Abstract
SEA (sea urchin sperm protein, enterokinase, agrin) domains, many of which possess autoproteolysis activity, have been found in a number of cell surface and secreted proteins. Despite high sequence divergence, SEA domains were also proposed to be present in dystroglycan based on a conserved autoproteolysis motif and receptor‐type protein phosphatase IA‐2 based on structural similarity. The presence of a SEA domain adjacent to the transmembrane segment appears to be a recurring theme in quite a number of type I transmembrane proteins on the cell surface, such as MUC1, dystroglycan, IA‐2, and Notch receptors. By comparative sequence and structural analyses, we identified dystroglycan‐like proteins with SEA domains in Capsaspora owczarzaki of the Filasterea group, one of the closest single‐cell relatives of metazoans. We also detected novel and divergent SEA domains in a variety of cell surface proteins such as EpCAM, α/ε‐sarcoglycan, PTPRR, collectrin/Tmem27, amnionless, CD34, KIAA0319, fibrocystin‐like protein, and a number of cadherins. While these proteins are mostly from metazoans or their single cell relatives such as choanoflagellates and Filasterea, fibrocystin‐like proteins with SEA domains were found in several other eukaryotic lineages including green algae, Alveolata, Euglenozoa, and Haptophyta, suggesting an ancient evolutionary origin. In addition, the intracellular protein Nucleoporin 54 (Nup54) acquired a divergent SEA domain in choanoflagellates and metazoans.
Keywords: SEA domain, autoproteolysis, cell surface proteins, dystroglycan, cadherin, Nup54
Introduction
The SEA domain was originally detected in a number of cell surface and secreted proteins such as sea urchin sperm protein 63 kDa, enterokinase (also called enteropeptidase), agrin, perlecan, and several membrane‐associated mucins.1 It was proposed to have functions related to sugar moieties since it often resides in heavily glycosylated multi‐domain proteins.1 Both agrin and perlecan are large proteoglycans responsible for interactions with numerous extracellular matrix and cell surface proteins.2 SEA domains are also present in two interphotoreceptor matrix proteoglycans (IMPG1 and IMPG2). Mutations located in the SEA domains of IMPG1 and IMPG2 have been associated with genetic disorders of vitelliform macular dystrophies3 and autosomal‐recessive retinitis pigmentosa,4 respectively. SEA‐domain‐containing mucins, such as MUC1 (Mucin‐1) and MUC16 (Mucin‐16), have extensive O‐linked glycosylation in their characteristic serine and threonine‐rich regions and have been linked to various cancers.5, 6 In addition to their roles in protection and lubrication of the epithelial surfaces of the internal ducts, some of these mucins such as MUC1 are involved in cell signaling that is regulated by multiple events of proteolysis.7 The SEA‐domain‐containing enterokinase is a type II single‐pass transmembrane protein (N‐terminus located in cytosol). It initiates intestinal digestion by proteolytically activating trypsin.8 Besides enterokinase, SEA domains were found in a number of other type II transmembrane serine proteases, including matriptase and matriptase‐2.9 Matriptase, overexpressed in numerous cancer cell lines, performs proteolysis on various cell surface proteins such as protease‐activated receptor 2 (PAR‐2) and the zymogen of the urokinase‐type plasminogen activator.10 Matriptase‐2 regulates iron homeostasis by proteolytic processing of hemojuvelin, the co‐receptor of bone morphogenetic protein.11 SEA domains were also observed in two adhesion‐type G‐protein coupled receptors (GPR110 and GPR116)12, 13 and the uromodulin like 1 protein.14
The SEA domain in the type I single‐pass transmembrane protein MUC1 (C‐terminus located in cytosol) was found to undergo autoproteolysis,15 creating two noncovalently bound α‐subunit and β‐subunit. Structural studies of MUC1 SEA domain revealed that it adopts a ferredoxin‐like fold, and the cleavage site is located in the middle of the β‐hairpin of the second and third β‐strands.16 The serine hydroxyl in the GSϕϕϕ consensus motif (ϕ: a hydrophobic residue) is responsible for the autoproteolysis that occurs at the glycine‐serine peptide bond. SEA domains with this motif also undergo proteolytic processing between the conserved glycine and serine in other proteins such as MUC3 and MUC12,17 enterokinase,18 matriptase,19 and the G‐protein coupled receptor Ig‐Hepta (the mouse ortholog of the human protein GPR116),20 presumably with the same autoproteolysis mechanism. However, this autoproteolysis motif is not conserved in all SEA domains. MUC16, for example, has multiple SEA domains, and none of them possesses this motif.21
A few divergent copies of SEA domains have been discovered by careful sequence and structure comparisons. One example is the SEA domains in the extracellular matrix receptor dystroglycan.22, 23 Like MUC1, dystroglycan was processed into the ligand‐binding α‐subunit and the membrane‐bound β‐subunit. Secondary structure and tertiary structure predictions coupled with the conservation of the autoproteolysis motif suggest that dystroglycan possesses a divergent SEA domain,23 which exhibits limited sequence similarity to previously identified SEA domains such as those in MUC1 and agrin.
Presence of an extracellular SEA domain in the membrane‐proximal stem region appears to be a recurring theme found in type I transmembrane proteins dystroglycan and MUC1 as well as some type II transmembrane serine proteases such as enterokinase, matriptase and matriptase‐2. Such a theme is also employed in another cell surface protein, the receptor‐type protein tyrosine phosphatase IA‐2.24, 25 IA‐2 is a type I transmembrane protein with a ferredoxin‐like extracellular domain adjacent to the transmembrane segment. This ferredoxin‐like domain was proposed to be a divergent SEA domain based on 3‐dimenional structural similarities and profile‐based sequence similarity searches.24 Moreover, Notch receptors possess an extracellular juxtamembrane domain (the heterodimeric (HD) domain) of the ferredoxin fold with noticeable structural similarity to mucin SEA domains and the IA‐2 SEA domains.26, 27 Like signaling mucins,7 the HD domains of Notch receptors are regulated by proteolysis events. The S1 furin cleavage site of the Notch HD domains is located in the same region as the autoproteolysis site of MUC1 SEA domain.26 Similarities in structures and active site locations suggest that the Notch HD domain is evolutionarily related to SEA domains.
Divergent SEA domains in dystroglycan, IA‐2, and Notch receptors suggest that SEA domain detection can be a challenging task. Here, we rely on comparative sequence and structural analyses to find new SEA domains and study their phylogenetic distributions. New SEA domains are proposed to be present in a number of cell surface proteins such as PTPRR, EpCAM, collectrin/Tmem27, Amnionless, CD34, KIAA0319, fibrocystin‐like protein, and several cadherins. SEA domains predate the last common ancestor of metazoans, as they were discovered in dystroglycan‐like proteins in Capsaspora owczarzaki (with conserved autoproteolysis motif) and fibrocystin‐like proteins beyond Holozoa. The functional regulation of SEA domain proteolysis could affect a much broader range of cell surface proteins than previously recognized. Interestingly, a new SEA domain was also found inside the cell in nucleoporin 54 (Nup54) in metazoans and choanoflagellates.
Results and Discussion
Sequence similarity searches of canonical SEA domains
Transitive PSI‐BLAST28 searches (see Materials and methods) starting from the SEA domain of MUC1 (NCBI GenBank accession: P15941.3, residues 1041‐1143) detected more than ten thousand proteins containing SEA domains in the non‐redundant protein database. They include all founding members of the SEA domains1 as well as numerous other SEA‐domain‐containing proteins. We name this large set of SEA domains related to MUC1 SEA domain as “canonical SEA domains” to distinguish them from the more divergent SEA domains that cannot be linked by PSI‐BLAST, such as those in dystroglycan23 and receptor‐type protein tyrosine phosphatase IA‐224 (described below). Canonical SEA domains are represented in the Pfam family SEA (PF01390). The NCBI GenBank accession numbers of canonical SEA domains and their residue ranges are listed in Supporting Information Table S1.
At least 26 proteins in the human genome were found to contain canonical SEA domains, including agrin (human gene official symbol: AGRN), perlecan (HSPG2), several mucins (MUC1, MUC3A, MUC12, MUC13, MUC16, and MUC17), two adhesion G‐protein coupled receptors (ADGRF1 (GPR110) and ADGRF5 (GPR116)), ten serine peptidases (TMPRSS11A, TMPRSS11B, TMPRSS11D, TMPRSS11E, TMPRSS11F, TMPRSS6 (matriptase‐2), TMPRSS7, TMPRSS9, ST14 (matriptase), and TMPRSS15 (enterokinase)), two interphotoreceptor matrix proteoglycans (IMPG1 and IMPG2), integrin beta‐4 (ITGB4), HEG1, C3orf52, and uromodulin like 1 (UMODL1). While most canonical SEA domains found by PSI‐BLAST were from metazoans, two proteins from choanoflagellates (GenBank: XP_001747329.1 of Monosiga brevicollis and GenBank: XP_004990163.1 of Salpingoeca rosetta) were also detected. These two proteins possess the autoproteolysis motif with the conserved glycine and serine, suggesting that both SEA domain and the autoproteolysis mechanism evolved before the advent of metazoans.
An alignment containing several canonical SEA domains including one from Monosiga is shown in Figure 1(A). The consensus autoproteolysis motif GSϕϕϕ is not present in all canonical SEA domains. Although the alignment contains positions with conserved hydrophobic residues (highlighted in yellow background) (Fig. 1), no positions with invariant residues were found. Canonical SEA domains were found to associate with a variety of extracellular domains [examples shown in Fig. 2(A)]. Many proteins with canonical SEA domains have serine/threonine‐rich regions that could undergo extensive O‐linked glycosylation, as observed in membrane‐bound mucins.29
Figure 1.
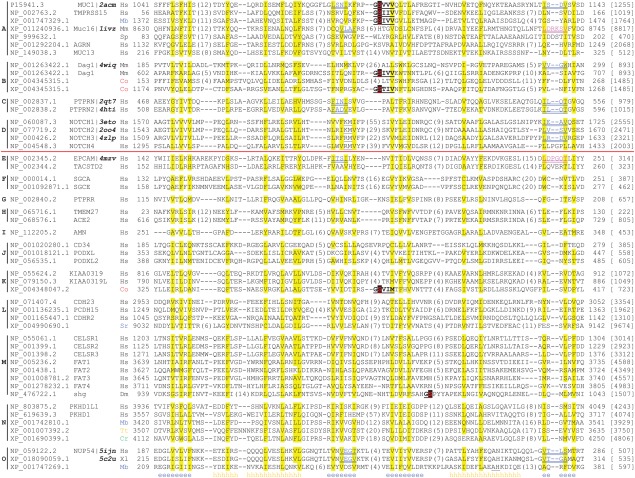
Multiple sequence alignment of SEA domains. Representative sequences of 15 SEA domain groups are shown: A – canonical SEA domains; B – dystroglycan group; C – receptor‐type protein tyrosine phosphatase IA‐2 group; D – Notch group; E – EpCAM group; F – α/ε‐sarcoglycan group; G – PTPRR group; H – collectrin group; I – amnionless group; J – CD34 group; K – KIAA0319 group; L – cadherin group 1; M – cadherin group 2; N – fibrocystin‐like group; O – Nup54 group. A red line separates known SEA domain groups (A–D) and newly discovered SEA domain groups (E–O). Official gene symbols are shown for human, mouse and fruit fly proteins, including some for the commonly used names in literature: enterokinase – TMPRSS15; Mucin‐1 – MUC1; agrin – AGRN; Mucin‐13 – MUC13; Mucin‐16 – Muc16; dystroglycan – Dag1; α‐sarcoglycan – SCGA; ε‐sarcoglycan – SCGE; IA‐2: PTPRN; IA‐2 β – PTPRN2; amnionless – AMN; TROP2 – TACSTD2; collectrin – TMEM27. For sequences with available structures, their four‐letter pdb IDs are shown as bold and italic letters after the accession numbers or gene symbols. In these sequences, β‐bulges with the ϕxxϕ motif are shown as double‐underlined blue letters. The corresponding regions with a small helix and the ϕxxxxϕ motif in 1ivz and 4mzv are shown as double‐underlined magenta letters. Autoproteolysis motifs are shown as underscored bold letters with the catalytic serine highlighted in black background. Noncharged residues in mainly hydrophobic positions are in yellow background. Long insertions are replaced with number of residues in parentheses. Residue insertions between two underlined residues are omitted, which occur in the second core β‐strand of Notch proteins (seven residues) and the last helix of Monosiga Nup54 (64 residues). Starting and ending residue numbers of the domains are shown before and after the sequences, respectively. Protein lengths are shown in brackets. Consensus secondary structure predictions are shown in the last line: “e” for β‐strand and “h” for α‐helix. Two‐letter organism name abbreviations shown after the accession numbers or official gene symbols are as follows: Co – Capsaspora owczarzaki; Cr – Chlamydomonas reinhardtii; Dm – Drosophila melanogaster; Hs – Homo sapiens; Mb – Monosiga brevicollis; Mm – Mus musculus; Sp – Strongylocentrotus purpuratus; Sr – Salpingoeca rosetta; Tt – Tetrahymena thermophila; Xl – Xenopus laevis. Organism name abbreviations are colored as follows: metazoa – black; choanoflagellate – blue; Filasterea – red; ciliate – orange; green algae – green.
Figure 2.
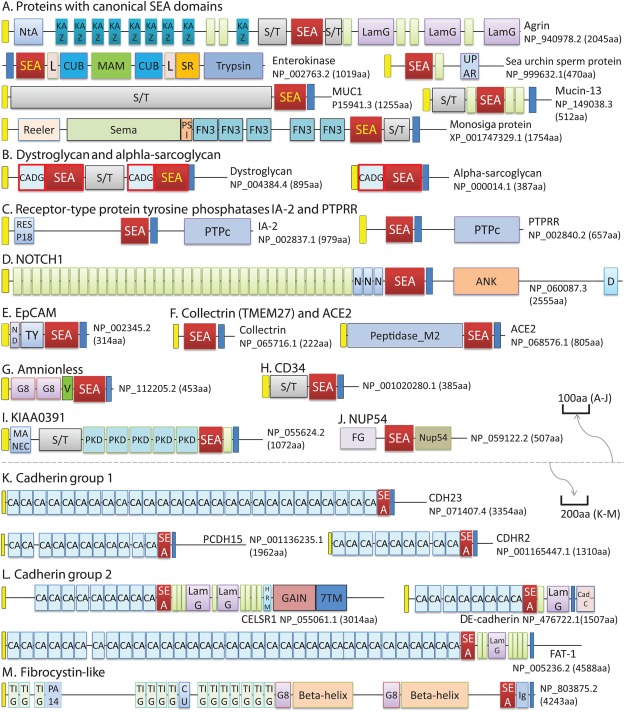
Domain diagrams of select SEA‐containing proteins. Domains or regions with sequence motifs are shown as rectangular boxes. N‐terminal signal peptides, transmembrane segments, and EGF domains are shown as yellow, blue and green unlabeled boxes, respectively. Other domains or regions are labeled with their names or name abbreviations in the boxes. The abbreviations are: 7TM – GPCR seven‐pass transmembrane domain; ANK – ankyrin repeats; CA – cadherin domain; Cad_C: cadherin cytoplasmic domain; CADG – cadherin‐like domain in dystroglycan; CU – Cupredoxin domain; D – DUF3454 domain; FG – FG repeat region in nucleoporins; KAZ – Kazal domain; L – LDLa domain; LamG – Laminin G domain; ND – N‐terminal domain of EpCAM; PTPc – protein phosphatase catalytic domain; S/T – serine and threonine rich region; UPAR – UPAR_LY6_2 domain; V – VWC domain. The CADG + SEA module are highlighted with thick red outlines. Names of SEA domains with the autoproteolysis motif are shown in yellow font. Domain diagrams above and below the dashed lines are shown in different length scales (suggested by the arrows) to accommodate several large proteins with more than 3,000 amino acid residues. The majority of these proteins are from human with the exceptions of the sea urchin sperm protein, a Monosiga protein with canonical SEA domains and the DE‐cadherin (shotgun) of D. melanogaster. GenBank accession numbers and protein lengths are shown for each protein.
Canonical SEA domains adopt a ferredoxin‐like fold with a βαββαβ sequential arrangement of core β‐strands (β) and α‐helices (α), as exemplified by one from mouse Muc16 [Fig. 3(A)]21 and one from human MUC1 [Fig. 3(B)].16 The first and third β‐strands in the middle of the β‐sheet exhibit the ϕxϕxϕ sequence pattern (Fig. 1), with the sidechains of the hydrophobic residues (ϕ) pointing to the core the structure. The β‐sheets in these two structures are curved, due in part to β‐bulges in the middle of the second and fourth core β‐strands, which are edge β‐strands. These β‐bulges create the ϕxxϕ hydrophobic pattern (double underlined blue letters in Fig. 1), with the two hydrophobic residues (ϕ) contributing to the core of the structure. MUC1 SEA domain and Muc16 SEA domain have the ϕxxϕxϕ and ϕxϕxxϕ patterns in the second core β‐strands respectively due to different locations of the β‐bulges [Fig. 1(A)]. The fourth core β‐strand of MUC1 SEA domain contains a β‐bulge with the ϕxxϕ motif, which is aligned to a region containing a short α‐helix with the ϕxxxxϕ motif in Muc16 SEA domain (shown as pink double‐underlined letters in Fig. 1). For both MUC1 and Muc16 SEA domain structures, a short α‐helix exists before the first core α‐helix that mainly interacts with the β‐hairpin of the second and third core β‐strands at an angle of about 45 degrees. The second core α‐helix is bended in MUC1 SEA domain [Fig. 3(A)], which allows it to interact with the fourth core β‐strand using mainly its N‐terminal half and to interact with the second core α‐helix using mainly its C‐terminal half. These structure and interaction features are largely conserved in Muc16, except that the C‐terminal part of the second core α‐helix in MUC1 is replaced by a loop in Muc16 [Fig. 3(B)].
Figure 3.
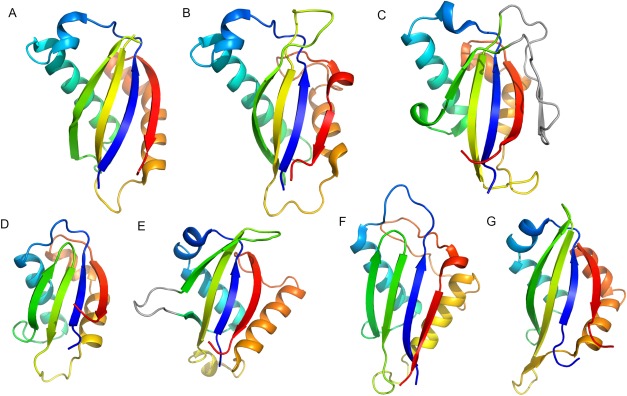
Structures of SEA domains. Cartoon representations are shown with rainbow coloring from N‐terminus (blue) to C‐terminus (red). These SEA domains are from: A. human MUC1 (pdb: 2acm); B. mouse Muc16 (pdb: 1ivz); C. N‐terminal region of mouse dystroglycan (pdb: 4wiq), with the long insertion between the second and third β‐strands colored grey; D. human IA‐2 (pdb: 2qt7); E. human NOTCH1 (pdb: 3eto); F. human EpCAM (pdb: 4mzv); G. Xenopus laevis Nup54 (pdb: 5c2u).
Nonmetazoan origin of SEA domains in dystroglycan
A divergent SEA domain was inferred to be present in the extracellular matrix receptor dystroglycan based on structure modeling and conservation of the autoproteolysis motif found in canonical SEA domains.23 This dystroglycan SEA domain exhibits limited sequence similarity to canonical SEA domains. Indeed, transitive PSI‐BLAST searches could not link this dystroglycan SEA domain to canonical SEA domains, and vice versa. In the Pfam database, this dystroglycan SEA domain is mapped to the DAG1 family (PF05454) and could not find canonical SEA domains (Pfam family: PF01390) by HMMER‐based searches.30 Like MUC1, the SEA domain with the autoproteolysis motif in dystroglycan lies adjacent to the transmembrane segment.23 Dystroglycan also possesses a cadherin‐like immunoglobulin domain (CADG domain in the SMART31 database)32 N‐terminally to this SEA domain.
Interestingly, the structure of the N‐terminal region of mouse dystroglycan33, 34 revealed a second divergent SEA domain [Fig. 3(C)] showing low sequence similarity to the dystroglycan SEA domain with autoproteolysis motif (sequence identity: 8%, based on the alignment shown in Fig. 1). A HHpred search35 using the mouse dystroglycan C‐terminal SEA domain (GenBank accession: NP_001263422.1, residues 602‐707) as the query found the dystroglycan N‐terminal SEA domain (pdb: 4wiq) with a statistically significant probability score of 97.2%, supporting the homology between the two SEA domains. However, transitive PSI‐BLAST could not link the dystroglycan N‐terminal SEA domain to canonical SEA domains and the dystroglycan C‐terminal SEA domain. A HMMER search using the mouse dystroglycan N‐terminal SEA domain as the query also could not detect the canonical SEA domain (Pfam: PF01390) and the dystroglycan C‐terminal SEA domain (Pfam: PF05454). Dystroglycan N‐terminal SEA domain does not possess the autoproteolysis motif, but instead has a long insertion in between the second and third core β‐strands [colored gray in Fig. 3(C)]. It is also preceded by a CADG domain, suggesting that it arose from a duplication [Fig. 2(B)]. In fact, the CADG + SEA module appears to have been duplicated one or more times in different metazoan lineages.36
Previous studies on the origin of dystroglycan only identified this protein within metazoans.36, 37 The CADG domain, on the other hand, has a much deeper evolutionary origin, as it was found in various eukaryotes outside metazoans such as fungi, various protists, and some bacteria.32 While our transitive PSI‐BLAST searches of the two SEA domains in dystroglycan found only proteins from metazoans, PSI‐BLAST searches of CADG domains identified three CADG‐containing proteins in Capsaspora owczarzaki of the Filasterea group, one of the closest single‐cell relatives of metazoans.37 We used HHpred to investigate if, like the metazoan dystroglycans, the regions after CADG in these Capsaspora proteins are SEA domains. Indeed, HHpred hits to dystroglycan SEA domains were found in all three CADG‐containing proteins of Capsaspora, suggesting that dystroglycan‐like proteins were present in the common ancestor of Filasterea and metazoans.
HHpred results suggest that one Capsaspora protein (GenBank: XP_004345315.1) has the same domain structure as the mouse dystroglycan [Fig. 2(B)], with two CADG + SEA modules and a serine/threonine‐rich region in between them. The C‐terminal SEA domain adjacent to the predicted transmembrane segment of this Capsaspora protein has the autoproteolysis motif with conserved glycine and serine that can be aligned to those in metazoan dystroglycan SEA domains [Fig. 1(B)]. Similar to the case of mouse dystroglycan, the N‐terminal SEA domain of this Capsaspora protein does not have the autoproteolysis motif [Fig. 1(B)]. The other two Caspaspora proteins (GenBank: XP_004365299.2 and XP_004365318.1) have one CADG + SEA module adjacent to a predicted transmembrane segment, a S/T‐rich region before CADG + SEA, and one (GenBank: XP_004365299.2) or two (GenBank: XP_004365318.1) discoidin domains (Pfam38 domain: F5_F8_type_C)39 in the N‐terminal region. The SEA domains in these two Capsaspora proteins also maintain the autoproteolysis motif with conserved glycine and serine, indicating that the autoproteolysis mechanism in dystroglycan evolved before the divergence of Filasterea and metazoans. We did not identify SEA domains in CADG‐containing proteins outside Filasterea, such as the fungi protein Axl2p.40 These findings suggest that the CADG + SEA module in dystroglycan could be a novel invention in Holozoa (Metazoa and its close single cell replatives Choanoflagellatea, Filasterea and Ichthyosporea).41 This invention could be an important event in the evolution of multicellularity in animals considering the multiple roles of dystroglycan in cell adhesion and the communication between extracellular matrix and cytoskeleton.42
Structural features of known SEA domains
Another example of a previously identified divergent SEA domain is in the receptor‐type protein tyrosine phosphatase IA‐2 (insulinoma‐associated protein 2, human gene official symbol: PTPRN).24, 25 IA‐2 is a type I transmembrane protein with an ectodomain adopting a ferredoxin‐like fold [Fig. 3(D)], which was proposed to be a SEA domain based on structural similarities.24 IA‐2 SEA domain undergoes autoproteolysis in vitro by reactive oxygen species.24 Our transitive PSI‐BLAST searches could not link the SEA domain in IA‐2 (Pfam family: Receptor_IA‐2 (PF11548)) with canonical SEA domains, indicating high sequence divergence between them. The human genome possesses a paralog of IA‐2, IA‐2β (also known as Phogrin, human gene official symbol: PTPRN2)43 [Fig. 1(C)].
A common theme found in type I transmembrane cell surface proteins MUC1, dystroglycan, and IA‐2 is the location of a SEA domain adjacent to the transmembrane segment. Such a recurring SEA + TM module (TM: transmembrane segment) was also identified in the cell surface Notch receptors [Fig. 2(D)], as they also possess a ferredoxin‐like domain26, 44 adjacent to the transmembrane segment. The ferredoxin‐like HD domains of Notch receptors exhibit significant structural similarity to other SEA domains, as previously reported.26, 27 For example, a DaliLite search45 using the human Notch1 ferredoxin‐like domain as query (pdb id: 3eto, chain A, residues 1572‐1727) [Fig. 3(E)] retrieved the ferredoxin‐like domains of Nup54 (another SEA domain described below) (e.g., pdb id: 5c2u, chain A, Z‐score: 10.4), IA‐2/IA‐2β (e.g., pdb id: 4hti, chain A, Z‐score: 7.8) and a canonical SEA domain (pdb id: 1ivz, chain A, Z‐score: 7.1) as the top hits.
While the Ferredoxin‐like βαββαβ fold has been observed in many protein domains, they could exhibit large structural differences in terms of secondary structure lengths, curvature of β‐sheet, and relative orientation of secondary structure elements. For example, the iron‐sulfur‐binding ferredoxins with the same fold usually have shorter β‐strands than those in the SEA domains. DaliLite comparisons of an iron‐sulfur‐binding ferredoxin structure (pdb: 2fdn, chain A) to SEA domain structures of MUC1 (pdb: 2acm), Muc16 (pdb: 1ivz), IA‐2 (pdb: 2qt7) and Notch1 (pdb: 3eto) all have Dali Z‐scores less than 2, suggesting large structural differences. On the other hand, the structures of Notch1, IA‐2, and dystroglycan exhibit a few common features with canonical SEA domain structures from MUC1 and Muc16, supporting the homology among them. The β‐sheets in all these structures have a concave surface on the side not interacting with the core α‐helices. A β‐bulge with the ϕxxϕ motif is located at the beginning of the fourth β‐strands in the structures of Notch1, IA‐2, dystroglycan, and MUC1. In addition, the first core α‐helix lies about 45 degrees relative to the β‐hairpin of the second and third core β‐strands in all these structures. A distinct structural feature for Notch SEA domains is the insertion of a loop [colored gray in Fig. 3(E) and omitted between the underlined KM or RM letters in Fig. 1(D)] in the region corresponding to the β‐bulges in the second core β‐strand of Muc16.
Compared to the autoproteolysis sites of SEA domains in MUC1 and dystroglycan, Notch SEA domain possesses a long insertion between the two corresponding β‐strands. Cleavage of Notch receptors at this site is not through autoproteolysis, but instead likely performed by furin‐like proteases.46, 47 Notch SEA domain is represented as two Pfam domains NOD (PF06816) and NODP (PF07684) that are separated at this insertion site. PSI‐BLAST and HHpred searches indicate that Notch and its SEA domain are restricted to metazoans, including organisms from basal metazoan groups such as Porifera and Ctenophora.
EpCAM has a novel divergent SEA domain
Structural comparison and domain architecture analysis suggest that the ferredoxin‐like domain in EpCAM (Epithelial cell adhesion molecule)48, 49 [Fig. 3(F)] is a SEA domain in yet another incidence of the SEA + TM module [Fig. 2(E)], which has been observed in known SEA‐containing proteins MUC1, IA‐2, dystroglycan, and Notch. Like the SEA domain structures of MUC1 and IA‐2, EpCAM SEA domain structure contains a β‐bulge with the ϕxxϕ motif in the second core β‐strand. It also harbors a short α‐helix in the ϕxxxxϕ motif at the start of the fourth core β‐strand that can be aligned with the one in Muc16 (Figs. 1 and 3). While the GSϕϕϕ autoproteolysis motif is not present in EpCAM, EpCAM is a target of proteolysis at multiple sites including those in the transmembrane segment and the ectodomain, one of which is mapped in the SEA domain.50 The homodimerization of EpCAM SEA domains could contribute to the forming of EpCAM cis‐dimers on the cell surface.48 A close homolog of EpCAM is TROP2 (trophoblast cell‐surface antigen‐2), also named TACSTD2 (tumor‐associated calcium signal transducer 2). TROP2 is a calcium signal transducer that shows differential expression in a variety of cancers.51 PSI‐BLAST searches suggest that close homologs of EpCAM and TROP2 are only present in vertebrates, suggesting a relatively late origin of them compared to other SEA‐domain containing proteins such as dystroglycan and Notch. EpCAM SEA domain has not been incorporated in the Pfam database (version 30.0).
Putative novel SEA domains in other cell surface proteins revealed by profile‐profile searches
We identified a number of additional cell surface proteins that potentially contain SEA domains through profile‐profile searches by HHpred. They include α/ε‐sarcoglycan, PTPRR, collectrin/Tmem27, amnionless, CD34, KIAA0319, fibrocystin‐like protein, and two groups of cadherins. Most of these proteins are type I transmembrane proteins. Like SEA domains in MUC1, dystroglycan, IA‐2, Notch, and EpCAM, the newly identified SEA domains often lie adjacent or close to the transmembrane segment (Fig. 2). SEA domains of α/ε‐sarcoglycan, collectrin/Tmem27, amnionless, and CD34 have been incorporated into the Pfam database (version 30.0) in the family entries of Sarcoglycan_2 (PF05510), Collectrin (PF16959), Amnionless (PF14828), and CD34_antigen (PF06365), respectively. On the other hand, newly discovered SEA domains in PTPRR, KIAA0319, fibrocystin‐like protein, and cadherins cannot be mapped to existing Pfam families by HMMER searches.
The α/ε‐sarcoglycan SEA domain group
Pofile‐profile based sequence similarity searches by HHpred35 suggest that a single copy of the CADG + SEA module is present in α‐sarcoglycan and ε‐sarcoglycan [Fig. 2(B)]. α‐sarcoglycan is mainly expressed in striated muscle tissues and forms the sarcoglycan subcomplex with β‐, γ‐, and δ‐sarcoglycans. Mutations in the subunits of the sarcoglycan subcomplex can lead to the limb‐girdle muscular dystrophy.52 Unlike α‐sarcoglycan that is a type I transmembrane protein, β‐, γ‐, and δ‐sarcoglycans are type II transmembrane proteins and do not possess SEA domains. Both the sarcoglycan subcomplex and dystroglycan are parts of the dystrophin‐associated glycoprotein complex that links the actin cytoskeleton to the extracellular matrix in muscles.53 ε‐sarcoglycan, mutations of which cause the Myoclonus dystonia syndrome, has a wider tissue distribution than α‐sarcoglycan and could be involved in dystrophin‐associated complex in tissues such as brain.54 The SEA domain of α/ε‐sarcoglycan cannot be linked by transitive PSI‐BLAST or HMMER to canonical SEA domains and dystroglycan SEA domains. They are only found in metazoans including Bilateria and Cnidaria.
The PTPRR SEA domain group
A previously unnoticed and highly divergent SEA domain was discovered in receptor‐type protein tyrosine phosphatase R (human gene official symbol: PTPRR)55 [Figs. 1(E) and 2(C)], which could not be linked by transitive PSI‐BLAST searches to canonical SEA domains or SEA domains from receptor‐type protein tyrosine phosphatase IA‐2. While IA‐2 SEA domains were found in metazoans beyond chordates, the PTPRR SEA domains appear to be restricted to vertebrates. The narrower phyletic distribution of PTPRR compared to IA‐2 and their shared domain structure suggest that PTPRR could have arisen from a gene duplication of IA‐2. A proteolysis site has been mapped to the SEA domain region of the mouse PTPRR protein.56
The collectrin/Tmem27 SEA domain group
Collectrin (human gene official symbol: TMEM27) and ACE2 share similarity in part of the extracellular region corresponding to the SEA domain, transmembrane segment and the cytosolic region.57 ACE2 has an additional N‐terminal peptidase domain similar to ACE (angiotensin‐converting enzyme) that is involved in the renin‐angiotensin system58 [Fig. 2(F)]. Collectrin/Tmem27 and ACE2 play important roles in renal and intestinal amino acid transport by acting as binding partners of amino acid transporters, regulating their trafficking and expression on the cell surface, and involving in their catalytic activities.59, 60, 61 Collectrin/Tmem27 has been shown to bind to protein complexes involved in intracellular and ciliary movement of vesicles and membrane proteins.62 Collectrin/Tmem27 in pancreatic beta cells was proteolytically processed in the extracellular region.63 Close homologs of collectrin/Tmem27 and ACE2 were only found in chordates including amphioxus and urochordates, suggesting a relatively late appearance of these proteins in evolution.
The amnionless SEA domain group
Amnionless is part of the multi‐ligand receptor (amnionless+ cubilin) responsible for absorption of vitamin B12.64, 65 As a type I transmembrane protein, Aminionless contains two G8 domains66 and a cysteine‐rich VWC domain67 N‐terminal to the SEA + TM module [Fig. 2(G)]. Amnionless SEA domain appears to be only present in metazoans. A few amnionless‐like proteins in choanoflagellates found by PSI‐BLAST have the N‐terminal G8 domains, but lack the VWC and SEA domains.
The CD34 SEA domain group
CD34 and its closely related proteins such as podocalyxin and podocalyxin‐like protein 2 (also named endoglycan)68 are cell surface glycoproteins with a heavily glycosylated mucin‐like serine/threoine‐rich region. These proteins also possess the SEA + TM module [CD34 domain structure shown in Fig. 2(H)]. Despite its use as a marker of various tissue‐specific stem cells including hematopoietic stem cells, the exact function of CD34 remains unclear.69 Close homologs of CD34 proteins were only found in vertebrates and Cephalochordata.
The KIAA0319 SEA domain group
The human dyslexia‐associated protein KIAA031970, 71 is a highly glycosylated type I plasma membrane protein with a MANEC (motif at the N terminus with eight cysteines) domain,72 five PKD (polycystic kidney disease) domains,73 and an EGF domain [Fig. 2(I)]. The SEA domain of KIAA0319 lies N‐terminally to the EGF domain near the predicted transmembrane segment. Like Notch receptors, KIAA0319 undergoes proteolysis in both extracellular region and the transmembrane segment.74 Close homologs of KIAA0319 SEA domain were mostly identified in metazoans, including KIAA0319‐like proteins (human gene official symbol: KIAA0319L) that are paralogs of KIAA0319 in vertebrates. A protein from Capsaspora owczarzaki (GenBank: XP_004348047.2) is the single nonmetazoan protein with the KIAA0319 SEA domain among the PSI‐BLAST hits. Interestingly, it possesses the GSϕϕϕ motif [Fig. 1(K)] that can be aligned to the autoproteolysis motifs of canonical SEA domains and dystroglycan SEA domains, suggesting that the Capasspora KIAA0319 homolog could possess autoproteolysis activity. On the other hand, none of the metazoan KIAA0319 homologs has the autoproteolysis motif.
The CDH23/PCDH15/CDHR2 cadherin SEA domain group
We found previously unnoticed SEA domains in a number of cell adhesion proteins belonging to the cadherin superfamily,75, 76 which consists of proteins with the cadherin domain. They can be divided in two groups. SEA domains in these two groups of cadherins exhibit quite large sequence diversity and could not be linked by transitive PSI‐BLAST searches. One group of SEA‐containing cadherins includes human proteins CDH23 (cadherin 23), PCDH15 (protocadherin related 15), and CDHR2 (cadherin related family member 2) [Fig. 1(L)]. CDH23 and PCDH15 mediate cell‐cell adhesion by interacting with each other in sensory hair cells.77 These human cadherin superfamily members have the SEA + TM module and contain no other domains except cadherin domains in the extracellular region [Fig. 2(K)]. Two cadherin proteins with Coherin domain and Dockerin domain from choanoflagellates78 were also found by transitive PSI‐BLAST searches (Monosiga brevicollis protein GenBank: XP_001750073.1 and Salpingoeca rosetta protein GenBank: XP_004990690.1), suggesting that the SEA domain in these cadherins originated before the advent of metazoans.
The CELSR/FAT cadherin SEA domain group
The other group of cadherins with SEA domains consists of the cadherin EGF LAG seven‐pass G‐type receptors (e.g., human CELSR1, CELSR2 and CELSR3),79 the Fat family of protocadherins80 (e.g., human FAT1, FAT2, FAT3 and FAT4) [Fig. 1(M)], and the invertebrate DN‐cadherins and DE‐cadherins. SEA domains in these cadherins are located C‐terminally to the cadherin domain (CA) repeats and N‐terminally to EGF repeat(s) and Lamimin G (LamG) domain(s) [Fig. 2(L)]. This SEA domain corresponds to the “Flamingo box” region in Flamingo, a cadherin EGF LAG seven‐pass G‐type receptor in Drosophila melanogaster.79 It also corresponds to the “primitive classical cadherin proteolytic site domain” (PCPS) in invertebrate DE‐ and DN‐cadherins,81 as this SEA domain in D. melanogaster DE‐cadherin (official gene symbol: shg, gene name: shotgun) has been found to undergo proteolysis.82 The cleavage site is also between a glycine and a serine in the SAHGSPYY segment,82 albeit this motif is located between the third core β‐strand and the second core α‐helix [Fig. 1(M)], unlike the location of autoproteolysis motif of canonical SEA domains (between the second and third core β‐strands). While the DE and DN‐cadherins and Fat proteins are type I transmembrane proteins, the CELSR proteins have the modular domains (CA + SEA + EGF + LamG) grafted to seven‐pass adhesion GPCRs83, 84 that also contain the HRM domain and the GAIN domain.85, 86 These SEA‐domain‐containing cadherins were only found in metazoans in PSI‐BLAST searches.
The fibrocystin‐like SEA domain group
A divergent SEA domain was also identified in a group of proteins that include human fibrocystin (encoded by the PKHD1 (polycystic kidney and hepatic disease 1) gene) and fibrocystin‐like protein (encoded by the PKHD1L1 gene). Mutations in fibrocystin are the cause of autosomal recessive polycystic kidney disease.87 Fibrocystin and fibrocystin‐like proteins possess several TIG domains,88 two G8 domains,66 and two regions of β‐helix repeats89 [Fig. 2(M)]. The SEA domain in the human Fibrocystin‐like protein lies in the C‐terminal part of the extracellular region before an Ig‐like domain (detected by HHpred) [Fig. 2(M)]. Like some of the Notch receptors, both human fibrocystin and fibrocystin‐like protein possess the Rx[KR]R motif in the loop region between the second and third core β‐strands of the predicted ferredoxin‐like fold. In fact, fibrocystin undergoes Notch‐like sequential proteolysis events including the processing by a probable proprotein convertase at the furin‐cleavage site, an ADAM metalloproteinase, and γ‐secretase.90 Fibrocystin‐like proteins were found beyond Holozoa in diverse eukaryotic lineages including green algae, Alveolata, Euglenozoa, and Haptophyta. The SEA domains are also present in these proteins [a few of them shown in Fig. 1(M)], suggesting that the SEA domains in fibrocystin‐like proteins have a deep eukaryotic origin.
Identification of SEA domain in the intracellular protein nucleoporin 54
All previously described SEA domains have extracellular localization. Interestingly, transitive PSI‐BLAST searches (e‐value inclusion threshold: 1e‐3) using cadherin SEA domains found a domain with ferredoxin‐like fold in Nup54 (nucleoprotein 54) with statistically significant e‐values (less than 1e‐3). Nup54 is a subunit of the Nup62•58•54 nuclear pore complex.91 Vertebrate Nup54 proteins contain an N‐terminal region with FG repeats, a ferredoxin‐like domain (not included in the current Pfam version 30.0 database) and a C‐terminal domain mainly consisting of coiled coils (Pfam family Nup54 (PF13874)) that mediates interactions with Nup62 and Nup58 [Fig. 2(J)].92
Evidence that the ferredoxin‐like domain in Nup54 is homologous to SEA domains also comes with structural comparisons. A DaliLite search using the ferredoxin‐like domain of a vertebrate Nup54 protein (pdb id: 5c2u, chain A, residues 214‐315)92 [Fig. 3(F)] as the query against the PDB database retrieved several SEA domain‐containing structures as the top hits. The best structural similarity hit is the SEA domain from mouse Muc16 (pdb: 1ivz)21 with a Z‐score of 7.7. The second best hit is the SEA domain from Notch3 (pdb: 4zlp)44 with a Z‐score of 7.6. The hits to the SEA domains of receptor‐type protein tyrosine phosphatases IA‐2 and IA‐2 β come next immediately after various hits to Notch SEA domains (e.g., pdb id: 4hti43 with a Z‐score of 6.9). Structural superpositions of Nup54 to the top two hits (1ivz and 4zlp) are included in Supporting Information Figure S1. Like other SEA domains with available structures, Nup54 SEA domain has a concave surface of the β‐sheet [Fig. 3(G)]. It contains a β‐bulge in the fourth core β‐strand that can be aligned with those in MUC1 (pdb: 2acm), dystroglycan (pdb: 4wiq), IA‐2 (pdb: 2qt7), IA‐2 β (pdb: 4hti), and Notch receptors (pdbs: 3eto, 2oo4, and 4zlp) (Fig. 1). It also has a β‐bulge in the second core β‐strand that can be aligned with the one in Muc16 (pdb: 1ivz). Like SEA domain structures of MUC1, IA‐2 and dystroglycan, the second core α‐helix in Nup54 is kinked, which allows it to interact with the fourth core β‐strand and the first core α‐helix in a similar fashion.
Nup54 homologs with the N‐terminal FG repeat region and the C‐terminal Nup54 domain (Pfam family PF13874) are found in most eukaryotic lineages including fungi, metazoans, plants, and various protists such as Naegleria gruberi, Phytophthora parasitica and Acanthamoeba castellanii, suggesting that it may be present in the common ancestor of eukaryotes. The ferredoxin‐like SEA domain of Nup54, on the other hand, were only found in metazoans in a previous study.92 Using HHpred, we were also able to locate the SEA domain in the Nup54 protein from the choanoflagellate M. brevicollis [Fig. 1(N)], but not in organisms outside Holozoa. Nup54 SEA domains do not possess the autoproteolysis motif observed in some canonical SEA domains and dystroglycan, nor do most of the newly identified SEA domains (one exception is the Capsaspora KIAA0319 protein). Whether Nup54 can undergo proteolysis by other proteases awaits experimental studies.
Conclusions
Since the first description of canonical SEA domains more than twenty years ago, a few divergent SEA domains have been revealed in dystroglycan, receptor‐type protein tyrosine phosphatase IA‐2, and Notch receptors. By comprehensive sequence and structural analyses, we further expanded the repertoire of SEA domains in a diverse array of cell surface proteins including EpCAM, α/ε‐sarcoglycan, PTPRR, collectrin/Tmem27, amnionless, CD34, KIAA0319, fibrocystin‐like protein, and two groups of cadherins. A SEA domain was also inferred to have transferred to nucleoporin 54 in the ancestor of choanoflagellates and metazoans. The homology among the divergent SEA domain groups is supported by profile‐based similarity searches, structure predictions and comparisons, domain structure analysis and sequence motif analysis. Known and newly discovered SEA domain groups exhibit distinct phyletic distributions (Supporting Information Table S2). SEA‐domain‐containing fibrocystin‐like proteins are present in various eukaryotic lineages outside Holozoa, suggesting an ancient evolutionary origin of SEA domains. On the other hand, the other SEA domain‐containing proteins appear to be restricted to metazoans and their closest single‐cell relatives such as choanoflagellates and Filasterea. SEA domains tend to occur in membrane proximal regions of cell surface proteins, and experimental studies revealed that many SEA domains serve as hotspots for proteolytic cleavage, either by autoproteolysis or through the action of other proteases. The proteolysis events occurring within or near the SEA domains could function in creating ligand‐receptor alliances,93 protecting cells from rupture,16 modulating ligand‐binding activities,23 or generating fragments that transduce signals from cell membrane to the nucleus.7 Identification of novel SEA domains has significant functional implications and could offer new research directions for proteins containing them. Nonmetazoan origin of SEA domains in proteins such as dystroglycan and fibrocystin‐like protein suggests their contribution to the expansion of functional modules in cell adhesion and extracellular matrix in the evolutionary process that led to animal multicellularity.
Materials and Methods
Sequence similarity searches
PSI‐BLAST (28) iterations were conducted to search for homologs of canonical SEA domain starting from the SEA domain of MUC1 (NCBI GenBank accession: P15941.3, residues 1041‐1143) against the NCBI non‐redundant (nr) protein database (e‐value inclusion cutoff: 1e‐3). To perform transitive searches, PSI‐BLAST hits were grouped by BLASTCLUST (with the score coverage threshold (−S, defined as the bit score divided by alignment length) set to 1, length coverage threshold (−L) set to 0.5, and no requirement of length coverage on both sequences (−bF)), and a representative sequence from each group was used to initiate new PSI‐BLAST searches. Such an iterative procedure was repeated until convergence. This transitive PSI‐BLAST procedure was also used for finding homologs of divergent SEA domains. HHpred web server35 was used for profile‐profile‐based similarity searches to identify distant homologous relationships of SEA domains (profile databases used: Pfam,38 pdb70 and the proteome databases of available eukaryotic organisms in the server).
Sequence alignment and domain architecture analysis
The multiple sequence alignment for select members of SEA domains was made by PROMALS3D94 and improved by manual adjustment. HMMER330 and HHpred web server35 were used to detect known Pfam domains in SEA‐domain‐containing proteins with default parameter settings. Phobius95 was used to predict transmembrane segments and N‐terminal signal peptides.
Supporting information
Supporting Information Figure 1.
Supporting Information Table 1.
Acknowledgments
We would like to thank Lisa Kinch for critical reading of the manuscript and helpful discussion. This work is supported by National Institutes of Health (GM094575 to NVG) and Welch Foundation (I‐1505 to NVG).
Conflict of Interest: None declared
References
- 1. Bork P, Patthy L (1995) The SEA module: a new extracellular domain associated with O‐glycosylation. Protein Sci 4:1421–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Iozzo RV (1998) Matrix proteoglycans: from molecular design to cellular function. Ann Rev Biochem 67:609–652. [DOI] [PubMed] [Google Scholar]
- 3. Manes G, Meunier I, Avila‐Fernandez A, Banfi S, Le Meur G, Zanlonghi X, Corton M, Simonelli F, Brabet P, Labesse G, Audo I, Mohand‐Said S, Zeitz C, Sahel JA, Weber M, Dollfus H, Dhaenens CM, Allorge D, De Baere E, Koenekoop RK, Kohl S, Cremers FP, Hollyfield JG, Senechal A, Hebrard M, Bocquet B, Ayuso Garcia C, Hamel CP (2013) Mutations in IMPG1 cause vitelliform macular dystrophies. Am J Human Genet 93:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bandah‐Rozenfeld D, Collin RW, Banin E, van den Born LI, Coene KL, Siemiatkowska AM, Zelinger L, Khan MI, Lefeber DJ, Erdinest I, Testa F, Simonelli F, Voesenek K, Blokland EA, Strom TM, Klaver CC, Qamar R, Banfi S, Cremers FP, Sharon D, den Hollander AI (2010) Mutations in IMPG2, encoding interphotoreceptor matrix proteoglycan 2, cause autosomal‐recessive retinitis pigmentosa. Am J Human Genet 87:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kufe DW (2009) Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer 9:874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bafna S, Kaur S, Batra SK (2010) Membrane‐bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 29:2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cullen PJ (2011) Post‐translational regulation of signaling mucins. Curr Opin Struct Biol 21:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Light A, Janska H (1989) Enterokinase (enteropeptidase): comparative aspects. Trends Biochem Sci 14:110–112. [DOI] [PubMed] [Google Scholar]
- 9. Bugge TH, Antalis TM, Wu Q (2009) Type II transmembrane serine proteases. J Biol Chem 284:23177–23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uhland K (2006) Matriptase and its putative role in cancer. Cell Mol Life Sci 63:2968–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramsay AJ, Hooper JD, Folgueras AR, Velasco G, Lopez‐Otin C (2009) Matriptase‐2 (TMPRSS6): a proteolytic regulator of iron homeostasis. Haematologica 94:840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fredriksson R, Lagerstrom MC, Hoglund PJ, Schioth HB (2002) Novel human G protein‐coupled receptors with long N‐terminals containing GPS domains and Ser/Thr‐rich regions. FEBS Lett 531:407–414. [DOI] [PubMed] [Google Scholar]
- 13. Lum AM, Wang BB, Beck‐Engeser GB, Li L, Channa N, Wabl M (2010) Orphan receptor GPR110, an oncogene overexpressed in lung and prostate cancer. BMC Cancer 10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shibuya K, Nagamine K, Okui M, Ohsawa Y, Asakawa S, Minoshima S, Hase T, Kudoh J, Shimizu N (2004) Initial characterization of an uromodulin‐like 1 gene on human chromosome 21q22.3. Biochem Biophys Res Commun 319:1181–1189. [DOI] [PubMed] [Google Scholar]
- 15. Levitin F, Stern O, Weiss M, Gil‐Henn C, Ziv R, Prokocimer Z, Smorodinsky NI, Rubinstein DB, Wreschner DH (2005) The MUC1 SEA module is a self‐cleaving domain. J Biol Chem 280:33374–33386. [DOI] [PubMed] [Google Scholar]
- 16. Macao B, Johansson DG, Hansson GC, Hard T (2006) Autoproteolysis coupled to protein folding in the SEA domain of the membrane‐bound MUC1 mucin. Nature Struct Mol Biol 13:71–76. [DOI] [PubMed] [Google Scholar]
- 17. Palmai‐Pallag T, Khodabukus N, Kinarsky L, Leir SH, Sherman S, Hollingsworth MA, Harris A (2005) The role of the SEA (sea urchin sperm protein, enterokinase and agrin) module in cleavage of membrane‐tethered mucins. FEBS J 272:2901–2911. [DOI] [PubMed] [Google Scholar]
- 18. Matsushima M, Ichinose M, Yahagi N, Kakei N, Tsukada S, Miki K, Kurokawa K, Tashiro K, Shiokawa K, Shinomiya K, H Umeyama, H Inoue, T Takahashi, K Takahashi (1994) Structural characterization of porcine enteropeptidase. J Biol Chem 269:19976–19982. [PubMed] [Google Scholar]
- 19. Cho EG, Kim MG, Kim C, Kim SR, Seong IS, Chung C, Schwartz RH, Park D (2001) N‐terminal processing is essential for release of epithin, a mouse type II membrane serine protease. J Biol Chem 276:44581–44589. [DOI] [PubMed] [Google Scholar]
- 20. Abe J, Fukuzawa T, Hirose S (2002) Cleavage of Ig‐Hepta at a “SEA” module and at a conserved G protein‐coupled receptor proteolytic site. J Biol Chem 277:23391–23398. [DOI] [PubMed] [Google Scholar]
- 21. Maeda T, Inoue M, Koshiba S, Yabuki T, Aoki M, Nunokawa E, Seki E, Matsuda T, Motoda Y, Kobayashi A, Hiroyasu F, Shirouzu M, Terada T, Hayami N, Ishizuka Y, Shinya N, Tatsuguchi A, Yoshida M, Hirota H, Matsuo Y, Tani K, Arakawa T, Carninci P, Kawai J, Hayashizaki Y, Kigawa T, Yokoyama S (2004) Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16). J Biol Chem 279:13174–13182. [DOI] [PubMed] [Google Scholar]
- 22. Henry MD, Campbell KP (1996) Dystroglycan: an extracellular matrix receptor linked to the cytoskeleton. Curr Opin Cell Biol 8:625–631. [DOI] [PubMed] [Google Scholar]
- 23. Akhavan A, Crivelli SN, Singh M, Lingappa VR, Muschler JL (2008) SEA domain proteolysis determines the functional composition of dystroglycan. FASEB J 22:612–621. [DOI] [PubMed] [Google Scholar]
- 24. Primo ME, Klinke S, Sica MP, Goldbaum FA, Jakoncic J, Poskus E, Ermacora MR (2008) Structure of the mature ectodomain of the human receptor‐type protein‐tyrosine phosphatase IA‐2. J Biol Chem 283:4674–4681. [DOI] [PubMed] [Google Scholar]
- 25. Lan MS, Lu J, Goto Y, Notkins AL (1994) Molecular cloning and identification of a receptor‐type protein tyrosine phosphatase, IA‐2, from human insulinoma. DNA Cell Biol 13:505–514. [DOI] [PubMed] [Google Scholar]
- 26. Gordon WR, Vardar‐Ulu D, Histen G, Sanchez‐Irizarry C, Aster JC, Blacklow SC (2007) Structural basis for autoinhibition of Notch. Nature Struct Mol BIol 14:295–300. [DOI] [PubMed] [Google Scholar]
- 27. Gordon WR, Roy M, Vardar‐Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC (2009) Structure of the Notch1‐negative regulatory region: implications for normal activation and pathogenic signaling in T‐ALL. Blood 113:4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jonckheere N, Skrypek N, Frenois F, Van Seuningen I (2013) Membrane‐bound mucin modular domains: from structure to function. Biochimie 95:1077–1086. [DOI] [PubMed] [Google Scholar]
- 30. Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR (2015) HMMER web server: 2015 update. Nucleic Acids Res 43:W30–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43:D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dickens NJ, Beatson S, Ponting CP (2002) Cadherin‐like domains in alpha‐dystroglycan, alpha/epsilon‐sarcoglycan and yeast and bacterial proteins. Curr Biol 12:R197–R199. [DOI] [PubMed] [Google Scholar]
- 33. Bozic D, Sciandra F, Lamba D, Brancaccio A (2004) The structure of the N‐terminal region of murine skeletal muscle alpha‐dystroglycan discloses a modular architecture. J Biol Chem 279:44812–44816. [DOI] [PubMed] [Google Scholar]
- 34. Bozzi M, Cassetta A, Covaceuszach S, Bigotti MG, Bannister S, Hubner W, Sciandra F, Lamba D, Brancaccio A (2015) The structure of the T190M mutant of murine alpha‐dystroglycan at high resolution: Insight into the molecular basis of a primary dystroglycanopathy. PloS One 10:e0124277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams JC, Brancaccio A (2015) The evolution of the dystroglycan complex, a major mediator of muscle integrity. Biol Open 4:1163–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suga H, Chen Z, de Mendoza A, Sebe‐Pedros A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sanchez‐Pons N, Torruella G, Derelle R, Manning G, Lang BF, Russ C, Haas BJ, Roger AJ, Nusbaum C, Ruiz‐Trillo I (2013) The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun 4:2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador‐Vegas A, Salazar GA, Tate J, Bateman A (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baumgartner S, Hofmann K, Chiquet‐Ehrismann R, Bucher P (1998) The discoidin domain family revisited: new members from prokaryotes and a homology‐based fold prediction. Protein Sci 7:1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao XD, Sperber LM, Kane SA, Tong Z, Tong AH, Boone C, Bi E (2007) Sequential and distinct roles of the cadherin domain‐containing protein Axl2p in cell polarization in yeast cell cycle. Mol Biol Cell 18:2542–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lang BF, O'Kelly C, Nerad T, Gray MW, Burger G (2002) The closest unicellular relatives of animals. Curr Biol 12:1773–1778. [DOI] [PubMed] [Google Scholar]
- 42. Moore CJ, Winder SJ (2010) Dystroglycan versatility in cell adhesion: a tale of multiple motifs. Cell Commun Signal 8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Noguera ME, Primo ME, Jakoncic J, Poskus E, Solimena M, Ermacora MR (2015) X‐ray structure of the mature ectodomain of phogrin. J Struct Funct Genomics 16:1–9. [DOI] [PubMed] [Google Scholar]
- 44. Xu X, Choi SH, Hu T, Tiyanont K, Habets R, Groot AJ, Vooijs M, Aster JC, Chopra R, Fryer C, Blacklow SC (2015) Insights into autoregulation of Notch3 from structural and functional studies of its negative regulatory region. Structure 23:1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holm L, Rosenstrom P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–54W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A (1998) The Notch1 receptor is cleaved constitutively by a furin‐like convertase. Proc Natl Acad Sci USA 95:8108–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Tetering G, Vooijs M (2011) Proteolytic cleavage of Notch: “HIT and RUN”. Curr Mol Med 11:255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pavsic M, Guncar G, Djinovic‐Carugo K, Lenarcic B (2014) Crystal structure and its bearing towards an understanding of key biological functions of EpCAM. Nat Commun 5:4764. [DOI] [PubMed] [Google Scholar]
- 49. Schnell U, Cirulli V, Giepmans BN (2013) EpCAM: structure and function in health and disease. Biochim Biophys Acta 1828:1989–2001. [DOI] [PubMed] [Google Scholar]
- 50. Schnell U, Kuipers J, Giepmans BN (2013) EpCAM proteolysis: new fragments with distinct functions? Bioscience Rep 33:e00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shvartsur A, Bonavida B (2015) Trop2 and its overexpression in cancers: regulation and clinical/therapeutic implications. Genes Cancer 6:84–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lim LE, Campbell KP (1998) The sarcoglycan complex in limb‐girdle muscular dystrophy. Curr Opin Neurol 11:443–452. [DOI] [PubMed] [Google Scholar]
- 53. Tarakci H, Berger J (2016) The sarcoglycan complex in skeletal muscle. Front Biosci 21:744–756. [DOI] [PubMed] [Google Scholar]
- 54. Waite AJ, Carlisle FA, Chan YM, Blake DJ (2016) Myoclonus dystonia and muscular dystrophy: varepsilon‐sarcoglycan is part of the dystrophin‐associated protein complex in brain. Movement Disord 31:1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hendriks WJ, Dilaver G, Noordman YE, Kremer B, Fransen JA (2009) PTPRR protein tyrosine phosphatase isoforms and locomotion of vesicles and mice. Cerebellum 8:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dilaver G, van de Vorstenbosch R, Tarrega C, Rios P, Pulido R, van Aerde K, Fransen J, Hendriks W (2007) Proteolytic processing of the receptor‐type protein tyrosine phosphatase PTPBR7. FEBS J 274:96–108. [DOI] [PubMed] [Google Scholar]
- 57. Zhang H, Wada J, Hida K, Tsuchiyama Y, Hiragushi K, Shikata K, Wang H, Lin S, Kanwar YS, Makino H (2001) Collectrin, a collecting duct‐specific transmembrane glycoprotein, is a novel homolog of ACE2 and is developmentally regulated in embryonic kidneys. J Biol Chem 276:17132–17139. [DOI] [PubMed] [Google Scholar]
- 58. Kobori H, Nangaku M, Navar LG, Nishiyama A (2007) The intrarenal renin‐angiotensin system: from physiology to the pathobiology of hypertension and kidney disease. Pharmacol Rev 59:251–287. [DOI] [PubMed] [Google Scholar]
- 59. Danilczyk U, Sarao R, Remy C, Benabbas C, Stange G, Richter A, Arya S, Pospisilik JA, Singer D, Camargo SM, Makrides V, Ramadan T, Verrey F, Wagner CA, Penninger JM (2006) Essential role for collectrin in renal amino acid transport. Nature 444:1088–1091. [DOI] [PubMed] [Google Scholar]
- 60. Camargo SM, Singer D, Makrides V, Huggel K, Pos KM, Wagner CA, Kuba K, Danilczyk U, Skovby F, Kleta R, Penninger JM, Verrey F (2009) Tissue‐specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology 136:872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fairweather SJ, Broer A, Subramanian N, Tumer E, Cheng Q, Schmoll D, O'Mara ML, Broer S (2015) Molecular basis for the interaction of the mammalian amino acid transporters B0AT1 and B0AT3 with their ancillary protein collectrin. J Biol Chem 290:24308–24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Y, Wada J, Yasuhara A, Iseda I, Eguchi J, Fukui K, Yang Q, Yamagata K, Hiesberger T, Igarashi P, Zhang H, Wang H, Akagi S, Kanwar YS, Makino H (2007) The role for HNF‐1beta‐targeted collectrin in maintenance of primary cilia and cell polarity in collecting duct cells. PloS One 2:e414. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63. Akpinar P, Kuwajima S, Krutzfeldt J, Stoffel M (2005) Tmem27: a cleaved and shed plasma membrane protein that stimulates pancreatic beta cell proliferation. Cell Metabol 2:385–397. [DOI] [PubMed] [Google Scholar]
- 64. Fyfe JC, Madsen M, Hojrup P, Christensen EI, Tanner SM, de la Chapelle A, He Q, Moestrup SK (2004) The functional cobalamin (vitamin B12)‐intrinsic factor receptor is a novel complex of cubilin and amnionless. Blood 103:1573–1579. [DOI] [PubMed] [Google Scholar]
- 65. Kozyraki R, Gofflot F (2007) Multiligand endocytosis and congenital defects: roles of cubilin, megalin and amnionless. Curr Pharmaceut Des 13:3038–3046. [DOI] [PubMed] [Google Scholar]
- 66. He QY, Liu XH, Li Q, Studholme DJ, Li XW, Liang SP (2006) G8: a novel domain associated with polycystic kidney disease and non‐syndromic hearing loss. Bioinformatics 22:2189–2191. [DOI] [PubMed] [Google Scholar]
- 67. Hunt LT, Barker WC (1987) von Willebrand factor shares a distinctive cysteine‐rich domain with thrombospondin and procollagen. Biochem Biophys Res Commun 144:876–882. [DOI] [PubMed] [Google Scholar]
- 68. Furness SG, McNagny K (2006) Beyond mere markers: functions for CD34 family of sialomucins in hematopoiesis. Immunol Res 34:13–32. [DOI] [PubMed] [Google Scholar]
- 69. Nielsen JS, McNagny KM (2008) Novel functions of the CD34 family. J Cell Sci 121:3683–3692. [DOI] [PubMed] [Google Scholar]
- 70. Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, Owen MJ, O'Donovan MC, Williams J (2005) Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Human Genet 76:581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Paracchini S, Steer CD, Buckingham LL, Morris AP, Ring S, Scerri T, Stein J, Pembrey ME, Ragoussis J, Golding J, Monaco AP (2008) Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am J Psychiatry 165:1576–1584. [DOI] [PubMed] [Google Scholar]
- 72. Guo J, Chen S, Huang C, Chen L, Studholme DJ, Zhao S, Yu L (2004) MANSC: a seven‐cysteine‐containing domain present in animal membrane and extracellular proteins. Trends Biochem Sci 29:172–174. [DOI] [PubMed] [Google Scholar]
- 73. Bycroft M, Bateman A, Clarke J, Hamill SJ, Sandford R, Thomas RL, Chothia C (1999) The structure of a PKD domain from polycystin‐1: implications for polycystic kidney disease. EMBO J 18:297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Velayos‐Baeza A, Levecque C, Kobayashi K, Holloway ZG, Monaco AP (2010) The dyslexia‐associated KIAA0319 protein undergoes proteolytic processing with {gamma}‐secretase‐independent intramembrane cleavage. J Biol Chem 285:40148–40162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hulpiau P, van Roy F (2009) Molecular evolution of the cadherin superfamily. Intl J Biochem Cell Biol 41:349–369. [DOI] [PubMed] [Google Scholar]
- 76. Angst BD, Marcozzi C, Magee AI (2001) The cadherin superfamily: diversity in form and function. J Cell Sci 114:629–641. [DOI] [PubMed] [Google Scholar]
- 77. Kazmierczak P, Sakaguchi H, Tokita J, Wilson‐Kubalek EM, Milligan RA, Muller U, Kachar B (2007) Cadherin 23 and protocadherin 15 interact to form tip‐link filaments in sensory hair cells. Nature 449:87–91. [DOI] [PubMed] [Google Scholar]
- 78. Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N (2012) Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/beta‐catenin complex. Proc Natl Acad Sci USA 109:13046–13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T (1999) Flamingo, a seven‐pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98:585–595. [DOI] [PubMed] [Google Scholar]
- 80. Tanoue T, Takeichi M (2005) New insights into Fat cadherins. J Cell Sci 118:2347–2353. [DOI] [PubMed] [Google Scholar]
- 81. Oda H, Takeichi M (2011) Evolution: structural and functional diversity of cadherin at the adherens junction. J Cell Biol 193:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Oda H, Tsukita S (1999) Nonchordate classic cadherins have a structurally and functionally unique domain that is absent from chordate classic cadherins. Dev Biol 216:406–422. [DOI] [PubMed] [Google Scholar]
- 83. Langenhan T, Aust G, Hamann J (2013) Sticky signaling–adhesion class G protein‐coupled receptors take the stage. Sci Signal 6:re3. [DOI] [PubMed] [Google Scholar]
- 84. Bjarnadottir TK, Fredriksson R, Hoglund PJ, Gloriam DE, Lagerstrom MC, Schioth HB (2004) The human and mouse repertoire of the adhesion family of G‐protein‐coupled receptors. Genomics 84:23–33. [DOI] [PubMed] [Google Scholar]
- 85. Arac D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Sudhof TC, Brunger AT (2012) A novel evolutionarily conserved domain of cell‐adhesion GPCRs mediates autoproteolysis. EMBO J 31:1364–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liao Y, Pei J, Cheng H, Grishin NV (2014) An ancient autoproteolytic domain found in GAIN, ZU5 and Nucleoporin98. J Mol Biol 426:3935–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Harris PC, Torres VE (2009) Polycystic kidney disease. Ann Rev Med 60:321–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bork P, Doerks T, Springer TA, Snel B (1999) Domains in plexins: links to integrins and transcription factors. Trends Biochem Sci 24:261–263. [DOI] [PubMed] [Google Scholar]
- 89. Jenkins J, Pickersgill R (2001) The architecture of parallel beta‐helices and related folds. Prog Biophys Mol Biol 77:111–175. [DOI] [PubMed] [Google Scholar]
- 90. Kaimori JY, Nagasawa Y, Menezes LF, Garcia‐Gonzalez MA, Deng J, Imai E, Onuchic LF, Guay‐Woodford LM, Germino GG (2007) Polyductin undergoes notch‐like processing and regulated release from primary cilia. Human Mol Genet 16:942–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hu T, Guan T, Gerace L (1996) Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol 134:589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chug H, Trakhanov S, Hulsmann BB, Pleiner T, Gorlich D (2015) Crystal structure of the metazoan Nup62*Nup58*Nup54 nucleoporin complex. Science 350:106–110. [DOI] [PubMed] [Google Scholar]
- 93. Wreschner DH, McGuckin MA, Williams SJ, Baruch A, Yoeli M, Ziv R, Okun L, Zaretsky J, Smorodinsky N, Keydar I, Neophytou P, Stacey M, Lin HH, Gordon S (2002) Generation of ligand‐receptor alliances by “SEA” module‐mediated cleavage of membrane‐associated mucin proteins. Protein Sci 11:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pei J, Grishin NV (2014) PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three‐dimensional structural information. Methods Mol Biol 1079:263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kall L, Krogh A, Sonnhammer EL (2004) A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.
Supporting Information Table 1.


