Abstract
We herein report a biomimetic technique to modify plastic substrates for bioassays. The method first places a polydopamine adhesion layer to plastic surface, and then grows conformal silica coating. As proof of principle, we coated plastic microbeads to construct a disposable filter for point-of-care nucleic acid extraction.
Thermoplastics often have ideal material properties to be used in biosensors and for lab-on-a-chip microfluidics. For example, plastics are transparent to allow optical measurements, rigid but not fragile, and solvent-resistant. Furthermore, plastic devices can be molded into complex shapes, and mass-produced at low cost.1 Plastics, however, are challenging materials for bioconjugation; the material surface is highly hydrophobic and inherently lacks active functional groups. A few methods of chemical modification are available. Proteins, for example, can be non-specifically adsorbed via hydrophobic interactions,2 but with little controllability.3 Oxygen plasma treatment or ultraviolet photoactivation can be used to generate carboxyl groups on plastic surfaces, although the effect is temporary and limited to specific plastic types (e.g., poly-methylmethacrylate, cyclic-olefin-copolymer, or polycarbonate) with ester or ami de bonds.4 Developing robust and generalizable surface chemistries, particularly for hydrocarbon-based plastics (e.g., polyethylene, polypropylene, polystyrene) would catalyze the more widespread use of cost-effective plastics in biosensing.5
Bio-inspired chemistries could be a new way to functionalize challenging materials in a scalable and environment-friendly manner.6-9 For example, DOPA (3,4-dihydroxyphenylalanin) and lysine residues in mussel foot proteins (mfps) have been identified to form water-resistant adhesion to various substrates via multimodal chemical interactions (e.g., hydrophobic, electrostatic/cation-π interactions, hydrogen bonding, π-π staking, Schiff-base formation, metal coordination).10-14 The observation has led to novel material-independent coating techniques. Catecholamines such as dopamine,15 norepinephrine,16 and pyrogallol 2-aminoethane17 have been used as a building block to polymerize into ‘artificial mfps’ that strongly bind to a wide range of materials.18 The biosilicification process in marine diatoms has also inspired novel biomimetic chemistries. Silaffin peptides, rich with polyamine moieties and phosphorylated serine residues, form supramolecular complexes with water-soluble silicate, leading to silica biomineralization in diatoms.19 These processes have been recently adopted to polymerize monosilicic acid into silica in aqueous, mild pH solution.20
We herein report a bio-inspired, generalizable approach to modify various plastic substrates for bioassays. We reasoned that i) a plastic surface can be effectively coated with an adhesion layer through catecholamine polymerization, and ii) a subsequent silica layer can then be layered on top. The latter can be further functionalized with biomolecules (e.g., antibodies, enzymes, and nucleic acids). As proof-of-concept, we modified different plastic beads with the bilayer method, and implemented a disposable filter for point-of-care (POC) nucleic-acid extraction. The bead-filters, irrespective of the plastic used, showed high extraction yields that were comparable to those of commercial chemical columns. Yet, the process was considerably cheaper and operated in an equipment-free manner. Using the system, we isolated RNA from cancer cells and their exosomes and compared mRNA expressions. Moreover, combined with an isothermal nucleic acid amplification method, we also detected antibiotic-resistance genes in bacterial samples. The developed method offers an effective, economically scalable mechanism for functionalizing plastic devices for biosensing purposes.
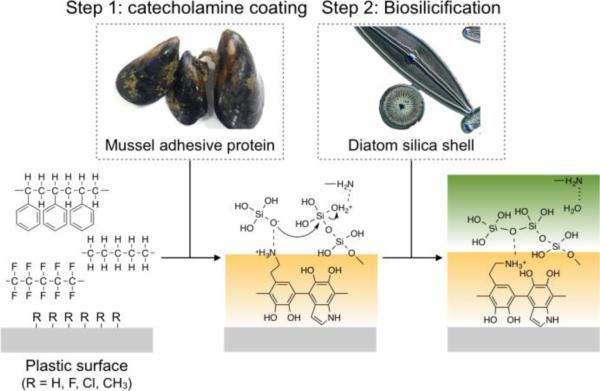
Scheme 1 shows the overall coating strategy. We first used mussel-inspired polydopamine (pDA) coating to generate a polyamine-based adhesion layer on the plastic surface. Following the pDA coating, we grew a biocompatible silica (SiO2) layer, mimicking the silicification process in diatom shell formation. The binding mechanism between pDA and plastics is likely hydrophobic interactions 21 due to the hydrophobic nature of plastics surfaces. The silicification process is the same as occurred in diatoms; i) electrostatic interaction triggering the chemisorption of monosilicic acid on positively charged pDA layer and ii) simultaneous polymerization of surface-bound monosilicic acid into SiO2. The SiO2 layer rendered the surface highly hydrophilic and facilitated bioconjugation through well-established silane chemistry.22,23
Scheme 1. Bioinspired plastic coating.
Mussel-inspired polydopamine (pDA) coating is used to make an adhesive layer on the hydrophobic plastic surface. Following the pDA coating, a biocompatible SiO2 layer can be grown by mimicking the silicification process in diatom shell formation.
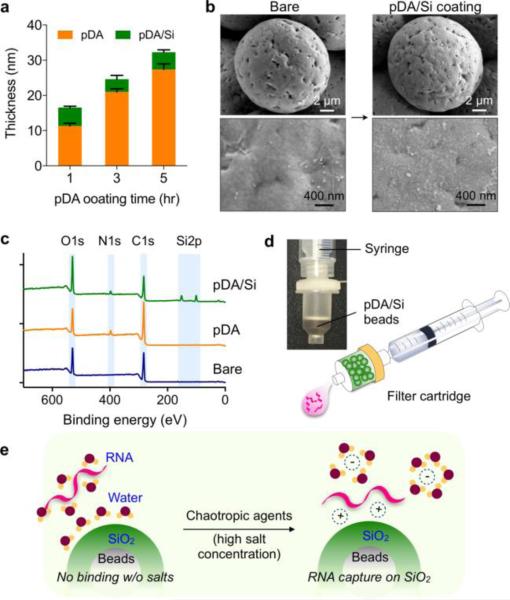
We initially used a planar Si wafer as a coating substrate, and measured the thickness of pDA and SiO2 layers via ellipsometry (Fig. 1a). We first grew a pDA layer varying the coating time. The layer thickness gradually increased up to 5 hrs. We then grew a SiO2 layer for an additional hour. Regardless of the thickness of the underlying pDA layers, the silica coating had the thickness of ~5 nm. We thus set the pDA coating to 1 hr, which would result in 17 nm of total thickness (pDA, 12 nm; SiO2, 5 nm). We next used different types of plastic microbeads (diameter range, 11-21 μm; see Methods for details) as a substrate, and sequentially coated them with pDA and SiO2 (pDA/Si). Fig. 1b shows an example, poly(methyl methacrylate) (PMMA) microbeads (14.7 μm in diameter), before and after the pDA/Si coating. The overall shape of beads was maintained, and no aggregates were formed after the surface modification (Fig. 1b, top). High magnification images revealed nanoscale SiO2 (<100 nm) covering the bead surface (Fig. 1b, bottom). We applied the Brunauer-Emmett-Teller (BET) method to analyze surface morphology (Fig. S1). From bare to pDA-coated beads, the surface area slightly increased (9%), whereas the increase was much more substantial (780%) with the silica coating (Fig. S1a). Such a large enhancement could be attributed to the formation of nanoporous structure on the bead surface. Barrett-Joyner-Halenda (BJH) analyses showed that bare and pDA-coated beads contained flat surfaces without detectable pores. On the pDA/Si-coated beads, however, the analysis indicated the existence of two portions of nanopores, below 10 nm and around 45 nm in diameter (Fig. S1b). pDA/Si-coated beads thus have nanoscale roughness, which increases the reactive surface area.
Fig. 1. Silicification of plastic microbeads.
(a) Thickness of polydopamine (pDA) and silica (SiO2) coated on a planar substrate (Si wafer) was measured via ellipsometry. The pDA layer gradually thickened with the coating time up to 5 hrs. The SiO2 layer, grown for 1 hr, was about 5 nm thick. (b) Scanning electron micrographs of microbeads before (left) and after (right) polydopamine/silica (pDA/Si) coating. The nano-thin layer of pDA/Si coating does not alter the surface roughness of pristine beads. (c) XPS analysis of modified surface showing a N1s peak, which appeared at around 400 eV after pDA coating, and two Si2p peaks at around 100 eV after Si coating. (d) Beads-loaded disposable filter for point-of-care (POC) operation. (e) The pDA/Si-coated beads can adsorb negatively charged nucleic acids in the presence of a chaotropic agent, and the adsorbed nucleic acid can be eluted in water by changing the salt concentration.
We used spectroscopic methods to verify pDA and SiO2 layer formations. Energy-dispersive X-ray spectroscopy (EDS) showed the Si peak at 1.74 keV (Fig. S2a), and the elemental mapping (Fig. S2b) confirmed homogeneous pDA (from N) and SiO2 (from Si) coatings on the bead surface. In-depth analyses were performed via X-ray photoelectron spectrum. Following the silicification, two peaks corresponding to Si2p1/2 and Si2p3/2 appeared (Fig. 1c). The high-resolution spectra (Fig. S3) confirmed that the chemical status of the top layer (~10 nm depth from the surface) changed after pDA and pDA/Si coating. For example, after pDA coating, a characteristic C=O peak from bare PMMA decreased (C1s spectra), and a new N1s peak emerged. Two peaks in O1s spectra, arising from ester bonds in PMMA (C-O and C=O), broadened into a single peak due to the presence of multiple oxygen species in pDA. After SiO2 coating, the O1s peak further increased and a new Si2p peak appeared. The C1s and N1s peaks decreased, but were still present, which indicated that the SiO2 thickness was likely <10 nm, the maximum depth measurable by XPS. 24 We finally used Fourier transform infrared (FT-IR) spectroscopy to double check the existence of SiO2 on the surface (Fig. S4). The pDA/Si-coated beads showed a shoulder peak at around 3500-3600 cm−1, which corresponds to the hydroxyl groups on the surface of SiO2. We also observed two characteristic peaks in the fingerprint region25: Si-O-Si and Si-O-C stretching at around 1050 cm−1 and bending at around 880 cm−1.
All coating processes were in solution phase, and could be performed in bulk scale. We used ca. 1 g of beads in a single batch. As a model device, we built disposable filters for nucleic-acid extraction by loading pDA/Si-coated beads into cartridges. Two filter types were prepared: a syringe attachment for POC operation (Fig. 1d) and an in-flow column for use with a bench-top centrifuge (Fig. S5). Fig. 1e shows the RNA binding mechanism on the surface of pDA/Si-modified beads in the presence of chaotropic agents such as guanidine thiocyanate. Chaotropic agents have shown to reduce the repulsion between the negatively charged nucleic acids and silica.26,27 These agents can dehydrate nucleic acids by disrupting their surrounding water molecules. As a result, non-covalent interactions (e.g., hydrogen bonding, van der Waals force, hydrophobic interactions) can be weakened, leading to denaturation of biomolecules. This effect has been exploited to lyse cells, reduce protein binding on silica, and inhibit enzymes (e.g, RNase) during nucleic acid extractions.28 Importantly, such dehydration could trigger cation-mediated binding between RNA and silica,27,29 which can be reversed through the control of salt concentrations.
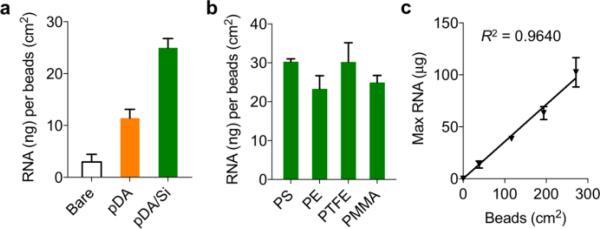
We prepared four different types of polymer microbeads for RNA extra ction: PMMA, polystyrene (PS), polyethylene (PE), and polytetrafluoroethylene (PTFE). Cell lysates (120 μL, 1 × 106 cells) were prepared and passed through the filters, followed by washing and elution steps. We used a commercial buffer set (RNeasy, Qiagen) optimized for silica membranes. For the syringe filter, the solution was manually injected; for the in-flow column, we applied a short centrifugation (9,000 g, 30 sec) to generate fluidic flow (detailed procedures in Table S1). We first tested the effect of surface coating by using bare, pDA-only, and pDA/Si-coated PMMA microbeads. As expected, pDA/Si-coated ones showed the highest yield, followed by pDA-coated beads (Fig. 2a). When different bead materials were compared, the extraction yields of pDA/Si-coated beads were similar with no statistically significant difference among bead types (p = 0.1901; one-way ANOVA) (Fig. 2b). The RNA capture capacity thus largely depended only on the amount of packed beads, and the maximum capacity of 0.357 μg RNA per beads surface (cm2) was achieved (Fig. 2c). The extraction performance of the bead filters, including the RNA amounts and purity, was comparable with that of a commercial kit (Table S2). The bead filters, however, enabled faster (a few seconds for the syringe type), near equipment-free, and more cost-effective operations.
Fig. 2. Bead-filter characterization.
(a) The amount of total RNA from A2780 cells (~106 cells) extracted by the unmodified (white), pDA-coated (orange), pDA/Si-coated (green) PMMA. (b) pDA/Si-coated microbeads showed similar extraction yields regardless of the core plastic types (p = 0.1901, one-way ANOVA) (c) Maximum binding capacity of a filter device containing pDA/Si-coated PE beads.
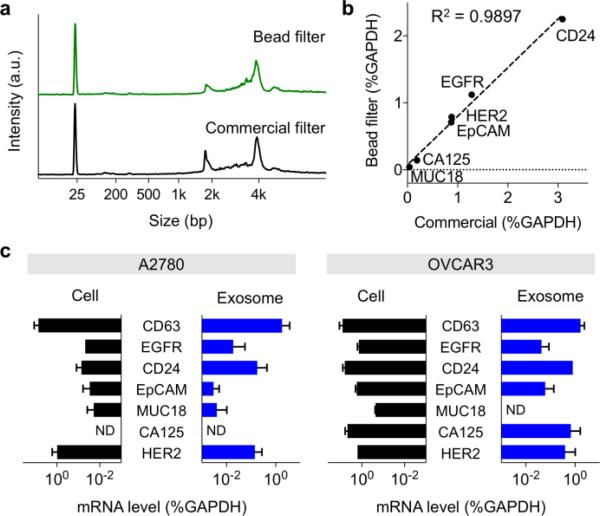
We applied the filter system to extract RNA from cancer cells and cell-derived exosomes. A panel of ovarian cancer cell lines (A2780, OVCAR3, OV420) and normal control (TIOSE4) were cultured. First, we used cell lysates and extracted RNA using the plastic-bead filter as well as a commercial column device. Electrophoresis analysis (Bioanalyzer, Agilent) showed that RNA collected by the bead filter retained structural integrity and quality similar to that of column-extracted RNA (Fig. 3a). RT-qPCR measurements further confirmed unbiased RNA extraction by the bead-filter; the levels of different mRNA targets matched well (R2 = 0.99) between bead-filtered and column-extracted samples (Fig. 3b and Fig. S6). We next examined exosomes, nanoscale membrane vesicles secreted by cells. These vesicles carry molecular cargos of parent cells, offering a timely peripheral window. We harvested exosomes from conditioned cell culture media. Exosome lysates were then processed by the bead-filter, and extracted RNA was amplified. Comparison between exosomal and cellular mRNA patterns revealed high correlation (98.6% for A2780 and 87% for OVCAR3, Fig. 3c). Similar observations have been made with other tumor types (e.g., Glioblastoma Multiforme)30,31; the current results further strengthen the use of exosomes as cellular surrogates.
Fig. 3. mRNA analysis of ovarian cancer cell lines and exosomes.
(a) Comparison of extracted RNA. RNA extracted by the bead-filter showed the structural integrity and quality profile similar to that extracted by a commercially available column. a.u., arbitrary unit. (b) Comparison of mRNA levels isolated by pDA/Si-coated PE and a commercial column from Qiagen. Note the excellent correlation between mRNA markers from plastic beads and the commercial column. (c) Exosome mRNA analysis (~1010 exosomes) compared with mRNA found in the parental cell (~106 cells). Marker expression levels correlated well between exosomal mRNAs and those found in cells (mRNA quantities were normalized to intrinsic GAPDH mRNA)
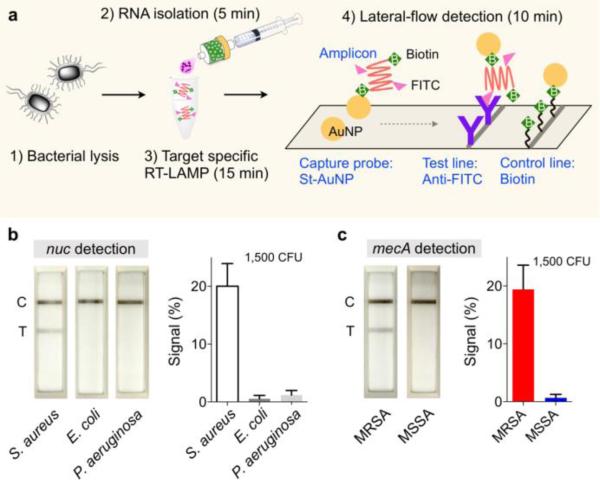
We further established an assay kit for nucleic acid detection, combining the bead-filter, a heating block, and lateral-flow strips (Fig. 4a). We used the syringe bead-filter for quick RNA extraction, and amplified target genes via loop-mediated isothermal amplification (LAMP). Amplicons were labeled with biotin and FITC during the LAMP reaction in the tube. Without any purification, we then loaded samples to the lateral-flow strip containing streptavidin-coated gold nanoparticles (AuNPs) which captured biotinylated amplicons. The complex further migrated and captured on the test line spotted with anti-FITC antibodies, creating a visible black line; excess AuNPs were collected on the control line. The assay was fast (30 min), cost-effective (~$1 per assay), and performed with minimal equipment (a heat block at constant temperature for LAMP). We prepared the kit to detect methicillin-resistant S. aureus (MRSA). We used primer sets (Table S3)32 for target-specific amplification of two genes33: nuc which is common to S. aureus; mecA which confers methicillin resistance to S. aureus. Four types of samples, each containing MRSA (nuc+, mecA+), methicillin-sensitive S. aureus (nuc+, mecA−), E. coli (nuc−, mecA−), and P. aeruginosa (nuc−, mecA−), were processed by the assay kit. The nuc assay correctly detected S. aureus species (Fig. 4b), and the antibiotic resistance status was confirmed by the mecA test (Fig. 4c).
Fig. 4. Point-of-care (POC) pathogen detection.
(a) A POC assay kit was assembled by combining bacterial RNA extraction, reverse transcription loop-mediated isothermal amplification (RT-LAMP) reaction, and lateral-flow detection. During the RT-LAMP, amplicons are labeled with biotin and FITC. Gold nanoparticles (AuNPs) are used to capture amplicons and generate optical signal. (b, c) Different strains of S. aureus (SA) were detected. S. aureus could be identified with positive nuc signal. The SA strains could further be genotyped for methicillin-resistance status by checking mecA signal. Signal level is the ratio between the test (T) and the control lines. MRSA, methicillin-resistant S. aureus; MSSA, methicillin-sensitive S. aureus
In summary, we have developed a universal, biomimetic technique for functionalizing various plastic types. The method allows coating of diverse plastics with silica, using polydopamine as an intermediate adhesive. Polydopamine is a nano-thin layer that binds to plastics and silica through various modes of chemical interactions that exhibit considerable strength. The resulting surface is highly hydrophilic and amenable to bioconjugation chemistries. We applied the method to coat plastic beads, and used them as an adsorbent for nucleic acid capture. The device achieved RNA capture yields comparable to those of a commercial product, and could be operated in an equipment-free manner. We envision that the developed coating technique could be applicable to various plastic devices of different shapes and complexity, particularly for creating complex bioactive microfluidic devices.
Supplementary Material
Acknowledgments
This work was supported in part by US NIH Grants R21-CA205322 (to H.L.), R01-HL113156 (to H.L.), R01-CA204019 (to R.W.), R01-EB010011 (to R.W.), K12CA087723-11A1 (to C.M.C.), MGH Physician Scientist Development Award (to C.M.C.) and Basic Science Research Program 2014R1A6A3A03059728 (K.S.P) through the National Research Foundation of Korea.
Footnotes
Electronic Supplementary Information (E SI) available: Experimental de tails. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Chantiwas R, et al. Chem. Soc. Rev. 2011;40:3677. doi: 10.1039/c0cs00138d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salas C, et al. ACS appl. mater. interfaces. 2013;5:6541. doi: 10.1021/am401065t. [DOI] [PubMed] [Google Scholar]
- 3.Trilling AK, et al. Analyst. 2013;138:1619. doi: 10.1039/c2an36787d. [DOI] [PubMed] [Google Scholar]
- 4.Jackson JM, et al. Lab chip. 2014;14:106. doi: 10.1039/c3lc50618e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D, Herr AE. Biomicrofluidics. 2013;7:041501. doi: 10.1063/1.4816934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu K, et al. Chem. Soc. Rev. 2010;39:3240. doi: 10.1039/b917112f. [DOI] [PubMed] [Google Scholar]
- 7.Liu K, Jiang L. Annu. Rev. Mater. Res. 2012;42:231. [Google Scholar]
- 8.Kirschner CM, Brennan AB. Ann. Rev. Mater. Res. 2012;42:211. [Google Scholar]
- 9.Liu Y, et al. Chem. Rev. 2014;114:5057. doi: 10.1021/cr400407a. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, et al. Proc. Natl. Acad. Sci. U. S. A. 2013;110:15680. doi: 10.1073/pnas.1315015110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim C, et al. Angew. Chem. Int. Ed. 2016;55:3342. doi: 10.1002/anie.201510319. [DOI] [PubMed] [Google Scholar]
- 12.Wilker JJ. Nat. Chem. Biol. 2011;7:579. doi: 10.1038/nchembio.639. [DOI] [PubMed] [Google Scholar]
- 13.Levine ZA, et al. Proc. Natl. Acad. Sci. U. S. A. 2016:201603065. [Google Scholar]
- 14.Lee H, et al. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12999. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H, et al. Science. 2007;318:426. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang SM, et al. J. Am. Chem. Soc. 2009;131:13224. doi: 10.1021/ja905183k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong S, et al. Adv. Mater. Interfaces. 2014;1:1400113. [Google Scholar]
- 18.Hong S, et al. Angew. Chem. Inter. Ed. 2013;52:9187. doi: 10.1002/anie.201301646. [DOI] [PubMed] [Google Scholar]
- 19.Kröger N, Lorenz S, Brunner E, Sumper M. Science. 2002;298:584. doi: 10.1126/science.1076221. [DOI] [PubMed] [Google Scholar]
- 20.Kang SM, et al. Chem. Mater. 2012;24:3481. [Google Scholar]
- 21.Zhou Q, et al. J. Sep. Sci. 2013;36:1516. doi: 10.1002/jssc.201201163. [DOI] [PubMed] [Google Scholar]
- 22.Haensch C, et al. Chem. Soc. Rev. 2010;39:2323. doi: 10.1039/b920491a. [DOI] [PubMed] [Google Scholar]
- 23.Ulman A. Chem. Rev. 1996;96:1533. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert JB, et al. Proc. Natl. Acad. Sci. U. S. A. 2013;110:6651. doi: 10.1073/pnas.1222325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung CJ, et al. Curr. Appl. Phys. 2010;10:428. [Google Scholar]
- 26.Melzak KA, et al. J. Colloid Interface Sci. 1996;181:635. [Google Scholar]
- 27.Iannone E. Labs on chip: Principles, design and technology. Vol. 3. Taylor & Francis Group; USA: 2014. p. 183. [Google Scholar]
- 28.Boom RCJA, et al. J. Clin. Microbiol. 1990;28:495. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogelstein B, Gillespie D. Proc. Natl. Acad. Sci. U. S. A. 1979;76:615. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skog J, et al. Nat. Cell Biol. 2008;10:1470. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao H, et al. Nat. Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudhaharan S, et al. J. Clin Diagn. Res. 2015;9:DC06. doi: 10.7860/JCDR/2015/13962.6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park KS, et al. Sci. Adv. 2016;2:e1600300. doi: 10.1126/sciadv.1600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.