Abstract
This study aimed to investigate the spread of methicillin-resistant coagulase-positive staphylococci (MRCoPS) among veterinary staff, hand-touch sites and surgical tissue during cystotomy operations on cats and dogs that were patients, and to analyze the genetic relatedness and antimicrobial resistance profiles of the isolates. Human and environmental samples were obtained from the nasal passageways of 12 surgeons and veterinary assistants and from 29 hand-touch sites of instruments in operative units and subjected to bacterial isolation and enumeration. Swab samples were collected in triplicate from 29 dogs and three cats at the site of incision, from the incision area, from the peritoneum during surgery and from the peritoneum before suture. MRCoPS were identified by mecA gene detection and characterized by their antibiogram profile, SCCmec type and pulsed-field gel electrophoresis. Twenty-four staphylococci were isolated, derived from one veterinary assistant, 12 operating room floor areas and hand-touch sites, three dogs and one cat. Methicillin-resistant S. pseudintermedius (MRSP) was found on an electric clipper and rebreathing circuits in the operating room. Three dogs were positive for MRSP during surgery, and one methicillin-resistant S. aureus (MRSA) was detected in a cat. All MRCoPS were resistant to doxycycline, erythromycin, clindamycin and enrofloxacin, but no patients developed surgical site infections. According to their genotypic patterns, the clones obtained from the environment and human sources differed from the animal clones. Despite intensive hygienic management, a variety of MRCoPS clones were present within the surgical unit and during surgery.
Keywords: dog, methicillin-resistant coagulase-positive staphylococci, Staphylococcus pseudintermedius, surgical patient, veterinary hospital
Staphylococcus aureus, S. pseudintermedius and S. schleiferi subsp. coagulans are the major coagulase-positive staphylococci (CoPS) found in dogs with and without dermatitis [5]. These bacteria are believed to be an important cause of canine pyoderma, surgical site infections (SSIs) and otitis externa in small animal patients [15, 24], and especially members of the methicillin-resistant coagulase-positive staphylococci (MRCoPS) group. MRCoPS isolates contain the mecA gene and play a significant role in nosocomial infections in human and veterinary hospitals [8, 25], with evidence of pet-to-client transmission [17].
In recent years, the high prevalence of MRCoPS in veterinary hospitals has become increasingly evident [3, 8, 25], but the clinical impact of MRCoPS remains unclear. Methicillin-resistant S. pseudintermedius (MRSP) has been found in dogs, veterinary staff and pet owners at rates of 45, 8 and 3%, respectively, while methicillin-resistant S. aureus (MRSA) is less common (0–1.5%) [16]. MRSP has been found on the skin and in the nasal passageways of pets, as well as on inanimate sites that are touched by hands, such as keyboards and weight scales, and the floors in animal hospitals [2, 13]. MRSP also is capable of causing large outbreaks that can be difficult to control. Prolonged hospital admission and length of anesthesia were found to increase the possibility of acquiring MRSP [12]. Moreover, a link between MRSP and MRSA in the environment and recurrent infections in surgical units and wards was suspected in an equine hospital [3]. Thus, a high prevalence of MRCoPS may be seen as a risk for SSIs for post-operative animals in veterinary hospitals [3, 4, 10, 23].
The main aim of this study was to establish the identity and likely sources of the bacteria responsible for frequent cases of surgical site infections after cystotomy in the Small Animal Teaching Hospital at Chulalongkorn University, Bangkok. Systemic surveillance was established to help define the source of contamination, in particular exploring the occurrence of MRCoPS in the environment of the surgical unit, and associated with veterinary staff and surgical patients from cystotomy to post-operation. To help track the source of the infection, MRSP was analyzed for their genetic relatedness, and antimicrobial resistance profiles were generated.
MATERIALS AND METHODS
Hospital background
Samples for bacterial culture were collected from the Small Animal Teaching Hospital of the Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand, which treats approximately 140,000 patients annually. About 250 surgical cases are routinely handled by the surgical unit. In this unit, the floors are cleaned with 2.5% quaternary ammonium compound (Laboratoire Huckert’s International, Wavre, Belgium) between 6:30 to 7:00 AM, while electric dog clippers, hematocrit centrifuges, the operating theater and surgical cabinets are cleaned with 0.5% quaternary ammonium compound when the unit is not in use. Scrub suits, endotracheal tubes and scissors are autoclaved after use. This surgical unit was chosen for the study in response to an incident of SSIs observed in cystotomy patients before the study began.
Sample collection
Hand-touch sites in the surgical unit.
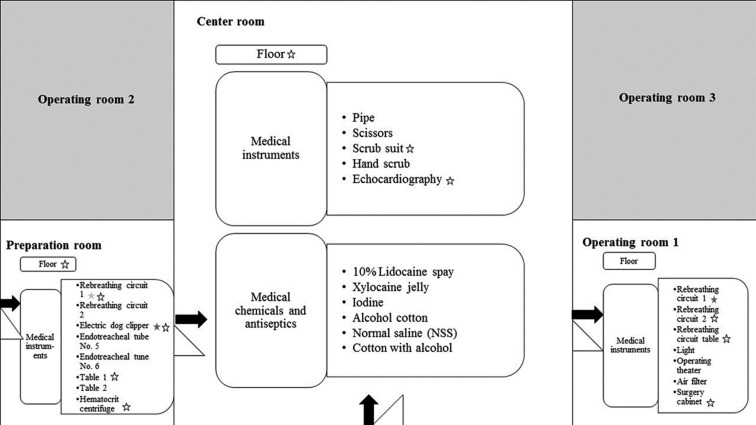
Following routine cleaning of the surgical unit between 7:00 to 8:00 AM, samples were obtained from 29 areas including floors and all sites frequently touched by hand (“hand-touch sites”). The surgical unit consisted of three subunit rooms: 1) the operating theater for abdominal surgery, including cystotomy; 2) the preparation room for hair shaving and pre-anesthetic medication; and 3) the central room where surgeons wash and dress for surgery (Fig. 1). Swab samples from the floors in the surgery (3 × 4 m2/room) and preparation room (3 × 4 m2/room) were obtained from three areas/room (left, middle and right) using 3 × 3 cm2/swabs; in the central room (6 × 8 m2), samples were collected from six areas/room: left north, left south, middle right, middle left, right north and right south. Prior to surgery, swab samples were obtained from 21 hand-touch sites on different pieces of equipment including electric dog clippers, rebreathing circuits, tables, a hematocrit centrifuge, scrub suits, endotracheal tubes, pipes, scissors, lights, operating theater, air filters, surgery cabinets and an electrocardiography unit, either as individual samples from hand-touch sites or pooled samples from floors, as previously described [14]. The samples were kept on ice for less than 2 hr before processing for culture. Medical chemicals and antiseptics, including lidocaine spray, xylocaine jelly, povidone-iodine, cotton with alcohol, normal saline and cotton with alcohol also were sampled. Each swab was separately cultured, and the colony numbers for each site of collection then were analyzed by determining the average number of colonies per area swabbed.
Fig. 1.
Presence of staphylococci divided by area and hand-touch sites according to floor
plan of the surgery unit in the veterinary teaching hospital. Star markers represent
subjects contaminated with staphylococci.  Methicillin-susceptible
coagulase-positive staphylococci (MSCoPS). ★Methicillin-resistant coagulase-positive
staphylococci (MRCoPS). ☆Coagulase-negative staphylococci.
Methicillin-susceptible
coagulase-positive staphylococci (MSCoPS). ★Methicillin-resistant coagulase-positive
staphylococci (MRCoPS). ☆Coagulase-negative staphylococci.
Veterinarians and nurses in the surgical unit
Twelve nasal passageway samples were obtained from healthy veterinarians and veterinary assistants in the surgical unit with the approval of the Ethical Review Committee for Research Involving Human Research Subjects, Health Science Group, Chulalongkorn University (081/54). All staff had worked full time at the surgical unit for at least 5 years. Staff histories, age, gender, previous illnesses, prior antimicrobial use and occupations were recorded. Each nasal swab was soaked with sterile peptone dilution saline (PDS) (0.09% NaCl and 0.1% peptone), then inserted at least 0.5 cm into the nasal cavities, rubbed against the epithelium and subsequently stored in transport tubes containing 1 ml of PDS [6]. The samples were kept on ice for less than 2 hr before processing for culture.
Pet patients in the surgical unit
Samples were collected in 2012–2013 from 32 surgical patients treated with a cystotomy for urolithiasis (29 dogs and 3 cats, aged from 3 months to 11 years). Sampling protocols and consent forms were approved by the Institutional Animal Care and Use Committee (IACUC) (113/56). Pre/post-operative antimicrobial therapies and previous illness histories were recorded thoroughly under veterinarian authorization. Urinalysis, with bacterial culture and antimicrobial susceptibility was performed prior to the operation for follow-up treatment planning. The choice of antimicrobial for administration was based on the results from the susceptibility tests on any isolates that were present.
After induction of anesthesia using 2% xylazine HCl (Bayer, Istanbul, Turkey), the surgical area of the animal patients was shaved using electrical clippers in the preparation room. The area then was scrubbed with 4% chlorhexidine gluconate scrub (Ecolab, Bangkok, Thailand) for 5 min and wiped with 95% ethyl alcohol. This process was conducted at least twice until all skin debris had been removed. Thirty minutes before the patient was moved to the operating room, they were injected intramuscularly with an appropriate dose of the most suitable antimicrobial compound identified from antimicrobial susceptibility testing of isolates from the urine.
In the operating room, the surgical site was cleaned three times with 10% povidone-iodine (LF Asia, Bangkok, Thailand) for 1 min and wiped with 95% ethyl alcohol before the initial incision. The first sample (T1) was collected using a sterile cotton swab at a 1 × 1 cm2 site at the incision line. Within 5 min after entering the abdomen, a second sample (T2) was swabbed from the peritoneum 0.5 cm under the incision line. After the cystotomy procedure, abdominal lavage was performed four to five times using approximately 200 ml of 37oC sterile normal saline. The third sample (T3) was obtained at the same site as T2. The skin incision line was stitched using monofilament sutures in both the inner and outer layers of the skin. The entire procedure was completed within less than 2 hr in all cases. The patients then were taken home for convalescence. At days 7–10 post-operation, the sutures were removed from the uncomplicated operation wound in the surgical unit. If SSI was present, a fourth sample (T4) was collected from the infected site.
Staphylococcal and MRCoPS identification
All swab samples were cultured within 2 hr after collection. A total of 100 µl sample suspension from floors, instruments and disinfectant agents was incubated on Baird-Parker agar at 37°C for 48 hr for enumeration [9] and on mannitol salt agar containing 0.5 µg/ml of oxacillin (MSA-O) at 35oC for 48 hr for isolation of methicillin-resistant S. pseudintermedius [18]. After counting the bacterial colonies, at least three staphylococcus-like colonies were picked for identification.
The staphylococci were identified using biochemical tests [5]. Species identification was confirmed by multiplex-PCR (M-PCR) [19]. The control strains were S. aureus ATCC 25923, S. pseudintermedius CVMC 0108, S. schleiferi subsp. coagulans CVMC 0208 (canine origin), S. intermedius CVMP 0309 and S. delphini CVMP 0109 [5].
The presence of MRCoPS amongst suspected colonies grown on MSA-O was confirmed by oxacillin disk diffusion following the Clinical Laboratory Standardization Institute procedure [7] and by looking for the presence of the mecA gene using PCR [22]. S. aureus ATCC 25923 and methicillin-resistant S. aureus (MRSA) N315 were used as negative and positive controls for the mecA gene, respectively.
Antibiograms
The antimicrobial susceptibility of MRCoPS to 11 antimicrobials (Oxoid, Hampshire, U.K.) [amoxicillin/clavulanic acid (20/10 µg), cefazolin (30 µg), cefoxitin (30 µg), clindamycin (2 µg), doxycycline (30 µg), enrofloxacin (5 µg), erythromycin (15 µg), gentamicin (30 µg), imipenem (5 µg), mupirocin (5 µg) and trimethoprim/sulfamethoxazole (1.25/23.75 µg)] was determined using the disk diffusion method [7].
Molecular typing
The SCCmec types of methicillin-resistant S. pseudintermedius (MRSP) were identified using a multiplex PCR for detection of the conserved fragments of the mec gene complex and ccr gene complex [16]. Pulsed-field gel electrophoresis (PFGE) illustrated the DNA fingerprint pattern of S. pseudintermedius using the Cfr9I restriction enzyme. DNA separation was achieved using 6 V/cm of voltage with a switch time of 0.5–5 sec for 18 hr and 20–25 sec for 5 hr in a CHEF-DRIII apparatus (Bio-Rad, Hercules, CA, U.S.A.) [21]. The genetic relatedness of the strains was analyzed by dendrogram construction using UPGMA in the GeneDirectory program (Syngene, Frederick, MD, U.S.A.) and setting 1.0% position tolerance. The DNA marker for gel normalization was XbaI-digested chromosomal DNA of Salmonella Braenderup H9812. PFGE clusters were grouped by more than 80% similarity of patterns.
Data analysis
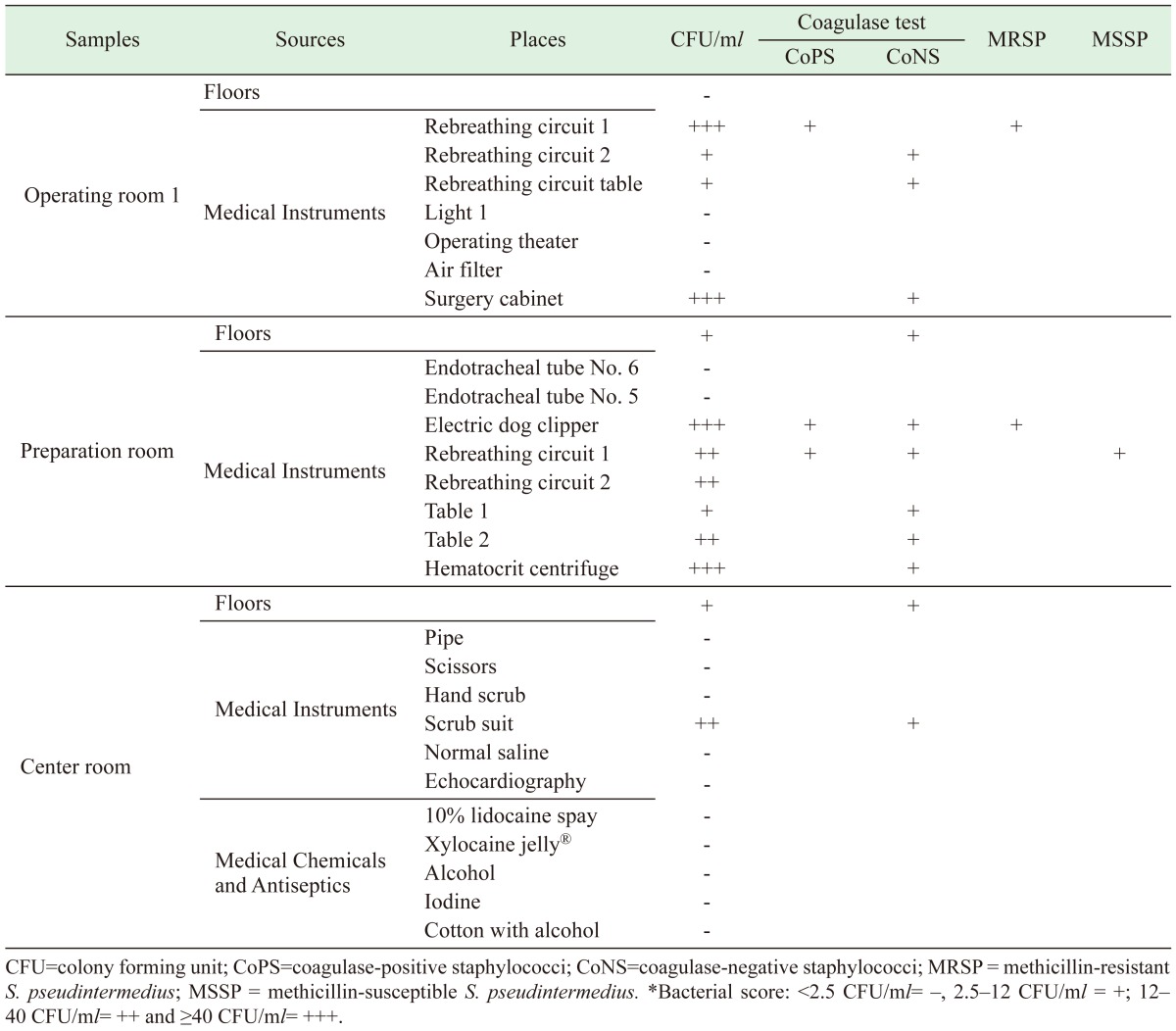
In this study, descriptive analysis was performed using the IBM SPSS Statistics Desktop version 22.0 (SPSS Inc.; Chicago, IL, U.S.A.). The populations of CoPS and MRCoPS were described by percentile. The criteria for bacterial growth on hand-touch sites and floors were those used by Dancer (2008): <2.5 CFU/ml= –, 2.5–12 CFU/ml = +; 12–40 CFU/ml = ++ and ≥ 40 CFU/ml = +++. GeneDirectory® software associated with the dice coefficient (1.5) was used to analyze the PFGE patterns.
RESULTS
Hand-touch sites and humans
Staphylococci isolated from the hand-touch sites consisted of 11/29 coagulase-negative staphylococci (CoNS) and 3/29 CoPS in all rooms. All rebreathing circuits in the operating room were positive for staphylococci, including MRSP. In operating room 1 and the preparation room either, both or one of CoPS and CoNS was found on the rebreathing circuit and electric clippers (Table 1 and Fig. 1). Two of three CoPS were MRSP with a high colony count. There was no detectable MRSP in the nasal passageways of staff in the surgical unit, but MRSA was found on one veterinarian. None of the disinfectant solutions or chemicals contained cultivable bacteria.
Table 1. Occurrence and number of staphylococci detected within three rooms of the surgical unit.
Animals
The 29 dogs and three cats had an average age of 6.78 ± 2.3 years. They all had follow-up examinations for MRCoPS detection during their cystotomy operation. Their histories pre-surgery based on urine culture and antibiotic use are presented in Table 2. Seven of 32 pets (nine observations) were found to have staphylococci in at least one sample, comprising four MSSP and four MRSP on dogs at T2 and T3, and one MRSA on a cat at T1. The number of bacterial colonies growing on the primary agar ranged from 0–7 CFU/swab. No staphylococci were detected on dogs at T1. Six of seven cases were treated with enrofloxacin during or after surgery. Only Dog 8 was positive for MRCoPS in at least two samples. There were no SSIs during post-operative care (T4).
Table 2. Staphylococcal detection and identification in surgical patients and their urine culture and antimicrobial histories.
| Pet No. | Sp. | Positive sample | Urine CULTURE | Pre-operative antimicrobial history (OP) | Pre-operative medicine (IM) | Post-operative antimicrobial history (OP) | ||
|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | ||||||
| Dog 6 | D | MSSP | CoNS | Enrofloxacin | Enrofloxacin | Enrofloxacin | ||
| Dog 8 | D | MRSP | MRSP | CoPS | Enrofloxacin | Enrofloxacin | Enrofloxacin | |
| Dog 11 | D | MSSP | Corynebacterium sp. | Amoxicillin/clavulanic acid | Cefazolin | Cephalexin | ||
| Cat 13 | C | MRSA | MSSP | ND | Enrofloxacin | Enrofloxacin | Enrofloxacin | |
| Dog 15 | D | MRSP | CoPS, Pseudomonas sp. | Enrofloxacin, Doxycycline | Enrofloxacin | Enrofloxacin | ||
| Dog 18 | D | MSSP | CoPS, Proteous sp. | Enrofloxacin, Amoxicillin/clavulanic acid | Enrofloxacin | Enrofloxacin | ||
| Dog 19 | D | MRSP | CoPS, Corynebacterium sp. | Marbofloxacin, Metronidazole | Enrofloxacin | Enrofloxacin | ||
Sp.=Species; C=cat; D=dogs; MRSP=Methicillin-resistant S. pseudintermedius; MRSSc=Methicillin-resistant S. schleiferi subsp. coagulans; MRSA=Methicillin-resistant S. aureus; MSSP=Methicillin-sensitive S. pseudintermedius, CoNS=Coagulase-negative staphylococci; CoPS=Coagulase-positive staphylococci; ND=not detected.
Antibiograms, SCCmec type and MRSP clone relatedness
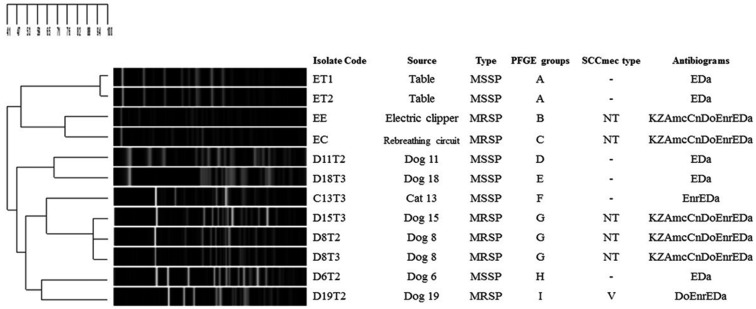
The antibiogram patterns and PFGE typing of S. pseudintermedius isolated from surgical patients and hand-touch sites are shown in Fig. 2. PFGE types G (D15T3, D8T2 and D8T3) and I (D19T2) belonging to MRSP were detected on dog patients at T2 and T3 of cystotomy, whereas B and C types were found on veterinary equipment. PGFE types A and D belonging to MSSP were detected on the tables, scrub suits and floor, but types E, F and H were found on surgical tissue. Feline MRSA was resistant to oxacillin, cefoxitin, erythromycin and clindamycin, while all MRSP were resistant to amoxicillin/clavulanic acid, cefazolin, cefoxitin, doxycycline, erythromycin, clindamycin and enrofloxacin. MSSP (PFGE types A, D, E and H) were resistant only to erythromycin and clindamycin, and one MSSP (F) also was resistant to enrofloxacin. By SCCmec typing, only D19T2 could be classified as V type, whereas the others were presented on an untypable cassette. Four identical PFGE types were distinguished: these included G type from Dog 8 and Dog 15, I type from Dog 19, B type from the electric clippers, C type from the rebreathing circuit and D type from Dog 19.
Fig. 2.
PGFE patterns, SCCmec types and antibiograms of S. pseudintermedius derived from patient and environmental sources. Kz=cefazolin, Do=doxycycline, E=erythromycin, Da=clindamycin, Mup=mupirocin, Sxt=co-trimoxazole, Cn=gentamicin, Amc=Amoxicillin/clavulanic acid, Enr=enrofloxacin, “-”=not detected, NT=non-typable.
DISCUSSION
The surgical unit in the veterinary teaching hospital was used as a model for monitoring bacterial distribution in a hospital unit. Its good hygienic management, antibiotic use protocols, high frequency of daily operations and restriction to outsiders meant that there were few confounders for the study. Despite the good hygienic conditions, staphylococci were found in the surgical unit and on equipment that remained within each room, including rebreathing circuits, surgical cabinets, electric clippers and a centrifuge. They also were present on animal patients and veterinary staff. Contamination on the scrub suits might have been transmitted by the reservoir user. As bacteria were not detected from diluted disinfectant solutions or chemicals associated with patients, they were not a source of bacterial distribution in this case. Interestingly, the higher CFU number found for hand-touch sites should be taken into reconsideration when considering hygienic manipulation and strategy in animal hospitals. CoNS, a low pathogenicity Staphylococcus, was commonly found in this study, as previously reported in a dairy environment [11].
Sample collection and cultures were performed using an aseptic technique to ensure that most of the cases were free from staphylococci. Surprisingly, MSSP, MRSP and MRSA were still found at the incision site and in the abdominal operative area, even though all patients underwent standard aseptic preparation [26]. Moreover, the result from urine cultures was not related to detection of staphylococci associated with surgery. It is speculated that staphylococci contamination might pass from adjacent areas to the incision site. However, staphylococci contamination at the surgical site was not the only factor associated with SSI, which might also result from underlying patient factors, such as immunological defects and household management [1].
DNA analysis showed the presence of a variety of MRSP and MSSP clones around the surgical unit and distinguished the animal clones found in surgical tissue from environmental clones. Thus, the most likely source of staphylococcal contamination of patients during the surgical procedure was themselves or other patients. The PFGE type G strain, with an identical antibiogram and SCCmec type, was abundant in this study. Nevertheless, this could not be considered a representative outbreak strain without performing long-term observations in relation to SSIs [3]. Using an antibiogram, enrofloxacin-resistant MRSP was detected at the surgical site, but there was no SSI; this finding confirms that low CoPS or MRCoPS contamination during surgery has an insignificant effect on SSIs [20]. Furthermore, results from in vitro susceptibility testing may not reflect the in vivo outcome, where host defense mechanism is involved. Likewise, MRSA was found on one veterinarian and one cat, confirming the low prevalence of MRSA on veterinary staff and animals [16]. The clonal relationship between the two MRSA isolates was not determined, since the cat had not been exposed to the positive veterinarian. Therefore, it would have been difficult to anticipate a causal role in transmission.
In a previous report, the risk factors for SSIs were related to pet health, pre-operative conditions, operating room environment, duration of operation, surgical instrument management, surgical attire and post-operative factors [24]. One incident of recurring infection derived from environmental surfaces was confirmed in a Swedish veterinary hospital [3]. However, most risk factors in the surgical unit were controllable, while this was not the case during convalescence as all patients convalesced in their home. Another study showed that SSIs might be caused by household contamination or wound management by clients [10]. Pet owners are likely to be the key persons involved in hygiene management in this case.
In conclusion, this report showed the presence of staphylococci, MSSP and MRCoPS, at hand-touch sites and on animal tissues during operations in a surgical unit. Electric clippers and rebreathing circuits might be the main source of contamination in this surgical unit. These results should be brought to the attention of individuals responsible for the hygiene policy of veterinary hospitals, particularly due to the multidrug-resistant properties of these bacteria.
Acknowledgments
This work was supported by the 90th Anniversary of Chulalongkorn University Fund, Ratchadaphiseksomphot Endowment Fund, F-31-GS-ES13, No. 49, the Chulalongkorn University Veterinary Science Research Fund 2014 and the National Research Council of Thailand (NRCT) 2014-2015. The authors would like to thank the surgical unit staff of the Small Animal Teaching Hospital, Faculty of Veterinary Science, Chulalongkorn University for assistance during sample collection. The Center of Excellence for Emerging, Re-emerging Infectious Diseases in Animals (EIDA) and STAR; Diagnosis and Monitoring Animal Pathogens also provided a student research scholarship. We thank Professor David J. Hampson of Murdoch University, Australia and the Office of Research Affairs, Chulalongkorn University for assistance during manuscript preparation.
REFERENCES
- 1.Bannoehr J., Guardabassi L.2012. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet. Dermatol.23: 253-266, e51-52. [DOI] [PubMed]
- 2.Bender J. B., Schiffman E., Hiber L., Gerads L., Olsen K.2012. Recovery of staphylococci from computer keyboards in a veterinary medical centre and the effect of routine cleaning. Vet. Rec. 170: 414. doi: 10.1136/vr.100508 [DOI] [PubMed] [Google Scholar]
- 3.Bergström K., Aspan A., Landén A., Johnston C., Grönlund-Andersson U.2012. b. The first nosocomial outbreak of methicillin-resistant Staphylococcus aureus in horses in Sweden. Acta Vet. Scand. 54: 11. doi: 10.1186/1751-0147-54-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergström A., Gustafsson C., Leander M., Fredriksson M., Grönlund U., Trowald-Wigh G.2012. a. Occurrence of methicillin-resistant Staphylococci in surgically treated dogs and the environment in a Swedish animal hospital. J. Small Anim. Pract. 53: 404–410. doi: 10.1111/j.1748-5827.2012.01238.x [DOI] [PubMed] [Google Scholar]
- 5.Chanchaithong P., Prapasarakul N.2011. Biochemical markers and protein pattern analysis for canine coagulase-positive staphylococci and their distribution on dog skin. J. Microbiol. Methods 86: 175–181. doi: 10.1016/j.mimet.2011.04.019 [DOI] [PubMed] [Google Scholar]
- 6.Chanchaithong P., Perreten V., Schwendener S., Tribuddharat C., Chongthaleong A., Niyomtham W., Prapasarakul N.2014. Strain typing and antimicrobial susceptibility of methicillin-resistant coagulase-positive staphylococcal species in dogs and people associated with dogs in Thailand. J. Appl. Microbiol. 117: 572–586. doi: 10.1111/jam.12545 [DOI] [PubMed] [Google Scholar]
- 7.CLSI2013. Vet01-S2 Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; Second Informational Supplement. In: Clinical and Laboratory Standards Institute. 26. [Google Scholar]
- 8.Coombs G. W., Nimmo G. R., Pearson J. C., Christiansen K. J., Bell J. M., Collignon P. J., McLaws M. L., Australian Group for Antimicrobial Resistance2009. Prevalence of MRSA strains among Staphylococcus aureus isolated from outpatients, 2006. Commun. Dis. Intell. Q. Rep. 33: 10–20. [PubMed] [Google Scholar]
- 9.Dancer S. J.2008. Importance of the environment in meticillin-resistant Staphylococcus aureus acquisition: the case for hospital cleaning. Lancet Infect. Dis. 8: 101–113. doi: 10.1016/S1473-3099(07)70241-4 [DOI] [PubMed] [Google Scholar]
- 10.Davis M. F., Iverson S. A., Baron P., Vasse A., Silbergeld E. K., Lautenbach E., Morris D. O.2012. Household transmission of meticillin-resistant Staphylococcus aureus and other staphylococci. Lancet Infect. Dis. 12: 703–716. doi: 10.1016/S1473-3099(12)70156-1 [DOI] [PubMed] [Google Scholar]
- 11.De Visscher A., Supré K., Haesebrouck F., Zadoks R. N., Piessens V., Van Coillie E., Piepers S., De Vliegher S.2014. Further evidence for the existence of environmental and host-associated species of coagulase-negative staphylococci in dairy cattle. Vet. Microbiol. 172: 466–474. doi: 10.1016/j.vetmic.2014.06.011 [DOI] [PubMed] [Google Scholar]
- 12.Grönthal T., Moodley A., Nykäsenoja S., Junnila J., Guardabassi L., Thomson K., Rantala M.2014. Large outbreak caused by methicillin resistant Staphylococcus pseudintermedius ST71 in a Finnish Veterinary Teaching Hospital--from outbreak control to outbreak prevention. PLOS ONE 9: e110084. doi: 10.1371/journal.pone.0110084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton E., Kaneene J. B., May K. J., Kruger J. M., Schall W., Beal M. W., Hauptman J. G., DeCamp C. E.2012. Prevalence and antimicrobial resistance of Enterococcus spp and Staphylococcus spp isolated from surfaces in a veterinary teaching hospital. J. Am. Vet. Med. Assoc. 240: 1463–1473. doi: 10.2460/javma.240.12.1463 [DOI] [PubMed] [Google Scholar]
- 14.Hoet A. E., Johnson A., Nava-Hoet R. C., Bateman S., Hillier A., Dyce J., Gebreyes W. A., Wittum T. E.2011. Environmental methicillin-resistant Staphylococcus aureus in a veterinary teaching hospital during a nonoutbreak period. Vector Borne Zoonotic Dis. 11: 609–615. doi: 10.1089/vbz.2010.0181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igimi S., Takahashi E., Mitsuoka T.1990. Staphylococcus schleiferi subsp. coagulans subsp. nov., isolated from the external auditory meatus of dogs with external ear otitis. Int. J. Syst. Bacteriol. 40: 409–411. doi: 10.1099/00207713-40-4-409 [DOI] [PubMed] [Google Scholar]
- 16.Kondo Y., Ito T., Ma X. X., Watanabe S., Kreiswirth B. N., Etienne J., Hiramatsu K.2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51: 264–274. doi: 10.1128/AAC.00165-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris D. O., Lautenbach E., Zaoutis T., Leckerman K., Edelstein P. H., Rankin S. C.2012. Potential for pet animals to harbour methicillin-resistant Staphylococcus aureus when residing with human MRSA patients. Zoonoses Public Health 59: 286–293. doi: 10.1111/j.1863-2378.2011.01448.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki T., Kikuchi K., Tanaka Y., Takahashi N., Kamata S., Hiramatsu K.2007. Methicillin-resistant Staphylococcus pseudintermedius in a veterinary teaching hospital. J. Clin. Microbiol. 45: 1118–1125. doi: 10.1128/JCM.02193-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki T., Tsubakishita S., Tanaka Y., Sakusabe A., Ohtsuka M., Hirotaki S., Kawakami T., Fukata T., Hiramatsu K.2010. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J. Clin. Microbiol. 48: 765–769. doi: 10.1128/JCM.01232-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmid-Hempel P., Frank S. A.2007. Pathogenesis, virulence, and infective dose. PLoS Pathog. 3: 1372–1373. doi: 10.1371/journal.ppat.0030147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soedarmanto I., Kanbar T., Ülbegi-Mohyla H., Hijazin M., Alber J., Lämmler C., Akineden Ö., Weiss R., Moritz A., Zschöck M.2011. Genetic relatedness of methicillin-resistant Staphylococcus pseudintermedius (MRSP) isolated from a dog and the dog owner. Res. Vet. Sci. 91: e25–e27. doi: 10.1016/j.rvsc.2011.01.027 [DOI] [PubMed] [Google Scholar]
- 22.Strommenger B., Kettlitz C., Werner G., Witte W.2003. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 41: 4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turk R., Singh A., Weese J. S.2015. Prospective surgical site infection surveillance in dogs. Vet. Surg. 44: 2–8. [DOI] [PubMed] [Google Scholar]
- 24.Weese J. S.2008. A review of post-operative infections in veterinary orthopaedic surgery. Vet. Comp. Orthop. Traumatol. 21: 99–105. [DOI] [PubMed] [Google Scholar]
- 25.Weese J. S., van Duijkeren E.2010. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet. Microbiol. 140: 418–429. doi: 10.1016/j.vetmic.2009.01.039 [DOI] [PubMed] [Google Scholar]
- 26.Zubrod C. J., Farnsworth K. D., Oaks J. L.2004. Evaluation of arthrocentesis site bacterial flora before and after 4 methods of preparation in horses with and without evidence of skin contamination. Vet. Surg. 33: 525–530. doi: 10.1111/j.1532-950X.2004.04074.x [DOI] [PubMed] [Google Scholar]