Abstract
Purpose
Eliciting and understanding patient and research participant preferences regarding return of secondary test results is a key aspect of genomic medicine. A valid instrument should be easily understood without extensive pre-test counseling, while still faithfully eliciting patients’ preferences.
Methods
We conducted focus groups with 110 adults to understand patient perspectives on secondary genomic findings and the role preferences should play. We then developed and refined a draft instrument, and used it to elicit preferences from parents participating in a genomic sequencing study in children with intellectual disabilities.
Results
Patients preferred filtering of secondary genomic results to avoid information overload and to avoid learning what the future holds, among other reasons. Patients preferred to make autonomous choices about which categories of results to receive and to have their choices applied automatically before results are returned to them and their clinicians. The Preferences Instrument for Genomic Secondary Results (PIGSR) is designed to be completed by patients or research participants without assistance and to guide bioinformatic analysis of genomic raw data. Most participants wanted to receive all secondary results, but a significant minority indicated other preferences.
Conclusions
Our novel instrument – PIGSR – should be useful in a wide range of clinical and research settings.
Introduction
When genomic sequencing is performed for a specific clinical or research purpose, it becomes necessary to develop a plan for managing and returning secondary results, i.e. results not related to the primary reason for testing. Some secondary results are generated incidentally through quality assurance procedures and other routine analyses.1 Others are “hidden” in raw sequencing data and can be identified manually and through automated analysis algorithms.2
While secondary findings are not novel to genomic sequencing, they do pose important ethical and practical challenges for clinical and research programs that perform genomic sequencing on human samples. In 2013, the American College of Medical Genetics and Genomics (ACMG) recommended that laboratories performing sequencing on clinical samples should routinely include additional analyses to identify highly actionable secondary results. Examples include variants in genes related to cancer or heart disease that could lead to beneficial clinical interventions.3 In this recommendation document, the authors argued that these analyses should be performed routinely without considering patient preferences. Their rationale for this recommendation was based on a proposed fiduciary duty for laboratories to seek and disclose secondary results that could prevent patient harm, as well as concern that eliciting patient preferences about secondary findings would be impractical because it would require genetic counseling on numerous conditions.3
In 2014, the ACMG revised this set of recommendations to stress that patients should have a right to decline receipt of secondary genomic findings. Despite this revision, the concern about the practicality of eliciting preferences remains important. It remains necessary to identify workable and effective methods to collect patient preferences about which genomic findings they would like to receive. This is complicated by the extremely large number of potential secondary findings that could be generated, and the potential for new findings to be uncovered over time. It is necessary to organize potential genomic results into a set of choices that patients can easily understand without extensive pre-test counseling, while still faithfully eliciting their granular preferences.
A number of possible approaches have been proposed for categorizing potential results to facilitate eliciting preferences.4 Many of these approaches utilize the distinction between results that are considered actionable and those that are not.5 This distinction is problematic for a number of reasons. First, the concept of actionability is ambiguous.6 Many define actionability on the basis of clinical interventions alone, while others highlight the possibility that patients will want to take other types of actions, such as changing their health behaviors or purchasing additional insurance.7,8 Another common approach organizes results using a range of categories, including organ systems.9
In this report, we provide evidence that patients’ personal experiences with specific medical conditions are an important driver of preferences for receiving secondary genomic findings. Building on this observation, we describe a new patient preferences instrument – the Preferences Instrument for Genomic Secondary Results (PIGSR) – that is designed to be responsive to patient experiences with specific conditions, to be completed rapidly and independently by a patient or research participant, and to support the development of analysis algorithms capable of extrapolating a brief set of patient responses to a wider range of potential secondary genomics results.
Methods
Overview of Study Components
PIGSR was developed using data from focus groups and semi-structured interviews undertaken as a part of the Pharmacogenomic Resource for Enhanced Decisions in Care & Treatment (PREDICT) effort at Vanderbilt University Medical Center.10 Once finalized, PIGSR was administered to parents participating in a clinical sequencing study at HudsonAlpha Institute to diagnose children with developmental delay. The HudsonAlpha project is a part of the Clinical Sequencing Exploratory Research (CSER) Consortium. This is a translational study designed to examine the effectiveness and clinical utility of whole genome sequencing to identify genetic causes for intellectual disability, developmental delay, and related phenotypes. Both the development (qualitative data) and the initial experience (quantitative data) with PIGSR are described here. The focus group and semi-structured interview elements of this study were approved by the Institutional Review Board (IRB) at Vanderbilt University. The HudsonAlpha CSER project was approved by Western IRB and the University of Alabama at Birmingham IRB. Informed consent was obtained from all participants.
Focus Groups
We conducted ten focus groups designed to inform the development of PREDICT, a clinical pharmacogenomic testing program at Vanderbilt University.10 Round 1 was comprised of five focus groups conducted in 2010, and focused on communication, consent, data security, and reporting of primary and secondary results. Round 2 was comprised of five focus groups conducted in 2011 and was undertaken to explore perspectives on the management of secondary results in greater detail.
The vignette-based discussion guides for these focus group sessions were developed iteratively by two authors (KBB and EWC) in collaboration with operational and administrative staff leading the development of the PREDICT program (JMP) (Supplemental Figures 1 and 2). Potential participants were invited by e-mail through ResearchMatch.org, an online research recruitment database.11 Those who expressed interest were screened by phone to meet purposive sampling aims, and in particular to generate focus group sessions that were diverse in terms of age, gender, race, ethnicity, employment status, income, and marital status. Each round was comprised of four focus group sessions conducted in English and one session conducted in Spanish.
Focus groups were conducted at a commercial facility, where sessions were both video recorded and observed by investigators. A moderator led each session utilizing standard moderating techniques to facilitate discussion. A professional transcription service transcribed the recordings.
Transcriptions were coded using a Framework Analysis methodology, a qualitative coding technique that starts with previously identified (deductive) themes that are then expanded by adding themes and subthemes inductively.12 A single coding framework was utilized across both rounds of focus groups. All transcriptions were coded independently by at least two investigators using Atlas.ti qualitative coding software (Version 7, Scientific Software Development GmbH), with disagreements resolved through review by a third investigator. Themes covering a wide range of topics were identified; this report focuses on only those themes related to analysis and management of secondary genomic results across both rounds of focus groups.
Instrument Development
Qualitative findings from these focus group sessions informed our insight that a brief instrument designed to elicit patient preferences on secondary genomic findings needed to be developed, and thus informed our work to develop PIGSR. Four investigators (KBB, EWC, MJW, and MFW) designed an initial draft of the instrument and received feedback from two independent experts on ethical issues in clinical genomics. We then recontacted focus group participants to complete the draft PIGSR and provide feedback. Participants were selected for recontact using purposive sampling; we used focus group transcripts to identify participants with diverse perspectives on the types and numbers of secondary genomics results they would prefer to receive, and to represent a diversity of perspectives based on gender, ethnic and racial background, educational attainment, and age.
Ten focus group participants returned to provide feedback on PIGSR. After completing the instrument, each participated in a 30-minute, face-to-face semi-structured interview focused on the questionnaire. Domains addressed in this interview included the success of PIGSR in allowing the respondent to thoroughly record his or her preferences, and the ease with which the instrument could be completed without guidance from a healthcare professional. Interviews were audio recorded and transcribed; transcriptions underwent qualitative coding as described above. Interview results were used to revise the instrument, generating the final version of the PIGSR described below.
Initial Experience in a Genomic Research Study
Families participating in the HudsonAlpha CSER project were enrolled from a pediatric neurology practice in Huntsville, Alabama. Parents completed PIGSR to record their preferences for receiving secondary genomic results generated in the analysis of their own genome. Secondary findings were only queried in children when it was necessary to determine whether the child had inherited a secondary finding identified in the parent. For this reason, preferences related to the child’s secondary findings were not elicited.
Fifty percent of families were randomized to record their preferences at the time of enrollment (Prospective Preferences Group), while those remaining were asked to record their preferences immediately prior to their visit to receive genomic results (Just-in-Time Preferences Group). This approach was designed to examine whether the timing for eliciting preferences influences outcomes such as anxiety and numbers of results requested. These outcomes will be reported in a future manuscript. In this report we focus on our interim experience with the first 100 families (49 in the Prospective group and 51 in the Just-in-Time group) in whom both biological parents were available to record their preferences and undergo testing.
Parents recorded their preferences using a paper version of PIGSR and were asked to complete the instrument without consulting their spouse or study personnel. Upon completion of the instrument, the genetic counselor (KME or WVK) reviewed parents’ responses, provided an opportunity for them to ask questions and provide feedback, and asked each parent whether he or she had additional preferences that were not captured using the instrument. Observations from these discussions were recorded in discursive research notes.
Responses to PIGSR were entered into a research database and used to guide the return of secondary findings uncovered through the genomic analysis conducted as a part of the research study. For this report, we performed descriptive analyses and tests for differences in preferences between mothers and fathers. Paired-sample t-tests were used to test for differences in continuous demographic variables. Chi-squared tests were used for categorical demographic variables, for differences in responses on individual preference items, and for differences in preference patterns between genders. Paired sample t-tests were used to test for differences in the number of genetic tests preferred between mothers and fathers. The categorical data met all the assumptions of the categorical analyses performed, and all continuous data were verified to be normally distributed using traditional statistics for verification.
Results
Focus Group Findings
The demographics of focus group participants are listed in Supplemental Table 1. In both rounds of focus groups, the concept of secondary findings was introduced to participants using a case vignette of a patient undergoing prospective pharmacogenomics testing prior to the placement of a coronary artery stent. Attention was drawn to the potential for using this test to generate other genetic findings. Participants were asked to discuss whether they would like these potential results “filtered.” Follow-up questions asked participants to discuss what criteria should be used for this filtering and, using a schematic of the testing pathway (Supplemental Figure 3), to identify where in this process filtering should be applied.
Some participants were initially opposed to the idea of “filtering” results. One respondent stated, “I would not want it to be filtered because it would potentially leave out information that the doctor or myself would need” (35yo male). Another respondent evoked genetic exceptionalism as a reason that filtering should not be performed, “Just because this is genetics we seem to be treating it as a special test… If I go to my doctor for one test and [he] sees that you’ve got another disease [than what] we were testing for, I think he’s got a responsibility to tell me” (44yo female).
Typically, however, respondents quickly generated reasons for not returning every possible result (Table 1). Many endorsed these reasons for themselves or affirmed their importance to others. One participant said, “I would want to know everything. But I know others would not, and I think that’s their right” (31yo male). For these reasons, many participants concluded that individual preferences should be the primary criteria used to filter results. One observed that, “They don’t want to know everything about themselves – just like hypothetically we’ve already spoken about Alzheimer’s… Other people want to know anything and everything. So I think it’s going to depend on the person” (21yo male). In a smaller number of cases, participants expressed a desire for the patient’s physician to make such decisions. According to one participant:
Table 1.
Reasons given for filtering genomic results or for not wanting to receive certain results
| Proposed Reason | Participant Information | Quote |
|---|---|---|
|
Prevent information
overload |
21yo Male | “I believe in the doctors but I think that sometimes we have too much information. I think that medical and technology has gone too far… So I think that it should just be kept really simple.” |
|
Desire not to know the
future |
68yo Male | “You go through life with a lot of expectations that you may not want to really know what lies in your future. Sometimes people wanna keep it as a surprise.” |
|
Concerns about quality
of information |
42yo Female | “I like how [participant] talked about the reliability. That is a fear. Whether it is.” |
| Religious beliefs | 47yo Male | “I know a lot of people who consider themselves very… religious… They believe it’s God’s way and that’s the way it is and man cannot change it and these tests are just interference if you will, in their beliefs.” |
|
Risk for workplace
discrimination |
23yo Female | “This person has a certain genetic predisposition for her to suffer from cancer in 2 years, and therefore she may suffer from a certain type of discrimination in the future, in the workplace or X situation.” |
|
Risk for psychological
harm |
61yo Female | “There are so many variables in people’s psychological makeups that to hear this kind of news no matter whether you ask for it or not. Sometimes you don’t even know whether you’ll be prepared.” |
|
Risk for adverse effect
on insurance coverage |
58yo Male | “Obama’s healthcare plan finding out about it and then jacking your rates up or dropping you.” |
|
Risk for revealing
sensitive information to unauthorized persons |
58yo Male | “The reason I disagree is… one of the pharmacy techs is a friend of my daughter’s… I don’t want that girl looking at my records and then saying ‘Gee, [daughter]’s dad is getting ready to drop dead.” |
I’m paying the doctor for his expertise… So if the doctor knows something about me that can affect me I think he has an obligation to tell me… Some people may not like that, but people get told they have cancer everyday… and they have to deal with it. (52yo male)
In discussing where in the analytic process filtering should be applied, many respondents expressed reservations about physician gatekeeping and selective disclosure: “I’d much rather prefer the information stay hidden, but everything that comes out I want to know” (24yo female). Filtering by physicians was also seen as discordant with patients’ ability to access their own results online: “If they see [in the online patient portal] that their doctor is withholding something from them, they’re going to feel betrayed” (38yo female).
Most participants seemed to prefer a filtering approach that would involve the use of personal preferences to direct computer-based analysis, such that only results desired by the patient would be analyzed and reported to the patient and provider. When asked to suggest ways that preferences could be elicited, participants suggested a wide variety of approaches. One participant, for example, proposed actionability, heritability, and disease severity criteria: “We can ask her: ‘Do you want to know only diseases that can be prevented? … Do you want to know diseases that can be passed on to your kids? Do you want to know things that have consequences of death’” (59yo man)? Quality of life, an aspect of disease severity, was raised as potentially important, “Each individual person is going to be looking for different types of things. Whether this disease can kill me… Another person might [be interested in whether a disease] will decrease my quality of life” (21yo male).
Other participants suggested that options should be organized around specific medical conditions: “You don’t want to list all the diseases… so maybe there’s a section for heart conditions… and then different type of blood diseases… Just some kind of way to keep it specific as opposed to listing every single thing” (32yo female). This focus on specific diseases was also emphasized because learning of risk for certain conditions could affect quality of life. Alzheimer’s disease was included in the discussion guide as an example of a risk result that could be generated, and was referenced frequently. “You may have a person who would worry tremendously about Alzheimer’s and you really affect their quality of life, knowing” (21yo male). Experience with a family member with Alzheimer’s also seemed important. “If that genetic testing [Alzheimer’s risk] was offered today and my mother has it, I don’t want to know” (35yo female).
Family and personal experience with individual diseases was considered so important that many respondents assumed that personal and family history would be used to generate the choices offered to patients. “I think also gathering information from personal previous history, health history and also if you look… [at] your mother and father, grandmother, grandfather, and on like that” (51yo female).
Instrument Development
The criteria we abstracted from our focus group findings to inform the design of PIGSR are summarized in Table 2. Using these criteria, we designed a draft instrument that was used to elicit feedback (see below).
Table 2.
Evidence-based criteria for a genomic secondary results preferences instrument
| Criterion | Rationale |
|---|---|
|
Preferences should be elicited
through structured choices rather than open-ended questions |
Patients prefer for preferences to be prospectively applied during the computer-based analysis of genomic raw data. They do not want providers to receive results that will not be reported to the patient. |
|
Preference items should be
organized around specific diseases or disease categories |
Patient preferences often seem to be driven by personal experience with a disease in a family member or other loved one. |
|
Preference items should be
immediately understandable by most patients |
Genomic testing will increasingly be delivered without the benefit of genetic counseling, including in the PREDICT program at Vanderbilt University. |
|
Preferences items should be limited
in number |
In many clinical and research settings, including the projects described here, a brief instrument is desirable to minimize respondent burden and support efficient clinical flow. |
|
Preference items should be carefully
selected to support inference about patients’ other preferences |
Genomic sequencing creates the potential to generate a wide range of secondary results. |
Feedback Interview Findings
Demographics of interview participants are listed in Supplemental Table 2. All ten interview participants endorsed that the language used in the instrument was clear and that all of the diseases and/or disease categories included in the instrument were familiar. The only exception to this was that one respondent reported that he was not familiar with “cystic fibrosis.” Participants were also asked if they would have responded differently for questions that were grouped into one item (such as “heart attack, heart rhythm problem, or stroke”); in all cases, respondents reported that groupings did not affect their ability to accurately record their preferences.
Participants were asked to identify conditions that were not included on the draft instrument but are important to their preferences. Cited conditions included autism, chronic fatigue syndrome, diabetes (two respondents), Down syndrome, and fibromyalgia. In revisions to the instrument, risk for developing diabetes and risk for having a child with autism were added. Based on feedback, questions were also reorganized to facilitate understanding. The final version of PIGSR is distributed freely online at www.PIGSR.org.
Initial Experience – Qualitative Findings
Demographics for the first 100 families with a biological mother and father enrolled in the HudsonAlpha CSER project are reported in Table 3. Among these, all parents reported to the genetic counselors (KME or WVK) that PIGSR accurately captured their preferences and that they understood the options being presented. Misunderstandings that were noted tended to be related to genetic concepts rather than the instrument itself. For example, on several occasions a white parent requested all secondary findings, but declined findings related to risk for having a child with sickle cell disease. When asked about this, the parent responded that they did not need that information because they did not see themselves as at risk for having a child with that condition because of their racial identity.
Table 3.
Demographics of mothers and fathers in the first 100 families enrolled in the HudsonAlpha CSER project
| Continuous Variables | Total Mean (SD) |
Fathers Mean (SD) |
Mothers Mean (SD) |
|---|---|---|---|
| Age** | 32.25 (11.1) | 34.31 (11.5) | 30.22 (10.5) |
| No. Typical Children | 1.71 (0.8) | 1.75 (0.9) | 1.67 (0.7) |
| No. Affected Children | 1.13 (0.4) | 1.15 (0.4) | 1.10 (0.3) |
| Categorical Variables |
Total
N=200 (%) |
Fathers
N=100 (%) |
Mothers
N=100 (%) |
|
Race
White Black Other |
163 (81.5%) 12 (6.0%) 25 (12.5%) |
79 (79.0%) 6 (6.0%) 15 (15.0%) |
84 (84.0%) 6 (6.0%) 10 (10.0%) |
|
Education
< High School High School Some College College Advanced |
19 (9.5%) 34 (17.0%) 67 (33.5%) 49 (24.5%) 31 (15.5%) |
11 (11.0%) 20 (20.0%) 31 (31.0%) 25 (25.0%) 13 (13.0%) |
8 (8.0%) 14 (14.0%) 36 (36.0%) 24 (24.0%) 18 (18.0%) |
| Employed** | 127 (63.5%) | 82 (82.0%) | 45 (45.0%) |
Significant difference between mothers and fathers, p<0.001
Levene’s test for equality of variance did not reach significance for any of the comparisons above. For this reason, traditional t-statistic and associated p-value was reported. No adjustment for unequal variances (e.g., Satterthwaite) was used.
Initial Experience – Quantitative Findings
Perhaps not surprisingly since all of the participating families had at least one child with an intellectual disability, mothers were significantly less likely to be employed than fathers (82% vs. 45%, p<0.001). Mothers were also significantly younger than fathers (30yo vs. 34yo, p<0.001).
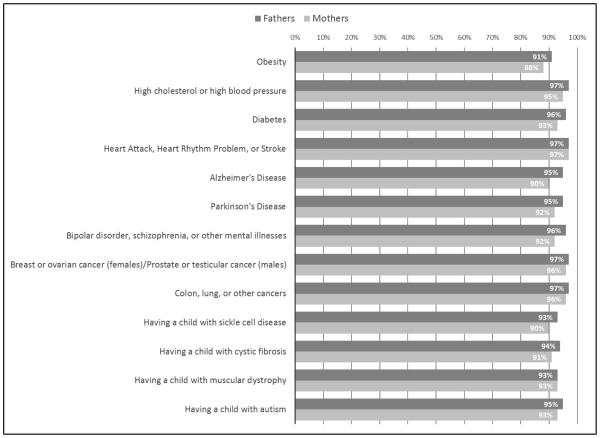
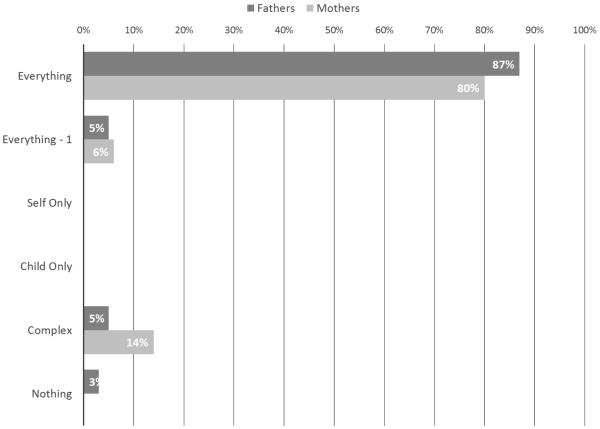
Most parents requested most secondary findings (Figure 1). The least frequently requested result was risk for developing obesity (n=179, 89.5%); the most frequently requested result was risk for having a heart attack, heart rhythm problem, or stroke (n=194, 97.0%). Combinations of preference responses can be categorized into “patterns,” which may inform extrapolation from items on PIGSR to secondary results not included in the instrument (described below). As shown in Figure 2, 87% of fathers and 80% of mothers want to receive all possible genomic secondary results, i.e. “Everything.” In this preliminary data we are underpowered to detect significant differences in preferences between genders for both individual preferences and patterns of preferences. However, it is interesting to note that no mothers responded that they wanted “Nothing,” while three fathers recorded this preference.
Figure 1.
Preferences of mothers and fathers for receiving categories of genomic secondary results from their own sequence data
Figure 2.
Patterns of genomic secondary result preferences for mothers and fathers
In this study, secondary findings for parents were limited to pathogenic variants in the 56 genes identified in the ACMG recommendations on secondary findings,3 carrier status in genes related to cystic fibrosis, sickle cell anemia, and Tay-Sachs disease, and carrier status for any condition listed in the Online Mendelian Inheritance in Man (OMIM) database for which both parents happen to be carriers. Given the focus of the study, variants conferring risk for intellectual disability, autism spectrum disorders, epilepsy, and related conditions were considered primary results and were returned to all families.
Among these first 100 families, secondary findings were identified in 10 mothers and 12 fathers. Because this is the first study to use PIGSR to collect preferences, we are not using automated analyses to apply participant preferences to secondary results; all potential secondary results are reviewed by a committee and participant preferences recorded on PIGSR are applied manually. In only one case was a secondary result identified but not reported due to participant preferences: a variant pathogenic for arrhythmogenic cardiomyopathy was identified in TMEM43 in one father. Because the participant had recorded a preference not to receive results related to “chance of having a heart attack, heart rhythm problem, or stroke,” the result was not returned. Interestingly, his wife was found to have a pathogenic variant for a related condition. This gave the genetic counselor an opportunity to confirm the father’s preference. Even knowing that his wife had a variant for a similar condition, he reiterated his preference not to receive this type of result.
Discussion
Comparison with other approaches for eliciting preferences
Two of the most well-known approaches for eliciting preferences share a great deal in common. Prior to a warning issued by the Food and Drug Administration (FDA) in 2014,13 the company 23andMe offered direct-to-consumer testing for genetic risk. 23andMe offered a number of disease-based choices that allowed clients to select which results they wished to view through their online portal. The online interface for my46 provides a similar set of choices that allows participants undergoing genomic testing to select the results they wish to view.14 Like PIGSR, both of these tools were originally designed to allow users to record their preferences without the assistance of a healthcare provider. In contrast with PIGSR, however, they provide very detailed choices about the results that are viewed, and allow users to opt-in to receiving additional results over time. By design, PIGSR includes only 13 items. It is intended to give participants prospective control over the results they will receive in clinic and research contexts where results are to be returned at a future clinic visit rather than through a web interface.
In this respect, PIGSR bears some similarity with the four approaches to preference-setting proposed recently by Bacon et al.4 These methods – which include a branching diagram model, an example-based model, a grid model with checklist, and a step-wise grid model – are also designed for prospective preference setting, but differ from PIGSR in that they organize preferences around disease preventability and severity. Laboratories that perform clinical exome or genome sequencing collect prospective preferences organized around similar criteria as well as age of disease onset.15 Important research, like that currently taking place in the CSER Consortium, will help uncover the strengths and weaknesses of these various approaches to eliciting preferences, and may inform refinements that will lead to the development of second generation tools.
Applying patterns of preferences to genomic secondary results
The current version of PIGSR elicits preferences on 13 diseases/disease categories, but these preferences are intended to be extrapolated to all potential secondary genomic results. The hypothesis behind this approach is that a small number of preference items can be used to reveal patterns of preferences, and that these patterns can be used to infer preferences about other results not included in PIGSR. The most common pattern in our CSER family members, as demonstrated in Figure 2, is the “Everything” pattern. This applies when a participant indicates that she wishes to receive every result listed in PIGSR. We hypothesize that when a respondent’s preferences reveal this pattern, she prefers to receive every possible secondary result generated. Our proposed patterns, along with their definitions and hypothesized implications, are provided in Supplemental Table 3. We are currently examining the effectiveness of this strategy by gathering quantitative and qualitative data on parents completing PIGSR and receiving secondary results, and anticipate that future iterations of PIGSR will incorporate refinements to this strategy.
Adapting PIGSR to new clinical or research genomics projects
PIGSR’s first item gives the respondent the opportunity to opt-out of receiving secondary results. If this option is selected, additional preferences on secondary findings are not elicited. However, those results that should be considered “primary” and “secondary” will necessarily differ depending on the clinical or research context. For example, for the PREDICT project, the primary reason for genomic testing is pharmacogenomics. For the HudsonAlpha CSER project, results relevant to intellectual disability and related conditions are considered “primary.” For these reasons, it will be necessary to modify the structure of PIGSR to accommodate changing genomic testing contexts. Suggestions for adapting PIGSR are provided at www.PIGSR.org. Future research in this area will help reveal whether these adaptations affect the ability of this instrument to accurately elicit respondent’s preferences.
Limitations
While we undertook purposive sampling techniques to maximize the diversity of perspectives in our focus groups and interviews, our findings would have benefited from an even broader set of perspectives. In particular, our focus group participants (enrolled to help guide the development of PIGSR) were patients at Vanderbilt University, thus excluding a number of perspectives that may exist both inside and outside the Nashville community. Our findings from the HudsonAlpha CSER project are preliminary; enrollment for genomic testing and quantitative and qualitative data collection is ongoing. This ongoing work, and future studies involving genomic sequencing among other populations, will shed additional light on the issues addressed in this report.
Like other methods for eliciting preferences on genomic secondary results, the development of PIGSR involved a number of trade-offs. In order to attain a brief and straightforward instrument, we knowingly excluded several items that are important to some persons undergoing genomic testing, including some participants in our focus groups and semi-structured interviews. Our ongoing research will help reveal the implications of these trade-offs.
Conclusion
PIGSR is a brief tool to allow adults undergoing genomic testing to record their preferences about getting incidental or secondary results. It is designed to be completed prospectively by patients or research participants without the assistance of a healthcare provider and to guide computer-based analysis of genomic raw data. Ongoing research will help reveal the strengths and weaknesses of this approach and identify contexts where this method can be used effectively to elicit preferences and guide the return of secondary genomic results. PIGSR is freely available online at www.PIGSR.org under a Creative Commons license.
Supplementary Material
Acknowledgements
The authors would like to thank staff members at the following organizations who supported this research: the Office of Research Support Services and the Center for Biomedical Ethics and Society (both at Vanderbilt University Medical Center in Nashville, TN), the Kosair Charities Pediatric Clinical Research Unit (University of Louisville in Louisville, KY), North Alabama Children's Specialists (Children’s of Alabama in Huntsville, AL), and HudsonAlpha Institute for Biotechnology (Huntsville, AL). We would also like to thank our colleagues in the Clinical Sequencing and Exploratory Research (CSER) Consortium who provided feedback and advice on this project, and Evan Brothers who designed the PIGSR.org website. This project was funded by the National Human Genome Research Institute (NHGRI) through UM1 HG007301 (HudsonAlpha Institute) and R21 HG006612 (Vanderbilt University and McGill University). It received additional funding support from the National Center for Advancing Translational Sciences (NCATS) through UL1 TR000445 (Vanderbilt University).
Funding:
This project was funded by the National Human Genome Research Institute (NHGRI) through UM1 HG007301 (to G.M.C. and R.M.M.; HudsonAlpha Institute) and R21 HG006612 (Vanderbilt University and McGill University). It received additional funding support from the National Center for Advancing Translational Sciences (NCATS) through UL1 TR000445 (Vanderbilt University). Its contents are solely the responsibility of the authors and do not necessarily represent official views of the NHGRI, NCATS, or the National Institutes of Health.
Abbreviations and Acronyms
- CSER
Clinical Sequencing Exploratory Research Consortium
- dbGaP
Database of Genotypes and Phenotypes
- eMERGE
Electronic Medical Records and Genomics Network
- HIPAA
Health Insurance Portability and Accountability Act
- IRB
Institutional Review Board
Footnotes
Conflict of Interest:
The authors report that they have no commercial association or other financial interest that might pose or create a conflict of interest with information presented in this manuscript.
References
- 1.Fullerton SM, Wolf WA, Brothers KB, et al. Return of individual research results from genome-wide association studies: experience of the Electronic Medical Records and Genomics (eMERGE) Network. Genetics in Medicine. 2012;14(4):424–431. doi: 10.1038/gim.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg JS, Adams M, Nassar N, et al. An informatics approach to analyzing the incidentalome. Genetics in Medicine. 2013 Jan;15(1):36–44. doi: 10.1038/gim.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in Medicine. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacon PL, Harris ED, Ziniel SI, et al. The Development of a Preference-Setting Model for the Return of Individual Genomic Research Results. Journal of Empirical Research on Human Research Ethics. 2015 Apr;10(2):107–120. doi: 10.1177/1556264615572092. [DOI] [PubMed] [Google Scholar]
- 5.Berg JS, Amendola LM, Eng C, et al. Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research Consortium. Genetics in Medicine. 2013 Nov;15(11):860–867. doi: 10.1038/gim.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett JR. Actionability in Contemporary Bioethics: A Conceptual Analysis and Critique. 2014 Unpublished. [Google Scholar]
- 7.Christensen KD, Green RC. How could disclosing incidental information from whole-genome sequencing affect patient behavior? Personalized medicine. 2013;10(4):377–386. doi: 10.2217/pme.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenbaum D. If you don't know where you are going, you might wind up someplace else: incidental findings in recreational personal genomics. American Journal of Bioethics. 2014;14(3):12–14. doi: 10.1080/15265161.2013.879946. [DOI] [PubMed] [Google Scholar]
- 9.My46. 2012 https://www.my46.org/. Accessed 7/12/2012. [Google Scholar]
- 10.Pulley J, Denny J, Peterson J, et al. Operational implementation of prospective genotyping for personalized medicine: The design of the Vanderbilt PREDICT project. Clinical Pharmacology & Therapeutics. 2012;92(1):87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris PA, Scott KW, Lebo L, Hassan N, Lightner C, Pulley J. ResearchMatch: a national registry to recruit volunteers for clinical research. Academic Medicine. 2012 Jan;87(1):66–73. doi: 10.1097/ACM.0b013e31823ab7d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pope C, Mays N. Qualitative Research in Health Care. Blackwell Publishing/BMJ Books; Malden, MA: 2006. [Google Scholar]
- 13.Annas GJ, Elias S. 23andMe and the FDA. New England Journal of Medicine. 2014 Jun 5;370(23):2248–2249. doi: 10.1056/NEJMc1404692. [DOI] [PubMed] [Google Scholar]
- 14.Yu J-H, Jamal SM, Tabor HK, Bamshad MJ. Self-guided management of exome and whole-genome sequencing results: changing the results return model. Genetics in Medicine. 2013;15(9) doi: 10.1038/gim.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamal SM, Yu JH, Chong JX, et al. Practices and policies of clinical exome sequencing providers: analysis and implications. American Journal of Medical Genetics Part A. 2013 May;161A(5):935–950. doi: 10.1002/ajmg.a.35942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.