SUMMARY
Somites form during embryonic development and give rise to unique cell and tissue types, such as skeletal muscles and bones and cartilage of the vertebrae. Using somitogenesis stage human embryos, we performed transcriptomic profiling of human presomitic mesoderm as well as nascent and developed somites. In addition to conserved pathways such as WNT/β-catenin, we also identified BMP and TGFβ signaling as major regulators unique to human somitogenesis. This information enabled us to develop an efficient protocol to derive somite cells in vitro from human pluripotent stem cells (hPSCs). Importantly, the in vitro differentiating cells progressively expressed markers of distinct developmental stages known during in vivo somitogenesis. Furthermore, when subjected to lineage-specific differentiation conditions, the hPSC-derived somite cells were multipotent in generating somite derivatives, including skeletal myocytes, osteocytes and chondrocytes. This work improves our understanding of human somitogenesis and may enhance our ability to treat diseases affecting somite derivatives.
Keywords: Human Pluripotent Stem Cells, Somite, Skeletal Myogenesis, Osteogenesis, Chondrogenesis, Differentiation, Development
Graphical Abstract
INTRODUCTION
Human pluripotent stem cells (hPSCs) hold great promise in understanding human biology, modeling diseases and generating cells and tissues for regenerative medicine. To fully harness their power, it is imperative to design robust and efficient protocols to differentiate hPSCs toward desired lineages, which often involves following cues seen during development of the tissue or organ system of interest (Murry and Keller, 2008). Somites are transient mesodermal structures formed during early embryonic development that are common ancestors to a plethora of cell and tissue types, including trunk and limb skeletal muscles, axial bones and cartilage, subtypes of smooth muscles and endothelial cells, brown fat as well as dermis of the back (Brent and Tabin, 2002). Thus, an effective protocol to derive somite cells in vitro from hPSCs would enable development of a wide range of targeted cell and tissue types that more closely recapitulate the endogenous lineages.
Somitogenesis progresses through a series of developmental stages. During early gastrulation, formation of the primitive streak (PS) initiates, and later on a subpopulation of PS cells give rise to presomitic mesoderm (PSM) alongside the forming anterior-posterior (A–P) axis. As the PSM expands, the anterior part (aPSM) segregates to generate pairs of somites flanking the A-P axis (Benazeraf and Pourquie, 2013). Research in model organisms has shown a decreasing posterior to anterior (P–A) gradient of WNT/β-catenin and FGF signaling as well as periodic activation of NOTCH signaling within the PSM. Accordingly, the clock and wavefront model has been shown to be the essential regulator of somitogenesis from aPSM cells when they reach subthreshold WNT/FGF activity with simultaneous activation of NOTCH signaling (Hubaud and Pourquie, 2014; Saga, 2012). Once the nascent somites form, they rapidly differentiate into sub-compartments, i.e., dermomyotome (DM) and sclerotome (Scl) that later on give rise to differentiated cells and tissues including skeletal muscles and the axial skeleton, respectively (Applebaum and Kalcheim, 2015; Brent and Tabin, 2002).
Extensive efforts have been made in the past few years to induce paraxial mesoderm (presomitic and somitic mesoderm) in vitro from hPSCs and derive downstream lineages (Borchin et al., 2013; Shelton et al., 2014; Umeda et al., 2012; Xu et al., 2013). A common theme of these protocols is activating WNT/β-catenin signaling, which successfully generates PSM cells. However, the transition from PSM to a somite stage in human in these reports is not well defined. Chal et al. recently performed a thorough microarray analysis on different regions of mouse paraxial mesoderm ranging from the most posterior tail bud to the newly formed somites (Chal et al., 2015). Based on their transcriptomic profiling, the authors developed a protocol to derive somite cells from mouse embryonic stem cells (mESCs) progressing through successive developmental stages. This protocol was adapted to hPSCs, which with further differentiation successfully generated skeletal muscles. Nevertheless, the progression of in vivo human or in vitro hPSC paraxial mesoderm development has not been characterized, and efficient differentiation into multiple lineages derived from hPSC-somites has not been shown.
Here, we carried out transcriptomic profiling of human PSM and somites obtained from early human embryos at somitogenesis stages (Carnegie stage (CS) 13–14; embryonic age 4.5–5 weeks of gestation). RNA sequencing (RNA-seq) analysis identified differentially regulated pathways in nascent somites compared to PSM, including the retinoic acid (RA) and NOTCH signaling (upregulated in nascent somites) as well as WNT, BMP and TGFβ signaling (downregulated in nascent somites). From this, we demonstrated that during hPSC differentiation, inhibition of BMP signaling following WNT/β-catenin activation robustly specifies pPSM cells toward the aPSM and somite fate. Moreover, we found that inhibition of TGFβ signaling, which has not been implicated in somitogenesis in model organisms, further enhanced hPSC somite specification efficiency. Additional RNA-seq analysis further identified upregulated WNT signaling in matured compared to nascent somites, thus enabling us to control the divergence of somite cells to distinct sub-compartment fates of DM and Scl. When subjected to further lineage-specific differentiation conditions, our in vitro hPSC-somite cells gave rise to three of the major derivatives of in vivo somites, i.e., skeletal myocytes, osteocytes and chondrocytes.
RESULTS
Transcriptomic profiling of developing paraxial mesoderm from somitogenesis stage human embryos
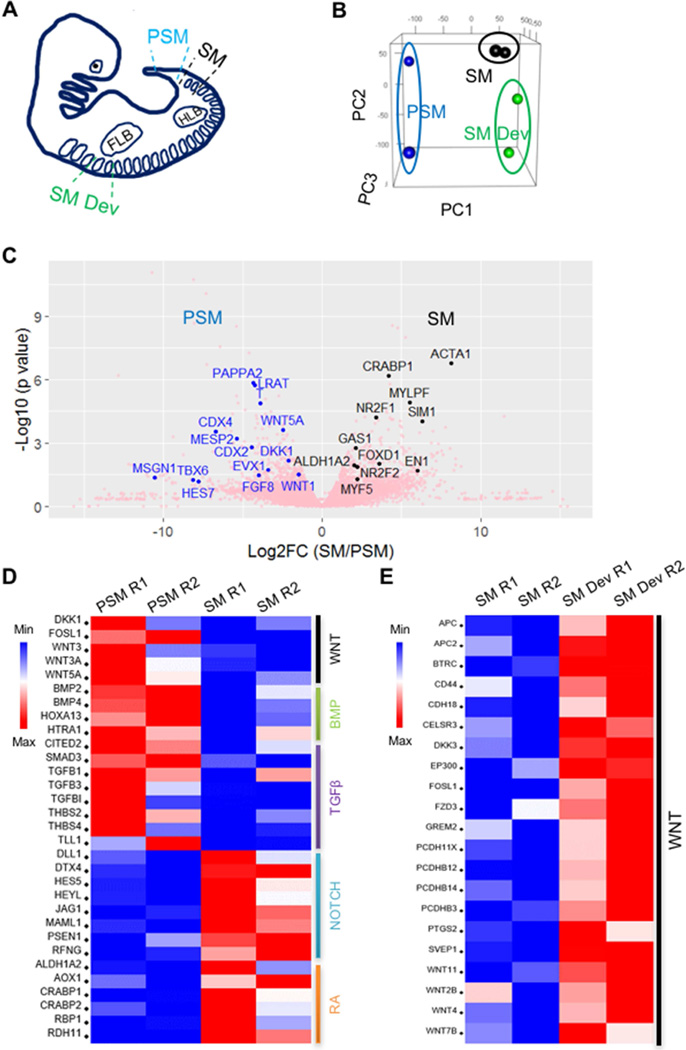
In order to improve our understanding of human somite development and to discover pathway(s) that can be modulated to efficiently drive human somite specification in vitro from hPSCs, we performed transcriptomic profiling of PSM, nascent somites (SM) as well as matured somites (SM Dev; more developed somites at the forelimb bud level) from CS 13–14 (embryonic age 4.5–5 weeks of gestation) human embryos (Table 1) undergoing somitogenesis (Figure 1A). Hierarchical clustering (Figure S1A) and principal component analysis (PCA) (Figure 1B) show that the PSM, SM and SM Dev replicates cluster with each other and form three distinct groups. Moreover, the human PSM or SM tissues are enriched in the respective marker genes well described in model organisms (Figure 1C).
Table 1.
The ID numbers, Carnegie stages, embryonic ages and tissue types of each human embryo used in this study.
| Embryo ID | Carnegie Stage | Embryonic Age | Tissues Used for RNA-seq |
|---|---|---|---|
| ID306 | CS 13–14 | 4.5–5 week | SM R1 & SM Dev R1 |
| ID332 | CS 14 | 5 week | PSM R1 & SM R2 |
| ID338 | CS 13 | 4.5 week | PSM R2 & SM Dev R2 |
PSM: presomitic mesoderm; SM: nascent somites; SM Dev: matured somites; R1 and R2: replicate 1 and 2 for RNA-seq samples.
Figure 1. Transcriptomic profiling of somitogenesis stage human embryos identifies differentially regulated pathways among PSM, SM and SM Dev.
(A) Illustration of human PSM, SM and SM Dev dissection. FLB and HLB: fore- and hind-limb bud. (B) PCA of PSM, SM and SM Dev replicates. (C) Volcano plot of PSM and SM gene expression profiles with selected PSM and SM markers highlighted in blue and black, respectively. (D) Heatmap showing RNA-seq expression of selected components of the differentially regulated signaling pathways between PSM and SM that were evaluated in this study. (E) Heatmap showing RNA-seq expression of selected components of the WNT signaling pathway that are differentially regulated between SM and SM Dev. See also Figure S1 and Tables S1–S3.
Next, we performed differential gene expression analysis in SM and PSM and found that there are 322 genes upregulated and 290 genes downregulated in SM compared to PSM (p < 0.05 and fold change > 2) (Table S1). Functional clustering of these differentially expressed genes (DEGs) reveals enrichment of certain biological processes and signaling pathways involved in anterior/posterior pattern formation, embryonic morphogenesis, cell motility, cell-matrix adhesion and cellular metabolism (Figure S1B and S1C). This reflects the dynamic nature of cell and tissue remodeling during human paraxial mesoderm development. NOTCH signaling and RA metabolism were found to be enriched amongst the DEGs upregulated in SM (downregulated in PSM) (Figures 1D and S1B), while WNT, BMP and TGFβ signaling were implicated in DEGs downregulated in SM (upregulated in PSM) (Figures 1D and S1C). Similar DEG (Table S2) and functional clustering analyses were also performed comparing SM Dev and SM. As expected, processes involved in muscle and skeletal system morphogenesis were found to be enriched in SM Dev, reflecting their more advanced developmental stage and lineage commitment compared to SM (Figure S1D). Interestingly, we also found that WNT signaling is upregulated in SM Dev vs. SM (Figures 1E and S1D).
To compare nascent somite formation in human and mouse, we utilized the recently published mouse paraxial mesoderm microarray gene expression dataset by Chal et al. as mentioned above (GEO database series#: GSE39613) (Chal et al., 2015). This dataset comprises 7 micro-dissected pieces of the posterior region from E8.0–9.5 mouse embryos representing the progression of somitogenesis from tail bud (Part 1) to the most newly formed somite pairs (Part 7). To mimic our human PSM and SM dissections, we grouped the mouse paraxial mesoderm Part 1–6 as PSM and assigned Part 7 as SM. DEG (Table S3). Functional clustering analyses revealed some conservation between the two species, such as the common enrichment of RA metabolism in SM and WNT signaling in PSM (H.X. and A.D.P., unpublished data). However, contrary to the downregulation of TGFβ signaling in human SM, this signaling pathway was found to be upregulated in mouse SM (Figure S1E and S1F). Overall, our transcriptomic analysis reveals candidate signaling pathways critical for human somite development, and indicates potential molecular differences between human and mouse somitogenesis.
BMP inhibition followed by WNT/β-catenin activation specifies hPSCs to a somite fate
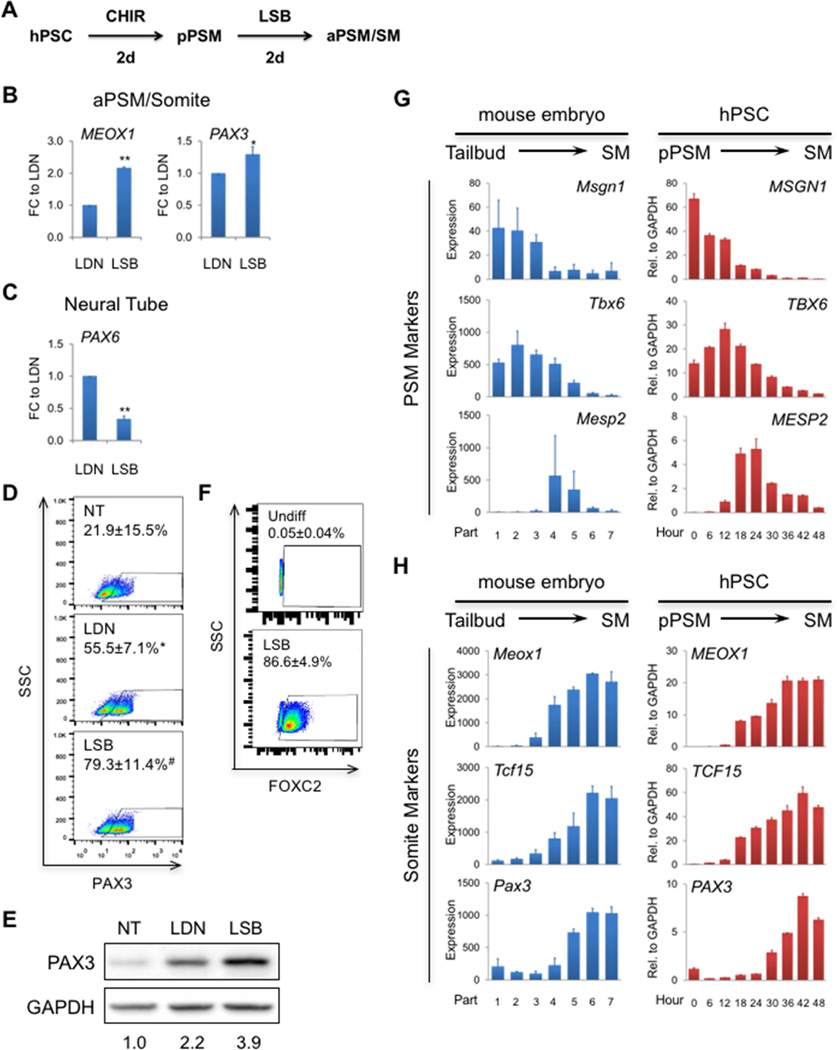
Our human profiling data reveal enriched WNT signaling in PSM, consistent with its pivotal role in paraxial mesoderm development in model organisms (Arnold and Robertson, 2009; Hubaud and Pourquie, 2014; Tam and Loebel, 2007). Accordingly, we initiated differentiation of H9 hESCs with the small molecule GSK3β inhibitor and WNT/β-catenin activator, CHIR99021 (CHIR) (Figure 2A). Compared to non-treatment (NT, treated with vehicle), CHIR significantly upregulated the PS markers T (also known as Brachyury) and MIXL1 and induced peak expression of these markers at day 1 of differentiation (Figure 2B). This was followed by the peak expression of the pPSM markers MSGN1 and TBX6 at day 2 (Figure 2C). Immunofluorescent (IF) staining showed that more than 80% of cells were TBX6+ at day 2 (Figure 2D), which correlated well with flow cytometric results for the pPSM populations defined by PDGFRα+KDR− and CDX2+ (Figures 2E and 2F).
Figure 2. Transient activation of canonical WNT/β-catenin signaling progressively specifies hPSCs to the PS and pPSM fate.
(A) Schematic of differentiation procedure in (B–F). Expression of the PS (B) and pPSM (C) markers by qRT-PCR shown as average ± SEM (n = 5). *, p<0.05, **, p<0.01, and ***, p<0.001 to the corresponding vehicle-treated (no treatment or NT) controls. Data were shown as relative expression to GAPDH levels. (D) Representative IF staining of TBX6 (top) and quantification of TBX6+ cells (bottom) shown as average ± SEM (n = 3). **, p<0.01 to Undiff (undifferentiated/day 0 samples). Scale bar: 100 µm. (E) Representative flow plots of PDGFRα and KDR (bottom) and quantification of PDGFRα+KDR− cell population (top) shown as average ± SEM (n = 3). **, p<0.01 to Undiff. (F) Representative flow plots of CDX2 and quantification of CDX2+ cell population shown as average ± SEM (n = 4). See also Figures S2 and S5, and Table S4.
We aimed to increase the efficiency of CHIR induction of pPSM cells by manipulating additional signaling pathways, including activating FGF (bFGF/FGF2 or FGF8), Activin (Activin A) or cAMP (Forskolin) signaling as well as inhibiting BMP (LDN-193189/LDN or Noggin), TGFβ (SB431542/SB) or PI3K (LY294002/LY) pathway. As assayed by qRT-PCR, none of these conditions we have tested consistently increased the expression of pPSM markers induced by CHIR, except for the combination of FGF2 and LY (Cheung et al., 2012) (Table S4, and Figures S2A and S2B). However, this combination did not increase the frequency of the TBX6+ nor PDGFRα+KDR− population (Figure S2C and S2D). Collectively, we confirmed that canonical WNT/β-catenin activation (CHIR) alone rapidly and potently induces the sequential PS and pPSM fate specification.
The BMP signaling pathway was identified to be downregulated in SM compared to PSM from our human profiling analysis. Therefore, we hypothesized that inhibition of this pathway would facilitate the specification of a somite fate of differentiating hPSCs in vitro. Indeed, the small molecule BMP inhibitor LDN significantly upregulated the aPSM/somite markers MEOX1 and TCF15 in CHIR-induced pPSM cells after 1 day of treatment (Figures 3A and 3B). The somite marker PAX3 was significantly increased after 2 days of LDN treatment (Figure 3B). Prolonged treatment of LDN did not further increase the expression of these markers. However, it gradually increased the expression of the Scl marker PAX1 (Figure 3C), indicating progression of some of the induced somite cells to a Scl state with prolonged culture. Nascent somites ubiquitously express PAX3, which is also highly expressed in the neighboring neural tube and neural crest tissues (Goulding et al., 1991). We found that the neural crest markers TFAP2A and SOX10 were not upregulated in LDN-treated cells in contrast to the NT control cells, whereas the neural tube marker PAX6 was slightly increased at day 3 and 4 and more robustly upregulated with prolonged incubation (Figure 3C). Notably, these observations were recapitulated with recombinant Noggin proteins (H.X. and A.D.P., unpublished data). In correlation with the qRT-PCR data, IF staining showed that TCF15+ cells started to appear at day 3 and PAX3+ cells could only be seen at day 4 (Figure 3D). Without LDN, there were no TCF15+ cells and only very few PAX3+ cells (Figure 3D). These findings indicate that inhibiting BMP signaling by LDN sequentially drives the CHIR-induced pPSM cells toward the aPSM and somite fate.
Figure 3. BMP inhibition drives pPSM cells to a somite fate.
(A) Schematic of differentiation procedure in (B–D). Expression of the aPSM/somite (B) as well as Scl, neural crest and neural tube (C) markers by qRT-PCR shown as average ± SEM (n = 6). *, p<0.05, **, p<0.01, and ***, p<0.001 to the corresponding NT controls. Data were shown as relative expression to GAPDH levels. (D) Representative IF staining of TCF15 and PAX3 of cells treated by LDN (left) or NT (right) (n = 5). Scale bar: 100 µm. See also Figure S3.
In addition to BMP signaling, we also found RA metabolism (upregulated in SM), NOTCH signaling (upregulated in SM) and TGFβ signaling (downregulated in SM) being differentially regulated in human PSM and SM (Figures 1D, and S1B and S1C). Thus, we activated the former two pathways by RA and DLL-1, respectively, and inhibited the latter one with SB (Figure S3A). However, none of these conditions alone consistently increased multiple aPSM and somite marker expression compared to the NT controls (Figure S3B–S3D). As low levels of FGF signaling and its downstream ERK activity have been shown in vivo to act as the “wavefront” to control somitogenesis (Hubaud and Pourquie, 2014; Saga, 2012), we also examined the effects of inhibition of these pathways in our in vitro system. The small molecules PD173074 and PD0325901 were used to block the FGF receptors and MEK-ERK signaling, respectively. Similar to the above results, none of the inhibitors enhanced the expression of aPSM and somite markers (Figure S3E).
Blocking TGFβ signaling enhances the efficiency of somite fate specification induced by BMP inhibition
Next, we tested whether manipulating the above mentioned pathways would augment hPSC somite specification induced by BMP inhibition. We found that the TGFβ receptor inhibitor SB significantly increased the expression of MEOX1 and PAX3 induced by LDN (Figures 4A and 4B). Interestingly, SB significantly decreased the expression of PAX6 compared to LDN alone (Figure 4C). To better quantify the effects of SB on the percentage of PAX3+ cells, we performed intracellular flow cytometric analysis. Under NT conditions, there were ∼20% PAX3+ cells, whereas treatment with LDN increased the percentage to ∼50%, and the addition of SB (LSB) further augmented it to ∼80% (Figure 4D). This finding is further supported by Western blot analysis of PAX3 protein expression in cells treated with NT, LDN or LSB (Figure 4E). Flow cytometric analysis of another somite marker FOXC2 also showed efficient generation of FOXC2+ population under LSB treatment (∼90%; Figure 4F).
Figure 4. Inhibition of TGFβ signaling synergizes with BMP inhibition to increase somite specification efficiency.
(A) Schematic of differentiation procedure. LSB: LDN+SB. Expression of the aPSM/somite (B) and neural tube (C) markers by qRT-PCR shown as average ± SEM (n = 5). GAPDH was used as the housekeeping gene and fold changes (FC) were compared to LDN only-treated samples (which were set to 1.0). *, p<0.05 and **, p<0.01 to LDN only. (D) Representative flow plots of PAX3 and quantification of PAX3+ population shown as average ± SEM (n = 7). *, p<0.05 to NT and #, p<0.05 to LDN only. (E) Representative Western blots of PAX3 protein expression (n = 2). Numbers under each treatment condition indicate intensity of PAX3 normalized to GAPDH. (F) Representative flow charts of FOXC2 and quantification of FOXC2+ population shown as average ± SEM (n = 3). (G and H) Expression of select PSM (G) and somite (H) markers from dissected mouse tail paraxial mesoderm and differentiating H9 cells. Mouse microarray data were obtained from GEO database (series#: GSE39613) and shown as average ± SEM (n = 3 biological replicates). Increasing part number represents posterior (tail bud; Part 1) to anterior (nascent somites; Part 7). Human data were obtained from cells pre-treated with 2 days of CHIR (labeled as Hour 0) followed by another 2 days of LSB (Hour 6–48) and analyzed by qRT-PCR shown as average ± SD of technical triplicates from a representative experiment (n = 3). Mouse data were absolute expression value from microarray, and human data were relative expression to GAPDH levels. See also Figures S4 and S5.
To examine the potential involvement of the canonical vs. non-canonical TGFβ signaling (Derynck and Zhang, 2003) in SB-enhanced somite specification, we performed Western blot analysis of these signaling pathways. To minimize the potential influence on pathway activities due to cell fate change, cells were treated by LDN or LSB for only 8 hrs when no overt cell fate conversion could be observed. At this time point, the addition of SB completely abolished the canonical TGFβ target pSMAD2/3 compared to LDN alone, whereas it did not change the level of pERK1/2, a non-canonical TGFβ target (Figure S4A). We have also examined other targets of non-canonical TGFβ signaling, i.e., pJNK, p-p38 and pAKT, but their levels were not detectable under basal conditions with LDN alone (H.X. and A.D.P., unpublished data). Furthermore, inhibition of another non-canonical TGFβ signaling target, ROCK, with Y-27632 during LDN treatment did not increase somite marker expression compared to LDN alone (H.X. and A.D.P., unpublished data). Thus, these data suggest that canonical, rather than non-canonical, TGFβ signaling pathway mediates SB-induced improvement on hPSC somite specification.
In addition to blocking TGFβ pathway, we also examined whether activating RA or NOTCH signaling, or inhibiting FGF or MEK/ERK pathway could improve LDN-induced somite specification. As shown in Figures S4C–S4F, none of the small molecule or growth factor modulators of the above signaling pathways consistently enhance aPSM and somite marker expression or the percentage of PAX3+ cell population.
Subsequently, we asked whether the dynamics of our in vitro somite derivation reflects that of in vivo somitogenesis. Due to the scarcity of somitogenesis stage human embryos, we turned to the mouse dataset by Chal et al. as a resource for detailed dynamics of in vivo somite development. Using this dataset, we compared the expression levels of selected common markers for PSM and somite in the mouse tissues to our in vitro differentiating cells during LSB-mediated somite specification. In mouse tissues, the pPSM markers Msgn1 and Tbx6 showed a high to low expression gradient from in vivo tailbud to nascent somites, while the somite markers Meox1, Tcf15 and Pax3 showed an opposite expression pattern (Figures 4G and 4H, left panels). Strikingly, this expression dynamic was well recapitulated during the LSB-mediated hPSC differentiation from in vitro pPSM (0 hr) to somite (48 hr) (Figures 4G and 4H, right panels). Notably, expression of the aPSM marker MESP2 during hPSC differentiation was only transiently upregulated at the intermediate time points around 24 hours post LSB incubation. This was well correlated with its narrow expression window in mouse Part 4 and 5 tissues, which reflects its strict in vivo expression pattern only limited to a sub-region of aPSM just posterior to the forming segmentation border (Hubaud and Pourquie, 2014; Saga, 2012).
Overall, our data demonstrate that a short 4-day protocol comprising 2 days of WNT/β-catenin activation (CHIR) followed by 2 days of combinatorial BMP (LDN) and TGFβ (SB) inhibition, robustly induces progressive PSM and somite marker expression from H9 hESCs, which correlates nicely with the dynamics of in vivo mouse somitogenesis. Without cell sorting or enrichment, this protocol generates a relatively pure population with ∼80% and ∼90% of the cells positive for the nascent somite markers PAX3 and FOXC2. To our best knowledge, this is the highest percentage of somite cell population obtained from any hPSC paraxial mesoderm differentiation scheme. Moreover, these findings are repeatable in an in-house derived 1002 iPSC line (Young et al., 2016) with increased initial CHIR concentration to 10 µM (Figure S5 and H.X. and A.D.P., unpublished data), suggesting slight optimizations may be required per hPSC line (Kattman et al., 2011).
In vitro hPSC-somite cells are functional in generating the major in vivo somite derivatives – myogenic, osteogenic and chondrogenic cells
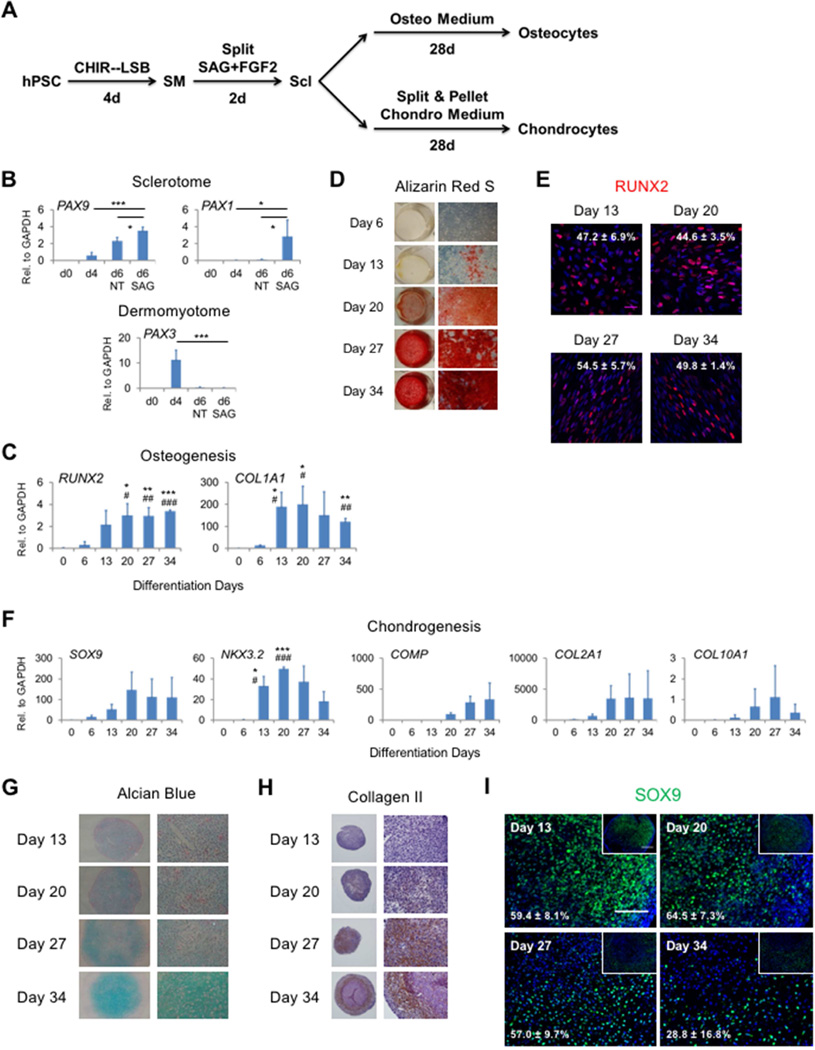
To test the functionality of hPSC-derived somite cells, we subjected these cells to further differentiation to assess their capacity to generate the three major somite-derived cell types, i.e., myocytes, osteocytes and chondrocytes.
As myogenic cells are derived from the DM sub-compartment of developed somites, we first examined whether we can efficiently guide hPSC-derived nascent somite cells to a DM fate. Our human embryo profiling analysis revealed upregulated WNT signaling in SM Dev vs. SM. Thus, we tested the effect of WNT activation using CHIR on the fate decision of somite cells. As shown in Figures 5A and 5B, 2 days of CHIR treatment after somite specification boosted the expression of the DM markers EN1 and SNAI1 while repressing that of the Scl markers PAX1 and PAX9, when compared to NT samples. Subsequently, cells were switched to a KSR/HGF/IGF1-based medium (Chal et al., 2015) for another 14–21 days to facilitate later stage myogenic differentiation (Figure 5A). As shown in Figure 5C, expression of myogenic markers was gradually increased toward day 20 of myogenic differentiation. At day 27, large areas of myosin heavy chain (MyHC) positive cells emerged throughout the culture, and the majority also expressed the sarcomeric protein Titin (Figures 5D and S6). The culture also contained a high proportion of PAX7+, MYOD+ and/or MYOG+ myogenic cells (Figures 5D and S6).
Figure 5. hPSC-derived somite cells can undergo skeletal myogenesis.
(A) Schematic of differentiation procedure in (B–D). (B) Expression of the DM and Scl markers by qRT-PCR shown as average ± SEM (n = 10). *, p<0.05, **, p<0.01 and ***, p<0.001. Data were shown as relative expression to GAPDH levels. (C) Expression of the myogenic markers by qRT-PCR shown as average ± SEM (n = 3). *, p<0.05 to d0 and #, p<0.05 to d6. Data were shown as relative expression to GAPDH levels. (D) Representative IF staining of the indicated myogenic markers at day 27 of differentiation (n = 3). DAPI is shown in blue in merged images. Scale bar: 200 µm. (E) Schematic of split, expansion (SkGM2+FGF2) and fusion (N2) of cells pre-differentiated as in (A). (F) Left: Representative IF staining of the indicated myogenic markers at the end of the culture (n = 2). A portion of PAX7+ cells (arrowhead) lie next to the MyHC+ myotubes containing multiple MYOD+ nuclei (arrow). Scale bar: 100 µm. Right: Quantification of nuclei within the MyHC+ myotubes, as well as those that were outside myotubes and either PAX7 and MYOD single or double positive. Total myogenic percentage was calculated as the sum of all the above populations. See also Figure S6.
To facilitate the quantification of myogenic cells, we dissociated the myogenic culture at day 29, replated and expanded the cells in SkGM2 medium containing FGF2 as monolayer culture (Figure 5E). Following fusion induction by N2 medium (Shelton et al., 2014), myogenic cells formed multinucleated myotubes with PAX7+ cells associated with some of these myotubes (Figure 5F, left). We quantified the cell nuclei within MyHC+ myocytes/myotubes (58.7±9.2%), as well as cells outside MyHC+ area that were PAX7+MYOD− (6.5±3.7%), PAX7−MYOD+ (9.1±3.3%) or PAX7+MYOD+ (4.9±0.1%). Overall, nearly 80% (79.2±9.7%; sum of the above 4 populations) of the cells were myogenic at the end of the differentiation protocol shown at Day 44 (Figure 5F, right).
Next, we assessed the ability of the hPSC-derived somite cells to undergo osteogenesis and chondrogenesis. The somite cells at day 4 of differentiation were split to mimic the less compact mesenchymal structure of Scl, the common precursor of axial bones and cartilage. We also activated the SHH signaling in these cells with a small molecule Smoothened agonist (SAG) (Figure 6A), as Shh from the ventral notochord and floor plate is a critical morphogen in specifying Scl in vivo (Brent and Tabin, 2002; Chal and Pourquié, 2009). Expression of the Scl markers PAX1 and PAX9 dramatically increased within as short as 2 days (Figure 6B). This was accompanied by a rapid decrease of the expression of the DM marker PAX3 (Figure 6B).
Figure 6. hPSC-derived somite cells are osteogenic and chondrogenic.
(A) Schematic of differentiation procedure in (B–I). (B) Expression of the Scl and DM markers by qRT-PCR shown as average ± SEM (n = 9). *, p<0.05 and ***, p<0.001. Data were shown as relative expression to GAPDH levels. (C) Expression of the osteogenic markers by qRT-PCR shown as average ± SEM (n = 4). *, p<0.05, **, p<0.01 and ***, p<0.001 compared to d0; #, p<0.05, ##, p<0.01 and ###, p<0.001 compared to d6. Data were shown as relative expression to GAPDH levels. (D) Alizarin Red S staining showing representative images of whole wells (left) and higher magnification (5X objective; right) (n = 6). (E) Representative confocal images of RUNX2 staining and quantification of RUNX2+ population shown as average ± SEM (n = 2). RUNX2 is shown in red and DAPI in blue. Scale bar: 50 µm. (F) Expression of the chondrogenic markers by qRT-PCR shown as average ± SEM (n = 3). *, p<0.05 and ***, p<0.001 compared to d0; #, p<0.05 and ###, p<0.001 compared to d6. Data were shown as relative expression to GAPDH levels. (G) Alcian Blue staining showing representative images of whole pellets (10X objective; left) and central areas under higher magnification (32X objective; right) (n = 3). (H) IHC of Collagen II showing representative images of whole pellets (10X objective; left) and lower-left areas under higher magnification (40X objective; right) (n = 2). (I) Representative images of chondrogenic pellets sectioned and stained with SOX9 and quantification of SOX9+ population shown as average ± SEM (n = 2; total of 4 pellets each time point). Insets showing lower magnification images capturing larger areas of the pellets. SOX9 is shown in green and DAPI in blue. Scale bar: 100 µm (insets 200 µm).
To differentiate the hPSC-derived Scl cells to an osteogenic lineage, we cultured them for another 28 days in StemPro osteogenic medium (Figure 6A). Expression of the osteogenic markers were upregulated (Figure 6C), and the cells also displayed progressively increased Alizarin Red S staining (Figure 6D). Moreover, RUNX2+ cells were observed throughout the osteogenic differentiation process and maintained relatively constant between 45–55% (Figure 6E). For chondrogenic differentiation, we subjected Scl cells to a pellet culture (Dar et al., 2012) in StemPro chondrogenic medium for 4 weeks (Figure 6A). During the pellet culture, expression of chondrogenic markers were upregulated (Figure 6F), along with gradually increased Alcian Blue (Figure 6G) and Collagen II staining (Figure 6H). The cells were ∼60% SOX9+ from day 13–27, and the percentage decreased to ∼30% at day 34 of differentiation. In summary, our hPSC-derived somite cells are multipotent in generating myogenic, osteogenic and chondrogenic cells, which are among the major derivatives of functional somites in vivo.
DISCUSSION
Tremendous efforts have been made to derive paraxial mesoderm cells from hPSCs in the past few years (Borchin et al., 2013; Chal et al., 2015; Mendjan et al., 2014; Shelton et al., 2014; Xu et al., 2013). However, the efficiency of paraxial mesoderm specification is generally suboptimal, resulting in highly heterogeneous populations or requiring sorting and purification of the mesoderm progenitors to perform downstream lineage differentiation (Sakurai et al., 2012; Umeda et al., 2012). To gain more insight into the regulatory mechanisms of human somitogenesis in vivo and increase hPSC somite specification and downstream lineage differentiation efficiency in vitro, we performed transcriptomic profiling of PSM, SM as well as SM Dev from somitogenesis stage human embryos (CS 13–14; embryonic age 4.5–5 weeks of gestation) (Figures 1 and S1).
Our RNA-seq analysis identified downregulated BMP signaling in SM compared to PSM. Based on this information, we inhibited the BMP signaling by LDN which rapidly and efficiently drove CHIR-induced pPSM cells toward the aPSM and somite fate (Figure 3) compared to published protocols that used CHIR followed by spontaneous differentiation (Borchin et al., 2013; Shelton et al., 2014). Consistent with the ectopic somite formation induced by exogenous Noggin in model organisms (Dias et al., 2014; Tonegawa and Takahashi, 1998), our findings that LDN dramatically induced aPSM/somite marker expression argue for a proactive role of BMP inhibition in somite fate specification rather than merely blocking the formation of the lateral plate mesoderm (LPM) (Umeda et al., 2012). This notion is further supported by the lack of LPM marker (KDR) upregulation (Figure 2E and H.X. and A.D.P., unpublished data), and the inability of LDN or Noggin to augment CHIR-induced pPSM specification (Table S4). Notably, our human paraxial mesoderm profiling also revealed a decrease of TGFβ signaling in SM vs. PSM. Whereas the role of the TGFβ signaling in somite formation is unclear in model organisms, inhibition of this pathway by SB significantly enhanced LDN-induced hPSC-somite specification, suggesting it may be a human specific feature of somite specification (Figure 4). In fact, similar transcriptomic analysis of mouse SM and PSM revealed increased TGFβ signaling in mouse SM (Figures S1E and S1F), which is the opposite to human, demonstrating the importance and uniqueness of our human specific somitogenesis profiling information in guiding optimized hPSC somite differentiation.
We did find that RA and NOTCH signaling were upregulated in SM over PSM, which is consistent with findings in model organisms (Hubaud and Pourquie, 2014; Saga, 2012). However, functionally we did not observe increased somite specification of hPSCs when these pathways were activated (Figures S3 and S4). Similarly, no increase of hPSC somite derivation was observed when FGF or MEK/ERK signaling were blocked to mimic their signaling gradients in model organism somitogenesis. These findings further indicate potential uniqueness in human paraxial mesoderm development and strengthen the importance of studying early human embryos to understand human somitogenesis and guide hPSC somite differentiation.
Compared to the protocol reported by Chal et al., which was devised based on mouse somitogenesis profiling (Chal et al., 2015), our somite specification protocol incorporates TGFβ inhibition based on our human somitogenesis analysis. Our protocol also implements transient instead of continuous WNT activation based on the detailed time course analysis showing decreased pPSM specification efficiency after longer WNT signaling stimulation (Figure 2). These optimizations generated a highly pure somite cell population with 80–90% of cells expressing somite markers within as short as 4 days (Figures 4D and 4F).
One of the drawbacks of the current work is the low replicate numbers limited by the rare week 4.5–5 human embryos available for study. Additionally, despite extensive efforts, we could not completely exclude the contaminating surrounding tissues during paraxial mesoderm dissections, which could dilute mild differences between sample groups. Another limitation results from the age of the embryos, which are not likely to be exactly the same due to the nature of the sample acquisition procedures. This sample age variation could contribute to variances between biological replicates, such as the relatively high variability on PC2 clustering of the PSM samples (Figure 1B). It also prevented us from performing more detailed sub-regional profiling as conducted in model organisms where ample embryos with synchronized ages could readily be obtained (Chal et al., 2015; Ozbudak et al., 2010). Thus, an alternative to the bulk RNA-seq approach would be to use the evolving single cell RNA-seq platforms and analysis tools (Grun and van Oudenaarden, 2015) to profile individual cells and unbiasedly identify distinct populations within the developing human paraxial mesoderm.
In addition to molecular characterizations, we also demonstrated the functional capacity of our hPSC-derived somite cells in giving rise to major somite derivatives including skeletal myocytes, osteocytes and chondrocytes. Guided by our discovery of upregulated WNT signaling in SM Dev vs. SM, we found that activating WNT in our hPSC-derived somite cells skewed their fate toward DM over Scl (Figure 5B). Combined with our efficient somite derivation protocol, it enabled us to achieve fairly robust hPSC skeletal muscle differentiation within a relatively short time frame, which does not require any cell purification or enrichment (Figures 5 and S6). Nevertheless, our proof-of-concept myogenesis protocol still generates heterogeneous cultures that contain myogenic cells at different stages as well as non-myogenic populations, a common feature of the currently available protocols (Rinaldi and Perlingeiro, 2014). Future work is required to achieve purer and more synchronized myogenic populations during hPSC differentiation or to determine if this heterogeneity is required for myogenic specification. A critical step to realize this goal is to improve our understanding and optimization of the lineage progression of nascent somite cells to DM cells and subsequently DM cells toward myogenic commitment and maturation of myogenic progenitor cells through different stages of embryonic and fetal development. In this regard, bulk RNA-seq using more pure progenitor populations or unbiased single cell RNA-seq of progenitor cells arising from developing human embryos should help address this challenge.
Contrary to skeletal muscles, which mostly originate from somites (Tajbakhsh and Cossu, 1997), bones and cartilage at different anatomical sites derive from distinct precursors (Chal and Pourquié, 2009). Thus, it is conceivable that it might be necessary to obtain bones and cartilage of different ontogeny to match the tissues of interest for disease modeling and cell-based therapy. Importantly, hPSC osteogenic and chondrogenic differentiation through a common somite/Scl stage has not been reported so far. Here, we show efficient specification toward both osteogenic and chondrogenic lineages from our hPSC-derived somite cells (Figure 6). Notably, robust upregulation of Scl markers along with downregulation of DM marker were found to precede osteogenesis and chondrogenesis. Together with the skeletal myogenic ability, our data strongly support the somite origin of the myogenic, osteogenic and chondrogenic cells obtained from our hPSC directed differentiation protocol.
One of the major roadblocks preventing the fulfillment of hPSCs’ tremendous potential lies in the lack of sufficient knowledge of early human development, and thus inefficient specification toward targeted lineages. Although studies on model organisms have provided great insights into potentially similar human developmental events, significant differences do exist (Risebro et al., 2015; van den Berg et al., 2015). This might at least partially explain the general difficulty in hPSC differentiation to somite-derived cell and tissue types. Here, we performed transcriptomic profiling of human paraxial mesoderm and identified differentially regulated pathways unique to human somitogenesis. This enabled us to develop an efficient protocol to differentiate hPSCs to functional somite cells, which could serve as a common starting population to obtain various cell and tissue types originated from somites. These cells and tissues could be beneficial for disease modeling, drug development or regenerative applications for conditions affecting somite derivatives, such as muscular dystrophies, spinal osteoporosis and fractures, as well as degenerative disc disease.
EXPERIMENTAL PROCEDURES
Human pluripotent stem cell differentiation
Somite differentiation
On day −1, hPSC colonies were dissociated into single cells with TrypLE Express (Thermo Fisher Scientific) and seeded on Matrigel-coated tissue culture plates at 25,000 cells/cm2 in mTeSR1 medium containing 10 µM of Y-27632 (Tocris). Somite differentiation was initiated the next day (day 0) when medium was switched to basal differentiation medium (BDM; DMEM/F12, 1% ITS-G and 0.5% Penicillin-Streptomycin; all from Thermo Fisher Scientific) supplemented with 3 µM of CHIR99021 (CHIR; Tocris) for 2 days. On day 2, cells were switched to BDM supplemented with 200 nM of LDN193189 (LDN; Stemgent) and 10 µM of SB431542 (SB; Tocris) for another 2 days. Cells were fed with fresh medium every day.
Myogenic differentiation
Cells were cultured under somite differentiation conditions for the first 4 days as described above. On day 4, culture medium was changed to BDM supplemented with 3 µM of CHIR and 20 ng/ml of FGF2 (bFGF; Thermo Fisher Scientific) for 2 days (FGF2 was included at this stage to support the survival of the dense culture). On day 6, medium was switched to a KSR/HGF/IGF1-based differentiation medium (DMEM, 0.5% Penicillin-Streptomycin and 15% KSR (Knockout Serum Replacement) (all from Thermo Fisher Scientific), as well as 10 ng/ml of HGF (PeproTech) and 2 ng/m of IGF1 (Sigma Aldrich)) for 14–21 days. Cells were fed with fresh medium every day until day 6 and every other day thereafter. At day 29, some cells were dissociated by Collagenase IV (2 mg/ml; Thermo Fisher Scientific) for ∼5 min followed by TrypLE Express for another ∼5 min. Cell suspension was filtered sequentially through 100 µm and 40 µm cell strainers to exclude cell aggregates and generate single cell suspension. Filtered cells were resuspended in SkGM2 medium (Lonza) supplemented with 20 ng/ml FGF2, and replated at 15,000–20,000 cells/cm2 onto Matrigel-coated tissue culture plates. Cells were cultured for 7–10 days until reach >70% confluency and then medium was switched to N2 medium (BDM containing 1% N2 supplement (Thermo Fisher Scientific)) (Shelton et al., 2014) to induce fusion for 5 days.
Osteogenic differentiation
Cells were cultured under somite differentiation conditions for the first 4 days as described above. On day 4, cells were split into single cells with TrypLE Express, seeded onto Matrigel-coated plates at 35,000 cells/cm2 in BDM containing 20 ng/ml of FGF2 and 300 nM of Smoothened Agonist (Santa Cruz Biotechnology), and cultured for 2 days. On day 6, cells were switched to StemPro Osteogenesis Differentiation medium (Thermo Fisher Scientific) and cultured for another 28 days. Cells were fed with fresh medium every day until day 6 and every other day thereafter.
Chondrogenic differentiation
Cells were cultured the same as osteogenic differentiation for the first 6 days as described above. On day 6, cells were split again into single cells with TrypLE Express, and 200,000 cells in 1 ml of StemPro Chondrogenesis Differentiation medium (Thermo Fisher Scientific) were pelleted at 300 g for 5 min in a 5 ml Eppendorf centrifuge tube. Pellets were cultured in the same medium for 28 days. Cells were fed with fresh medium every day until day 6 and every three days thereafter.
Human embryonic tissues
Human fetal tissue was obtained from legally terminated embryos (Carnegie stage 13–14; embryonic age 4.5–5 weeks of gestation) following informed consent and de-identification, in accordance with institutional guidelines. Procedures were approved by the research ethics committee of University of Tübingen (IRB #710/2012BO1 and #312/2016BO1). Subsequent use of the above tissues by researchers at UCLA was deemed IRB exempt by the UCLA Office of the Human Research Protection Program (IRB #15-000959). Tissues were immediately washed in sterile PBS and placed in PBS supplemented with 5% fetal bovine serum (Thermo Fisher Scientific), 1% Penicillin-Streptomycin and 2.5 µg/mL Amphotericin B (Sigma Aldrich). Within 48 hours, targeted tissues were micro-dissected from the embryos and RNA extracted using the QIAGEN RNeasy Plus Micro Kit. Nascent somites (SM) were defined as the two pairs of somites just anterior to the most caudal segmentation border that was visible. Presomitic mesoderm (PSM) was dissected as the tail region mesoderm posterior to SM (PSM tissues were not separated into pPSM and aPSM due to variability in developmental stages from the human embryos available for this study). Two pairs of more developed somites at the forelimb bud level were used as SM Dev. Embryos used in this study are listed in Table 1.
RNA-sequencing and analysis
The Ovation RNA-Seq System V2 kit (Nugen) was used to make and amplify cDNA from up to 100 ng of human RNA inputs, and sequencing was performed using an Illumina HiSeq 3000 sequencer. Raw sequencing reads were mapped to the hg19 reference genome via Tophat. Gene expression values were obtained through Cufflinks and DEG analysis was performed through Cuffdiff. DEGs were chosen based on p < 0.05 and an absolute value of log2FC > 1. The DAVID Bioinformatics Resources was used for functional annotation analysis. Clustering, PCA and Volcano plot was performed using RStudio. RNA-seq data were deposited to the GEO database with accession number GSE90876. See also Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank the UCLA Technology Center for Genomics & Bioinformatics (TCGB) for consultation and execution of RNA-seq, and the UCLA Institute for Quantitative and Computational Biosciences (QCBio) Collaboratory Workshop for general guidance on RNA-seq analysis. We are grateful to the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research (BSCRC) Flow Cytometry Core. We acknowledge the UCLA Translational Pathology Core Laboratory (TPCL) for chondrogenic cell pellet embedding and sectioning. We also thank Drs. Ayelet Dar (UCLA) and Denis Evseenko (USC) for suggestions on osteogenesis and chondrogenesis experiments. We thank Drs. Carrie Miceli, Melissa Spencer, Leanne Jones, Alex Hoffman, Steven Bensinger and Jeff Miller (UCLA) for their support on equipment. This work was supported by NIH/NIAMS (R01AR064327), the BSCRC, and a Rose Hills Foundation Research Award to A.D.P. The study was also supported by a UCLA Muscular Dystrophy P30 Core Center grant (NIH/NIAMS P30AR057230), the UCLA Clinical and Translational Science Institute (CTSI) (UL1TR000124), and the UCLA Center for Duchenne Muscular Dystrophy (CDMD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The accession number for the RNA-seq data generated in this study is GEO: GSE90876
AUTHOR CONTRIBUTIONS
Conceptualization and Methodology: H.X., B.V.H and A.D.P.; Validation, Formal Analysis, Visualization and Writing – Original Draft: H.X.; Investigation: H.X., W.F., K.G., M.J., S.L. and B.V.H.; Resources and Funding Acquisition: K.S-L and A.D.P.; Writing – Review & Editing: H.X., M.J., B.V.H., K.S-L. and A.D.P.; Supervision: A.D.P.; Project Administration: H.X. and A.D.P.
REFERENCES
- Applebaum M, Kalcheim C. Mechanisms of myogenic specification and patterning. Results Probl. Cell Differ. 2015;56:77–98. doi: 10.1007/978-3-662-44608-9_4. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Benazeraf B, Pourquie O. Formation and segmentation of the vertebrate body axis. Annu. Rev. Cell Dev. Biol. 2013;29:1–26. doi: 10.1146/annurev-cellbio-101011-155703. [DOI] [PubMed] [Google Scholar]
- Borchin B, Chen J, Barberi T. Derivation and FACS-Mediated Purification of PAX3+/PAX7+ Skeletal Muscle Precursors from Human Pluripotent Stem Cells. Stem Cell. Reports. 2013;1:620–631. doi: 10.1016/j.stemcr.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent AE, Tabin CJ. Developmental regulation of somite derivatives: muscle, cartilage and tendon. Curr. Opin. Genet. Dev. 2002;12:548–557. doi: 10.1016/s0959-437x(02)00339-8. [DOI] [PubMed] [Google Scholar]
- Chal J, Oginuma M, Al Tanoury Z, Gobert B, Sumara O, Hick A, Bousson F, Zidouni Y, Mursch C, Moncuquet P, et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 2015;33:962–969. doi: 10.1038/nbt.3297. [DOI] [PubMed] [Google Scholar]
- Chal J, Pourquié O. 3 Patterning and Differentiation of the Vertebrate Spine. Cold Spring Harbor Monograph Archive. 2009;53 [Google Scholar]
- Cheung C, Bernardo AS, Trotter MW, Pedersen RA, Sinha S. Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nat. Biotechnol. 2012;30:165–173. doi: 10.1038/nbt.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar A, Domev H, Ben-Yosef O, Tzukerman M, Zeevi-Levin N, Novak A, Germanguz I, Amit M, Itskovitz-Eldor J. Multipotent vasculogenic pericytes from human pluripotent stem cells promote recovery of murine ischemic limb. Circulation. 2012;125:87–99. doi: 10.1161/CIRCULATIONAHA.111.048264. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Dias AS, de Almeida I, Belmonte JM, Glazier JA, Stern CD. Somites without a clock. Science. 2014;343:791–795. doi: 10.1126/science.1247575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, van Oudenaarden A. Design and Analysis of Single-Cell Sequencing Experiments. Cell. 2015;163:799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]
- Hubaud A, Pourquie O. Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 2014;15:709–721. doi: 10.1038/nrm3891. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell. Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Mendjan S, Mascetti VL, Ortmann D, Ortiz M, Karjosukarso DW, Ng Y, Moreau T, Pedersen RA. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell. Stem Cell. 2014;15:310–325. doi: 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Tassy O, Pourquie O. Spatiotemporal compartmentalization of key physiological processes during muscle precursor differentiation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4224–4229. doi: 10.1073/pnas.0909375107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi F, Perlingeiro RC. Stem cells for skeletal muscle regeneration: therapeutic potential and roadblocks. Transl. Res. 2014;163:409–417. doi: 10.1016/j.trsl.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risebro CA, Vieira JM, Klotz L, Riley PR. Characterisation of the human embryonic and foetal epicardium during heart development. Development. 2015;142:3630–3636. doi: 10.1242/dev.127621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y. The mechanism of somite formation in mice. Curr. Opin. Genet. Dev. 2012;22:331–338. doi: 10.1016/j.gde.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Sakaguchi Y, Shoji E, Nishino T, Maki I, Sakai H, Hanaoka K, Kakizuka A, Sehara-Fujisawa A. In vitro modeling of paraxial mesodermal progenitors derived from induced pluripotent stem cells. PLoS One. 2012;7:e47078. doi: 10.1371/journal.pone.0047078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton M, Metz J, Liu J, Carpenedo RL, Demers SP, Stanford WL, Skerjanc IS. Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell. Reports. 2014;3:516–529. doi: 10.1016/j.stemcr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Cossu G. Establishing myogenic identity during somitogenesis. Curr. Opin. Genet. Dev. 1997;7:634–641. doi: 10.1016/s0959-437x(97)80011-1. [DOI] [PubMed] [Google Scholar]
- Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Tonegawa A, Takahashi Y. Somitogenesis controlled by Noggin. Dev. Biol. 1998;202:172–182. doi: 10.1006/dbio.1998.8895. [DOI] [PubMed] [Google Scholar]
- Umeda K, Zhao J, Simmons P, Stanley E, Elefanty A, Nakayama N. Human chondrogenic paraxial mesoderm, directed specification and prospective isolation from pluripotent stem cells. Sci. Rep. 2012;2:455. doi: 10.1038/srep00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg CW, Okawa S, Chuva de Sousa Lopes SM, van Iperen L, Passier R, Braam SR, Tertoolen LG, del Sol A, Davis RP, Mummery CL. Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development. 2015;142:3231–3238. doi: 10.1242/dev.123810. [DOI] [PubMed] [Google Scholar]
- Xu C, Tabebordbar M, Iovino S, Ciarlo C, Liu J, Castiglioni A, Price E, Liu M, Barton ER, Kahn CR, Wagers AJ, Zon LI. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell. 2013;155:909–921. doi: 10.1016/j.cell.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, Karumbayaram S, Kumagai-Cresse C, Wang D, Zack JA, et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell. Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.