Abstract
Runt-related transcription factor 2 (Runx2) is essential for osteogenesis. This study aimes at identification of the genomic region differentially methylated in DNA for regulation of Runx2 expression. In the proximal promoter of mouse Runx2, DNA methylation was frequent at the region further than 3 kb relative to the transcription start site, in contrast to lower methylation status of the closer locus within 2 kb from the transcription start site. At the intermediate part, we identified a novel differentially methylated region in the Runx2 promoter region (Runx2-DMR): from −2.7 to −2.2 kb relative to the start site of Runx2 transcription in mice. In this region, the DNA methylation rate correlated negatively with Runx2 expression among mouse organs as well as among primary cultures of bone marrow from different dogs. Induction of mouse and dog mesenchymal-like cells into osteoblastic differentiation decreased the methylation rate of Runx2-DMR. Thus, in this study, we identified a novel genomic region in which DNA methylation status is related to Runx2 expression and detected demethylation of Runx2-DMR during osteoblastic differentiation in mouse and dog.
Keywords: correlation analysis, DNA methylation, osteogenesis, promoter, Runx2
Runt-related transcription factor 2 (Runx2, also known as Cbfα1) is essential for osteogenesis and chondrogenesis. By regulating transcription of osteogenic genes including Osterix (Sp7), osteocalcin (Spp1) and bone sialoprotein (Ibsp) [4, 7, 19], Runx2 exerts its action on osteoblastic differentiation [4], chondrocyte maturation and vascular invasion into the cartilage [6, 9]. In addition to the absence of bone formation and a cartilage defect in Runx2-deficient mice [10,11,12], mutations in Runx2 are associated with cleidocranial dysplasia in humans [18, 21]. Forced expression of Runx2 promotes osteogenic differentiation of mesenchymal lineage cells [2, 4]; therefore, Runx2 is considered the potent activator of osteoblastic differentiation.
Two distinct promoters initiate Runx2 transcription from different start sites. In mice, the proximal promoter yields the type I isoform (Runx2-I) with the length of 6,475 bp that encodes 514 amino acid residues, whereas Runx2-II (coding for additional 19 N-terminal amino acid residues) is produced by the distal promoter 78 kb upstream of the proximal promoter. The use of these promoters depends on developmental timing and cell type [1, 7]. Targeted deletion of Runx2-II results in impairment of endochondral bone formation in mice, suggesting that Runx2-II has a function in osteogenesis, whereas Runx2-I is sufficient for early skeletogenesis and intramembranous bone formation [26].
In eukaryotes, most of DNA methylation occurs on cytosines within CpG sites and has been implicated in formation of heterochromatin regions. Proper distribution of DNA methylation in the genome is essential for embryogenesis [16], and aberrant methylation is associated with pathologies including carcinogenesis [8]. Some research groups reported that expression of osteogenic genes including Runx2 is affected by DNA methylation [3, 13, 23,24,25, 27]. The transcription start site of Runx2-I locates on a CpG island, which contains abundant cytosine bases that can be methylated, while the distal promoter for Runx2-II is composed of a CpG-poor sequence. Using an ex vivo experimental model, Uehara et al. identified a differentially DNA methylated CpG site at upstream of Runx2-I locus in which hypermethylation is associated with down-regulation of Runx2 expression [23]. However, a concrete responsible region that controls Runx2 expression via its DNA methylation remains unclear, as is its physiological function. Here, we report a novel differentially DNA methylated region located on the proximal promoter of Runx2 (Runx2-DMR), in which DNA methylation is related to Runx2 expression and osteoblastic differentiation.
MATERIALS AND METHODS
Animal experiments
All animal procedures were approved by the Animal Care and Use Committee of the University of Miyazaki (approval number: 2013–001). For molecular biological analyses, cerebellum, salivary glands, lung, heart, stomach, small intestine, liver, spleen, kidneys, testes, bone (femur), skeletal muscle of the thigh and skin of the trunk were excised from 8-week-old male C57BL/6N mice (Charles River, Wilmington, MA, U.S.A.) after euthanasia. A piece of kidney and an epididymal fat were used for cell culture.
Cell culture
Among the dogs brought to the Veterinary Teaching Hospital of Miyazaki University by their owners, bone marrow was collected by bone puncture for clinical use (Supplementary Table S1). A part of the marrow was used in this study with the informed consent of their owners. Murine kidney and adipose tissue, and canine marrow samples were cultured in plastic dishes in Dulbecco’s modified Eagle medium (Sigma-Aldrich, St. Louis, MO, U.S.A.) containing 10% of fetal bovine serum (Biofill, Victoria, Australia), 50 µM β-mercaptoethanol (Life Technologies, Carlsbad, CA, U.S.A.), 10 U/ml penicillin, 10 µg/ml streptomycin and 250 ng/ml amphotericin B (antibiotic-antimycotic, Life Technologies) at 37°C in 5% in air. Tissue pieces were removed from the culture dishes 4 days later, and monolayer cells were allowed to proliferate in the dishes. After the first passage, amphotericin B was not added to the medium (Penicillin-Streptomycin, Life Technologies). The mesenchymal-like cell line derived from mouse adipose tissue was established after 20 passages.
Analysis of DNA methylation
To assess the methylation rate of arbitrary CpG sites, combined bisulfite restriction analysis (CoBRA) was conducted. Genomic DNA was isolated from tissues and cultured cells using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The bisulfite reaction was run using an EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, U.S.A.). PCR was conducted using specific primers (Supplementary Table S2) and BIOTAQ HS DNA Polymerase (Bioline, London, U.K.). The amplified product was reacted with the restriction enzyme HpyCH4IV (New England Biolabs, Ipswich, MA, U.S.A.) and was also subcloned and subjected to bisulfite sequencing. Fragments produced by HpyCH4IV were loaded onto a 2% agarose gel for electrophoresis or quantified by means of a microchip electrophoresis system (MultiNA, Shimadzu, Kyoto, Japan). The degree of methylation in each sample was calculated by means of the formula IMe(IU + IMe) ×100, where IMe and IU represent intensity of restriction fragments and intact fragments, respectively, in each sample [20].
Real-time RT-PCR
To study gene expression, real-time RT-PCR was performed. Total RNA was extracted from organs and cultured cells using the RNeasy Plus Mini Kit (Qiagen). Complementary DNA was synthesized using SuperScript III Reverse Transcriptase (Life Technologies) with random primers. PCR (Power SYBR Green PCR Master Mix, Life Technologies) was performed using an Applied Biosystems 7300 Real Time PCR system (Life Technologies) with specific primers (Supplementary Table S2). Relative expression was calculated by the ddCT method using mouse Gapdh or dog Rpl13a as endogenous standards.
Induction of osteoblastic differentiation
Osteoblastic differentiation was induced as described previously [14, 27]. Briefly, the established mouse mesenchymal-like cells (5.0 × 104 cells per well) were seeded in 24-well plates in the medium described above and cultured at 37°C in 5% in air. Two days later, 10 mM β-glycerophosphate (Sigma-Aldrich), 50 µg/ml ascorbic acid 2-phosphate (Sigma-Aldrich), 100 nM dexamethasone (Sigma-Aldrich) and 100 ng/ml recombinant human BMP-2 (Wako, Osaka, Japan) were added to the medium of confluent cells, but not to the control group. The medium was replaced every other day. For analysis of DNA methylation, genomic DNA was isolated from three distinct wells per group at day 7 of the induction, and we calculated the methylation rate in each well. To detect alkaline phosphatase (ALP)-positive osteoblasts, cells in the other three wells per group were stained with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) substrates (MK300, TaKaRa, Otsu, Japan). The image analysis was carried out with an inverted microscope (BZ-9000, Keyence, Osaka, Japan) and the optional software (BZ-analysis application, Keyence). The proportion of osteoblasts was calculated by means of AALP/Awhole ×100, where AALP and Awhole represent area covered with ALP-positive osteoblasts and whole cells, respectively. For osteoblastic differentiation of canine marrow cells, similar experiments as mouse cells were conducted, but the cells were pre-cultured for four days followed by fourteen days of osteoblastic induction, because, in this study, the canine cells proliferated slower than the mouse cell.
Statistics
The χ2 test was applied to statistical evaluation of DNA methylation at each CpG site examined by bisulfite sequencing. Spearman’s rank correlation coefficient was calculated to estimate the relation between DNA methylation and gene expression. Student’s or paired t test was applied to statistical evaluation in the experiment on osteoblastic induction of mouse or canine cells, respectively.
RESULTS
Identification of Runx2-DMR in mice
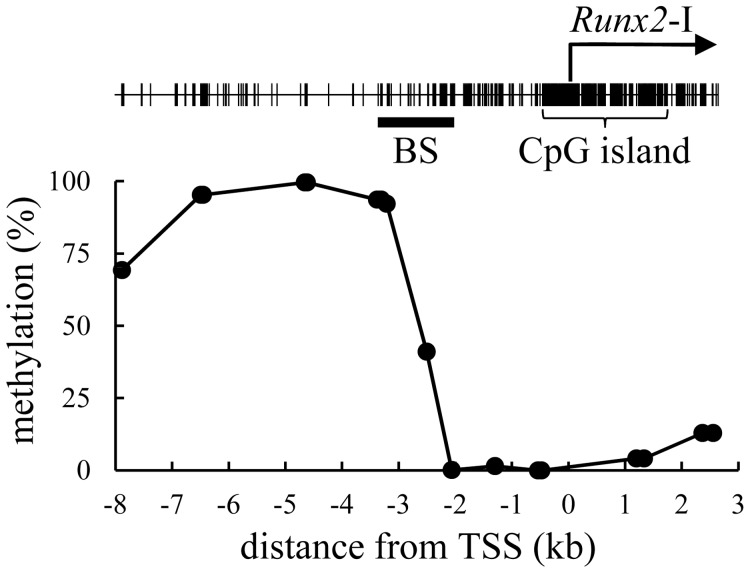
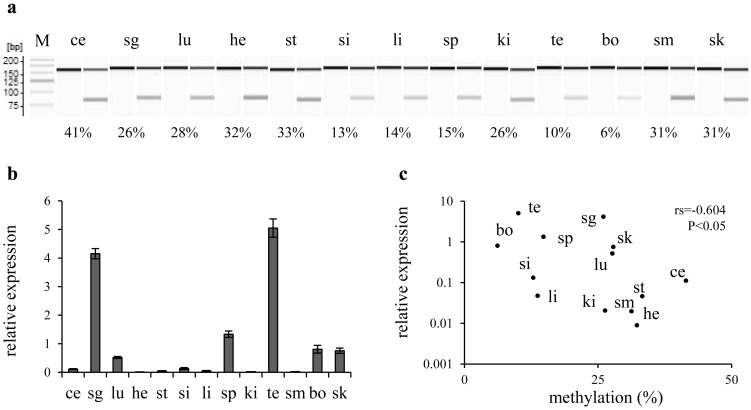
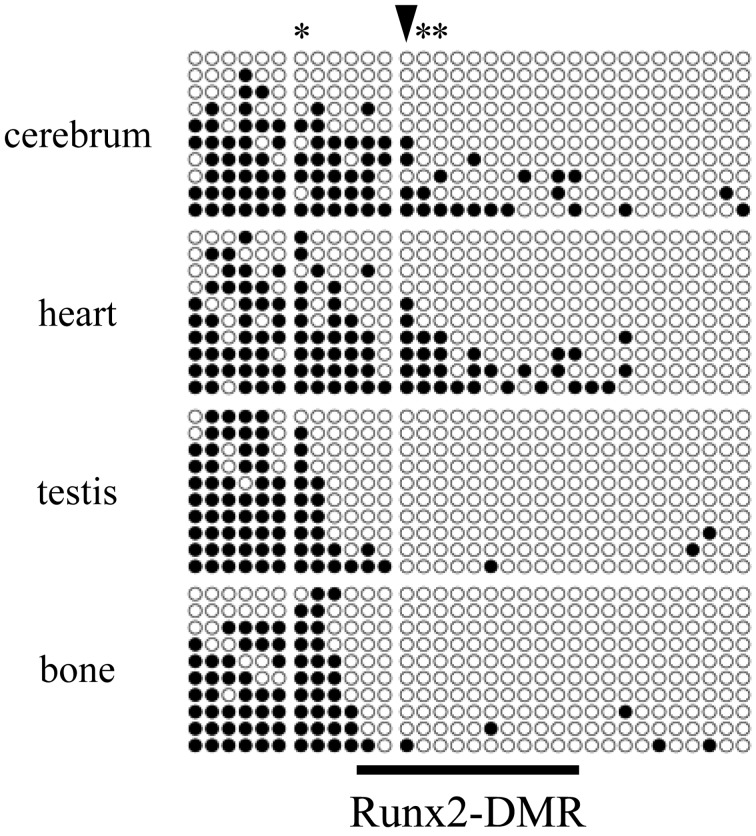
The methylation status of arbitrary CpG sites located between −8 kb and +3 kb relative to the transcription start site of Runx2-I (TSS) was first screened by combined bisulfite restriction analysis (CoBRA) in primary culture cells derived from a mouse kidney. DNA methylation was rare in the region between −2 and +3 kb relative to the TSS (Fig. 1). In contrast, the CpG sites located at −8 to −3 kb were frequently methylated. The CpG site at −2,505 bp relative to the TSS (CpG-2,505), which is located in an intermediate part between highly and weakly methylated regions, was methylated in 41% of genomes in the cultured cells. We analyzed the methylation rate of CpG-2,505 in various organs, including the cerebellum, salivary glands, lungs, heart, stomach, small intestine, liver, spleen, kidneys, testes, bone, skeletal muscle and skin. The methylation rate ranged from a minimum of 6% in bone to a maximum of 41% in the cerebellum (Fig. 2). Expression of Runx2 mRNA correlated negatively with the methylation rate of CpG-2,505 among the mouse organs (rs=−0.604, n=13, P<0.05). CpG-3,210 was highly methylated in almost all organs, and methylation of CpG-1,297 was not detected (Supplementary Figure). We further surveyed the methylation state of all CpG sites between −3,371 and −2,039 bp by bisulfite sequencing. The result indicated again that CpG sites were frequently methylated at the distal location from TSS in contrast to rare methylation at the proximal region in each organ (Fig. 3). Although testes and bone, which highly expressed Runx2 mRNA, were rarely methylated in the region between −2,658 and −2,039 bp, a certain level of methylation was detected between −2,658 and −2,226 bp in the cerebellum and heart, which expressed Runx2 mRNA weakly. Notably, a statistical association between DNA methylation and the organ type was confirmed at CpG-2,505 (P<0.01) and at CpG-2,487 and CpG-2,412 (P<0.05). In light of these results, we defined the region from −2.7 to −2.2 kb relative to the TSS as Runx2-DMR in mice.
Fig. 1.
Distribution of DNA methylation within the proximal Runx2 promoter. The line plot indicates the methylation rate of arbitrary CpG sites in primary cells derived from the mouse kidney. Location of CpG sites is marked as vertical lines in the upper panel. BS indicates the range studied by bisulfite sequencing in Fig. 3.
Fig. 2.
Correlation between DNA methylation and Runx2 expression among mouse organs. a) A gel image as output from the results of combined bisulfite restriction analysis (CoBRA) targeting CpG-2,505 in mouse organs. Fragments digested with HpyCH4IV were loaded onto odd lanes, except for the leftmost lane (showing a 25-bp DNA ladder; M). In the lanes immediately on the left, the fragment without the digestion is loaded as the experimental control. The PCR product 150 bp long is digested into fragments with the length of 73 and 77 bp when CpG-2,505 is methylated. Various rates of methylation are shown as indicating each score below their lanes. b) A bar chart shows the results of real-time RT-PCR. Columns and their error bars indicate mean and standard error, respectively, of relative expression levels of Runx2 mRNA in each organ to the sample mean. c) Each organ is plotted on the scatter diagram. Vertical and horizontal scales indicate relative expression levels of Runx2 mRNA and the methylation rate of CpG-2,505, respectively. A negative correlation was detected between Runx2 expression and CpG-2,505 methylation among mouse organs. ce: cerebellum, sg: salivary glands, lu: lungs, he: heart, st: stomach, si: small intestine, li: liver, sp: spleen, ki: kidneys, te: testes, bo: bone, sm: skeletal muscle and sk: skin.
Fig. 3.
Bisulfite sequencing of the Runx2 promoter in mouse organs. Each row of circles indicates whether CpG sites are methylated (black) or unmethylated (white) in each read from −3,371 to −3,148 (left), −2,985 to −2,649 (middle) and from −2,505 to −2,039 bp (right) relative to the transcription start site (TSS). Three divided ranges cover all CpG sites from −3,371 to −2,039 bp. A gradient of methylation rates is detectable along the genomic sequence. CpG sites in the cerebrum and heart are methylated more frequently in the intermediate region than those in testes and bone. Asterisks and an arrowhead that marks CpG-2,505 are shown at statistically significantly dependent CpG sites during methylation (P<0.05 and P<0.01, respectively). A horizontal line at the bottom of the figure indicates the defined Runx2-DMR extending from −2,658 to −2,226 bp.
Identification of Runx2-DMR in dog
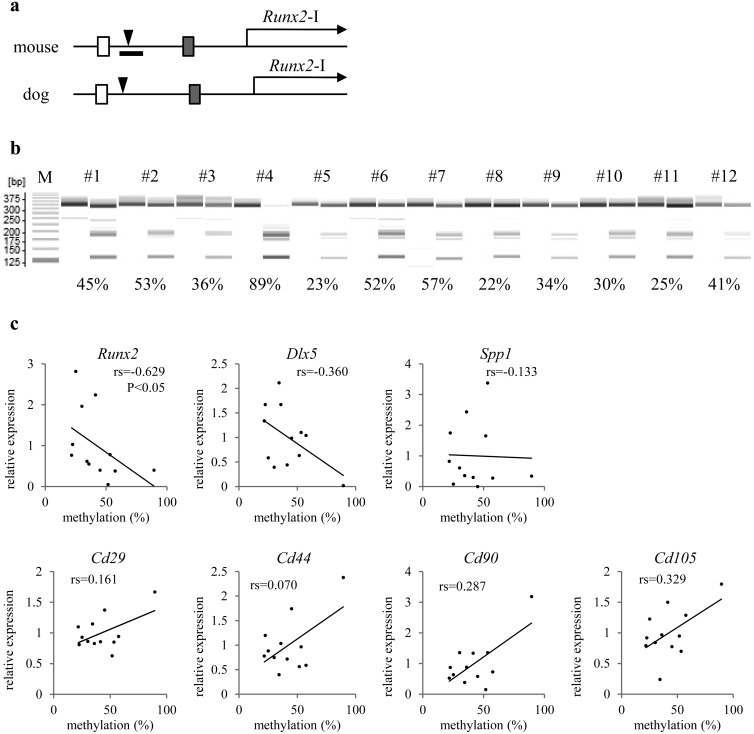
To firmly validate the observed relation between Runx2 expression and DNA methylation in Runx2-DMR using multiple lines of evidence, assays similar to those above were conducted on primary cultures derived from canine marrow (12 dogs; Supplementary Table S1). This set of samples with a relatively common type of cellular population containing Runx2-expressing osteoblasts enabled us to evaluate the cross-species relations via comparison to the results of our mouse experiments. According to the open genome database, the upstream genomic sequences (relative to the TSS) from −3,409 to −3,171 bp and from −1,387 to −1,169 bp in dogs showed more than 90% similarity with the sequences from regions −3,161 to −2,924 bp and −1,347 to −1,126 bp in mice, respectively (Fig. 4a). The former region in mice also showed 91% similarity with the region −3,712 to −3,475 bp relative to the human TSS. The sequence of Runx2-DMR is, however, not conserved between dogs and humans. We estimated that canine Runx2-DMR lies near −2.95 to −2.35 kb and examined canine CpG-2,829 in the methylation assay. The methylation rates at canine CpG-2,829 showed a broad range, from a minimum of 22% to a maximum of 89% (Fig. 4b) and correlated negatively with Runx2 expression among marrow cultures (Fig. 4c, rs=−0.629, n=12, P<0.05). Therefore, this result suggests that Runx2-DMR is shared at least between mice and dogs. We could not find significant correlation between the methylation status of canine CpG-2,829 and expression of the other osteogenic genes, Dlx5 and Spp1, and marker genes of mesenchymal stem cells (MSCs), CD29, CD44, CD90 and CD105. But, high expression of all of the MSC markers was detected in the culture consisting of abundant cells carrying methylated CpG-2,829, raising the possibility that differentiation of MSCs into osteoblasts is accompanied with demethylation of Runx2-DMR.
Fig. 4.
Correlation between DNA methylation and gene expression among primary cultures of canine marrow (from different dogs). a) Structure of the proximal Runx2 promoter in the mouse genome and dog genome is shown. White and shaded boxes indicate conserved regions corresponding to each color. Arrowheads mark mouse CpG-2,505 and canine CpG-2,829. The range of defined Runx2-DMR is indicated as a horizontal bar in the mouse panel. b) A gel image as output from the results of combined bisulfite restriction analysis (CoBRA) targeting CpG-2,829 in canine marrow cultures. Fragments digested with HpyCH4IV are loaded onto odd lanes, except for the leftmost lane (showing a 25-bp DNA ladder; M). In the lanes immediately on the left, the fragment without the digestion is loaded as the experimental control. Each number on the lanes is assigned to each individual. The PCR product 315 bp long is digested into fragments 128 and 187 bp long when canine CpG-2,829 is methylated. Various rates of methylation are shown as indicating each score below their lanes. c) Marrow cells derived from each individual are plotted on the scatter diagrams. Vertical and horizontal scales indicate relative expression levels of genes headlining each diagram to the sample mean and the methylation rate of canine CpG-2,829, respectively. Statistical significance of correlation is shown only between CpG-2,829 methylation and Runx2 expression.
Dynamics of DNA methylation during osteoblastic differentiation
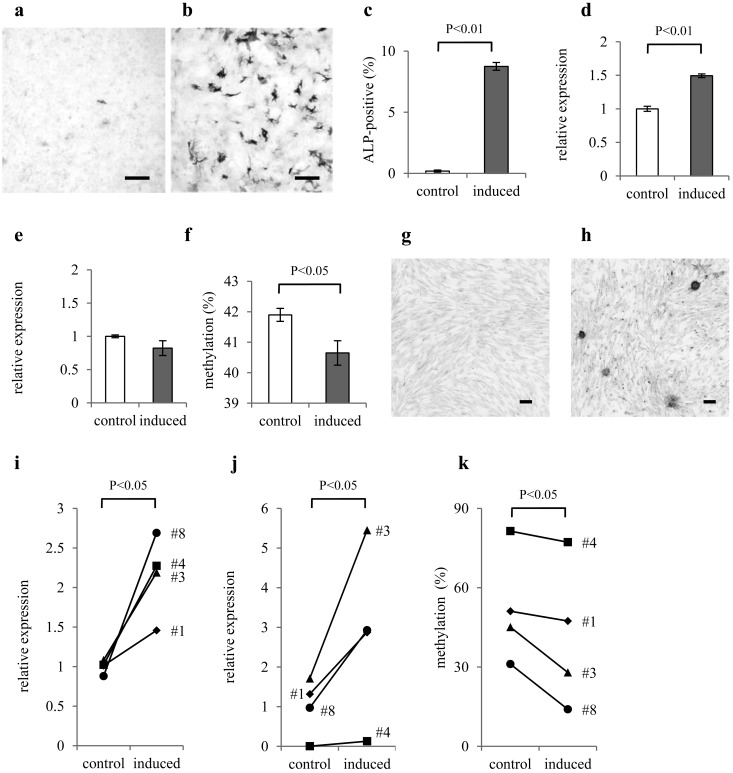
To detect demethylation of Runx2-DMR during osteoblastic differentiation, we studied the effect of osteoblastic induction on CpG-2,505 methylation in a mouse mesenchymal cell line, which we had established. At day 7 of osteoblastic induction, 8.75 ± 0.32% of ALP-positive cells appeared in contrast to 0.19 ± 0.08% of ALP-positive cells in the control group (Fig. 5a–5c). The induced group significantly showed higher Runx2 expression than the control group (Fig. 5d), but Dlx5 expression was comparable (Fig. 5e). CpG-2,505 methylation in the induced group was significantly lower than that in the control group (Fig. 5f), indicating that Runx2-DMR is demethylated during osteoblastic differentiation. We further investigated the effect of osteoblastic induction on demethylation of canine Runx2-DMR. As raised colony of ALP-positive osteoblasts appeared by the induction of canine marrow cells (Fig. 5g and 5h), we could not quantify the abundance of osteoblasts by our method that is based on microscopic observation. Osteoblastic genes, Runx2 and Dlx5, were increased by the induction at fourteen days (Fig. 5i and 5j). Compared to the control group, canine CpG-2,829 methylation was significantly lower in the induced group (Fig. 5k), supporting the demethylation of Runx2-DMR accompanied with osteoblastic differentiation.
Fig. 5.
Demethylation of Runx2-DMR in mesenchymal-like cells under the influence of osteoblastic induction. The results of ALP-staining for the mouse mesenchymal-like cell culture a) without and b) with osteoblastic induction were under inverted microscopy. Scale bars are 200 µm. c) The proportions of ALP-positive cells, d) relative expression levels of Runx2 and e) Dlx5 mRNAs to the mean of control group, and f) methylation ratio of CpG-2,505 are presented as bar charts. The percentage of ALP-positive cells and Runx2 expression are increased by osteoblastic induction. Mouse CpG-2,505 is demethylated under the influence of osteoblastic induction. Columns and their error bars indicate mean and standard error of the methylation rate of CpG-2,505. The results of ALP-staining for the canine marrow-derived cell culture g) without and h) with osteoblastic induction were under inverted microscopy. Scale bars are 200 µm. i) The relative expression levels of Runx2 and j) Dlx5 mRNAs to the mean of control group, and k) methylation ratio of canine CpG-2,829 are presented as line graphs. Expression of Runx2 and Dlx5 is significantly increased by osteoblastic induction. Canine CpG-2,829 is demethylated under the influence of osteoblastic induction.
DISCUSSION
Because Runx2 exerts a potent osteogenic action, some researchers have investigated the relation between DNA methylation and Runx2 expression. One research group reported that DNA methylation of the region −384 to −101 bp relative to the TSS does not correlate with a change in expression of Runx2 associated with the passage number of MSCs derived from the human placenta [17]. The other group demonstrated hypermethylation of CpG sites located at −1.9 kb but not −0.7 and −0.4 kb, relative to the TSS in human periodontal ligament cells exposed to lipopolysaccharide extracted from Porphyromonas gingivalis; the latter treatment downregulates Runx2 expression by altering DNA methylation [23]. They also revealed an accelerant effect of DNA methyltransferase inhibitor, 5-aza-2’-deoxycytidine, on human Runx2 expression as well as the previous observation from mouse experiments [23, 27]. Thus, epigenetic regulation of Runx2 expression would be similar in mouse and human. In the present study, we surveyed a wider region of the mouse Runx2 promoter and then clarified the genomic region closely related to Runx2 expression via its DNA methylation status at upstream of the locus previously investigated. The fact that the correlation between methylation rate in Runx2-DMR and Runx2 expression was applicable across species supports the importance of Runx2-DMR for regulating Runx2 expression in mammals.
The Runx2 promoter was clearly divided into a proximal ummethylated and a distal methylated region. Notably, the border located at appropriately −2.5 kb was closer to TSS in organs that do not express Runx2, suggesting that an expansion of the continuous methylated region toward the TSS rather than an emerging hypermethylation at a specific site surrounded by the unmethylated locus is related to Runx2 silencing. This insight is consistent with the previous observation that the hypermethylation was detected only at −1.9 kb site but not the more proximal part in the human Runx2 promoter [17, 23], implying preserved dynamics of DNA methylation in mouse and human Runx2 promoter. Because the previous reported hypermethylation was achieved by exposure of lipopolysaccharides, it might be affected by expansion of the distal methylated region beyond the physiologically variable region as human Runx2-DMR.
The canine culture conducted in this study is considered as heterogeneous population of bone marrow cells, although these cells share adhesiveness to the plastic dish. The variation in methylation rate of Runx2-DMR among twelve individuals would be due to difference in cell composition of each marrow culture. We had formerly hypothesized that the methylation rate of Runx2-DMR in marrow culture would be associated with contained amount of MSCs that possess the ability to differentiate into osteoblasts, but we could not find any evidences of the relationship (unpublished data). The strongest correlation was observed between DNA methylation of Runx2-DMR and expression of Runx2 itself, and methylation rate of Runx2-DMR was decreased by the osteoblastic induction. Therefore, the DNA demethylation of Runx2-DMR would proceed just before the activation of Runx2 transcription during osteoblastic differentiation.
In general, a genomic region receiving frequent methylation at CpG sites forms heterochromatin, which makes it difficult to recruit factors participating in gene transcription, e.g., RNA polymerase. As Runx2-DMR is 2 kb away from a CpG island that surrounds the TSS, the repressive effect of DNA methylation on Runx2 expression would not be mediated by heterochromatin formation at the TSS under physiological conditions. The conserved sequence that exists immediately upstream of Runx2-DMR seems to be important for Runx2 regulation. If this sequence acts as an enhancer of Runx2 expression, Runx2-DMR may control the chromatin conformation upstream of the TSS to activate the predicted enhancer. While, a promoter assay performed previously did not work under any conditions, even though the construct contained a complete sequence from −4,056 to +246 bp relative to the TSS [15]. Thus, activation of the proximal Runx2 promoter would also require the other remote enhancer working under the specific conformation of chromatin controlled by epigenetic modifications including DNA methylation of Runx2-DMR.
In conclusion, we first identified a genomic region in which DNA methylation status is responsible for Runx2 expression and detected demethylation in this region during osteoblastic differentiation. Because elevated expression of Runx2 is associated with vascular calcification during atherosclerosis [2, 5, 22], orderly activation of Runx2 is necessary for maintenance of physical health. Runx2-DMR may participate in locking up the Runx2 promoter via its DNA methylation to prevent unexpected activation of Runx2 that triggers pathological calcification. The uncovered mechanism should act as a key that unlocks the Runx2 promoter during a physiological process driven by Runx2 action, e.g., osteogenesis.
Supplementary Material
Acknowledgments
We thank Dr. Nobuyuki Kanno, formerly of the Laboratory of Veterinary Surgery, Faculty of Agriculture, University of Miyazaki, for providing canine marrow samples. This study was supported by a grant-in-aid from the University of Miyazaki to SW and by a Terumo Foundation grant to KN.
REFERENCES
- 1.Banerjee C., Javed A., Choi J. Y., Green J., Rosen V., van Wijnen A. J., Stein J. L., Lian J. B., Stein G. S.2001. Differential regulation of the two principal Runx2/Cbfa1 n-terminal isoforms in response to bone morphogenetic protein-2 during development of the osteoblast phenotype. Endocrinology 142: 4026–4039. doi: 10.1210/endo.142.9.8367 [DOI] [PubMed] [Google Scholar]
- 2.Byon C. H., Javed A., Dai Q., Kappes J. C., Clemens T. L., Darley-Usmar V. M., McDonald J. M., Chen Y.2008. Oxidative stress induces vascular calcification through modulation of the osteogenic transcription factor Runx2 by AKT signaling. J. Biol. Chem. 283: 15319–15327. doi: 10.1074/jbc.M800021200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho Y. D., Yoon W. J., Kim W. J., Woo K. M., Baek J. H., Lee G., Ku Y., van Wijnen A. J., Ryoo H. M.2014. Epigenetic modifications and canonical wingless/int-1 class (WNT) signaling enable trans-differentiation of nonosteogenic cells into osteoblasts. J. Biol. Chem. 289: 20120–20128. doi: 10.1074/jbc.M114.558064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G.1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89: 747–754. doi: 10.1016/S0092-8674(00)80257-3 [DOI] [PubMed] [Google Scholar]
- 5.Engelse M. A., Neele J. M., Bronckers A. L., Pannekoek H., de Vries C. J.2001. Vascular calcification: expression patterns of the osteoblast-specific gene core binding factor alpha-1 and the protective factor matrix gla protein in human atherogenesis. Cardiovasc. Res. 52: 281–289. doi: 10.1016/S0008-6363(01)00375-3 [DOI] [PubMed] [Google Scholar]
- 6.Enomoto H., Enomoto-Iwamoto M., Iwamoto M., Nomura S., Himeno M., Kitamura Y., Kishimoto T., Komori T.2000. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J. Biol. Chem. 275: 8695–8702. doi: 10.1074/jbc.275.12.8695 [DOI] [PubMed] [Google Scholar]
- 7.Håkelien A. M., Bryne J. C., Harstad K. G., Lorenz S., Paulsen J., Sun J., Mikkelsen T. S., Myklebost O., Meza-Zepeda L. A.2014. The regulatory landscape of osteogenic differentiation. Stem Cells 32: 2780–2793. doi: 10.1002/stem.1759 [DOI] [PubMed] [Google Scholar]
- 8.Hattori N., Ushijima T.2014. Compendium of aberrant DNA methylation and histone modifications in cancer. Biochem. Biophys. Res. Commun. 455: 3–9. doi: 10.1016/j.bbrc.2014.08.140 [DOI] [PubMed] [Google Scholar]
- 9.Himeno M., Enomoto H., Liu W., Ishizeki K., Nomura S., Kitamura Y., Komori T.2002. Impaired vascular invasion of Cbfa1-deficient cartilage engrafted in the spleen. J. Bone Miner. Res. 17: 1297–1305. doi: 10.1359/jbmr.2002.17.7.1297 [DOI] [PubMed] [Google Scholar]
- 10.Inada M., Yasui T., Nomura S., Miyake S., Deguchi K., Himeno M., Sato M., Yamagiwa H., Kimura T., Yasui N., Ochi T., Endo N., Kitamura Y., Kishimoto T., Komori T.1999. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev. Dyn. 214: 279–290. doi: [DOI] [PubMed] [Google Scholar]
- 11.Kim I. S., Otto F., Zabel B., Mundlos S.1999. Regulation of chondrocyte differentiation by Cbfa1. Mech. Dev. 80: 159–170. doi: 10.1016/S0925-4773(98)00210-X [DOI] [PubMed] [Google Scholar]
- 12.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T.1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764. doi: 10.1016/S0092-8674(00)80258-5 [DOI] [PubMed] [Google Scholar]
- 13.Lee J. Y., Lee Y. M., Kim M. J., Choi J. Y., Park E. K., Kim S. Y., Lee S. P., Yang J. S., Kim D. S.2006. Methylation of the mouse DIx5 and Osx gene promoters regulates cell type-specific gene expression. Mol. Cells 22: 182–188. [PubMed] [Google Scholar]
- 14.Lee M. H., Kim Y. J., Kim H. J., Park H. D., Kang A. R., Kyung H. M., Sung J. H., Wozney J. M., Kim H. J., Ryoo H. M.2003. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-β 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J. Biol. Chem. 278: 34387–34394. doi: 10.1074/jbc.M211386200 [DOI] [PubMed] [Google Scholar]
- 15.Lee M. H., Kim Y. J., Yoon W. J., Kim J. I., Kim B. G., Hwang Y. S., Wozney J. M., Chi X. Z., Bae S. C., Choi K. Y., Cho J. Y., Choi J. Y., Ryoo H. M.2005. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J. Biol. Chem. 280: 35579–35587. doi: 10.1074/jbc.M502267200 [DOI] [PubMed] [Google Scholar]
- 16.Li E., Bestor T. H., Jaenisch R.1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926. doi: 10.1016/0092-8674(92)90611-F [DOI] [PubMed] [Google Scholar]
- 17.Li Z., Liu C., Xie Z., Song P., Zhao R. C., Guo L., Liu Z., Wu Y.2011. Epigenetic dysregulation in mesenchymal stem cell aging and spontaneous differentiation. PLoS ONE 6: e20526. doi: 10.1371/journal.pone.0020526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R.1997. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89: 773–779. doi: 10.1016/S0092-8674(00)80260-3 [DOI] [PubMed] [Google Scholar]
- 19.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B.2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108: 17–29. doi: 10.1016/S0092-8674(01)00622-5 [DOI] [PubMed] [Google Scholar]
- 20.Nishino K., Hattori N., Tanaka S., Shiota K.2004. DNA methylation-mediated control of Sry gene expression in mouse gonadal development. J. Biol. Chem. 279: 22306–22313. doi: 10.1074/jbc.M309513200 [DOI] [PubMed] [Google Scholar]
- 21.Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J.1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771. doi: 10.1016/S0092-8674(00)80259-7 [DOI] [PubMed] [Google Scholar]
- 22.Sun Y., Byon C. H., Yuan K., Chen J., Mao X., Heath J. M., Javed A., Zhang K., Anderson P. G., Chen Y.2012. Smooth muscle cell-specific runx2 deficiency inhibits vascular calcification. Circ. Res. 111: 543–552. doi: 10.1161/CIRCRESAHA.112.267237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uehara O., Abiko Y., Saitoh M., Miyakawa H., Nakazawa F.2014. Lipopolysaccharide extracted from Porphyromonas gingivalis induces DNA hypermethylation of runt-related transcription factor 2 in human periodontal fibroblasts. J. Microbiol. Immunol. Infect. 47: 176–181. doi: 10.1016/j.jmii.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 24.Vaes B. L., Lute C., van der Woning S. P., Piek E., Vermeer J., Blom H. J., Mathers J. C., Müller M., de Groot L. C., Steegenga W. T.2010. Inhibition of methylation decreases osteoblast differentiation via a non-DNA-dependent methylation mechanism. Bone 46: 514–523. doi: 10.1016/j.bone.2009.09.033 [DOI] [PubMed] [Google Scholar]
- 25.Villagra A., Gutiérrez J., Paredes R., Sierra J., Puchi M., Imschenetzky M., Wijnen Av A., Lian J., Stein G., Stein J., Montecino M.2002. Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. J. Cell. Biochem. 85: 112–122. doi: 10.1002/jcb.10113 [DOI] [PubMed] [Google Scholar]
- 26.Xiao Z. S., Hjelmeland A. B., Quarles L. D.2004. Selective deficiency of the “bone-related” Runx2-II unexpectedly preserves osteoblast-mediated skeletogenesis. J. Biol. Chem. 279: 20307–20313. doi: 10.1074/jbc.M401109200 [DOI] [PubMed] [Google Scholar]
- 27.Zhang R. P., Shao J. Z., Xiang L. X.2011. GADD45A protein plays an essential role in active DNA demethylation during terminal osteogenic differentiation of adipose-derived mesenchymal stem cells. J. Biol. Chem. 286: 41083–41094. doi: 10.1074/jbc.M111.258715 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.