Abstract
Large-scale mutagenesis of target DNA sequences comprehensively assesses the effects of single nucleotide changes. Here we demonstrate the construction of a systematic allelic series (SAS) using massively parallel single nucleotide mutagenesis with reversibly-terminated deoxyinosine triphosphates (rtITP). We created a mutational library containing every possible singleton nucleotide mutation surrounding the active site of the TEM-1 β-lactamase gene. When combined with high-throughput functional assays, SAS mutational libraries have the ability to expedite the functional assessment of genetic variation.
The difficulty of assessing the effects of genetic variants in the human population is a major obstacle confronting precision medicine. The power and utility of rapid, accurate mutation library production has been recently demonstrated (1–5). Several large-scale mutagenesis methods have been utilized to screen both genes and non-coding loci. Mutagenesis libraries have been created using a variety of methods, including error-prone or inosine containing PCR, or by using synthetic oligonucleotides (1–10). However, each of these approaches has their limitations. While error-prone PCR is inexpensive, products often have multiple mutations and high transition:transversion ratios, whereas the production of oligonucleotides requires substantial up-front financial investment.
Here we demonstrate the construction of a systematic allelic series (SAS) by performing mutagenesis of segments of DNA using reversibly-terminated deoxyinosine triphosphates (rtITPs). By combining cycle termination used in Sanger sequencing, reversible termination used in Illumina sequencing, and the ability of inosine to base pair with each of the four bases, we have systematically incorporated single inosine molecules into DNA molecules allowing the introduction of one and only one mutation per molecule during PCR amplification.
Briefly, we generated reversibly-terminated deoxyinosine triphosphates (rtITP) by sequential enzymatic reaction of reversibly-terminated deoxyadenosine triphosphates (Supplementary Fig. 1 and Supplementary Note). This multistep synthesis was required due to the inability of adenosine deaminase to deaminate adenosine triphosphate and the lack of commercially available rtITP. A linear amplification was then performed using a biotinylated forward primer, a special polymerase (Firebird475, Firebird Biomolecular Sciences, LLC) required for the incorporation of reversibly-terminated nucleotides, and a dNTP pool containing >50% w/v rtITP (Fig. 1a). Incorporation of rtITP molecules and their termination of extension was confirmed by amplification with a 5’-ROX-labeled primer (Supplementary Fig. 2). For short products, bands consisting of products >20bp and <full length were gel extracted to remove unwanted full length product and unextended primers. Linear amplification products were subsequently isolated by hybridization to streptavidin-coated magnetic beads. The 3’-O-NH2 termination moiety on the rtITPs was then removed by exposure to sodium nitrite (0.7M, pH 5.5) and products extended using a high-fidelity polymerase. Beads were then washed to remove template DNA and amplified to produce PCR products containing each of the 4 alternative nucleotides at each site where inosines had been incorporated.
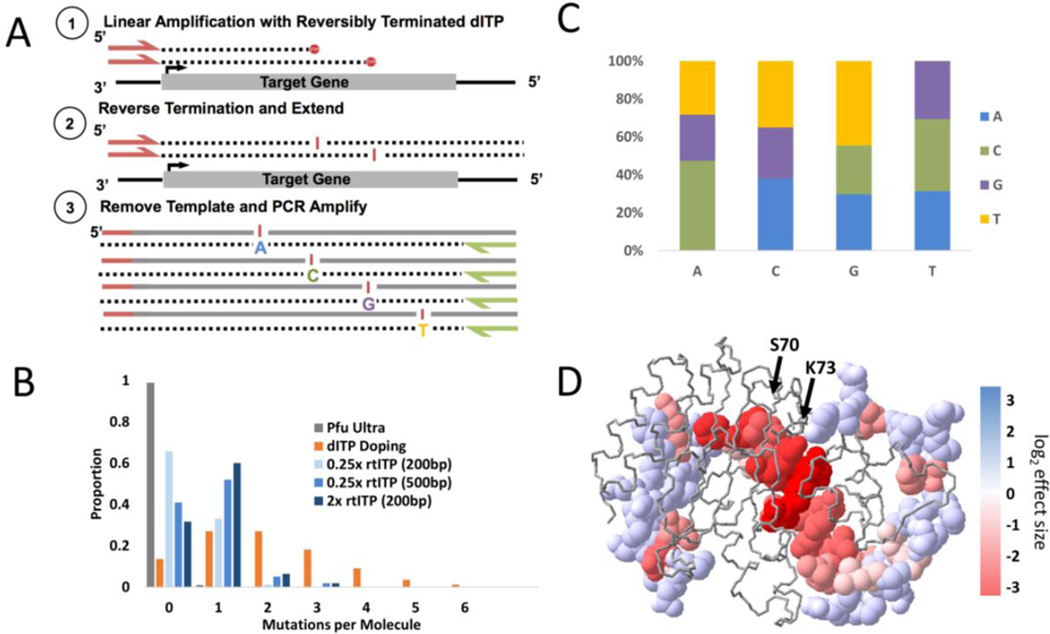
Figure 1. Schematic and library characteristics of SAS mutagenesis.
(a) Schematic of SAS mutagenesis (b) Histogram of mutations per molecule for libraries under various reaction conditions (c) Average proportions of nucleotide changes observed in the two SAS libraries (217 and 687bp) (d) Crystal structure of TEM-1 β-lactamase with mutated sites pictured with space filled molecular model and non-tested sites with backbone only. Sites are colored according to their log2 effect size after ampicillin selection with red indicating a greater negative effect size.
We utilized SAS mutagenesis to create a library of the ampicillin resistance gene (AmpR) encoding TEM-1 β-lactamase. First, 217bp consisting of a portion of the active site of the ampicillin resistance gene was amplified and cloned in-frame into a wild-type β-lactamase plasmid also containing the kanamycin resistance gene, allowing the plasmid library to replicate efficiently without ampicillin selection. Completely overlapping paired-end sequencing of this SAS library unselected for ampicillin resistance revealed that 33% of clones contained one and only one mutation when created with a 1:1 ratio of rtITP:dNTPs and >50% with a 4:1 ratio of rtITP:dNTPs. Importantly, only ~1% of clones contained >1 mutation for the 1:1 ratio and ~5% for the 4:1 ratio (Figure 1b). This is a marked reduction in secondary mutations compared to previous methods, the most effective of which produced a similar rate of single mutations (33–47%) but produced a substantial proportion of molecules with secondary mutations (21–35%) (2). We initially limited our mutagenesis reactions to < 250bp so that they could be fully sequenced from both directions to distinguish true mutations from sequencing errors. We observed good correlation between true mutation counts and observed mutation counts, however, suggesting that shotgun sequencing can suffice to calculate mutation effect sizes (Supplementary Figure 3). The nucleotide composition of generated mutations within the SAS library was similar at each nucleotide resulting in an average transition:transversion ratio of 0.48 (Figure 1c and Supplementary Table 1) compared to error-prone PCR which showed the expected inflated transition:transversion ratio of 3.2.
Using this SAS mutagenized library, we assessed the functional impact of mutations present within the first 217bp of the AmpR gene. Upon submitting the SAS library to selection, we observed strong depletion of mutations resulting in non-conservative (i.e. hydrophobic to polar, etc) non-synonymous amino acid substitutions (p<2x10−20, Mann-Whitney U) (Supplementary Figure 4) with specific residues within the β-lactamase active site (S70 and K73) among the most strongly depleted with effect sizes dropping off further away from the active site (Figure 1d). We also observed a strong correlation of our observed enrichment scores with those described previously (11) (r2=0.31–0.68) (Supplementary Figure 5).
Lastly, we sequenced the distal 687bp segment of the AmpR gene to demonstrate the ability of SAS mutagenesis to create long mutational libraries. We appended unique molecular identifiable (UMI) sequences via primers with 20-mer random sequences followed by a universal 5’ overhang during the final PCR amplification in order to uniquely identify mutant DNA molecules during sequencing (Supplementary Figure 6). Similar rates of singleton mutations (~50% total or ~8% singleton mutant molecules per 100bp mutated) were observed within this long SAS library with slight drop-off along the length of the SAS library out to approximately 1kb (Supplementary Figure 7). The rate of incorporation of each nucleotide was also comparable to that observed for the first 200bp of the SAS library (Supplementary Table 1). These data suggest that long SAS libraries can be created without substantial change in mutagenic properties along the length of the product.
Mutagenesis using rtITP enables the rapid construction of comprehensive libraries containing all possible single nucleotide changes within a region of DNA for a fraction of the cost of current methods. While saturation mutagenesis of splice-sites and coding genes can now be performed at their native loci using CRISPR-Cas9 technologies (1), these experiments have thus far been limited due to size restrictions of oligonucleotide arrays. SAS mutagenesis will enable rapid, cost-effective production of homology-directed repair template pools to simultaneously assess the functional impact of a library of mutations within coding and non-coding loci. Overall, integration of SAS mutagenesis with high-throughput functional testing will enable the rapid assessment and fine-scale understanding of the millions of DNA variants present within the human population.
Online Methods
Production of reversibly-terminated deoxyinosine triphosphate
3’-O-NH2 -deoxyadenosine triphosphate (Firebird Biomolecular Sciences, LLC) was enzymatically converted to 3’-O-NH2 -deoxyinosine triphosphate by the sequential addition and heat inactivation of recombinant shrimp alkaline phosphatase (rSAP) (New England Biolabs) to create 3’-O-NH2–deoxyadenosine, deamination with adenosine deaminase (Sigma) to create 3’-O-NH2 –deoxyinosine and a mixture of T4 PNK, pyruvate kinase and myokinase (adenylate kinase) (Sigma) to create 3’-O-NH2 –deoxyinosine triphosphate. This product is then used directly in the SAS mutagenesis protocol. The ability of rtITP molecules to be incorporated into linear amplification products and terminate polymerase extension was testing using a 5’-ROX-labeled primer identical in sequence to the biotinylated primer used to perform SAS mutagenesis on the AmpR gene. Fluorescently labeled linear amplification products were produced using varying concentrations of either rtATP or rtITP (Supplementary Figure 2).
SAS mutagenesis
Linear amplification of target DNA was performed using a biotinylated primer, a 1:1 or 1:4 ratio of dNTPs and rtITP and Firebird Taq 475 (Firebird Biomolecular Sciences, LLC). This polymerase was specifically developed to incorporate 3’-O-NH2 linked nucleotides. The products were then bound to streptavidin beads and washed. Beads were then exposed to 70mM Sodium Nitrite, pH 5.5 to reverse the termination, washed and cycle extended in the presence of template DNA. Upon extension, beads were washed and DNA eluted using 0.1M NaOH followed by neutralization with 1M Tris-HCl. Beads with bound DNA were then used as the DNA template for a PCR to produce final mutagenized products with randomly inserted nucleotides (A, C, G, and T) in place of each inosine. A detailed protocol can be found in Supplementary Information.
Cloning and functional selection of TEM-1 β-lactamase
A dual selection plasmid (plasmid pGH1) was created by cloning the full TEM-1 β-lactamase gene amplified from plasmid pCMV6-XL6 into the plasmid pCR-Blunt-II-TOPO which contains the kanamycin resistance gene. A 217bp segment containing the active site was then amplified from plasmid pGH1 containing both the AmpR and the KanR genes and SAS mutagenesis was performed on it as described above. The inverse of 217bp segment within plasmid pGH1 was then amplified using primers that were the reverse complement of those used to amplify the 217bp fragment. This PCR product was then Gibson assembled with the SAS library derived from the 217bp fragment. This plasmid library was then transformed into XL10-Gold ultracompetent cells and grown in LB to saturation in the presence of 50ug/ml kanamycin. For ampicillin selection, kanamycin outgrowth was diluted 1:1000 in 100ml of selective media containing 200ug/ml ampicillin. Cells were cultured for 2 hours, centrifuged, washed by resuspending in 1ml LB without antibiotic, centrifuged and washed again, then resuspended in 5ml LB media without antibiotic and grown overnight to saturation. Plasmids were purified using Qiagen Mini Plasmid purification kit.
Sequencing and enrichment analysis
Mutated segments were sequenced in both directions with 250bp paired-end reads using an Illumina MiSeq. The consensus sequence of each read-pair was aligned using Novoalign. Mutation counts were obtained from pileups of aligned consensus sequencing reads. Enrichment
scores were determined using the following formula for each observed mutation.
For validation, enrichment scores were compared to those observed previously for mutations in the TEM-1 β-lactamase gene using linear regression of enrichments scores compared to scores obtained previously. Projection of enrichment scores onto the crystal structure of TEM-1 β-lactamase (PBD:1FQG) was performed using Swiss-PBD viewer 4.1.0.
Data Availability
The data that support the findings of this study are available from the corresponding author upon request
Supplementary Material
Acknowledgments
We thank Carlos Cruchaga and John Budde and members of the Gurnett/Dobbs Lab for helpful discussion. This work was supported by a postdoctoral research fellowship (84291-STL) from the Shriners Hospital for Children (G.H.), a US National Institutes of Health (NIH) grant (R01AR067715-01) (C.G and M.D.) and Shriners Hospital for Children research grant (85200-STL) (C.G and M.D.)
Footnotes
We declare that there are no competing interests.
Editorial summary:
The incorporation of reversibly-terminated deoxyinosine triphosphates during linear amplification allows the incorporation of one mutation per molecule and the generation of a systematic allelic series.
Accession Numbers
Protein Database ID for TEM-1 Beta-lactamase: 1FQG
Contributions
G.H., D.A., R.M., M.D. and C.G. designed the study and wrote the manuscript. G.H. and K.M. performed experiments. All authors contributed to and approved the final manuscript.
Competing financial interests
Washington University in St. Louis has filed a provisional patent application on this method, with G.H, D.A., C.G. and M.D. as inventors.
References
- 1.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513(7516):120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kitzman JO, Starita LM, Lo RS, Fields S, Shendure J. Massively parallel single-amino-acid mutagenesis. Nature methods. 2015;12(3):203–206. doi: 10.1038/nmeth.3223. 4 p following 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nature biotechnology. 2012;30(3):265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nature biotechnology. 2009;27(12):1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starita LM, Young DL, Islam M, Kitzman JO, Gullingsrud J, Hause RJ, et al. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics. 2015;200(2):413–422. doi: 10.1534/genetics.115.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nature genetics. 2013;45(9):1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirino PC, Mayer KM, Umeno D. Generating mutant libraries using error-prone PCR. Methods in molecular biology. 2003;231:3–9. doi: 10.1385/1-59259-395-X:3. [DOI] [PubMed] [Google Scholar]
- 8.Copp JN, Hanson-Manful P, Ackerley DF, Patrick WM. Error-prone PCR and effective generation of gene variant libraries for directed evolution. Methods in molecular biology. 2014;1179:3–22. doi: 10.1007/978-1-4939-1053-3_1. [DOI] [PubMed] [Google Scholar]
- 9.McCullum EO, Williams BA, Zhang J, Chaput JC. Random mutagenesis by error-prone PCR. Methods in molecular biology. 2010;634:103–109. doi: 10.1007/978-1-60761-652-8_7. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y, Zhao H, Lv M, Sun G, Yang X, Wang H. A simple error-prone PCR method through dATP reduction. Wei sheng wu xue bao = Acta microbiologica Sinica. 2014;54(1):97–103. [PubMed] [Google Scholar]
- 11.Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM-1 beta-lactamase. Cell. 2015;160(5):882–892. doi: 10.1016/j.cell.2015.01.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request