Abstract
Nucleotide excision repair (NER) is a pivotal life process for repairing DNA nucleotide mismatch caused by chemicals, metal ions, radiation, and other factors. As the initiation step of NER, the xeroderma pigmentosum complementation group A protein (XPA) recognizes damaged DNA molecules, and recruits the replication protein A (RPA), another important player in the NER process. The stability of the Zn2+-chelated Zn-finger domain of XPA center core portion (i.e., XPA98–210) is the foundation of its biological functionality, while the displacement of the Zn2+ by toxic metal ions (such as Ni2+, a known human carcinogen and allergen) may impair the effectiveness of NER and hence elevate the chance of carcinogenesis. In this study, we first calculated the force field parameters for the bonded model in the metal center of the XPA98–210 system, showing that the calculated results, including charges, bonds, angles etc., are congruent with previously reported results measured by spectrometry experiments and quantum chemistry computation. Then, comparative molecular dynamics simulations using these parameters revealed the changes in the conformation and motion mode of XPA98–210 Zn-finger after the substitution of Zn2+ by Ni2+. The results showed that Ni2+ dramatically disrupted the relative positions of the four Cys residues in the Zn-finger structure, forcing them to collapse from a tetrahedron into an almost planar structure. Finally, we acquired the binding mode of XPA98–210 with its ligands RPA70N and DNA based on molecular docking and structural alignment. We found that XPA98–210’s Zn-finger domain primarily binds to a V-shaped cleft in RPA70N, while the cationic band in its C-terminal subdomain participates in the recognition of damaged DNA. In addition, this article sheds light on the multi-component interaction pattern among XPA, DNA, and other NER-related proteins (i.e., RPA70N, RPA70A, RPA70B, RPA70C, RPA32, and RPA14) based on previously reported structural biology information. Thus, we derived a putative cytotoxic mechanism associated with the nickel ion, where the Ni2+ disrupts the conformation of the XPA Zn-finger, directly weakening its interaction with RPA70N, and thus lowering the effectiveness of the NER process. In sum, this work not only provides a theoretical insight into the multi-protein interactions involved in the NER process and potential cytotoxic mechanism associated with Ni2+ binding in XPA, but may also facilitate rational anti-cancer drug design based on the NER mechanism.
Keywords: Cytotoxic mechanism, Nickel ion, Nucleotide excision repair, XPA, Zn-finger
Introduction
Nucleotide mismatch is a ubiquitous phenomenon in all life processes that may result in the cessation of cell cycles. After the excision of the mismatched nucleotides, the DNA polymerase can synthesize new segments and fill in the gap, which is subsequently ligated to the original chain by the DNA ligase, a process referred to as “nucleotide excision repair” (NER) [1, 2]. Should the NER pathway be impaired, a variety of human genetic diseases may emerge, such as xeroderma pigmentosum (XP), Cockayne syndrome, and trichothiodystrophy etc [3, 4]. As a pivotal protective mechanism in advanced living organisms, the NER process repairs UV- and cisplatin-induced intra-strand crosslinks in the DNA, as well as a wide variety of single base bulky DNA adducts formed by environmental carcinogens. It is therefore the most versatile DNA repair system known to humans [5]. In this complicated process, XP complementation group A (XPA) protein and replication protein A (RPA) family (including RPA70, RPA32, and RPA14) interact and cooperate with each other to assist in the recognition and elongation of DNA [6–8].
As a key step in the NER reaction, the XPA protein initiates the NER via recognizing the damaged DNA due to its high affinity for preferentially binding with damaged DNA. The XPA constitutes three structurally independent domains: the N-terminal portion (NTP, residues 1–97), the center core portion (CCP, residue 98–219), and the C-terminal portion (CTP, residue 220–273). The primary function of XPA1–97 is to bind with RPA34 and ERCC1, whereas that of RPA220–273 is to bind with TFIIH; XPA98–219, the key functional region in XPA, serves as an adaptor between damaged DNA and RPA70 [9–11]. Ikegami and Buchko et al. both reported NMR solution CCP structures of XPA, and their results elucidated that the CCP of XPA consists of a Zn-finger domain (102–129) and a C-terminal subdomain (138–209), connected by an eight amino acid linker sequence (130–137) [12, 13]. In the Zn-finger domain, the Zn ion is chelated with four residues, namely Cys105, Cys108, Cys126, and Cys129. The four sulfur atoms in these four Cys residues basically form a regular tetrahedron, hereinafter referred to as the CCCC pyramid. A key factor in maintaining the stability of XPA98–210 structure is hydrophobic interactions [12, 13]. The XPA98–210 contains three hydrophobic cores: the first core is formed by the encircling of loop1 (98–102) and loop3 (113–129) and it mainly functions to stabilize the Zn-finger domain. The second hydrophobic core consists of the loop3 (113–129), linker (130–137), the trailing end of loop4 (148–163), and β4 (164–165), and it mainly sustains the relative position and linkage of the Zn-finger domain with the C-terminal domain. The last hydrophobic core, which is the largest one, is formed by the β3 (138–140), α1 (141–147), the leading end of loop4 (148–163) and α2 (183–194), and it maintains the stability of the C-terminal domain (see Fig. S1).
Following its specific recognition of DNA, the XPA subsequently recruits RPA, and the elongation steps for the DNA repair ensue. The human homolog of RPA is a heterotrimer with subunits of 70, 32, and 14 kDa (RPA70, RPA32, and RPA14, respectively). Bochkareva et al. solved the X-ray structure of the RPA70C-RPA32D-RPA14 trimer, based on which they proposed the interaction mechanism between the three components, and determined the three DNA binding sites [14]. Li et al. carried out a series of pull-down experiments with different mutants of RPA, and the result indicated that more than one subunit in the RPA protein had interactions with XPA at different sites, but only that of RPA70 is essential for NER [3]. Affinity chromatography and immunoprecipitation biological experiments [15, 16] also showed that RPA’s direct binding with XPA at the very early stage of NER may aid the recognition between endonuclease and damaged DNA. Furthermore, Hey et al.’s fluorescence anisotropy assay and Wang et al.’s surface plasmon resonance (SPR) assay both investigated the interaction of XPA and RPA with two major types of UV-damaged DNA, and their results consistently demonstrated that both proteins were able to bind to DNA independently; however, when XPA and RPA were incubated together, the stability of the XPA-damaged DNA complex was significantly enhanced [17, 18].
Zinc as an essential trace metal is known to play important roles in life processes [19]. In biological systems, most Zn2+ exists in Zn-fingers, which is a common structural motif in DNA binding proteins. The Zn-finger is a finger-like structure formed by a Zn2+ coordinating with either four Cys residues (C4 type) or two Cys and two His residues (C2H2 type) located on an approximately 30-residue sequence. It bears great importance for gene transcription, enzyme activity, and protein conformational stability. Naturally, the substrate for the Zn-finger domain is Zn2+, whereas in the presence of reductive agents, the Zn-finger domain can also bind with Ni2+ and other ions [20, 21]. Nickel is classified by the World Health Organization as a cytotoxic element, many of whose compounds are confirmed as human carcinogens and allergens. In vitro and ex vivo studies demonstrated that soluble Ni compounds can damage or distort the conformation of Zn-finger domains and inhibit Zn-finger containing DNA repair proteins (such as XPA) [22]. Asmuss et al. conducted polyacrylamide gel electrophoreses (PAGE) followed by Southern blotting to investigate the effect of various toxic metals on the Zn-finger structures in both bacterial Fpg protein and mammalian XPA protein [23, 24]. Their results showed that Ni2+ specifically decreased the binding affinity between XPA and damaged DNA while not affecting that of Fpg, implying that the sensitivity of different Zn-finger domains to toxic metal compounds may be dependent on what proteins they reside in. On the other hand, the atomic absorption and UV-CD spectra results by Cleaver and Asahina et al. indicated that XPA binds preferentially to UV-induced DNA, and loss of XPA function leads to a severe human disorder characterized by UV hypersensitivity and enhanced cancer risk [25, 26]. This finding is supported by the systematic review by Hartwig et al., which suggested that the NER-related proteins are very sensitive targets for compounds of Ni2+, and the consequence of this interaction could lead to a diminished removal of endogenous DNA lesions, which in turn may increase the risk of tumor formation [27, 28]. Moreover, the review also pointed out that the displacement of Zn2+ by Ni2+ in the Zn-finger structures of NER systems may be one of the underlying mechanisms for the cytotoxicity of these compounds.
The Zn2+ ion in the XPA98–210 structure forms a complex with the CCCC pyramid in the Zn-finger domain and plays an essential role in the stabilization of protein structure and subsequent biological functions. Three methodologies have been universally adopted to study the interaction between metals and protein residues in metalloproteins: the non-bonded model, the cationic dummy atom model and the bonded model. In the non-bonded model, we only use two types of non-covalent bonds (i.e., van deer Walls and electrostatic interactions) to depict the interactions between the metal and the surrounding residues in metalloproteins. Therein, the metal charges are usually set as positive integers, and the charge distributions of the surrounding residues do not change at all. Therefore, the non-bonded model is simplistic and easy to implement, but it suffers from a low accuracy, and the result relies heavily on the initial structure of the metal center [29]. The cationic dummy atom model places the charges between the metal ions and surrounding residues to mimic covalent bonds and offers a more sophisticated electrostatic model. The cationic dummy atom model requires an intricate parameterization process due to the inclusion of numerous empirical parameters, which severely restricts the application of this method [30]. In the bonded model, the force field parameters for the metal and key atoms in its surrounding residues, including bond lengths, bond angles, dihedral angles, van deer Walls, and coulomb interactions, can either be computed based on quantum mechanics or determined by experiments. The interactions between metal and its surrounding residues are deemed as covalent bonds, and their force field parameters can all be utilized for subsequent molecular dynamics (MD) simulations [31]. Although in bonded models, quantum chemistry is required to calculate the force field constants, it is nevertheless a more accurate way to model the ions that form coordination bonds with surrounding residues. In our work, we adopted the package of Metal Center Parameter Builder (MCPB) based on the bonded model to provide the metal center parameters in XPA98–210, for the following considerations: 1) the computation is more accurate; 2) our system does not involve changes in coordination number (CN) and chemical reactions; 3) MCPB is a semi-automatic method that can provide point charges and other force field parameters, which has been extensively used in previous studies [32–34].
There have been numerous studies discussing the important role of XPA in the NER pathway and the potential cytotoxicity mechanism of Ni2+ by influencing the Zn-finger of XPA [22–24]. However, four meaningful scientific challenges remain to be further elucidated: 1) the mechanisms of both the multiprotein complex interactions among the many proteins participating in the NER process and the DNA’s transmission among these proteins are still not fully understood; 2) the XPA-RPA70-DNA complex structure, the crucial structure for studying the initial DNA recognition step of XPA in the NER process, has not been included in the PDB; 3) Ni metal may exhibit cytotoxic effect through its influence on the Zn-finger structure of XPA, but the mechanism of which remains unclear; 4) the bonded model of Zn in the XPA’s Zn-finger domain has not yet been studied, and successful determination of its force field constants can provide important insights into its structural properties as well as high-accuracy parameters for future MD simulation research. In this work, we first used the MCPB package to calculate the force field parameters of the metal center in the XPA98–210 Zn-finger domain, and then performed 100 ns comparative molecular dynamics (MD) simulations on the three XPA98–210 systems: Zn-bound, Ni-bound, and metal-free. By analyzing the impact of metal on the conformation of the protein, especially the Zn-finger domain, we summarized and discussed the differential motion modes of the three systems and proposed a potential cytotoxic mechanism of Ni. Finally, we combined the data from molecular docking, structural biology information, as well as structural alignments and came up with the multi-protein interactions involving XPA98–210, RPA70, RPA32, and RPA14, which demonstrated the binding pattern of DNA to the multi-protein complex in NER. This scheme, as we believed, will substantially aid insight into the design of anti-cancer drugs.
Systems and methods
Construction of the initial structures for MD simulations
The system discussed in this study is the center core portion (CCP, residue 98–219) of XPA, including the Zn-finger domain. Previously, Ikegami and Buchko et al. used solution NMR to obtain the CCP structure of XPA–XPA98–210 (PDB code: 1XPA) [12], and XPA98–208 (PDB code: 1D4U), respectively [13]. Both structures share a high similarity in terms of secondary structure and the detailed conformation in the Zn-finger domain. The only difference lies in the angle formed by the two domains of XPA98–210 (Zn-finger domain: 98–129; C-terminal subdomain: 138–219) around the linker (130–137), and this discrepancy can be overcome by sufficient sampling during the long time MD simulation. Here, we selected 1XPA structure as the initial structure for the MD simulation for two reasons: 1) between the two solution NMR PDB structures, 1XPA is more rigorous than 1D4U and presumably more reasonable, based on their selection criteria and conformers calculated; 2) 1XPA is two residues longer than 1D4U. Since the Zn-finger of 1XPA NMR structure contains a Zn2+, it is defined as XPA98–210-Zn in this article. Then we simply deleted Zn2+ or replaced it with Ni2+ (based on the same coordinates, considering the same valence and similar space of Zn2+ and Ni2+), and acquired two systems, namely XPA98–210-apo and XPA98–210-Ni. These three systems will be further used for long-time comparative MD simulations, which will investigate the influence of Ni2+ on the conformation of XPA98–210 and discover the potential mechanism for the cytotoxicity of Ni2+.
Metal center parameter builder (MCPB)
The MCPB method [31] was developed by Peters et al. and embedded into the MTK++ software package [35] in AmberTools version 1.5. The MCPB method has been widely used to facilitate the modeling of metal effects of the metalloprotein under the AMBER force field. In the XPA98–210-Zn and XPA98–210-Ni systems, we adopted the MCPB method based on bonded models for representing the interactions between metal and the surrounding residues. The major steps for MCPB include: 1) prepare the PDB by completing residues and adding hydrogens; 2) substitute the protein backbone C atoms with methyl groups, and then use the B3LYP/6-31G* method [36, 37] with the Gaussian 03 software [38] to obtain the stable and convergent sidechain model. This model is then used to calculate the force field parameters; 3) establish a large model based on the sidechain model by capping each of the methyl groups with acetylamide (ACE) and N-methylamide (NME) group, and apply the same quantum chemistry strategy to yield the restrained electrostatic potential (RESP) charge parameters of the large model; 4) replace the standard amino acid force field information in the ff99SB-ILDN with the new force field parameters calculated from the 2nd and 3rd step, including the bond length, bond angle, dihedral angle and RESP charge, and then integrate all the metal bond information. This will generate all the parameter files required for the following AMBER MD simulation.
Molecular dynamics simulation
Molecular dynamics (MD) simulation is a method based on Newtonian mechanics, which is widely used to investigate the dynamics and structural properties of protein systems [39–41]. In this work, three 100 ns independent MD simulations were carried out for the Ni2+-bound XPA98–210 (XPA98–210-Ni) and metal-free XPA98–210 (XPA98–210-apo) systems using the AMBER 12 suite of programs [35, 42]. The Amber ff99SB-ILDN all atom force field was adopted [43]. ff99SB-ILDN force field contains some modifications and updates to ff99SB, improving the amino acid side-chain torsion potentials, which exhibits considerably better agreement with the NMR data. In each simulation, the solute was solvated in a truncated periodic box with a 1.2 nm solute-wall distance using the TIP3P water model [44]. The simulation boxes (XPA98–210-Zn/XPA98–210-Ni, XPA98–210-apo) contained the XPA98–210 protein (113 residues), 9828/9828/10512 water molecules, 8/8/6 sodium ions for a total of 31357/31357/33410 atoms per periodic cell. Before MD simulations, two stages of minimization were performed for the initial structures. First, the solute atoms were restrained by the force constant 2.09 × 105 kJ mol−1 · nm−2. Water molecules were minimized for 10,000 steps with the steepest descent method followed by 10,000 steps of the conjugate gradient. Then, the restraint for the solute was removed and 10,000 steps energy minimization of the steepest descent followed by 10,000 steps of the conjugate gradient were carried out for the total system. The convergence criterion was 4.182 kJ · mol−1 · nm−1.
MD simulations were set to two stages. First, we restrained the solutes (the restraining force constant was 4.182 × 103 kJ mol−1 · nm−2) and slowly heated the systems up from 0 to 300 K over 1 ns. Then, three nonrestraint MD simulations at 300 K were performed for 99 ns. In these simulations, the system structures were monitored simultaneously with the VMD 1.9.1 package [45]. The SHAKE algorithm [46] was applied to constrain covalent bonds involving hydrogen atoms and a 1.2 nm cut-off was used for all non-bonded interactions. The particle mesh Ewald (PME) method was utilized for long-range electrostatic interactions. The MD time step was taken as 2 fs, and one snapshot was sampled every 5000 steps (i.e. 10 ps). Thus, total 10,000 conformations were obtained during each 100 ns MD simulation.
It is worth mentioning that, AMBER 12 provides different protonation states for titrating side chains of acidic and basic amino acids as well as cysteines. Since the pH in the simulated environment of this study was set at 7.0, all histidines were not protonated due to the low protonation ratio at this pH. On the other hand, the side chains of Asp, Glu, Arg, and Lys were charged (as Asp−, Glu−, Arg+, and Lys+) in all simulations. For the metal-containing systems (XPA98–210-Zn and XPA98–210-Ni), the cysteines in the zinc-finger domain (i.e., Cys105/Cys108/Cys126/Cys129) lose their hydrogen atoms and become Cys− (referred to as CYM in AMBER 12), whereas other cysteines were all treated as CYS.
Principal component analysis
Principal component analysis (PCA) is a standard method for finding global, correlated motions from MD trajectories through dimensionality reduction [47–49]. PCA has been successfully applied to some functional motion studies in many biological systems [50–52]. The definition of PCA is based on the construction and diagonalization of the covariance matrix. The element Cij in this covariance matrix is given by [53]:
| (1) |
where xi (xj) is the coordinate of the ith (jth) atom and · · · represents an ensemble average. The eigenvectors of the matrix describe the directions of the motions. Meanwhile, the eigenvalues give the magnitude of the motion for each direction. The first few principal components (PCs) are the eigenvectors with the highest eigenvalues, which usually contain the most important conformational changes of a biomolecular system. While for the rest of the few PCs with lower eigenvalues, the corresponding motions are composed of high frequency local vibrations and contain less biological functional motion information [54, 55].
To find out the functional motions from an MD simulation, it is useful to filter out all other motions by projecting the whole MD trajectory along the directions described by the first few selected eigenvectors. Then through superimposing the two extreme projection structures, the detailed functional motion information can be distinctly revealed by the porcupine map, where the length of cone in each atom represents the motion magnitude and the orientation of the cone describes the motion direction. In this work, PCA was performed for the Cα atoms in the XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo systems based on all their MD trajectories (each having a total of 10,000 conformations) with Gromacs 4.5 package [56].
Free energy landscape
The free-energy landscape (FEL) can provide a quantitative description of protein dynamics [53]. The FEL is obtained from the joint-probability distribution of reaction coordinates. It can be estimated by:
| (2) |
where KB is the Boltzmann constant, T is the temperature of the simulation, and P(X) is the probability distribution of conformations along the reaction coordinate X. In the FEL, the local basins populated with high probability usually represent low-energy conformations in stable states. And the barriers populated with low probability indicate the high-energy transient states. In our study, the first and second principal components were chosen to be the reaction coordinates, and the two-dimensional FEL was computed to identify the dominant conformations with lower energy.
Results and discussion
Force field parameters
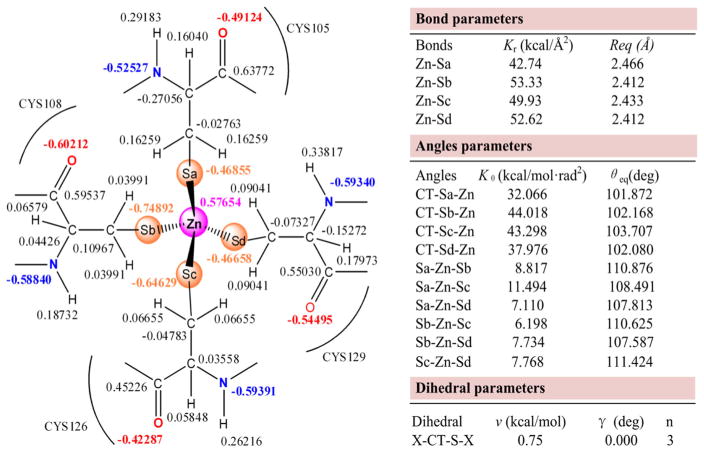
As is well known, in metalloproteins, the metal can affect the electronic charge distributions of its adjacent residues, and the charge distributions of these residues would differ significantly from metal-free residues. In this work, we obtained the bonding parameters of the Zn and Ni metal centers using the MCPB method [31], and used them to characterize the binding modes with the Zn-finger. Here, we adopted the RESP algorithm [57, 58] for the partial charge distribution in the metal center.
Figures 1 and S2 show all force field parameters of the XPA98–210-Zn and XPA98–210-Ni systems, respectively. When the metal is chelated in the CCCC pyramid, the partial charges of Zn and Ni ions in XPA98–210 were +0.57654 and +0.56223 |e|, and those of the four Cys-S’s in direct contact with Zn and Ni ranged from −0.75686 to −0.41725 |e|. Notably, the Zn and Ni in the metal center no longer maintained at +2 |e|, but went through a phenomenon where the electrons in the CCCC were transferred to the metal. Hou et al. [32] used the DFT method to compute the force field parameters of the Zn-tetra-coordination center in the Gelatinase-A protein, where the valence of Zn converged at +0.5493 |e|. Sindhikara et al. [59] studied the structure-activity relationship of the apo-and Ni-bound NikR protein using molecular dynamics simulation, where the bonding parameters between Ni and the CHHH pyramid were calculated using the DFT method. In their results, the charge of Ni was +0.66275 |e|, and that of Cys97S was −0.52432 |e|. Our partial charge data agreed with the relevant studies. As for bond lengths, those of Zn-S and Ni-S ranged within 2.412–2.466 and 2.281–2.293 Å, respectively. The force constant of Ni-S was also greater than that of Zn-S, suggesting a greater bond energy for Ni-S than Zn-S. The Ni-S bond, which possesses a stronger attractive force than Zn-S, may be the cause for the distortion of the residues around and within the CCCC pyramid. Sakharov et al. [19] have developed a novel potential energy function (PEF) for protein simulations of Zn2+ bound to Cys− and/or His0. Their Zn-Cys4 model suggested that the Zn-S bond length ranged within 2.37–2.43 Å. In the study on the magnetic behavior of a series of Ni compounds reported by Ni et al. [60], the Ni-S bond length ranged within 2.133–2.153 Å. The same bond length in Sindhikara et al.’s paper on nickel-bound NikR protein [59] was 2.206 Å. Although the bond lengths in these reports are smaller than our result from the CCCC-Ni model, it is indubitable that the bond length of Zn-S is slightly greater than that of Ni-S. In terms of bond angle, the bond angles around the S atom remained around the sp3 hybridization angle (109°28′), but those around the metal varied. The S-Zn-S basically maintained around the sp3 hybridization angle, while the S-Ni-S changed greatly; for instance, the Sa-Ni-Sd and the Sb-Ni-Sc each twisted to 164.389 and 154.097 degrees. The alteration of the surroundings of the CCCC pyramid in XPA98–210 can lead to a different chelating site of Ni from that of Zn. The S-Zn-S bond angle determined by Sakharov et al. using the PEE method was 107 ± 5 degrees [19], while the S-Ni-S bond angles in the series of metal-chelating small molecules reviewed by Liu et al. all deviated from the sp3 hybridization angle [61]. It can be speculated that, to adapt to the force field after the displacement of Zn2+ by Ni2+, the four Cys residues changed their relative positions to each other.
Fig. 1.
Force field parameters of Zn center in the XPA98–210-Zn system
To sum up, we successfully obtained the force field parameters by the MCPB method, and used them on subsequent MD simulations. All of these parameters readily agreed with the previous results of the numerous spectrometry experiments and quantum chemistry computations [19, 32, 59–61].
Overall structural convergence parameter
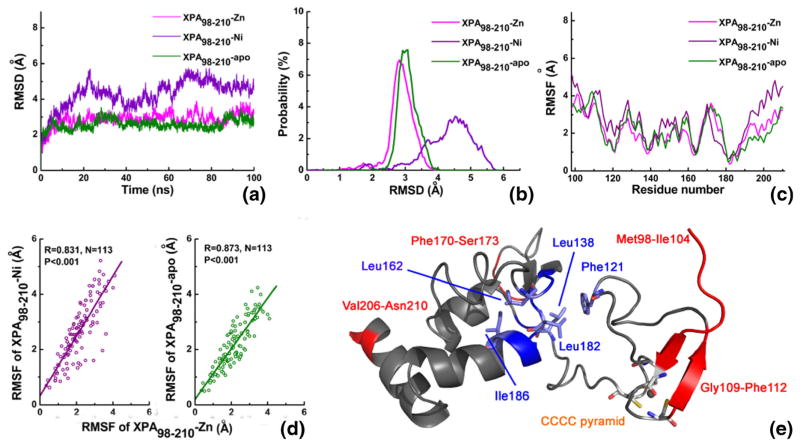
We performed 100 ns explicit water MD simulations on XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo systems. First, we calculated the root mean standard deviation (RMSD) of all the Cα atoms in the three protein systems, in order to monitor the fluctuation of the protein structure from the initial structure (see Fig. 2a). The XPA98–210-Zn and XPA98–210-apo systems essentially reached their equilibria after 10 ns, whereas the RMSD fluctuation of the XPA98–210-Ni backbone was obviously greater than the former two. Figure 2b shows the probability distribution of RMSD in the three systems. It can be clearly seen from both Fig. 2a and b that, when Zn ion was displaced by Ni ion, the stability of the system declined, while in the absence of metal, the stability was not significantly different from that of XPA98–210-Zn. In other words, the rank of molecular flexibility for the three systems is: XPA98–210-Ni > XPA98–210-apo ≈ XPA98–210-Zn. It is worth mentioning that XPA98–210 is a small protein with only 113 amino acids, however its RMSD value at equilibrium remained at 3 Å, such a large RMSD level is mainly for two reasons: 1) since the structure of the system comes from an NMR structure, the starting point of the molecular simulation is in a complete solution state, rather than a rigid structure formed by crystal stacking; 2) rigid secondary structures are middle flexibility regions rare in the XPA98–210 system, while random coils comprise a large proportion, especially in the long linker that connects the two large domains, contributing to a very large flexibility of the molecule. This generally highly-flexible structure of XPA98–210 will be further studied using secondary structure analysis.
Fig. 2.
Comparative MD analysis of the XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo systems. (a) RMSD values of all the Cα atoms during the simulation time. (b) Probability distribution of RMSDs. (c) RMSF values of all the Cα atoms. (d) RMSFs’ correlation of XPA98–210-Zn with XPA98–210-Ni and XPA98–210-apo. (e) Flexibility distribution of XPA98–210 system. Red, blue, and gray colors represent the high, low, and
The root mean standard fluctuation (RMSF) has been widely used for estimating the protein flexibility at residue level. The choice of trajectories at different time periods may have an influence on the results of RMSF analyses. In this work, considering the obviously differential motion modes of the three systems, we based our RMSF analysis upon the entire MD simulation trajectory, in order to better contrast molecular flexibility information across the three systems (i.e., 0~100 ns, total 10,000 conformations). Figure 2c shows the RMSF distributions of all Cα atoms in the XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo systems. The RMSF distribution trends of the three systems appear to be fairly similar. Generally, when the initial structure of an MD simulation comes from X-ray crystallography experiment, the correlation between experimental B-factor values and calculated RMSF values can be used to validate the simulation. Since the initial structure adopted for MD simulation in this work was an NMR structure, B-factor was not available, we alternatively analyzed the correlations between the Cα RMSF values of XPA98–210-Ni, XPA98–210-apo, and those of XPA98–210-Zn. The correlation coefficient was 0.831 and 0.873, respectively, which indicated highly significant correlations for a sample size of 113 (see Fig. 2d). The high correlations between the RMSF values suggest high consistency and reasonableness of the three MD simulation datasets. Based on the data given in Fig. 2c, we defined the high flexibility region (RMSF > 3.0 Å), medium flexibility region (1.0 Å < RMSF ≤ 3.0 Å), and low relatively rigid region (RMSF ≤ 1.0 Å). Figure 2e shows the residue flexibility distributions of the three systems, and the high flexibility regions are: (1) the flanking residues, such as Met98-Ile104 and Val206-Asn210; (2) spatially independent and long loop regions, such as Phe170-Ser173; (3) polar residues at the surface of the molecule, which have extensive contact with water, such as Lys110 and Glu111. The low flexibility regions are: Phe121, Leu138-Ile139, Leu162-Phe164, and Tyr181-Ile186, all of which are key residues constituting the hydrophobic core 2 and 3. It can be seen that, as an important factor for maintaining the stability of XPA98–210, the hydrophobic core 2 and 3 remained stable during the relatively long MD simulation. The hydrophobic core 1 in the Zn-finger domain, however, had higher motility. On the other hand, the CCCC pyramid (Cys105, Cys108, Cys126, and Cys129, as an important component for the Zn-finger) exhibited a moderate flexibility during simulation.
To investigate the outward extension of the system structure from the center, we also analyzed the change of the radius of gyration with time during the MD simulation (see Fig. S3). From Fig. S3, the XPA98–210-Zn system remained generally steady and had the least fluctuation. It also manifested a slight trend of shrinkage, unlike the slight expansion that is typically observed for normal crystal structures in water. This is because the initial structure for the simulation was an NMR structure and had already been sufficiently solubilized. In contrast, the XPA98–210-Ni had the greatest fluctuation of the radius of gyration and the worst stability, which agrees with the preceding RMSD analysis. This is mainly due to the shorter bond length of Ni-S than that of Zn-S (see Figs. 1 and S2): the defined bond length forced the system to reach a shrunk status after energy optimization, but with the elapse of the simulation time, the global adjustment of the protein conformation gradually made the system stretched and relaxed. The XPA98–210-apo system, meanwhile, had a medium fluctuation of gyration radius and had an overall tendency of relaxation, which is the result of the loss of metal chelation.
For a more intuitive view into the detailed change of the radii of gyration in the three systems, we superimposed the final conformations of the MD simulations (at 100 ns, see Fig. S4). It can be seen from the figure that the structural difference after the MD simulations of the three systems primarily lay in the Met98-Ser115, Asp122-Leu138, and Lys163-Lys179 segments. It is also conspicuous that the Ni2+ in XPA98–210-Ni moved more outwardly and further away from the C-terminal subdomain than the Zn2+ in XPA98–210-Zn did, which may explain why XPA98–210-Ni had a larger radius compared to XPA98–210-Zn to some extent.
Residue contact and second structure analysis
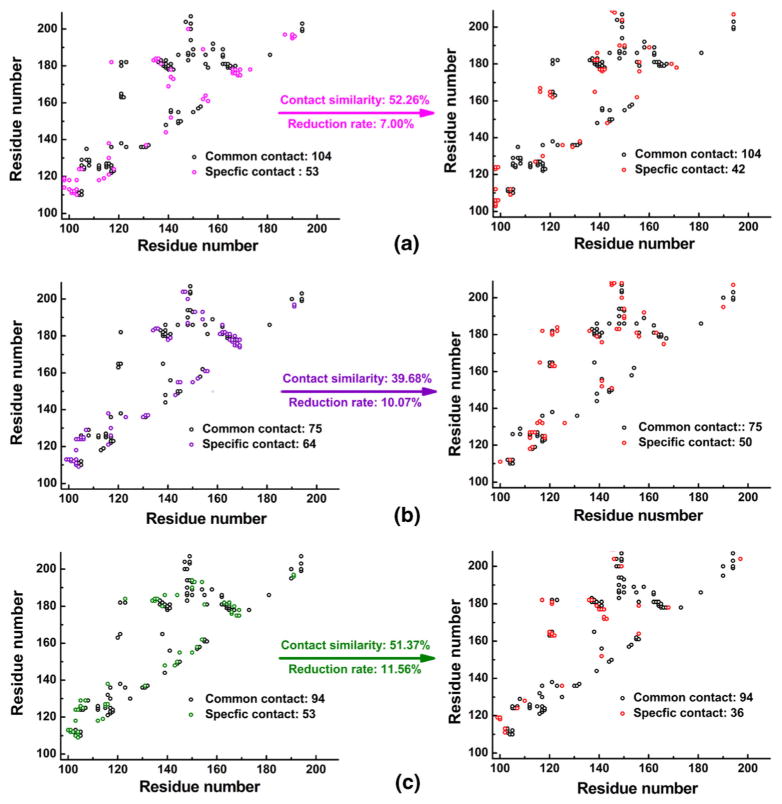
Residue contact analysis is an effective method for describing the intramolecular conformational change [62]. We defined contact between two residues as having a distance less than 4.5 Å, a threshold for residue interaction. Here we analyzed the differential residue contacts between the initial (at 0 ns) and final structures (at 100 ns) of the three systems (XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo).
In the XPA98–210-Zn system, the initial structure of MD simulation (at 0 ns) had 157 residue contacts, while the final structure had 146. In both structures 104 contacts were present, while 53 and 42 contacts were specific to either structure, respectively (see Fig. 3). Here, we defined two parameters: contact similarity and reduction rate. The contact similarity demonstrates the conservativeness of the contacts, and is calculated by the common contacts in both initial and final structures divided by the total number of contacts (including common contact and specific contacts in both the initial and final structures of MD simulation). The reduction rate, meanwhile, characterizes the extent of expansion and relaxation of the system, and is computed by dividing the difference between the number of specific contacts in each structure by the total number of contacts in the initial conformation (including common and specific contacts). We can see that in XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo systems, the contact similarities and reduction rates were 52.26/39.68/51.37 % and 7.00/10.07/11.56 %, respectively. The XPA98–210-Zn system showed the highest stability throughout the simulation. When Zn ion was displaced by Ni ion, the XPA98–210-Ni system had the lowest contact similarity, implying the greatest structural change and the least residue contact conservativeness. The metal-free XPA98–210-apo system, due to the expansion of the system, had the greatest reduction rate, indicating the sharpest decrease in residue contacts.
Fig. 3.
Residue contact map of the three systems. (a) The XPA98–210-Zn system. (b) The XPA98–210-Ni system. (c) The XPA98–210-apo system
It can be seen from Fig. 2a that the three systems were still undergoing fluctuation at 100 ns. To better reveal the global structural change, we also compared the contact residues in the initial structures and respective averaged structures in the three systems (see Fig. S5). This analysis also yielded similar results, which is that the XPA98–210-Ni had a greater reduction rate and a lower contact similarity compared to that of XPA98–210-Zn. This clearly suggested that the substitution of Ni2+ by Zn2+ lowered the conformational stability of XPA98–210.
Detailed analysis of the residue contacts in different regions of XPA98–210 revealed four characteristics: (1) the residue contact distribution of XPA98–210-Zn, XPA98–210-Ni and XPA98–210-apo shared a high similarity; (2) the internal residue contacts in loop5 (166–179) of the three systems all went through a sharp decline, which suggests that this loop became very relaxed during the MD simulation, congruent with the previous RMSF analysis results (see Fig. 2e); (3) in all three systems, Leu116-Leu123 (part of the loop3), Leu162-Leu165 (the terminus of loop4 plus β4), and Tyr181-Ile186 (β5 and part of α2) all had increasing numbers of contacts as the simulation went on. This can be explained by the strong hydrophobic interactions in hydrophobic core 2 and 3, the key driving force that sustains the global stability of the XPA98–210 structure; (4) the total number of contacts within the Zn-finger domain was well maintained in the XPA98–210-Zn system, but those numbers in both XPA98–210-Ni and XPA98–210-apo significantly decreased.
To describe the overall protein conformational changes, we also analyzed the stability of protein secondary structure. Based on the three MD simulation trajectories (XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo), we collected a total of 100 snapshots at each 1 ns interval. The secondary structures in each snapshot, as well as their changes over time, were analyzed with the DSSP program [63, 64]. Among these three systems, the color of C-terminal subdomain (138–210) remained coherent during the MD simulation process, which indicated a high stability of the overall protein secondary structure (see Fig. S6). However in the XPA98–210-Ni system, the Zn-finger domain (98–129) experienced significant conformational changes. Moreover, residues 127–129 region formed constant α-helix structure in the XPA98–210-Zn system, while in the other two systems they stayed as bends or turns. Surprisingly, the β1 (residues 103–104) and β2 (residues 111–112) kept stable in the metal-free XPA98–210-apo system, which suggested that the stacking effect between β1 and β2 contributed greatly to the maintenance of Zn-finger domain structure stability in the absence of metal chelation.
In sum, the analysis of residue contacts and secondary structures indicates that the absence of and the displacement of Zn2+ by Ni2+ did not exert a major impact on the structure of the C-terminal domain (138–210), but drastically disrupted the residue contacting in the Zn-finger domain (98–129), altering the conformation of the Zn-finger domain.
Hydrogen bonds and domain motion analysis
Typically, hydrogen bonds play a crucial role in maintaining the stability of protein structures. We compared the changes in the intra-molecular hydrogen bonds in XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo. Herein, the geometric criteria we adopted for the formation of hydrogen bonds were: 1) the angle formed by the donor atom, the hydrogen atom, and the receptor atom was larger than 120° and 2) the distance between donor atom and receptor atom was shorter than 3.0 Å. It is the same as RMSF analysis in that all 10,000 MD simulation conformations are adopted to hydrogen bond analysis to ensure the comparability of analysis. When the Zn2+ in the XPA98–210-Zn system was absent (XPA98–210-apo) or displaced by Ni2+ (XPA98–210-Ni), conspicuous changes have occurred in important intra-molecular hydrogen bonds during the simulation (see Table 1). In the XPA98–210-Zn system, there were six hydrogen bonds with occupancies over 40 %, among which four were formed at the Zn-finger domain (98–129), while the other two were located at the C-terminal subdomain (138–210). Notably, no hydrogen bonds were found between the two regions. The higher hydrogen bond frequency in the Zn-finger domain suggested that hydrogen bonds were one of the dominant forces that maintained the Zn-finger domain. After losing the chelating metal (XPA98–210-apo), the system became more relaxed, which dramatically disrupted the hydrogen bonds that originally existed in the XPA98–210-Zn structure: the number of hydrogen bonds with over 40 % occupancy declined to four. In the XPA98–210-Ni system, meanwhile, there were five hydrogen bonds with >40 % occupancy. The hydrogen bonds in the C-terminal subdomain remained virtually unchanged, while the only significant change happened in the Zn-finger domain. It can be inferred that Ni2+ exhibited its cytotoxic effect mainly through its impact on the conformation of Zn-finger domain and thereby on the NER process.
Table 1.
Hydrogen bonds in the three systems with frequency over 40 %
| XPA98–210-Zn
|
XPA98–210-apo
|
XPA98–210-Ni
|
||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| Donor | Acceptor | Freq. | Donor | Acceptor | Freq. | Donor | Acceptor | Freq. |
| Ser115-OG-HG | Thr125-O | 98.99 % | Ser115-OG-HG | Thr 125-O | 94.48 % | Ser115-OG-HG | Thr 125-O | 57.74 % |
| His136-ND1-HD1 | Cys129-O | 82.75 % | Ser190-OG-HG | Tyr148-O | 53.59 % | Arg158-NH1-HH12 | Asp154-OD1 | 54.68 % |
| His120-ND1-HD1 | Tyr116-O | 59.18 % | Arg130-NH2-HH21 | Asp127-OD2 | 42.73 % | Ser190-OG-HG | Tyr148-O | 52.60 % |
| Ser190-OG-HG | Tyr148-O | 54.74 % | Arg158-NH1-HH12 | Asp154-OD2 | 42.18 % | Thr125-OG1-HG1 | Leu123-O | 48.80 % |
| Arg158-NH1-HH12 | Asp154-OD2 | 45.41 % | Asn119-ND2-HD22 | Ser115-O | 40.61 % | |||
| Asn119-ND2-HD22 | Ser115-O | 40.13 % | ||||||
The analysis results of the hydrogen bond show that the hydrogen bond was also one of the key driving forces for the stability of Zn-finger domain. In sum, when Zn2+ was displaced by Ni2+ or simply absent, the hydrogen bonds in the C-terminal subdomain were almost unchanged. Nevertheless, when Zn2+ was displaced by Ni2+, the hydrogen bonds in Zn-finger domain were partially disrupted and weakened.
From the previous hydrogen bond analysis, we observed no hydrogen bond contacts between the C-terminal subdomain and Zn-finger in XPA98–210, demonstrating the independent motion modes of the two domains connected by the linker (130–137). In short, we treated the mass centers of Zn-finger (ZF), linker (L), and C-terminal subdomain (CT) as three points, and analyzed the change of the ZF-CT distance and the ZF-L-CT angle over time (see Fig. S7). From Fig. S7, the ZF-CT distances for the XPA98–210-Zn/XPA98–210-Ni/XPA98–210-apo systems were 23.49 ± 0.59/24.26 ± 0.79/24.25 ± 0.64 Å, and the corresponding ZF-L-CT angles were 100.76 ± 4.06/101.70 ± 5.89/102.74 ± 3.36 degrees. In terms of movement scales, the rank of the weak open-and-close movement between the Zn-finger and C-terminal domain in the three systems was: XPA98–210-Ni > XPA98–210-apo ≈ XPA98–210-Zn. When Zn2+ was displaced by Ni2+, the open-and-close movement between the C-terminal subdomain and Zn-finger domain became more intense, which destabilized the hydrophobic core 2 and hence impaired the stability of the XPA98–210 system. This result is consistent with the result of the preceding RMSD and RMSF analyses.
The structure of Zn-finger domain
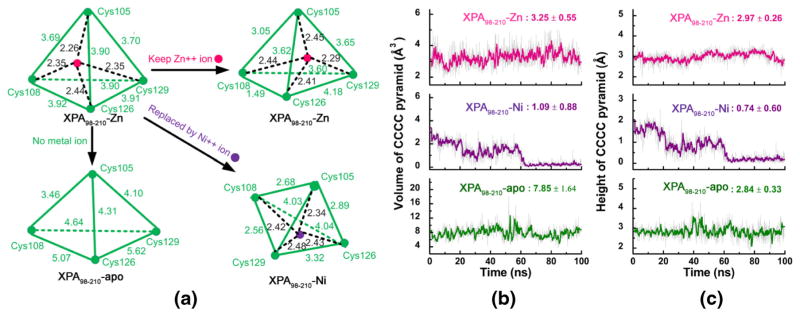
First, we analyzed the change of potential energy and presented the potential energy plot (see Fig. S8). It can be seen from Fig. S8 that the XPA98–210-Zn and XPA98–210-Ni molecules generally remained stable and reached an equilibrium, except that XPA98–210-Ni had a very slight decrease in energy at around 60 ns and shifted to a new plateau of lower energy. In addition, when compared to Fig. 2a, its RMSD also had an increase after 60 ns. In order to illustrate the structural factors behind the changes in energy and RMSD, we superimposed the averaged structures before and after 60 ns. The result showed that the structure of the C-terminal domain was very stable and had a nice conformational overlap, while the Zn-finger domain was relatively unstable. Detailed analysis focusing on the four S atoms in the CCCC pyramid and the Ni2+ revealed that Ni2+ had a tendency of escaping from the inside of the CCCC pyramid; in other words, the CCCC moved in one direction, while the Ni2+ moved in another, disrupting the original pyramidal structure. The detailed geometrical data will be further discussed in the next paragraph. The result here has already suggested that the shift basically occurred after 60 ns, when the CCCC pyramid in the XPA98–210-Ni system collapsed, and its four apex sulfur atoms nearly fell into a plane, shifting from one initially stable conformation to another stable conformation, and remained unchanged to the end of this simulation.
To fully explore the structure of Zn-finger domain, we selected every tenth conformation from the 10,000 conformations in the trajectories of the three systems (XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo), and analyzed the change of the geometric measures of the CCCC (i.e., Cys105, Cys108, Cys126, and Cys129) pyramid in the Zn-finger structure over time (see Fig. 4). Figure 4a shows the spatial scheme of the CCCC pyramids in the three systems, the vertices of which represent the sulfur atoms in the four cysteine residues. Compared with the CCCC pyramid in the 1XPA crystal structure, all six distances between the sulfur atoms in the XPA98–210 apo increased by an average rate (the sum of all percent increments divided by 6) of +17.86 %, due to the loss of metal chelation. Meanwhile, in the XPA98–210-Zn and XPA98–210-Ni systems which contained metal ions, the distances experienced overall reduction by −14.78 % and −15.35 %, respectively. Figure 4b showed the volume changes of the CCCC pyramids in the three systems over simulation time. It can be seen that the pyramid volume in XPA98–210-Zn remained relatively stable at 3.25 ± 0.55 Å3, with a fluctuation of 16.92 %. When the Zn2+ was displaced by Ni2+, the mean volume went down to 1.09 ± 0.88 Å3 (fluctuation = 80.73 %), exhibiting a stepwise trend of shrinkage. After 60 ns, the volume almost went down to zero. As anticipated, the CCCC pyramid volume in XPA98–210-apo was relatively larger, with mean value and fluctuation being 7.85 ± 1.64 Å3/20.89 %, respectively. What exactly caused the CCCC pyramid in XPA98–210-Ni to dramatically shrink, while that in XPA98–210-apo to expand? To address this problem, we analyzed the change of distance from the apex sulfur atom in Cys105 to the base plane formed by the sulfur atoms in Cys108, Cys126, and Cys129 over simulation time. It can be seen from Fig. 4c that the pyramid height in XPA98–210-Ni went close to zero, which indicates that the four sulfur atoms were almost coplanar. The pyramid height in XPA98–210-apo, meanwhile, was still comparable with that in XPA98–210-Zn, therefore the change of the pyramid volume in XPA98–210-apo mainly lies in the stretching of the base area formed by Cys108, Cys126, and Cys129, a result of the lack of constraint by the metal ion.
Fig. 4.
The schematic structure, volume, and height of CCCC pyramid in the XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo systems. (a) All the geometry distances of CCCC pyramid; (b) volume of CCC pyramid over simulation time; (c) height of CCCC pyramid over simulation time
Noticeably, the Cys108 and Cys126 in the XPA98–210-Zn system were drawn close to each other. This situation is likely attributed to two reasons: 1) the system used in our simulation (Met98-Asn210) is not a complete protein, and the missing parts may influence the Zn-finger domain structure; 2) the RESP charges of the Zn center calculated by MCPB may require further correction. Notwithstanding this issue, the XPA98–210-Zn structure, or the relative positions among the four Cys residues in particular, experienced low fluctuation throughout the simulation. This is mainly because the force field parameters including the lengths of the Zn-S bonds rendered Zn the most suitable metal for stabilizing the conformation of the Zn-finger domain, presumably the consequence of natural selection in evolution. When Zn2+ is displaced by Ni2+, the stronger chelating effect (with larger Kr and shorter Req, see Fig. 1 and Fig. S2) drove the four Cys residues close to each other; yet, the global hydrophobic effect of the XPA98–210 molecule tends to maintain the overall conformation: this conflict ultimately led to the breakdown of the folded structure of the CCCC pyramid, with the four S atoms in the four Cys residues virtually collapsing onto the same plane. In the metal-free XPA98–210-apo system, despite that the pyramidal spatial structure was maintained, the CCCC pyramid became more relaxed due to the net effect of the solvent effect and the dispelling force among the charges on the four Cys residues. This also reflected the vital stabilizing effect of Zn’s presence on the Zn-finger structure.
Motion mode and conformational change
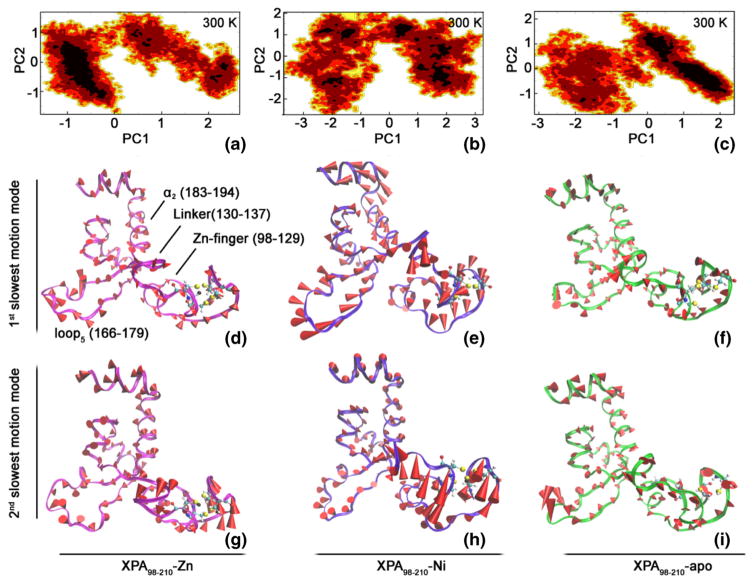
To further analyze the impact of metals on the motion mode and conformational change of XPA98–210, principal component analysis (PCA) was carried out based on the MD simulation trajectories of the three systems (i.e., XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo). PCA is an effective method for extracting the major motion mode from complex MD trajectories, and has been widely used in protein structure-activity relationship studies [50–52]. Generally speaking, the top principal components carry the most important conformational change information of the molecule, and can reveal the slow motion modes that are closely related to its functions. Figure S9 shows the distribution of the top 20 eigenvalues of the MD trajectories of XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo systems. Apparently, the tendencies of the eigenvalue distributions in the three systems were similar. Then, the ratios taken up by the top two eigenvalues were calculated by dividing each of the eigenvalues by the sum of all eigenvalues. These ratios reflected the proportions of the motion modes corresponding to the eigenvectors in the overall motion. It can be seen that the proportions of the top two motion modes in the three systems (XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo) were 55.34 %, 62.79 %, and 62.70 %, respectively. Therefore, it is PC1 and PC2 that represented the functional slow motion modes in our case.
To further investigate the conformational fluctuation in the direction of each primary component, Fig. 5a–c respectively shows the free energy contour maps of the XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo systems at 300 K on the basis of their MD trajectories. Lower energy levels are represented by deeper shades of color. It can be seen from Fig. 5a–c that, the fluctuation range of PC1 is wider than that of PC2. For any given PCn, bigger n reasonably implicates a narrower fluctuation range of motion, and the corresponding conformational fluctuation further resembles a Gaussian distribution, a general characteristic of intra-system, high-frequency, and non-functional fast motion modes. In the low free energy areas of the XPA98–210-Zn system, PC1 varies from −1 to 0, and PC2 varies from −1.2 to 0.7 (see Fig. 5a). In the XPA98–210-Ni system, in contrast, the PC1 and PC2 in the low free energy areas distribute more dispersedly, and conformational fluctuations are greater, indicating lower structural stability. In the same area of the XPA98–210-apo system, PC1 varies from 0 to 2.0 and PC2 varies from −1.0 to 1.2 (see Fig. 5c). The distribution range of PC1 in XPA98–210-apo happened to be exactly the contrary to the PC1 distribution in XPA98–210-Zn, and the width is also larger, indicating a major difference in the motion direction and motion magnitude.
Fig. 5.
Free energy contour map by motion modes, as well as the 1st and 2nd slowest motion modes in the XPA98–210-Zn (a, d, and g), XPA98–210-Ni (b, e, and h), XPA98–210-apo (c, f and i) systems. In the free energy contour maps (a, b, and c), deeper color indicates lower energy, corresponding to free-energy minima. For d–i, the length of the cone is positively correlated with motive magnitude, and the orientation of the cone describes motive direction
In addition, in order to further illustrate the result of the PCA analysis, we also concatenated the three trajectories, and performed an overall PCA, and the projected each trajectory along the first two PCs of XPA98–210-Zn (see Fig. S10). As shown in Fig. S10, both XPA98–210-Ni and XPA98–210-apo had only one low-energy basin. It can also be seen that XPA98–210-Ni exhibited the fiercest functional motion amongst the three systems.
Figure 5d–i shows the 1st and 2nd slowest motion modes of the three systems with cone model. For the 1st slowest motion mode of XPA98–210-Zn, the scope of global motion was small and the Zn-finger domain remained stable; and the motion of its α2 (183–194) and loop5 (166–179) primarily faced outward; the motion of the linker (130–137) was primarily rotation with no observable distortion (see Fig. 5d). The 2nd slowest motion mode of XPA98–210-Zn had a slightly increased scope, especially in its Zn-finger domain (see Fig. 5g). Compared with the 1st and 2nd slowest motion modes of XPA98–210-Zn, those of XPA98–210-Ni were dramatically enhanced, which was most significant in its Zn-finger domain. Meanwhile, both loop5 (166–179) and Zn-finger had an enclosing tendency; and the linker (130–137) went through a major distortion. The 1st and 2nd motion modes of XPA98–210-Ni were basically the opposite and complement of each other, and together they coordinated the movement of the molecule as a whole (see Fig. 5e and h). The 1st slowest motion mode of XPA98–210-apo was similar to that of XPA98–210-Zn, which is consistent with the comparable stabilities suggested by the RMSD result. Detailed features of the motion modes include: 1) the linker (130–137) of XPA98–210-Zn mainly exhibited rotational movement, whereas the XPA98–210-apo mainly went through translational movement, and the latter also had a greater scope; 2) the motion direction of the Zn-finger of XPA98–210-apo mainly pointed outward, which indicated a loss of metal restriction, and the volume of the Zn-finger domain enlarged, which was consistent with the previous calculation of CCCC pyramid volume (Fig. 4b); 3) unlike those of XPA98–210-Zn and XPA98–210-Ni, the loop5 in the 2nd slowest motion mode of XPA98–210-apo had a large motion scope, whereas the Zn-finger region in the same motion mode still remained a low motion scope, representing the diversity and complexity of biological macromolecules’ motion.
To summarize the above analysis, we can observe three characteristics of the motion modes of XPA98–210-Ni, XPA98–210-apo, and XPA98–210-Zn: (1) the motion scopes can be sorted descending as: XPA98–210-Ni > XPA98–210-apo ≈ XPA98–210-Zn, which is perfectly congruent with the results of RMSD, hydrogen bonds, and residue contact analyses; (2) the direction of the 2nd slowest motion mode was inversely correlated with that of the 1st slowest motion mode, especially in XPA98–210-Ni and XPA98–210-Zn, which is likely to contribute to the coordination of molecular global movement; 3) in the primary functional slow motion mode (i.e., the 1st slowest motion mode), the movement of the linker (130–137) in XPA98–210-Ni, XPA98–210-apo, and XPA98–210-Zn behaved differently, exhibiting slight gyration/significant torsion/moderate translation, respectively. It can be inferred that the difference in the global motion scope of the entire molecule and in motion mode of the linker (130–137) may be associated with the cytotoxic mechanism of Ni2+ in the NER process.
From the aforementioned analysis on the RMSF and the superimposition of the final conformations, it can be seen that there existed a distinguishable difference in the linker regions (residues 130–137) among the three systems. The motion modes of this region also appeared to be different. Notably, the linker connects the C-terminal subdomain and the Zn-finger domain, two substructures that are functionally independent, and it participates in binding with RPA70N (see Fig. 5d–i). Therefore, we analyzed the differences in the dihedrals around the eight residues in the linker region (see Table S1). Compared with XPA98–210-Zn and XPA98–210-apo, the residues in XPA98–210-Ni that had the largest fluctuations in dihedrals were Arg130, Asp131, His136, and Lys137. This clearly indicates that the substitution of Ni2+ had an impact on the conformation of the residues flanking the linker, and thus on the relative positions of the C-terminal subdomain and the Zn-finger domain. The reason why the residues flanking the linker in XPA98–210-Zn and XPA98–210-apo had relatively smaller fluctuation was the presence of stable H-bonds between the ND1-HD1 atom in His136 and the O atom in Cys129 in XPA98–210-Zn, and between the NH2-HH21 atom in Arg130 and the OD2 atom in Asp127 in XPA98–210-apo. It is also worth mentioning that we clearly observed a plateau shift in the XPA98–210-Ni’s Arg130-Φ, His136- Ψ, and Lys137- Ψ dihedrals. For example, the Arg130-Φ maintained around −50 degrees during 0~60 ns, whereas it changed to around +50 degrees during 60~100 ns. This indicates that there was a dramatic conformational change at 60 ns during the MD simulation, which is consistent with the time for the potential energy change as well as for the collapse of the CCCC pyramid.
Putative binding modes of XPA98–210 with DNA and RPA70N
XPA98–210 has been known to interact with DNA and some other NER-related proteins such as RPA70 [3, 65], suggesting XPA’s role as a functional hub in the NER mechanism [66]. To date, there have been no reports on the binding mode among DNA, RPA70, and human XPA. Previous structural biology studies [12, 67] indicate a long, narrow cationic bend lying across the C-terminal subdomain of the human XPA98–210, which is formed by Lys141, Lys145, Lys151, Lys179, Lys204, and Arg207. Structurally speaking, this region is ideal for binding both single-strand and double strand DNA. Mutation study by Maltseva et al. [68] indicated that, K179E and K141E mutants of XPA exhibited significantly weakened binding affinity with DNA compared to the wild-type XPA. Sugitani et al. suggested via a fluorescence anisotropy DNA-binding assay that, Lys204, Arg207, as well as the unresolved Phe219, Ala229, Trp235, and Lys236, participated in DNA binding [69]. In 2015, Koch et al. acquired the crystal structure of DNA binding with the yeast XPA (PDB code: 5A3D) [6]. The yeast XPA188–301 shares a high homology with human XPA98–210, the sequence comparison of which is shown in Fig. S11. In yeast XPA, the four Cys residues that form a Zn-finger pyramid structure with Zn2+ are Cys191/Cys194/Cys213/Cys216, which exactly correspond to the four Cys residues in human XPA98–210 (i.e., Cys105/Cys108/Cys126/Cys129). The key residues in yeast XPA188–301 that participate in DNA recognition are Lys229, Lys233, Thr239, His258, and R294. Through sequence alignment and structural superimposition, these residues correspond to Lys141, Lys145, Lys151, His171, Lys179, and Arg207 in human XPA98–201 (see Fig. S11).
The above mentioned analyses on residue contact, second structure, and hydrogen bonds indicated that the conformation of the C-terminal domain (98–129) of XPA98–210 was not significantly influenced by Zn2+ ion’s absence or displacement, and hence the direct recognition of DNA by human XPA was unaffected. Another role of XPA98–210 apart from binding DNA is to form a complex with RPA70 [12]. Then, what regions in both XPA98–210 and RPA70 are responsible for this interaction, and how exactly do they bind? The answer to these questions may hold the key to unraveling the mechanism for the functional change of XPA98–210 caused by the absence or displacement of Zn2+ by Ni2+.
Table S2 lists all the available structural biology data of RPA70 from PDB in descending order of time. In the PDB database there have already been 24 reports on the structure of RPA70, as well as 21 resolved structures of the N-terminal domain of RPA70 (RPA70N, residues 1–120), 20 of which were co-crystal structures that had mutual RMSD values less than 0.5 Å; the only one left was an NMR solution structure published in 1999, probably due to the difference in methodology and environment. The structures of C-terminal domain (439–616) [14] and the central domain (183–420) [70, 71] were reported by Bochkareva et al. and Bochkarev et al., respectively. These two regions form a trimer with RPA14 and RPA32 and have the ability to form a complex with DNA. Notably, the conformation of the RPA70 central domain experienced a major alteration after binding with DNA. The N-terminal domain of RPA70 (RPA70N), based on the structural biology analysis by Bochkareva and Bochkarev et al., primarily participates in binding with XPA.
By analyzing all the available PDB structures of RPA70N in complex with its substrates (see Table S2), we summarized the substrate-binding regions of RPA70N as follows: Arg31-Arg43, Ser54-Thr60, and Ile83-Met97 (which are located exactly in the V-shaped pocket formed by Arg31-Arg41 and Thr86-Arg92). We also analyzed the amino acid types in the small peptide bound in RPA70N, and it was found that acidic amino acids took up a large percentage [72, 73], for instance the small peptide substrate D5F6T7A8D9D10L11E12E13W14 in crystal structure of PDB 4NB3. NMR experiment [12] has discovered that the C-terminal subdomain (138–210) in XPA98–210 primarily functions for DNA binding, while the Zn-finger region (98–129) for recognition of RPA70N. A series of helix-rich small peptides were designed by Frank et al. [72], and they could strongly bind to RPA70N at the V-shaped pocket formed by Arg31-Arg41 and Thr86-Arg92. All these structural biology studies suggest that the V-shaped pocket in RPA70N is optimal for protein binding. The structural characteristics of the Zn-finger domain in XPA98–210, on the other hand, indicate that the regions most likely to be recognized by the V-shaped pocket in RPA70N are Tyr116-Asp122 and Asp127-Asp134, both of which have: 1) most helix-like secondary structures; 2) optimal lengths to be anchored in the V-shaped pocket.
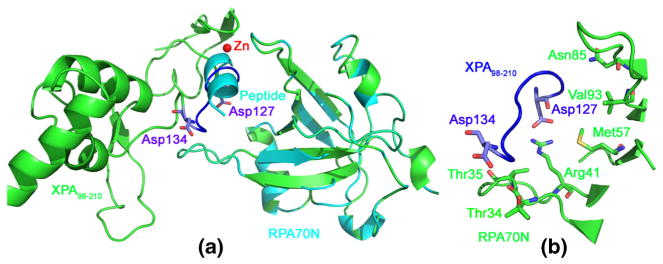
Based on the structural biology information, we obtained the binding mode between XPA98–210-Zn and RPA70N (see Fig. 6) using Z-dock server [74, 75]. In all the docking modes of XPA98–210-Zn-RPA70N, it was Asp127-Asp134 in XPA98–210-Zn that bound to the V-shaped pocket of RPA70N. In contrast, Tyr116-Asp122 refused to bind to the V-shaped pocket, mainly because of the significant steric hindrance from the adjacent Ile165-Lys167 loop. From Fig. 6, the position of Asp127-Asp134 (of XPA98–210) in the XPA98–210-Zn-RPA70N docking modes is similar to that of the small peptide in the RPA70N-peptide crystal structure (PDB ID: 4NB3) [72]. Therefore, this XPA98–210-Zn-RPA70N docking mode is reasonable. Here, the contact residues in RPA70N that participated in binding Asp127-Asp134 of XPA98–210 were: Thr34, Thr35, Arg41, Asn85, Val93, and Met157, which were consistent with the structure biology information given by Table S2. Analysis of the XPA98–210-Zn-RPA70N complex model showed that, apart from Asp127-Asp134, the contact residues in XPA98–210 which had less than 4 Å distances to RPA70N also included Val166-Lys168. Our result is consistent with the pull-down assay by Li et al. [3] which showed that the Cys153-Gly176 residues may be involved in RPA70N binding.
Fig. 6.
The binding modes between XPA98–210-Zn and RPA70N. (a) Superimposition of the docked XPA98–210-Zn-RPA70N model (solid ribbon in green) with RPA70N-peptide crystal structure (solid ribbon in cyan, PDB ID: 4NB3). (b) The contact resides of RPA70N (stick model in green) binding with XPA98–210-Zn (ribbon model in blue)
Similar molecular docking strategy was adopted to yield the binding modes of XPA98–210-Ni and RPA70N (data not shown). The result showed that the V-shaped pocket mainly binds to Met98-Ile104 in XPA98–210-Ni, instead of Asp127-Asp134. The difference of the binding sites in XPA98–210-Zn and XPA98–210-Ni to RPA70N may be attributed to two reasons: 1) Secondary structure analysis (see Fig. S6) indicated that, the Met98-Ile104/Asp127-Asp134 residue regions were predominantly bend-coil/helix-turn and helix-turn/bend-turn in the XPA98–210-Zn and XPA98–210-Ni systems. In comparison with the same structures in XPA98–210-Zn, the Met98-Ile104 in XPA98–210-Ni had greatly increased helicity, while the helix in the Asp127-Asp134 almost disappeared; the V-shaped pocket of RPA70N preferentially bind with more stable α-helixes. 2) Based on the preceding motion mode analysis (see Fig. 5d–f) results, the linker (130–137) in both XPA98–210-Zn and XPA98–210-Ni had differential motion modes (slight gyration vs. significant distortion), which also prevents RPA70N from binding the Asp128-Asp134 region in XPA98–210-Ni. It can be speculated that the differential binding modes of XPA98–210-Ni with RPA70N compared to that of XPA98–210-Zn, in addition to the weakening of the binding affinity, may explain the cytotoxicity of Ni2+.
We have already acquired the structures of both XPA98–210-DNA and XPA98–210-RPA70N through super-imposition and molecular docking, respectively. Based on the almost perfectly superimposed structures of XPA98–210 in both XPA98–210-DNA and XPA98–210-RPA70N, we constructed the structure of the XPA98–210-DNA-RPA70N complex structure (see Fig. S12). The figure shows that the C-terminal subdomain (139–210) in XPA98–210-Zn primarily participates in DNA recognition, while the Zn-finger domain (98–129) and linker (130–137) mainly contribute to the binding with RPA70N. Here, the Cys126 residue in the CCCC pyramid of the Zn-finger was right adjacent to the Asp127-Asp134 region which directly binds to RPA70N, so this complex structure model agrees with the result from previous the NMR experiment [12].
The importance of Zn-finger domain in the NER process
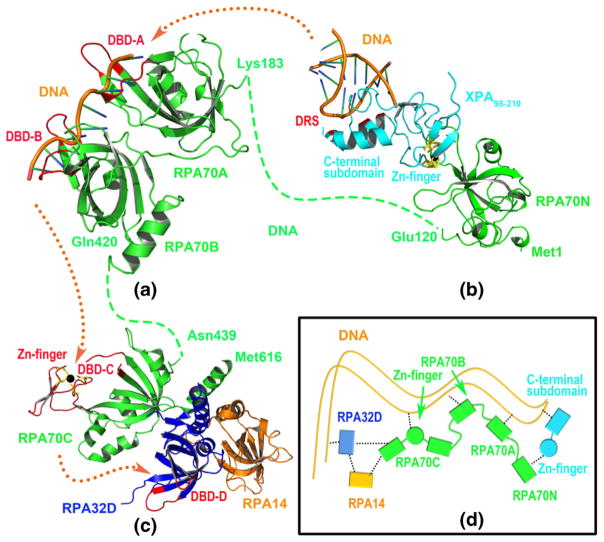
The co-crystal structures of RPA70183–300 (RPA70A) and RPA70301–420 (RPA70B) in complex with DNA have been obtained by Bochkarev et al. [71] using X-ray crystallography. The DNA binding regions in RPA70A and RPA70B were two V-shaped clefts consisting of Ser213-Glu218/Ile263-Lys267 (referred to as DNA binding domain A, DBD-A, see Fig. 7a) and Val334-Arg339/Ser384-Arg389 (referred to as DNA binding domain B, DBD-B, see Fig. 7a), respectively. Bochkareva et al. [14] reported the structure of the RPA trimer at 2.8 angstrom resolution. This structure included the C-terminal domain of RPA70 (RPA70C), the DNA binding region in RPA32 (RPA32D) and the whole RPA14 subunit. Interestingly, there are also two V-shaped cleft in RPA70C and RPA32D for DNA binding, which consist of Cys481-Tyr512/Tyr581-Lys588 (referred to as DBD-C, see Fig. 7c) and E84-N89/F135-K138 (referred to as DBD-D, see Fig. 7c), respectively. Notably, the Zn-finger motif is a core structure that constitutes DBD-C. The molecular simulation result and data from the structural biology experiment indicated that, the DNA is first recognized by the C-terminal subdomain (138–210) in XPA98–210, or more specifically, by Lys141, Lys145, Lys151, His171, Lys179, and Arg207, a region referred to as DNA recognition site (DRS, see Fig. 7b). Then, the Zn-finger region (98–129) of XPA98–210 is recognized by RPA70N. Obviously, the correct and stable recognition of XPA by RPA70N is essential for the entire NER process.
Fig. 7.
Extension of the DNA in NER process. (a) The 3D-structure of RPA70A-RPA70B-DNA complex based on PDB 1JMC; (b) The 3D-structure of RPA70N-XPA98–210-DNA complex model; (c) The 3D-struccture of RPA70C-RPA32D-RPA14 trimer from PDB 1L1O. Blue and origin dotted lines represent the unresolved part of RPA70 proteins and the extension direction of DNA. Five interaction regions (i.e., DRS, DBD-A, DBD-B, DBD-C, and DBD-D) with DNA are shown with red ribbon mode. (d) Schematic diagram of the interactions between a series of associated proteins and DNA in the NER process, and the black dotted line indicates the existing interactions for the multi-proteins NER system
Combining the crystal structures of RPA70A-RPA70B-DNA, RPA70C-RPA32D-RPA14 trimer, and our simulation result, the transmission process of the DNA in the NER pathway is shown in Fig. 7. The DNA has five binding sites in the NER process, and it is recognized and transmitted in such order: first by DRS of XPA98–210, then DBD-A of RPA70A, DBD-B of RPA70B, DBD-C of RPA70C, and finally DBD-D of RPA32. Intriguingly, two Zn-finger domains are present in the entire NER process: one of them is located in XPA98–210 and is responsible for binding RPA70N, and the other one lies in RPA70C and directly participates in DNA binding. Apparently, the zinc metal exhibits dual biological functions in the complicated NER machinery. The cytotoxic mechanism proposed in this work, where Ni2+ decreases the effectiveness of the NER process by replacing Zn2+ in XPA98–210 and weakening its recognition by RPA70N, may be one of the many artifices adopted by Ni2+.
Conclusions
In this paper, we studied the impact of Ni2+ on XPA98–210 structure using comparative MD simulation, and also explored the potential mechanism for Ni2+-mediated cytotoxicity in the NER process. The force field parameters of Zn2+ and Ni2+ ion center in XPA98–210 were calculated by MCPB method, where the ranges of Zn-S and Ni-S bond length are 2.412–2.466 and 2.281–2.293 Å, respectively. The bond energy of the latter is higher. Three 100 ns explicit water MD simulations were carried out for XPA98–210-Zn, XPA98–210-Ni, and XPA98–210-apo using these parameters. The RMSF of the three simulations showed high consistency and correlation, suggesting that the methodology is valid. Regular MD simulation metrics, including RMSD and radius of gyration, all clearly showed that the conformational fluctuation of XPA98–210-Ni was greater than that of XPA98–210-apo and XPA98–210-Zn, with the order of molecular stability being XPA98–210-Zn ≈ XPA98–210-apo > XPA98–210-Ni. Further analysis of residue contacts, secondary structures, and hydrogen bonding suggested that, even though the C-terminal domain (138–210) structure are generally not affected by the absence of or the displacement of Zn2+ by Ni2+, such conditions obviously reduced the number of residue contacts and hydrogen bonds, and altered the distribution of secondary structures, indicating a large conformational change in the Zn-finger domain.
To quantitatively depict such changes, we used the volume and height of the CCCC pyramid as measures of the shape for Zn-finger pyramid during the simulation process. In XPA98–210-Zn, the volume and height both remained steady through-out the simulation, suggesting a relatively higher stability of the Zn-finger structure. In the absence of metal chelation, i.e., XPA98–210-apo, the overall shape of the CCCC pyramid in the Zn-finger domain remained basically unchanged, whereas the mutual distances of the four Cys residues were enlarged, leading to an increase in the volume and height of the pyramid and a more relaxed structure. When the Zn2+ is displaced by Ni2+, however, strong attractive forces by the Ni-S bonds, in addition to the high flexibility of the adjacent loop regions, drove the four Cys residues to fall on a plane, leading to the collapse of the pyramid. To analyze the impact of metal ion on the overall motion mode of XPA98–210, principal component analysis was performed on the MD trajectories of the three systems, and the results demonstrate that the order of motion amplitude was XPA98–210-Ni > XPA98–210-apo ≈ XPA98–210-Zn. In addition, an obvious difference in the motion mode of the linker (130–137) among the three systems was observed, with XPA98–210-Ni, XPA98–210-apo, and XPA98–210-Zn, showing small rotation, large distortion, and moderate translational movement, respectively.
To unravel the biological implications of such conformational and motional change of XPA98–210 induced by the displacement of Zn2+ by Ni2+, we combined the results from molecular docking, structural biology information, and structural alignment, and obtained the model for XPA98–210-RPA70N-DNA complex, and a possible interaction pattern during NER process involving multiple proteins (including XPA, RPA70N, RPA70A, RPA70B, RPA70C, RPA32D, and RPA14). The alterations of Zn-finger domain in XPA induced by displacement of Zn2+ by Ni2+ could impair its binding to RPA70N, hence reducing the NER efficiency. Considering that there are more than one type of protein containing Zn-finger domain during the NER process, we speculate that the proposed mechanism in this study could be one of the contributing factors for Ni2+ cytotoxicity. In sum, our study investigated the influence of Ni2+ on the structure and functions of XPA98–210, and suggested a putative molecular mechanism for metal-induced cytotoxicity, which will facilitate novel anti-cancer drug design targeting the NER signaling pathway, and will provide insight into the multiple protein interactions in the context of NER.
Supplementary Material
Acknowledgments
This work was supported by the funding from NIDA (P30 DA035778A1) and NIH (R01 DA025612), and in part by the National Natural Science Foundation of China (11147175, 11247018), the Key Project of Sichuan Provincial Education Bureau (12ZA066), the Project of Leshan Science and Technology Administration (14GZD022).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00894-016-3017-x) contains supplementary material, which is available to authorized users.
Disclaimer The findings and conclusions in this paper have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any agency determination or policy. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by Department of Health and Human Services.
References
- 1.Sanear A. Mechanisms of DNA excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- 2.Gillet LC, Schärer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Lu X, Peterson CA, Legerski RJ. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol Cell Biol. 1995;15:5396–5402. doi: 10.1128/mcb.15.10.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feltes BC, Bonatto D. Overview of xeroderma pigmentosum proteins architecture, mutations and post-translational modifications. Mutat Res Rev Mut Res. 2015;763:306–320. doi: 10.1016/j.mrrev.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 6.Koch SC, Kuper J, Gasteiger KL, Simon N, Strasser R, Eisen D, Geiger S, Schneider S, Kisker C, Carell T. Structural insights into the recognition of cisplatin and AAF-dG lesion by Rad14 (XPA) Proc Natl Acad Sci U S A. 2015;112:8272–8277. doi: 10.1073/pnas.1508509112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oei AL, Vriend LE, Crezee J, Franken NA, Krawczyk PM. Effects of hyperthermia on DNA repair pathways: one treatment to inhibit them all. Radiat Oncol. 2015;10:165–179. doi: 10.1186/s13014-015-0462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugitani N, Chazin WJ. Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Prog Biophys Mol Biol. 2015;117:206–211. doi: 10.1016/j.pbiomolbio.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li CL, Golebiowski FM, Onishi Y, Samara NL, Sugasawa K, Yang W. Tripartite DNA lesion recognition and verification by XPC, TFIIH, and XPA in nucleotide excision repair. Mol Cell. 2015;59:1025–1034. doi: 10.1016/j.molcel.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilton B, Shkriabai N, Musich PR, Kvaratskhelia M, Shell S, Zou Y. A new structural insight into XPA-DNA interactions. Biosci Rep. 2014;34:831–840. doi: 10.1042/BSR20140158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones CJ, Wood RD. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 12.Ikegami T, Kuraoka I, Saijo M, Kodo N, Kyogoku Y, Morikawa K, Tanaka K, Shirakawa M. Solution structure of the DNA-and RPA-binding domain of the human repair factor XPA. Nat Struct Mol Biol. 1998;5:701–706. doi: 10.1038/1400. [DOI] [PubMed] [Google Scholar]
- 13.Buchko GW, Daughdrill GW, De Lorimier R, Sudha Rao B, Isern NG, Lingbeck JM, Taylor JS, Wold MS, Gochin M, Spicer LD. Interactions of human nucleotide excision repair protein XPA with DNA and RPA70ΔC327: Chemical shift mapping and 15N NMR relaxation studies. Biochemistry. 1999;38:15116–15128. doi: 10.1021/bi991755p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 16.Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protić M, Hübscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 17.Hey T, Lipps G, Krauss G. Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy. Biochemistry. 2001;40:2901–2910. doi: 10.1021/bi002166i. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Mahrenholz A, Lee SH. RPA stabilizes the XPA-damaged DNA complex through protein-protein interaction. Biochemistry. 2000;39:6433–6439. doi: 10.1021/bi000472q. [DOI] [PubMed] [Google Scholar]
- 19.Sakharov DV, Lim C. Zn protein simulations including charge transfer and local polarization effects. J Am Chem Soc. 2005;127:4921–4929. doi: 10.1021/ja0429115. [DOI] [PubMed] [Google Scholar]
- 20.Michalek JL, Besold AN, Michel SL. Cysteine and histidine shuffling: mixing and matching cysteine and histidine residues in zinc finger proteins to afford different folds and function. Dalton Trans. 2011;40:12619–12632. doi: 10.1039/c1dt11071c. [DOI] [PubMed] [Google Scholar]
- 21.Besold AN, Lee SJ, Michel SL, Sue NL, Cymet HJ. Functional characterization of iron-substituted neural zinc finger factor 1: metal and DNA binding. J Biol Inorg Chem. 2010;15:583–590. doi: 10.1007/s00775-010-0626-1. [DOI] [PubMed] [Google Scholar]
- 22.Witkiewicz-Kucharczyk A, Bal W. Damage of zinc fingers in DNA repair proteins, a novel molecular mechanism in carcinogenesis. Toxicol Lett. 2006;162:29–42. doi: 10.1016/j.toxlet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Asmuss M, Mullenders LH, Eker A, Hartwig A. Differential effects of toxic metal compounds on the activities of Fpg and XPA, two zinc finger proteins involved in DNA repair. Carcinogenesis. 2000;21:2097–2104. doi: 10.1093/carcin/21.11.2097. [DOI] [PubMed] [Google Scholar]
- 24.Hartwig A, Asmuss M, Ehleben I, Herzer U, Kostelac D, Pelzer A, Schwerdtle T, Bürkle A. Interference by toxic metal ions with DNA repair processes and cell cycle control: molecular mechanisms. Environ Health Perspect. 2002;110:797–799. doi: 10.1289/ehp.02110s5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleaver J. The DNA damage-recognition problem in human and other eukaryotic cells: the XPA damage binding protein. Biochem J. 1997;328:1–12. doi: 10.1042/bj3280001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asahina H, Kuraoka I, Shirakawa M, Morita EH, Miura N, Miyamoto I, Ohtsuka E, Okada Y, Tanaka K. The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat Res/DNA Repair. 1994;315:229–237. doi: 10.1016/0921-8777(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 27.Hartwig A. Carcinogenicity of metal compounds: possible role of DNA repair inhibition. Toxicol Lett. 1998;102:235–239. doi: 10.1016/s0378-4274(98)00312-9. [DOI] [PubMed] [Google Scholar]
- 28.Hartwig A. Zinc finger proteins as potential targets for toxic metal ions: differential effects on structure and function. Antioxid Redox Signal. 2001;3:625–634. doi: 10.1089/15230860152542970. [DOI] [PubMed] [Google Scholar]
- 29.Li P, Roberts BP, Chakravorty DK, Merz KM., Jr Rational design of particle mesh Ewald compatible Lennard-Jones parameters for + 2 metal cations in explicit solvent. J Chem Theory Comput. 2013;9:2733–2748. doi: 10.1021/ct400146w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang YP. Successful molecular dynamics simulation of two zinc complexes bridged by a hydroxide in phosphotriesterase using the cationic dummy atom method. Proteins: Struct Funct Bioinform. 2001;45:183–189. doi: 10.1002/prot.1138. [DOI] [PubMed] [Google Scholar]
- 31.Peters MB, Yang Y, Wang B, Füsti-Molnár L, Weaver MN, Merz KM., Jr Structural survey of zinc-containing proteins and development of the zinc AMBER force field (ZAFF) J Chem Theory Comput. 2010;6:2935–2947. doi: 10.1021/ct1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou TJ, Zhang W, Xu XJ. Binding affinities for a series of selective inhibitors of gelatinase-A using molecular dynamics with a linear interaction energy approach. J Phys Chem B. 2001;105:5304–5315. [Google Scholar]
- 33.Tounge BA, Reynolds CH. Calculation of the binding affinity of β-secretase inhibitors using the linear interaction energy method. J Med Chem. 2003;46:2074–2082. doi: 10.1021/jm020513b. [DOI] [PubMed] [Google Scholar]
- 34.Zhou M, Du K, Ji P, Feng W. Molecular mechanism of the interactions between inhibitory tripeptides and angiotensin-converting enzyme. Biophys Chem. 2012;168:60–66. doi: 10.1016/j.bpc.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Pa K. AMBER 12. University of California; San Francisco: 2012. [Google Scholar]
- 36.Becke AD. Density-functional thermochemistry. III. the role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 37.Dill JD, Pople JA. Self-consistent molecular orbital methods. XV. extended Gaussian-type basis sets for lithium, beryllium, and boron. J Chem Phys. 1975;62:2921–2923. [Google Scholar]
- 38.Frisch M, Trucks G, Schlegel H, Scuseria G, Robb M, Cheeseman J, Montgomery J, Jr, Vreven T, Kudin K, Burant J, et al. Gaussian 03. Gaussian Inc; Wallingford: 2004. [Google Scholar]
- 39.Feng Z, Kochanek S, Close D, Wang L, Srinivasan A, Almehizia AA, Iyer P, Xie X-Q, Johnston PA, Gold B. Design and activity of AP endonuclease-1 inhibitors. J Chem Biol. 2015:1–15. doi: 10.1007/s12154-015-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Z, Pearce LV, Xu X, Yang X, Yang P, Blumberg PM, Xie X-Q. Structural insight into tetrameric hTRPV1 from homology modeling, molecular docking, molecular dynamics simulation, virtual screening, and bioassay validations. J Chem Inf Model. 2015;55:572–588. doi: 10.1021/ci5007189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Z, Alqarni MH, Yang P, Tong Q, Chowdhury A, Wang L, Xie X-Q. Modeling, molecular dynamics simulation, and mutation validation for structure of cannabinoid receptor 2 based on known crystal structures of GPCRs. J Chem Inf Model. 2014;54:2483–2499. doi: 10.1021/ci5002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct Funct Bioinform. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 45.Shen R, Han W, Fiorin G, Islam SM, Schulten K, Roux B. Structural refinement of proteins by restrained molecular dynamics simulations with non-interacting molecular fragments. PLoS Comput Biol. 2015;11:e1004368. doi: 10.1371/journal.pcbi.1004368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 47.Amadei A, Linssen A, De Groot B, Van Aalten D, Berendsen H. An efficient method for sampling the essential subspace of proteins. J Biomol Struct Dyn. 1996;13:615–625. doi: 10.1080/07391102.1996.10508874. [DOI] [PubMed] [Google Scholar]
- 48.Hu JP, Wang CX. Molecular dynamics simulation of HIV-1 integrase dimer complexed with viral DNA. Chin J Chem. 2010;28:33–40. [Google Scholar]
- 49.Wan H, Chang S, Hu JP, Tian YX, Tian XH. Molecular dynamics simulations of ternary complexes: comparisons of LEAFY protein binding to different DNA motifs. J Chem Inf Model. 2015;55:784–794. doi: 10.1021/ci500705j. [DOI] [PubMed] [Google Scholar]
- 50.Srikumar P, Rohini K, Rajesh PK. Molecular dynamics simulations and principal component analysis on human laforin mutation W32G and W32G/K87A. Protein J. 2014;33:289–295. doi: 10.1007/s10930-014-9561-2. [DOI] [PubMed] [Google Scholar]
- 51.Nagai T, Mitsutake A, Takano H. Principal component relaxation mode analysis of an all-atom molecular dynamics simulation of human lysozyme. J Phys Soc Jpn. 2013;82:023803. [Google Scholar]
- 52.Ivanov PM. Conformations of some large-ring cyclodextrins derived from conformational search with molecular dynamics simulations and principal component analysis. J Phys Chem B. 2010;114:2650–2659. doi: 10.1021/jp9110426. [DOI] [PubMed] [Google Scholar]
- 53.Maisuradze GG, Liwo A, Scheraga HA. Relation between free energy landscapes of proteins and dynamics. J Chem Theory Comput. 2010;6:583–595. doi: 10.1021/ct9005745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan H, Hu JP, Li KS, Tian XH, Chang S. Molecular dynamics simulations of DNA-free and DNA-bound TAL effectors. PLoS One. 2013;8:e76045. doi: 10.1371/journal.pone.0076045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan H, Hu JP, Tian XH, Chang S. Molecular dynamics simulations of wild type and mutants of human complement receptor 2 complexed with C3d. Phys Chem Chem Phys. 2013;15:1241–1251. doi: 10.1039/c2cp41388d. [DOI] [PubMed] [Google Scholar]
- 56.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 57.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 58.Woods RJ, Khalil M, Pell W, Moffat SH, Smith VH. Derivation of net atomic charges from molecular electrostatic potentials. J Comput Chem. 1990;11:297–310. [Google Scholar]
- 59.Sindhikara DJ, Roitberg AE, Merz KM., Jr Apo and nickel-bound forms of the Pyrococcus horikoshii species of the metalloregulatory protein: NikR characterized by molecular dynamics simulations. Biochemistry. 2009;48:12024–12033. doi: 10.1021/bi9013352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ni C, Dang D, Song Y, Gao S, Li Y, Ni Z, Tian Z, Wen L, Meng Q. An interesting magnetic behavior in molecular solid containing one-dimensional Ni (III) chain. Chem Phys Lett. 2004;396:353–358. [Google Scholar]
- 61.Liu S, Han YF, Jin GX. Formation of direct metal–metal bonds from 16-electron “pseudo-aromatic” half-sandwich complexes Cp “M [E 2 C 2 (B 10 H 10)] Chem Soc Rev. 2007;36:1543–1560. doi: 10.1039/b701869j. [DOI] [PubMed] [Google Scholar]
- 62.Feig M, Karanicolas J, Brooks CL. MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graphics Modell. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 63.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 64.Andersen CA, Palmer AG, Brunak S, Rost B. Continuum secondary structure captures protein flexibility. Structure. 2002;10:175–184. doi: 10.1016/s0969-2126(02)00700-1. [DOI] [PubMed] [Google Scholar]
- 65.Tripsianes K, Folkers GE, Zheng C, Das D, Grinstead JS, Kaptein R, Boelens R. Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization. Nucleic Acids Res. 2007;35:5789–5798. doi: 10.1093/nar/gkm503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair. 2011;10:673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 67.Buchko GW, Isern NG, Spicer LD, Kennedy MA. Human nucleotide excision repair protein XPA: NMR spectroscopic studies of an XPA fragment containing the ERCC1-binding region and the minimal DNA-binding domain (M59-F219) Mutat Res/DNA Repair. 2001;486:1–10. doi: 10.1016/s0921-8777(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 68.Maltseva E, Krasikova Y, Naegeli H, Lavrik O, Rechkunova N. Effect of point substitutions within the minimal DNA-binding domain of xeroderma pigmentosum group a protein on interaction with DNA intermediates of nucleotide excision repair. Biochem Mosc. 2014;79:545–554. doi: 10.1134/S000629791406008X. [DOI] [PubMed] [Google Scholar]
- 69.Sugitani N, Shell SM, Soss SE, Chazin WJ. Redefining the DNA-binding domain of human XPA. J Am Chem Soc. 2014;136:10830–10833. doi: 10.1021/ja503020f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bochkareva E, Belegu V, Korolev S, Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 72.Frank AO, Vangamudi B, Feldkamp MD, Souza-Fagundes EM, Luzwick JW, Cortez D, Olejniczak ET, Waterson AG, Rossanese OW, Chazin WJ. Discovery of a potent stapled helix peptide that binds to the 70N domain of replication protein A. J Med Chem. 2014;57:2455–2461. doi: 10.1021/jm401730y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.