Abstract
SIRT6, a member of the Sirtuin family of NAD+-dependent enzymes, has established roles in chromatin signaling and genome maintenance. Through these functions, SIRT6 protects against aging-associated pathologies including metabolic disease and cancer, and can promote longevity in mice. Research from the last few years has revealed that SIRT6 is a complex enzyme with multiple substrates and catalytic activities, and uncovered novel SIRT6 functions in maintenance of organismal health span. Here, we review these new discoveries and models of SIRT6 biology in four areas: heterochromatin stabilization and silencing, stem cell biology, cancer initiation and progression, and regulation of metabolic homeostasis. We discuss possible implications of these findings for therapeutic interventions in aging and aging-related disease processes.
Keywords: SIRT6, sirtuin, aging, chromatin, metabolism, cancer
SIRT6: an enzyme that protects against aging and disease pathologies
SIRT6 is a chromatin regulatory protein in the Sirtuin family of NAD+-dependent enzymes, whose members play diverse roles in aging, metabolism, and disease [1]. Studies in mice have uncovered key roles for SIRT6 in promoting organismal health: SIRT6-deficient mice have shortened lifespan and phenotypes associated with aging, cancer, and metabolic disorders. Conversely, increased levels of SIRT6 may have beneficial effects. For instance, SIRT6 over-expression in mice protects against metabolic pathologies associated with diet-induced obesity. Moreover, SIRT6 overexpression led to a subtle increase in lifespan in male mice, at least in a specific mouse background [2-5]. In addition, SIRT6 expression is induced in mice on calorie restriction, a dietary regimen that protects against many aging-related changes [6].
SIRT6 has been best characterized as a histone deacetylase that targets specific sites on histone H3, namely the acetylated lysines K9, K56 and K18 (H3K9ac, H3K56ac, and H3K18ac) [7-10]. Deacetylation of these chromatin marks by SIRT6 is important for chromatin compaction, transcriptional repression, and DNA damage responses. Thus, SIRT6 acts as a co-repressor of several transcription factors implicated in aging, cancer, and metabolism (e.g., NF-κB, HIF-1, and c-Myc); promotes DNA damage-dependent chromatin changes that are essential for DNA repair (See Box 2); and maintains telomeric chromatin structure to prevent genomic instability and cellular senescence [4]. Notably, SIRT6 is a complex enzyme, and has additional enzymatic mechanisms and substrates beyond histone deacetylation. It deacetylates several non-histone proteins involved in DNA repair and glucose homeostasis, promotes mono-ADP-ribosylation of certain DNA repair and chromatin silencing factors; and intriguingly, catalyzes deacylation of long-chain fatty-acyl-groups – protein modifications whose physiologic functions are just beginning to be elucidated (see Table 1). Advances in proteomic approaches promise to uncover more SIRT6 substrates, and elucidation of their importance for SIRT6 biology in cellular and organismal physiology are exciting goals of ongoing studies.
Box 2. SIRT6 function in DNA repair and chromatin signaling during DNA damage responses.
A role for SIRT6 in DNA repair was first suggested by observations that SIRT6-deficient mouse cells exhibit genomic instability and DNA damage hypersensitivities consistent with defects in DNA double strand break (DSB) repair or Base Excision Repair (BER) [2]. Subsequently, SIRT6 was shown to associate dynamically with chromatin at sites of DNA damage, and a growing body of work indicates that it coordinates multiple aspects of DNA repair and chromatin signaling during the cellular DNA damage response (DDR) [17, 81, 83]. In response to DSBs, SIRT6 interacts with the DSB repair factor DNA-dependent protein kinase (DNA-PK) and the chromatin-remodeling factor SNF2H, and promotes their recruitment to chromatin flanking the break [81, 83]. SIRT6 also promotes H3K56ac deacetylation and chromatin remodeling at DSBs, and this may be important for creating accessibility or generating binding sites for downstream DDR factors. Indeed, inactivation of SIRT6 leads to impaired recruitment of p53-binding protein 1 (p53BP1) and BRCA1 to DSB foci, and reduced DSB repair efficiency by both Non-Homologous End-Joining (NHEJ) and Homology-Directed Repair (HDR) [81]. Notably, overexpression of a mutant histone H3 protein that mimics constitutive H3K56 acetylation phenocopies the DNA repair defects in SIRT6-deficient cells, suggesting that SIRT6-dependent H3K56 deacetylation is important for DSB repair efficiency [81].
SIRT6 impacts on DNA repair through additional mechanisms beyond histone deacetylation. It promotes DNA end-resection, an important step in HDR, by deacetylating the DSB-resection factor C-terminal binding protein interacting protein (CtIP) [85]. In response to oxidative stress, SIRT6 is also reported to promote mono-ADP-ribosylation and activation of the PARP1 poly(ADP-ribose) polymerase, which is important for DSB repair by NHEJ and HDR, and repair of single stranded lesions by BER [17].
Thus, SIRT6 is important for several aspects of DNA damage signaling and repair, and recent work suggests that defects in these functions may contribute to aging, cancer, or other disease processes. For example, the DNA repair functions of SIRT6 can be activated by Lamin A, which is mutated in Hutchinson Gilford Progeria Syndrome (HGPS), suggesting that defective SIRT6-dependent DNA repair may contribute to the accelerated aging in HGPS [33]. SIRT6 expression is reduced in brains of Alzheimer’s Disease (AD) patients, and overexpression of SIRT6 can prevent DNA damage in a mouse model of AD [96]. A decline in SIRT6 levels is also observed in aged human fibroblasts, and overexpression of SIRT6 can rescue a decline in BER efficiency in these cells [78].
Table 1.
Identified substrates of SIRT6
| Activity | Targets | In vitro |
In cells |
SIRT6-linked cellular functions | Ref | |
|---|---|---|---|---|---|---|
| Deacetylation | Histones | H3K9ac | V | V | Telomere stability, transcriptional regulation, DNA damage response |
[7, 83, 84] |
| H3K56ac | V | V | Telomere stability, transcriptional regulation, DNA damage response |
[5, 8, 9, 81] |
||
| H3K18ac | V | V | Heterochromatin silencing | [10, 14] |
||
| H3K27ac | V | [14] | ||||
| Non- histones |
CtIP | V | V | Regulation of DSB processing and Homologous Directed Repair |
[85] | |
| NPM1 | V | Potential link to cellular senescence |
[86] | |||
| PKM2 | V | V | Regulation of PKM2 localization and oncogenic functions |
[58] | ||
| GCN5 | V | V | Regulation of GCN5 acetyltransferase activity |
[69] | ||
| FoxO3a | V | Modulation of chemotherapy resistance in breast cancer |
[87] | |||
| De-fatty- acylation |
Histones | H3K9myristoyl | V | [15] | ||
| H3K9dodecanoyl | V | [15] | ||||
| H3K9decanoyl | V | [15] | ||||
| H3K9octanoyl, H3K18octanoyl, H3K27 octanoyl |
V | [14, 15] |
||||
| H3K9hexanoyl | V | [15] | ||||
| Non- histones |
TNFαK19myr, TNFαK20myr |
V | V | Regulation of TNFα secretion | [23] | |
| Mono-ADP- ribosylation |
Non- histones |
SIRT6 | V | [16] | ||
| PARP1 | V | V | Regulation of PARP1 poly-ADP- ribosyltransferase activity and DSB repair |
[17] | ||
| KAP1 | V | Regulation of KAP1 interaction with HP1α, and L1 silencing |
[18] | |||
Several excellent reviews provide comprehensive accounts of the evolution of the SIRT6 field [4, 11]. In this article, we begin with a review of the diverse catalytic activities and substrates of SIRT6 identified to date, and the cellular functions to which they have so far been linked (See Table 1). We then focus on four emerging areas of discovery that implicate SIRT6 in novel mechanisms of aging and disease biology. First, we discuss findings demonstrating that SIRT6 has essential roles in heterochromatin silencing, and that in the absence of SIRT6 heterochromatin de-repression contributes to cellular dysfunction associated with aging and disease. We then review recently discovered functions of SIRT6 in adult and embryonic stem cell biology, and their implications for aging and stem cell-based therapeutic strategies. We also discuss studies of SIRT6 function in human cancers and their underlying mechanisms, and emerging insights into SIRT6 biology in the complex regulation of glucose and lipid metabolism and their interplay with circadian rhythms.
Insights into biochemical substrates and physiologic functions of SIRT6 enzymatic activities
Characterizing the biochemical and molecular functions of SIRT6 is challenging, particularly given its three distinct enzymatic activities and diverse deacetylase, ADP-ribosyltransferase, and defatty-acylase substrates. Moreover, SIRT6 exerts these activities in specialized cellular contexts, such as packaged heterochromatin, which may not be appropriately reproduced in standard in vitro enzymatic reactions. Nonetheless, deciphering which activities and substrates of SIRT6 mediate its different cellular and physiologic functions is an important area of ongoing study, and may be critical for achieving specificity in therapeutic strategies based on SIRT6 modulation.
Much evidence indicates that deacetylation by SIRT6 is highly substrate-specific, and only a handful of histone acetylation sites and non-histone substrates of SIRT6 have been reported. Importantly, while SIRT6 catalyzes robust histone deacetylation on nucleosomes [10, 12], it has much weaker activity on histone peptides, suggesting that its efficiency depends on physiologic chromatin context [13]. Recently, using a novel approach to generate nucleosomes that are specifically acetylated on a single defined site, Wang and colleagues definitively confirmed the high efficiency and selectivity of SIRT6 in deacetylating its previously described substrates H3K9ac and H3K18ac. In addition, they showed that SIRT6 can also deacetylate nucleosomal H3K27ac, a chromatin mark associated with transcriptional enhancer elements, suggesting potential functions of SIRT6 in enhancer-regulated gene expression programs [14]. Interestingly, the in vitro deacetylation of histone peptides by SIRT6 can be activated up to 35-fold fold by free fatty acids (FFAs; e.g., myristic acid), suggesting that in vivo, efficient deacetylation of known or novel SIRT6 substrates could depend on endogenous FFA activators that fluctuate with metabolic conditions [15].
SIRT6 was first observed to undergo auto-mono-ADP-ribosylation when incubated with NAD in vitro, in an intramolecular reaction [16]. Subsequently, Gorbunova and colleagues reported SIRT6-dependent mono-ADP-ribosylation of the PARP1 polymerase [17] and KAP1 co-repressor in trans [18]. Of note, several biochemical studies reported observing weak or negligible mono-ADP-ribosylation activity of SIRT6 in vitro [19-22]. It is possible that, as for histone deacetylation, efficient in vitro ADP-ribosylation by SIRT6 could depend critically on use of specific physiologic substrates or conditions.
One of the most intriguing new areas in SIRT6 biology stems from the finding that in addition to deacetylating lysines, SIRT6 also efficiently catalyzes removal of certain long chain fatty-acyl groups from lysine residues in vitro, suggesting that regulation of such modifications may have important physiologic functions. Studies from the Lin and Denu labs first detected the defatty-acylation activity of SIRT6 using in vitro assays on acylated peptide substrates, such as TNFα peptides myristoylated on lysine K19 or K20, and histone H3 peptides with 6- to 16-carbon chain fatty acyl groups on lysine K9 [15, 23]. Crystal structure of SIRT6 in complex with a H3K9-myristoylated peptide revealed that the long fatty acid chain is accommodated in a hydrophobic binding cavity, likely conferring a more catalytically competent conformation to SIRT6 [23]. Indeed, a similar conformation change could be induced by FFAs, and explain their dramatic activating effect on the in vitro deacetylation activity of SIRT6. By contrast, free myristic acid does not activate, and in fact inhibits, the demyristoylation activity of SIRT6, because it competes with the myristoylated peptide substrate for the same binding pocket. Based on these and other findings, Feldman and colleagues suggested the intriguing model that increased cellular levels of specific FFAs (which could occur in response to changes in food intake) could direct a “switch” in SIRT6 activities, from defatty-acylation to deacetylation [15].
Very recently, Wang and colleagues developed a novel approach to detect defatty-acylation by SIRT6 using more physiologic substrates than isolated acyl peptides. They assembled acyl-nucleosomes using histone H3 proteins engineered to harbor long-chain acyl groups on single defined sites, and developed a chemical biology approach to visualize deacylation. They showed that SIRT6 removes long-chain octenoyl-groups on H3K9, H3K18, and H3K27 (and with weaker activity, on H3K4 and H3K23), but was unable to de-octenoylate these residues when acylated free H3 histones or acylated H3-H4 tetramers were used as substrates. Thus, both the deacetylase and long-chain defatty-acylase activities of SIRT6 appear to be much more active on histones in the physiologic context of nucleosomal structure [14].
Unraveling the biologic importance of the different SIRT6 enzymatic activities and substrates for specific cellular and physiologic functions of SIRT6 is a challenging and important goal. The physiologic importance of lysine deacetylation by SIRT6 has been underscored by the many cellular functions that have been linked to this activity over the past decade, including regulation of DNA damage responses (Box 2) and many of the transcriptional programs highlighted in the sections below. Indeed, Kugel et al recently showed that several recurrent SIRT6 point mutations in human tumors selectively impair SIRT6-dependent histone deacetylation without affecting defatty-acylation [22], and they directly linked the deacetylase activity of SIRT6 to its tumor suppressive function. In similar separation-of-functions studies, Zhang and colleagues recently concluded that the transcriptional functions of SIRT6 depend predominantly on its deacetylase activity [21].
Separation-of-function mutations of SIRT6 were also instrumental in studies of the physiologic relevance of mono-ADP-ribosylation by SIRT6 for specific cellular functions. Mao et al used mutant SIRT6 proteins, which they observed to retain only deacetylase activity (SIRT6 G60A) or only ADP-ribosyltransferase activity (SIRT6 R65A), to provide evidence that mono-ADP-ribosylation of PARP1 by SIRT6 increases PARP1 activity in enhancing double-strand-break (DSB) repair under oxidative stress (Box 2) [17]. Similarly, the study from Van Meter et al used these mutant proteins to implicate SIRT6-dependent mono-ADP-ribosylation of KAP1 in heterochromatin silencing of LINE1 retroelements [18]. These findings constitute some of the first evidence for the physiologic importance of mono-ADP-ribosylation by SIRT6 in regulating nuclear protein functions.
By contrast to SIRT6-dependent deacetylation and ribosylation, the de-fatty-acylation activities of SIRT6 were only discovered quite recently, and their physiologic substrates and cellular functions are just starting to be uncovered. The Lin Lab provided important first insights in this context. They showed that de-myristoylation of the tumor necrosis factor-αDDTNFαD by SIRT6 is important for promoting TNFα secretion [23]. More recently, they further reported that the mutant SIRT6 G60A protein is fully active in defatty-acylation, but contrary to the previous findings, lacks both deacetylation and ADP-ribosylation activities [21]. They used this mutation in separation-of-function studies to show that de-fatty-acylation by SIRT6 is sufficient for its promotion of TNFα secretion in cells. Moreover, proteomic analyses comparing SIRT6-knockout and control MEFs revealed changes in the secretion of many other proteins (some showing increased secretion, others showing reduced secretion), and reconstitution with the de- fatty-acylation-specific SIRT6 G60A enzyme was sufficient to restore their secretion to wild-type levels. Among the proteins showing increased secretion in SIRT6 KO MEFs were 41 ribosomal proteins (RPs, not known as classical secreted proteins). Intriguingly, treatment of NIH3T3 cells with RP-containing exosomes that were secreted by SIRT6 KO MEFs led to increased cell proliferation. Zhang and colleagues propose that the delivery of proliferation-promoting material via exosomes to surrounding cells could contribute to the reported tumorigenicity of immortalized SIRT6 KO MEFs [5]. Together, these findings highlight the importance of elucidating novel fatty acyl substrates of SIRT6 in vivo, and the relevance of long-chain fatty acyl-groups for protein functions and dynamics.
The identification of additional mutations that selectively inactivate distinct enzymatic activities of SIRT6 can provide powerful tools to delineate which activities contribute to the diverse cellular and organismal functions of SIRT6. However, the discrepancies in detected activities of mutant SIRT6 proteins between different studies [17, 21] suggest that careful quantitative analysis of the mutant proteins will be important. Regardless, development of novel approaches to study specific SIRT6 activities and substrates, link them to cellular functions is an important future goal.
Heterochromatin silencing and genome maintenance in aging and disease
There is growing appreciation that aging is associated with epigenetic chromatin changes that impact the stability and proper expression of the genome. The hypothesis that heterochromatin loss contributes to aging dates back to the 1990s, with the findings that silent chromatin regions are de-repressed during aging of budding yeast, and that Sir2, the founding member of the sirtuin family, counteracts yeast aging by promoting heterochromatin silencing [24]. Recently, genome-wide studies have revealed that mammalian aging is also accompanied by chromatin de-compaction and impaired silencing at repetitive DNA elements, such as centromeres, telomeres, and retrotransposons. Indeed, aberrant transcription and instability of these elements is observed in aging cells and tissues. Notably, similar defects in heterochromatin silencing are also observed in cancer, suggesting underlying mechanistic links between aging-associated heterochromatin dysfunction and cancer pathogenesis [18, 25-28].
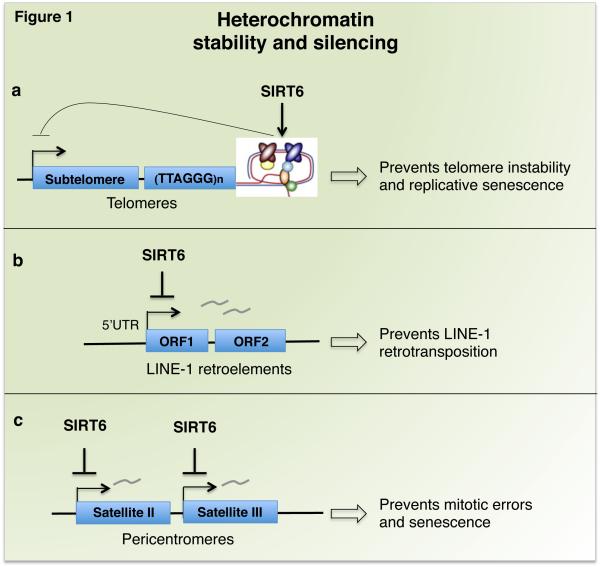
The first reported chromatin-regulatory activity of SIRT6 was uncovered at telomeres (Figure 1a). SIRT6-dependent histone deacetylation at telomeres establishes a specialized chromatin state, which is important for binding of Werner syndrome ATP-dependent helicase (WRN), the DNA-processing factor that is mutated in the premature aging disorder Werner syndrome. In the absence of SIRT6, telomere dysfunction leads to genomic instability and premature cellular senescence of primary human fibroblasts [7]. SIRT6 also stabilizes repressive heterochromatin at sub-telomeric regions to silence transcription of telomere-proximal genes, and de-regulation of such genes could in theory contribute to cellular changes in aging [29].
Figure 1. Heterochromatin silencing by SIRT6 maintains genomic stability and transcriptional silencing of repetitive DNA.
SIRT6 has activating (arrows) or repressing (head-line) activities. a) SIRT6 binds to and stabilizes telomeric chromatin, an activity important for preventing genomic instability and replicative senescence. Stability of telomeric repeats also contributes to maintaining the silence of sub-telomeric genes (telomere position effect). b) SIRT6 maintains heterochromatin at LINE-1 retroelements, which suppresses their transcription and prevents retrotransposition events that can destabilize the genome. c) SIRT6 maintains heterochromatin at Satellite II and Satellite III pericentric repeats; this prevents accumulation of pathologic satellite transcripts that trigger mitotic errors and cellular senesce.
Recent studies have now identified novel mechanisms of SIRT6 in promoting heterochromatin silencing at other genomic loci, which prevent genomic instability and aging-related cellular dysfunction. Retrotransposable elements (RTEs), especially Long Interspersed Element-1 (LINE-1) RTEs, comprise a significant fraction of mammalian genomes, and their transposition can give rise to genomic instability, mutagenesis, and transcriptional de-regulation [30]. Accumulating evidence suggests that heterochromatic silencing of such elements is compromised in aging cells and tissues [26]. Recently, Van Meter and colleagues showed that SIRT6 promotes heterochromatin packaging of LINE-1 RTEs to suppress transposition, and this function may be compromised during aging (Figure 1b) [18]. SIRT6 binds LINE-1 regulatory elements, but is depleted from these sequences both in aging and in response to DNA damage (during which SIRT6 is recruited to DNA breaks). These findings suggested the model that mobilization of SIRT6 to DNA damage that accumulates during aging may lead to depletion of SIRT6 at LINE-1 sites, with resulting heterochromatin de-repression and activation of transposition. Interestingly, studies of separation-of-function mutations in SIRT6 suggest that its effects on LINE-1 silencing are mediated predominantly by ADP-ribosylation of the heterochromatin-regulatory factor KAP1, rather than histone deacetylation.
Growing evidence indicates that during mammalian aging and in cancer, heterochromatin silencing also breaks down at pericentric regions of centromeres, but the functional relevance of this has been unclear [26]. Recent findings revealed that SIRT6 maintains pericentric heterochromatin silencing by selectively deacetylating a previously unreported histone substrate, H3K18ac, and accumulation of pathologic pericentric transcripts in SIRT6-deficient cells has a causal role in triggering aging- and cancer-associated defects including mitotic errors, genomic instability, and cellular senescence (Figure 1c) [10]. Like at LINE-1 RTEs, pericentric heterochromatin silencing by SIRT6 involves KAP1, but surprisingly, is mediated by a distinct mechanism that may be independent of traditional heterochromatin regulatory factors. Specifically, the data suggest a model in which deacetylation of H3K18ac by SIRT6 is important for KAP1 retention at pericentric satellite repeats, and in the absence of SIRT6, H3K18 hyperacetylation triggers KAP1 release and transcriptional derepression. It will be interesting to further compare the distinct mechanisms of functional interplay between SIRT6 and KAP1 at LINE-1 elements versus pericentric repeats, and potentially at other genomic loci.
Intriguingly, recent studies have suggested that impaired heterochromatin maintenance due to reduced SIRT6 activity could contribute to pathogenesis of Hutchinson-Gilford Progeria Syndrome (HGPS), a human premature aging disorder [31]. HGPS patient cells express an abnormal Lamin A protein, a scaffold protein that is important for heterochromatin maintenance. Like SIRT6-deficient cells, HGPS cells exhibit defects in pericentric heterochromatin, aberrant satellite transcription, and telomere dysfunction [32]. Recently, Lamin A was found to activate SIRT6-dependent histone deacetylation and ADP-ribosylation (see Box 1), but this function is lost in the HGPS mutant Lamin A protein [33]. Moreover, other work revealed that SIRT6 levels were reduced in HGPS cell lines, and SIRT6 re-expression in these cells attenuated their premature senescence [31]. It will be interesting to determine whether the underlying mechanisms involve SIRT6-dependent re-silencing of heterochromatin at pericentric or other genomic regions.
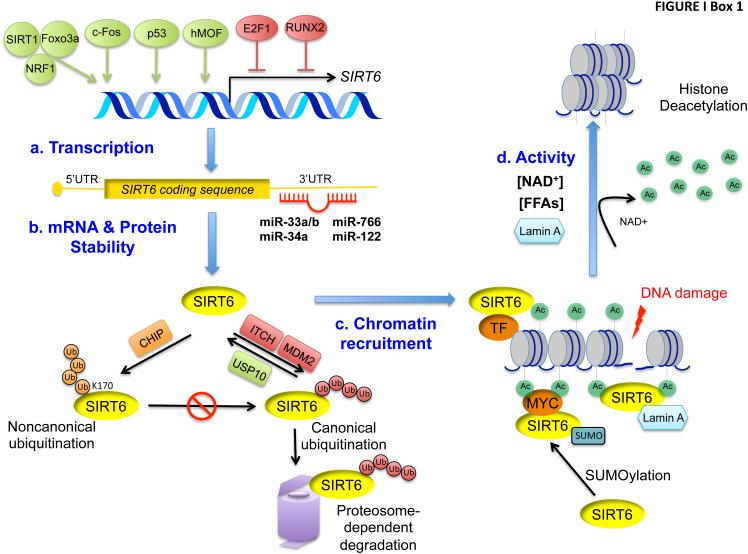
Box 1. Molecular mechanisms of SIRT6 regulation.
Elucidation of mechanisms through which SIRT6 levels or activity are regulated has potential to provide important insights for understanding SIRT6 physiology and developing pharmacologic strategies to modulate SIRT6 activity. SIRT6 gene expression is positively and negatively regulated by several transcription factors (Figure Ia). The SIRT1 deacetylase, in complex with the FoxO3a and NRF1 transcription factors, promotes SIRT6 transcription in liver cells upon nutritional stress [65]. c-Fos activates SIRT6 transcription in mouse models of liver cancer initiation [59], the p53 tumor suppressor activates SIRT6 transcription in primary mouse fibroblasts and cancer cells [68], and the histone acetyltransferase hMOF (human Males-absent on the first) binds to the SIRT6 promoter to promote SIRT6 transcription in hepatocellular carcinoma cells [88]. The transcription factors E2F1 and RUNX2 repress SIRT6 transcription in response to hypoxia [56] and low glucose conditions respectively [55].
SIRT6 is regulated post-transcriptionally at the RNA and protein levels (Figure Ib). Several microRNAs, including miR-33a/b, −34a, −766, and -122, reduce SIRT6 mRNA stability [45, 73, 76, 89, 90]. The E3 ubiquitin ligases ITCH and MDM2 ubiquitylate and target SIRT6 protein for protease-dependent degradation [91, 92], and this is suppressed by the ubiquitin-specific peptidase USP10 [93]. Conversely, non-canonical ubiquitylation of SIRT6 at lysine 170 by the ubiquitin ligase CHIP leads to reduced proteasome-dependent degradation and increased SIRT6 protein stability [94].
SIRT6 activity is also affected by regulated localization of SIRT6 at chromatin (Figure Ic). SIRT6 is targeted to specific gene promoters by interactions with transcription factors (TFs) such as NF-κB and MYC [5, 84]. DNA damage triggers SIRT6 recruitment to chromatin surrounding DNA double strand breaks [17, 81, 83] and recent work suggests that this is augmented by Lamin A [33]. SUMOylation (small ubiquitin-like modification) of SIRT6 was also recently shown to be important for SIRT6 binding to MYC and deacetylation of H3K56ac at MYC-target genes [95].
The intrinsic enzymatic activities of SIRT6 can be regulated by trans-acting factors (Figure Id). Like other NAD+-dependent enzymes, changes in NAD+ availability may influence SIRT6 catalytic activity, though contexts where this is physiologically relevant have yet to be described. Intriguingly, the histone deacetylase activity of SIRT6 is activated in vitro by biologically relevant FFAs, which may increase the affinity of SIRT6 binding to H3 [15]. Lamin A was also reported to enhance SIRT6 intrinsic catalytic activity [33].
Together, these findings suggest that activating SIRT6 to silence and stabilize repetitive regions of the genome could be a promising therapeutic approach to counteract cellular dysfunction in premature or physiologic aging. Moreover, because telomere attrition has been associated with increased risk of cancer, and de-regulated transcription of pericentric and L1 sequences is observed in many tumors, heterochromatin maintenance by SIRT6 may also contribute to tumor suppressive effects of SIRT6. Notably, intense efforts have been made over the last decade to develop sirtuin-activating compounds, or STACs. Dozens of SIRT1 activators have been tested in animal models of diabetes, neurodegeneration, and other aging-related pathologies, and one STAC, SRT2104, has progressed to Phase 2 human patient clinical trials [34, 35]. By contrast, the development of SIRT6-selective compounds is at an earlier stage, and no SIRT6-activating small molecules are as yet available. However, the advances in elucidating SIRT6 enzymatic activities and substrates have already led to development of new small molecule screening strategies based on SIRT6-dependent H3K56 deacetylation and lysines de-myristoylation, which can be useful tools for future drug discovery efforts for SIRT6 activation [36, 37]. Moreover, the expanding understanding of SIRT6 regulation – by post-translational modifications, stress conditions, cellular metabolites, or miRNAs (Box 1) – could suggest additional approaches to modulate SIRT6 levels or activity.
Epigenetic regulation of stem cell homeostasis and differentiation
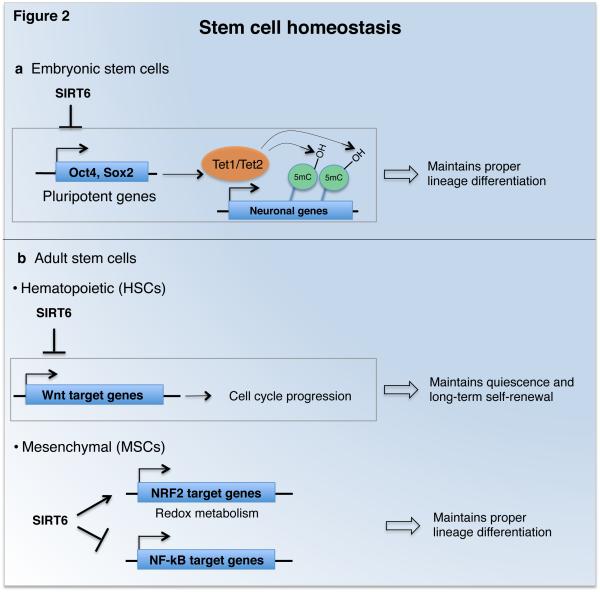
Embryonic stem cells (ESCs) differentiate during development into somatic ectodermal, endodermal, and mesodermal lineage cells. ESC pluripotency is maintained by master transcriptional regulators (Oct4, Sox2, and Nanog), which must be repressed for ESC differentiation along lineage-specific transcriptional programs. Chromatin modification and DNA methylation patterns undergo extensive reprogramming during ESC differentiation, but the underlying mechanisms and their effects on lineage-specific epigenetic programs are still being elucidated [38]. Etchegaray and colleagues recently uncovered a role for SIRT6-dependent histone deacetylation in ESC differentiation (Figure 2a). They showed that SIRT6 represses expression of Oct4, Sox2, and Nanog through H3K9ac and H3K56ac deacetylation. This in turn represses expression of Tet1 and Tet2, enzymes that modulate cell lineage choice during ESC differentiation by promoting 5-hydroxymethylation (5-hmC) of DNA [39]. In SIRT6-deficient ESCs, increased Tet1 and Tet2 expression and 5-hmC levels lead to up-regulation of neuronal transcription programs and skewing of differentiation towards neuroectodermal lineages [40]. Thus, SIRT6 has a pivotal role in the interplay between chromatin modifications and epigenetic DNA methylation during ESC differentiation.
Figure 2. SIRT6 maintains embryonic and adult stem cell homeostasis.
a) SIRT6 maintains proper differentiation of embryonic stem cells by suppressing expression of pluripotent genes and associated increased levels of Tet1 and Tet2, enzymes that promote 5-hmC DNA methylation at neuronal genes. b) SIRT6 maintains long-term self-renewal and multi-lineage reconstitution capacity of adult hematopoietic stem cells (HSCs) by repressing Wnt target genes involved in cell cycle progression. Maintenance of proper differentiation of mesenchymal stem cells (MSCs) by SIRT6 has been linked to both altered redox metabolism through co-activation of NRF2 transcription factor, and to co-repression of NF-κB target gene expression programs.
Like ECSs, adult tissue-specific stem cells are pluripotent and can self-renew indefinitely. These cells allow for replenishment of somatic cells that are lost during physiologic cell turnover or in response to pathologic insults, and a decline in stem cell properties may contribute to aging-associated tissue dysfunction. Accumulating evidence indicates that SIRT6 has key roles in regulating stem cell homeostasis and function in different types of adult stem cells (Figure 2b). New work from Wang and colleagues has revealed that SIRT6 is important for preserving the long-term self-renewal capacity of hematopoietic stem cells (HSCs). Through H3K56 deacetylation, SIRT6 represses transcription of genes in the Wnt pathway, a key regulator of HSC homeostasis. Deletion of SIRT6 in HSCs led to reduced quiescence, a non-cycling state critical for long-term maintenance of stem cell pools, and to impaired multi-lineage reconstitution and self-renewal in competitive hematopoetic-transplantation studies [41]. It will be interesting to examine whether repression of Wnt signaling by SIRT6 contributes to homeostasis of other stem cell types, or could be involved in cancers in which Wnt signaling is de-regulated.
SIRT6-dependent histone deacetylation is also implicated in homeostasis of mesenchymal stem cells (MSCs), which give rise to bone, cartilage, and fat cell lineages. MSC exhaustion can contribute to degenerative attrition of mesenchymal tissues in premature or physiologic aging, and interestingly, SIRT6-deficient mice exhibit degenerative defects in these tissues [2]. Now, new studies have shown that SIRT6-deficient MSCs exhibit compromised osteoblast and chondrocyte differentiation, signs of cellular senescence, and accelerated cellular attrition when transplanted in vivo [42, 43]. These phenotypes are associated with altered redox metabolism, oxidative stress hypersensitivity, and down-regulation of several targets of the NRF2 transcription factor, a master regulator of antioxidant responses. Pan et al propose that SIRT6 coactivates transcription of NRF2 target genes, through deacetylation of H3K56ac. However, H3K56ac deacetylation is generally linked to transcriptional repression, so it remains possible that the activating effects of SIRT6 in this context are due to additional mechanisms. Interestingly, a modest increase in SIRT6 expression is observed with age in MSCs, and might be a compensatory mechanism to alleviate aging-dependent changes in MSC functions [43]. Indeed, SIRT6 overexpression improved osteogenic differentiation of rat MSCs partially by attenuating NF-κB signaling, and enhanced new bone formation and bone repair [44].
These findings in HSCs and MSCs suggest that loss of SIRT6 activity in specific stem cell compartments could contribute to aging-associated tissue dysfunction. In addition, activation of SIRT6 could be useful for stem cell-based therapeutic strategies and tissue engineering. Moreover, work from Sharma and colleagues revealed an important role for SIRT6 in maintaining the epigenetic plasticity of primary human cells for reprogramming into induced pluripotent stem cells (iPSCs). iPSCs-based therapies could have tremendous benefit for treatment of degenerative diseases, by allowing tissue transplantation from a patient’s own somatic cells. Sharma et al showed that SIRT6 expression and the efficiency of iPSC reprogramming is over 3-fold lower in cells from older donors, and restoration of SIRT6 levels in these cells restores reprogramming efficiency [45]. Thus, SIRT6 activation could be useful for iPSC-based therapeutic strategies, particularly for degenerative disorders associated with aging.
Mechanisms of tumor suppression and regulation of cancer progression
Early observations of genomic instability and DNA repair defects in SIRT6 deficient mice suggested that SIRT6 could have protective roles in cancer (See Box 2, Figure 3e) [2]. Subsequently, Sebastian et al directly showed that inactivation of SIRT6 in immortalized mouse embryonic fibroblasts (MEFs) leads to oncogenic transformation, and SIRT6 deficiency increases tumor growth and invasiveness in a mouse colorectal cancer model [5]. They also observed greatly reduced levels of SIRT6 expression in human pancreatic and colorectal cancer tissues, and deletion of the Sirt6 locus was observed in 20% of human patient tumors analyzed [5]. Reduced SIRT6 expression was also reported in breast, ovarian, hepatocellular, lung and other tumors, and in some cases was associated with increased tumor progression and poor clinical prognosis ([5, 46], and reviewed in [11]). Importantly, recent work identified multiple loss-of-function SIRT6 point mutations in human cancers that selectively impair the deacetylase activity of SIRT6, solidifying the role of SIRT6-dependent histone deacetylation in tumor suppression (as discussed earlier) [22].
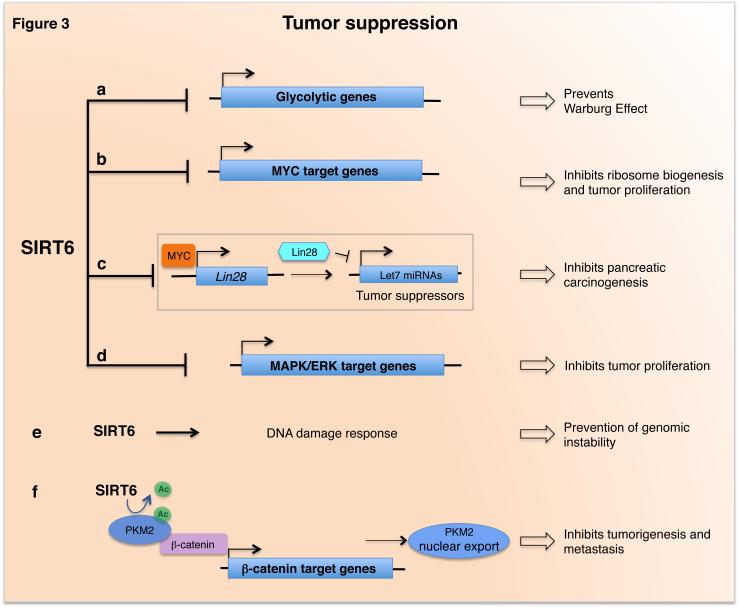
Figure 3. Role and mechanisms of SIRT6 in tumor suppressive pathways.
SIRT6 acts as a tumor suppressor by repressing several transcription programs: a) SIRT6 represses glycolytic genes, which in turn prevents a metabolic switch required for supporting the rapid growth of cancer cells (the Warburg Effect); b) SIRT6 co-represses MYC target genes involved in ribosome biogenesis and proliferation; c) SIRT6 inhibits pancreatic carcinogenesis by transcriptional repression of Myc-target oncofetal protein Lin28b, an inhibitor of the tumor suppressive let-7 family of miRNAs; d) SIRT6 represses MAPK/ERK-target genes involved in tumor proliferation. e) SIRT6 functions in DNA damage responses through a variety of pathways that protect cells from DNA damage and contribute to tumor suppression. f) SIRT6 directly deacetylates PMK2, allowing its nuclear export and subsequent lost of co-activation of β-catenin, impairing its nuclear oncogenic and pro-metastatic functions.
Other studies, however, have revealed that SIRT6 is up-rather than down-regulated in a subset of human cancers (reviewed in [11]). SIRT6 copy number is increased in some hematopoietic cancer cell lines [5]. Moreover, elevated SIRT6 expression occurs in cases of Multiple Myeloma (MM) and some solid tumors, and in some instances is associated with poor clinical outcome [47-52]. Thus, SIRT6 could have oncogenic rather than tumor suppressive functions in certain contexts. Alternatively, SIRT6 overexpression in these cancers could represent a compensatory mechanism to oppose tumor progression. Indeed, increased SIRT6 expression in MM cells inhibits tumor cell proliferation via H3K9 deacetylation and transcriptional repression of pro-proliferative MAPK/ERK target genes, and enhances SIRT6-dependent DNA damage response functions (See Figure 3d-e, Box 2) [47]. These findings suggest that modulation of SIRT6 activity in cancer cells varies with tumor type and cancer stage, and elucidating the complex mechanisms and pleiotropic effects of SIRT6 in cancer cell biology will be crucial for attempts to target SIRT6 for cancer therapy.
A key mechanism underlying the tumor suppressor activity of SIRT6 involves preventing a glycolytic metabolic shift, known as the Warburg effect, that is important for supporting the rapid growth of cancer cells (Figure 3a) [5, 53]. Sebastian and colleagues showed, in both cellular and mouse model studies, that SIRT6 promotes tumor suppression by inhibiting glycolytic cancer cell metabolism [5]. This could be mediated by SIRT6 co-repression of HIF-1α, a master transcriptional regulator of glycolytic genes and mediator of the Warburg effect [54]. Indeed, down-regulation of SIRT6 correlated with increased glycolytic gene expression in human pancreatic and colorectal carcinomas [5]. Other studies confirmed the role of SIRT6 in regulating glycolysis during tumor progression in additional cancers (breast, bladder, and prostate), and showed that the RUNX2 and E2F1 transcription factors can facilitate tumor progression by repressing SIRT6 (See Box 1) [55, 56].
SIRT6 also impacts on cancer cell biology through additional mechanisms, which can be specific to tumor type or stage. Loss of histone deacetylation in SIRT6-deficient colon cancer cells leads to increased transcription of ribosome biogenesis genes by the MYC oncogene, which may be important for meeting the high energetic demands of tumor growth (Figure 3b) [5]. Interestingly, new work from Kugel et al. has uncovered a novel mechanism of tumor suppression by SIRT6 that is lost in pancreatic ductal adenocarcinomas (PDACs) – tumors with dismal prognosis that are among the most lethal of human cancers. They showed that in mouse and human PDAC tumors, the major tumorigenic effects of SIRT6 loss depend on de-repression of the Myc-target oncofetal protein Lin28b. Lin28b inhibits the tumor suppressive let-7 family of miRNAs, and reactivation of Lin28b in SIRT6-deficient PDACs leads to up-regulation of let-7 target genes that promote PDAC survival and malignancy (Figure 3c). Strikingly, in a subset (30-40%) of human PDAC patients with extremely poor prognosis, PDAC cell proliferation and malignancy depend upon reduced SIRT6 expression and high Lin28 expression. Thus, epigenetic regulation of the Lin28b/let-7 axis by SIRT6 is a novel pathway in pancreatic cancer progression, and targeting this pathway could provide a much-needed therapeutic option for a subset of PDAC tumors [57].
In hepatocellular carcinoma cells, SIRT6 may also promote tumor suppression by deacetylating pyruvate kinase M2 (PKM2), a rate-limiting glycolytic enzyme that is up-regulated in many cancers. In the nucleus, PKM2 has additional oncogenic effects as a kinase and transcriptional co-activator of β-catenin, but deacetylation by SIRT6 results in nuclear export of PKM2 and loss of these tumorigenic functions [58] (Figure 3f). Anti-proliferative and pro-apoptotic effects of SIRT6 have also been observed in lung, ovarian, glioma, and other cancer cell types, and likely contribute to its tumor suppressive activity [11, 59].
Importantly, however, recent studies uncovered contexts in which inhibition of SIRT6 activity can augment cancer cell apoptosis or inhibit tumor growth [51]. SIRT6 overexpression in hepatocellular carcinoma promotes cell survival by inhibiting the pro-apoptotic protein Bax, and in lung carcinoma cells, degradation of SIRT6 can augment radiation-induced apoptosis [51, 60]. In addition, certain pro-inflammatory functions of SIRT6 can promote the development of a chronic inflammatory microenvironment that contributes to cancer cell survival, proliferation, and invasiveness. In human skin cancer cells, SIRT6 up-regulation is proposed to increase the mRNA stability of the pro-inflammatory protein cyclooxygenase-2 (COX-2) [48]. In pancreatic cancer cells, SIRT6 enzymatic activity increases intracellular levels of ADP-ribose, which activates the Ca2+ channel TRPM2, and the resulting increase in Ca2-dependent signaling enhances expression of pro-inflammatory cytokines TNFα and interleukin 8 (IL8) [61]. In hepatocellular carcinoma cells, SIRT6 may contribute to an increase in tumorigenicity associated with reduced tumor cell senescence that is observed in response to TGF-β1 or H2O2/HOCl reactive oxygen species [62]. Thus, therapeutic attempts to activate SIRT6 for tumor suppression could be complicated by unwanted effects on cancer cell survival and proliferation in certain contexts.
An important goal of future work is to continue deciphering the molecular mechanisms of tumor suppression by SIRT6 in both initiation and progression of many cancers, and determine the potential of SIRT6 as a biomarker or therapeutic target for malignancy. At the same time, potential tumor-promoting effects of SIRT6 should be carefully considered, and it may be essential to study specific effects of SIRT6 in different tumor types, and even in different patient sub-groups. Indeed, inhibition of SIRT6 could be explored for treatment of certain cancers in which up-regulated SIRT6 expression may contribute to malignancy. Moreover, inhibitors of non-sirtuin histone deacetylase enzymes (HDACs) are already in the clinics for certain cancers [63], and SIRT6 inhibition could conceivably be useful in the context of combination therapy with these agents.
Regulation of glucose and lipid homeostasis in metabolism and disease
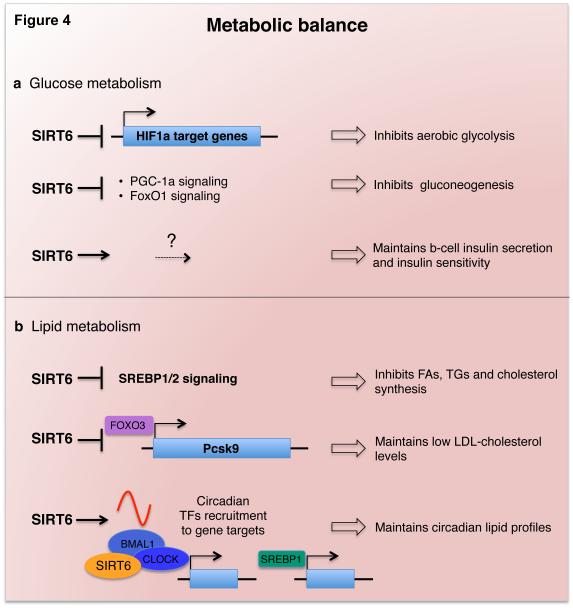
Glucose homeostasis is important for meeting tissue energy demands in response to physiologic and environmental conditions, and its de-regulation contributes to diabetes and other metabolic disorders. SIRT6 is essential for maintaining glucose homeostasis (Figure 4a), as first revealed by the severe hypoglycemia of SIRT6-deficient mice, associated with increased glucose uptake and insulin signaling [2, 54, 64]. Through histone deacetylation, SIRT6 co-represses transcription by HIF1α, a master glycolysis regulator, which channels glucose flux towards glycolysis and away from more energy-efficient mitochondrial respiration. In SIRT6-deficient mice, glucose flux through glycolysis is dramatically increased, and this may trigger pathologic glucose uptake, insulin signaling, and lethal hypoglycemia as compensatory mechanisms [2, 54]. Moreover, liver-specific inactivation of SIRT6 in mice leads to fatty liver formation due to increased glycolysis and triglyceride synthesis [65].
Figure 4. SIRT6 maintains glucose and lipid metabolic balance.
a) SIRT6 regulates glucose metabolism through several mechanisms. It inhibits glycolysis through HIF-1α co-repression, inhibits gluconeogenesis by repressing PGC-1α and FoxO1 signaling, and positively regulates β-cell insulin secretion and insulin sensitivity by mechanisms yet to be elucidated. b) SIRT6 represses SREBP1/2- and FoxO3-mediated transcriptional programs involved in fatty acid, triglyceride, and cholesterol homeostasis. SIRT6 also regulates circadian lipid profiles in the liver by maintaining a proper cyclic recruitment of CLOCK/BMAL1 and SREBP1 transcription factors.
Mounting evidence indicates that activation of SIRT6 can have beneficial effects on glucose homeostasis in pathologic conditions. In mice on high-fat or high-calorie diets, overexpression of SIRT6 protects against defects in glucose tolerance, glucose-stimulated insulin secretion (GSIS), and insulin sensitivity [66, 67]. SIRT6 also inhibits hepatic gluconeogenesis through FoxO1- and PGC1α-dependent pathways, and prevents hyperglycemia in a diabetic/obese mouse model [68, 69]. Notably, inactivation of SIRT6 in a pancreatic beta cell line and in beta cell-specific SIRT6 knockout mice leads to impaired GSIS, demonstrating a cell-intrinsic role of SIRT6 in promoting beta cell insulin secretion [70]. In addition, mice with tissue specific inactivation of SIRT6 in the brain had low growth hormone (GH) and insulin-like growth factor-1 (IGF-1) levels, similar to whole body SIRT6 knockout mice [71]. Thus, although serum glucose levels in these mice were within normal, it is possible that in some physiologic contexts, SIRT6 may also impact on glucose metabolism and insulin sensitivity through GH/IGF-1 signaling.
In addition to promoting glucose homeostasis, SIRT6 also regulates lipid metabolism (Figure 4b). It represses triglyceride synthesis and fatty acid uptake, promotes fatty acid β-oxidation, and maintains low levels of LDL-cholesterol. Accordingly, SIRT6 overexpressing mice are protected from diet-induced obesity, and liver-specific SIRT6-deficient mice develop fatty liver [65, 66, 72, 73]. Mechanistically, histone deacetylation by SIRT6 represses transcription of key lipid metabolism factors such as PCSK9 and the SREBP1 and SREBP2 transcriptional regulators [72-74]. Notably, SIRT6 regulation of SREBP1/2 is complex and occurs at multiple levels, including post-translational modification and chromatin targeting [73, 75]. Thus, activation of SIRT6 might have therapeutic applications for glucose and lipid normalization in the contexts of obesity, metabolic syndrome, and insulin-resistant diabetes.
Recently, a new study has suggested that many of these functions of SIRT6 may be impacted by a complex interplay with miR-122, the most abundant hepatic microRNA [76]. Like SIRT6 overexpressing mice, miR-122-deficient mice have improved lipid profiles and are resistant to pathologies of diet-induced obesity [66, 77]. Elhanati et al now provide evidence that SIRT6 and mi-R122 negatively regulate each other and have opposite effects on fatty acid metabolism and expression of β-oxidation regulatory enzymes [76]. These findings suggest that modulation of miR-122 levels could be useful for treatment of metabolic disorders associated with reduced SIRT6 activity in aging, obesity, or diabetes [45, 69, 78]. In addition, the functional interplay between SIRT6 and miR-122 in lipid metabolism may have important pathological effects in hepatocellular carcinomas (HCC), because a strong negative correlation between their expression is predictive of poor survival in HCC patients [76].
Finally, work by Masri and colleagues has revealed an important function of SIRT6 in coordinating circadian rhythms of specific lipid regulatory gene expression programs [75]. Circadian oscillation of gene expression is essential for many physiologic functions, and disruption of this rhythmicity contributes to metabolic disorders, cancer, and aging-related pathologies [79, 80]. Circadian pacemakers present in peripheral tissues are driven by the core clock transcription factor CLOCK:BMAL1, which generates oscillations in up to 10% of genes in any given tissue. Masri et al. showed that deletion of SIRT6 in the liver leads to deregulation of the hepatic circadian transcriptome, with consequent perturbed oscillation of lipid-related metabolites [75]. The transcriptional changes are due to loss of SIRT6 function in controlling the chromatin recruitment of CLOCK:BMAL1 at promoters of clock-controlled genes (CCGs), and of SREBP1 at promoters of its target lipid and cholesterol regulatory genes.
The involvement of SIRT6 in fatty acid metabolism is intriguing given the recent findings that SIRT6 can be activated by FFAs in vitro. Thus, SIRT6 could act as an in vivo sensor of FFA levels, which peak at specific circadian phases [75], and increased SIRT6 activity at those moments could direct timed recruitment of transcription factors to genes involved in fatty acid metabolism. SIRT6 would thus act as a key translator of metabolic cues into epigenetic signals that control circadian rhythmicity.
Future studies should aim to elucidate whether SIRT6 controls circadian physiology in other tissues, or in the master circadian pacemaker of behavioral rhythms in the hypothalamus. In addition, studies of SIRT6 function in circadian biology could suggest ways to combat aging-associated diseases by preventing a decline in circadian rhythms with age.
Concluding Remarks and Future Perspectives
A decade of research has established SIRT6 as a key epigenetic regulator of chromatin and nuclear signaling programs important for mammalian health and disease prevention. The role of SIRT6 in site-specific histone deacetylation has now been linked to regulation of many processes that are deregulated in aging, cancer, and metabolic syndromes, including DNA repair, heterochromatin silencing, genomic instability, glucose and lipid homeostasis, circadian regulation, and stem cell function. Early concerns about the weak histone deacetylase activity of SIRT6 observed in vitro have been resolved by detection of efficient deacetylase activity using improved assays that better mimic physiologic conditions at chromatin, and by development of approaches with optimized detection of substrate-specific deacetylation [10, 12, 14, 23]. Such assays also provide tools for the identification of novel SIRT6 substrates in the future.
Recent chromatin and genomic studies showing that SIRT6 selectively affects different histone substrates depending on genomic or cellular contexts (e.g., H3K18ac in pericentric heterochromatin silencing, and H3K9ac and H3K56ac in distinct transcription and DNA damage response programs), has also led to new testable paradigms for how site-specific histone deacetylation is translated into downstream cellular functions [5, 10, 81]. Notably, studies using point mutations in SIRT6 (including human cancer mutations) that selectively inactivate deacetylase activity have reinforced the fundamental importance of SIRT6-dependent deacetylation for tumor suppression and transcriptional regulation [21, 22].
At the same time, the discovery of novel functions for SIRT6-mediated mono-ADP-ribosylation and deacylation of long-chain fatty acyl groups has opened up new avenues of research into these less understood modifications and raised many important questions [17, 18, 21, 23, 82]. What are the “ribosylome” or “fatty-acylomes” that are regulated by such modifications? How do the modifications affect protein function, cellular signaling, or epigenetic networks? How are these or other SIRT6-dependent modifications linked to cellular energy states or metabolic fluctuations? And can new separation-of-function mutations in SIRT6 be identified and used to further parse the unique or overlapping links of specific substrates and activities to physiologic and pathologic effects of SIRT6 [17, 18, 21, 22]? Answering these and other questions will be crucial for assessing the potential specificity and efficacy of SIRT6 modulation as a therapeutic strategy (see Outstanding Questions).
Box 1 Figure I. Mechanisms of SIRT6 regulation.
Regulation of SIRT6 at the levels of transcription (a), mRNA and protein stability (b), chromatin recruitment (c), and enzymatic activity (d).
Outstanding questions.
Given the importance of SIRT6 in preventing heterochromatin defects associated with aging, is down-regulation of SIRT6 a general phenomenon during aging, and can activation of SIRT6 in such contexts alleviate degenerative changes of aging?
What are the mechanisms of SIRT6 function in diverse tissue stem cells, and are these de-regulated in aging? Can activation of SIRT6 be used to rejuvenate aging stem cells for therapeutic purposes?
How does the interplay between the pleiotropic effects of SIRT6 impact tumor biology, and which mechanisms are most relevant in particular cancers or patient sub-groups? In tumors in which down-regulation of SIRT6 is linked to poor clinical outcome, can activation of SIRT6 be used for therapeutic treatment? Conversely, in cancers where SIRT6 is upregulated, is this a compensatory tumor suppressive response or does it have pathologic effects?
SIRT6 enzymatic reactions are NAD-dependent and can be activated by FFAs. Does SIRT6 serve as a redox or nutrient sensor in chromatin and nuclear signaling programs, and are these pathways altered in diabetes, obesity, and other metabolic disorders? Can dietary regimens be used to modulate SIRT6 function?
Which catalytic activities and substrates of SIRT6 mediate its different cellular and physiologic effects, and what mechanisms regulate these activities in specific contexts? How does SIRT6 selectively affect acetylation at specific histone residues, and how is this translated into distinct downstream signaling pathways? Similarly, are deacetylation, mono-ADP-ribosylation, and de-fatty-acylation by SIRT6 selectively regulated by specific cues? Can small molecule SIRT6 regulators be developed that affect one catalytic activity but not the others?
What are the physiologic targets of de-fatty-acylation and mono-ADP-ribosylation by SIRT6? Can improved proteomic strategies better define the “fatty acylome” and “mono-ADP-ribosylome” in different tissues and cell types, and their regulation by SIRT6?
Trends Box.
SIRT6 promotes heterochromatin silencing at repetitive DNA sequences. Decreased SIRT6 activity may underlie heterochromatin defects that contribute to cellular dysfunction in aging and cancer.
New studies link chromatin regulation by SIRT6 to maintenance of stem cell homeostasis and function, and suggest that SIRT6 may oppose aging-related stem cell dysfunction.
SIRT6-dependent deacetylation promotes tumor suppression through diverse mechanisms. Reduced SIRT6 activity or levels may contribute to cancer progression in many types of human tumors. However, elevated SIRT6 levels in some cancers may have complex effects on tumor progression.
Activation of SIRT6 could have beneficial effects on glucose and lipid metabolism for control of obesity and diabetes. SIRT6 regulation of circadian metabolic programs sheds new light on how the enzyme couples chromatin dynamics to metabolism.
Acknowledgements:
We thank Tie-Mei Li and John Coan for useful discussions and comments on the manuscript.
Glossary
- β-oxidation
Mitochondrial catabolic processes by which fatty acid molecules are broken down to acetyl-CoA
- c-Fos
proto-oncogene that regulates transcription of genes involved in cell proliferation, differentiation and survival
- Cellular senescence
State of permanent cell-cycle arrest caused by telomere shortening during prolonged replication of primary mammalian cells, or by genotoxic and other stress. It is associated with aging in mammalian tissues and can contribute to changes in tissue function
- Circadian rhythm
Any biological process that displays ~24 hour oscillations. They are driven by an endogenous circadian clock, and can be modulated by external cues including light, temperature, and nutrient availability
- E2 transcription factor 1 (E2F1)
member of the E2F family of transcription factors that regulates cell cycle and tumor suppressor proteins
- Fatty acylation
Covalent attachment of fatty acids to proteins (e.g. myristoylation), which can regulate protein function, localization, or interactions
- Forkhead box protein O1 (FoxO1)
transcription factor that regulates gluconeogenesis, glycogenolysis, and adipogenesis through insulin signaling pathways
- Forkhead box O3a (FoxO3a)
tumor suppressor and transcription factor implicated in regulation of apoptosis and protection against oxidative stress
- Heterochromatin
Highly compacted regions of the chromatin in which transcription is silenced. Constitutive heterochromatin is present in all somatic cells and consists mainly of pericentric and telomeric regions, whereas facultative heterochromatin is established in a developmentally or environmental regulated manner
- Hypoxia inducible factor-1 alpha (HIF-1α)
subunit of the heterodimeric transcription factor HIF-1, a master transcriptional regulator of cellular responses to hypoxia and cancer cell metabolism
- Krüppel-associated box associated protein-1 (KAP1)
chromatin-binding protein involved in transcriptional regulation, cellular differentiation, proliferation, and DNA repair
- Long-interspersed element 1 (LINE-1) retrotransposable elements (LINE-1 RTEs)
a class of endogenous retrotransposon sequences
- Mono-ADP-ribosylation
Reversible post-translational modification involving transfer of an ADP-ribose moiety from NAD+ to amino acids of proteins
- Nicotinamide Adenine Dinucleotide (NAD)
molecule that functions as cofactor in redox reactions in cellular metabolism and is required for catalysis by sirtuin deacylase enzymes
- Proprotein convertase subtilisin/kexin type 9 (PCSK9)
protein involved in cholesterol and fatty acid metabolism; mutated in human hypercholesterolemia syndromes
- Peroxisome proliferator-activated receptor gamma PPAR-γ coactivator 1α (PGC1α)
transcriptional coactivator in mitochondrial biogenesis and cholesterol homoeostasis gene expression pathways
- Runt-related transcription factor 2 (RUNX2)
transcription factor involved in cartilage and bone remodeling
- Silent-Information Regulator-2 (Sir2)
S. cerevisiae gene encoding a histone deacetylase that maintains silent heterochromatin
- Sterol regulatory element-binding protein 1 and 2 (SREBP1/2)
transcription factors involved in cholesterol and fatty acid biosynthesis
- Ten-Eleven Translocation methylcytosine dioxygenase 1 and 2 (Tet1 and Tet2)
enzymes that promote 5-hydroxymethylation (5-hmC) of DNA, a process thought to promote DNA demethylation; implicated in modulating cell lineage choice during ESC differentiation
- Warburg effect
Metabolic abnormality of most cancer cells to rely on high rates of glycolysis, rather than mitochondrial oxidative phosphorylation, to generate energy needed for cellular processes and support rapid cell proliferation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nature reviews. Cancer. 2015;15:608–624. doi: 10.1038/nrc3985. [DOI] [PubMed] [Google Scholar]
- 2.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 3.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 4.Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends in biochemical sciences. 2014;39:72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sebastian C, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanfi Y, et al. Regulation of SIRT6 protein levels by nutrient availability. FEBS letters. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michishita E, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang B, et al. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tasselli L, et al. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nature structural & molecular biology. 2016 doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerrer B, et al. The complex role of SIRT6 in carcinogenesis. Carcinogenesis. 2016;37:108–118. doi: 10.1093/carcin/bgv167. [DOI] [PubMed] [Google Scholar]
- 12.Gil R, et al. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic acids research. 2013;41:8537–8545. doi: 10.1093/nar/gkt642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan PW, et al. Structure and biochemical functions of SIRT6. The Journal of biological chemistry. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang WW, et al. A Chemical Biology Approach to Reveal Sirt6-targeted Histone H3 Sites in Nucleosomes. ACS chemical biology. 2016 doi: 10.1021/acschembio.6b00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman JL, et al. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. The Journal of biological chemistry. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liszt G, et al. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. The Journal of biological chemistry. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 17.Mao Z, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Meter M, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nature communications. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du J, et al. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- 20.Feldman JL, et al. Sirtuin catalysis and regulation. The Journal of biological chemistry. 2012;287:42419–42427. doi: 10.1074/jbc.R112.378877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, et al. Identifying the functional contribution of the defatty-acylase activity of SIRT6. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kugel S, et al. Identification of and Molecular Basis for SIRT6 Loss-of-Function Point Mutations in Cancer. Cell reports. 2015;13:479–488. doi: 10.1016/j.celrep.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaeberlein M, et al. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & development. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, et al. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell cycle. 2011;10:3016–3030. doi: 10.4161/cc.10.17.17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Cecco M, et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging cell. 2013;12:247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Cecco M, et al. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging. 2013;5:867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tennen RI, et al. SIRT6 is required for maintenance of telomere position effect in human cells. Nature communications. 2011;2:433. doi: 10.1038/ncomms1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kemp JR, Longworth MS. Crossing the LINE Toward Genomic Instability: LINE-1 Retrotransposition in Cancer. Frontiers in chemistry. 2015;3:68. doi: 10.3389/fchem.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endisha H, et al. Restoring SIRT6 Expression in Hutchinson-Gilford Progeria Syndrome Cells Impedes Premature Senescence and Formation of Dysmorphic Nuclei. Pathobiology : journal of immunopathology, molecular and cellular biology. 2015;82:9–20. doi: 10.1159/000368856. [DOI] [PubMed] [Google Scholar]
- 32.Arancio W, et al. Epigenetic involvement in Hutchinson-Gilford progeria syndrome: a mini-review. Gerontology. 2014;60:197–203. doi: 10.1159/000357206. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S, et al. Lamin A Is an Endogenous SIRT6 Activator and Promotes SIRT6-Mediated DNA Repair. Cell reports. 2015;13:1396–1406. doi: 10.1016/j.celrep.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Carafa V, et al. Sirtuin functions and modulation: from chemistry to the clinic. Clin Epigenetics. 2016;8:61. doi: 10.1186/s13148-016-0224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nature reviews. Molecular cell biology. 2016 doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kokkonen P, et al. Studying SIRT6 regulation using H3K56 based substrate and small molecules. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences. 2014;63:71–76. doi: 10.1016/j.ejps.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Hu J, et al. A fluorogenic assay for screening Sirt6 modulators. Organic & biomolecular chemistry. 2013;11:5213–5216. doi: 10.1039/c3ob41138a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell stem cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Etchegaray JP, et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nature cell biology. 2015;17:545–557. doi: 10.1038/ncb3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, et al. SIRT6 Controls Hematopoietic Stem Cell Homeostasis through Epigenetic Regulation of Wnt Signaling. Cell stem cell. 2016;18:495–507. doi: 10.1016/j.stem.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Pan H, et al. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell research. 2016;26:190–205. doi: 10.1038/cr.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhai XY, et al. Knockdown of SIRT6 Enables Human Bone Marrow Mesenchymal Stem Cell Senescence. Rejuvenation research. 2016 doi: 10.1089/rej.2015.1770. [DOI] [PubMed] [Google Scholar]
- 44.Sun H, et al. SIRT6 regulates osteogenic differentiation of rat bone marrow mesenchymal stem cells partially via suppressing the nuclear factor-kappaB signaling pathway. Stem cells. 2014;32:1943–1955. doi: 10.1002/stem.1671. [DOI] [PubMed] [Google Scholar]
- 45.Sharma A, et al. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. The Journal of biological chemistry. 2013;288:18439–18447. doi: 10.1074/jbc.M112.405928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marquardt JU, et al. Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. Hepatology. 2013;58:1054–1064. doi: 10.1002/hep.26413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cea M, et al. Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood. 2015 doi: 10.1182/blood-2015-06-649970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ming M, et al. SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer research. 2014;74:5925–5933. doi: 10.1158/0008-5472.CAN-14-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, et al. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein & cell. 2013 doi: 10.1007/s13238-013-3054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu CT, et al. The potential of SIRT6 and SIRT7 as circulating markers for head and neck squamous cell carcinoma. Anticancer research. 2014;34:7137–7143. [PubMed] [Google Scholar]
- 51.Ran LK, et al. SIRT6 Overexpression Potentiates Apoptosis Evasion in Hepatocellular Carcinoma via BCL2-Associated X Protein-Dependent Apoptotic Pathway. Clin Cancer Res. 2016;22:3372–3382. doi: 10.1158/1078-0432.CCR-15-1638. [DOI] [PubMed] [Google Scholar]
- 52.Bai LH, et al. Upregulation of SIRT6 predicts poor prognosis and promotes metastasis of non-small cell lung cancer via the ERK1/2/MMP9 pathway. Oncotarget. 2016 doi: 10.18632/oncotarget.9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu LE, et al. Geroncogenesis: metabolic changes during aging as a driver of tumorigenesis. Cancer Cell. 2014;25:12–19. doi: 10.1016/j.ccr.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong L, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choe M, et al. The RUNX2 Transcription Factor Negatively Regulates SIRT6 Expression to Alter Glucose Metabolism in Breast Cancer Cells. J Cell Biochem. 2015;116:2210–2226. doi: 10.1002/jcb.25171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu M, et al. E2F1 enhances glycolysis through suppressing Sirt6 transcription in cancer cells. Oncotarget. 2015;6:11252–11263. doi: 10.18632/oncotarget.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kugel S, et al. SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b. Cell. 2016;165:1401–1415. doi: 10.1016/j.cell.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhardwaj A, Das S. SIRT6 deacetylates PKM2 to suppress its nuclear localization and oncogenic functions. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E538–547. doi: 10.1073/pnas.1520045113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Min L, et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nature cell biology. 2012;14:1203–1211. doi: 10.1038/ncb2590. [DOI] [PubMed] [Google Scholar]
- 60.Kim EJ, Juhnn YS. Cyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (Raf/mitogen-activated extracellular signal-regulated kinase/extracellular signal-regulated kinase) pathway. The Journal of biological chemistry. 2015;290:9604–9613. doi: 10.1074/jbc.M114.633198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bauer I, et al. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. The Journal of biological chemistry. 2012;287:40924–40937. doi: 10.1074/jbc.M112.405837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng XX, et al. Sirtuin 6 promotes transforming growth factor-beta1/H2O2/HOCl-mediated enhancement of hepatocellular carcinoma cell tumorigenicity by suppressing cellular senescence. Cancer science. 2015;106:559–566. doi: 10.1111/cas.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 64.Xiao C, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. The Journal of biological chemistry. 2010;285:36776–36784. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim HS, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell metabolism. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanfi Y, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 67.Anderson JG, et al. Enhanced insulin sensitivity in skeletal muscle and liver by physiological overexpression of SIRT6. Molecular metabolism. 2015;4:846–856. doi: 10.1016/j.molmet.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang P, et al. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10684–10689. doi: 10.1073/pnas.1411026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dominy JE, Jr., et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Molecular cell. 2012;48:900–913. doi: 10.1016/j.molcel.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong X, et al. Sirtuin 6 regulates glucose-stimulated insulin secretion in mouse pancreatic beta cells. Diabetologia. 2015 doi: 10.1007/s00125-015-3778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwer B, et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tao R, et al. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J Lipid Res. 2013;54:2745–2753. doi: 10.1194/jlr.M039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elhanati S, et al. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell reports. 2013;4:905–912. doi: 10.1016/j.celrep.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 74.Tao R, et al. FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. The Journal of biological chemistry. 2013;288:29252–29259. doi: 10.1074/jbc.M113.481473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masri S, et al. Partitioning Circadian Transcription by SIRT6 Leads to Segregated Control of Cellular Metabolism. Cell. 2014;158:659–672. doi: 10.1016/j.cell.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elhanati S, et al. Reciprocal Regulation between SIRT6 and miR-122 Controls Liver Metabolism and Predicts Hepatocarcinoma Prognosis. Cell reports. 2016;14:234–242. doi: 10.1016/j.celrep.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 77.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell metabolism. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 78.Xu Z, et al. SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell cycle. 2015;14:269–276. doi: 10.4161/15384101.2014.980641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maury E, et al. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circulation research. 2010;106:447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nature reviews. Neuroscience. 2012;13:325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toiber D, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Molecular cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feldman JL, et al. Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation. Biochemistry. 2015;54:3037–3050. doi: 10.1021/acs.biochem.5b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McCord RA, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging. 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaidi A, et al. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Lee N, et al. Comparative interactomes of SIRT6 and SIRT7: Implication of functional links to aging. Proteomics. 2014;14:1610–1622. doi: 10.1002/pmic.201400001. [DOI] [PubMed] [Google Scholar]
- 87.Khongkow M, et al. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis. 2013;34:1476–1486. doi: 10.1093/carcin/bgt098. [DOI] [PubMed] [Google Scholar]
- 88.Zhang J, et al. The histone acetyltransferase hMOF suppresses hepatocellular carcinoma growth. Biochemical and biophysical research communications. 2014;452:575–580. doi: 10.1016/j.bbrc.2014.08.122. [DOI] [PubMed] [Google Scholar]
- 89.Lefort K, et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. The EMBO journal. 2013;32:2248–2263. doi: 10.1038/emboj.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davalos A, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stohr R, et al. ITCH modulates SIRT6 and SREBP2 to influence lipid metabolism and atherosclerosis in ApoE null mice. Scientific reports. 2015;5:9023. doi: 10.1038/srep09023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thirumurthi U, et al. MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis and trastuzumab resistance in breast cancer. Science signaling. 2014;7:ra71. doi: 10.1126/scisignal.2005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin Z, et al. USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell reports. 2013;5:1639–1649. doi: 10.1016/j.celrep.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ronnebaum SM, et al. The ubiquitin ligase CHIP prevents SirT6 degradation through noncanonical ubiquitination. Molecular and cellular biology. 2013;33:4461–4472. doi: 10.1128/MCB.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cai J, et al. A crucial role of SUMOylation in modulating Sirt6 deacetylation of H3 at lysine 56 and its tumor suppressive activity. Oncogene. 2016 doi: 10.1038/onc.2016.24. [DOI] [PubMed] [Google Scholar]
- 96.Jung ES, et al. p53-dependent SIRT6 expression protects Abeta42-induced DNA damage. Scientific reports. 2016;6:25628. doi: 10.1038/srep25628. [DOI] [PMC free article] [PubMed] [Google Scholar]