Abstract
Irisin is secreted by skeletal muscle during exercise and influences energy and metabolic homeostasis. This hormone is a cleaved and secreted fragment of fibronectin type III domain-containing 5 (FNDC5). Elucidation of the FNDC5 gene regulation mechanism is necessary to clarify the function of irisin as a potential therapeutic target in human metabolic diseases. Thus, we investigated the genetic and epigenetic mechanisms that regulate expression of the FNDC5 gene. FNDC5 mRNA was strong expressed in major energy-dependent human tissues, including heart, brain, liver, and skeletal muscle. Promoter analysis of the FNDC5 gene revealed that the core promoter region of the FNDC5 gene contained one CpG island that was located just upstream of the transcriptional start site for variants 2 and 3. Treatment with the histone deacetylase inhibitor sodium butyrate and the demethylating agent 5-azacytidine increased mRNA expression of FNDC5 in Huh7 cells. Prediction of transcription factor binding sites suggested that the glucocorticoid receptor was involved in the regulation of FNDC5 expression, and indeed, cortisol treatment increased mRNA expression of FNDC5 in Huh7 cells. Collectively, these findings offer insight into the genetic and epigenetic regulation of FNDC5, providing the initial steps required for understanding the role of irisin in the metabolic homeostasis.
Recently, a new muscle-derived hormone, irisin, was reported. This protein is secreted from skeletal muscle and induced by overexpression of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α)1. Irisin is produced as a result of cleavage of the membrane protein fibronectin type III domain-containing protein 5 (FNDC5) and contributes to maintenance of metabolic homeostasis via induction of the “browning” of white adipocytes through increased expression of uncoupling protein 1 (UCP1), leading to increased energy expenditure1,2,3. Over the past 2 years, multiple studies on FNDC5 and the novel hormone in both rodents and humans have been undertaken3,4,5,6,7. Recently, the diverse role of FNDC5/irisin in diseases, such as diabetes8,9, hypothyroidism10, atherosclerosis11, nonalcoholic fatty liver disease12, and preeclampsia13, has been investigated. Especially, serum irisin level was tightly related with metabolic diseases and activation of FNDC5 showed beneficial clinical effects in animal and human8,9,14,15,16,17. In the animal study, FNDC5 knockout mice showed severe hepatic steatosis with impaired autophagy and fatty acid oxidation. In contrast, FNDC5 overexpression prevented hyperlipidemia, hepatic lipid accumulation and autophagy impairment in the high fat dieted mouse16. In the human study, the patients with type 2 diabetes (T2D) had decreased serum irisin level and T2D drug metformin or glucagon-like peptide-1(GLP-1) treatments increased serum irisin level in the T2D patients8,9.
Although accumulated evidences suggest that FNDC5 and irisin play important roles in the regulation of energy metabolism in multiple tissues, the detailed mechanisms for the regulation of expression of these factors remain unknown. Therefore, we investigated the genetic and epigenetic regulation mechanisms of this hormone by using several human tissue samples and hepatocellular carcinoma cell lines that differentially express FNDC5 variants genes.
Results
Differential expression of FNDC5 in human tissues and cell lines
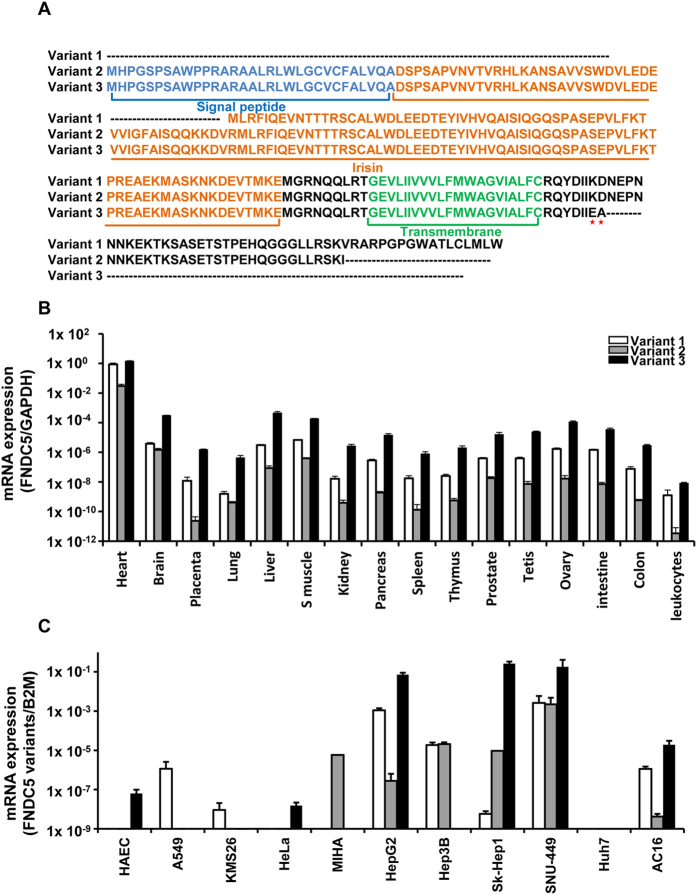
In humans, the FNDC5 gene has three variants that are distinguished by the signal peptide and C-terminal amino acids (Table 1). Multiple protein sequence alignments of human FNDC5 gene (Fig. 1A) were used to design unique primers to distinguish the variants in real-time PCR analysis. To determine the tissue specific expression patterns of FNDC5 variant genes, we performed auantitative real-time PCR of 16 human tissues and 11 human normal or cancer cell lines such as HAEC (human aortic endothelial cell), A549 (adenocarcinomic human alveolar basal epithelial cells), KMS26 (plasma cell myeloma cell), HeLa (cervical adenocarcinoma cell line), MIHA (nontumorigenic immortalized human hepatocyte cell line) HepG2, Hep3B, Sk-Hep1, SNU449, and Huh7(human hepatoma cell lines) and AC16 (human cardiomyocyte cell line).
Table 1. Human FNDC5 variants information.
| Variant | NCBI | Amino Acid | NCBI Description |
|---|---|---|---|
| FNDC5-1 | NP_001165412.1 | 153 | This variant (1) represents the longest transcript and encodes the shortest isoform (1). |
| FNDC5-2 | NP_715637.2 | 212 | This variant (2) contains multiple differences in the UTRs and coding region, compared to variant 1. It initiates translation from an alternate “AUA” start codon that is conserved as an “AUG” start codon in orthologs. The encoded isoform (2) is longer and has distinct N- and C-termini, compared to isoform 1. |
| FNDC5-3 | NP_001165411.2 | 181 | This variant (3) contains multiple differences in the UTRs and coding region, compared to variant 1. It initiates translation from an alternate “AUA” start codon that is conserved as an “AUG” start codon in orthologs. The encoded isoform (3) is longer and has distinct N- and C-termini, compared to isoform 1. |
Figure 1. Schematic sequence of human FNDC5 variants and mRNA expression of FNDC5 variants in human tissues and cell lines.
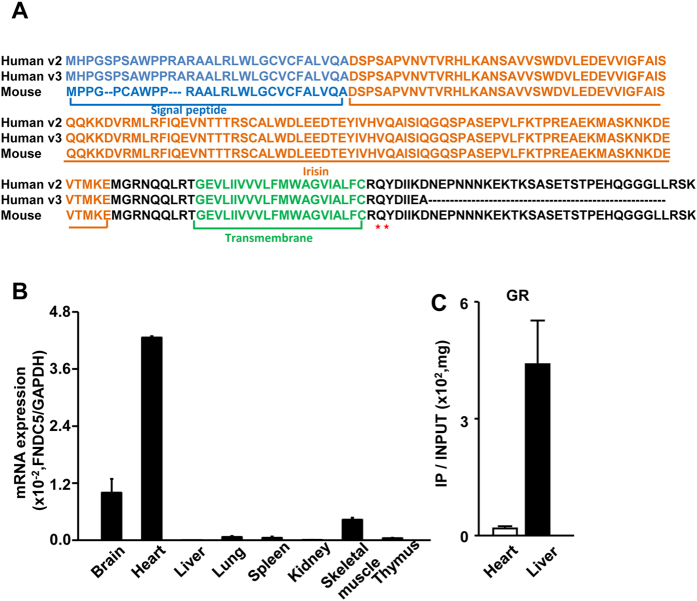
(A) Multiple amino acid sequence alignment of the variants of the human FNDC5 gene. Variant 1 contains no signal peptide and is cleaved to the irisin sequence. Variants 2 and 3 have a different start codon (ATA) than variant 1, and the C-terminal amino acids are KD → EA in variant 3 (blue = signal peptide, orange = irisin, green = transmembrane sequence). (B) The mRNA levels of the three FNDC5 variants in human tissues are shown. (C) The mRNA levels of the three FNDC5 variants are shown for human cell lines. The expression level of each sample was measured by quantitative real-time RT-PCR using GAPDH or B2M for normalization. HAEC; human aortic endothelial cell, A549; adenocarcinomic human alveolar basal epithelial cells, KMS26; Plasma cell myeloma cell, HeLa; cervical adenocarcinoma cell line, MIHA; nontumorigenic immortalized human hepatocyte cell line, HepG2, Hep3B, Sk-Hep1, SNU449, and Huh7: human hepatoma cell lines, AC16: human cardiomyocyte cell line.
The real-time PCR results showed different mRNA expression levels for the FNDC5 gene in several tissues and cell lines (Fig. 1B and C). The highest expression of FNDC5 mRNA was found in the heart, with higher expression in the brain, liver, skeletal muscle, and ovary compared to other tissues. Expressions of FNDC5 variants were diverse in different types of normal and cancer cell lines. The human adult cardiomyocyte AC16 and the human hepatocellular carcinoma cell lines including HepG2, Sk-Hep1 and SNU449 showed high mRNA expression level of FNDC5 variants which were observed in normal heart and liver tissues, while Huh7 cells exhibited extremely low levels of the three FNDC5 variants. We tested MIHA, an immortalized cell line established from human hepatocytes, as a model of non-tumorigenic normal human hepatocytes. However, the gene expression pattern of FNDC5 in the cell line was quite different from liver tissues. Based on this screening result, we selected hepatocellular carcinoma cell lines in the proceeding studies to discriminate the transcriptional regulation mechanism of FNDC5.
Modification of the CpG island region within the FNDC5 promoter correlates with regulation of FNDC5 expression
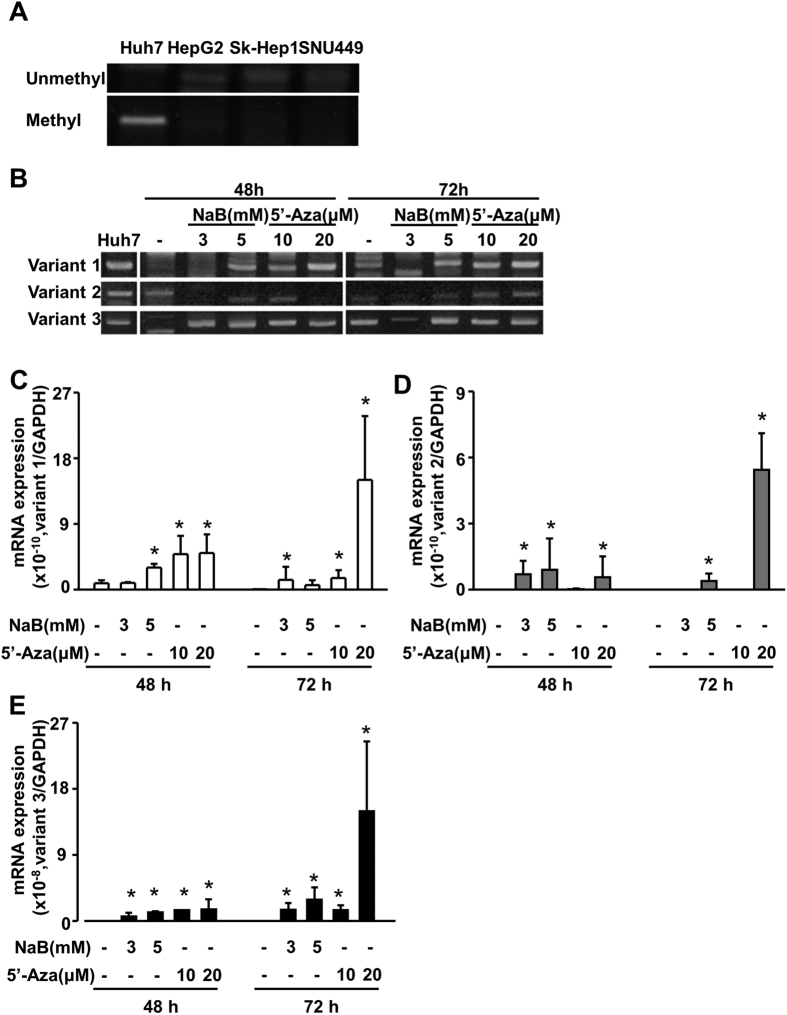
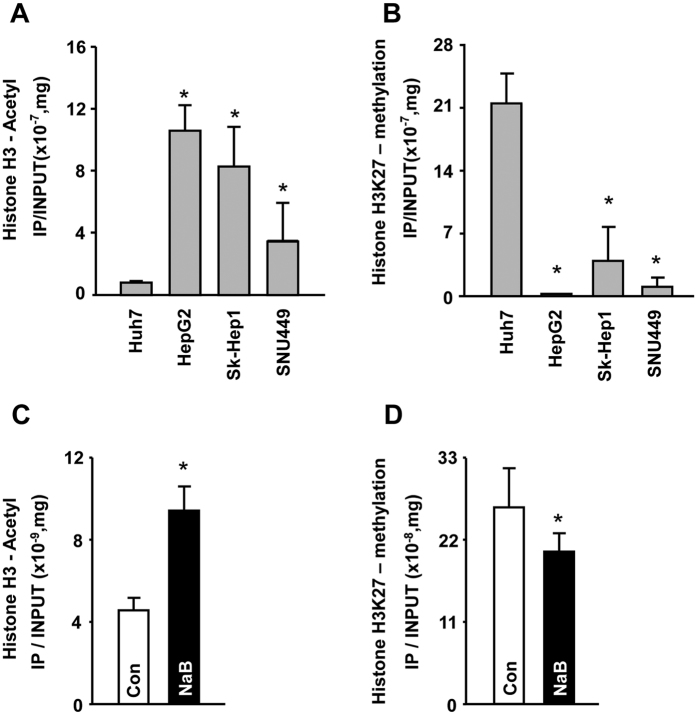
To further elucidate the epigenetic regulation mechanisms involved in FNDC5 gene expression, we predicted the presence of CpG islands using Meth primer site (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). According to this prediction program, a single CpG island region is located at −52~−442 bp of the FNDC5 variant 2/3 promoter. We designed primers for methylation-specific PCR and chromatin-immunoprecipitation (ChIP) analysis (Fig. 2) to examine the epigenetic modification at the CpG island in four cell lines (Huh7, HepG2, Sk-Hep1, and SNU449). Huh7 cells were highly methylated compared to the other cell lines (Fig. 3A). Next, we determined the mRNA expression levels of the three variants of FNDC5 following treatment of cells with the histone deacetylase (HDAC) inhibitor sodium butyrate (NaB) or the DNA demethylation agent 5-azacytidine (5-Aza). In Huh7 cells, the mRNA expression of all variants was increased by NaB or 5-Aza treatment (Fig. 3B–E). Based on our previous results, we investigated histone H3 modification using a ChIP assay with H3Ac and H3K27me2 antibodies. Levels of histone H3 acetylation were significantly suppressed in Huh7 cells in comparison to levels in other cell types (Fig. 4A). In contrast to the acetylation levels, Huh7 cells showed significantly greater histone H3 K27 di-methylation levels than the other cell types (Fig. 4B). These data indicate that the epigenetic regulation at histone H3 in the CpG island regulates the mRNA expression of FNDC5. Thus, for confirmation, we treated Huh7 cells with 3 mN NaB for 72 h and then performed a ChIP assay with histone H3Ac and H3K27me2 antibodies. NaB-treated cells exhibited higher levels of histone H3 acetylation and lower H3 K27 methylation levels (Fig. 4C and D) in contrast with the first set of ChIP results. These findings confirm that histone H3 modification at the CpG island contribute to the transcriptional regulation of FNDC5.
Figure 2. Structure of human FNDC5 variants promoter.
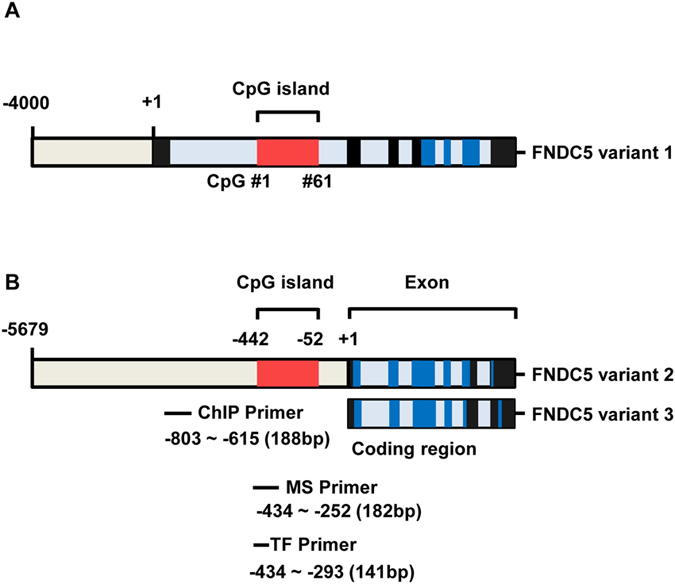
(A and B) The human FNDC5 variants have different promoter regions. The CpG island in the FNDC5 gene promoter was predicted by “Methprimer” (http://www.urogene.org/cgi-bin/methprimer/Methprimer.cgi). The CpG island is located at −52 to −442 bp in the FNDC5 promoter, and this region has 61 CpG sites. The CpG island is close to the variant 2 and 3 transcription start sites. Red = CpG island, blue = translational region, black = untranslational region, ChIP primer = chromatin immunoprecipitation primer, MS primer = methylation-specific PCR primer set, TF primer = transcription factor-binding region-specific primer set.
Figure 3. Sodium butyrate (NaB) and 5-azacytidine (5-Aza) increase mRNA expression of FNDC5 in human hepatocellular carcinoma cell lines.
(A) Methylation in the CpG island region was measured by methylation-specific PCR. (B–E) Huh7 cells were treated with NaB or 5-, and mRNA expression levels of the three FNDC5 variants were measured by RT-PCR and quantitative real-time PCR.
Figure 4. Histone H3 modification at the CpG island in the FNDC5 promoter is related to the transcriptional regulation of the FNDC5 gene.
(A and B) ChIP was performed in four cell lines (Huh7, HepG2, Sk-Hep1, and SNU449) using histone H3 acetylation and K27 di-methylation antibodies. (C and D) ChIP analysis of the CpG island was performed as for (A) and (B) following treatment of cells with/without 3 mM NaB for 72 h, and expression was measured by quantitative real-time PCR using ChIP primers.
The core promoter of the FNDC5 gene spans the region from −1 bp to −1 kb
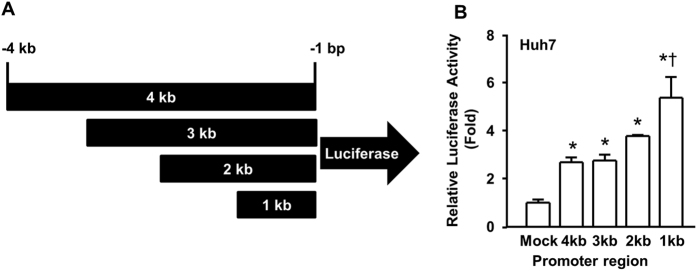
To further probe the genetic regulation of FNDC5 gene expression, we generated FNDC5 deletion mutants in a luciferase vector (Fig. 5A) for luciferase activity assays in Huh7 cells. Interestingly, the promoter region from −1 bp to −1 kb exhibited high activity compared to that of the other deletion constructs (Fig. 5B). According to these data, the region between −1 bp and −1 kb in the promoter serves as the core regulatory region.
Figure 5. Analysis of FNDC5 gene expression using an FNDC5 promoter-driven luciferase reporter plasmid.
(A) Depiction of the FNDC5 gene promoter deletion mutants that were used for gene expression analysis. (B) Luciferase activity assay with FNDC5 promoter constructs was performed in Huh7 cells.
Glucocorticoid receptor (GR) regulates FNDC5 gene expression
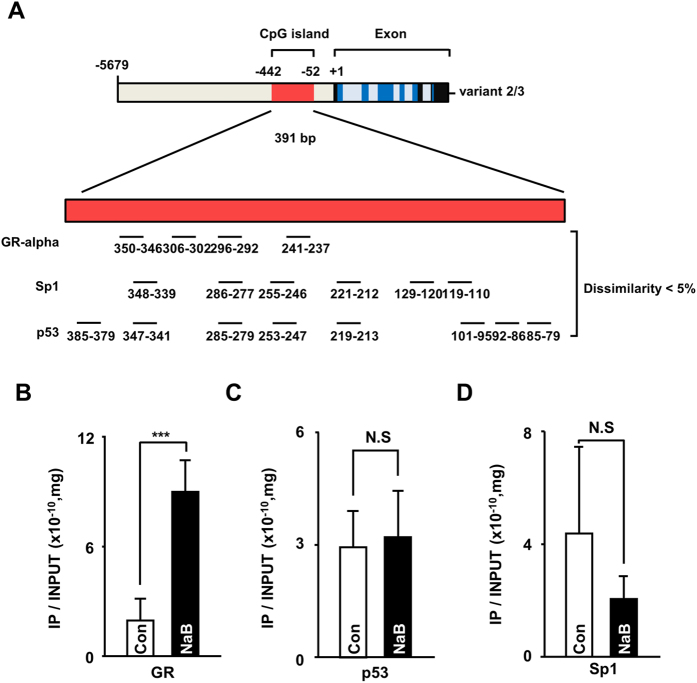
To examine the regulation mechanism associated with the CpG island within the core promoter region (1 bp - 1 kb), we screened transcription factors that were predicted to bind the CpG island in the FNDC5 gene promoter with the PROMO and transcription factor binding site (TFBS) search analysis tool18. Of the identified potential factors, we selected three transcription factors: GR-alpha, Sp1, and p53. The binding site for these transcription factors was abundant and yielded high search accuracies in the CpG island (Fig. 6A). Next, we performed ChIP analysis with these transcription factors in Huh7 cells in the presence or absence of NaB. Interestingly, the GR-alpha binding sites in the CpG island were significantly increased by NaB treatment; however, neither the p53 nor Sp1 binding sites responded to NaB stimulation (Fig. 6B–D). For confirmation, we examined mRNA expression levels following treatment of cells with different doses of cortisol to activate the GR signaling pathway. Cortisol treatment induced increased mRNA expression of FNDC5 variant 1 and 2 in Huh7 cells (Fig. 7). The effective dose of cortisol required to enhance transcription of FNDC5 differed between variant 1 and variant 2 and was 5 μM and 20 μM, respectively (Fig. 6A,B). The effect of cortisol on FNDC5 variant 3, however, was not significant for any treatment dose (Fig. 6C). Thus, the GR-alpha appears to play a role in regulation of FNDC5 variant expression.
Figure 6. GR is a regulator of FNDC5 gene expression.
(A) Schematic of the predicted transcription factor binding sites in the FNDC5 gene promoter. Prediction of these binding sites was performed using PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) and TF search (http://www.cbrc.jp/research/db/TFSEARCH.html). Each transcription factor has <5% dissimilarity. (B–D) ChIP assay in NaB-treated and untreated Huh7 cells with GR-α, p53, and SP1 antibodies. N.S; none significant, ***p < 0.001.
Figure 7. Cortisol increased mRNA expression of FNDC5 variants 1 and 2 in Huh7 cells.
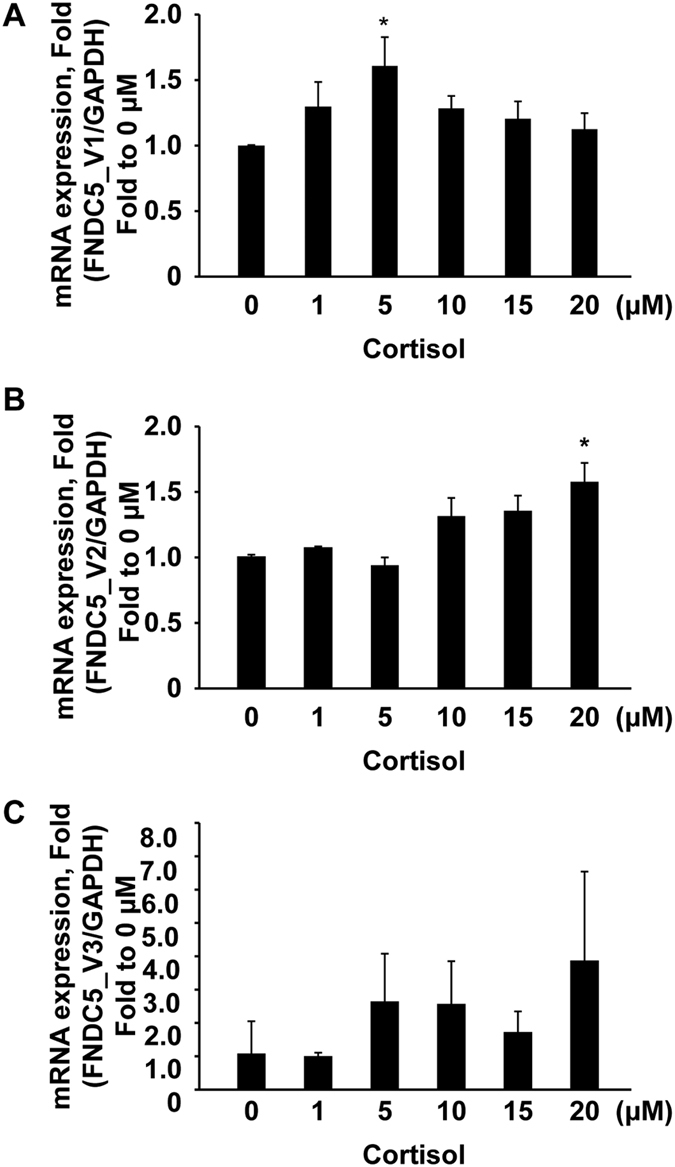
Different doses of cortisol (1–20 μM) treatment significantly increased mRNA expression of FNDC5 variant 1 (A) and variant 2 (B) after 24 h. (C) Cortisol treatment did not significantly alter FNDC5 variant 3 expression at any dose.*p < 0.05.
FNDC5 expression is regulated similarly in mice
To explore the similarities in FNDC5 regulation between human cell lines and rodents, we first compared the amino acid sequences of FNDC5 in humans and rodents (Fig. 8A). In mice, no variants were observed, and the sequence showed remarkable similarity with human variant 2. Tissue-dependent mRNA levels of FNDC5 in mice were investigated using qRT-PCR (Fig. 8B). The observed pattern of mRNA expression in mice is similar to that observed for human variant 2. A mouse tissue ChIP assay was performed to test whether GR also binds FNDC5 genes in murine heart and liver tissue (C57BL/6, 8w, male, n = 3). ChIP with an anti-GR antibody revealed that GR binds FNDC5 in liver more strongly than that from heart (Fig. 8C). These findings in mice reflect our findings in human cell lines.
Figure 8. Expression level and regulation of the mouse FNDC5 gene.
(A) Amino acid sequence of mouse FNDC5 was compared with human variants 2 and 3. (B) The mRNA expression level of FNDC5 in mouse tissues. (C) ChIP of mouse heart and liver tissue with GR antibody.
Discussion
Irisin is generated by cleavage of FNDC5 and has recently become the focus of much research. Irisin is a myokine that is secreted by skeletal muscle and may be involved in energy and metabolic homeostasis especially in diabetes and obesity1,2,3,5,6,7. Numerous reports have suggested that irisin exerts beneficial effects on metabolic disease and have uncovered the mechanism of the involved pathway; however, these results remain controversial and have not definitely demonstrated the regulation mechanisms19. In this study, we investigated the genetic and epigenetic regulation of FNDC5 gene expression. As an essential finding of the present study, we found that FNDC5 has 3 different variants, and these variants showed various expression patterns in several types of tissues and cell lines. Remarkably, high expression of FNDC5 was detected in heart and skeletal muscle tissues. Clearly, the high expression of FNDC5 in skeletal muscle is well matched with previous reports that FNDC5 is highly expressed in skeletal muscles and in response to exercise2,6. Although the latest study showed that FNDC5 level in rat heart is comparable with skeletal muscle20, the high expression of FNDC5 gene in the human heart tissues was not previously reported, with the exception in ‘The Human Protein Atlas (http://www.proteinatlas.org)’. Thus, our result provided valuable information regarding the expression pattern of FNDC5 variant genes in the human heart. Aside from heart and skeletal muscles, the expression of FNDC5 was also high in the brain, liver, and ovary. Coincidentally, recent studies have suggested that FNDC5 is an important mediator for the beneficial effect of exercise on the brain via regulation of brain-derived neurotrophic factor21,22 and is also a mediator of hepatic glucose and lipid metabolism23. The gene expression of FNDC5 in human ovary has not been reported yet. It only had been reported that the serum irisin level of polycystic ovary syndrome (PCOS) patients were higher compared to the normal control group, however it had been uncertain whether increased irisin level is only a molecular marker for the metabolic syndrome associated with PCOS or whether irisin directly plays a role in ovarian abnormality24. The high expression of FNDC5 gene in our result might give a clue regarding the function of FNDC5 in reproductive organs. In the energy metabolism aspect, FNDC5 mRNA expression levels were higher in high energy-dependent tissues (Fig. 1). This distribution of FNDC5 mRNA expression in energy-dependent tissues suggests that not only the cleaved irisin protein but also the FNDC5 protein is involved in energy metabolism in humans.
The expression patterns of FNDC5 variants were diverse in the normal and cancer cell lines. Similar with human tissues, cell lines originating from the heart and liver showed higher expression of FNDC5 than others. However, liver cell lines (HepG2, Hep3B, Sk-Hep1 and SNU449) expressed much higher FNDC5 gene expression than heart cell lines (AC16). We also found that the FNDC5 expressions were different among tissues and matched non-tumor cell lines of heart (AC16) and liver (MIHA), which might be due to transcriptional remodeling during immortalization and adaptation of cells in in vitro environments.
Among the tested cell lines, most human hepatocellular carcinoma cells (HepG2, Hep3B, Sk-Hep1, and SNU449) exhibited high mRNA levels of the FNDC5 variants, although Huh7 cells did not. We therefore determined whether an epigenetic difference in the FNDC5 gene exists between Huh7 cells and the other cell lines.
CpG islands are short interspersed DNA sequences that play an important role in epigenetic gene transcription initiation. The DNA or histone methylation status in CpG islands is an important regulation factor that determines activation or inactivation of promoters25. We investigated the structure of the promoter regions of FNDC5 variants using the UCSC genome browser, Methprimer, and PROMO sites. We found a single CpG island in the promoter region (−1 bp to −1 kb region), and this CpG island is close to the start codon of variants 2 and 3 (Fig. 2). Analysis of DNA methylation in this CpG island using MS-PCR revealed post-modification of histone H3 or DNA using NaB/5-Aza (Fig. 3). Interestingly, Huh7 DNA was highly methylated compared to the other cell lines, and treatment with the HDAC inhibitor NaB or demethylating agent 5-Aza significantly enhanced the mRNA expression of FNDC5 in Huh7 cell line. These data indicate that different FNDC5 mRNA expression levels in human hepatocellular carcinoma cells result from methylation of DNA or histones in the CpG island of the promoter (Fig. 3)26. These epigenetic modifications were further tested by comparison of histone H3 acetylation and H3K27 di-methylation levels between Huh7 and the other cells. Both deacetylation and methylation have repressive effects on the transcription of a gene in cancer cells27. FNDC5 expression was clearly lower in Huh7 cells concomitant with a higher level of histone H3K27 methylation and a lower level of H3 acetylation in the CpG island. Moreover, treatment with the HDAC inhibitor NaB significantly increased the acetylation level and decreased the methylation level of the CpG island in the Huh7 cells (Fig. 4).
The role of H3K27me2 has not been as thoroughly studied as that of H3K27me3, which is known to be important for epigenetic regulation. Trimethylation of H3K27 induces inactivation of gene expression. In addition, H3K27me1 is associated with activation of promoters25,28,29. A recent study demonstrated that H3K27me2 has a broad effect on promoters and a role in silencing non-cell type-specific enhancers28,29,30. In another study, H3K27me2 was found to be associated with hypo-acetylation30 and that acetylation of H3K27 exerts opposite and antagonistic effects to H3K27me330,31. These results suggest that FNDC5 gene transcription is epigenetically regulated by histone H3 acetylation and H3K27 di-methylation.
To identify the major transcription factor promoter binding site, we constructed FNDC5 promoter deletion mutants in a luciferase vector to detect promoter activity in Huh7 cells (Fig. 5A). Interestingly, the −1 kb region displayed the highest activity compared with the other deletion constructs (Fig. 5B). Subsequently, we searched for key transcription factors that bind to the CpG island within the identified promoter region (−1 kb region). Three transcription factors, GR-alpha, Sp1, and p53, were predicted with a probability >95% to bind to this region and were most frequently observed (Fig. 6A). ChIP assay analysis of these three key transcription factors in the presence or absence of NaB indicated that GR exhibited a higher binding affinity in the NaB-treated group (Fig. 6B–D). In confirmation of this result, we found that cortisol, which can boost GR signaling in Huh7 cells, increased the mRNA expression of FNDC5 variants 1 and 2. Variant1 mRNA expression was highest at a cortisol dose of 5 μM but decreased at higher doses, while variant 2 mRNA expression gradually increased and then was significantly increased by a cortisol treatment dose of 20 μM. Interestingly, variant 3 did not respond to cortisol treatment at all. These results suggest that different FNDC5 variants respond differently to cortisol or corticosteroids at the transcriptional level.
Collectively these results indicated that GR-α is a positive regulator of FNDC5 in human hepatocellular cells and that FNDC5 transcription may be regulated by cortisol levels. Circulating cortisol levels are increased only via high-intensity exercise training with >60% VO2 max levels32, and irisin response is also greater during high-intensity exercise than during low-intensity exercise33. These studies strongly support the hypothesis that high-intensity exercise enhances circulating cortisol levels and increases GR binding to the FNDC5 promoter site, resulting in increased transcription of the FNDC5 gene and consequently increased irisin levels.
Glucocorticoids and the GR are an important genomic regulator that controls the transcription of essential genes involved in development, energy metabolism, immune system, cardiovascular system, and neuronal systems34,35. Also, GR engages in cross talk with estrogen receptors to reprogram the chromatin configuration36. The GR complex binds to target genes and regulates gene expression via glucocorticoid response element (GRE) complex with p300, CBP, PCAF, and SRC that induce histone acetylation37,38,39,40. PGC-1a is well known as a master regulator of energy metabolism and a key regulator of irisin and FNDC51,41. In addition, PGC-1a is a co-activator of nuclear receptors, including GR, and PGC-1α elevates the binding of GR to its target binding sites41,42. These results suggest the existence of an inter-relationship among GR, PGC-1α, and FNDC5 regulation. Taken together, these data suggest that GR and cortisol may be potential regulating factors that are involved in exercise-induced PGC-1a and FNDC5/irisin regulation (Fig. 8D).
Although we demonstrated the GR is a possible transcriptional regulator of FNDC5 in human cell lines and mouse liver tissue, we did not yet test whether exogenous cortisol treatment increases FNDC5 expression and circulating irisin levels in an animal model. The inter-relationship between cortisol, GR, and FNDC5/irisin requires additional investigations in such an in vivo setting. Furthermore, the role of cortisol in the exercise-induced irisin elevation should be examined in specific GR-knockout mice in future studies.
Conclusion
In conclusion, this study revealed a 1-kb core promoter region of the FNDC5 gene and identified GR as the potential transcription factor of FNDC5, which may control the histone acetylation and methylation of the gene. Overall, these finding provide the foundation for understanding the molecular regulatory mechanisms of FNDC5/irisin and may lay the groundwork for therapeutic targeting of these factors for treatment of metabolic disease.
Methods
Cell culture
Human adult cardiomyocyte AC16 cells, human hepatocellular carcinoma Huh7, HepG2, Sk-Hep1, and SNU449 cells (ATCC), adenocarcinomic human alveolar basal epithelial cells A549, Plasma cell myeloma cell KMS26, cervical adenocarcinoma cell HeLa, nontumorigenic human aortic endothelial cell (HAEC) and nontumorigenic immortalized human hepatocyte cell line (MIHA) were used for this study. All cells were maintained in DMEM/F12 or DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. All cells were incubated at 37 °C in an atmosphere of 5% CO2 in air and sub-cultured when reaching at least 80% confluences. For experiments, cells were cultured with NaB (Sigma, #B5887) or 5-Aza (Sigma, #A2385) for 24 to 72 h.
Quantitative Real-time PCR
Total RNA from human cell lines and mouse tissues was extracted using the Trizol® reagent (Ambion, #15596-018) and the RNease Mini kit (Qiagen, #74104), and 1 μg of total RNA of the human first-strand cDNA (Clontech, Human MTC Panel I and II) were transcribed using a first-strand cDNA synthesis kit (Permentas, #K1621). Real time PCR was performed on cDNA with gene-specific primers pairs (Table 2). Reactions were performed in triplicate. The qPCR products were detected with SYBR Green connect Real-Time PCR platform. Results were quantified using the absolute quantitative analysis method via normalization with GAPDH or β-2-mitcroglobulin.
Table 2. Quantitative real-time PCR gene-specific primer sequences.
| Gene | Primer sequence |
|---|---|
| Human FNDC5 variant 1 3′ | TACCAGAGCATGAGGCACAG |
| Human FNDC5 variant 2 3′ | TTTCATATCTTGCTGCGGAGA |
| Human FNDC5 variant 3 3′ | ACAGGCAGTCACGCTTCAAT |
| Human FNDC5 5′ | CCTCCAAGAACAAAGATGAGG |
| Human B2M 5′ | CTCGCTCCGTGGCCTTAG |
| Human B2M 3′ | CAAATGCGGCATCTTCAA |
| Rat/Mouse FNDC5 3′ | GGCTCGTTGTCCTTGATGAT |
| Rat/Mouse FNDC5 5′ | GACCTGGAGGAGGACACAGA |
| Mouse GAPDH 5′ | CACCATCTTCCAGGAGCGAG |
| Mouse GAPDH 3′ | CCTTCTCCATGGTGGTGAAGAC |
Luciferase activity assay
FNDC5 promoter sequences were obtained from UCSC genome browser (https://genome.ucsc.edu/). FNDC5 promoters were amplified from human genomic DNA (Clontech) with specific primers (Table 3) and inserted into the pGL4.14 vector (Promega) using the KpnI/HindIII enzyme sites and the In-Fusion HD cloning kit (Clontech). The plasmids containing the FNDC5 promoter and renilla luciferase genes for control were co-transfected using Lipofectamine® LTX with Plus™ Reagent (Life Technologies). Luciferase activity was measured with a dual luciferase assay kit (Promega). Luciferase activity was normalized to the renilla luciferase activity.
Table 3. Primers used for FNDC5 promoter construction.
| Cloning region | Primer sequence |
|---|---|
| FNDC5 V1 prom1.5F | CTAACTGGCCGGTACCAAAAGAATGAAACTCCGTCTC |
| FNDC5 V1 prom2.5F | CTAACTGGCCGGTACCTCTCAAGTTAATTACCTTTTC |
| FNDC5 V1 prom1F | CTAACTGGCCGGTACCGGAAGAATGATTTTTTTTCCA |
| FNDC5 V1 prom2F | CTAACTGGCCGGTACCGTGAGCCCTGAGCCCTGGATC |
| FNDC5 V1 prom3F | CTAACTGGCCGGTACCACCCAGCCACTCTCCACCACA |
| FNDC5 V1 prom4F | CTAACTGGCCGGTACCGCAGCTGAGAATCTGTCTCCA |
| FNDC5 V1 promR | CCGGATTGCCAAGCTTCTTTCTCTCCCCTTATTATCT |
| FNDC5 prom1DR | CCGGATTGCCAAGCTTTGGAAAAAAAATCATTCTTCC |
| FNDC5 prom2DR | CCGGATTGCCAAGCTTGATCCAGGGCTCAGGGCTCAC |
| FNDC5 prom3DR | CCGGATTGCCAAGCTTTGTGGTGGAGAGTGGCTGGGT |
Methylation-specific PCR
The genomic DNA from cells was extracted using a genomic DNA extraction kit (DNeasy Blood & Tissue kit, Qiagen). As much as 3 μg of gDNA was used for CT conversion with bisulfate treatment (DNA Methylation-Gold kit, Zymo Research) and then amplified with non-methylation- and methylation-specific primers. FNDC5 nonmethyled primer set (forward 5′–TGGTAGTTAAGTTAGGGTTGTGTATG-3′, reverse 5′–AATAAAAACCAAAAAACACC-3′) and methylation primer set (forward 5′–CGGTAGTTAAGTTAGGGTTGTGTAC-3′, reverse 5′-AAATAAAAACCGAAAAACGC-3′).
ChIP analysis
The antibodies utilized in ChIP were HitoneH3Ac (Millipore, 06-599), H3K27me2 (Abcam, ab24684), GR (CST, 3660), Sp1 (Santa Cruz, SC-420), and p53 (Santa Cruz, SC-126). ChIP was carried out using a ChIP kit (Active Motif) according to the manufacturer’s specifications, and then the precipitated material was used in quantitative analysis. ChIP was performed with the following CpG island-specific primer pairs: ChIP primer set for human cells (forward 5′-GCAAAGAAAGCTCAGCATAGTATC-3′, reverse 5′-GAGCAGAGAGTGCTAAGTGGAC-3′), ChIP-TF primer set for human cells (forward 5′-CTGTGCACGGGAGAGAGAG-3′, reverse 5′-CTATTAGGTCCTCTGCCGGG-3′), and the ChIP primer set for mouse cells (forward 5′-GATGGGGTACAGATGGGCAT-3′, reverse 5′-GTCCCCGCTCTTCACTCTG -3′).
Statistical analysis
Data are presented as means ± standard error of the mean (SEM). Differences between the control and treatment groups were evaluated using a one-way analysis of variance (ANOVA), and the control-to-treatment comparison over time or dose was tested using a two-way ANOVA with Origin 8.0 software (Origin Lab). Differences with a P-value ≤ 0.05 were considered significant.
Additional Information
How to cite this article: Kyu Kim, H. et al. Glucocorticoid receptor positively regulates transcription of FNDC5 in the liver. Sci. Rep. 7, 43296; doi: 10.1038/srep43296 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
These investigators were supported by a grant (2010-0020224, 2015R1A2A1A13001900, and 2015R1D1A1A01057937) from the Priority Research Centers Program of the National Research Foundation of Korea (NRF), which is funded by the Ministry of Education, Science and Technology of the Republic of Korea.
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.J.J. and H.K.K. designed experiments and performed ChIP analysis and PCR experiments. I.-S.S and Y.H.N. performed Histone acetylation and methylation analysis. K.W.S. and M. K performed cortisol-induced FNDC5 mRNA expression analysis. J.H. supervised and coordinated the study. Y.J.J. and H.K.K. wrote the manuscript. All authors reviewed the manuscript.
References
- Bostrom P. et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481, 463–468, doi: 10.1038/nature10777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Navarrete J. M. et al. Irisin is expressed and produced by human muscle and adipose tissue in association with obesity and insulin resistance. The Journal of clinical endocrinology and metabolism 98, E769–778, doi: 10.1210/jc.2012-2749 (2013). [DOI] [PubMed] [Google Scholar]
- Timmons J. A., Baar K., Davidsen P. K. & Atherton P. J. Is irisin a human exercise gene? Nature 488, E9-10; discussion E10-11, doi: 10.1038/nature11364 (2012). [DOI] [PubMed] [Google Scholar]
- Besse-Patin A. et al. Effect of endurance training on skeletal muscle myokine expression in obese men: identification of apelin as a novel myokine. International journal of obesity 38, 707–713, doi: 10.1038/ijo.2013.158 (2014). [DOI] [PubMed] [Google Scholar]
- Kurdiova T. et al. Effects of obesity, diabetes and exercise on Fndc5 gene expression and irisin release in human skeletal muscle and adipose tissue: in vivo and in vitro studies. The Journal of physiology 592, 1091–1107, doi: 10.1113/jphysiol.2013.264655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkala S. et al. Are skeletal muscle FNDC5 gene expression and irisin release regulated by exercise and related to health? The Journal of physiology 591, 5393–5400, doi: 10.1113/jphysiol.2013.263707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke S. et al. Evidence against a beneficial effect of irisin in humans. PloS one 8, e73680, doi: 10.1371/journal.pone.0073680 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Hu Y., Zhang H., Xu Y. & Wang G. Exenatide treatment increases serum irisin levels in patients with obesity and newly diagnosed type 2 diabetes. J Diabetes Complications, doi: 10.1016/j.jdiacomp.2016.07.020 (2016). [DOI] [PubMed] [Google Scholar]
- Du X. L., Jiang W. X. & Lv Z. T. Lower Circulating Irisin Level in Patients with Diabetes Mellitus: A Systematic Review and Meta-Analysis. Horm Metab Res, doi: 10.1055/s-0042-108730 (2016). [DOI] [PubMed] [Google Scholar]
- Zybek-Kocik A., Sawicka-Gutaj N., Wrotkowska E., Sowinski J. & Ruchala M. Time-dependent irisin concentration changes in patients affected by overt hypothyroidism. Endokrynol Pol, doi: 10.5603/EP.a2016.0030 (2016). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Protective Effect of Irisin on Atherosclerosis via Suppressing Oxidized Low Density Lipoprotein Induced Vascular Inflammation and Endothelial Dysfunction. PLoS ONE 11, e0158038, doi: 10.1371/journal.pone.0158038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyzos S. A., Kountouras J., Anastasilakis A. D., Geladari E. V. & Mantzoros C. S. Irisin in patients with nonalcoholic fatty liver disease. Metabolism 63, 207–217, doi: 10.1016/j.metabol.2013.09.013 (2014). [DOI] [PubMed] [Google Scholar]
- Garces M. F. et al. Irisin levels during pregnancy and changes associated with the development of preeclampsia. J Clin Endocrinol Metab 99, 2113–2119, doi: 10.1210/jc.2013-4127 (2014). [DOI] [PubMed] [Google Scholar]
- Yang Z., Chen X., Chen Y. & Zhao Q. Decreased irisin secretion contributes to muscle insulin resistance in high-fat diet mice. Int J Clin Exp Pathol 8, 6490–6497 (2015). [PMC free article] [PubMed] [Google Scholar]
- Xiong X. Q. et al. FNDC5 overexpression and irisin ameliorate glucose/lipid metabolic derangements and enhance lipolysis in obesity. Biochim Biophys Acta 1852, 1867–1875, doi: 10.1016/j.bbadis.2015.06.017 (2015). [DOI] [PubMed] [Google Scholar]
- Liu T. Y. et al. FNDC5 Alleviates Hepatosteatosis by Restoring AMPK/mTOR-Mediated Autophagy, Fatty Acid Oxidation and Lipogenesis in Mice. Diabetes, doi: 10.2337/db16-0356 (2016). [DOI] [PubMed] [Google Scholar]
- Bang H. S. et al. Ursolic Acid-induced elevation of serum irisin augments muscle strength during resistance training in men. Korean J Physiol Pharmacol 18, 441–446, doi: 10.4196/kjpp.2014.18.5.441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer X. et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 (2002). [DOI] [PubMed] [Google Scholar]
- Erickson H. P. Irisin and FNDC5 in retrospect: An exercise hormone or a transmembrane receptor? Adipocyte 2, 289–293, doi: 10.4161/adip.26082 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Rodriguez B. M. et al. FNDC5 expression and circulating irisin levels are modified by diet and hormonal conditions in hypothalamus, adipose tissue and muscle. Sci Rep 6, 29898, doi: 10.1038/srep29898 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrann C. D. et al. Exercise induces hippocampal BDNF through a PGC-1alpha/FNDC5 pathway. Cell Metab 18, 649–659, doi: 10.1016/j.cmet.2013.09.008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrann C. D. FNDC5/Irisin–their role in the nervous system and as a mediator for beneficial effects of exercise on the brain. Brain Plasticity 1, 55–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo L. et al. Irisin Is Regulated by CAR in Liver and Is a Mediator of Hepatic Glucose and Lipid Metabolism. Mol Endocrinol 30, 533–542, doi: 10.1210/me.2015-1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostanci M. S. et al. Serum irisin levels in patients with polycystic ovary syndrome. Eur Rev Med Pharmacol Sci 19, 4462–4468 (2015). [PubMed] [Google Scholar]
- Deaton A. M. & Bird A. CpG islands and the regulation of transcription. Genes & development 25, 1010–1022, doi: 10.1101/gad.2037511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J. Y. et al. Epigenetic modification regulates both expression of tumor-associated genes and cell cycle progressing in human colon cancer cell lines: Colo-320 and SW1116. Cell Res 14, 217–226, doi: 10.1038/sj.cr.7290222 (2004). [DOI] [PubMed] [Google Scholar]
- Jaenisch R. & Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33 Suppl, 245–254, doi: 10.1038/ng1089 (2003). [DOI] [PubMed] [Google Scholar]
- Barski A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837, doi: 10.1016/j.cell.2007.05.009 (2007). [DOI] [PubMed] [Google Scholar]
- Ferrari K. J. et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Molecular cell 53, 49–62, doi: 10.1016/j.molcel.2013.10.030 (2014). [DOI] [PubMed] [Google Scholar]
- Miao F. & Natarajan R. Mapping global histone methylation patterns in the coding regions of human genes. Molecular and cellular biology 25, 4650–4661, doi: 10.1128/MCB.25.11.4650-4661.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F. et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141, doi: 10.1242/dev.037127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E. E. et al. Exercise and circulating cortisol levels: the intensity threshold effect. J Endocrinol Invest 31, 587–591, doi: 10.1007/BF03345606 (2008). [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y. et al. High-intensity exercise causes greater irisin response compared with low-intensity exercise under similar energy consumption. Tohoku J Exp Med 233, 135–140 (2014). [DOI] [PubMed] [Google Scholar]
- Lee S. R. et al. Non-genomic effect of glucocorticoids on cardiovascular system. Pflugers Arch 464, 549–559, doi: 10.1007/s00424-012-1155-2 (2012). [DOI] [PubMed] [Google Scholar]
- Lee S. R. et al. Glucocorticoids and their receptors: Insights into specific roles in mitochondria. Prog Biophys Mol Biol 112, 44–54, doi: 10.1016/j.pbiomolbio.2013.04.001 (2013). [DOI] [PubMed] [Google Scholar]
- Miranda T. B. et al. Reprogramming the chromatin landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res 73, 5130–5139, doi: 10.1158/0008-5472.CAN-13-0742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. & Cidlowski J. A. The human glucocorticoid receptor: one gene, multiple proteins and diverse responses. Steroids 70, 407–417, doi: 10.1016/j.steroids.2005.02.006 (2005). [DOI] [PubMed] [Google Scholar]
- Ramamoorthy S. & Cidlowski J. A. Exploring the molecular mechanisms of glucocorticoid receptor action from sensitivity to resistance. Endocrine development 24, 41–56, doi: 10.1159/000342502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadmiel M. & Cidlowski J. A. Glucocorticoid receptor signaling in health and disease. Trends in pharmacological sciences 34, 518–530, doi: 10.1016/j.tips.2013.07.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley R. H. & Cidlowski J. A. The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. The Journal of allergy and clinical immunology 132, 1033–1044, doi: 10.1016/j.jaci.2013.09.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck B. N. & Kelly D. P. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest 116, 615–622, doi: 10.1172/JCI27794 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W. G. et al. Glucocorticoid receptor mediated repression of human insulin gene expression is regulated by PGC-1alpha. Biochem Biophys Res Commun 352, 716–721, doi: 10.1016/j.bbrc.2006.11.074 (2007). [DOI] [PubMed] [Google Scholar]