Abstract
The complex multi-chain architecture of antibodies has spurred interest in smaller derivatives that retain specificity but can be more easily produced in bacteria. Domain antibodies consisting of single variable domains are the smallest antibody fragments and have been shown to possess enhanced ability to target epitopes that are difficult to access using multidomain antibodies. However, in contrast to natural camelid antibody domains, human variable domains typically suffer from low stability and high propensity to aggregate. This review summarizes strategies to improve the biophysical properties of heavy chain variable domains from human antibodies with an emphasis on aggregation resistance. Several protein engineering approaches have targeted antibody frameworks and complementarity determining regions to stabilize the native state and prevent aggregation of the denatured state. Recent findings enable the construction of highly diverse libraries enriched in aggregation-resistant variants that are expected to provide binders to diverse antigens. Engineered domain antibodies possess unique advantages in expression, epitope preference and flexibility of formatting over conventional immunoreagents and are a promising class of antibody fragments for biomedical development.
Keywords: human domain antibody, variable heavy domain, antibody engineering, synthetic antibodies, phage display, aggregation
1. INTRODUCTION
1.1 Antibody fragments, in vitro display and synthetic antibody libraries
A conventional monoclonal antibody (mAb) is composed of two heavy and two light polypeptide chains connected via multiple disulfide bonds (Figure 1). The two antibody arms (antigen binding fragments, Fabs) can independently bind antigens and the constant stem region (fragment crystallizable, Fc) is responsible for effector functions. Immunoglobulin G (IgG) is the most abundant antibody class in human serum and in therapeutic development. The heavy chains of IgGs consist of three constant domains (CH1, CH2 and CH3) and one variable domain (VH), and each domain consists of a characteristic β-sandwich fold [1, 2]. The light chains contain one constant [3] and one variable domain (VL). The complementarity determining regions (CDRs) are located in loops that connect the β-sheets of the VH and VL domains. Sequence diversity in the CDRs produces a contiguous paratope capable of recognizing diverse molecular surfaces. Five of these loops adopt one of the canonical structures defined by specific loop and framework interactions [4]. No canonical structures have been identified for the third heavy chain CDR (CDRH3), which varies significantly in length, sequence and conformation [5, 6].
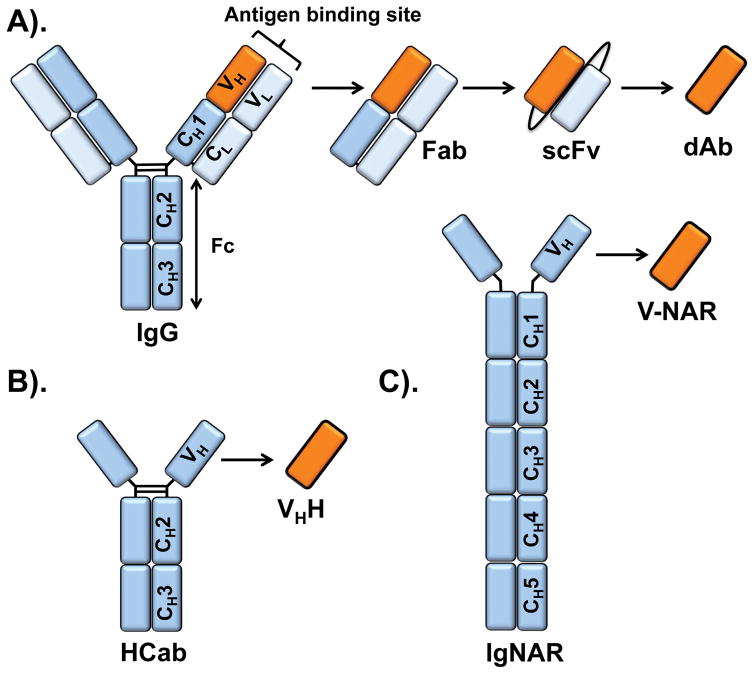
Figure 1. Immunoglobulins from various species.
(A) Schematic representation of a human IgG and related fragments [fragment crystallizable (Fc), fragment antigen binding (Fab), single-chain fragment variable (scFv) and VH domain antibody (dAb)]. (B) Camelid heavy chain only antibody (HCab) and its autonomous antigen-binding domain (VHH). (C) Immunoglobulin new antigen receptor (IgNAR), found in cartilaginous fish, and its autonomous antigen-binding V-NAR domain.
The success of antibodies as affinity reagents in research and diagnostic applications as well as therapeutics is due to their extraordinary specificity, high affinity, long serum half-life and amenability to engineering [7, 8]. The modular nature of immunoglobulins can also be exploited to engineer smaller antibody fragments such as Fabs [9], which are heterodimers consisting of the variable (VH and VL) and constant (CH1 and CL) domains. Other important antibody fragments include the fragment variable (Fv) [10], which consists of the VH and VL domains and the single-chain fragment variable (scFv) [11] where the VH and VL domains are joined by a peptide linker. Single domain antibodies (dAbs) consisting of only the variable region from either the heavy or light chain are the smallest antigen-binding fragments of antibodies (11–15 kDa) [12].
Antibody fragments can retain the affinity and specificity of their parent antibodies while enabling the use of bacterial expression systems, which are simpler and less costly than the mammalian expression systems used to produce full-length antibodies [8]. However, due to the lack of the Fc region, antibody fragments have fewer modes of action than full-length mAbs. The Fc domain recruits cytotoxic effector functions through complement activation and binding to Fcγ receptors, and endows long serum half-life via binding to the neonatal Fc receptor (FcRn) [13]. Antibody fragments can also elicit therapeutic action by binding a ligand or receptor or be used in applications where small size or lack of effector functions is desired. Autonomous constant (CH2) domains derived from human IgG have also been engineered as antigen-binding scaffolds [14]. An attractive feature of engineered CH2 domains is their potential for both antigen and FcRn binding, the later of which prolongs serum half-life [15, 16]. Soluble autonomous CH3 domains have been described [17] and loops on CH3 have been recruited for antigen binding in so-called Fcabs (Fc antigen binding) [18–20]. There are also structurally related non-immunoglobulin scaffolds such as the fibronectin type III domain (FN3), which has been extensively characterized and shown to be a robust platform for generating novel binders [21, 22].
Since the introduction of in vitro display technologies some 30 years ago [23], many antibodies and antibody fragments have been selected and improved by using these methods [24–27]. These technologies provide a physical linkage between the phenotype (displayed antibody) and the genotype (encapsulated DNA) and enable amplification, selection and manipulation of recombinant antibodies in vitro. Display technologies permit the use of controlled selection conditions and provide direct access to the gene sequence encoding the antibody. Numerous studies have demonstrated successful selection of antibodies from in vitro repertoires that rival or surpass antibodies obtained by immunization in terms of functionality [28, 29]. In fact, the general features of natural antibody structures and functions are now so well understood that sophisticated design strategies can be employed to encode and display fully synthetic human antibody libraries optimized for molecular recognition in vitro [30–32]. Precise control over design enables the use of highly optimized human frameworks and the introduction of defined chemical diversity at positions that are most likely to contribute to antigen recognition [33]. Our work has demonstrated that remarkably diverse antibody functions can be supported by a single scaffold based on a highly stable human framework originally used for the humanization of therapeutic antibodies [34, 35]. Starting from initial minimalist designs that used two or four amino acid codes [36, 37], synthetic antibody designs have been progressively optimized by incrementally expanding the size of the diversified paratope and by increasing chemical diversity [32]. In a recent optimized library design of a Fab fragment, binary diversity was introduced in CDRH1 and CDRH2 whereas CDRH3 and CDRL3 contained more chemical and length diversity [38]. These synthetic repertoires are truly naïve because they have not been subjected to the restrictions imposed by self-tolerance of natural repertoires [33]. Furthermore, initial clones can be rapidly affinity-matured using secondary libraries and in vitro display and can be reformatted into the natural IgG format or into various non-natural antibody fragment or bispecific formats for specialized downstream applications [28, 29, 39].
1.2 Natural autonomous variable domains can access restricted epitopes
Early experiments suggested that antibody heavy chains alone could sometimes bind antigen in the absence of a light chain partner [40]. Twenty-five years later, antigen-specific dAbs were isolated from a cloned repertoire of antibody fragments derived from mice immunized with lysozyme [41]. Although it might seem surprising that three CDR loops are sufficient to confer antigen binding affinities comparable to those of conventional antibodies, evolution has itself arrived at that very solution in camelids and cartilaginous fish [42]. These species naturally produce heavy chain antibodies with binding sites contained within a single unpaired variable domain (Figure 1). These unusual variable domains from heavy chain antibodies are referred to as VHH domains in camelids [43] and V-NAR (variable domain of new antigen receptor) in cartilaginous fish [44, 45]. Several structures have shown that isolated human and camelid dAbs adopt a β-sheet structure similar to variable domains in IgGs and function as independent antigen-binding units in vitro [46–49]. Upon loss of the associated light chain, VHH domains have acquired framework substitutions that are not observed in paired VH domains. These substitutions are thought to have evolved to accommodate the absence of the VL and CH1 domains. In contrast to VHHs that are believed to have originated from an IgG molecule that underwent selection and permanently lost its light chain, V-NARs appear to have evolved convergently from a cell-surface receptor [50].
Two fundamental limitations of full-length IgGs are their poor penetration into tissues (e.g. solid tumors) and limited ability to target certain antigens (e.g. viral and bacterial antigens) that are more accessible to antibody fragments of smaller size [8, 14, 51]. The small size of dAbs has potential to provide better access to such sterically restricted epitopes. The paratope of Fvs can form a flat surface, a groove or a cleft to recognize flat protein epitopes, linear peptides or haptens, respectively [52]. The absence of the VL means that the paratope of an autonomous VH is often smaller and more convex, which allows for penetration of concave epitopes and clefts that are poorly accessible to conventional paratopes. The convex shape of many VHH domains originates from an unusually long CDRH3 loop, which averages 17 residues for camelid antibodies versus 12 or 9 residues for human and mouse antibodies, respectively [6, 48, 53]. The CDRH3 of VHH domains often packs against hydrophobic framework residues and sequesters them from solvent [54, 55]. Furthermore, the entropic penalty imposed by fixation of a long CDRH3 loop upon binding is sometimes minimized by the presence of a disulfide bond connecting CDRH3 to CDRH1, FR2 or FR4 [42, 48, 56].
Several examples emphasize the unique ability of dAbs to target epitopes that are difficult to access with conventional antibodies (Figure 2). Long protruding loops can insert into catalytic sites of enzymes [45, 46, 57, 58], which are often located at the largest cavity on the enzyme surface [59]. Restricted epitopes on the surface of pathogens have also been targeted using dAbs, including an immunosilent malarial surface protein canyon and a trypanosome surface glycoprotein [60, 61]. VHH domains have been shown to engage epitopes generally inaccessible to conventional antibodies on G-protein coupled receptors (GPCRs), which are a class of medically important protein targets. For example, VHHs have been used as chaperones to facilitate structural analysis of conformational states of membrane proteins [62, 63]. The limited ability of the human antibody response to target such proteins may be due to the limited diversity of CDR loop length, the large size of the antigen-binding units and/or the fact that the paratopes have mostly flat or concave topologies. Although CDRH3 is typically primarily responsible for antigen binding in VHHs and V-NARs, the buried surface area at the antibody-antigen interface is often comparable to that for conventional antibodies [64].
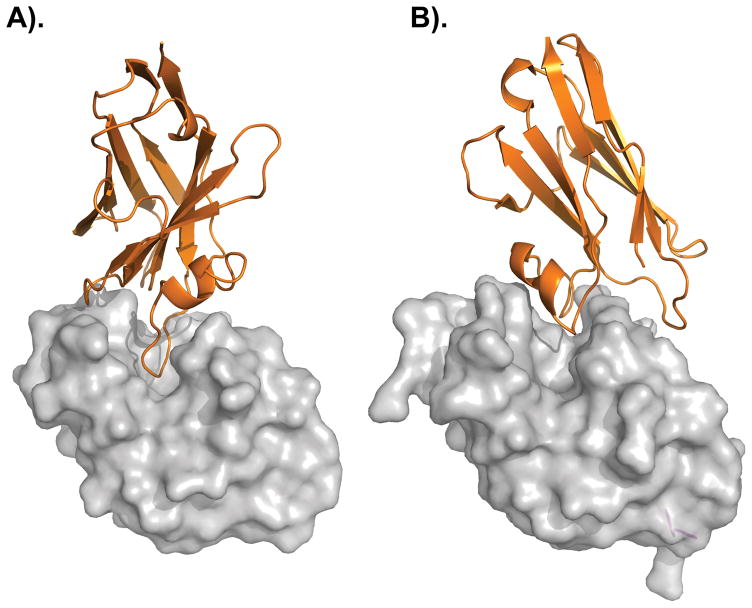
Figure 2. dAbs binding to the catalytic cleft of lysozyme.
(A) VHH and (B) V-NAR domains with the dAbs colored in orange and lysozyme shown in gray. Pictures were generated in PyMOL using PDB 1MEL [46] and PDB 1SQ2 [45].
Around the same time that heavy-chain only antibodies were identified, it was found that bovines could produce antibodies with exceptionally long CDRH3 loops [65]. In some bovine antibodies, a β-ribbon stalk protrudes from the surface to display a disulfide bonded knob domain, which provides a unique paratope that can potentially engage concave epitopes [48, 66]. The substantial antigen recognition adaptations in camelids, cartilaginous fish and cows suggest that a capability of binding epitopes via a protruding paratope may provide an important evolutionary advantage.
1.3 Aggregation propensities of human domain antibodies
Improving the biophysical properties of antibodies has important implications for clinical, diagnostic and biotechnological applications. Some human mAbs display low stability and/or are prone to aggregation. Lack of an Fc domain in antibody fragments can further increase the risk of aggregation during production and purification [8, 67], which in turn decreases purification yields, shortens shelf-life and increases the risk of immunogenicity [68–70]. The biophysical properties of full-length antibodies are often limited by the stability of their variable domains [71, 72], which can be prone to aggregation and/or poorly soluble in isolation. This is a result of exposure of a large hydrophobic surface (~700 Å2) in the absence of an interacting chain [2, 73] and a lack of the domain-domain interactions that stabilize larger antibody formats [67, 74]. The thermodynamic stability of human VHs is comparable to that of VHHs, but most human VHs do not refold reversibly without aggregating. In contrast, many VHHs and V-NARs refold reversibly, resist aggregation even when unfolded and retain activity after being heated above their melting temperatures [75, 76]. Both colloidal stability (solubility) and conformational (folding) stability contribute to antibody solution properties [77]. Conformational stability is primarily governed by solvent-shielded residues within the domains or at domain interfaces and is critical for long-term antibody activity. The colloidal stability of human dAbs, which is arguably the main obstacle holding back the field [78, 79], arises from intermolecular self-interactions mediated by solvent-exposed residues that are commonly located in CDRs [80]. Strategies for improving colloidal stability include altering solution properties such as pH and ionic strength, and the use of excipients [67, 77]. Proteins with low conformational stability are more vulnerable to aggregation upon exposure to environmental stress (e.g. heating, freezing, low pH), which is due to attractive self-interactions between unfolded molecules [3, 78]. If the interactions between unfolded dAbs are insufficiently attractive to mediate aggregation, the proteins may refold.
Considering the beneficial properties of naturally occurring single variable domains in heavy chain only antibodies, it is desirable to generate human dAbs that would have similar properties but would be potentially less immunogenic. Human variable domains are divided into families based on their sequence homology and different families exhibit distinct biophysical properties. The VH3 family has been identified as having the best biophysical properties among human subtypes [73]. Furthermore, VH3 is the largest germline family of the human repertoire, it is the most frequently used human acceptor framework for CDR grafts and it is overrepresented in exceptionally stable antibody constructs [81]. Notably, the VH3 family is also most closely related to VHH domains from camelid heavy chain only antibodies [82].
Sequence comparison of human and camelid variable domains led to the process of camelization, which involves the transfer of a few hallmark residues of aggregation-resistant VHH domains to human VH domains [83]. These solvent-exposed residues (most of which are hydrophilic) are located in the former light chain interface and are assumed to impede the association with a VL domain. Analogous mutational approaches to humanize VHHs have also been attempted [84, 85]. However, these strategies generally only partially eliminate the tendency to aggregate and the modified VH domains may display poor expression levels, low solubility, low stability, increased stickiness on size-exclusion columns and/or impaired antigen binding [84–88]. Similar strategies have been applied to V-NARs, a process that is further complicated by the lower sequence homology and larger structural differences compared to human VH domains [89].
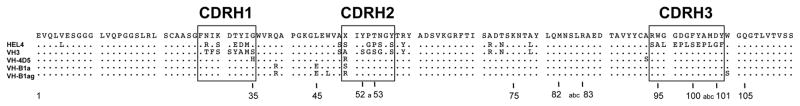
Studies of human dAbs have identified key sequence and structural determinants that differentiate aggregation-prone and aggregation-resistant antibodies [79, 90, 91]. A striking example is the aggregation resistant hen egg lysozyme binder HEL4, which displays properties similar to those of VHH domains while the hydrophobic framework residues of the former VL interface are maintained [92]. HEL4 only differs from its aggregation-prone germline VH parent (Dp47d) in its CDRs (Figure 3A). Structural analysis revealed an unusual flipping of a hydrophobic framework tryptophan side chain into a cavity facilitated by a glycine residue in CDRH1, which increases the hydrophilicity of the surface that is normally covered by the light chain. However, in spite of its monomeric state in solution, HEL4 crystallizes as a dimer where the crystal packing contacts resemble the interactions observed at a VH/VL interface. Interestingly, mutational dissection of HEL4 demonstrated that grafting of CDRH1 from HEL4 to Dp47d could confer aggregation-resistance (Figure 3B) [93]. The aggregation hotspot was further pinpointed to a charged triad of residues in the middle of CDRH1. In contrast, transfer of CDRH2 or CDRH3 from HEL4 to Dp47d or incorporation of charged mutations outside of the CDRs of Dp47d failed to suppress aggregation. Taken together, these findings indicate that modification of CDR sequences alone might alter aggregation behavior.
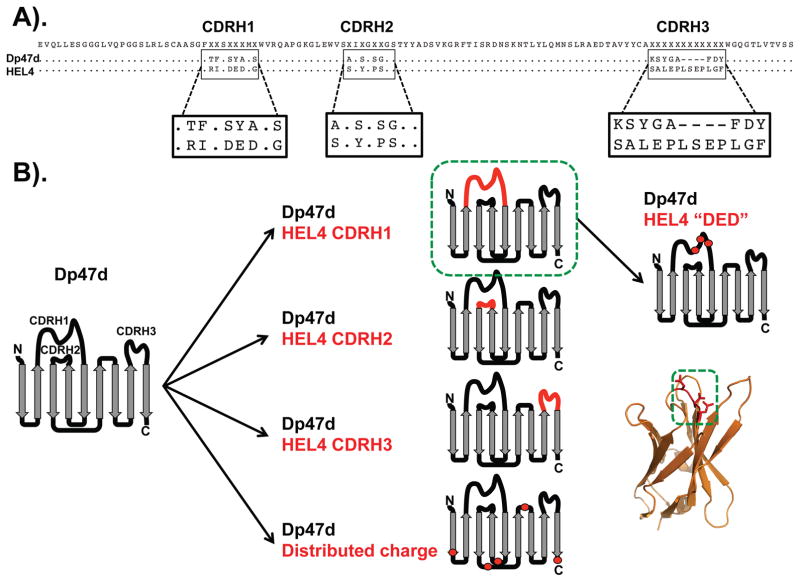
Figure 3. Role of CDR sequences in dAb aggregation.
(A) Amino acid alignment of the aggregation-prone germline VH segment Dp47d and the aggregation-resistant dAb HEL4. The sequences only differ in the CDRs. Sequence identities are shown as dots. The figure was prepared using Geneious version 7.1.9 [94]. (B) Grafting of CDRs from HEL4 to Dp47d or incorporation of charged substitutions in non-CDR regions. Grafting of CDRH1 from HEL4 to Dp47d conferred resistance to aggregation (green box) whereas all of the other dAb constructs aggregated. Aggregation resistance was attributed to a charged triad of residues (Asp-Glu-Asp; DED) within CDRH1, which is also shown in the HEL4 crystal structure (green box) (image was prepared from PDB 1OHQ [92] using PyMOL).
1.4 Rational engineering of aggregation-resistant human dAbs
The findings based on CDR-swapping between HEL4 and Dp47d suggested that incorporation of charged residues in the CDRs might improve antibody solubility. Moreover, a charged substitution for a large hydrophobic residue adjacent in CDRH1 (F29D; Kabat numbering [5]) could by itself inhibit aggregation of Dp47d [93]. This hydrophobic residue is conserved across diverse human antibodies [95] and therefore typically not involved in the antibody paratope. This finding demonstrates that highly beneficial mutations are rarely explored by natural mechanisms that create antibody diversity. Negatively charged residues, as observed in HEL4, have previously been shown to contribute favorably to protein solubility [96]. Aggregation resistance of antibody fragments can also be mediated by attaching negatively charged peptide sequences, as demonstrated by fusion of the acidic tail of α-synuclein [97], a penta glutamic acid tag [98] or a Glu-Ala dipeptide [99]. Similar solubilizing effects have been reported for the commonly used acidic FLAG-peptide [100, 101].
The potential solubilizing effects of negatively-charged CDR insertions were examined using so called Grafted AMyloid-Motif AntiBODIES (gammabodies) [102, 103], which are VH domains that display hydrophobic peptide segments within CDRH3. VH domains grafted with peptides derived from the Alzheimer Aβ peptide bind to Aβ-oligomers and fibrils but their hydrophobic CDRs make them prone to aggregate. Intriguingly, insertion of an increasing number of negatively charged residues at the edges of the hydrophobic CDRH3 gradually prevented aggregation while maintaining binding activity [91]. Three charged residues at each edge of CDRH3 could completely suppress aggregation while charged mutations outside this loop failed to achieve this. Furthermore, it was shown that charge insertion at the CDRH3 edge closest to the aggregation hotspot alone could eliminate aggregation whereas insertion at the CDRH3 edge opposite to the aggregation hotspot was ineffective. Hence, the solubilizing effect of charged residues in CDRs may be directly related to the proximity to clusters of hydrophobic residues [91]. Detailed investigation of polar mutations at the edges of the grafted CDR loop demonstrated that negatively-charged or asparagine insertions are more effective than positively-charged or other polar residues [104]. Moreover, the negatively-charged insertions did not compromise the conformational specificity of gammabodies for Aβ-fibrils relative to monomers while other insertions often impacted on this specificity. Similar results were obtained using a VH domain grafted with a long hydrophobic peptide derived from islet amyloid polypeptide (IAPP). However, this strategy is more difficult to apply in cases where more than one CDR is involved in hydrophobic patches that mediate binding.
Although the overall charge can have a dramatic effect on antibody aggregation [105], results from gammabodies and mutational analysis of HEL4 demonstrate the importance of localized charge. Moreover, the net charge of the antibody scaffold will influence how effective positively- or negatively-charged substitutions are on preventing aggregation [106]. In summary, attempts to predict dAb aggregation propensity must consider both the net charge and the spatial distribution of charge and hydrophobicity.
1.5 Combinatorial engineering of aggregation-resistant human dAbs
The power of directed evolution allows the selection of molecules with desired properties even in the absence of detailed mechanistic insights. This allows dAb libraries to be constructed and subjected to selection conditions designed to identify variants that have incorporated desired properties. Such technologies are particularly useful to screen for aggregation-resistant variants since this trait is highly complex and difficult to rationally link to sequence information. Moreover, in the absence of the VH-VL combinatorial diversity, there is increased need for constructing highly diverse dAb libraries for antigen selection. The diversity is inherently limited to only three CDRs compared to six in conventional mAbs and large libraries are typically required to yield binders with high affinities [25, 107].
One strategy that has proven to be a powerful tool to explore the influence of CDR-sequences on aggregation resistance involves displaying multiple copies of dAbs on the surface of phage (Figure 4) [108]. Transient heating of the phage pool at a temperature above the melting temperature is used to induce unfolding and aggregation of poorly soluble dAbs at the phage tip upon cooling. Aggregation-resistant dAbs that are able to refold without aggregating are selected using Protein A, a bacterial superantigen that binds folded VH3 domains in a CDR-independent manner [109]. This strategy was applied to isolate several aggregation-resistant dAbs from a library based on HEL4 in which all three CDRs were diversified. Importantly, the selected dAbs often have similar or lower stabilities than their aggregation-prone counterparts, which suggest that the results are not simply due to stabilization of the native state. In general, a lack of correlation between conformational stability and aggregation propensity is a common observation for dAbs [55, 91].
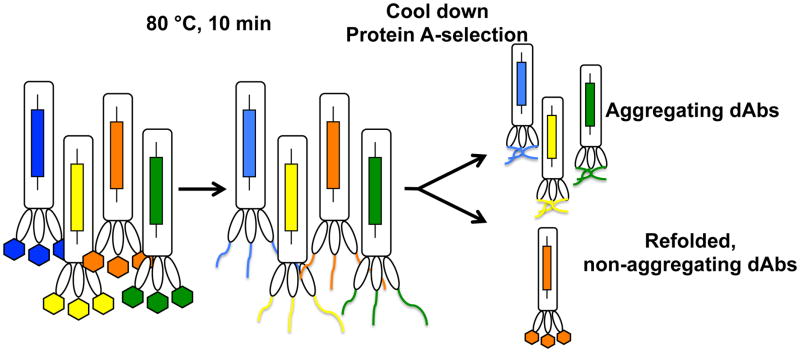
Figure 4. A phage display selection for aggregation resistance [108].
Phages displaying multiple copies of dAbs are heated to denature the displayed proteins. After cooling, aggregation-resistant dAbs refold and can be selected by binding to Protein A whereas aggregation-prone dAbs remain unfolded and are washed away.
By recombining heat-tolerant CDR sequences isolated from the first HEL4-library [108] into a new library, the frequency of aggregation-resistant clones could be increased 10-fold [110]. Furthermore, this new repertoire could be used to select antigen-specific dAbs, some of which retained the beneficial properties of the selected CDR building blocks. Another approach [111] aimed to increase the fraction of aggregation resistant dAbs in the library by using a modified HEL4 sequence as the scaffold. The solubilizing CDRH1 sequence of the template was retained and combined with an additional aggregation-preventing Asp29 residue adjacent to it [93], and diversity was introduced in CDRH2 and CDRH3. Previous libraries based on the same framework have restricted diversity at position 29 to particular hydrophobic residues [108] whereas camelid VHH sequences typically encode Tyr or Ser [55]. However, analysis of selected clones showed that the sequence motifs that were retained in the library design did not confer a general resistance to aggregation [111].
An interesting observation from the phage-based heat denaturation system was that a majority of the identified aggregation resistant dAbs exhibit an acidic isoelectric point [108, 112]. Similar results have been reported for dAbs isolated from another human VH library exposed to comparable selection conditions [113] as well as for other non-antibody proteins [114–117]. A more systematic mutagenesis approach of human variable domains combined with the phage-based aggregation method revealed a significant preference for negatively charged residues in several positions clustered in CDRH1 of a VH3 scaffold [118]. Moreover, the effects of the charged substitutions were additive, with maximal effects observed for two or more substitutions in this region, and also appeared to be independent of sequence diversity at other positions. These general rules for charged mutations are in agreement with HEL4 and other aggregation-resistant dAbs isolated from HEL4-based libraries. Interestingly, the same approach applied to a human VL domain using Protein L for selection identified a similar aggregation hotspot centered in the antigen-binding site in CDRL2 [118].
A related study used a library based on another autonomous VH domain (m0) that contained diversity from natural sources incorporated in CDRH2 and CDRH3 and degenerate four amino acid codons (A/D/S/Y) in CDRH1 [119]. Selected clones had a significant increase in Asp residues at several positions in CDRH1, one of which overlaps with the positions described in the study by Dudgeon et al. [118]. Furthermore, the heat-denaturation method on phage can be used as a predictor of expression and refolding yields of dAbs [120]. This study was facilitated by the use of a panel of dAbs with varying aggregation propensities based on the rules for charged substitutions in CDRH1 [118]. Since the sequences of those model domains are highly similar yet have varying yields of soluble protein, the results point to protein aggregation as a major limiting step in expression of human VH domains. It is relatively easy to select mutations that confer improved stability or antigen binding. However, it is more challenging to select sets of rare mutations that improve binding but are not destabilizing. The importance of simultaneously optimizing affinity and stability was recently demonstrated in selections of yeast-displayed gammabodies where simultaneous selection for antigen binding and Protein A binding was employed [121].
In an effort to extend the multivalent phage display method [108] to select thermodynamically stable dAbs, the phage pool was incubated in acid in place of heating [122]. Capture of non-denatured dAbs using Protein A yielded variants with higher melting temperatures. Although no general stabilizing sequence features were identified, a common substitution (T28R) was found to increase aggregation resistance at low pH without compromising thermodynamic stability.
More recent libraries that exclusively focus diversity to CDRs have incorporated some of the characteristics of aggregation-resistant dAbs in their design criteria [123]. In one library (Garvan I), the CDRH1 of HEL4 was retained in order to maintain its aggregation resistant properties while seven or nine positions in a fixed length CDRH3 were randomized. Interestingly, analysis of sequences that retained Protein A binding after heating demonstrated that a large portion of randomly selected clones displayed considerable resistance to heat-induced aggregation on phage. However, the median levels were well below that observed for HEL4 and only moderate affinities were observed for binders obtained from antigen selections using this repertoire. A following design (Garvan II) retained two negatively charged residues in CDRH1 and encoded limited diversity in two adjacent positions. This strategy fulfilled the previously demonstrated requirement of at least two negatively charged residues in this region [118] and allowed some variety within the encoded CDRH1 sequences. The library also included limited diversity in five positions in CDRH2. CDRH3 diversity allowed two loop lengths and was biased towards Tyr, Gly and Ser but also contained small fractions of all other residues except cysteine. In addition, two previously analyzed mutations that are partly responsible for the beneficial properties of HEL4 were preserved (Arg28 and Gly35) [92, 122, 124].
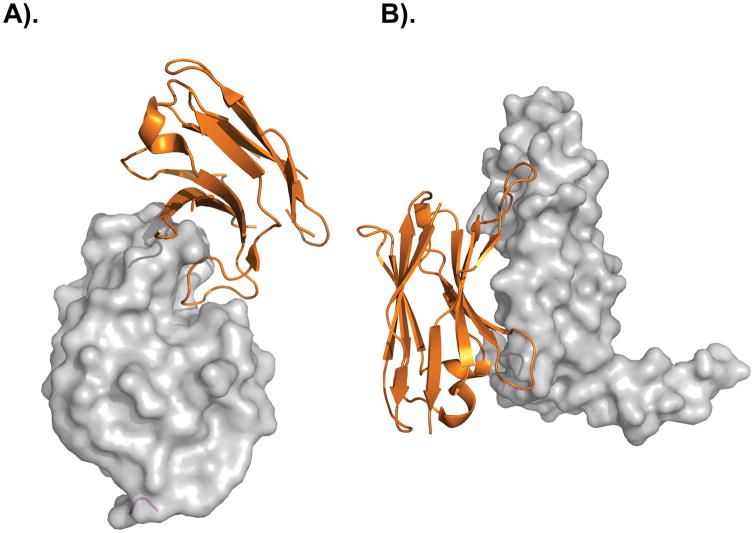
The lack of high-affinity binders obtained from the Garvan I repertoire may be a result of the multivalent phage display format, which is a prerequisite for the phage-based heat denaturation method but is also known to promote the enrichment of low affinity clones due to avidity effects. To overcome this limitation, a phagemid system was used to allow monovalent display of dAbs in the Garvan II library. Compared to the Garvan I repertoire, the median level of resistance to heat-induced aggregation on phage was further decreased. This result was most likely a consequence of the increased repertoire diversity (more randomized positions) in the Garvan II repertoire. Affinities for five antigens were generally in the high nanomolar range. Several of the selected dAbs demonstrated favorable biophysical properties, showing that CDR composition alone can mediate aggregation resistance also in antigen specific clones from a naïve library. The most striking result was the preference of selected lysozyme binders to bind epitopes within or in proximity to the active site cleft, which is the dominant epitope for VHH domains but an uncommon epitope for human antibodies. Structural analysis of one dAb in complex with lysozyme revealed that the binding interface was dominated by CDRH3, which protrudes deeply into the active site cleft (Figure 5A). Interestingly, the degree of shape complementarity was higher and more in the range of complexes of antigens with VHH or V-NAR domains compared to reported structures of antigen complexes with human dAbs [123].
Figure 5. Different modes of antigen recognition by human dAbs.
(A) Human dAb (orange) binding to lysozyme [105] via the insertion of CDRH3 into the catalytic cleft. (B) Human dAb (orange) binding to a flat epitope on vascular endothelial growth factor [105] via an extended CDRH3 loop and the former light chain interface. Pictures were generated in PyMOL using PDB 4PGJ [123] and PDB 3P9W [125].
In summary, these libraries demonstrate that it is possible to generate human dAbs that rival the solubility and cleft binding properties of domains derived from heavy chain only antibodies. However, it is difficult to construct libraries where a large fraction of variants benefit from sequence characteristics of CDRH1 that have been linked to aggregation resistance. The remaining CDRs need to be highly diversified in order to yield competitive antigen-binding affinities, which in turn might offset the solubilizing effects of mutations in or near CDRH1.
1.6 Systematic analysis of factors that contribute to autonomous behavior in a human VH domain
A systematic approach based on combinatorial phage-displayed libraries and conventional biophysical methods has been used to generate autonomous human VH domains [124]. This strategy aimed to identify structurally compatible hydrophilic substitutions in the former light chain interface that promote autonomous behavior. In contrast to most previous attempts to elucidate features contributing to the autonomous nature of VHH or VH domains, this approach was unbiased since diversity was not restricted to natural sequence variation. The VH domain of 4D5, a therapeutic monoclonal antibody [34], was displayed on phage as a starting point to engineer autonomous domains (Figure 6). The Fab and scFv forms of this antibody have been used as scaffolds to construct synthetic antibody libraries [126, 127], and the VH domain belongs to the VH3 family that binds to Protein A through conformation-specific interactions involving framework regions opposite to the light chain interface. This VH domain has potential to function as a universal human scaffold for the generation of dAb reagents. However, the shape of the paratope may impose restrictions on which epitopes and antigens that can be targeted (e.g. binding to linear peptides or haptens).
Figure 6. Amino acid alignment of selected VH3 domains.
HEL4 is an aggregation-resistant human dAb isolated from a library by panning against lysozyme [92]. A consensus VH3 domain [128] is shown as a representative of this biophysically favorable VH family. The consensus sequence is shown with a CDRH3 derived from hu4D5-8 [34]. VH-B1a [124] and VH-B1ag [125] are autonomous VH domains derived from VH-4D5. Boxes denote CDRs, sequence identities are shown as dots, and numbering is according to Kabat. The figure was prepared using Geneious version 7.1.9 [94].
An initial VH-4D5 library [124] was designed around residues that have been implicated in the stabilization of VHH domains and the autonomous human VH domain HEL4 [92]. Three hydrophobic residues of the VHH tetrad (Val37, Leu45 and Trp47) were randomized with degenerate codons that encode all 20 amino acids. The fourth tetrad residue (Gly44) was already hydrophilic and was thus left unchanged. In addition, position 35 in CDRH1 was targeted for randomization because it has been associated with stabilization of HEL4. To account for possible stabilization of VH domains through interactions between CDRH3 and the former light chain interface, CDRH3 was replaced by random loops of diverse lengths. The library was selected for binding to Protein A and sequences of selected clones were analyzed for conserved motifs. In contrast to similar analyses of VHH domains [54, 129], no bias for hydrophobic CDRH3 residues was observed suggesting that the VH domain is not dependent on stabilizing interactions between the former light chain interface and CDRH3. The hydrophobic character of the wild-type residues in positions 37 and 45 was conserved. Many substitutions were observed in position 47, which is occupied by a large tryptophan residue in the wild-type sequence, and position 35 was biased towards small residues. This may indicate that interactions between small residues in position 35 and hydrophobic residues in position 47 enhance hydrophilicity in a manner similar to what has been observed for HEL4. Alternatively, substitution of Trp47 with a smaller or charged residue may promote autonomous behavior.
Analysis of six purified VH domains from the first selection identified four variants with improved properties compared to the aggregation prone wild-type domain [124]. A second-generation library was designed using the most promising first generation variant (VH-A1) as a template. Analysis of the structure of the wild-type domain identified ten positions involved in the light chain interface that were simultaneously targeted for randomization. A soft randomization approach was employed to bias the library towards template residues since the template already exhibited many of the desired characteristics. CDRH3 was also targeted for diversification but was retained at the same length as in VH-A1. Following selection for binding to Protein A, many clones were sequenced to determine the amino acid distribution at each randomized position. Expression of 40 variants showed that most gave higher yields than VH-4D5 but only one domain (VH-B1) exhibited improved yield over VH-A1. VH-B1 exhibited a high melting temperature (73°C), monomeric behavior and essentially quantitative refolding.
To thoroughly assess stabilizing effects of individual residues within CDRH3, VH-A1 and ten variants from the second-generation library were subjected to shotgun alanine scanning analysis [130]. Briefly, this strategy diversified every residue in CDRH3 using a degenerate codon that encodes alanine and the wild-type residue. The wt/Ala ratio was determined for each scanned position by sequencing clones after two rounds of selection for binding to Protein A. Wild-type/Ala ratios greater than one imply a stabilizing effect of the wild-type residue whereas a value below one implies a destabilizing effect. This analysis demonstrated that the stabilities of many domains were independent of the CDRH3 loops.
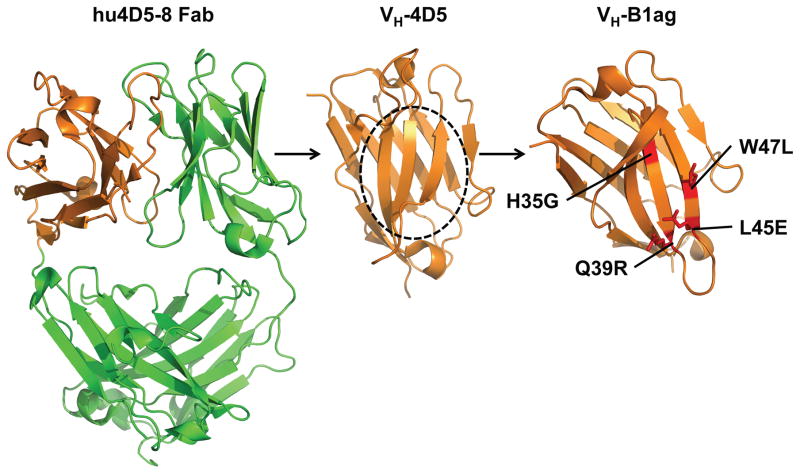
Next, quantitative saturation scanning mutagenesis [132] was performed on nine positions in VH-B1 to provide a more complete picture of the sequence requirements in the autonomous former light chain interface. Modest biases were observed at most positions and the data indicated that the stabilization of the domain was due to the cumulative effects of contributions from several substitutions. To confirm that the CDRH3 sequence of VH-B1 did not influence stability, a new variant (VH-B1a) was constructed by replacing this loop with the CDRH3 of VH-4D5. The resulting protein only differs from the wild-type domain by four substitutions in the former light chain interface (H35G, Q39R, L45E and R50S).
The crystal structure of VH-B1a (Figure 7) was solved to visualize how the novel mutations contribute to autonomous behavior. Occurrence of Gly35 suggested that the VH domains might be stabilized by a similar mechanism as observed in HEL4 [92]. However, in contrast to the HEL4 structure where Trp47 occupies the cavity formed by Gly35, Trp95 located at the beginning of CDRH3 occupies this space in VH-B1a. This interaction brings CDRH3 closer to the framework while leaving Trp47 exposed to solvent. However, CDRH3 adopts different conformations in the four molecules of the asymmetric unit, which indicates that it is flexible in solution. Arg39 and Glu45 increase the hydrophilicity and the opposite charges can presumably stabilize the folded structure via electrostatic interactions [78]. The single back mutations R39Q or E45L reduced the monomeric fraction or increased the elution time from a size-exclusion column, respectively, which indicated that the hydrophobic Leu45 promoted interactions with the chromatography matrix. Ser50 is in close proximity to Gly35 and its shorter side chain may facilitate formation of the cavity that reorientates the adjacent aromatic residue. However, it was unclear if Ser50 contributes favorably to solubility.
Figure 7. Engineering of autonomous VH domains.
VH-4D5, derived from hu4D5-8 [34], is prone to aggregation. Systematic combinatorial mutagenesis identified key substitutions in the former light chain interface (circled) that promote autonomous behavior [124]. Four substitutions in this region (shown in red) transformed the template VH domain into the autonomous VH-B1ag. Pictures were generated from PDB 1N8Z [131] and 3B9V [124] using PyMOL.
Because the hydrophobic Trp47 was exposed to solvent in the crystal structure and has previously been implicated in the autonomous behavior of VH and VHH domains [55, 92], variants were generated in which Trp47 was replaced by residues that were commonly observed in the saturation scanning data set [124]. Moreover, additional substitutions were evaluated in the W47L background because this substitution reduced gel filtration elution time and led to increased thermostability. Analysis of 20 single- and double mutants of VH-B1 demonstrated that most of them were monomeric with a high melting temperature and a high degree of refolding after thermal denaturation.
As a final validation of the four substitutions in the former light chain interface that differ between VH-B1 and VH-4D5 they were individually backmutated in three different sequence backgrounds [VH-B1a, VH-B1a (W47L) or VH-B1a (W47T)][124]. In all cases the back mutations G35H, R39Q and E45L had detrimental effects while the back mutation at R50S improved monomeric behavior. Taken together, additive effects of mutations that decrease the hydrophobicity of positions 39, 45 and 47 improved the solubility of this VH domain. Incorporation of Gly35 was beneficial when Trp47 was retained but not necessary when Trp47 was substituted with a less hydrophobic residue. Many of the mutations that were most effective at improving autonomous behavior of VH-4D5 are rare in natural antibody repertoires, which highlights the importance of a comprehensive and unbiased mutational approach. Unlike VHH domains, these engineered autonomous VH domains contain only a few framework changes and these are independent of CDRH3. This demonstrates that highly conserved antibody sequences can be further optimized by synthetic antibody technology and in vitro selections. Taken together, these data indicated that VH-B1a is an attractive scaffold for synthetic dAb libraries.
As a first step towards synthetic antibody libraries based on VH-B1a, its capacity to support diversity within its antigen-binding site was compared to that of the wild-type domain VH-4D5 paired with a VL domain in a conventional Fab format [125]. Each of the three CDRs and the third framework region (FR3) were targeted for randomization in a total of four libraries. FR3 was included since it is located close to the CDRs and sometimes contributes to antigen contacts. The libraries were selected for binding to Protein A and selected clones were analyzed for mutational tolerance at each position. Taken together, the results indicated that the autonomous VH domain and the Fab were very similar in terms of regions in the antigen-binding site that can tolerate diversity without compromising stability. Although FR3 was less tolerant to mutations than the CDRs, six contiguous positions in this region were fairly tolerant, which suggested that this region might be recruited for antigen binding. The tolerance for length variation in each of the three CDRs of VH-B1a was investigated in a set of new libraries. The data indicated that CDRH3 was highly tolerant to length variation while CDRH1 displayed a more modest tolerance.
Two synthetic dAb libraries were constructed based on the results from this analysis [125]. A new VH domain (VH-B1ag) was generated from VH-B1a to serve as a scaffold for these libraries. VH-B1ag differs from VH-B1a in three framework positions (W47L, S50R and W103S), which have been evaluated in detail in a previous study [124]. Only CDRH3 was diversified in the first library where it was allowed to vary between 4 and 24 residues in length. The same CDRH3 randomization was used in the second library, which also incorporated diversity in a CDRH2 of fixed length and some length variation in CDRH1. Selection on human vascular endothelial growth factor (VEGF) using both libraries identified several binders, some of which exhibited high affinities. Affinity maturation of a first generation clone yielded a high-affinity variant (KD = 16 nM) and the structure of this dAb was solved in complex with VEGF (Figure 5B). The dAb binds VEGF via an extended CDRH3 loop but, unexpectedly, also makes contacts via the former light chain interface and the overall paratope is flat. This unusual mode of binding shows that exploitation of a combination of loops and framework regions can expand the antigen recognition capacity of VH domains. Interestingly, similar modes of binding have been observed for engineered fibronectin domains [133] and have also been used to guide new library designs [134].
CONCLUSIONS
Human domain antibodies are a promising class of antibody fragments for therapeutic and diagnostic applications. dAbs with affinities comparable to those of larger antibody fragments can be generated and optimized by using in vitro directed evolution. However, the advancement of human dAbs as next-generation antibody therapeutics has been hampered by poor biophysical properties. Recent advances in antibody engineering and selection show promise to overcome these barriers using systematic mutational strategies to maximize antibody solubility and affinity. dAbs can now be endowed with high stability, aggregation resistance, reversible folding and high protein yields in microbial hosts. Although human VH sequences are used, the described mutational approaches may increase immunogenicity of engineered dAbs, which needs to be evaluated on a case-by-case basis. Their small size provides distinct advantages compared to larger antibody fragments including superior biodistribution well suited for tumor targeting applications and may also facilitate new routes of administration and novel mechanisms of action. Furthermore, the modular nature of dAbs enables flexible construction of various bi- and multivalent antibody formats. Importantly, this strategy overcomes the inherent chain association problem that arises when heterodimeric bispecific IgG molecules are produced [51, 135]. Human dAbs have been produced with the ability to target cryptic epitopes or antigens in obstructed locations, which often elude recognition by classical antibody paratopes [119, 123]. In spite of a growing arsenal of small alternative scaffold binders, the observed epitope preference appears to be a unique property of dAbs.
The commercial potential of dAbs is reflected by interests in development of single domain antibody agents in the therapeutic and diagnostic fields, including VHH domains (Ablynx), v-NARs (Wyeth/Pfizer), FN3-based binders (Adnexus/Bristol-Myers Squibb), human dAbs (Domantis/GlaxoSmithKline, Arana/Cephalon) and Humabody® VH domains from transgenic mice (Crescendo Biologics). Several additional companies and academic labs are involved in development of dAb libraries and reagents. Given the beneficial attributes of dAbs and current engineering abilities, we envision a bright future for these agents.
Acknowledgments
This work was supported by funds from the Swedish Research Council (637-2013-468 to J.N.), National Institutes of Health (R01GM104130 to P.M.T.) and National Science Foundation (CBET grants 0954450 and 1159943 to P.M.T.), and the Canadian Institutes of Health Research (operating grant MOP-93725 to S.S.S.).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no competing financial interests.
References
- 1.Lesk AM, Chothia C. Evolution of proteins formed by beta-sheets. II. The core of the immunoglobulin domains. J Mol Biol. 1982;160:325–42. doi: 10.1016/0022-2836(82)90179-6. [DOI] [PubMed] [Google Scholar]
- 2.Padlan EA. Anatomy of the antibody molecule. Mol Immunol. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan A, Clementel V, Woods R, Harn N, Bowen MA, Mo W, et al. Engineering a therapeutic IgG molecule to address cysteinylation, aggregation and enhance thermal stability and expression. MAbs. 2013;5:255–62. doi: 10.4161/mabs.23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–83. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 5.Kabat EA, Wu TT, Perry H, Gottesman K, Foeller C. Sequences of Proteins of Immunological Interest. 5. NIH; 1991. [Google Scholar]
- 6.Wu TT, Johnson G, Kabat EA. Length distribution of CDRH3 in antibodies. Proteins. 1993;16:1–7. doi: 10.1002/prot.340160102. [DOI] [PubMed] [Google Scholar]
- 7.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10:345–52. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- 8.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9:767–74. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 9.Better M, Chang CP, Robinson RR, Horwitz AH. Escherichia coli secretion of an active chimeric antibody fragment. Science. 1988;240:1041–3. doi: 10.1126/science.3285471. [DOI] [PubMed] [Google Scholar]
- 10.Skerra A, Pluckthun A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science. 1988;240:1038–41. doi: 10.1126/science.3285470. [DOI] [PubMed] [Google Scholar]
- 11.Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A. 1988;85:5879–83. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt LJ, Herring C, Jespers LS, Woolven BP, Tomlinson IM. Domain antibodies: proteins for therapy. Trends Biotechnol. 2003;21:484–90. doi: 10.1016/j.tibtech.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Andersen JT, Sandlie I. The versatile MHC class I-related FcRn protects IgG and albumin from degradation: implications for development of new diagnostics and therapeutics. Drug Metab Pharmacokinet. 2009;24:318–32. doi: 10.2133/dmpk.24.318. [DOI] [PubMed] [Google Scholar]
- 14.Ying T, Gong R, Ju TW, Prabakaran P, Dimitrov DS. Engineered Fc based antibody domains and fragments as novel scaffolds. Biochim Biophys Acta. 2014;1844:1977–82. doi: 10.1016/j.bbapap.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ullman C, Mathonet P, Oleksy A, Diamandakis A, Tomei L, Demartis A, et al. High Affinity Binders to EphA2 Isolated from Abdurin Scaffold Libraries; Characterization, Binding and Tumor Targeting. PLoS One. 2015;10:e0135278. doi: 10.1371/journal.pone.0135278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao X, Feng Y, Vu BK, Ishima R, Dimitrov DS. A large library based on a novel (CH2) scaffold: identification of HIV-1 inhibitors. Biochem Biophys Res Commun. 2009;387:387–92. doi: 10.1016/j.bbrc.2009.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying T, Chen W, Feng Y, Wang Y, Gong R, Dimitrov DS. Engineered soluble monomeric IgG1 CH3 domain: generation, mechanisms of function, and implications for design of biological therapeutics. J Biol Chem. 2013;288:25154–64. doi: 10.1074/jbc.M113.484154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobner E, Traxlmayr MW, Obinger C, Hasenhindl C. Engineered IgG1-Fc - one fragment to bind them all. Immunol Rev. 2016;270:113–31. doi: 10.1111/imr.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Traxlmayr MW, Lobner E, Antes B, Kainer M, Wiederkum S, Hasenhindl C, et al. Directed evolution of Her2/neu-binding IgG1-Fc for improved stability and resistance to aggregation by using yeast surface display. Protein Eng Des Sel. 2013;26:255–65. doi: 10.1093/protein/gzs102. [DOI] [PubMed] [Google Scholar]
- 20.Wozniak-Knopp G, Bartl S, Bauer A, Mostageer M, Woisetschlager M, Antes B, et al. Introducing antigen-binding sites in structural loops of immunoglobulin constant domains: Fc fragments with engineered HER2/neu-binding sites and antibody properties. Protein Eng Des Sel. 2010;23:289–97. doi: 10.1093/protein/gzq005. [DOI] [PubMed] [Google Scholar]
- 21.Bloom L, Calabro V. FN3: a new protein scaffold reaches the clinic. Drug Discov Today. 2009;14:949–55. doi: 10.1016/j.drudis.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–51. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 23.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–7. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 24.Boder ET, Raeeszadeh-Sarmazdeh M, Price JV. Engineering antibodies by yeast display. Arch Biochem Biophys. 2012;526:99–106. doi: 10.1016/j.abb.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Lofblom J. Bacterial display in combinatorial protein engineering. Biotechnol J. 2011;6:1115–29. doi: 10.1002/biot.201100129. [DOI] [PubMed] [Google Scholar]
- 27.Pluckthun A. Ribosome display: a perspective. Methods Mol Biol. 2012;805:3–28. doi: 10.1007/978-1-61779-379-0_1. [DOI] [PubMed] [Google Scholar]
- 28.Bradbury AR, Sidhu S, Dubel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol. 2011;29:245–54. doi: 10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geyer CR, McCafferty J, Dubel S, Bradbury AR, Sidhu SS. Recombinant antibodies and in vitro selection technologies. Methods Mol Biol. 2012;901:11–32. doi: 10.1007/978-1-61779-931-0_2. [DOI] [PubMed] [Google Scholar]
- 30.Adams JJ, Sidhu SS. Synthetic antibody technologies. Curr Opin Struct Biol. 2014;24:1–9. doi: 10.1016/j.sbi.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Miersch S, Sidhu SS. Synthetic antibodies: concepts, potential and practical considerations. Methods. 2012;57:486–98. doi: 10.1016/j.ymeth.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Sidhu SS, Fellouse FA. Synthetic therapeutic antibodies. Nat Chem Biol. 2006;2:682–8. doi: 10.1038/nchembio843. [DOI] [PubMed] [Google Scholar]
- 33.Rajan S, Sidhu SS. Simplified synthetic antibody libraries. Methods Enzymol. 2012;502:3–23. doi: 10.1016/B978-0-12-416039-2.00001-X. [DOI] [PubMed] [Google Scholar]
- 34.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A. 1992;89:4285–9. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6:343–57. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 36.Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular recognition by a binary code. J Mol Biol. 2005;348:1153–62. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 37.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci U S A. 2004;101:12467–72. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persson H, Ye W, Wernimont A, Adams JJ, Koide A, Koide S, et al. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. J Mol Biol. 2013;425:803–11. doi: 10.1016/j.jmb.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kontermann RE, Brinkmann U. Bispecific antibodies. Drug Discov Today. 2015;20:838–47. doi: 10.1016/j.drudis.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Utsumi S, Karush F. The Subunits of Purified Rabbit Antibody. Biochemistry. 1964;3:1329–38. doi: 10.1021/bi00897a024. [DOI] [PubMed] [Google Scholar]
- 41.Ward ES, Gussow D, Griffiths AD, Jones PT, Winter G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature. 1989;341:544–6. doi: 10.1038/341544a0. [DOI] [PubMed] [Google Scholar]
- 42.Flajnik MF, Deschacht N, Muyldermans S. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol. 2011;9:e1001120. doi: 10.1371/journal.pbio.1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–8. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 44.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–73. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 45.Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science. 2004;305:1770–3. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 46.Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol. 1996;3:803–11. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 47.Krah S, Schroter C, Zielonka S, Empting M, Valldorf B, Kolmar H. Single-domain antibodies for biomedical applications. Immunopharmacol Immunotoxicol. 2016;38:21–8. doi: 10.3109/08923973.2015.1102934. [DOI] [PubMed] [Google Scholar]
- 48.Muyldermans S, Smider VV. Distinct antibody species: structural differences creating therapeutic opportunities. Curr Opin Immunol. 2016;40:7–13. doi: 10.1016/j.coi.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riechmann L, Davies J. Backbone assignment, secondary structure and protein A binding of an isolated, human antibody VH domain. J Biomol NMR. 1995;6:141–52. doi: 10.1007/BF00211778. [DOI] [PubMed] [Google Scholar]
- 50.Streltsov VA, Varghese JN, Carmichael JA, Irving RA, Hudson PJ, Nuttall SD. Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor. Proc Natl Acad Sci U S A. 2004;101:12444–9. doi: 10.1073/pnas.0403509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–36. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 52.Webster DM, Henry AH, Rees AR. Antibody-antigen interactions. Curr Opin Struct Biol. 1994;4:123–9. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 53.Vu KB, Ghahroudi MA, Wyns L, Muyldermans S. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol Immunol. 1997;34:1121–31. doi: 10.1016/s0161-5890(97)00146-6. [DOI] [PubMed] [Google Scholar]
- 54.Bond CJ, Marsters JC, Sidhu SS. Contributions of CDR3 to V H H domain stability and the design of monobody scaffolds for naive antibody libraries. J Mol Biol. 2003;332:643–55. doi: 10.1016/s0022-2836(03)00967-7. [DOI] [PubMed] [Google Scholar]
- 55.Ewert S, Cambillau C, Conrath K, Pluckthun A. Biophysical properties of camelid V(HH) domains compared to those of human V(H)3 domains. Biochemistry. 2002;41:3628–36. doi: 10.1021/bi011239a. [DOI] [PubMed] [Google Scholar]
- 56.Govaert J, Pellis M, Deschacht N, Vincke C, Conrath K, Muyldermans S, et al. Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J Biol Chem. 2012;287:1970–9. doi: 10.1074/jbc.M111.242818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A. 2006;103:4586–91. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lauwereys M, Arbabi Ghahroudi M, Desmyter A, Kinne J, Holzer W, De Genst E, et al. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998;17:3512–20. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein clefts in molecular recognition and function. Protein Sci. 1996;5:2438–52. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henderson KA, Streltsov VA, Coley AM, Dolezal O, Hudson PJ, Batchelor AH, et al. Structure of an IgNAR-AMA1 complex: targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure. 2007;15:1452–66. doi: 10.1016/j.str.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 61.Stijlemans B, Conrath K, Cortez-Retamozo V, Van Xong H, Wyns L, Senter P, et al. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J Biol Chem. 2004;279:1256–61. doi: 10.1074/jbc.M307341200. [DOI] [PubMed] [Google Scholar]
- 62.Shukla AK, Gupta C, Srivastava A, Jaiman D. Antibody fragments for stabilization and crystallization of G protein-coupled receptors and their signaling complexes. Methods Enzymol. 2015;557:247–58. doi: 10.1016/bs.mie.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 63.Steyaert J, Kobilka BK. Nanobody stabilization of G protein-coupled receptor conformational states. Curr Opin Struct Biol. 2011;21:567–72. doi: 10.1016/j.sbi.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacCallum RM, Martin AC, Thornton JM. Antibody-antigen interactions: contact analysis and binding site topography. J Mol Biol. 1996;262:732–45. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 65.Saini SS, Allore B, Jacobs RM, Kaushik A. Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur J Immunol. 1999;29:2420–6. doi: 10.1002/(SICI)1521-4141(199908)29:08<2420::AID-IMMU2420>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 66.Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, Bazirgan O, et al. Reshaping antibody diversity. Cell. 2013;153:1379–93. doi: 10.1016/j.cell.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lowe D, Dudgeon K, Rouet R, Schofield P, Jermutus L, Christ D. Aggregation, stability, and formulation of human antibody therapeutics. Adv Protein Chem Struct Biol. 2011;84:41–61. doi: 10.1016/B978-0-12-386483-3.00004-5. [DOI] [PubMed] [Google Scholar]
- 68.Ratanji KD, Derrick JP, Dearman RJ, Kimber I. Immunogenicity of therapeutic proteins: influence of aggregation. J Immunotoxicol. 2014;11:99–109. doi: 10.3109/1547691X.2013.821564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tessier PM, Wu J, Dickinson CD. Emerging methods for identifying monoclonal antibodies with low propensity to self-associate during the early discovery process. Expert Opin Drug Deliv. 2014;11:461–5. doi: 10.1517/17425247.2014.876989. [DOI] [PubMed] [Google Scholar]
- 70.Wu J, Schultz JS, Weldon CL, Sule SV, Chai Q, Geng SB, et al. Discovery of highly soluble antibodies prior to purification using affinity-capture self-interaction nanoparticle spectroscopy. Protein Eng Des Sel. 2015;28:403–14. doi: 10.1093/protein/gzv045. [DOI] [PubMed] [Google Scholar]
- 71.Garber E, Demarest SJ. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem Biophys Res Commun. 2007;355:751–7. doi: 10.1016/j.bbrc.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 72.Wang X, Das TK, Singh SK, Kumar S. Potential aggregation prone regions in biotherapeutics: A survey of commercial monoclonal antibodies. MAbs. 2009;1:254–67. doi: 10.4161/mabs.1.3.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ewert S, Huber T, Honegger A, Pluckthun A. Biophysical properties of human antibody variable domains. J Mol Biol. 2003;325:531–53. doi: 10.1016/s0022-2836(02)01237-8. [DOI] [PubMed] [Google Scholar]
- 74.Rothlisberger D, Honegger A, Pluckthun A. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J Mol Biol. 2005;347:773–89. doi: 10.1016/j.jmb.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 75.Barelle C, Porter A. VNARs: An Ancient and Unique Repertoire of Molecules That Deliver Small, Soluble, Stable and High Affinity Binders of Proteins. Antibodies. 2015;4:240–58. [Google Scholar]
- 76.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–97. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 77.Tiller KE, Tessier PM. Advances in Antibody Design. Annu Rev Biomed Eng. 2015;17:191–216. doi: 10.1146/annurev-bioeng-071114-040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perchiacca JM, Tessier PM. Engineering aggregation-resistant antibodies. Annu Rev Chem Biomol Eng. 2012;3:263–86. doi: 10.1146/annurev-chembioeng-062011-081052. [DOI] [PubMed] [Google Scholar]
- 79.Rouet R, Lowe D, Christ D. Stability engineering of the human antibody repertoire. FEBS Lett. 2014;588:269–77. doi: 10.1016/j.febslet.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 80.Sule SV, Cheung JK, Antochshuk V, Bhalla AS, Narasimhan C, Blaisdell S, et al. Solution pH that minimizes self-association of three monoclonal antibodies is strongly dependent on ionic strength. Mol Pharm. 2012;9:744–51. doi: 10.1021/mp200448j. [DOI] [PubMed] [Google Scholar]
- 81.Honegger A, Malebranche AD, Rothlisberger D, Pluckthun A. The influence of the framework core residues on the biophysical properties of immunoglobulin heavy chain variable domains. Protein Eng Des Sel. 2009;22:121–34. doi: 10.1093/protein/gzn077. [DOI] [PubMed] [Google Scholar]
- 82.Riechmann L, Muyldermans S. Single domain antibodies: comparison of camel VH and camelised human VH domains. J Immunol Methods. 1999;231:25–38. doi: 10.1016/s0022-1759(99)00138-6. [DOI] [PubMed] [Google Scholar]
- 83.Davies J, Riechmann L. ‘Camelising’ human antibody fragments: NMR studies on VH domains. FEBS Lett. 1994;339:285–90. doi: 10.1016/0014-5793(94)80432-x. [DOI] [PubMed] [Google Scholar]
- 84.Conrath K, Vincke C, Stijlemans B, Schymkowitz J, Decanniere K, Wyns L, et al. Antigen binding and solubility effects upon the veneering of a camel VHH in framework-2 to mimic a VH. J Mol Biol. 2005;350:112–25. doi: 10.1016/j.jmb.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 85.Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem. 2009;284:3273–84. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- 86.Davies J, Riechmann L. Antibody VH domains as small recognition units. Biotechnology (N Y) 1995;13:475–9. doi: 10.1038/nbt0595-475. [DOI] [PubMed] [Google Scholar]
- 87.Riechmann L. Rearrangement of the former VL interface in the solution structure of a camelised, single antibody VH domain. J Mol Biol. 1996;259:957–69. doi: 10.1006/jmbi.1996.0373. [DOI] [PubMed] [Google Scholar]
- 88.Tanha J, Nguyen TD, Ng A, Ryan S, Ni F, Mackenzie R. Improving solubility and refolding efficiency of human V(H)s by a novel mutational approach. Protein Eng Des Sel. 2006;19:503–9. doi: 10.1093/protein/gzl037. [DOI] [PubMed] [Google Scholar]
- 89.Kovalenko OV, Olland A, Piche-Nicholas N, Godbole A, King D, Svenson K, et al. Atypical antigen recognition mode of a shark immunoglobulin new antigen receptor (IgNAR) variable domain characterized by humanization and structural analysis. J Biol Chem. 2013;288:17408–19. doi: 10.1074/jbc.M112.435289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim DY, Hussack G, Kandalaft H, Tanha J. Mutational approaches to improve the biophysical properties of human single-domain antibodies. Biochim Biophys Acta. 2014;1844:1983–2001. doi: 10.1016/j.bbapap.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 91.Perchiacca JM, Ladiwala AR, Bhattacharya M, Tessier PM. Aggregation-resistant domain antibodies engineered with charged mutations near the edges of the complementarity-determining regions. Protein Eng Des Sel. 2012;25:591–601. doi: 10.1093/protein/gzs042. [DOI] [PubMed] [Google Scholar]
- 92.Jespers L, Schon O, James LC, Veprintsev D, Winter G. Crystal structure of HEL4, a soluble, refoldable human V(H) single domain with a germ-line scaffold. J Mol Biol. 2004;337:893–903. doi: 10.1016/j.jmb.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 93.Perchiacca JM, Bhattacharya M, Tessier PM. Mutational analysis of domain antibodies reveals aggregation hotspots within and near the complementarity determining regions. Proteins. 2011;79:2637–47. doi: 10.1002/prot.23085. [DOI] [PubMed] [Google Scholar]
- 94.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tomlinson IM, Walter G, Marks JD, Llewelyn MB, Winter G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol. 1992;227:776–98. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- 96.Trevino SR, Scholtz JM, Pace CN. Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. J Mol Biol. 2007;366:449–60. doi: 10.1016/j.jmb.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goldman ER, Brozozog-Lee PA, Zabetakis D, Turner KB, Walper SA, Liu JL, et al. Negative tail fusions can improve ruggedness of single domain antibodies. Protein Expr Purif. 2014;95:226–32. doi: 10.1016/j.pep.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 98.Tan PH, Chu V, Stray JE, Hamlin DK, Pettit D, Wilbur DS, et al. Engineering the isoelectric point of a renal cell carcinoma targeting antibody greatly enhances scFv solubility. Immunotechnology. 1998;4:107–14. doi: 10.1016/s1380-2933(98)00011-6. [DOI] [PubMed] [Google Scholar]
- 99.Schaefer JV, Pluckthun A. Engineering aggregation resistance in IgG by two independent mechanisms: lessons from comparison of Pichia pastoris and mammalian cell expression. J Mol Biol. 2012;417:309–35. doi: 10.1016/j.jmb.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 100.Kvam E, Sierks MR, Shoemaker CB, Messer A. Physico-chemical determinants of soluble intrabody expression in mammalian cell cytoplasm. Protein Eng Des Sel. 2010;23:489–98. doi: 10.1093/protein/gzq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Waugh DS. Making the most of affinity tags. Trends Biotechnol. 2005;23:316–20. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 102.Ladiwala AR, Bhattacharya M, Perchiacca JM, Cao P, Raleigh DP, Abedini A, et al. Rational design of potent domain antibody inhibitors of amyloid fibril assembly. Proc Natl Acad Sci U S A. 2012;109:19965–70. doi: 10.1073/pnas.1208797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Perchiacca JM, Ladiwala AR, Bhattacharya M, Tessier PM. Structure-based design of conformation- and sequence-specific antibodies against amyloid beta. Proc Natl Acad Sci U S A. 2012;109:84–9. doi: 10.1073/pnas.1111232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee CC, Julian MC, Tiller KE, Meng F, DuConge SE, Akter R, et al. Design and Optimization of Anti-amyloid Domain Antibodies Specific for beta-Amyloid and Islet Amyloid Polypeptide. J Biol Chem. 2016;291:2858–73. doi: 10.1074/jbc.M115.682336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miklos AE, Kluwe C, Der BS, Pai S, Sircar A, Hughes RA, et al. Structure-based design of supercharged, highly thermoresistant antibodies. Chem Biol. 2012;19:449–55. doi: 10.1016/j.chembiol.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perchiacca JM, Lee CC, Tessier PM. Optimal charged mutations in the complementarity-determining regions that prevent domain antibody aggregation are dependent on the antibody scaffold. Protein Eng Des Sel. 2014;27:29–39. doi: 10.1093/protein/gzt058. [DOI] [PubMed] [Google Scholar]
- 107.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10:866–76. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jespers L, Schon O, Famm K, Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat Biotechnol. 2004;22:1161–5. doi: 10.1038/nbt1000. [DOI] [PubMed] [Google Scholar]
- 109.Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Charbonnier JB, et al. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U S A. 2000;97:5399–404. doi: 10.1073/pnas.97.10.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Christ D, Famm K, Winter G. Repertoires of aggregation-resistant human antibody domains. Protein Eng Des Sel. 2007;20:413–6. doi: 10.1093/protein/gzm037. [DOI] [PubMed] [Google Scholar]
- 111.Mandrup OA, Friis NA, Lykkemark S, Just J, Kristensen P. A novel heavy domain antibody library with functionally optimized complementarity determining regions. PLoS One. 2013;8:e76834. doi: 10.1371/journal.pone.0076834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dudgeon K, Famm K, Christ D. Sequence determinants of protein aggregation in human VH domains. Protein Eng Des Sel. 2009;22:217–20. doi: 10.1093/protein/gzn059. [DOI] [PubMed] [Google Scholar]
- 113.Arbabi-Ghahroudi M, To R, Gaudette N, Hirama T, Ding W, MacKenzie R, et al. Aggregation-resistant VHs selected by in vitro evolution tend to have disulfide-bonded loops and acidic isoelectric points. Protein Eng Des Sel. 2009;22:59–66. doi: 10.1093/protein/gzn071. [DOI] [PubMed] [Google Scholar]
- 114.Chiti F, Calamai M, Taddei N, Stefani M, Ramponi G, Dobson CM. Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16419–26. doi: 10.1073/pnas.212527999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lawrence MS, Phillips KJ, Liu DR. Supercharging proteins can impart unusual resilience. J Am Chem Soc. 2007;129:10110–2. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shaw BF, Schneider GF, Bilgicer B, Kaufman GK, Neveu JM, Lane WS, et al. Lysine acetylation can generate highly charged enzymes with increased resistance toward irreversible inactivation. Protein Sci. 2008;17:1446–55. doi: 10.1110/ps.035154.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilkinson DL, Harrison RG. Predicting the solubility of recombinant proteins in Escherichia coli. Biotechnology (N Y) 1991;9:443–8. doi: 10.1038/nbt0591-443. [DOI] [PubMed] [Google Scholar]
- 118.Dudgeon K, Rouet R, Kokmeijer I, Schofield P, Stolp J, Langley D, et al. General strategy for the generation of human antibody variable domains with increased aggregation resistance. Proc Natl Acad Sci U S A. 2012;109:10879–84. doi: 10.1073/pnas.1202866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen W, Zhu Z, Feng Y, Xiao X, Dimitrov DS. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J Mol Biol. 2008;382:779–89. doi: 10.1016/j.jmb.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dudgeon K, Rouet R, Christ D. Rapid prediction of expression and refolding yields using phage display. Protein Eng Des Sel. 2013;26:671–4. doi: 10.1093/protein/gzt019. [DOI] [PubMed] [Google Scholar]
- 121.Julian MC, Lee CC, Tiller KE, Rabia LA, Day EK, Schick AJ, 3rd, et al. Co-evolution of affinity and stability of grafted amyloid-motif domain antibodies. Protein Eng Des Sel. 2015;28:339–50. doi: 10.1093/protein/gzv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Famm K, Hansen L, Christ D, Winter G. Thermodynamically stable aggregation-resistant antibody domains through directed evolution. J Mol Biol. 2008;376:926–31. doi: 10.1016/j.jmb.2007.10.075. [DOI] [PubMed] [Google Scholar]
- 123.Rouet R, Dudgeon K, Christie M, Langley D, Christ D. Fully Human VH Single Domains That Rival the Stability and Cleft Recognition of Camelid Antibodies. J Biol Chem. 2015;290:11905–17. doi: 10.1074/jbc.M114.614842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Barthelemy PA, Raab H, Appleton BA, Bond CJ, Wu P, Wiesmann C, et al. Comprehensive analysis of the factors contributing to the stability and solubility of autonomous human VH domains. J Biol Chem. 2008;283:3639–54. doi: 10.1074/jbc.M708536200. [DOI] [PubMed] [Google Scholar]
- 125.Ma X, Barthelemy PA, Rouge L, Wiesmann C, Sidhu SS. Design of synthetic autonomous VH domain libraries and structural analysis of a VH domain bound to vascular endothelial growth factor. J Mol Biol. 2013;425:2247–59. doi: 10.1016/j.jmb.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 126.Lee CV, Liang WC, Dennis MS, Eigenbrot C, Sidhu SS, Fuh G. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol. 2004;340:1073–93. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 127.Sidhu SS, Li B, Chen Y, Fellouse FA, Eigenbrot C, Fuh G. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J Mol Biol. 2004;338:299–310. doi: 10.1016/j.jmb.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 128.Ewert S, Honegger A, Pluckthun A. Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods. 2004;34:184–99. doi: 10.1016/j.ymeth.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 129.Bond CJ, Wiesmann C, Marsters JC, Jr, Sidhu SS. A structure-based database of antibody variable domain diversity. J Mol Biol. 2005;348:699–709. doi: 10.1016/j.jmb.2005.02.063. [DOI] [PubMed] [Google Scholar]
- 130.Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 2000;97:8950–4. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–60. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 132.Pal G, Kouadio JL, Artis DR, Kossiakoff AA, Sidhu SS. Comprehensive and quantitative mapping of energy landscapes for protein-protein interactions by rapid combinatorial scanning. J Biol Chem. 2006;281:22378–85. doi: 10.1074/jbc.M603826200. [DOI] [PubMed] [Google Scholar]
- 133.Gilbreth RN, Koide S. Structural insights for engineering binding proteins based on non-antibody scaffolds. Curr Opin Struct Biol. 2012;22:413–20. doi: 10.1016/j.sbi.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koide A, Wojcik J, Gilbreth RN, Hoey RJ, Koide S. Teaching an old scaffold new tricks: monobodies constructed using alternative surfaces of the FN3 scaffold. J Mol Biol. 2012;415:393–405. doi: 10.1016/j.jmb.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Klein C, Sustmann C, Thomas M, Stubenrauch K, Croasdale R, Schanzer J, et al. Progress in overcoming the chain association issue in bispecific heterodimeric IgG antibodies. MAbs. 2012;4:653–63. doi: 10.4161/mabs.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]