Abstract
Mammalian SIRT7 is a member of the sirtuin family that regulate multiple biological processes including genome stability, metabolic pathways, stress responses, and tumorigenesis. SIRT7 has been shown to be important for ribosome biogenesis and transcriptional regulation. SIRT7 knockout mice exhibit complications associated with fatty liver and increased aging in hematopoietic stem cells. However, the molecular basis for its biological function remains unclear, in part due to the lack of efficient enzymatic activity in vitro. Previously we have demonstrated that double-stranded DNA could activate SIRT7’s deacetylase activity in vitro, allowing it to deacetylate H3K18 in the context of chromatin. Here we show that RNA can increase the catalytic efficiency of SIRT7 even better and that SIRT7 can remove long chain fatty acyl groups more efficiently than removing acetyl groups. Truncation and mutagenesis studies revealed residues at both the amino and carboxyl termini of SIRT7 that are involved in RNA-binding and important for activity. RNA immunoprecipitation-sequencing (RIP-seq) identified ribosomal RNA (rRNA) as the predominant RNA binding partners of SIRT7. The associated RNA was able to effectively activate the deacetylase and defatty-acylase activities of SIRT7. Knockdown of SIRT7 increased the lysine fatty acylation of several nuclear proteins based on metabolic labeling with an alkyne-tagged fatty acid analog, supporting that the defatty-acylase activity of SIRT7 is physiologically relevant. These findings provide important insights into the biological functions of SIRT7, as well as an improved platform to develop SIRT7 modulators.
Sirtuins are a family of enzymes possessing nicotinamide adenine dinucleotide (NAD)-dependent protein lysine deacetylase activities1–3. There are seven mammalian sirtuins (SIRT1-SIRT7)4, which regulate a variety of biological processes such as metabolism, gene transcription and lifespan extension5, 6. Although all sirtuins have a conserved catalytic core domain, emerging evidence demonstrates that they exhibit varying catalytic activities towards different acyl lysine modifications. SIRT1-3 display robust deacetylase activities, whereas SIRT4-7 have only weak deacetylase activities in vitro. SIRT5 and SIRT6, two of the four sirtuins with weak deacetylase activities, preferentially hydrolyze succinyl/malonyl/glutaryl7, 8 and long-chain fatty acyl lysine9, respectively. These studies demonstrate that sirtuins are able to hydrolyze a variety of lysine acyl modifications and regulate a broad range of cellular events.
As one of the least understood human sirtuins, SIRT7 is normally concentrated in nucleoli where it activates transcription of rRNA genes10, 11. Under certain stress, SIRT7 can translocate from nucleoli to nucleoplasm resulting in hyperacetylation of PAF53 (a subunit of Pol I) and thus inhibits Pol I transcription12. By deacetylating histone H3 Lys18 (H3K18), SIRT7 also suppresses mRNA transcription mediated by the Pol II machinery13. Moreover, SIRT7 regulates the transcription of nucleus-encoded mitochondrial biogenesis genes by deacetylating and activating a transcription factor, GABPβ114. Interestingly, three recent studies link SIRT7 to the regulation of liver function in mouse, although different effects of SIRT7 on fatty liver formation are reported14–16.
So far, all the biological functions of SIRT7 have been attributed to its deacetylase activity. However, the in vitro deacetylase activity of SIRT7 in many cases cannot be detected. We have previously shown that double-stranded DNA (dsDNA) can significantly improve SIRT7’s deacetylase activity and allows it to deacetylate H3K18 in the context of chromatin17. However, the in vitro activity on peptide substrates is still rather weak compared to other sirtuins with efficient deacetylase activities. Here we report that RNA can increase the catalytic efficiency of SIRT7 even better. Furthermore, upon RNA activation, SIRT7 can remove long chain fatty acyl groups more efficiently than removing acetyl groups. Both ribosomal RNA (rRNA) and transfer RNA (tRNA) are potent activators of SIRT7 in vitro. The predominant endogenous RNA binding partners of SIRT7 are rRNA, which can efficiently activate SIRT7 in vitro. Mutagenesis study identified key residues at the N- and C- domains of SIRT7 that are important for the interaction with RNA and enzymatic activity. We believe these results will help to better understand the biological functions of SIRT7 and to develop small molecule modulators of SIRT7.
Results and Discussion
SIRT7 deacetylase activity is dramatically enhanced by RNA
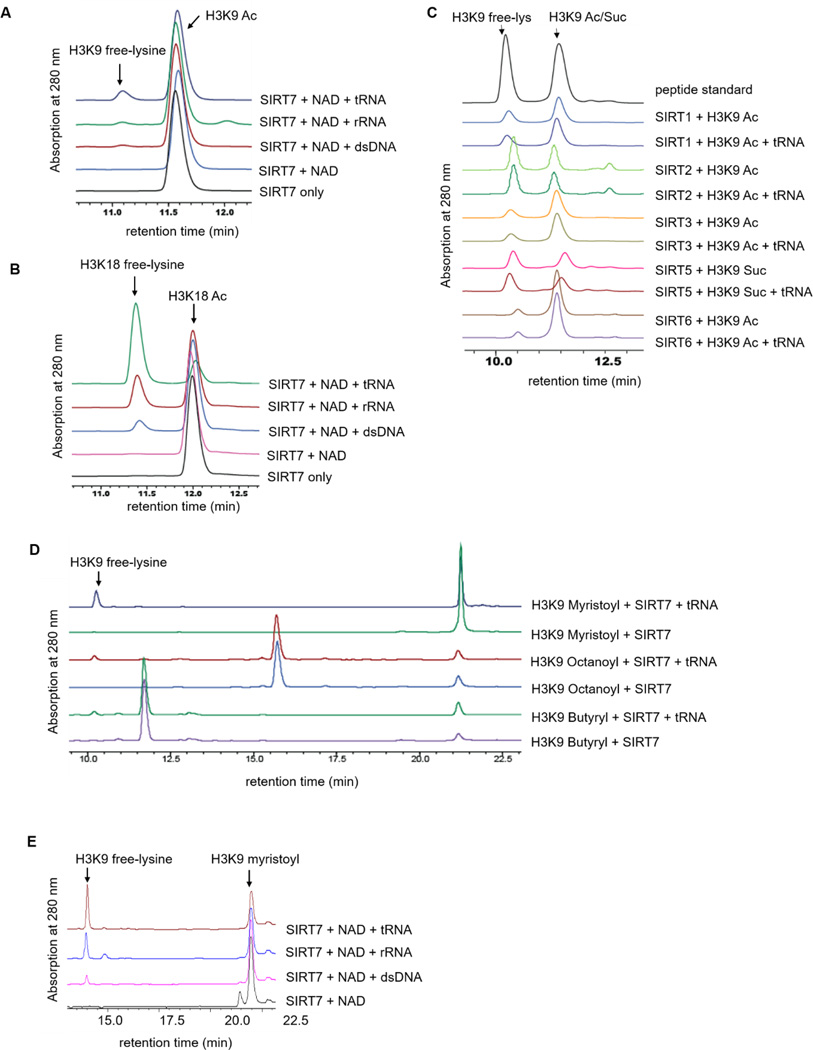
We have recently reported that SIRT7 can be activated by dsDNA17, which explains its role as a H3K18 deacetylase in regulating transcription. SIRT7 is also involved in regulating rRNA gene transcription, ribosome biogenesis, and protein synthesis10, 18, 19, which are also thought to be mediated by its deacetylase activity. It is not clear how SIRT7’s catalytic activity would be stimulated when functioning to regulate ribosome biogenesis and protein synthesis. We wondered whether RNA species could also activate SIRT7. To test this hypothesis, we detected the deacetylase activity of SIRT7 on H3K9 and H3K18 acetyl (H3K9 Ac and H3K18 Ac) peptides with different nucleic acids (dsDNA, rRNA, and tRNA). In the absence of nucleic acid, no product was formed. In the presence of either DNA or RNA, the deacetylated H3K18 peptide was detected (Figure 1A & 1B). Interestingly, among all the nucleic acids we tested in vitro, tRNA turned out to be the most potent activator of SIRT7. Interestingly, RNA was unable to activate any of the other sirtuin proteins (Figure 1C). Therefore, among all the seven human sirtuins, SIRT7 is unique in that its enzymatic activity can be activated by both DNA and RNA on peptide substrates.
Figure 1.
SIRT7 deacylase activity is significantly increased by RNA. (A) Overlaid HPLC traces showing SIRT7-catalyzed hydrolysis of H3K9 acetyl (Ac) peptide with different nucleic acids. (B) HPLC traces showing SIRT7-catalyzed hydrolysis of H3K18 acetyl (Ac) peptide with nucleic acids. (C) Overlaid HPLC traces showing that other sirtuin proteins cannot be activated by tRNA. We cannot acquire recombinant SIRT4 from E.coli. (D) SIRT7 can hydrolyze fatty acyl groups with different carbon chain length from lysine residues. (E) SIRT7 demyristoylase activity is dramatically activated by rRNA and tRNA.
SIRT7 exhibits efficient defatty-acylase activity upon activation by RNA
It was recently reported that defatty-acylation (removing long chain fatty acyl groups) is an intrinsic activity of several sirtuins9, 20, 21. In particular, SIRT6, which is also a class IV sirtuin like SIRT7, can remove long chain fatty acyl groups more efficiently in vitro. Therefore, we investigated whether SIRT7 could also remove long chain fatty acyl groups. We tested the activity of recombinant human SIRT7 on H3K9 peptides with different acyl groups (butyryl, octanoyl, and myristoyl). In the absence of RNA, no deacylation product was detected (except very little product that was observed when using the H3K9 myristoyl peptide) (Figure 1D). In the presence of tRNA, SIRT7 was able to catalyze the hydrolysis of all three H3K9 acyl peptides. Among the three acyl peptides, the hydrolysis of myristoyl lysine was the most efficient (Figure 1D & 1E), suggesting that SIRT7 has a preference for long chain fatty acyl groups similar to its closest sirtuin member, SIRT69.
To quantitatively compare the effects of tRNA activation, we determined the turnover number (kcat) and Michaelis constant (Km) of SIRT7 on H3K9 peptides with different acyl groups (Table 1). The molar ratio of tRNA to SIRT7 was fixed at 3:1 (Figure S1). The kcat/Km for demyristoylation with tRNA (41 M−1s−1) was approximately 40-fold better than that for deacetylation (1.1 M−1s−1) on H3K9 peptides. tRNA improved the catalytic efficiency of SIRT7 for demyristoylation nearly 40-fold, which was achieved mainly by increasing kcat (25-fold), as well as decreasing Km slightly (1.6-fold). Similarly, tRNA improved the catalytic efficiency of SIRT7 on H3K18 acyl peptides (Table 1).
Table 1.
Catalytic efficiencies of SIRT7 on H3 acyl peptides with different nucleic acids
| Acyl peptide | activator | kcat (min−1) | Km (µM) | kcat/Km (M−1s−1) |
|---|---|---|---|---|
| H3K18 Ac | - | NP | NP | NP |
| tRNA | 1.2 ± 0.2 | 309 ± 59 | 65 | |
| H3K18 Myr | - | NP | NP | NP |
| tRNA | 0.02 ± 0.002 | <2 | >167 | |
| H3K9 Ac | - | NP | NP | NP |
| dsDNA | ND | ND | 1.0 | |
| tRNA | ND | ND | 1.0 | |
| H3K9 Myr | - | 0.004 ± 0.001 | 65 ±26 | 1.1 |
| dsDNA | 0.04 ± 0.005 | 32 ±10 | 21 | |
| tRNA | 0.1 ± 0.01 | 41 ± 7 | 41 | |
| H3K9 But | tRNA | ND | ND | 2.0 |
| H3K9 Oct | tRNA | ND | ND | 2.0 |
ND: Not determined because V~[S] was linear. Only kcat/Km value could be obtained.
NP: No product formed (product formation was below HPLC detection limit).
Ac: acetyl; But: butyryl; Oct: octanoyl; Myr: myristoyl.
Sirt7 knockdown increased global lysine fatty-acylation level
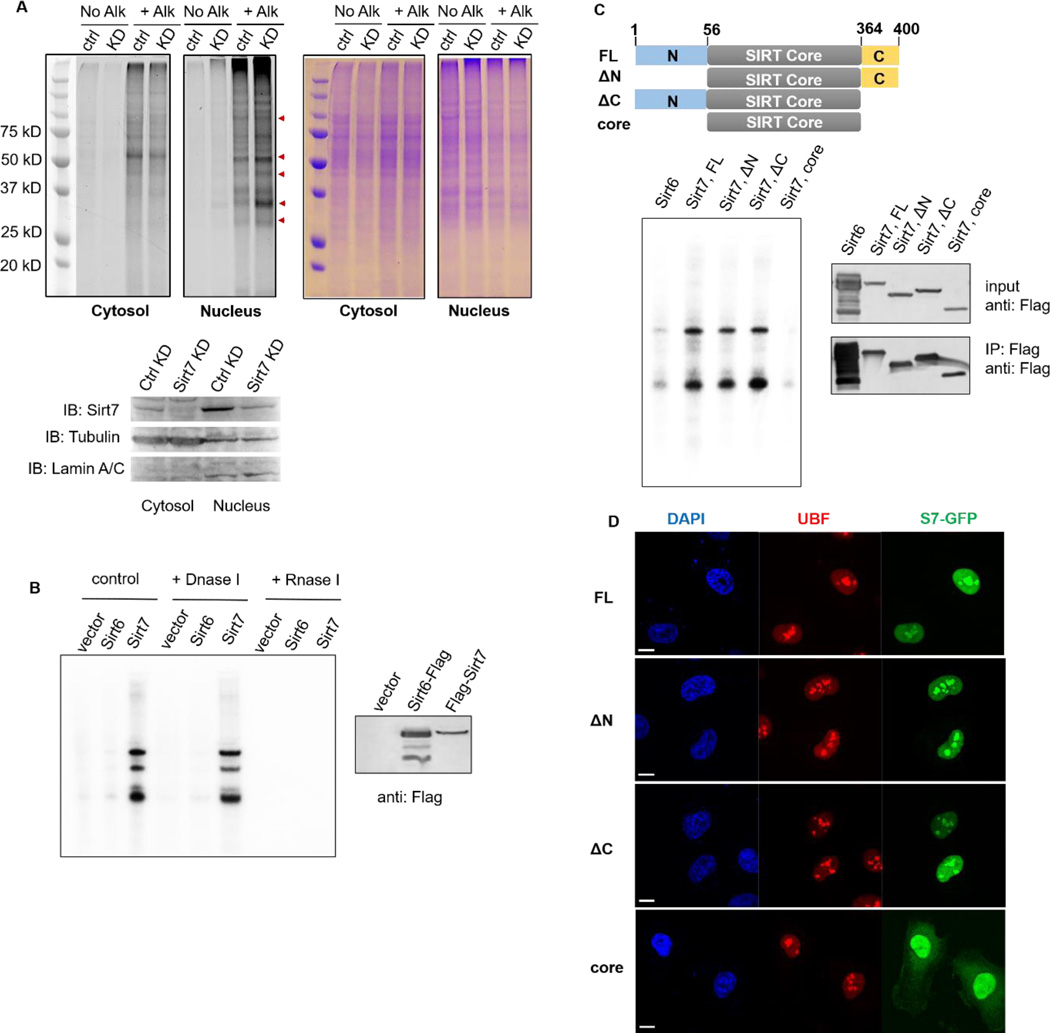
To investigate whether the defatty-acylase activity of SIRT7 is physiologically relevant, we examined whether global lysine fatty-acylation is regulated by SIRT7. We transiently knocked down Sirt7 expression in HEK 293T cells using short hairpin RNA (shRNA) (Figure 2A). Both control and Sirt7 knockdown (Sirt7 KD) cells were cultured in the presence of Alk14, an alkyne-tagged fatty acid analogue. Then equal amounts of cell lysates were conjugated to BODIPY-azide (BODIPY-N3) using click chemistry to detect proteins modified by Alk14. Hydroxylamine was used to remove cysteine fatty acylation before resolving samples by gel electrophoresis. As shown in Figure 2A, nuclear fractions of Sirt7 KD cells had increased global lysine fatty-acylation level (red arrows) compared to control cells, whereas cytosolic fractions of Sirt7 ctrl and KD cells exhibited similar levels of global lysine fatty-acylation. The data suggested that SIRT7 regulates global lysine fatty acylation level, especially in the nucleus where SIRT7 is primarily located.
Figure 2.
SIRT7 regulates global lysine fatty acylation level and associates with RNA in mammalian cells. (A) Sirt7 KD increased global lysine fatty acylation level in HEK 293T cells. Alk: Alkyne-14; ctrl: control; KD: knockdown. (B) SIRT7 pulled down endogenous RNA species. SIRT6 was used as a negative control. (C) SIRT7 core domain did not bind endogenous RNA. Scheme showing SIRT7 full-length (FL: 1-400), N-terminal deletion (ΔN: 56-400), C-terminal deletion (ΔC: 1-364), and core domain (56-364). (D) SIRT7 core domain was no longer localized in nucleoli and dispersed into nucleoplasm and cytoplasm. Scale bar: 10 μm.
SIRT7 associates with RNA in mammalian cells
Previous studies have demonstrated that SIRT7 interacts with transcription factors, such as ELK4 and MYC, and almost exclusively resides in a chromatin-enriched fraction13, 16. We believe in this scenario, DNA acts as the activator of SIRT7, which hydrolyzes acetylated H3K18 and suppresses transcription. We had thus far revealed that RNA species (rRNA and tRNA) can serve as effective activators for SIRT7 in vitro. Therefore, we asked whether SIRT7 directly binds RNA in mammalian cells. To address this question, we stably overexpressed Flag-tagged SIRT7 in HEK 293T cells and performed RNA immunoprecipitation (RIP). Empty vector and Flag-tagged SIRT6 were used as negative controls. Co-immunoprecipitated RNA was labeled with [32P]pCp at the 3’-end and further resolved on the denaturing gel. As shown in Figure 2B, Flag-SIRT7 was able to pull down several major RNA species, which were resistant to DNase treatment but were completely degraded by RNase. In contrast, very little RNA was pulled down by Flag-SIRT6.
To dissect which domain(s) of SIRT7 interacts with RNA, we expressed Flag-tagged SIRT7 truncation mutants (ΔN 56-400, ΔC 1-364, and core domain 56-364, Figure 2C) and conducted CLIP (UV-crosslinking and immunoprecipitation of RNA) experiments and further labeled the 3’-end of immunoprecipitated RNA with [32P]pCp (Figure 2C). Interestingly, SIRT7 core domain (56-364) was incapable of pulling down any RNA species while both ΔN and ΔC were still able to pull down RNA. Thus, both the N- and C-termini of SIRT7 contribute to its interaction with endogenous RNA.
We also transiently overexpressed GFP-tagged SIRT7 truncation proteins and examined their localization in Hela cells. FL, ΔN, ΔC were all localized in the nucleus with enrichment in nucleoli as indicated by co-localization with UBF. In contrast, the core domain of SIRT7 did not show nucleoli-enriched localization and some SIRT7 molecules were also localized in the cytosol (Figure 2D). These results support that the amino and carboxyl termini are needed for both RNA binding and proper localization of SIRT7.
SIRT7 mainly interacts with rRNA in vivo
In order to characterize the repertoire of RNAs that are associated with SIRT7 in vivo, we sequenced the RNAs co-immunoprecipitated with Flag-tagged SIRT7 using RIP-seq (RNA immunoprecipitation sequencing). As summarized in Figure S2A, in the cytosol, 99% of the RNA binding partners of SIRT7 turned out to be rRNA and similarly in the nucleus, 97% of the RNAs associated with SIRT7 were rRNAs, consistent with the reported roles of SIRT7 involved in pre-rRNA transcription and processing10, 19. We further mapped all the rRNA reads to human pre-rRNA sequences (Figure S3). The majority of SIRT7-interacting rRNA reads corresponded to the transcribed regions of mature rRNA (18S, 5.8S and 28S rRNA), constant with our RIP-32P-labeling results (Figure 2B & Figure S5). Noticeably, we also identified 5S rRNA in the RIP-seq result (Figure S4). The 5S rRNA is transcribed by Pol III. Our result is in agreement with a previous SIRT7 interactome study, which demonstrates that SIRT7 modulates Pol III function by interacting directly with components of the Pol III transcription complex18.
To further analyze the 3% non-rRNA binding partners of SIRT7, we removed rRNA and sequenced the non-rRNA repertoire (from both SIRT-7 RIP RNA and pre-RIP input RNA). The results show that 82% of the non-rRNA pool was represented by mRNA while 11% was non-coding RNA (ncRNA) (Figure S2B). Gene Ontology analysis of SIRT7-bound mRNA suggested that the major biological processes included translation (ribosome protein mRNA cluster) and chromatin assembly (histone mRNA cluster), and the top KEGG pathway was ribosome (Figure S2C). The interaction between SIRT7 and identified mRNA was validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) using gene-specific primers (Figure S2D).
We were also able to identify some tRNA species in the SIRT7 RIP samples (Table S1). The reads were very low probably due to difficulties associated with sequencing of tRNA. The percentage of tRNAs in the SIRT7 RIP sample decreased compared to that in the input RNA repertoire. Therefore, we think that tRNAs are unlikely to be major binding partners of SIRT7 in cells.
SIRT7 can bind to and be activated by rRNA in vitro
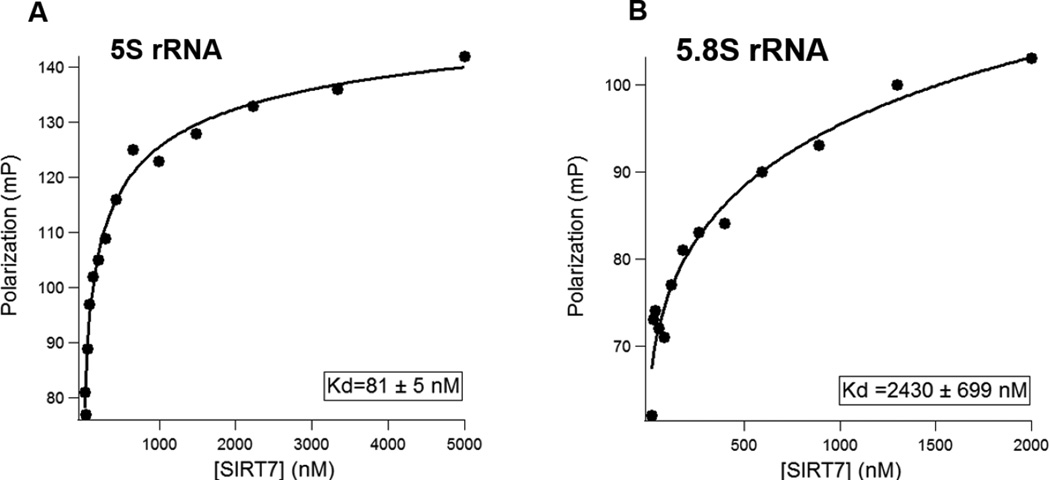
To confirm that SIRT7 directly binds to rRNA, we in vitro transcribed 5S and 5.8S rRNAs, and determined the binding affinities of SIRT7 for 5S and 5.8S rRNA using a fluorescence polarization (FP) assay. In vitro transcribed 5S and 5.8S rRNA were gel-purified and subsequently labeled with fluorescein 5-thiosemicarbazide (FTSC) at the 3’ end. By fixing the rRNA fluorescent probe at 5 nM, we titrated SIRT7 protein concentration from 25 nM to 5000 nM. As plotted in Figure 3A & 3B, the dissociation constants (Kd) of SIRT7 were determined to be 81 nM for 5S rRNA and 2.4 μM for 5.8S rRNA.
Figure 3.
Fluorescence Polarization (FP) assay to determine binding affinity of SIRT7 for 5S and 5.8S rRNA. (A) The dissociation constant (Kd) of SIRT7 for 5S rRNA. (B) The Kd of SIRT7 for 5.8S rRNA. Human full-length 5S and 5.8S rRNA were in vitro transcribed.
We then measured the catalytic efficiencies of SIRT7 on H3K18 Ac and H3K9 myristoyl peptides in the presence of 5S or 5.8S rRNA (Table 2). With H3K18 Ac as the substrate, the catalytic efficiencies of SIRT7 with 5S and 5.8S rRNA were 6–7-fold lower than those with tRNA (11.4 and 9.4 M−1s−1 versus 65 M−1s−1). In contrast, with H3K9 myristoyl as the substrate, the catalytic efficiencies of SIRT7 with 5S and 5.8S rRNA were 2–4-fold better than those with tRNA (163 and 88 M−1s−1 versus 41 M−1s−1). Thus, 5S rRNA and 5.8S rRNA are competent activators for the defatty-acylase activity of SIRT7 in vitro. The fact that 5S rRNA served as a better activator of SIRT7 than 5.8S rRNA is consistent with the higher binding affinity of 5S rRNA for SIRT7.
Table 2.
Catalytic efficiencies of SIRT7 on H3 acyl peptides with 5S and 5.8S rRNA
| Acyl peptide | activator | kcat (min−1) | Km (µM) | kcat/Km (M−1s−1) |
|---|---|---|---|---|
| H3K18 Ac | - | NP | NP | NP |
| 5.8S rRNA | 0.3 ± 0.02 | 532 ± 55 | 9.4 | |
| 5S rRNA | 0.3 ± 0.05 | 440 ± 122 | 11.4 | |
| H3K9 Myr | - | 0.004 ± 0.001 | 65 ±26 | 1.1 |
| 5.8S rRNA | 0.03 ± 0.009 | 5.7 ± 0.6 | 88 | |
| 5S rRNA | 0.05 ± 0.004 | 5.1 ± 1.5 | 163 | |
NP: No product formed (product formation was below HPLC detection limit).
Ac: acetyl; Myr: myristoyl.
Both amino (N-) and carboxyl (C-) termini of SIRT7 are important for its activities
We showed that both N- and C-terminal extensions of SIRT7 are important for its full activation by dsDNA in vitro17. Therefore, we tested whether the N- and C-termini of SIRT7 were also important for its activation by RNA. We purified WT and truncated SIRT7 proteins from E. coli: full-length (FL, 1-400), N-terminal truncation (ΔN, 56-400), C-terminal truncation (ΔC, 1-364), and core domain (56-364) (Figure 2C). Using tRNA as the in vitro activator, we measured the activities of the four SIRT7 constructs on H3K18 Ac peptide. SIRT7 ΔC and the core domain, which lack the C-terminal domain, exhibited no detectable deacetylase activity (Table 3), suggesting that the C-terminus is essential for SIRT7’s deacetylase activity. SIRT7 ΔN still showed some activity in the presence of tRNA, but the turnover number kcat (0.002 s−1) was only 1/10 of FL SIRT7 (kcat=0.02 s−1). Thus, the N-terminus of SIRT7 is also important for the deacetylase activity.
Table 3.
Catalytic efficiencies of SIRT7 truncates on H3K18 Ac and H3K9 Myr peptides
| SIRT7 | Acyl Peptide |
activator | kcat (min−1) | Km (µM) |
kcat/Km (M−1s−1) |
relative catalytic efficiency to FL |
|---|---|---|---|---|---|---|
| FL | H3K18 Ac | - | NP | NP | NP | NA |
| H3K18 Ac | tRNA | 1.2 ± 0.2 | 309 ± 59 | 65 | 1 | |
| H3K9 Myr | - | 0.004 ± 0.001 | 65 ±26 | 1.1 | 1 | |
| H3K9 Myr | tRNA | 0.1 ± 0.01 | 41 ± 7 | 41 | 1 | |
| ΔN | H3K18 Ac | - | NP | NP | NP | NA |
| H3K18 Ac | tRNA | 0.1 ± 0.003 | 294 ± 15 | 5.7 | 0.09 | |
| H3K9 Myr | - | 0.002 ± 0.0002 | 90 ± 16 | 0.4 | 0.36 | |
| H3K9 Myr | tRNA | 0.02 ± 0.002 | 18 ± 4.2 | 19 | 0.46 | |
| ΔC | H3K18 Ac | - | NP | NP | NP | NA |
| H3K18 Ac | tRNA | NP | NP | NP | NA | |
| H3K9 Myr | - | 0.003 ± 0.0003 | 18 ± 4.7 | 2.8 | 2.5 | |
| H3K9 Myr | tRNA | 0.04 ± 0.006 | 25 ± 8.6 | 27 | 0.66 | |
| core | H3K18 Ac | - | NP | NP | NP | NA |
| H3K18 Ac | tRNA | NP | NP | NP | NA | |
| H3K9 Myr | - | 0.001 ± 0.0001 | 89 ± 22 | 0.2 | 0.2 | |
| H3K9 Myr | tRNA | 0.005 ± 0.0003 | 39 ± 4.5 | 2.1 | 0.05 | |
NP: No product formed (product formation was below HPLC detection limit).
NA: Not available.
FL: 1–400; ΔN: 56–400; ΔC: 1–364; core: 56–364.
We also measured the activities of the different SIRT7 constructs on the H3K9 myristoyl peptide (Table 3). Deletion of either N- or C-terminal of SIRT7 decreased the demyristoylase activity of SIRT7, and the core domain had only about 5% of the activity of full length, again suggesting that both the N- and C-termini of SIRT7 are important for SIRT7’s catalytic activities.
Mutagenesis revealed residues of SIRT7 important for RNA-binding and activity
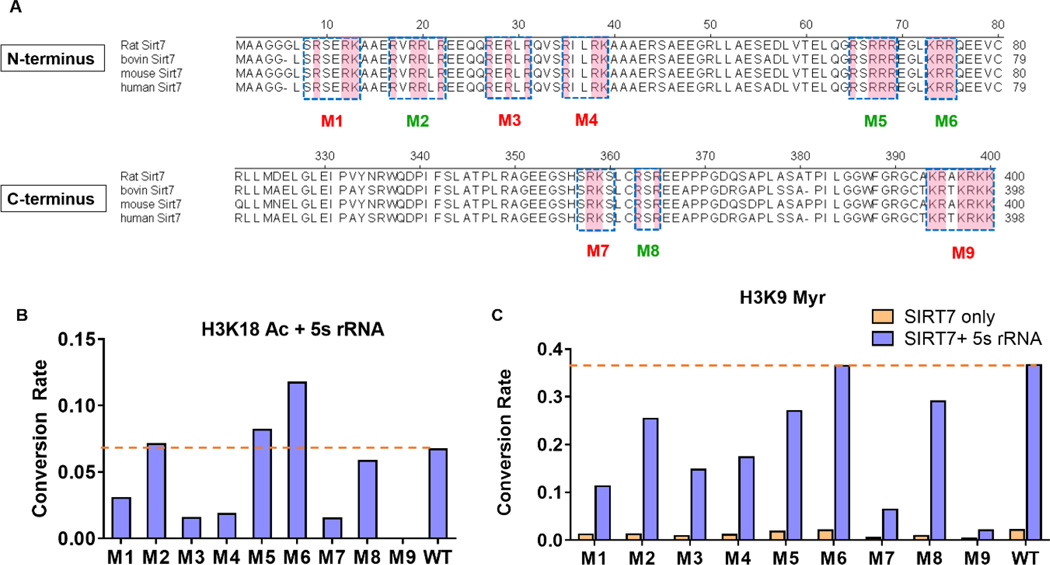
To further identify key residues at the N- and C-termini of SIRT7 that contribute crucially to SIRT7’s interaction with RNA, we first utilized a bioinformatics tool, Bind N (http://bioinfo.ggc.org/bindn/), and identified nine clusters of amino acids located at the N- and C- termini that might be central for binding to nucleic acids. Each cluster contains at least one basic amino acid (Lys or Arg, highlighted in pink in Figure 4A) and is conserved among different species (except for M9, in which one Thr residue is replaced with Ala in some species). We then mutated all the residues in each cluster to alanine to generate mutants M1-M9 (Figure 4A).
Figure 4.
Mutagenesis to identify key residues of SIRT7 involved in binding to RNA. (A) Schematics showing the residues mutated to alanines in the nine SIRT7 mutants. Basic amino acids are highlighted in pink. Red mutants: mutants with significantly decreased enzymatic activity (less than 50% of WT activity); Green mutants: mutants with more than 10-fold decrease in binding affinity but relatively unchanged enzymatic activity; (B) Conversation rates of SIRT7 mutants on H3K18 acetyl (Ac) peptide in the presence of 5S rRNA. SIRT7 (2 μM) was incubated with 100 μM of H3K18 acetyl peptide at 37 °C for 45 minutes with 6 μM of 5S rRNA. (C) Conversation rates of SIRT7 mutants on H3K9 myristoyl (Myr) peptide with 5S rRNA. SIRT7 (2 μM) was incubated with 20 μM of H3K9 myristoyl peptide at 37 °C for 45 minutes with 6 μM of 5S rRNA.
We measured the dissociation constants (Kd) between 5S rRNA and the different SIRT7 mutants using the FP assay (Table 4). All the mutants had increased Kd values. In particular, the Kd values of M2, M3, M4, M5, M6, M8 and M9 for 5S were 20~80-fold higher than that of WT SIRT7. Thus, these clusters are important for binding to 5S RNA.
Table 4.
Summary of SIRT7 WT and mutants’ activities and RNA-binding affinities in the presence of 5S rRNA
| SIRT7 | Kd (nM) | Relative Deacetylation (H3K18 Ac) |
Relative Demyristoylation (H3K9 Myr) |
Fraction of SIRT7 bound with 5SRNA in the assay ** |
|---|---|---|---|---|
| WT | 81 ± 5 | 1.00 | 1.00 | 0.98 |
| M1* | 520 ± 130 | 0.45 | 0.31 | 0.89 |
| M2 | 1600 ± 250 | 1.06 | 0.69 | 0.74 |
| M3* | 3000 ± 570 | 0.23 | 0.40 | 0.62 |
| M4* | 4200 ± 600 | 0.28 | 0.47 | 0.54 |
| M5 | 3800 ± 230 | 1.22 | 0.74 | 0.56 |
| M6 | 4000 ± 1500 | 1.74 | 1.00 | 0.55 |
| M7* | 540 ± 40 | 0.23 | 0.17 | 0.89 |
| M8 | 1600 ± 50 | 0.87 | 0.79 | 0.74 |
| M9* | 6800 ± 1000 | 0.00 | 0.06 | 0.43 |
Mutants with flawed catalytic activities (less than 50% of WT SIRT7) in the presence of 5S rRNA. Conversion Rate of WT SIRT7 was set at 1.00, all the mutants’ conversion rates were normalized using that of WT SIRT7.
SIRT7 (2 µM) was incubated with 6 µM of 5S rRNA. The fraction of SIRT7 bound with 5S rRNA was calculated assumpting that one molecule of SIRT7 associates with one molecule of 5S rRNA.
Using the H3K18 Ac and H3K9 Myr peptides, we screened the activities of these mutants with and without 5S rRNA. With H3K18 Ac as the substrate, since we could not determine the basal activity of SIRT7 without RNA, we only compared the substrate conversion rates of SIRT7 WT and the mutants in the presence of 5S rRNA. With H3K9 Myr as the substrate, we determined the substrate conversion rates with and without 5S RNA. Among the nine mutants, M9 (392KRTKRKK398 mutated to 392AAAAAAA398) showed no detectable deacetylase activity and very little demyristoylase activity, while M1, M3, M4, and M7 exhibited significantly reduced deacetylase and demyristoylase activities in the presence of 5S rRNA (Figure 4B–C & Table 4). In contrast, M2, M5, M6, and M8 largely retained the enzymatic activities in the presence of 5S rRNA. To rule out the possibility that the mutants had decreased protein stability, we examined the stability of SIRT7 WT and mutants at 4°C and 37°C. As summarized in Figure S7, no significant difference in stability was seen for the SIRT7 mutants and WT, suggesting that the defective deacylase activities of M1, M3, M4, M7 and M9 are primarily due to the alteration of residues important for enzymatic activity rather than protein stability.
Considering the fact that mutants with high Kd values might not fully bind with RNA in the assay condition (Figure 4B—C), we calculated the fraction of SIRT7 bound to 5S rRNA (Table 4). For M3 and M4, the decreased activities (~40% of WT) could be explained by the decreased “active fraction” bound with 5S rRNA (50%–60%). In contrast, for M1 and M7 (~90% bound to 5S rRNA under assay conditions), the decreased enzymatic activities (20%-30% of WT) could not be fully explained by decreased binding to 5S rRNA. One possible explanation is that the M1 and M7 mutants may impede SIRT7’s conformational change (which is important for activities) induced by RNA binding. The M9 mutant, with almost no detectable deacetylase and demyristoylase activities in the presence of 5S rRNA, was defective in binding to 5S rRNA (85 fold lower than WT) and also impeded RNA-induced conformational alteration. In contrast, the M5 and M6 mutants, after correcting for the fractions bound to 5S rRNA, had higher activities than WT. Altogether, these results reveal a delicate relationship between the binding of SIRT7 to RNA and the activation by RNA.
In summary, we have identified RNA as a more potent activator of SIRT7 and SIRT7 can efficiently remove long chain fatty acyl groups in vitro. SIRT7 plays important roles in the maintenance of oncogenic transformation13, non-alcoholic fatty liver formation14–16, ribosome biogenesis, and protein synthesis18. Despite the reported functions, the enzymatic activity and physiological substrates of SIRT7 are largely underexplored. SIRT7 has been shown to associate with chromatin to deacetylate histone H313 or transcription factors to regulate transcription16. However, no reports have established the connection between SIRT7’s physiological functions and its interaction with RNA molecules. Here we have demonstrated that SIRT7 interacts predominantly with mature regions of RNA polymerase I-transcribed 45S pre-rRNA (28S, 18S, and 5.8S) and RNA polymerase III-transcribed 5S rRNA. We have further shown that SIRT7 can be effectively activated by in vitro transcribed human 5S and 5.8S rRNA. Recent studies have revealed a multifaceted role of SIRT7 in regulating the transcription and processing of pre-rRNA19. Our finding that a variety of RNA species can dramatically enhance the catalytic efficiency of SIRT7 explains how SIRT7 could function as an efficient deacylase in vivo despite that no appreciable amount of product could be formed in vitro in the absence of nucleic acids in the assay. Our findings suggest that SIRT7 is only activated at the right locations where corresponding DNA or RNA molecules are bound to SIRT7.
Both the N- and C- termini are important for SIRT7’s activity. Subsequent systematic mutagenesis studies revealed that the polybasic sequences (residue 392–398, KRTKRKK) at the very end of SIRT7’s C-terminal region contributed crucially to its binding and activation by RNA. Interestingly, this stretch of polybasic amino acids (residue 392–398, KRTKRKK) has also been previously characterized as a nucleolar localization signal22. Furthermore, SIRT7 nucleoli localization has been linked to its interaction with newly synthesized pre-rRNA23. Thus, this polybasic sequence seems to play a central role in controlling SIRT7’s interaction with RNA, its dynamic subcellular localization, and its catalytic activity.
The regulation of SIRT7 activity by DNA and RNA may be important for further investigation of its role in various stress responses. For example, under normal conditions, SIRT7 may predominantly bind to rRNA and regulate ribosome biogenesis. Under certain stress conditions where rRNA production is inhibited, more SIRT7 may be released from rRNA to bind to other nucleic acids (e. g. DNA or mRNA. Note that certain mRNAs were pulled down by SIRT7) and be activated to work on other substrate proteins, which serves to regulate transcription (when SIRT7 binds to DNA) or translation (when SIRT7 binds to mRNA) in response to stress stimuli. In this way, SIRT7 may act to sense the stress and coordinate changes in ribosomal biogenesis, transcription, and translation. This model needs to be tested in future studies.
Upon RNA activation, SIRT7 shows robust defatty-acylase activity that is more efficient than its deacetylase activity. Thus, similar to SIRT6 which has physiologically relevant deacetylase and defatty-acylase activities, SIRT7 is also likely to be multifunctional. Our global labeling data demonstrated that upon Sirt7 knockdown, lysine fatty acylation levels of several nuclear proteins increased. Protein lysine fatty acylation was first reported over two decades ago, but to date, only a few proteins are reported to have this modification, and the only known biological function is the regulation of protein secretion9,24. However, given that five of the seven human sirtuins possess defatty-acylase activities in vivo or in vitro20, 21, we believe protein lysine fatty acylation is more abundant and important than currently appreciated. Future studies to identify the substrate proteins of SIRT7’s defatty-acylation activity will broaden our understanding of protein lysine fatty acylation and the biological function of SIRT7.
The finding that DNA17 and RNA can significantly activate SIRT7 also opens up opportunities to develop small molecule modulators of SIRT7. Sirtuin inhibitors have been reported to be potential therapeutic candidates for several human diseases.25, 26 SIRT7 inhibitors in particular could be useful for treating cancer.13 So far, no effective SIRT7 inhibitors are available due to the absence of efficient activity assays. With the more efficient activities of SIRT7 in the presence of DNA or RNA, it is now possible to use the activity assay to develop SIRT7 inhibitors.
Methods
SIRT7 purification
Nine full length SIRT7 mutants in pET28a vector were constructed using two-step PCR mutagenesis and all the sequences were confirmed by DNA sequencing. Purification procedures of SIRT7 WT, truncates (FL, ΔN, ΔC and core), and other mutants were the same as previously described17.
Synthesis of acyl peptides
Acyl peptides representing residues 4–13 and 12–24 of Histone H3 (NH2-KQTARKSTGGWW-COOH and NH2-GGKAPRKQLATKAWW-COOH) were synthesized as previously described17.
Acyl modifications (acetyl, butyryl, octanoyl, myristoyl) on lysine residues were introduced using Fmoc-Lys(acyl)-OH. Butyryl, octanoyl and myristoyl peptides were dissolved in DMSO whereas acetyl and free-lysine peptides were dissolved in water.
Activity assays and kinetics measurement for SIRT7
SIRT7 activity assay was set up and detected as previously described17. SUPERase-In™ RNase inhibitor (Thermo) was added to the reaction to prevent RNA degradation. Different nucleic acids were added as the in vitro activators: Salmon sperm DNA (Sigma, cat # D1626), ribosomal RNA from bioPLUS (cat # 11020001), and yeast tRNA from Thermo (cat# AM-7119). Mass ratio of RNA/DNA to SIRT7 was fixed at 3:1 in the assays shown in Figure 1A, 1B and 1E. For all the other experiments, molar ratio of RNA (tRNA, 5S rRNA and 5.8S rRNA) to SIRT7 was maintained at 3:1.
Kinetics experiments were performed as previously depicted17. For kinetics experiment using acetyl peptides as substrates, 2 μM SIRT7 (FL) or 8 μM SIRT7 (ΔN 56-400) was added to the reaction. The acetyl peptide concentrations was 10, 30, 50, 100, 150, 250, 500 μM and the reaction was proceeded for 45 mins. For Kinetics measurement with other acyl peptides (H3K9 butyryl, octanoyl and myristoyl), 2 μM SIRT7 (FL) or 5 μM SIRT7 deletion mutants (ΔN 56-400, ΔC 1-364, core 56-364) was used in the assay. The acyl peptide concentrations were 2, 5, 10, 15, 20, 30, 50, 80 μM and the reaction time was 45 mins. Reactions were quenched with 10% (v/v) TFA in methanol and then analyzed by HPLC.
Protein stability assay
SIRT7 WT and mutants were incubated with H3K18 acetyl or H3K9 myristoyl peptide at 4 °C or 37 °C for 20 mins respectively. The reaction consisted of 50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM NAD, 1 mM DTT, 50 μM H3K18 acetyl peptide or 20 μM H3K9 myristoyl peptide, 3 μM SIRT7, 9 μM tRNA, 0.5 μL SUPERase-In™ RNase inhibitor (20 U/L), protease inhibitor cocktail (1:100 dilution). After centrifugation at 17,000 g for 20 mins, equal volume of the supernatant of each reaction was resolved on SDS-PAGE and proteins were visualized by Coomassie blue staining.
Cell culture and viral transfection
Human 293T and Hela cell lines were acquired from the American Type Culture Collection (ATCC). Cells were cultured in DMEM (Thermo Fisher) supplemented with penicillin-streptomycin (Thermo Fisher) and 10% fetal bovine serum (FBS). Lentivirus transfection was performed as previously described9. Briefly, coding sequences (CDS) of SIRT7 (FL, ΔN, ΔC and core) were cloned into pCDH-CMV-MCS-EF1-Puro vector with an N-terminal Flag tag. For lentiviral packaging, 293T cells were co-transfected with pCMV-dR8.2, pMD2.G (obtained from Addgene) and corresponding SIRT7 pCDH plasmid or empty pCDH vector. The medium supernatant containing the lentiviruses was collected and filtered after 48 hrs. For virus infection, cells were incubated with virus-containing supernatant in the presence of 8 μg/mL polybrene. After 72 hrs, infected cells were selected using 1.5 μg /mL puromycin for one week. To generate transient Sirt7 knockdown cell lines, the pLKO.1-puro lentiviral shRNAs constructs toward luciferase and Sirt7 were purchased from Sigma-Aldrich. Luciferase shRNA (SHC007 and Sirt7 shRNA1 (TRCN0000359663)) were used. Lentivirus particles were generated as described above. HEK 293T cells were infected with lentivirus in the presence of 8 μg/ml polybrene for 6 hrs, and then incubated with fresh DMEM medium supplemented with 10% FBS for 48 hrs before subsequent global labeling experiments.
Global fluorescent labeling of fatty-acylated proteins in Sirt7 ctrl and KD cells
Prior to harvest, Cells were cultured in the presence of 50 μM Alk-14 for 6 hrs in DMEM medium supplemented with 10% FBS. The cell pellet was re-suspended in Buffer A (20 mM Tris, pH 8.0, 10 mM KCl, 1.5 mM MgCl2, 1mM DTT, 5% glycerol, 0.1% NP-40) with EDTA-free protease inhibitor cocktail (Sigma) freshly added and incubated on ice for 5 mins. After centrifugation at 1300 g for 10 mins at 4 °C, the supernatant was collected as crude cytosol fraction, which was further clarified at 17,000 g for 20 mins. The pellet was re-suspended in Buffer C (20 mM Tris, pH 8.0, 500 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 5% glycerol, 1% NP-40) with fresh EDTA-free protease inhibitor, held on ice for another 20 mins, then incubated with 1 µl Benzonase nuclease (Sigma) at 37 °C for 3 mins to release chromatin-associated proteins. Cell debris was removed by spinning at 17,000 g for 20 mins and the supernatant was collected as the nuclear fraction. Equal amounts of total protein (~100 µg) from the nuclear and cytoplasmic fractions were dispensed into 1.5 ml Eppendorf tube respectively. BODIPY-N3 (5 mM stock in DMF) was added to the reaction to a final concentration of 100 µM, followed by the addition of Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, 100 mM stock in DMF, final concentration 500 µM), CuSO4 (100 mM stock in water, final concentration 500 µM), and TCEP (40 mM stock in water, final concentration 1 mM). TCEP (40 mM) and CuSO4 stock (100 mM) solutions were prepared fresh. The click chemistry reactions were allowed to proceed at room temperature for 30 mins. The proteins were then precipitated with methanol/chloroform/water (4:1.5:3 volume ratio with the reaction volume set as 1) and the protein pellets were washed twice with 1 ml of ice-cold methanol. After drying the protein pellet in the air for 10~15 mins, 20 µl of a solution containing 4% (w/v) sodium dodecyl sulfate (SDS), 150 mM NaCl, 50 mM triethanolamine, pH 7.4 was added to completely dissolve the protein pellet. Protein loading dye with 100 mM DTT was added and the samples were heated at 95 °C for 5 mins. To remove cysteine fatty acylation, 500 mM NH2OH (final concentration) was added and the samples were heated at 95 °C for 10 mins. Finally, protein samples were resolved by SDS-PAGE and visualized by Typhoon 9400 Variable Mode Imager (GE Healthcare Life Sciences) with setting of Green (532 nm)/520BP40 PMT 600~800 V (normal sensitivity), and analyzed by ImageQuant TL v2005.
Fluorescence microscopy
Coding sequences (CDS) of human SIRT7 (FL, ΔN, ΔC and core) were inserted in-frame into the 5’-end of EGFP cDNA (pEGFP-N1 vector). Hela cells were transfected with EGFP-tagged SIRT7 using Fugene 6 transfection reagent following manufacturer’s instruction for 24 hrs. For immunofluorescence, cells were permeabilized with 0.3% triton X-100 in PBS for 10 mins at room temperature (RT). After washing three times with PBS, cells were fixed with 4% paraformaldehyde in PBS for 15 mins at RT. Cells were then blocked in PBS containing 5% BSA and 0.3% triton X-100 for 1 hr. Primary antibody (UBF, Santa Cruz Biotechnology cat # sc13125) was diluted at 1:50 in PBS containing 1% BSA and 0.3% triton X-100 and incubated with the cells for 1 hr. After rinsing three times with PBS, fluorophore-conjugated secondary antibody (Goat anti-mouse, Alexa Fluor® 594, Thermo) was diluted at 1:500 and applied for 1 hr at RT in the dark. Samples were washed three times with PBS and stained with prolong gold anti-fade reagent (Cell signaling technology) with DAPI. Confocal images were taken using a 63×, 1.4 NA objective, on an inverted microscope (Leica), and acquired with ZEN software. Image resizing, cropping, and brightness were uniformly adjusted in Photoshop (Adobe).
RIP to pull down RNA that binds to SIRT7
RNA immunoprecipitation (RIP) experiment was conducted following a previous report27. Two 15-cm plates of HEK 293T cells stably overexpressing Flag-tagged SIRT7 or Flag-tagged SIRT6 were grown to 80% confluency. For CLIP (crosslinking and immunoprecipitation of in vivo RNA) experiments, 293T cells stably overexpressing Flag-SIRT7 (FL, ΔN, ΔC and core) were UV-irradiated (254 nm, 0.3 J/cm2). Cells were washed twice with ice-cold PBS and collected. Cell pellets were lysed in 1 mL WCE buffer (1% NP-40, 30 mM Tris, pH 7.5, 100 mM NaCl, 66 mM KCl, 1 mM MgCl2, 1 mM DTT, 500 U SUPERase-In™ RNase inhibitor, complete protease inhibitor cocktail) on ice for 20 mins. Then 2 μL of TURBO DNase (2U/µL, Thermo) was added to each sample. The mixture was incubated at 37°C for 3 mins and left on ice for at least 1 min. Then the mixture was spun at 16,000 g in a tabletop centrifuge for 15 mins to pellet insoluble cell debris.
For subcellular fractionation, the protocol was adapted as previously described28. Briefly, the cell pellet was re-suspended in 500 μL Buffer A (20 mM Tris pH 8.0, 10 mM KCl, 1.5 mM MgCl2, 1mM DTT, 5% glycerol, 0.1% NP-40, 500 U of SUPERase-In™ RNase inhibitor, complete protease inhibitor cocktail) and incubated on ice for 5 mins. After centrifugation at 1300 g for 10 mins at 4 °C, the supernatant was collected as the cytosol fraction, which was further clarified at 17,000 g for 20 mins. The pellet was washed twice with 1 mL of Buffer A (no NP-40) and then re-suspended in 500 μL Buffer C (20 mM Tris, pH 8.0, 300 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 5% glycerol, 1% NP-40, 500 U of SUPERase-In™ RNase inhibitor, complete protease inhibitor cocktail), held on ice for another 20 mins, then incubated with 1 μL of Benzonase nuclease (Sigma) at 37 °C for 3 mins. Cell debris was removed by spinning at 17,000 g for 20 mins and the supernatant was collected as the nuclear fraction.
The supernatant (total cell lysate or fractionated cytosolic and nuclear lysates) was then incubated with anti-Flag M2 affinity gel (50 µL for two 15-cm plates of HEK 293T cells, pre-washed twice with PBS and four times with WCE buffer before incubation) for 2 hrs at 4 °C with gentle shaking. The beads were washed six times with 1 mL of WCE buffer, changing to new tubes for the final wash. For the CLIP experiment, beads were washed four times with 1 mL of high-salt buffer (50 mM Tris-HCl, pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate), followed by washing twice with 1 mL of WCE buffer. The beads were re-suspended in 200 µL of proteinase K solution and incubated at 37 °C for 20 mins with gentle shaking. Then, 200 µL of PK buffer (no proteinase K) with 2% SDS was added to the beads suspension and further incubated at 37 °C for 20 mins to completely digest proteins. Proteinase K solution (4 µg/µL) in PK buffer (100 mM Tris, pH 7.5, 10 mM EDTA, 50 mM NaCl) was pre-incubated at 37 °C for 20 mins to inactivate any RNase.
RNA was extracted using phenol-chloroform extraction followed by ethanol precipitation. Typically, 400 µL of sample was mixed with 400 µL of acid-phenol:chloroform:isoamyl alcohol (125:24:1, v/v/v, pH 4.5, Thermo) and vortex vigorously. Samples were spun at 16,000 g for 15 mins at 4 °C. Upper aqueous layer was transferred to a new tube, mixed with 400 µL of chloroform, and vortexed vigorously. After spinning at 16,000 g for 15 mins, the aqueous layer was transferred and mixed with 40 µL of 3 M NaOAC (pH 5.3), 1 mL of pure ethanol, and 2 µL of RNase-free glycogen (Thermo, cat # 10814010). RNA was precipitated at −20 °C for 2 hrs. The mixture was spun at 16,000 g for 30 mins at 4 °C. Supernatant was removed and RNA pellet was washed with 1 mL of 70% ethanol twice. After air drying the RNA pellet for 5 mins at RT, RNA pellet was dissolved in 20–40 µL of DEPC (Diethylpyrocarbonate)-treated H2O and stored at −80 °C.
Total RNA isolation
Total input RNA was purified using TRI reagent (Sigma) according to manufacturer’s instruction. For each isolation, 50 µL of total cell lysate was incubated with 500 µL of TRI reagent and vortexed briefly to mix. Then 100 µL (1/5 volume of TRI reagent) of chloroform was added to the sample, vortexed, and incubated at room temperature for 5 mins. Samples were spun at 16,000 g for 15 mins at 4 °C. The upper aqueous layer was transferred to a new tube and 250 µL of (1/2 volume of TRI reagent) isopropanol, 25 µL of NaOAC (pH 5.3) and 1 µL of RNase-free glycogen were added to the sample and incubated at −20 °C for 2 hrs. The samples were spun at 16,000 g for 30 mins at 4 °C. The RNA pellet was washed twice with 1 mL of 70% ethanol and re-solubilized in DEPC-H2O.
RNA 3’-end labeling using [32P] pCp
RNA (2–5 µg) were 3’-end labeled with T4 RNA ligase 1 (NEB) and homemade 5’-α-32P 5’-3’ cytidine bis-phosphate overnight at 4 °C in 1× RNA ligase buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 mM ATP, 10% v/v DMSO). Unincorporated radioactive materials were removed using NucAway™ Spin Columns (Ambion), and one half of the labeling reaction was resolved on a 12% denaturing polyacrylamide gel containing 7 M urea. The gel was sealed with plastic wrap and imaged on a PhosphoImager immediately.
RIP-seq and data analysis
The RIP-seq experiment was conducted using two biological replicates for each sample. For the total repertoire of SIRT7 RIP-RNA, we sequenced both cytosolic and nuclear RNAs that were associated with Flag-SIRT7 respectively. In order to deep sequence the non-rRNA repertoire, rRNA was subtracted from both pre-RIP input RNA and RNA co-immunoprecipitated with Flag-SIRT7 using the RiboZero Magnetic Gold Human/Mouse/Rat Kit (Illumina). All RNA samples were treated with DNase I in solution to completely remove genomic DNA. RNA samples were quantified with a Qubit 2.0 (RNA HS kit; Thermo) and were then analyzed on the BioAnalyzer High Sensitivity DNA chip (Agilent) to determine RNA integrity before library construction.
Stranded RNA-seq libraries were generated with the NEB Next Ultra RNA library Prep kit (NEB) using 25–100ng RNA. All samples were sequenced on an Illumina NextSeq500 to generate single-end 75-bp reads. Following removal of rRNA-matching reads with bowtie2, the deep sequencing data was mapped to the Human genome (UCSC hg19) by Tophat version 2.0. Cufflinks v2.2.1 was used to generate FPKM (Fragments Per Kilobase of Exon Per Million fragments mapped) values and statistical analysis of differential gene expression for annotated genes. Enrichment fold was calculated as log2 (FPKMIP/FPKMinput) against SIRT7 target genes with sufficient expression level (FPKM>5). For analysis of rRNA sequences in non-rRNA-subtracted libraries, depth of coverage counts was generated with SAM tools pileup and normalized to total reads per pre-rRNA in order to compare distribution of read depth over assembled rRNA transcript to indicate relative binding preference. Gene ontology (GO) annotations were performed using DAVID v6.7.
Reverse transcription and RT-qPCR
RNA templates in solution were treated with DNase I to remove any contaminant genomic DNA, and then were reversely transcribed to cDNA using a primer mixture consisting of oligo dT and random hexamers by Superscript III First-strand synthesis kit (Thermo). Real-time quantitative PCR (RT-qPCR) was performed to access the relative abundance of mRNA using iTaq Universal SYBR Green Supermix (Biorad) according to manufacturer’s instruction with 200–500 ng total cDNA as the template. β-Actin was used as an internal control for the following reasons: (1) β-actin mRNA was not bound by SIRT7 from RIP sequencing data; (2) β-actin expression is not affected by SIRT7. The primers used for RT-PCR are listed as below:
| Gene | NCBI Gene ID | Forward | Reverse |
|---|---|---|---|
| beta actin | 60 | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
| HIST1H4A | 8359 | AAGGGTTTGGGTAAGGGGG | TAGATCAGACCAGAGATCCGC |
| HIST1H4H | 8365 | GAGGAGCTAAGCGTCATCGC | ACACCTTCAGAACACCACGAG |
| HIST4H4 | 121504 | TGCTGCGGGACAATATCCAAG | AACCGCCGAAACCATAAAGGG |
| HIST1H2AG | 8969 | GCTAAGGCCAAGACTCGCTC | GACCCGCTCGGCATAGTTG |
| HIST3H2A | 92815 | GGTCGTGGTAAGCAGGGTG | CGCTCCGAATAGTTG CCCTT |
| HIST1H3A | 8350 | ACTGCTCGGAAGTCTACTGGT | GCGCTGGAAAGGTAGTTTACGA |
| RPS19 | 6223 | AAGCTGAAAGTCCCCGAATGG | AGTTCTCATCGTAGGGAGCAAG |
| FAM 195B | 348262 | AACGTCCGCTTCATTTACGAA | GGTTAGGGACCTTCTCCACATAC |
| PSMB10 | 5699 | TCCTTCGAGAACTGCCAAAGA | ATCGTTAGTGGCTCGCGTATC |
| PARP-1 | 142 | CGGAGTCTTCGGATAAGCTCT | TTTCCATCAAACATGGGCGAC |
| EIF5B | 9669 | AGGGCTTATGACAAAGCAAAACG | CCCAAGTACGCAGATAATAGGGG |
| NCL | 4691 | GGTGGTCGTTTCCCCAACAAA | GCCAGGTGTGGTAACTGCT |
| GRP78 | 3309 | CATCACGCCGTCCTATGTCG | CGTCAAAGACCGTGTTCTCG |
| PRPF6 | 24148 | CACCACGCGGTCAGACATT | TCCCCAACGGTTCTCTTGC |
T7 in vitro transcription
Full-length mature human 5S and 5.8S rRNAs were made by in vitro transcription using HiScribe T7 high yield RNA synthesis kit (NEB) according to manufacturer’s instruction. Briefly, 50 ng of linear dsDNA was used as the template (IDT), and mixed with 2 µL 10X T7 reaction buffer, rNTP mix (final concentration of each rNTP was 10 mM), 2 µL T7 RNA polymerase mix and DEPC water up to 20 µL. Transcription reaction was proceeded at 37 °C overnight in the presence of 0.5 µL SUPERase-In™ RNase inhibitor (20 U/µL, Thermo). Then 2 µL DNase I was added to the reaction mixture to remove template DNA. RNA transcripts were purified using canonical phenol-chloroform extraction followed by ethanol precipitation. Final RNA pellets were re-solubilized using DEPC-H2O and stored at −80°C.
Fluorescence polarization (FP)
FP assay was performed as previously described29. In vitro transcribed 5S and 5.8S rRNAs were PAGE purified and eluted into 10 mM Tris pH 7.5 buffer. Purified RNAs were then 3’-end labeled with fluorescein 5-thiosemicarbazide (Thermo Fisher) as previously described29. The purified 5S rRNA and 5.8S rRNA were diluted to 6.33 nM (1.3X) using EMSA buffer (20 mM Tris pH 8.0, 100 mM NaCl, 5 mM MgCl2, 1mM DTT, 2.5% glycerol, 0.01% triton X-100) with 0.5 µL SUPERase-In™ RNase inhibitor. The mixtures were heated to 65°C for 5 mins and then placed at room temperature to anneal for ~20 mins. Binding reactions were prepared with labeled RNA (final concentration 5 nM) and varying concentrations of recombinant SIRT7 or its mutants (SIRT7 WT: from 0 to 5000 nM, SIRT7 mutants: from 0 to 15,000 nM) in EMSA buffer to a final volume of 50 µL in black flat-bottom 96-well half-area microplate (Corning). All solutions were prepared using DEPC-H2O to prevent RNA degradation. Reactions were equilibrated for 2 hrs at room temperature in the dark before taking FP measurements on a Synergy H1 Microplate Reader (BioTek) with the appropriate filter for fluorescein (Excitation: 485/20, Emission: 528/20). Data was fit to a Hill equation using Igorpro software (Wavemetrics), which includes the Levenberg-Marquadt algorithm and statistical analysis tools.
Supplementary Material
Acknowledgments
This work is supported in part by grants from NIH GM086703 (H.L.), GM105933, CA152870 (Q.H.), and HK-RGC C7037-14G (Q.H.).
References
- 1.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 2.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 5.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin H. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.015156. 015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Seiler J, Santiago-Reichelt M, Felbel K, Grummt I, Voit R. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol Cell. 2013;52:303–313. doi: 10.1016/j.molcel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, Struhl K, Garcia BA, Gozani O, Li W, Chua KF. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, Lee JU, Zeviani M, Romand R, Hottiger MO, Schoonjans K, Auwerx J. A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab. 2014;20:856–869. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Yoshizawa T, Karim MF, Sato Y, Senokuchi T, Miyata K, Fukuda T, Go C, Tasaki M, Uchimura K, Kadomatsu T, Tian Z, Smolka C, Sawa T, Takeya M, Tomizawa K, Ando Y, Araki E, Akaike T, Braun T, Oike Y, Bober E, Yamagata K. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014;19:712–721. doi: 10.1016/j.cmet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, Li P, Cheng HL, Murphy AJ, Valenzuela DM, Luo H, Kapahi P, Krauss R, Mostoslavsky R, Yancopoulos GD, Alt FW, Chua KF, Chen D. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 2013;5:654–665. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong Z, Wang Y, Zhang X, Kim DD, Sadhukhan S, Hao Q, Lin H. SIRT7 Is Activated by DNA and Deacetylates Histone H3 in the Chromatin Context. ACS Chem Biol. 2016;11:742–747. doi: 10.1021/acschembio.5b01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai YC, Greco TM, Cristea IM. Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol. Cell Proteomics. 2014;13:73–83. doi: 10.1074/mcp.M113.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Blank MF, Iyer A, Huang B, Wang L, Grummt I, Voit R. SIRT7-dependent deacetylation of the U3-55k protein controls pre-rRNA processing. Nat Commun. 2016;7:10734. doi: 10.1038/ncomms10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, Lin H, Hao Q. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci Rep. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiran S, Chatterjee N, Singh S, Kaul SC, Wadhwa R, Ramakrishna G. Intracellular distribution of human SIRT7 and mapping of the nuclear/nucleolar localization signal. FEBS J. 2013;280:3451–3466. doi: 10.1111/febs.12346. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Seiler J, Santiago-Reichelt M, Felbel K, Grummt I, Voit R. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol Cell. 2013;52:303–313. doi: 10.1016/j.molcel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Khan S, Jiang H, Antonyak MA, Chen X, Spiegelman NA, Shrimp JH, Cerione RA, Lin H. Identifying the functional contribution of the defatty-acylase activity of SIRT6. Nat Chem Biol. 2016;12:614–620. doi: 10.1038/nchembio.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villalba JM, Alcaín FJ. Sirtuin activators and inhibitors. BioFactors. 2012;38:349–359. doi: 10.1002/biof.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J, Jing H, Lin H. Sirtuin inhibitors as anticancer agents. Future Med Chem. 2014;6:945–966. doi: 10.4155/fmc.14.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagano JM, Kwak H, Waters CT, Sprouse RO, White BS, Ozer A, Szeto K, Shalloway D, Craighead HG, Lis JT. Defining NELF-E RNA binding in HIV-1 and promoter-proximal pause regions. PLoS Genet. 2014;10:e1004090. doi: 10.1371/journal.pgen.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.