Abstract
Chronic Obstructive Pulmonary Disease (COPD) is characterised by reduced lung function and is the third leading cause of death globally. Through genome-wide association discovery in 48,943 individuals, selected from extremes of the lung function distribution in UK Biobank, and follow-up in 95,375 individuals, we increased the yield of independent signals for lung function from 54 to 97. A genetic risk score was associated with COPD susceptibility (odds ratios per standard deviation of the risk score (~6 alleles) (95% confidence interval) 1.24 (1.20-1.27), P=5.05x10-49) and we observed a 3.7 fold difference in COPD risk between highest and lowest genetic risk score deciles in UK Biobank. The 97 signals show enrichment in development, elastic fibres and epigenetic regulation pathways. We highlight targets for drugs and compounds in development for COPD and asthma (genes in the inositol phosphate metabolism pathway and CHRM3) and describe targets for potential drug repositioning from other clinical indications.
Maximally attained lung function and subsequent lung function decline together determine the risk of developing Chronic Obstructive Pulmonary Disease (COPD)1,2. COPD, characterised by irreversible airflow obstruction and chronic airway inflammation, is the third leading cause of death globally3. Smoking is the primary risk factor for COPD but not all smokers develop COPD and more than 25% of COPD cases occur in never-smokers4. Patients with COPD exhibit variable presentation of symptoms and pathology, with or without exacerbations, with variable amounts of emphysema and with differing rates of progression. Although risk factors for COPD are known, including smoking and environmental exposures in early5,6 and later life, the causal mechanisms are not well understood7. Disease-modifying treatments for COPD are required7.
Understanding genetic factors associated with reduced lung function and COPD susceptibility could inform drug target identification, risk prediction and stratified prevention or treatment. Previous genome-wide association studies (GWAS) of COPD identified several independent COPD-associated variants8–10 but the rate and scale of discovery has been limited by available sample sizes. We conducted a powerful GWAS for lung function, and followed up the robustly-associated variants in COPD case-control studies. Although previous GWAS have reported genome-wide significant associations with lung function11–16, there has not been a comprehensive study confirming the effect of these variants on COPD susceptibility. In this study, we hypothesised that: (i) undertaking GWAS of lung function of unprecedented power and scale would detect novel loci associated with quantitative measures of lung function; (ii) collectively these variants would be associated with the risk of developing COPD, and (iii) aggregate analyses of all novel and previously-reported signals of association, and the identification of genes through which their effects are mediated, would reveal further insight into biological mechanisms underlying the associations. Together these findings could provide potential novel targets17 for therapeutic intervention and pinpoint existing drugs which could be candidates for repositioning18 for the treatment of COPD.
Results
43 new signals for lung function
For stage 1, genome-wide association analyses of forced expired volume in 1 second (FEV1), forced vital capacity (FVC) and FEV1/FVC were undertaken in 48,943 individuals from the UK BiLEVE study16 who were selected from the extremes of the lung function distribution in UK Biobank (total n=502,682). From analysis of 27,624,732 variants, 81 independent variants associated with one or more traits with P<5x10-7 were selected for follow-up in stage 2, consisting of a further 95,375 independent individuals from UK Biobank, the SpiroMeta consortium and UK Households Longitudinal Study (UKHLS) (Supplementary Table 1). No evidence of sample overlap between stage 1 and stage 2 studies or between stage 2 studies was identified using LD score regression (Supplementary Table 2). Following meta-analysis of stage 1 and stage 2 results, 43 signals showed genome-wide significant (P<5x10-8) association with one or more of FEV1, FVC or FEV1/FVC (Table 1, Supplementary Table 3 and Supplementary Figure 1). We report these 43 signals as novel independent signals (Figure 1), almost doubling the number of confirmed independent genomic signals for lung function to 97 (Supplementary Table 4). Of the 43 novel signals, 33 represented novel loci whilst 10 were statistically independent signals (conditional P<5x10-7) within 500kb of another association signal. Based on an assumed heritability of 40%19,20 for each lung function trait, the novel signals explained 4.3% of the heritability of FEV1, 3.2% for FVC and 5.2% for FEV1/FVC bringing the total heritability explained by the 97 signals to 9.6%, 6.4% and 14.3%, respectively. The estimated effect sizes of lung function associated variants in children were correlated with those in adults (r=0.65, 73 variants with high imputation quality, Supplementary Figure 2). A genetic risk score based on these 73 variants, was also significantly associated with FEV1 and FEV1/FVC in children, (per risk allele β (s.e.) = -0.0177 (0.0040), P=1.03x10-5 and per risk allele β (s.e.) = -0.0213 (0.0037), P=1.27x10-8, respectively), but not with FVC (per risk allele β (s.e.) = -0.0037 (0.0041), P=0.366).
Table 1. Stage 1 and stage 2 association results for the 43 novel signals of association with lung function.
Where the discovery variant was not available in replication cohorts but a proxy with r2 > 0.8 was available, the proxy was used for replication in all cohorts (proxies are marked with * in rsid column). For discovery the standard errors and P values are genomic controlled except for conditional analyses (“Conditioned on” SNP is given in rsid column) where unadjusted standard errors and P values are given. Genomic controlled results were used for SpiroMeta. Unadjusted results were used for UK Biobank or UKHLS where genome-wide inflation factors were not available. Values are missing from stage 2 studies where there was quality control failure due to poor imputation (info < 0.5) or low minor allele count (MAC < 3). In the meta-analysis of the Stage 2 replication cohorts the 39 variants showing independent replication (Bonferroni correction for 81 tests: P <6.17×10-4) have P value in bold. Nearest gene gives either the nearest genes either side or the gene and location within the gene. Stage 1 association results (FEV1, FVC and FEV1/FVC) for the 54 signals of association that have been previously reported are given in Supplementary Table 4b.
| Stage 1 (discovery in UK BiLEVE) | Stage 2 | Meta-analysis of stage 1 and stage 2 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Top trait | Rsid (conditioned on) | Position b37 | Nearest gene(s) | Non /coding allele | Effect allele frequency | beta | se | P | UK Biobank beta | UK Biobank se | SpiroMeta beta | SpiroMeta se | UKHLS beta | UKHLS se | Meta beta | Meta se | Meta P | beta | se | Meta P |
| FEV1/FVC | rs17513135 | 1:40035686 | LOC101929516 (intron) | C/T | 23.15% | -0.047 | 0.008 | 1.25E-09 | -0.034 | 0.008 | -0.025 | 0.009 | -0.030 | 0.020 | -0.033 | 0.006 | 1.17E-08 | -0.038 | 0.005 | 2.31E-16 |
| FEV1/FVC | rs1192404 (rs12140637) |
1:92068967 | CDC7/TGFBR3 | A/G | 16.21% | -0.046 | 0.009 | 1.10E-07 | -0.047 | 0.009 | -0.046 | 0.010 | -0.033 | 0.023 | -0.050 | 0.007 | 9.31E-14 | -0.048 | 0.005 | 6.09E-20 |
| FEV1/FVC | rs12140637 | 1:92374517 | TGFBR3/BRDT2 | C/T | 31.30% | -0.036 | 0.007 | 3.49E-07 | -0.014 | 0.008 | -0.019 | 0.008 | -0.042 | 0.018 | -0.020 | 0.005 | 1.46E-04 | -0.026 | 0.004 | 1.18E-09 |
| FVC | rs200154334 | 1:118862070 | SPAG17/TBX15 | AT/- | 24.79% | 0.054 | 0.008 | 9.70E-12 | 0.025 | 0.008 | 0.023 | 0.009 | 0.001 | 0.020 | 0.024 | 0.006 | 1.69E-05 | 0.034 | 0.005 | 8.20E-14 |
| FEV1/FVC | rs6688537 | 1:239850588 | CHRM3 (intron) | C/A | 50.60% | -0.037 | 0.007 | 2.74E-08 | -0.042 | 0.006 | -0.023 | 0.008 | -0.049 | 0.017 | -0.039 | 0.005 | 4.05E-15 | -0.038 | 0.004 | 6.72E-22 |
| FEV1/FVC | rs61332075 | 2:239316560 | TRAF3IP1/ASB1 | G/C | 12.30% | 0.060 | 0.010 | 2.93E-09 | 0.025 | 0.010 | 0.021 | 0.012 | 0.029 | 0.025 | 0.026 | 0.007 | 5.11E-04 | 0.038 | 0.006 | 2.55E-10 |
| FEV1/FVC | rs1458979 | 3:55150677 | CACNA2D3/WNT5A | A/G | 50.11% | -0.035 | 0.007 | 1.52E-07 | -0.021 | 0.006 | -0.010 | 0.008 | -0.031 | 0.017 | -0.019 | 0.005 | 1.07E-04 | -0.025 | 0.004 | 4.42E-10 |
| FVC | rs1490265 | 3:67452043 | SUCLG2 (intron) | C/A | 70.79% | 0.039 | 0.007 | 1.03E-07 | 0.022 | 0.007 | 0.008 | 0.008 | 0.036 | 0.018 | 0.019 | 0.005 | 3.27E-04 | 0.026 | 0.004 | 1.58E-09 |
| FEV1/FVC | rs2811415 | 3:127991527 | EEFSEC (intron) | A/G | 84.04% | -0.057 | 0.009 | 2.64E-10 | -0.017 | 0.009 | -0.023 | 0.010 | -0.041 | 0.022 | -0.023 | 0.007 | 4.53E-04 | -0.035 | 0.005 | 5.52E-11 |
| FEV1/FVC | rs56341938* | 3:168715808 | LOC100507661/MECOM | A/G | 51.34% | 0.034 | 0.007 | 3.38E-07 | 0.037 | 0.006 | 0.013 | 0.008 | - | - | 0.027 | 0.005 | 1.97E-08 | 0.029 | 0.004 | 4.52E-14 |
| FEV1/FVC | rs13110699 (rs2045517) |
4:89815695 | FAM13A (intron) | T/G | 82.51% | -0.045 | 0.008 | 1.29E-07 | -0.037 | 0.008 | -0.030 | 0.009 | -0.014 | 0.024 | -0.035 | 0.006 | 7.80E-09 | -0.038 | 0.005 | 7.86E-15 |
| FVC | rs91731 | 5:33334312 | LOC340113/TARS | C/A | 90.53% | -0.070 | 0.011 | 8.10E-10 | -0.031 | 0.011 | -0.047 | 0.013 | 0.000 | 0.028 | -0.038 | 0.008 | 7.88E-06 | -0.049 | 0.007 | 4.31E-13 |
| FEV1/FVC | rs1551943 | 5:52195033 | ITGA1 (intron) | G/A | 23.01% | -0.052 | 0.008 | 3.12E-11 | -0.041 | 0.008 | -0.019 | 0.009 | -0.031 | 0.020 | -0.035 | 0.006 | 2.35E-09 | -0.041 | 0.005 | 1.92E-18 |
| FVC | rs2441026 | 5:53444498 | ARL15 (intron) | C/T | 46.27% | 0.034 | 0.007 | 4.59E-07 | 0.023 | 0.006 | 0.025 | 0.008 | 0.006 | 0.017 | 0.024 | 0.005 | 6.59E-07 | 0.027 | 0.004 | 2.75E-12 |
| FEV1/FVC | rs7713065 | 5:131788334 | C5orf56 (intron) | A/C | 73.67% | 0.039 | 0.007 | 2.21E-07 | 0.029 | 0.007 | 0.014 | 0.008 | 0.017 | 0.019 | 0.024 | 0.005 | 8.29E-06 | 0.029 | 0.004 | 2.77E-11 |
| FEV1 | rs3839234 | 5:148596693 | ABLIM3 (intron) | G/- | 47.01% | -0.038 | 0.007 | 8.87E-09 | -0.023 | 0.006 | -0.014 | 0.008 | 0.001 | 0.017 | -0.019 | 0.005 | 7.71E-05 | -0.026 | 0.004 | 4.48E-11 |
| FEV1/FVC | rs10515750 (rs1990950) | 5:156810072 | CYFIP2 (intron) | C/T | 7.18% | -0.063 | 0.012 | 2.61E-07 | -0.050 | 0.012 | -0.040 | 0.014 | -0.033 | 0.032 | -0.048 | 0.009 | 2.62E-07 | -0.054 | 0.007 | 5.26E-13 |
| FEV1/FVC | rs28986170 (rs2070600 rs9272528*) |
6:31556155 | LST1 (intron) | -/AA | 7.52% | 0.075 | 0.013 | 2.30E-08 | 0.034 | 0.014 | - | - | 0.096 | 0.036 | 0.048 | 0.014 | 6.49E-04 | 0.063 | 0.010 | 1.56E-10 |
| FEV1 | rs114229351 (rs34864796) |
6:32648418 | HLA-DQB1/HLA-DQA2 | T/C | 17.53% | -0.046 | 0.009 | 1.15E-07 | -0.026 | 0.008 | - | - | -0.045 | 0.026 | -0.030 | 0.008 | 1.78E-04 | -0.037 | 0.006 | 2.12E-10 |
| FEV1/FVC | rs141651520 | 6:73670095 | KCNQ5 (intron) | TTCTAT/- | 20.10% | 0.042 | 0.008 | 3.38E-07 | 0.049 | 0.008 | 0.026 | 0.009 | 0.025 | 0.020 | 0.042 | 0.006 | 5.49E-12 | 0.042 | 0.005 | 9.93E-18 |
| FEV1/FVC | rs10246303 | 7:7286445 | C1GALT1 (3’ UTR) | A/T | 41.74% | -0.034 | 0.007 | 4.42E-07 | -0.013 | 0.006 | -0.016 | 0.008 | -0.019 | 0.017 | -0.016 | 0.005 | 1.29E-03 | -0.022 | 0.004 | 2.35E-08 |
| FEV1/FVC | rs72615157 | 7:99635967 | ZKSCAN1 (3’ UTR) | G/A | 16.73% | 0.046 | 0.009 | 2.68E-07 | 0.015 | 0.009 | 0.030 | 0.010 | 0.030 | 0.022 | 0.024 | 0.007 | 2.56E-04 | 0.032 | 0.005 | 1.98E-09 |
| FEV1 | rs12698403 | 7:156127246 | LOC389602/LOC285889 | G/A | 44.36% | -0.036 | 0.007 | 7.43E-08 | -0.025 | 0.006 | -0.025 | 0.008 | -0.012 | 0.017 | -0.026 | 0.005 | 1.43E-07 | -0.029 | 0.004 | 1.11E-13 |
| FEV1 | rs7872188 | 9:4124377 | GLIS3 (intron) | C/T | 40.17% | -0.038 | 0.007 | 1.80E-08 | -0.019 | 0.007 | -0.020 | 0.008 | 0.005 | 0.017 | -0.019 | 0.005 | 1.41E-04 | -0.026 | 0.004 | 1.59E-10 |
| FVC | rs10870202 (rs10858246) |
9:139257411 | DNLZ (intron) | T/C | 50.01% | -0.033 | 0.006 | 3.25E-07 | -0.016 | 0.006 | -0.017 | 0.008 | -0.027 | 0.017 | -0.019 | 0.00 | 1.54E-04 | -0.024 | 0.004 | 9.32E-10 |
| FEV1/FVC | rs3847402 | 10:30267810 | SVIL/KIAA1462 | G/A | 40.57% | -0.036 | 0.007 | 1.00E-07 | -0.017 | 0.007 | -0.027 | 0.008 | -0.007 | 0.017 | -0.021 | 0.005 | 3.84E-05 | -0.027 | 0.004 | 7.72E-11 |
| FVC | rs7095607 | 10:69957350 | MYPN (intron) | G/A | 49.52% | -0.037 | 0.007 | 3.93E-08 | -0.021 | 0.006 | -0.029 | 0.008 | -0.030 | 0.017 | -0.027 | 0.005 | 2.26E-08 | -0.031 | 0.004 | 8.67E-15 |
| FEV1 | rs2509961 | 11:62310909 | AHNAK (intron) | T/C | 38.21% | 0.036 | 0.007 | 1.68E-07 | 0.030 | 0.007 | 0.017 | 0.008 | 0.025 | 0.017 | 0.027 | 0.005 | 1.07E-07 | 0.030 | 0.004 | 1.49E-13 |
| FEV1 | rs145729347* | 11:86442733 | ME3/PRSS23 | G/C | 15.08% | -0.056 | 0.009 | 1.67E-09 | -0.020 | 0.009 | -0.016 | 0.010 | - | - | -0.018 | 0.007 | 5.36E-03 | -0.031 | 0.005 | 8.58E-09 |
| FEV1 | rs567508 | 11:126008910 | CDON/RPUSD4 | G/A | 84.96% | 0.050 | 0.009 | 1.11E-07 | 0.029 | 0.009 | 0.013 | 0.010 | 0.053 | 0.024 | 0.026 | 0.007 | 1.08E-04 | 0.034 | 0.005 | 4.77E-10 |
| FEV1 | rs1494502 | 12:65824670 | MSRB3 (intron) | A/G | 36.20% | 0.036 | 0.007 | 2.72E-07 | 0.020 | 0.007 | 0.012 | 0.008 | 0.030 | 0.017 | 0.019 | 0.005 | 1.33E-04 | 0.025 | 0.004 | 9.80E-10 |
| FEV1/FVC | rs113745635 | 12:95554771 | FGD6 (intron) | C/T | 21.20% | -0.050 | 0.008 | 3.47E-10 | -0.039 | 0.008 | -0.018 | 0.009 | -0.061 | 0.020 | -0.036 | 0.006 | 1.41E-09 | -0.041 | 0.005 | 8.46E-18 |
| FVC | rs35506 | 12:115500691 | TBX3/MED13L | T/A | 71.25% | 0.037 | 0.007 | 4.31E-07 | 0.021 | 0.007 | 0.019 | 0.008 | 0.011 | 0.018 | 0.021 | 0.005 | 1.08E-04 | 0.027 | 0.004 | 9.87E-10 |
| FEV1/FVC | rs1698268 | 14:84309664 | LINC01467/LINC00911 | A/T | 29.44% | -0.039 | 0.007 | 1.12E-07 | -0.023 | 0.007 | -0.003 | 0.010 | 0.000 |
0.018 | -0.016 | 0.006 | 4.20E-03 | -0.025 | 0.005 | 3.19E-08 |
| FEV1/FVC | rs72724130 | 15:41977690 | MGA (intron) | A/T | 5.70% | -0.075 | 0.014 | 2.05E-07 | -0.046 | 0.014 | -0.039 | 0.021 | 0.007 | 0.035 | -0.043 | 0.012 | 2.62E-04 | -0.056 | 0.009 | 9.58E-10 |
| FEV1/FVC | rs12591467 (rs10851839) |
15:71788387 | THSD4 (intron) | C/T | 68.38% | 0.037 | 0.007 | 6.45E-08 | 0.021 | 0.007 | 0.011 | 0.008 | 0.030 | 0.018 | 0.019 | 0.005 | 2.17E-04 | 0.026 | 0.004 | 5.65E-10 |
| FEV1/FVC | rs66650179 | 15:84261689 | SH3GL3 (intron) | A/- | 81.34% | -0.048 | 0.009 | 2.60E-08 | -0.030 | 0.008 | - | - | -0.035 | 0.021 | -0.036 | 0.008 | 1.79E-05 | -0.042 | 0.006 | 3.71E-12 |
| FEV1/FVC | rs62070270* | 17:28263980 | EFCAB5 (intron) | A/G | 45.65% | -0.041 | 0.007 | 6.71E-10 | -0.036 | 0.006 | -0.021 | 0.008 | - | - | -0.030 | 0.005 | 8.00E-10 | -0.034 | 0.004 | 7.29E-18 |
| FEV1/FVC | rs11658500 | 17:36886828 | CISD3 (intron) | G/A | 14.16% | -0.051 | 0.009 | 4.70E-08 | -0.031 | 0.009 | -0.011 | 0.011 | -0.069 | 0.025 | -0.029 | 0.007 | 5.11E-05 | -0.037 | 0.006 | 7.22E-11 |
| FVC | rs6140050 | 20:6632901 | CASC20/BMP2 | C/A | 63.34% | 0.040 | 0.007 | 5.95E-09 | 0.026 | 0.007 | 0.028 | 0.008 | -0.011 | 0.017 | 0.026 | 0.005 | 5.23E-07 | 0.031 |
0.004 | 6.39E-14 |
| FEV1 | rs72448466 | 20:62363640 | ZGPAT (intron) | GT/- | 67.28% | -0.038 | 0.007 | 1.09E-07 | -0.020 | 0.007 | -0.029 | 0.008 | -0.032 | 0.017 | -0.027 | 0.005 | 3.68E-07 | -0.030 |
0.004 | 4.31E-13 |
| FEV1 | rs11704827 | 22:18450287 | MICAL3 (intron) | A/T | 23.14% | 0.049 | 0.008 | 6.08E-10 | 0.021 | 0.008 | 0.021 | 0.009 | 0.047 | 0.020 | 0.025 | 0.006 | 1.44E-05 | 0.033 | 0.005 | 8.32E-13 |
| FEV1 | rs2283847 | 22:28181399 | MN1 (intron) | C/T | 55.51% | -0.038 | 0.007 | 2.40E-08 | -0.026 | 0.007 | -0.014 | 0.008 | -0.003 | 0.021 | -0.021 | 0.005 | 3.65E-05 | -0.027 | 0.004 | 3.41E-11 |
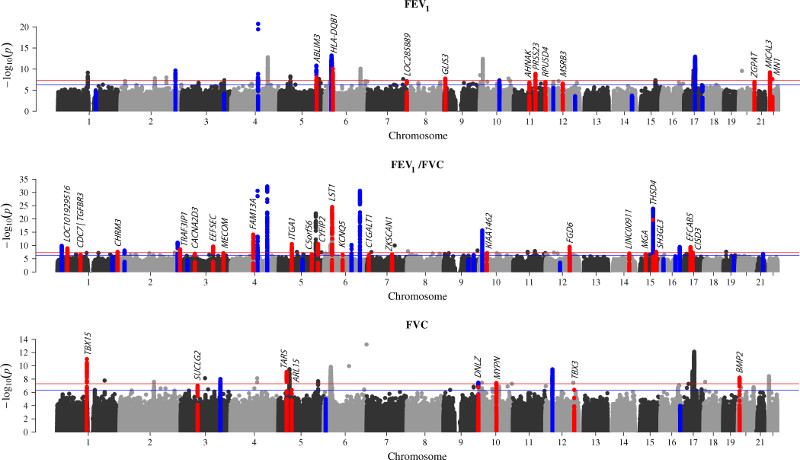
Figure 1.
Manhattan plots of genome-wide association results for FEV1 (top), FEV1/FVC (middle) and FVC (bottom). Previously reported signals are highlighted in dark blue (except signals with P>5x10-4in this study); and novel signals are coloured in red. Signals are highlighted for the trait with which they showed strongest association only. The red and blue lines correspond to the genome-wide significance level (P=5x10-8, -log10P=7.3) and the threshold used to select signals for follow up in stage 2 (P=5x10-7, -log10P=6.3) respectively. Labels show the nearest gene to the novel sentinel variants. There were 2 independent novel signals near CDC7 and TGFBR3 on chromosome 1 (labelled as CDC7/TGFBR3). See Supplementary Table 3 for full results. Image was created using a modified version of the R package qqman.
Using the stage 1 results, a 95% ‘credible set’ of variants (i.e. the set of variants that were 95% likely to contain the underlying causal variant, based on Bayesian refinement) was defined for all (novel and previously reported) association signals for which this was feasible (67 signals, Online Methods Supplementary Figures 3, 4 and 5 and Supplementary Table 5); 13 of these signals were fine-mapped to <=10 plausible causal variants and for 63 of the 67 signals fine-mapped, the sentinel (lowest P value) variant was also the top ranked variant by posterior probability. In addition, by refining six chromosome 6 MHC region association signals using imputation of classical alleles and amino acid changes (Online methods), we identified the MHC class II HLA-DQB1 gene product, HLA-DQβ1, amino acid change at position 57 (alanine compared to non-alanine) as the main driver of signals in the MHC region for both FEV1 (β (s.e.) = 0.048 (0.007), P=5.71×10-13, Supplementary Figure 6a) and FEV1/FVC (β (s.e.) = 0.062 (0.007), P=1.17×10-20, Supplementary Figure 6c) with secondary non-HLA gene signals in the MHC region remaining after conditioning on the HLA-DQβ1 position 57 variant for rs34864796:G>A (near ZKSCAN3, FEV1; conditional β (s.e.) = -0.058 (0.01), P=1.26x10-9, Supplementary Figure 6b) and rs2070600:C>T (in AGER, FEV1/FVC; conditional β (s.e.) = 0.120 (0.013), P=4.23x10-20, Supplementary Figure 6d), (Supplementary Table 6).
We identified that 29 of the lung function-associated signals had previously shown genome-wide significant association in GWAS of traits other than lung function or COPD. This included associations with inflammatory bowel disease (Crohn’s disease and/or ulcerative colitis, 3 signals) and height (9 signals, 3 of which showed a consistent direction of effect on height and the lung function measure with which they were most strongly associated) (Supplementary Table 7). With the exception of KANSL116, there was no significant (P<5.15x10-4) association with smoking for any of the signals (Supplementary Table 8).
95 variants and COPD susceptibility
The disease-relevance of lung function-associated variants has been questioned21. Therefore we tested association with COPD susceptibility for variants representing 95 of the 97 lung function associated signals in up to 20,086 COPD cases and 215,630 controls (data were unavailable for further study for the X-chromosome variant, rs7050036:A>T near AP1S2, and a rare variant, chr12:114743533:C>T) (Supplementary Table 9). These cases and controls comprised the COPD study at deCODE Genetics22, (COPD cases defined using spirometry, population-based controls excluding known cases, up to 1,964 moderate-severe cases, up to 142,262 controls), three lung resection cohorts23–25 (COPD definition based on spirometry, 310 moderate-severe cases, 332 controls), four case-control studies employing post-bronchodilator spirometry8–10,26–29 (5,778 moderate-severe cases, 3,950 controls), two studies within which COPD was determined from electronic medical records30 (eMR, total 1,487 cases, 15,138 controls), additional UK Biobank samples (COPD definition based on spirometry, 984 moderate-severe31 cases and 26,561 controls) and UK BiLEVE (COPD definition based on spirometry, 9,563 moderate-severe cases, 27,387 controls). UK BiLEVE COPD cases and controls were only used for single variant COPD association tests for the subset of 47 variants discovered independently from UK BiLEVE (that is excluding the 43 variants discovered using the UK BiLEVE data described in this paper and 5 variants reported in our previous study in the UK BiLEVE population16). Across all 95 variants, 51 showed nominal COPD association (P<0.05) and 30 showed associations with COPD susceptibility reaching a Bonferroni corrected threshold for 95 tests (P<5.26x10-4, Supplementary Table 10). Of these 30, 27 were variants discovered independently from UK BiLEVE and 3 were from the 48 lower powered association tests not including UK BiLEVE cases and controls.
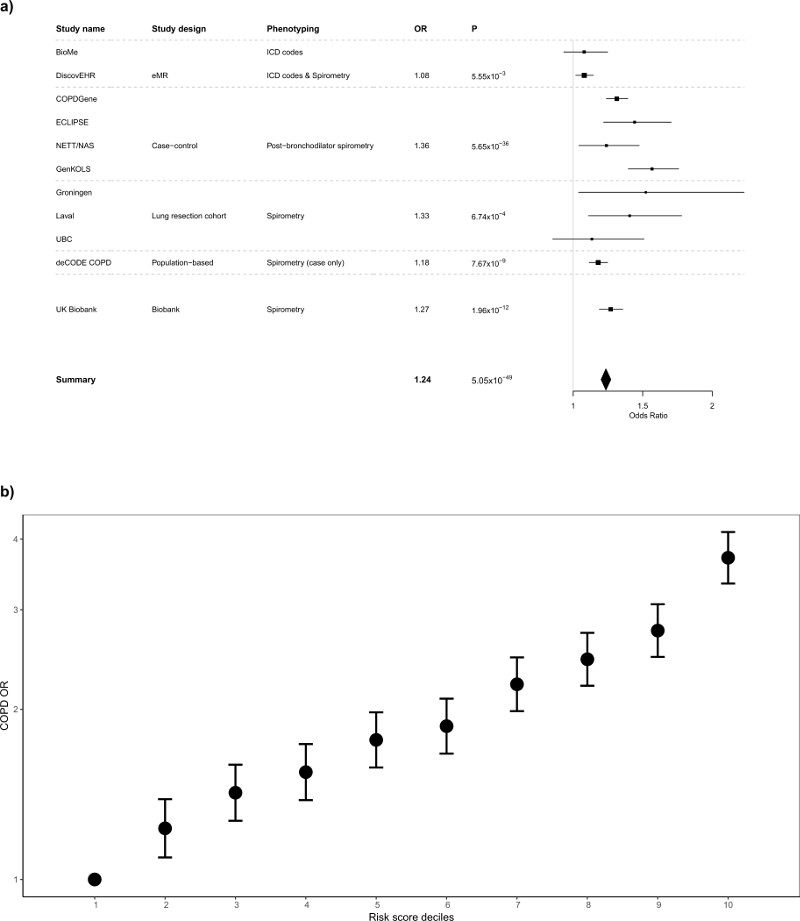
Using a risk score based on the available 95 sentinel variants or their best proxies, and using data from up to 9791 COPD cases and 120,462 controls (Online Methods), for the meta-analysis the OR (95% CI) per standard deviation change in risk score (~6 alleles) was 1.24 (1.20-1.27), P=5.05x10-49 (Figure 2a, Supplementary Table 11). We observed considerable heterogeneity in effect estimates between the different COPD studies (I2=92%) which had different approaches to ascertainment of COPD cases and variable disease severity. In UK Biobank (including UK BiLEVE) we found broadly similar effect size estimates of moderate-severe COPD to those in COPD case-control studies employing post-bronchodilator spirometry (OR=1.42 versus 1.36) and therefore we undertook further modelling showing a gradation in susceptibility to moderate-severe COPD across deciles of allelic risk score (Online Methods). The risk of moderate-severe COPD was more than three times higher in the top decile than the bottom decile (OR 3.71, 95% CI 3.34 to 4.12, Figure 2b). The estimated proportion of COPD cases attributable to allelic risk scores above the first decile (population attributable risk fraction) was 48.0% (95% CI 43.6 to 52.2%).
Figure 2.
Genetic Risk Score associations with COPD susceptibility (a) Forest plot of COPD results for the risk score analysis. Odds ratios per standard deviation of the risk score (~6 alleles) are presented for each study. Studies are grouped according to study design and phenotyping: “eMR”, electronic medical records, which used ICD codes to define COPD (DiscovEHR also used spirometry to refine the COPD definition); “case-control”, COPD case-control, which used post-bronchodilator spirometry to define COPD; “lung resection cohort”, which used a combination of pre and post-bronchodilator spirometry to define COPD; the Icelandic Biobank, deCODE, where cases were selected from a population based study and a study of COPD patients and defined using a spirometric definition, controls were selected as individuals within the cohort that were not known cases (no spirometric definition was used for controls); and UK Biobank (excluding UK BiLEVE), which used spirometry to define both COPD cases and controls. Further details are provided in the Supplementary Note. (b) Odds ratios for spirometrically-defined COPD for weighted genetic risk score deciles in UK Biobank (10,547 cases, pre-bronchodilator % predicted FEV1<80% and FEV1/FVC<0.7, and 53,948 controls, FEV1/FVC>0.7 and % predicted FEV1>80%, weights derived from non-discovery populations). For each decile, odds ratios were obtained using a logistic regression adjusted for age, age2, sex, height, smoking status, pack-years and the first 10 ancestry principal components. The OR comparing the 10th and the 1st decile in ever-smokers only was 3.35 (95% CI 2.93 to 3.84) and in never-smokers only was 4.27 (95% CI 3.61 to 5.06).
We tested association of individual variants and the 95-variant risk score with COPD exacerbations in subsets of individuals from UK Biobank, deCODE, four COPD case-control studies and two eMR studies (total 2,462 COPD exacerbation cases, 15,288 COPD non-exacerbation controls) and the Lung Health Study (100 exacerbation cases, 4,002 controls). There was no association of individual variants or genetic risk score with acute exacerbations of COPD (Supplementary Tables 12 and 13).
To evaluate whether these variants showed disease-relevant associations in a non-European population, we studied 71 variants for which data were available in 7,116 COPD cases (20,919 controls) and 5,292 exacerbation cases (1,824 controls) from the China Kadoorie Biobank cohort (CKB) (Supplementary Tables 10 to 13). The allelic risk score was associated with COPD susceptibility (OR per standard deviation change in risk score (95% CI) = 1.08 (1.04-1.11), P=4.2x10-6) suggesting some shared genetic contributions to COPD in European and East Asian descent populations. Thirty-nine of the variants showed a consistent direction of effect on COPD in European and Chinese samples and seven of these were significant (P<0.05). Two signals were significant after correction for multiple testing (Supplementary Table 10c).
To assess the impact of including individuals with asthma in a COPD case-control analysis, we tested for association with COPD in UK Biobank both before and after excluding individuals with self-reported doctor-diagnosed asthma and show that the effect size estimates were similar (Supplementary Figure 7).
Implicated genes highlight pathways and druggable targets
Gene expression and genotype data from lung, blood and multi-tissue resources were queried to identify whether the top variant at each of the 97 signals, or a proxy, were significantly associated with changes in expression of any gene (i.e. were an eQTL for any gene). Using this approach, and identification of deleterious variants within the association signal (Online methods, Supplementary Table 14), we implicated 234 genes with potentially causal effects on lung function (Supplementary Table 15). These 234 genes were enriched (False Discovery Rate (FDR) ≤5%) in elastic fibre pathways and in “signalling events mediated by the Hedgehog family”, the latter including CDON implicated by a novel intergenic signal (rs567508, between CDON and RPUSD4) on chromosome 11. We narrowed this group of 234 genes to 68 high-priority genes which were implicated via a deleterious variant or on stricter criteria for gene expression co-localisation (sentinel variant and top expression variant r2≥0.9, Table 2). We found that the 68 high-priority genes were overrepresented (FDR≤5%) among a number of gene ontology terms including SH3 domain binding, GTPase binding, actin binding and fibroblast migration (Supplementary Table 16). Alternative approaches to pathway analyses, which instead use all genome-wide association results, supported previous reports of enrichment of histone and systemic lupus erythematosus pathways14–16 and additional autoimmune and inflammatory pathways (Supplementary Table 17). Tests for tissue-specific enrichment of lung function signals overlapping histone marks identified enrichment in fetal lung, fetal heart and fibroblasts (H3K4me1), and stomach smooth muscle (H3K4me1 and H3K4me3) (Supplementary Table 18).
Table 2.
Genes implicated as high-priority genes for novel genome-wide significant and previously-reported signals using expression data and functional annotation. #Variant did not reach P<5.15x10-4 (Bonferroni corrected P value for 97 tests) in this study for any trait. *Gene implicated as it contained a deleterious variant (Supplementary Table 14); all other genes implicated by co-localisation of GWAS and eQTL signal. (*) implicated by both co-localisation of eQTL and GWAS, and a deleterious variant. All 234 genes implicated are listed in Supplementary Table 15.
| Genome-wide significant trait (additional traits with P<5.15x10-4) | Variant ID (position b37) | Nearest gene(s) | High-priority genes |
|---|---|---|---|
| Novel signals | |||
| FEV1/FVC (FVC) | rs17513135 (chr1:40,035,686) | LOC101929516 (intron) | PABPC4 |
| FEV1/FVC (FEV1) | rs6688537 (chr1:239,850,588) | CHRM3 (intron) | CHRM3 |
| FEV1/FVC (FEV1) | rs2811415 (chr3:127,991,527) | EEFSEC (intron) | RUVBL1 |
| FEV1/FVC (-) | rs13110699 (chr4:89,815,695) | FAM13A (intron) | FAM13A |
| FEV1 (FVC, FEV1/FVC) | rs3839234 (chr5:148,596,693) | ABLIM3 (intron) | GRPEL2, ABLIM3 |
| FEV1/FVC (FEV1) | rs10515750 (chr5:156,810,072) | CYFIP2 (intron) | ADAM19 |
| FEV1/FVC (FEV1) | rs200003338 (chr6:31,556,155) | LST1 (intron) | MICB*, MICA* |
| FEV1/FVC (FEV1) | rs10246303 (chr7:7,286,445) | C1GALT1 (3’ UTR) | C1GALT1 |
| FVC (FEV1) | rs10870202 (chr9:139,257,411) | DNLZ (intron) | INPP5E, CARD9 |
| FVC (FEV1) | rs7095607 (chr10:69,957,350) | MYPN (intron) | MYPN* |
| FEV1 (FVC) | rs2509961 (chr11:62,310,909) | AHNAK (intron) | ROM1, EML3, MTA2, GANAB, C11orf83* |
| FEV1/FVC (-) | rs59835752 (chr17:28,265,330) | EFCAB5 (intron) | EFCAB5, CRYBA1, SSH2, SLC6A4 |
| FEV1/FVC (FEV1) | rs11658500 (chr17:36,886,828) | CISD3 (intron) | CISD3* |
| FEV1 (FVC) | rs72448466 (chr20:62,363,640) | ZGPAT (intron) | LIME1 |
| Previously-reported signals | |||
| FEV1 (FVC) | rs6681426 (chr1:150,586,971) | MCL1/ENSA | GOLPH3L |
| FEV1/FVC (-) | rs4328080 (chr1:219,963,088) | LYPLAL1/RNU5F-1 | SLC30A10 |
| FEV1 (FVC, FEV1/FVC) | rs2571445 (chr2:218,683,154) | TNS1 (exon) | TNS1* |
| FEV1/FVC (-) | rs10498230 (chr2:229,502,503) | SPHKAP/PID1 | SPHKAP |
| FVC (FEV1) | rs1595029 (chr3:158,241,767) | RSRC1 (intron) | RSRC1 |
| FEV1 (FVC, FEV1/FVC) | rs10516526 (chr4:106,688,904) | GSTCD (intron) | INTS12, GSTCD, NPNT |
| FEV1/FVC (FEV1, FVC) | rs34712979 (chr4:106,819,053) | NPNT (intron) | NPNT |
| FEV1/FVC (FEV1) | rs138641402 (chr4:145,445,779) | GYPA/HHIP-AS1 | HHIP |
| FEV1/FVC (-) | rs153916 (chr5:95,036,700) | SPATA9/RHOBTB3 | RHOBTB3 |
| FEV1/FVC (FEV1) | rs1990950 (chr5:156,920,756) | ADAM19 (intron) | ADAM19 |
| FEV1 (FVC, FEV1/FVC) | rs34864796 (chr6:27,459,923) | ZNF184/LINC01012 | OR2B2* |
| FEV1/FVC (FEV1) | rs2857595 (chr6:31,568,469) | NCR3/AIF1 | MICB* |
| FEV1/FVC (-) | rs2070600 (chr6:32,151,443) | AGER (exon) | AGER(*) |
| FEV1 (FVC, FEV1/FVC) | rs114544105 (chr6:32,635,629) | HLA-DQB1/HLA-DQA2 | HLA-DQB1*, APOM, RNF5 |
| FEV1/FVC (FEV1) | rs113096699 (chr6:142,745,883) | GPR126 (intron) | GPR126 |
| FEV1/FVC (-) | rs148274477 (chr6:142,838,173) | GPR126/LOC153910 | GPR126* |
| FVC (FEV1) | rs10858246 (chr9:139,102,831) | QSOX2 (intron) | QSOX2 |
| FVC (FEV1) | rs2348418 (chr12:28,689,514) | CCDC91 (intron) | FLJ35252 |
| FEV1/FVC# (-) | rs11172113 (chr12:57,527,283) | LRP1 (intron) | LRP1 |
| FEV1# (-) | rs7155279 (chr14:92,485,881) | TRIP11 (intron) | ATXN3 |
| FEV1# (-) | rs117068593 (chr14:93,118,229) | RIN3 (exon) | RIN3(*) |
| FEV1/FVC (FEV1) | rs10851839 (chr15:71,628,370) | THSD4 (intron) | THSD4 |
| FEV1/FVC (-) | rs12447804 (chr16:58,075,282) | MMP15 (intron) | MMP15 |
| FEV1/FVC (FEV1) | rs3743609 (chr16:75,467,021) | CFDP1 (intron) | TMEM170A, BCAR1, CFDP1 |
| FEV1 (FVC, FEV1/FVC) | rs35524223 (chr17:44,192,590) | KANSL1 (intron) | KANSL1(*), MAPT(*), ARL17B, ARL17A, LRRC37A4, NUDT1, LRRC37A, CRHR1, LRRC37A2, ARHGAP27, FMNL1, PLEKHM1, WNT3, NSF, SPPL2C* |
| FEV1 (FVC) | rs7218675 (chr17:73,513,185) | TSEN54 (intron) | CASKIN2, TSEN54* |
| FEV1/FVC (-) | rs113473882 (chr19:41,124,155) | LTBP4 (intron) | LTBP4* |
Approved drugs, or drugs in development, target the protein products of 7 of the 234 genes (Supplementary Table 19a). This includes 3 high-priority genes CHRM3, SLC6A4 and CRHR1. CHRM3 and SLC6A4 were both implicated by novel signals (rs6688537:C>A in an intron of CHRM3 and rs59835752:-/A in an intron of EFCAB5, respectively) and encode targets for drugs approved for the treatment of asthma and COPD (CHRM3, muscarinic acetylcholine receptor M3) and anxiety and depression (SLC6A4, serotonin transporter). CRHR1 (implicated by rs35524223:T>A in an intron of KANSL1) encodes the corticotropin releasing factor receptor 1 which is a target for compounds in development for the treatment of anxiety, depression and irritable bowel syndrome. The other 4 genes include NDUFA12 (implicated by rs113745635:C>T in an intron of FGD6) encoding an NADH dehydrogenase which is a target for metformin hydrochloride, primarily used to treat type 2 diabetes, and ITK (implicated by rs10515750 in an intron of CYFIP2) encoding a tyrosine-protein kinase, a target for the cancer drug Pazopanib.
Using STRING32 to find proteins that interact with the proteins encoded by the high priority genes, we highlighted further druggable targets (Supplementary Table 19b). These included the PI3-kinase p110-delta subunit (part of the inositol phosphate metabolism pathway with INPP5E, which was implicated as a high-priority gene by rs10870202 in an intron of DNLZ, and a target for compounds in development for the treatment of COPD and asthma), and matrix metalloproteinases 1, 8 and 7 (targets for doxycycline, which is an antibiotic and anti-malarial).
Discussion
In this study, the power gained by sampling from the extremes of a large biobank whilst retaining the power of a quantitative trait analysis, coupled with strategies to improve coverage of the genome and extensive follow-up, enabled a near-doubling of the number of signals of association with lung function identified to date. We further explored 95 variants, representing 43 novel signals and 52 previously reported signals, and showed that collectively these variants are strongly associated with COPD susceptibility.
Using functional evidence from eQTL studies and deleterious variants to link signals to genes, we identified that 41 of the 97 lung function signals are also the strongest signals of association for expression of, or contain deleterious variants within, 68 genes (which we term “high-priority genes”). Amongst these, novel signals in or near FAM13A and ADAM19, both previously associated with lung function and COPD susceptibility9,33, along with evidence that these signals are themselves eQTLs for FAM13A and ADAM19, provide further evidence for FAM13A and ADAM19 themselves being the drivers of those signals. There was significant enrichment amongst the 68 genes for SH3 domain (including ADAM19), GTPase and actin binding, and fibroblast migration, highlighting the potential importance of pathways relating to the cytoskeleton.
The 68 genes identified as high-priority included genes at novel signals encoding targets for which there are approved drugs or drugs in development (Supplementary Table 19). Of note, the muscarinic acetylcholine receptor M3, encoded by CHRM3, is a well-characterised drug target for which many approved drugs exist, including for the treatment of asthma and obstructive lung disease. SLC6A4 encodes a serotonin transporter, a target for a number of drugs approved for treating depression and anxiety disorders, one of which (nortriptyline hydrochloride) has been trialed for use in inflammatory skin disorders (psoriasis and eczema); HTR4, which encodes a serotonin receptor, was identified in one of the earliest lung function GWAS13. INPP5E, identified as a high-priority gene for a novel signal of association with FVC (and FEV1) on chromosome 9, encodes inositol polyphosphate-5-phosphatase E, a component of the inositol phosphate metabolism pathway. Another component of the same pathway, phosphoinositide 3-kinase (PI3K) delta is a target of drugs under development for the treatment of a range of indications including COPD and asthma. Mutations in INPP5E cause ciliopathy (Joubert and MORM syndromes).
Protective genetic variants that reduce the function or expression of a target protein could be mimicked by drugs and so are of particular interest. The minor allele (MAF 17%) at the novel signal in an intron of FAM13A was associated with decreased expression of FAM13A in lung tissue and reduced risk of COPD. This, together with recent evidence from a study of the Fam13a knockout mouse34, suggests that pharmacological inhibition of FAM13A may be protective.
Extending our pathway analyses to all 234 genes implicated by gene expression or deleterious variants, we observed enrichment of genes related to “signalling events mediated by the Hedgehog family” pathway. Hedgehog signalling plays a crucial role in early development. Three members of this pathway, PTCH1, TGFB2 and HHIP, have been previously reported as likely causal genes underlying lung function association signals35. In this study, we additionally report PTHLH, encoding a parathyroid hormone-like hormone, and CDON¸ encoding a Hedgehog co-receptor, as likely causal genes (the latter at a novel signal). Of the 73 well-imputed variants available in children, we show correlation (r=0.62) between variant effect size estimates with those in adults. Should this pattern of correlation apply across all 97 lung-function-associated variants, then this would suggest that many of these variants may act, at least in part, via effects on lung development. Elastic fibre pathways were over-represented; products of elastin degradation have been shown to be elevated during acute exacerbations of COPD 36,37. In addition, degradation of elastin by excess neutrophil-released elastase in the lung leads to emphysema in individuals with alpha-1 antitrypsin deficiency. CARD9, another high-priority gene at a novel signal, encodes an adaptor protein involved in neutrophil recruitment in respiratory fungal infection38. Tissue-specific enrichment of lung function signals overlapping H3K4me1 was seen in stomach smooth muscle. Although comparable H3K4me1 data were not available for airway smooth muscle, similar findings have been reported previously for rectal smooth muscle39.
The 17q21.31 inversion has previously been associated with lung function. Custom imputation of additional structural variation at the locus, along with eQTL evidence and deleterious variants in the gene, suggested that KANSL1 may drive the association. Amongst the novel signals reported in this study, SNPs in an intron of EEFSEC on chromosome 3 are correlated with expression of nearby gene RUVBL1. Both KANSL1 and RUVBL1 encode members of histone modification complexes.
A novel signal on chromosome 20 (rs72448466, intronic in ZGPAT), which showed association with FVC almost as strong as its association with FEV1, is an eQTL for the telomere gene, RTEL1. Although rs72448466:->GT was not the strongest eQTL for RTEL1 (r2=0.6 with the top eQTL variant), RTEL1 is of interest as it has recently been implicated in familial pulmonary fibrosis40. Variant rs72448466 has also been associated with inflammatory bowel disease, prostate cancer and atopic dermatitis.
Our implication of genes of potential functional relevance to the 97 signals was based on gene expression data (eQTL) and associated deleterious variants within a gene. Although eQTL evidence currently gives the best in silico indication of which gene (or genes) might be functionally relevant to a signal, conclusive evidence for a causal relationship between SNP genotype and gene expression can only be obtained through direct molecular experiments.
Six signals of association have been previously identified within the HLA region. Using a custom imputation approach, we identified the presence of alanine (compared to aspartic acid, valine or serine) at amino acid position 57 in HLA-DQβ1 as associated with decreased lung function and the main driver of signals in this region. The presence of alanine is also strongly associated with risk of type 1 diabetes41.
The three lung function traits we studied are correlated. The overall and genetic correlations were: 0.88 and 0.87 between FEV1 and FVC; 0.46 vs 0.35 between FEV1 and FEV1/FVC and; 0.038 and -0.17 between FVC and FEV1/FVC (transformed traits, as studied in UK Biobank and SpiroMeta15, respectively). One might expect variants showing strongest association with FEV1 and FEV1/FVC to be of greatest relevance for COPD and genetic correlations of -0.76 and -0.9 have been reported between COPD and FEV1 and FEV1/FVC, respectively42. We show, however, that variants associated with one of these traits also tend to be associated with one of the other two lung function traits studied (for example, all but 2 signals for FVC are also associated (P<0.05) with FEV1, Supplementary Table 4). Although classification of COPD in UK Biobank was based on pre-bronchodilator spirometry, we have previously shown that this leads to minimal misclassification of moderate-severe (GOLD 2-4) COPD43. The effect size estimates for COPD associations could be influenced by differences in case ascertainment between the follow-up studies. Motivated by avoidance of potential winner’s curse bias for the 48 variants discovered using UK BiLEVE, we excluded UK BiLEVE from individual variant analyses. However, this excluded 9,563 moderate to severe COPD cases, and therefore the significance of COPD association tests for these variants should be interpreted with caution. Notably, we found effect size estimates only slightly smaller in deeply-characterised COPD case-control studies than in UK Biobank (OR per SD change in allelic risk score 1.36 compared to 1.42). Whilst we show an appreciable proportion of COPD cases could be attributable to allelic risk scores above the first decile, great caution must be exercised in interpretation of population attributable risk fraction estimates given considerations of shared etiologic responsibility44. The lung function-associated variants we report were not associated with acute exacerbations of COPD. Although more powerful studies of exacerbations will be required, this suggests that different genetic mechanisms could underlie risk of acute exacerbations.
A threshold of P<5x10-8 is a valid threshold for genome-wide significance in GWAS analyses of common variants45. Our genotyping and imputation strategy resulted in testing of 27.6 million variants of which 21.6 million had MAF<5% and 18.2 million had MAF<1%. Although all of our 43 signals were common, had we adopted a stricter threshold for genome-wide significance, for example, P<1x10-8 (recommended in a recent report of significance thresholds in whole genome sequencing45), only two of our signals (rs10246303:A>T in the 3’ UTR of C1GALT1 on chromosome 7, and rs1698268:A>T near LINC00911 on chromosome 14) would not have reached significance. Thirty-nine of the 43 signals were additionally supported by statistically significant independent replication in stage 2 (P<0.05/43, Supplementary Table 3).
In summary, our study provides the most comprehensive evidence yet regarding genetic variants associated with lung function and their association with susceptibility to COPD, with a more than threefold difference in COPD risk between highest and lowest allelic risk score deciles. Whilst translation of GWAS findings can take some years and requires extensive additional work, selecting genetically supported targets could double the drug development success rate17. The future clinical relevance of our findings include contributions towards understanding of disease pathogenesis, identification of drug targets for targeting or repositioning of drugs18, and potentially improved prediction of COPD or its subtypes.
Data Availability Statement
The stage 1 (UK BiLEVE) genome-wide association results for FEV1, FVC and FEV1/FVC are available from UK Biobank at http://www.ukbiobank.ac.uk/. The sources of all other data utilised in this study can be found in the Online Methods and Supplementary Note.
Online Methods
Study Governance
UK Biobank has ethical approval from the NHS National Research Ethics Service (Ref 11/NW/0382). Informed consent was obtained from all participants. All other studies were approved by an appropriate ethics committee or data protection authority (Supplementary Note).
Stage 1 study sample selection
A genome-wide discovery study for variants associated with lung function measures was performed in 48,943 individuals from the UK BiLEVE16 subset of UK Biobank (UK BiLEVE, stage 1). In brief, UK Biobank comprised 502,682 individuals of whom 275,939 were of self-reported European-ancestry and had ≥2 Forced Expired Volume in 1s (FEV1) and Forced Vital Capacity (FVC) measures (Vitalograph Pneumotrac 6800, Buckingham, UK) passing ATS/ERS criteria46. Based on the best (highest) available FEV1 measurement, 50,008 individuals from groups with extreme low (n=10,002), near-average (n=10,000) and extreme high (n=5,002) % predicted FEV1 were selected from amongst never-smokers (total n=105,272) and the same numbers from amongst the heavy-smokers (mean 35 pack-years of smoking, total n=46,758). FEV1, FVC and FEV1/FVC distributions are summarised in Supplementary Figure 8. Genotyping was undertaken using the Affymetrix Axiom UK BiLEVE array16 and imputed to the 1000 Genomes Project Phase 147 and UK10K48,49 combined panel. A total of 27,624,732 imputed or directly genotyped autosomal variants with imputation quality (info) >0.5 and minor allele count (MAC) ≥3 were included in the analysis. In total, 48,943 unrelated individuals passed all quality control steps and were used in this analysis.
Association testing and selection of signals from stage 1 for follow-up in stage 2
Power calculations were undertaken using Quanto (see URLs) (Supplementary Figure 9). For stage 1, genome-wide association studies of FEV1, FVC and FEV1/FVC were undertaken separately in heavy-smokers and never-smokers and then meta-analysed for each trait. Linear regression of age, age2, sex, height, the first 10 principal components of genetic ancestry and pack years of smoking (in smokers) on each trait was undertaken and residuals were ranked and transformed to inverse normally distributed Z-scores. For the first 26 lung function variants reported11,13,14,50 we showed Stage 2 effect size estimates14 were comparable with those from inverse normally distributed Z-scores in UK BiLEVE (Supplementary Figure 10). Subsequently these Z-scores were used for genome-wide association testing using an additive genetic model (SNPTEST v2.5). The full genome-wide stage 1 results are available via UK Biobank (see URLs).
From each of the three discovery GWAS, signals were selected for follow-up in stage 2 if they met an initial threshold of P<5x10-7. Low MAC variants (MAC between 3 and 20), were selected for follow-up only if the imputation quality (info) exceeded 0.8. Independence of signals was determined as follows: the most strongly associated (P<5x10-7) variant within a 1Mb region was selected as a putative signal and then the analysis repeated for that 1Mb region conditioning on the most strongly associated variant. Any variant which then had a conditional P<5x10-7 was then assigned as a secondary putative signal and also included in the conditional analysis. This was repeated until no variants with P<5x10-7 remained within the 1Mb region. Results were confirmed using a joint conditional analysis (GCTA51) and visual inspection of region plots. Previously reported signals were not included in the final list of putative signals to be taken for follow-up in stage 2. Where novel signals for different traits were in linkage disequilibrium (r2 > 0.2), the variant for the trait with the most significant association was followed up. Due to the extended LD structure in the MHC region, conditional analyses and GCTA were run over a 9Mb region (chr6:24,126,750-33,126,689). Two pairs of signals previously reported as being independent (rs16909859:G>A11 and rs16909898:A>G14 in PTCH1, and rs34712979:G>A16 and rs6856422:T>G15, in NPNT) were found to be correlated in our data.
Stage 2 – follow-up in independent studies (quantitative lung function)
Putative novel signals of association from stage 1 were followed up in three independent sets of samples (stage 2): (i) an independent subset of UK Biobank participants (UK Biobank, n=49,727), (ii) a population-based consortium (SpiroMeta, n=38,199)15 and (iii) UK Households Longitudinal Study (UKHLS, n=7,449). We did not include these studies in Stage 1 as: (ii) was to be utilised for independent replication and; (i) and (iii) were not yet available when Stage 1 was undertaken. Each signal was followed-up only for the trait with which it was most strongly associated in Stage 1. The first tranche of genotype data and imputation output (merged 1000 Genomes Project Phase 3 and UK10K imputation panel) from UK Biobank was released May 2015 (see URLs) and comprised the 49,979 individuals originally genotyped for UK BiLEVE (an unrelated subset of 48,943 of which were used as discovery in this study) and an additional 102,757 individuals selected at random from the entire UK Biobank. From these 102,757 individuals, we initially selected 51,117 samples that had lung function measurements (FEV1 and FVC) meeting ATS/ERS criteria and had covariates age, sex, height, principal components and smoking status recorded. Following further exclusion of individuals with sex mismatches (n=41), individuals of non-European ancestry (based on k-means clustering of principal components 1 and 2 with 4 clusters, n=124) and one individual from each pair of related samples (KING relatedness > 0.088 [2nd degree], n=1,225), a total of 49,727 individuals remained for analysis.
The details of the SpiroMeta consortium analysis (including contributing studies, spirometry details and methods) are described elsewhere15. In brief, this was an inverse variance weighted fixed effects meta-analysis of 17 studies with imputation to 1000 Genomes Project Phase 1 reference panel. Within each study, FEV1, FVC and FEV1/FVC were adjusted for age, age2, sex, height and population structure, separately for ever and never-smokers. Inverse normal transformed residuals were then tested for association within each smoking stratum assuming an additive genetic effect and then meta-analysed. Genomic control was applied to account for residual population structure. We only included SpiroMeta meta-analysis results in the meta-analysis in this study if Neffective > 70% (i.e. >70% of 38,199), where Neffective is the effective sample size after scaling for imputation quality15.
Summary statistics of a GWAS of FEV1, FVC and FEV1/FVC in 7,449 individuals were available from UKHLS (Supplementary Note). SNPs were genotyped using the Illumina Infinium HumanCoreExome BeadChip Kit and imputed against the same 1000 Genomes Project + UK10K combined imputation panel as used in discovery in this study. Association testing was performed separately for ever and never-smokers with covariates age, age2, sex height and ancestry principal components, as for Stage 1. We only included UKHLS results in the meta-analysis in this study if imputation info >0.5 and MAC >=3.
Meta-analysis of stage 1 and stage 2
All meta-analyses were undertaken using fixed effects inverse variance weighting which takes directionality of association into account. Effect estimates for all variants followed up in stage 2 were meta-analysed across the three stage 2 studies and then the combined result was meta-analysed with stage 1 results. Where the discovery variant was not present in any stage 2 study, a proxy (r2>0.8) that was available in all stage 1 and stage 2 studies was used. We report signals with association P<5x10-8 in the meta-analysis of stages 1 and 2 as novel signals of association with lung function.
Assessment of stage 1 and stage 2 sample overlap by LD score regression
LD score regression was used to assess the extent of confounding. Absence of significant confounding indicates that factors such as sample overlap and/or population stratification are not evident. Pre-computed LD scores from a European population were used (see URLs), based on genotypes for 1,293,150 HapMap3 SNPs in samples from the 1000 Genomes Project EUR population. Association results were filtered (info > 0.9 and MAF > 1%) before running LD score regression on (i) 3 pairwise meta-analyses of results from UK BiLEVE (stage 1) and UK Biobank (stage 2), UK BiLEVE and SpiroMeta and UK Biobank and SpiroMeta; (ii) bivariate analyses of the 3 pairs of cohorts.
Effect sizes in adults and children
The effects of variants on lung function in children were also tested in 5,062 children from ALSPAC (mean age 8.6) and 1,220 children from the Raine study (mean age 8.1). Data were available for 81 of the 97 variants (a proxy variant with r2>0.7 was used for 11 signals) with imputation quality >0.5 of which 73 had imputation quality >0.8 (71 variants in ALSPAC and 35 in the Raine study). Association results from the two cohorts were combined using inverse variance weighted meta-analysis. A weighted risk score was approximated using pooled single SNP results, as described in Dastani et al52, and weights obtained using estimated effect sizes from either SpiroMeta15 summary data (for SNPs discovered in UK Biobank), or from UK Biobank (for SNPs discovered elsewhere). The risk score was tested for the three lung function traits: FEV1, FVC and FEV1/FVC.
Refinement of signals
A Bayesian method53 was used to fine-map lung function-associated signals to the set of variants that were 95% likely to contain the underlying causal variant (assuming that the causal variant has been analysed). This was undertaken for novel signals and for previously-reported signals which reached P<10-5 in the stage 1 results. Following van de Bunt et al.54 we set the value of a prior W=0.4 in the approximate Bayes Factor formula. Signals in the HLA were not included.
We re-imputed our 48,943 discovery samples across the HLA (chr6:29,607,078-33,267,103 (b37)) using IMPUTE2 v2.3.1 with a reference panel incorporating classical HLA alleles and amino acid changes55. The reference panel contained haplotypes for 5,225 samples from the type 1 diabetes genetics consortium (T1DGC) across 8,961 biallelic variants comprised of 5,863 directly genotyped biallelic SNPs and 3,098 surrogate biallelic variants encoding multiallelic SNPs, indels, classical HLA alleles and amino acid changes. Association testing was then undertaken as described for stage 1 for FEV1 and FEV1/FVC.
Effects of lung function associated variants on other traits
To identify whether the novel and previously reported lung function-associated variants had been reported in previous GWAS as associated with traits other than lung function and COPD, we queried the GWAS Catalog56 (last update: 13/03/2016, downloaded on 17/03/16) and GRASP57 (v2.0, downloaded on 17/03/16) for genome-wide significant (P<5x10-8) signals using the 95% credible set (if calculated) or all proxy SNPs (r2>0.8) within 2Mb of the top variant in our data.
Clinical relevance – COPD susceptibility and risk of COPD exacerbations in European and Chinese populations
The effect on COPD susceptibility of up to 95 out of the 97 lung function-associated signals was tested in the COPD study at deCODE Genetics (deCODE COPD study) (1,964 COPD cases and 142,262 controls for single-variant analyses and 1,248 COPD cases and 74,700 controls for risk score analyses), in three lung resection studies: Groningen, Laval and UBC (310 COPD cases and 332 controls), in the following COPD case-control studies: COPDGene Study (2,812 COPD cases and 2,534 controls), Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) (1,736 COPD cases and 176 controls), National Emphysema Treatment Trial (NETT) and Normative Aging Study (NAS) (NETT/NAS, 376 COPD cases and 435 controls) and the Norway GenKOLS study (Genetics of Chronic Obstructive Lung Disease) (854 cases and 805 controls), in the following eMR studies: Mount Sinai BioMe Biobank (BioMe, 207 COPD cases and 1,817 controls) and Geisinger-Regeneron DiscovEHR Study (DiscovEHR, 1,280 COPD cases and 13,321 controls for single-variant analyses and 1,264 COPD cases and 13,032 controls for risk score analyses), and in UK Biobank (not including UK BiLEVE samples, 984 cases and 26,561 controls in total) and UK BiLEVE (9,563 moderate-severe cases, 27,387 controls). rs7050036, located in chromosome X, and chr12:114743533, with MAF= 0.15%, were not present in most studies and therefore were excluded from these analyses, bringing the 97 signals to 95. Of the 95 signals, 47 signals were previously discovered independently of UK BiLEVE and were tested for association using all available COPD cases and controls (20,086 COPD cases and 215,630 controls). The remaining 48 signals were discovered using UK BiLEVE data and so were tested for association using 10,523 COPD cases and 188,243 controls (UK BiLEVE excluded). The effect on risk of COPD exacerbation was additionally tested in the Lung Health Study (LHS) (100 COPD exacerbation cases and 4,002 COPD controls) as well as subsets of UK Biobank (including UK BiLEVE, 647 cases and 9,900 controls), COPDGene (557 cases and 2,255 controls), ECLIPSE (278 cases and 1,458 controls), NETT/NAS (87 cases and 277 controls), GenKOLS (120 cases and 734 controls), BioMe (8 cases and 199 controls) and DiscovEHR (774 cases and 472 controls). Analyses of the effect of lung function variants on COPD susceptibility and on risk of COPD exacerbations in a Chinese ancestry population were undertaken using the China Kadoorie Biobank prospective cohort (CKB) within which data were available for 71 (single variant analyses) or 70 (risk score analyses) of the 95 variants (or proxies) for analyses of COPD susceptibility (7,116 COPD cases and 20,919 controls) and risk of COPD exacerbation (5,292 cases and 1,824 controls). Further details of all studies, including case and control definitions are in the Supplementary Note and Supplementary Table 20.
To test the single variant associations with COPD susceptibility and risk of exacerbation, logistic regression using age, age2, sex, and height as covariates (unless otherwise indicated, Supplementary Note) and assuming an additive genetic effect was used. To test the joint effect of these variants, risk alleles in the subset of the 95 signals with data available in each study (from 86 to 95) were summed to create an unweighted genetic risk score and logistic regression was used to test the effect of the risk score, as a continuous variable, on COPD status and COPD exacerbation status (adjusted for age, age2, sex and height, unless otherwise indicated, Supplementary Note). Results, both from single variant and risk scores, were meta-analysed separately for studies where similar study design and phenotyping was used: eMR, case-control and lung resection, and results were also meta-analysed across studies. Inverse variance weighted meta-analysis was used. In CKB, analyses were adjusted for sex, age, age2, height, region (n=10) and disease status (n=5) and final results were GC-corrected based on genome-wide inflation estimates. Heterogeneity was tested using I2(ref 58).
We calculated odds ratios for spirometrically-defined COPD for weighted risk score deciles in UK Biobank (incorporating UK BiLEVE, 10,547 cases, pre-bronchodilator % predicted FEV1<80% and FEV1/FVC<0.7, and 53,948 controls, FEV1/FVC>0.7 and % predicted FEV1>80%). The weighting of the risk score was undertaken using COPD logOR calculated in studies free of winner’s curse bias (Supplementary Table 21). We scaled the logOR, so that the weights added up to 95.
Population attributable risk fraction calculation
The population attributable risk fraction (PARF) was calculated using the formula below
where P(E) is the probability of the exposure, in this case the probability of having more risk alleles than those in the lowest decile of the risk score (P(E) =0.9), and the OR refers to the odds of having COPD for individuals in deciles 2 to 10 of the risk score compared to the odds of having COPD for individuals in the lowest decile (decile 1) of the risk score. The ORs were calculated separately in ever and heavy-smokers using a logistic regression adjusted for age, age2, sex, height and the first 10 ancestry principal components, and an additional pack-years adjustment for heavy-smokers, and were then meta-analysed using inverse variance weighting. Confidence intervals were estimated using the formula above with the lower and upper bound of the meta-analysed OR estimated by logistic regression. These analyses were run using UK Biobank data and the COPD case definition described above: individuals with % predicted FEV1<80% and FEV1/FVC<0.7 were selected as COPD cases and those with FEV1/FVC>0.7 and % predicted FEV1>80% were selected as controls.
Implication of causal genes
In order to implicate the likely causal gene (or genes) for each of the novel and previously-reported signals (97 in total), we employed functional annotation and analysis of gene expression data. All variants within 25kb, variants within 500kb and with r2>0.5 of the top SNP at each signal and variants within 1Mb and with r2>0.8 with the top SNP were annotated using ENSEMBL’s Variant Effect Predictor (VEP). A variant was labelled as deleterious if it was a missense coding variant that was annotated as ‘deleterious’ by SIFT, ‘probably damaging’ or ‘potentially damaging’ by PolyPhen-2, had a CADD scaled score ≥ 20 (CADD_PHRED ≥ 20), or had a GWAVA score > 0.5. The deleterious variants were each, in turn, included as a covariate in the association analysis for the top SNP. If inclusion of the deleterious variant as a covariate reduced the association signal for the top SNP such that P>0.01, that deleterious variant was deemed to explain part of the signal. If annotation (e.g. a coding variant) implicated a specific gene, then the gene was classified as a high-priority gene for the relevant signal.
At each signal, the sentinel SNP and top proxies with r2>0.4 and within 2Mb, no limit on number of proxies, were used to query 3 eQTL resources; lung eQTL23,24,59, blood eQTL60 and GTEx61 (artery (aorta and tibia), adrenal gland, colon sigmoid, esophagus (gastroesophageal junction and mucosa), transformed fibroblasts, lung, spleen, skin (sun exposed lower leg), stomach, testis, thyroid, whole blood). A False Discovery Rate (FDR) of 10% was used as a threshold for significance in the lung and blood eQTL datasets and 5% in GTEx (due to large number of different tissues and cells, and small sample size). A gene was classified as a potential causal gene if the sentinel SNP or proxy (r2>0.4) showed significant evidence of being an eQTL signal for that gene. Genes were further classified as high-priority genes if the variant most strongly associated with the lung function traits (or a proxy with r2>0.9) was also the variant most strongly associated with expression of the gene in one or more of the eQTL datasets (i.e. there was co-localisation of the lung function associated SNP and the gene expression associated SNP). Due to extended linkage disequilibrium across the MHC region, only high-priority genes were identified for the signals in the MHC.
Pathway analyses
The genes implicated for each signal (high-priority genes only and all genes) were tested for enrichment of gene sets and pathways using ConsensusPathDB62. Pathways or gene sets represented entirely by genes implicated by the same association signal were excluded. Pathways or gene sets represented by 2 or more genes from the same association signal were flagged. Pathway enrichment using all genome-wide P values was undertaken using MAGENTA63 as previously described15. Gene sets/pathways with FDR<5% either including the HLA region or excluding the HLA region were reported.
Tissue specific enrichment of overlap of histone marks
Two methods were used to test for enrichment of the 97 signals of association with lung function for H3K4me1 and H3K4me3 histone marks in up to 127 different tissue and cell types from the ENCODE and RoadMap projects39.
First, enrichment was investigated using a hypergeometric test (as previously described39) using SNPs from the GWAS Catalog (hg19, downloaded 02/11/2015) as background. The GWAS Catalog was pruned within each contributing GWAS study to retain only SNPs that were at least 1Mb apart within that study resulting in 18,202 SNPs for further analysis. BEDtools was used to calculate overlap with precomputed “gapped peaks” for H3K4me1 and H3K4me3 histone marks and a hypergeometric test was used to test the significance of enrichment of the 97 lung function variants compared to the background of GWAS Catalog SNPs. Control for multiple testing was undertaken by picking 97 random variants from the pruned GWAS Catalog and repeating the enrichment computation. FDR was calculated from 10,000 randomizations and FDR=10% was used as a threshold.
The second method used, GoShifter, calculates overlap enrichment against a null distribution generated by locally shifting annotations64. Linkage disequilibrium was calculated using the stage 1 population. Precomputed “narrow peaks” for H3K4me1 and H3K4me3 histone marks from the Roadmap project were used. Tissues/cell types with overlap enrichment P<0.05 are reported.
Druggability
We searched the ChEMBL database (v21, last update: 01/02/2016, downloaded on 11/02/16) to identify whether any of the implicated genes encoded proteins that were targets for approved drugs, or drug compounds in development. We additionally searched for genes predicted to interact (parameters: STRING score ≥0.90; maximum of 10 interactions per gene) with each of the high-priority genes32.
Supplementary Material
Acknowledgements
This work was funded by a Medical Research Council (MRC) strategic award to M.D. Tobin, I.P. Hall, D. Strachan and L.V. Wain (MC_PC_12010). This research has been conducted using the UK Biobank Resource under Application Number 648. This article presents independent research funded partially by the National Institute for Health Research (NIHR). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. This research used the ALICE and SPECTRE High Performance Computing Facilities at the University of Leicester. Additional acknowledgements and funding can be found in the Supplementary Information.
Footnotes
URLs
UK Biobank genetic data release http://www.ukbiobank.ac.uk/scientists-3/genetic-data/
LD Score regression, Broad Institute http://www.broadinstitute.org/~bulik/eur_ldscores/
Author Contributions
L.V.W., D.J.P., M.J., A.L.J., N.J.W., J.F.W., B.S., H.S., N.M.P., S.K., C.G., I.J.D., I.Rudan, S.M.K., O.P., M.K., C.H., T.L., O.T.R., A.J.H., C.E.P., P.D.S., A.G., P.S.B., J.D.C., T.H.B., N.N.H., R.A.M., I.Ruczinski, K.C.B., Y.B., P.J., P.D.P., D.D.S., K.H., E.P.B., R.JF.L., R.G.W., Z.C., I.Y.M., L.L., E.Z., I.Sayers, D.P.S., I.P.H. and M.D.T. contributed to the conception and study design.
L.V.W., N.S., M.S., A.M.E., B.N., L.B., M.O., A.P.H., M.A.P., R.J.H., C.K.B., T.L.R., A.G.F., C.J., T.B., V.E.J., R.J.A., B.P.P., A.C., M.W., J.H., J.Z., P.K.J., B.S., R.R., M.I., N.M.P., S.E.H., J.M., S.E., I.Surakka, V.V., C.H., T.L., D.M.E., C.A.W., E.S.W., R.B., B.D.H., A.A.L., D.W.S., M.v.d.B, C.Brandsma, D.C.N., O.G., F.E.D., S.E.B., D.J.C., H.L.K., S.J., G.Thorleifsson, I.J., T.G., K.S., C.S., G.N., R.G.W., J.V., O.P.K., M.H.C., E.K.S., G.Trynka and D.P.S. contributed to data analysis.
L.V.W., N.S., M.S., A.M.E., B.N., M.O., A.P.H., M.A.P., R.J.H., C.K.B., T.L.R., A.G.F., C.J., V.E.J., A.C., M.J., B.S., R.R., H.S., M.I., N.M.P., S.K., C.G., C.H., A.G., C.S., G.N., R.JF.L., A.L.H., C.Brightling, I.Sayers, A.P.M., D.P.S., I.P.H. and M.D.T. contributed to data interpretation.
Competing Financial Interests Statement
Frederick E Dewey and Shannon E Bruse are employed by Regeneron Pharmaceuticals. David C Nickle is employed by Merck. In the past three years, Edwin K. Silverman received honoraria and consulting fees from Merck, grant support and consulting fees from GlaxoSmithKline, and honoraria and travel support from Novartis. Stefan Jonsson, Gudmar Thorleifsson, Ingileif Jonsdottir, and Kari Stefansson are employed by deCODE Genetics/Amgen. Michael Cho receives grant funding from GlaxoSmithKline.
References
- 1.Fletcher C, Peto R. The natural history of chronic airflow obstruction. Br Med J. 1977;1:1645–8. doi: 10.1136/bmj.1.6077.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lange P, Celli B, Agusti A. Lung-Function Trajectories and Chronic Obstructive Pulmonary Disease. N Engl J Med. 2015;373:1575. doi: 10.1056/NEJMc1510089. [DOI] [PubMed] [Google Scholar]
- 3.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salvi SS, Barnes PJ. Chronic obstructive pulmonary disease in non-smokers. Lancet. 2009;374:733–43. doi: 10.1016/S0140-6736(09)61303-9. [DOI] [PubMed] [Google Scholar]
- 5.Melen E. Bridging genetics, epidemiology, and respiratory medicine. Am J Respir Crit Care Med. 2014;190:716–8. doi: 10.1164/rccm.201408-1524ED. [DOI] [PubMed] [Google Scholar]
- 6.Svanes C, et al. Early life origins of chronic obstructive pulmonary disease. Thorax. 2010;65:14–20. doi: 10.1136/thx.2008.112136. [DOI] [PubMed] [Google Scholar]
- 7.ATS/ERS Task Force for COPD Research. An official American Thoracic Society/European Respiratory Society statement: research questions in COPD. Eur Respir J. 2015;45:879–905. doi: 10.1183/09031936.00009015. [DOI] [PubMed] [Google Scholar]
- 8.Pillai SG, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho MH, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–2. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho MH, et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–25. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock DB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loth DW, et al. Genome-wide association analysis identifies six new loci associated with forced vital capacity. Nat Genet. 2014;46:669–77. doi: 10.1038/ng.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Repapi E, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soler Artigas M, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–90. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soler Artigas M, et al. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat Commun. 2015;6:8658. doi: 10.1038/ncomms9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wain LV, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–81. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–60. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 18.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol. 2012;30:317–20. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 19.Palmer LJ, et al. Familial aggregation and heritability of adult lung function: results from the Busselton Health Study. European Respiratory Journal. 2001;17:696–702. doi: 10.1183/09031936.01.17406960. [DOI] [PubMed] [Google Scholar]
- 20.Wilk JB, et al. Evidence for major genes influencing pulmonary function in the NHLBI family heart study. Genet Epidemiol. 2000;19:81–94. doi: 10.1002/1098-2272(200007)19:1<81::AID-GEPI6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Weiss ST. Lung function and airway diseases. Nat Genet. 2010;42:14–6. doi: 10.1038/ng0110-14. [DOI] [PubMed] [Google Scholar]
- 22.Gudbjartsson DF, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47:435–44. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 23.Hao K, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamontagne M, et al. Refining susceptibility loci of chronic obstructive pulmonary disease with lung eqtls. PLoS One. 2013;8:e70220. doi: 10.1371/journal.pone.0070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obeidat M, et al. GSTCD and INTS12 Regulation and Expression in the Human Lung. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0074630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regan EA, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vestbo J, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) Eur Respir J. 2008;31:869–73. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 28.Fishman A, et al. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348:2059–73. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 29.Bell B, Rose CL, Damon H. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. Aging Hum Dev. 1972;3:5–17. [Google Scholar]
- 30.Dewey FE, et al. Inactivating Variants in ANGPTL4 and Risk of Coronary Artery Disease. N Engl J Med. 2016;374:1123–33. doi: 10.1056/NEJMoa1510926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Global Initiative for Chronic Obstructive Lung Disease. Global Strategy for the Diagnosis Management and Prevention of COPD. 2015 http://goldcopd.org/
- 32.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilk JB, et al. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. American Journal of Respiratory and Critical Care Medicine. 2012;186:622–632. doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Z, et al. A Chronic Obstructive Pulmonary Disease Susceptibility Gene, FAM13A, Regulates Protein Stability of beta-catenin. Am J Respir Crit Care Med. 2016 doi: 10.1164/rccm.201505-0999OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obeidat M, et al. Molecular mechanisms underlying variations in lung function: A systems genetics analysis. The Lancet Respiratory Medicine. 2015;3:782–795. doi: 10.1016/S2213-2600(15)00380-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter RI, Ungurs MJ, Mumford RA, Stockley RA. Aalpha-Val360: a marker of neutrophil elastase and COPD disease activity. Eur Respir J. 2013;41:31–8. doi: 10.1183/09031936.00197411. [DOI] [PubMed] [Google Scholar]
- 37.Sand JM, et al. Accelerated extracellular matrix turnover during exacerbations of COPD. Respir Res. 2015;16:69. doi: 10.1186/s12931-015-0225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jhingran A, et al. Compartment-specific and sequential role of MyD88 and CARD9 in chemokine induction and innate defense during respiratory fungal infection. PLoS Pathog. 2015;11:e1004589. doi: 10.1371/journal.ppat.1004589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cogan JD, et al. Rare variants in RTEL1 are associated with familial interstitial pneumonia. Am J Respir Crit Care Med. 2015;191:646–55. doi: 10.1164/rccm.201408-1510OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 42.Hobbs B, et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017 doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soler Artigas M, et al. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am J Respir Crit Care Med. 2011;184:786–95. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poole C. A history of the population attributable fraction and related measures. Ann Epidemiol. 2015;25:147–54. doi: 10.1016/j.annepidem.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 45.Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur J Hum Genet. 2016;24:1202–5. doi: 10.1038/ejhg.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller MR, et al. Standardisation of spirometry. Eur Respir J. 2005;26:319–38. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 47.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nature Communications. 2015;6 doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walter K, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–89. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilk JB, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. S1–3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dastani Z, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wakefield J. A Bayesian Measure of the Probability of False Discovery in Genetic Epidemiology Studies. The American Journal of Human Genetics. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van de Bunt M, et al. Evaluating the Performance of Fine-Mapping Strategies at Common Variant GWAS Loci. PLoS Genet. 2015;11:e1005535. doi: 10.1371/journal.pgen.1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia X, et al. Imputing Amino Acid Polymorphisms in Human Leukocyte Antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eicher JD, et al. GRASP v2.0: an update on the Genome-Wide Repository of Associations between SNPs and phenotypes. Nucleic Acids Res. 2015;43:D799–804. doi: 10.1093/nar/gku1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Obeidat M, et al. GSTCD and INTS12 regulation and expression in the human lung. PLoS One. 2013;8:e74630. doi: 10.1371/journal.pone.0074630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Segrè AV, et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trynka G, et al. Disentangling the Effects of Colocalizing Genomic Annotations to Functionally Prioritize Non-coding Variants within Complex-Trait Loci. Am J Hum Genet. 2015;97:139–52. doi: 10.1016/j.ajhg.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The stage 1 (UK BiLEVE) genome-wide association results for FEV1, FVC and FEV1/FVC are available from UK Biobank at http://www.ukbiobank.ac.uk/. The sources of all other data utilised in this study can be found in the Online Methods and Supplementary Note.