecular Ecology. 2005;14:1377–1390. doi: 10.1111/j.1365-294X.2005.02489.x. [DOI] [PubMed] [Google Scholar]
- Baums, Miller & Hellberg (2006).Baums IB, Miller MW, Hellberg ME. Geographic variation in clonal structure in a reef-building Caribbean coral, Acropora palmata. Ecological Monographs. 2006;76:503–519. doi: 10.1890/0012-9615(2006)076[0503:GVICSI]2.0.CO;2. [DOI] [Google Scholar]
- Belkhir et al. (1996).Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05 logiciel Sous windows Pour La génétique des populations. 1996. Laboratoire génome, populations, interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier, France.
- Bellwood et al. (2004).Bellwood DR, Hughes TP, Folke C, Nyström M. Confronting the coral reef crisis. Nature. 2004;429:827–833. doi: 10.1038/nature02691. [DOI] [PubMed] [Google Scholar]
- Boissin et al. (2016).Boissin E, Micu D, Janczyszyn-Le Goff M, Neglia V, Bat L, Todorova V, Panayotova M, Kruschel C, Macic V, Milchakova N, Keskin C, Anastasopoulou A, Nasto I, Zane L, Planes S. Contemporary genetic structure and post-glacial demographic history of the black scorpionfish, Scorpaena porcus, in the Mediterranean and the Black Seas. Molecular Ecology. 2016;25:2195–2209. doi: 10.1111/mec.13616. [DOI] [PubMed] [Google Scholar]
- Boschma (1948).Boschma H. The species problem in Millepora. Zoologische Verhandelingen. 1948;1:1–115. [Google Scholar]
- Boschma (1956).Boschma H. Milleporina and Stylasterina. In: Moore RC, editor. Treatise on invertebrate paleontology. Geological Society of America, University of Kansas; Lawrence: 1956. pp. 90–106. [Google Scholar]
- Bourmaud et al. (2013).Bourmaud C, Leung JKL, Bollard S, Gravier-Bonnet N. Mass spawning events, seasonality and reproductive features in Milleporids (Cnidaria, Hydrozoa) from Reunion Island. Marine Ecology. 2013;34:14–24. doi: 10.1111/maec.12024. [DOI] [Google Scholar]
- Cardinale et al. (2012).Cardinale BJ, Duffy E, Gonzalez A, Hooper DU, Perrings C, Venail P, Narwani A, Mace GM, Tilman D, Wardle DA, Kinzig AP, Daily GC, Loreau M, Grace JB, Larigauderie A, Srivastava D, Naeem S. Biodiversity loss and its impact on humanity. Nature. 2012;486:59–67. doi: 10.1038/nature11148. [DOI] [PubMed] [Google Scholar]
- Carreras-Carbonell, Macpherson & Pascual (2008).Carreras-Carbonell J, Macpherson E, Pascual M. Utility of pairwise mtDNA genetic distances for predicting cross-species microsatellite amplification and polymorphism success in fishes. Conservation Genetics. 2008;9:181–190. doi: 10.1007/s10592-007-9322-2. [DOI] [Google Scholar]
- Cheng et al. (2012).Cheng HL, Lee YH, Hsiung DS, Tseng MC. Screening of new microsatellite DNA markers from the genome of Platyeriocheir formosa. International Journal of Molecular Sciences. 2012;13:5598–5606. doi: 10.3390/ijms13055598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham & Buss (1993).Cunningham CW, Buss LW. Molecular evidence for multiple episodes of paedomorphosis in the family Hydractiniidae. Biochemical Systematics and Ecology. 1993;21:57–69. doi: 10.1016/0305-1978(93)90009-G. [DOI] [Google Scholar]
- Davies et al. (2013).Davies SW, Rahman M, Meyer E, Green EA, Buschiazzo E, Medina M, Matz MV. Novel polymorphic microsatellite markers for population genetics of the endangered Caribbean star coral, Montastraea faveolata. Marine Biodiversity. 2013;43:167–172. doi: 10.1007/s12526-012-0133-4. [DOI] [Google Scholar]
- Davies, Treml & Matz (2015).Davies SW, Treml EA, Matz MV. Exploring the role of Micronesian islands in the maintenance of coral genetic diversity in the Pacific Ocean. Molecular Ecology. 2015;24:70–82. doi: 10.1111/mec.13005. [DOI] [PubMed] [Google Scholar]
- Dawson et al. (2010).Dawson DA, Horsburgh GJ, Küpper C, Stewart IRK, Ball AD, Durrant KL, Hansson B, Bacon I, Bird S, Klein A, Krupa AP, Lee J-W, Martín-Gálvez D, Simeoni M, Smith G, Spurgin LG, Burke T. New methods to identify conserved microsatellite loci and develop primer sets of high cross-species utility—as demonstrated for birds. Molecular Ecology Resources. 2010;10:475–494. doi: 10.1111/j.1755-0998.2009.02775.x. [DOI] [PubMed] [Google Scholar]
- Dubé, Boissin & Planes (2016).Dubé CE, Boissin E, Planes S. Overgrowth of living scleractinian corals by the hydrocoral Millepora platyphylla in Moorea, French Polynesia. Marine Biodiversity. 2016;46:329–330. doi: 10.1007/s12526-015-0392-y. [DOI] [Google Scholar]
- Edmunds (1999).Edmunds PJ. The role of colony morphology and substratum inclination in the success of Millepora alcicornis on shallow coral reefs. Coral Reefs. 1999;18:133–140. doi: 10.1007/s003380050167. [DOI] [Google Scholar]
- Estoup et al. (1993).Estoup A, Presa P, Krieg F, Vaiman D, Guyomard R. (CT)n and (GT)n microsatellites: a new class of genetic markers for Salmo truttaL. (brown trout) Heredity. 1993;71:488–496. doi: 10.1038/hdy.1993.167. [DOI] [PubMed] [Google Scholar]
- Frankham (2005).Frankham R. Genetics and extinction. Biology Conservation. 2005;126:131–140. doi: 10.1016/j.biocon.2005.05.002. [DOI] [Google Scholar]
- Fukami et al. (2004).Fukami H, Budd AF, Levitan DR, Jara J, Kersanach R, Knowlton N. Geographic differences in species boundaries among members of the Montastraea annularis complex based on molecular and morphological markers. Evolution. 2004;38:324–337. [PubMed] [Google Scholar]
- Gilmour, Smith & Brinkman (2009).Gilmour JP, Smith LD, Brinkman RM. Biannual spawning, rapid larval development and evidence of self-seeding for scleractinian corals at an isolated system of reefs. Marine Biology. 2009;156:1297–1309. doi: 10.1007/s00227-009-1171-8. [DOI] [Google Scholar]
- Harrison (2011).Harrison PL. Sexual reproduction of scleractinian corals. In: Dubinsky Z, Stambler N, editors. Coral reefs: an ecosystem in transition. Springer; Berlin: 2011. pp. 59–85. [Google Scholar]
- Heckenhauer et al. (2014).Heckenhauer J, Schweinsberg M, Elbrecht V, John U, Tollrian R, Lampert KP. Isolation, characterization and cross amplification of eleven novel microsatellite loci for the hydrozoan coral Millepora. Conservation Genetic Resources. 2014;7:215–217. doi: 10.1007/s12686-014-0337-y. [DOI] [Google Scholar]
- Hendrix et al. (2010).Hendrix R, Hauswaldt JS, Veiths M, Steinfartz S. Strong correlation between cross-amplification success and genetic distance across all members of ‘True Salamanders’ (Amphibia: Salamandridae) revealed by Salamandra salamandra-specific microsatellite loci. Molecular Ecology Resources. 2010;10:1038–1047. doi: 10.1111/j.1755-0998.2010.02861.x. [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg et al. (2007).Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K, Knowlton N, Eakin CM, Iglesias-Prieto R, Muthiga N, Bradbury RH, Dubi A, Hatziolos ME. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- Hoffmann & Sgrò (2011).Hoffmann AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- Jan et al. (2012).Jan C, Dawson DA, Altringham JD, Burke T, Butlin RK. Development of conserved microsatellite markers of high cross-species utility in bat species (Vespertilionidae, Chiroptera, Mammalia) Molecular Ecology Resources. 2012;12:532–548. doi: 10.1111/j.1755-0998.2012.03114.x. [DOI] [PubMed] [Google Scholar]
- Kayal & Kayal (2016).Kayal M, Kayal E. Colonies of the fire coral Millepora platyphylla constitute scleractinian survival oases during Acanthaster outbreaks in French Polynesia. Marine Biodiversity. 2016 doi: 10.1007/s12526-016-0465-6. Epub ahead of print Mar 05 2016. [DOI] [Google Scholar]
- Keller & Waller (2002).Keller LF, Waller DM. Inbreeding effects in wild populations. Trends in Ecology & Evolution. 2002;17:19–23. doi: 10.1016/S0169-5347(02)02489-8. [DOI] [Google Scholar]
- Kuffner et al. (2015).Kuffner IB, Lidz BH, Hudson JH, Anderson JS. A century of ocean warming on Florida Keys coral reefs: historic in situ observations. Estuaries and Coasts. 2015;38:1085–1096. doi: 10.1007/s12237-014-9875-5. [DOI] [Google Scholar]
- Lagercrantz, Ellegren & Adersson (1993).Lagercrantz U, Ellegren H, Adersson L. The abundance of various polymorphic microsatellite motifs differ between plants and vertebrates. Nucleic Acids Research. 1993;21:1111–1115. doi: 10.1093/nar/21.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler et al. (2012).Leffler EM, Bullaughey K, Matute DR, Meyer WK, Ségurel L, Venkat A, Andolfatto P, Przeworski M. Revisiting an old riddle: what determines genetic diversity levels within species? PLOS Biology. 2012;10(9):e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis (2006).Lewis JB. Biology and ecology of the hydrocoral Millepora on coral reefs. Advances in Marine Biology. 2006;50:1–55. doi: 10.1016/S0065-2881(05)50001-4. [DOI] [PubMed] [Google Scholar]
- Lukoschek, Riginos & Van Oppen (2016).Lukoschek V, Riginos C, Van Oppen MJH. Congruent patterns of connectivity can inform management for broadcast spawning corals on the Great Barrier Reef. Molecular Ecology. 2016;25:3065–3080. doi: 10.1111/mec.13649. [DOI] [PubMed] [Google Scholar]
- Maduna et al. (2014).Maduna SN, Rossouw C, Roodt-Wilding R, Bester-van der Merwe AE. Microsatellite cross-species amplification and utility in southern African elasmobranchs: a valuable resource for fisheries management and conservation. BMC Research Notes. 2014;7:352. doi: 10.1186/1756-0500-7-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malausa et al. (2011).Malausa T, Gilles A, Maglécz E, Blanquart H, Duthoy S, Costedoat C, Dubut V, Pech N, Castagnone-Sereno P, Délye C, Feau N, Frey P, Gauthier P, Guillemaud T, Hazard L, Le Corre V, Lung-Escarmant B, Malé P-JG, Ferreira S, Martin J-F. High-throughput microsatellite isolation through 454 GS-FLX Titanium pyrosequencing of enriched DNA libraries. Molecular Ecology Resources. 2011;11:638–644. doi: 10.1111/j.1755-0998.2011.02992.x. [DOI] [PubMed] [Google Scholar]
- Martin et al. (2010).Martin J-F, Pech N, Meglécz E, Ferreira S, Costedoat C, Dubut V, Malausa T, Gilles A. Representativeness of microsatellite distributions in genomes, as revealed by 454 GS-FLX titanium pyrosequencing. BMC Genomics. 2010;11:560. doi: 10.1186/1471-2164-11-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meglécz et al. (2010).Meglécz E, Costedoat C, Dubut V, Gilles A, Malausa T, Pech N, Martin J-F. QDD: a user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics. 2010;26:403–404. doi: 10.1093/bioinformatics/btp670. [DOI] [PubMed] [Google Scholar]
- Mieog et al. (2009).Mieog JC, Van Oppen MJH, Berkelmans R, Stam WT, Olsen JL. Quantification of algal endosymbionts (Symbiodinium) in coral tissue using real-time PCR. Molecular Ecology Resources. 2009;9:74–82. doi: 10.1111/j.1755-0998.2008.02222.x. [DOI] [PubMed] [Google Scholar]
- Mokhtar-Jamaï et al. (2013).Mokhtar-Jamaï K, Coma R, Wang J, Zuberer F, Féral JP, Aurelle D. Role of evolutionary and ecological factors in the reproductive success and the spatial genetic structure of the temperate gorgonian Paramuricea clavata. Ecology and Evolution. 2013;3:1765–1779. doi: 10.1002/ece3.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley et al. (2015).Moodley Y, Masello JF, Cole TL, Calderon L, Munimanda GK, Thali MR, Alderman R, Cuthbert RJ, Marin M, Massaro M, Navarro J, Phillips RA, Ryan PG, Suazo CG, Cherel Y, Weimerskirch H, Quillfeldt P. Evolutionary factors affecting the cross-species utility of newly developed microsatellite markers in seabirds. Molecular Ecology Resources. 2015;15:1046–1058. doi: 10.1111/1755-0998.12372. [DOI] [PubMed] [Google Scholar]
- Mourier & Planes (2013).Mourier J, Planes S. Direct genetic evidence for reproductive philopatry and associated fine-scale migrations in female blacktip reef sharks (Carcharhinus melanopterus) in French Polynesia. Molecular Ecology. 2013;22:201–214. doi: 10.1111/mec.12103. [DOI] [PubMed] [Google Scholar]
- Nagelkerken & Nagelkerken (2004).Nagelkerken I, Nagelkerken WP. Loss of coral cover and biodiversity on shallow Acropora and Millepora reefs after 31 years on Curaçao, Netherlands Antilles. Bulletin of Marine Science. 2004;74:213–223. [Google Scholar]
- Nair (2014).Nair P. Conservation genomics. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:569. doi: 10.1073/pnas.1323086111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima et al. (2016).Nakajima Y, Zayasu Y, Shinzato C, Noriyuki S, Mitarai S. Genetic differentiation and connectivity of morphological types of the broadcast-spawning Galaxea fascicularis in the Nansei Islands, Japan. Ecology and Evolution. 2016;6:1457–1469. doi: 10.1002/ece3.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noreen, Harrison & Van Oppen (2009).Noreen AME, Harrison PL, Van Oppen MJH. Genetic diversity and connectivity in a brooding reef coral at the limit of its distribution. Proceedings of the Royal Society London: Biological Sciences. 2009;276:3927–3935. doi: 10.1098/rspb.2009.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetkau & Strobeck (1995).Paetkau D, Strobeck C. The molecular-basis and evolutionary history of a microsatellite null allele in bears. Molecular Ecology. 1995;4:519–520. doi: 10.1111/j.1365-294X.1995.tb00248.x. [DOI] [PubMed] [Google Scholar]
- Pandolfi et al. (2011).Pandolfi JM, Connolly SR, Marshall DJ, Cohen AL. Projecting coral reef futures under global warming and ocean acidification. Science. 2011;333:418–422. doi: 10.1126/science.1204794. [DOI] [PubMed] [Google Scholar]
- Peakall & Smouse (2006).Peakall R, Smouse P. Genalex 6: genetic analysis in excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzón et al. (2012).Pinzón JH, Reyes-Bonilla H, Baums IB, Lajeunesse TC. Contrasting clonal structure among Pocillopora (Scleractinia) communities at two environmentally distinct sites in the Gulf of California. Coral Reefs. 2012;3:765–777. doi: 10.1007/s00338-012-0887-y. [DOI] [Google Scholar]
- Pirog et al. (2016).Pirog A, Jaquemet S, Blaison A, Soria M, Magalon H. Isolation and characterization of eight microsatellite loci from Galeocerdo cuvier (tiger shark) and cross-amplification in Carcharhinus leucas, Carcharhinus brevipinna, Carcharhinus plumbeus and Sphyrna lewini. PeerJ. 2016;4:e2041. doi: 10.7717/peerj.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primmer et al. (1997).Primmer CR, Raudsepp T, Chowdhary BP, Møller AP, Ellegren H. Low frequency of microsatellites in the avian genome. Genome Research. 1997;7:471–482. doi: 10.1101/gr.7.5.471. [DOI] [PubMed] [Google Scholar]
- Reid, Hoareau & Bloomer (2012).Reid K, Hoareau TB, Bloomer P. High-throughput microsatellite marker development in two sparid species and verification of their transferability in the family Sparidae. Molecular Ecology Resources. 2012;12:740–752. doi: 10.1111/j.1755-0998.2012.03138.x. [DOI] [PubMed] [Google Scholar]
- Romiguier et al. (2014).Romiguier J, Gayral P, Ballenghien M, Bernard A, Cahais V, Chenuil A, Chiari Y, Dernat R, Duret L, Faivre N, Loire E, Lourenco JM, Nabholz B, Roux C, Tsagkogeorga G, Weber AA-T, Weinert LA, Belkhir K, Bierne N, Glémin S, Galtier N. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature. 2014;515:261–263. doi: 10.1038/nature13685. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ramos & Baums (2014).Ruiz-Ramos D, Baums I. Microsatellite abundance across the Anthozoa and Hydrozoa in the phylum Cnidaria. BMC Genomics. 2014;15:939. doi: 10.1186/1471-2164-15-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ramos, Weil & Schizas (2014).Ruiz-Ramos DV, Weil E, Schizas NV. Morphological and genetic evaluation of the hydrocoral Millepora species complex in the Caribbean. Zoological Studies. 2014;53:4. doi: 10.1186/1810-522X-53-4. [DOI] [Google Scholar]
- Schlötterer (2000).Schlötterer C. Evolutionary dynamics of microsatellite DNA. Chromosoma. 2000;109:365–371. doi: 10.1007/s004120000089. [DOI] [PubMed] [Google Scholar]
- Selkoe & Toonen (2006).Selkoe KA, Toonen RJ. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecology Letters. 2006;9:615–629. doi: 10.1111/j.1461-0248.2006.00889.x. [DOI] [PubMed] [Google Scholar]
- Shearer, Porto & Zubilaga (2009).Shearer TL, Porto I, Zubilaga AL. Restoration of coral populations in light of genetic diversity estimates. Coral Reefs. 2009;28:727–733. doi: 10.1007/s00338-009-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva & Gardner (2015).Silva CNS, Gardner JPA. Emerging patterns of genetic variation in the New Zealand endemic scallop. Pecten Novaezelandiae. Molecular Ecology. 2015;24:5379–5393. doi: 10.1111/mec.13404. [DOI] [PubMed] [Google Scholar]
- Stanley (2006).Stanley GD. Photosymbiosis and the evolution of modern coral reefs. Science. 2006;312:857–858. doi: 10.1126/science.1123701. [DOI] [PubMed] [Google Scholar]
- Tamura et al. (2013).Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth, Gáspári & Jurka (2000).Tóth G, Gáspári Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Research. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trench (1979).Trench RK. The cell biology of plant-animal symbiosis. Annual Review of Plant Physiology. 1979;30:485–531. doi: 10.1146/annurev.pp.30.060179.002413. [DOI] [Google Scholar]
- Van der Ven et al. (2016).Van der Ven RM, Triest L, De Ryck DJR, Mwaura JM, Mohammed MS, Kochzius M. Population genetic structure of the stony coral Acropora tenuis shows high but variable connectivity in East Africa. Journal of Biogeography. 2016;43:510–519. doi: 10.1111/jbi.12643. [DOI] [Google Scholar]
- Van Oosterhout et al. (2004).Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Microchecker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- Wahle (1980).Wahle CM. Detection, pursuit, and overgrowth of tropical gorgonians by milleporid hydrocorals: perseus and Medusa revisited. Science. 1980;229:689–691. doi: 10.1126/science.209.4457.689. [DOI] [PubMed] [Google Scholar]
- Waits, Luikart & Taberlet (2001).Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Molecular Ecology. 2001;10:249–256. doi: 10.1046/j.1365-294X.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Warner, Willis & Van Oppen (2016).Warner PA, Willis BL, Van Oppen MJH. Sperm dispersal distances estimated by parentage analysis in a brooding scleractinian coral. Molecular Ecology. 2016;25:1398–1415. doi: 10.1111/mec.13553. [DOI] [PubMed] [Google Scholar]
- Weir & Cockerham (1984).Weir B, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- Wilkinson (2008).Wilkinson C. Status of coral reefs of the world: 2008. Global Coral Reef Monitoring Network and Reef and Rainforest Research Center; Townsville: 2008. [Google Scholar]
- Zhang et al. (2011).Zhang J, Chiodini R, Badr A, Zhang G. The impact of next-generation sequencing on genomics. Journal of Genetics and Genomics. 2011;38:95–109. doi: 10.1016/j.jgg.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.nsm.buffalo.edu/Bio/burr/ for more details on the Symbiodinium strains.
TA, primer temperature annealing; Sp, Species; N, sample size; Namp, number of individuals with reliable amplification; Null, proportion of null alleles; LD, proportion of allele comparisons showing significant linkage disequilibrium (P < 0.05); Na, number of alleles; Ho, observed heterozygosity; He, expected heterozygosity; FIS, inbreeding coefficient. Significant values of FIS are indicated by bold values with * P < 0.05, ** P < 0.01 and *** P < 0.001. Clonal replicates were removed from our dataset for the measures of genetic diversity.
Each pie represents one 16S haplotype (with its area proportional to the number of individuals in which it was detected). The lengths of the grey lines connecting the 16S haplotypes are proportional to the number of mutations separating them with the number of mutations shown in red on each line. This haplotype network was reconstructed using the median joining algorithm (Bandelt, Forster & Rohl, 1999) in Network v5.0.0.0 (www.fluxus-engineering.com).
Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16:37–48.
Assignment analyses based on Bayesian clustering analysis using STRUCTURE (Pritchard, Stephens & Donnelly, 2000) for five of the six studied species: (1) M. platyphylla, (2) M. exaesa, (3) M. intricata, (4) M. dichotoma and (5) M. tenera. The x-axis shows species identification and y-axis shows the cluster membership ( K = 2). Initial STRUCTURE runs were used to determine the most likely number of clusters (K). Runs were performed with the default setting, a burn-in period of 50000, 50000 MCMC repeats and 10 iterations per K. The results were uploaded to STRUCTURE HARVESTER (Earl & vonHoldt, 2011) and the most likely K was retained for a second run in STRUCTURE with a burn-in period of 500000, 500000 MCMC repeats, 10 iterations and uniform prior setting.
Earl DA, vonHoldt BM. 2011. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361. Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959.
— indicates no amplification.
Data Availability Statement
The following information was supplied regarding data availability:
The raw data has been supplied as Data S1.