Abstract
Electron cryotomography (ECT) enables the 3D reconstruction of intact cells in a near-native state. Images produced by ECT have led to the proposal that an ancient sporulation-like event gave rise to the second membrane in diderm bacteria. Tomograms of sporulating monoderm and diderm bacterial cells show how sporulation can lead to the generation of diderm cells. Tomograms of Gram-negative and Gram-positive cell walls and purified sacculi suggest that they are more closely related than previously thought and support the hypothesis that they share a common origin. Mapping the distribution of cell envelope architectures onto a recent phylogenetic tree of life indicates that the diderm cell plan, and therefore the sporulation-like event that gave rise to it, must be very ancient. One explanation for this model is that during the cataclysmic transitions of the early Earth, cellular evolution may have gone through a bottleneck in which only spores survived, which implies that the last bacterial common ancestor was a spore.
Historically, bacteria were classified by their ability to retain the Gram stain1. Following the development of electron microscopy, the actual structure of the bacterial cell envelope became apparent. Gram-positive bacteria were seen to be monoderms (having a single membrane) with a relatively thick layer of peptidoglycan. Gram-negative bacteria were seen to be diderms (having two membranes) with a thin layer of peptidoglycan between the two membranes. Typical Gram-positive and Gram-negative cell envelope architectures are exhibited by the members of the Firmicutes and Proteobacteria phyla and are represented by the model organisms Bacillus subtilis and Escherichia coli, respectively2,3.
The cytoplasmic membrane of all cells (the innermost membrane in the case of diderm bacteria) contains α-helical integral membrane proteins and sustains a chemical gradient, which usually involves protons and is used to produce ATP. The structure and function of the outer membrane in diderm bacteria are very different. Whereas the inner membrane is a symmetrical lipid bilayer, the outer membrane of typical Gram-negative bacteria is asymmetric, with the outer leaflet being composed primarily of lipopolysaccharide (LPS). Outer membranes are also rich in outer-membrane proteins (OMPs; mostly β-barrel proteins) that allow the free diffusion of small molecules into and out of the periplasm2,4. Interesting exceptions to the typical Gram-positive and Gram-negative envelope architectures exist. For example, members of the class Mollicutes (in the Tenericutes phylum) lack peptidoglycan and members of the Corynebacterineae, a suborder of the Actinobacteria phylum that includes mycobacteria, have an unusual outer membrane that is rich in mycolic acids.
It is unclear how the different architectures of monoderm and diderm bacterial envelopes have evolved. However, recent insights provided by electron cryotomography (ECT) suggest that the cell walls of Gram-negative and Gram-positive bacteria are more closely related than previously assumed and that sporulation can lead to the generation of diderm cells from a monoderm progenitor. In this Opinion article, we review the evidence that supports this hypothesis and outline a possible scenario that puts sporulation at the onset of evolution of all bacteria.
Biogenesis through sporulation
Various theories exist about the origin of life on Earth. Some theories imagine larger and larger molecules replicating in rich organic pools, with lipid vesicles washing out of cavities in rocks to enclose primordial cells5. Assuming that the earliest primordial cells were monoderm3,6–9, the acquisition of an outer membrane must have been a major evolutionary step. Prior to the characterization of sporulation in diderm cells in 2011 (REF. 10), the most prominent, but highly criticized11,12, hypothesis for how outer membranes evolved was based on the distribution of protein families and proposed that a symbiosis between an ancient actinobacterium and an ancient clostridium produced the last common ancestor of all diderms11,13. Based on conserved protein inserts, Gupta proposed that diderms that lack LPS preceded the formation of diderms that contain LPS12. Although the mechanism by which the second membrane developed was not addressed, Gupta argued that antibiotic selection pressure was the main evolutionary driver.
We propose that the outer membrane probably first arose through the formation of endospores, a common process that is executed by many species in the Firmicutes phylum. The formation of endospores begins with genome segregation and asymmetric cell division. Next, in a process that is similar to phagocytosis in eukaryotic cells, the larger compartment (mother cell) engulfs the smaller compartment (future spore). Several protective layers are formed around the immature spore and then lysis of the mother cell releases a mature spore into the environment. Notably, spores have a complete copy of the genomic DNA. Later, germination returns the dormant spore to a vegetative state14,15.
Although the well-studied model endospore-forming species (Bacilli and Clostridia classes) are all monoderms, we studied Acetonema longum, a member of a lesser-known class of Firmicutes, the Negativicutes16,17, that forms endospores but is diderm10,18. The class Negativicutes contains more than 30 genera in two families, the Veillonellaceae and Acidaminococcaceae, and outer membrane and LPS biosynthesis genes are readily detectable in the genomes of these bacteria2,19. Biochemical characterization of the outer membrane also revealed that A. longum contains LPS similarly to model diderms20. Homology between many OMPs of A. longum and their counterparts in proteobacteria and other diderm species suggested that these proteins share an ancient common heritage (rather than being the result of convergent evolution)10. Moreover, phylogenetic analysis of several omp85 and omp87 family genes, which are involved in the insertion and assembly of OMPs, revealed a close relationship between mitochondria and α-proteobacteria, but no close relationship between A. longum and any other species in diderm phyla, which argues against recent horizontal gene transfer10. Therefore, A. longum and other members of the Veillonellaceae are candidate ‘missing links’ between monoderm and diderm bacteria. They have characteristics of both monoderms and diderms: an outer membrane and the ability to form endospores.
Mechanistic clues about how the formation of endospores may have given rise to bacterial outer membranes came from comparing images of sporulating cells of B. subtilis (monoderm) and A. longum (diderm)10,21 (FIG. 1). Cells from both species were imaged with ECT, which provides 3D reconstructions of intact cells to ‘macromolecular’ (3–4 nm) resolution22. Images of vegetative, sporulating and germinating cells revealed that both monoderm (B. subtilis; FIG. 1, left panels) and diderm (A. longum; FIG. 1, right panels) bacteria produced spores that were surrounded by two membranes. Furthermore, in both cases, the two membranes originated from the inner (or cytoplasmic) membrane of the mother cell. At some time between mid-to-late spore development and germination, B. subtilis loses its outer spore membrane to become a monoderm, ‘Gram-positive’ vegetative cell, whereas A. longum retains both spore membranes and the outer spore membrane remains and is remodelled to an outer membrane. Therefore, the formation of endospores provides a novel hypothesis for how the bacterial outer membrane could have evolved: a primordial monoderm cell may have first developed the ability to form endospores, and then this process could have given rise to diderm vegetative cells10,23,24 (FIG. 2).
Figure 1. Sporulation in Gram-positive and Gram-negative bacteria.
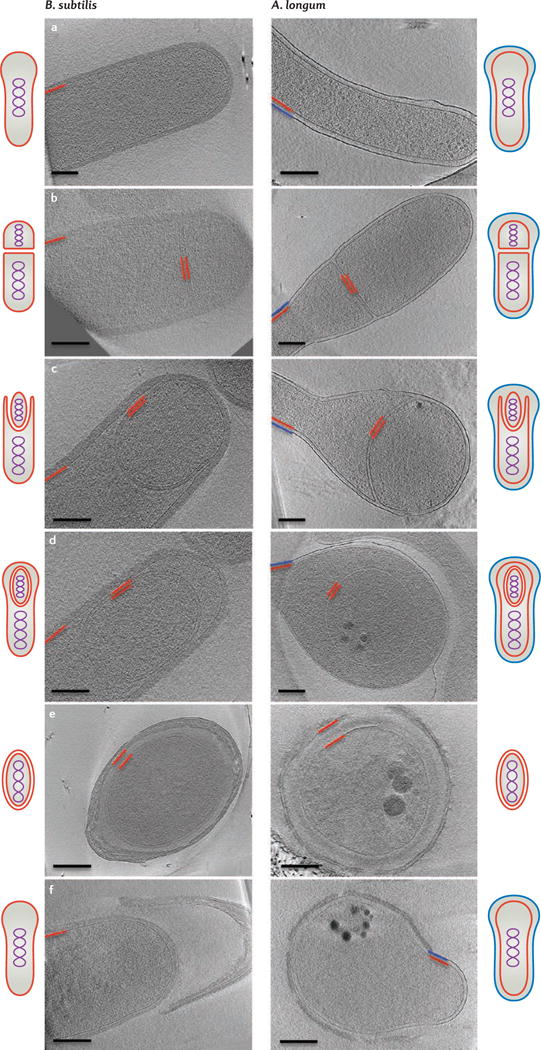
Overview of bacterial sporulation in Bacillus subtilis (left) and Acetonema longum (right) by electron cryotomography (ECT). Each panel represents a tomographic slice through a bacterial cell at a different stage of sporulation. Part a shows vegetative cells, part b shows the formation of the septum, part c shows engulfment, part d shows forespores, part e shows mature spores and part f shows germinating cells. The inner membrane and cytoplasmic membrane are depicted in red and the outer membrane of A. longum is depicted in blue. Schematic representations of all sporulation stages are shown next to the tomographic slices. Scale bar 200 nm. Images in the left panels of parts d–f are adapted with permission from REF. 21, Wiley. Images in the right panels of parts a, b, d, e and f are adapted with permission from REF.10, Elsevier.
Figure 2. Model for how the outer membrane arose as a byproduct of sporulation and how losses then led to the diversity of modern bacterial cell plans.
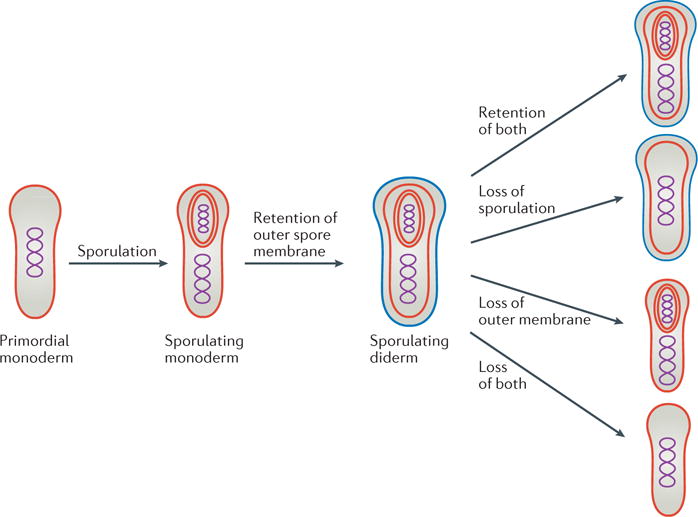
At some point during early evolution, the cell division and nutrient-uptake processes of a primordial cell were combined into a sporulation-like process. Retention of the second spore membrane led to sporulating diderm species. Losses of the outer membrane and/or the ability to sporulate in various lineages can explain the distribution of cell envelope architectures that is observed in modern bacteria. An alternative hypothesis (not shown) is that some monoderms evolved directly from a monoderm last common ancestor.
Peptidoglycan and sporulation
Further support for this hypothesis came from imaging the cell walls of both Gram-positive and Gram-negative bacteria. Typical Gram-negative peptidoglycan is a single-layer polymer that is composed of long glycan strands that are formed by repeating units of N-acetyl glucosamine–N-acetyl muramic acid crosslinked with peptide bonds25. Peptidoglycan is synthesized by transglycosylases and transpeptidases and other proteins that are influenced by cytoskeletal filaments26. The architecture of Gram-negative peptidoglycan was initially unclear27–29. However, direct visualization of the cell wall architecture of E. coli and Caulobacter crescentus by ECT revealed that the glycan strands of Gram-negative peptidoglycan run along the cell surface perpendicular to the long axis of the cell and parallel to the cell surface (in a ‘layered’, or ‘circumferential’ model)30.
Gram-positive peptidoglycan is thicker (~40 nm), but is closely related to Gram-negative peptidoglycan in two ways. First, the basic chemical structure of peptidoglycan is very similar among Gram-positive and Gram-negative bacteria — notable differences are mostly related to modifications in the composition of peptides, the extent of peptide crosslinks and the length of the peptidoglycan chains31. In fact, members of the Proteobacteria and Firmicutes phyla, in particular, share the same chemotype of peptidoglycan (A1γ), with a meso-diaminopimelic acid (meso-A2pm) residue at position 3 of the peptide and a direct crosslink to a D-alanine at position 4 of the neighbouring peptide24,32. Second, many biochemical and genetic studies have shown homology between the enzymes that are responsible for synthesizing peptidoglycan in Gram-negative and Gram-positive bacteria33–35; therefore, the cell walls that they build are likely to be similar.
Various models for the architecture of Gram-positive peptidoglycan have been proposed including circumferential, scaffold and coiled cable models28,36–39. Unfortunately, direct visualization of the architecture of Gram-positive peptidoglycan by ECT was not possible owing to its thickness and rigidity40. However, ECT imaging of sheared purified B. subtilis sacculi and coarse-grain molecular dynamics simulations supported the circumferential model40. Thus, the basic architecture of both Gram-negative and Gram-positive cell walls is similar.
Interestingly, the similarities in peptidoglycan architecture are supported by the observation that thick peptidoglycan and thin peptidoglycan can be interconverted21. At the onset of sporulation in the Gram-positive monoderm bacterium B. subtilis, thick peptidoglycan (~40 nm) is present between the two septal membranes (FIG. 3; top). Prior to engulfment, this thick peptidoglycan layer is remodelled into a thin layer that resembles Gram-negative peptidoglycan. The thin layer is extended by the synthesis of new peptidoglycan at the leading edges of engulfing membranes as they progress around the immature spore41. At the end of engulfment, a thin layer of peptidoglycan is found between the two spore membranes and probably acts as a foundation for the synthesis of the spore cortex (thick protective layers of glycan strands that are crosslinked by peptide bonds). Similar transitions of peptidoglycan thickness were observed in A. longum (FIG. 3; bottom). Thus, both A. longum and B. subtilis transform a thick layer of peptidoglycan into a thin, Gram-negative-like layer that eventually surrounds the immature spore during engulfment, and then expand this thin layer into a thick cortex during spore maturation. Both Gram-negative and Gram-positive bacteria can synthesize both thin and thick peptidoglycan and can gradually remodel one into the other, which strongly supports the concept that the cell walls have the same circumferential architecture and differ mainly in the number of layers that are present21. As the synthesis of peptidoglycan probably drives the engulfing membranes during sporulation10,41, the evolution of peptidoglycan probably preceded the evolution of sporulation. Appreciation of the diversity of cell wall thicknesses across the bacterial world is growing42. For example, the existence of medium-thick peptidoglycan in cyanobacteria43 and deinococci44 further supports the model that the basic peptidoglycan architecture is similar in Gram-negative and Gram-positive bacteria and just varies in thickness.
Figure 3. Peptidoglycan remodelling during sporulation.
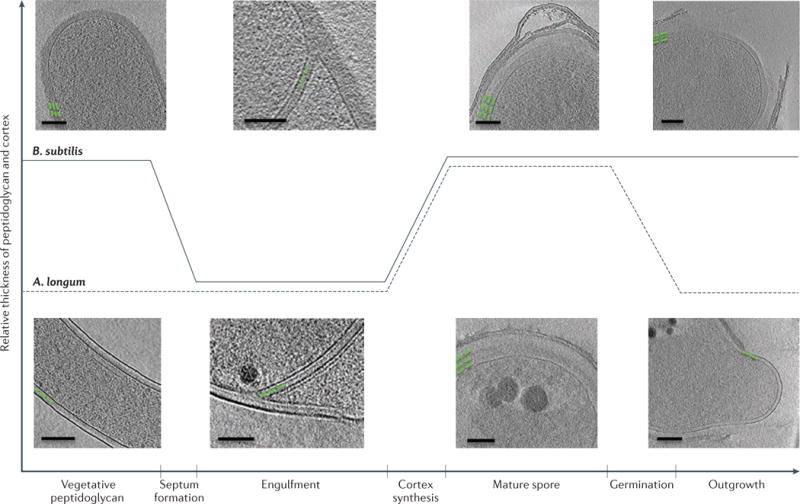
Peptidoglycan and cortex remodelling during sporulation in Bacillus subtilis and Acetonema longum show that both Gram-positive and Gram-negative bacterial cells can synthesize thick and thin peptidoglycan and can remodel one into the other, which supports the notion that all bacterial cell walls have the same basic architecture that is inherited from a common ancestor. Green lines represent the placement and thickness of peptidoglycan. Scale bar 100 nm. Adapted with permission from REF. 21, Wiley.
Because Gram-negative and Gram-positive cell walls have the same basic architecture (circumferential glycan strands) and can be interconverted during the sporulation cycle, we should not think of these two major divisions of bacteria as completely separate branches of the bacterial phylogenetic tree, but instead as potentially closely related or even phylogenetically intermixed (as they are in the Firmicutes phylum)17. This supports the hypothesis that the sporulation process in a primordial cell may have led to both the Gram-positive and Gram-negative cell plans.
Evolutionary implications
As the number of sequenced genomes has increased, more and more sophisticated phylogenetic analyses of bacteria have been possible45–48. Although this has made the relationships between phyla increasingly clear, unfortunately there is no agreement on how to root the tree of life. Therefore, it remains a mystery as to which modern species most closely resembles the last universal common ancestor49. To explore the evolutionary implications of our hypothesis we consider three different roots for the tree of life: between Archaea and Bacteria50,51 (FIG. 4a; although historically most favoured, it is not supported by recent findings)52; at the Chloroflexi phylum53,54 (FIG. 4b); and at the Firmicutes phylum45,55 (FIG. 4c; the last two roots are suggested by current methodologies in systematics). In each case we use a recently published and comprehensive tree of life46 to assert the relationships between the established phyla for which the envelope architecture is known. By simply rooting the tree of life in different places, we map the gains and losses for the ability to sporulate and the presence of an outer membrane in a way that minimizes the number of evolutionary events required (FIG. 4).
Figure 4. Rooted phylogenetic trees that represent relationships between bacterial phyla.
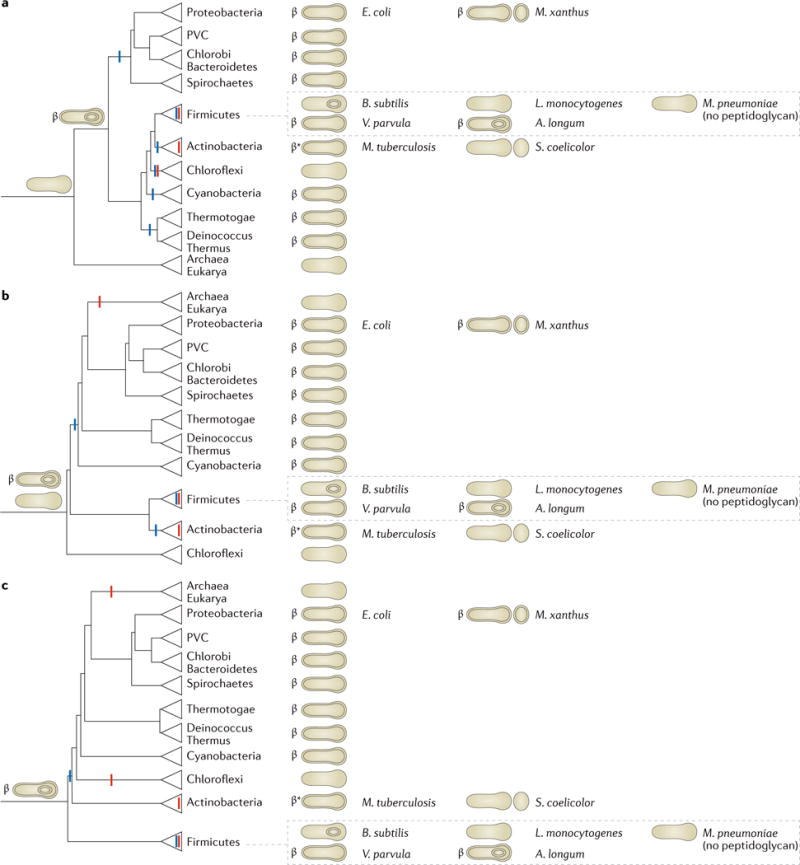
Three different schematic reproductions are shown of a recent phylogenetic tree of life based on 42 gene markers that are conserved throughout Bacteria and Archaea rooted in three different ways, following leading proposals in the field45,50,51,53–55. Cell envelope structures that are present in members of these phyla are shown on the right. Reported abilities to sporulate are represented by small circles within the cells (for endospores) or next to them (for exospores). The presence of the conserved Omp85 or Omp87 is denoted by the ‘β’ symbol and the presence of other conserved outer membrane proteins (OMPs) is denoted by the ‘β*’ symbol. The deduced basic cell plans of ancient ancestors are depicted on early branches. Losses of sporulation (red) and the outer membrane (blue) are marked, which together with the hypothesized cell plans of ancient ancestors minimize the number of evolutionary events that are required to explain the modern distributions of cell plans and sporulation abilities. In all cases, an endospore-forming diderm preceded all or at most one major branch point. A. longum, Acetonema longum; B. subtilis, Bacillus subtilis; E. coli, Escherichia coli; L. monocytogenes, Listeria monocytogenes; M. pneumoniae, Mycoplasma pneumoniae; M. tuberculosis, Mycobacterium tuberculosis; M. xanthus, Myxococcus xanthus; PVC, Planctomycetes–Verrucomicrobia–Chlamydiae; S. coelicolor, Streptomyces coelicolor; V. parvula, Veillonella parvula.
We begin with the first scenario (that the root of the tree of life lies between the three major kingdoms), as suggested by Gogarten and Woese50,51. Mapping the basic cell envelope structures of different species to the right of the corresponding tree (FIG. 4a) presents an interesting surprise: the monoderm phyla (Firmicutes, Tenericutes, Actinobacteria and Chloroflexi) are surrounded by diderm phyla. In addition, the Firmicutes and Actinobacteria phyla comprise both monoderm and diderm species. Possible explanations for this observation are that the diderm cell plan evolved several times independently or was horizontally transferred. However, as OMPs are homologous among all diderm phyla and the assembly, structure and function of the outer membrane depend on hundreds of genes, we propose that the last bacterial common ancestor was a diderm. The diversity of known phyla could then be explained by losses of the outer membrane and/or sporulation properties: members of diderm phyla, such as Proteobacteria, Deinococcus–Thermus, Thermotogae, Cyanobacteria, Spirochaetes, Chlorobi–Bacteriodetes and Planctomycetes–Verrucomicrobia–Chlamydiae (PVC superphylum), lost their ability to sporulate but retained their outer membrane, which explains why they all share common β-barrel OMPs2,10. In the Firmicutes phylum, most bacilli and clostridia (such as B. subtilis and Clostridium difficile) retained their ability to form endospores but discarded their outer membrane in the vegetative state, perhaps for reasons of increased efficiency. Other clostridia, such as A. longum, retained both properties, whereas others species such as Veillonella parvula lost the ability to sporulate but retained an outer membrane. Members of the Chloroflexi and most members of the Firmicutes (such as Listeria monocytogenes) lost both the ability to sporulate and their outer membrane, as did members of the Tenericutes (Mycoplasma spp.), which further discarded their peptidoglycan (as in FIG. 2).
The Actinobacteria phylum presents a particularly interesting case. Some actinobacteria such as Streptomyces coelicolor are monoderm. Other actinobacteria such as mycobacteria are diderm, but have a unique outer membrane that is linked to a thin layer of peptidoglycan by a network of arabinogalactan56,57. The lipid composition of the outer membrane is rich in mycolic acids, which is a notable difference from the outer membrane of members of the Proteobacteria phylum. Although different lipids comprise the outer membrane in mycobacteria and no omp85 or omp87 genes (which encode homologues of the conserved outer-membrane protein BamA) have been identified, a mycobacterial OmpA homologue has been found58, and bioinformatics and experimental approaches have identified over 100 putative OMPs59 that have β-barrel structure60–62. Again, this suggests that members of the Actinobacteria phylum descend from an ancient ancestor with an outer membrane and established OMPs but that the outer-membrane lipids in the mycobacterial branch of the phylum were exchanged for mycolic acids and there were accompanying adaptations. Alternatively, of course, the mycolic envelope is different enough that it may have evolved independently and received conserved OMPs through horizontal gene transfer. In either case, if the tree is rooted between kingdoms, the distribution of diderms throughout the rest of the tree suggests that the last bacterial common ancestor was a sporulating diderm (FIG. 4a).
The second evolutionary scenario we will consider arises from the proposals of Cavalier-Smith and Valas and Bourne40,41,63 that modern members of the Chloroflexi phylum are most closely related to the root of the tree of life (FIG. 4b). In this scenario, the Chloroflexi may have never been diderm64, but for similar reasoning as above (diderms are not monophyletic in this tree, and it is unlikely that the diderm cell plan evolved more than once or was laterally transferred), we hypothesize that a very early sporulation process led to a diderm that is the last common ancestor of all other cells.
A third scenario (FIG. 4c) is that modern members of the Firmicutes phylum are closest to the root of the tree of life (Lake and Bork45,55). Although the Firmicutes phylum is, by sequence analysis, monophyletic (all branches are clearly more related to each other than any other bacterial branch), five diverse cell forms are seen within the phylum (monoderm sporulating, monoderm non-sporulating, diderm sporulating, diderm non-sporulating and monoderm non-sporulating without a cell wall). If the monoderm members of the Firmicutes were directly related to a monoderm last bacterial common ancestor, but the diderm members of the Firmicutes descended from a different branch that also gave rise to all other diderm phyla, these two parts of the Firmicutes phylum would not cluster together. Thus, if the diderm plan evolved only once and was not later horizontally transferred, it must have preceded the branching of the Firmicutes phylum, which leads to the conclusion that the last universal common ancestor was most likely a sporulating diderm. The diversity seen within the Firmicutes phylum could then be explained by losses, again as in FIG. 2.
Other theories have been put forward over the years. For example, without proposing a mechanism whereby the outer membrane first arose, Gupta suggested that current monoderm phyla never went through a diderm stage — the Chloroflexi (monoderms) and Deinococcus–Thermus (diderms that lack LPS) phyla branched before all of the traditional LPS-containing Gram-negative phyla, and Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia obtained their outer membranes by lateral gene transfer or independent evolution12. However, phylogenetic analysis suggests that the OMPs and LPS in A. longum are ancient and are therefore not likely the result of recent gene transfer10. The sequence homology of these features with those in traditional Gram-negative bacteria such as E. coli further suggests a common ancestor and argues against convergent evolution. Finally, the Chloroflexi (monoderm) and Deinococcus–Thermus phyla do not cluster together in the most recent phylogenetic trees46, but rather are separated by LPS-containing diderms, which raises further problems with the theory of early branching of monoderm phyla without a diderm stage. Other theories also depend on the notion that the outer membrane could have evolved twice — for example, once leading to the majority of known diderms and once leading to the formation of the mycolate membrane65. Considering the need for hundreds of genes to encode all of the components of a membrane and their correct assembly, a single ancient origin seems more likely66.
Future work on cell envelope structure, phylogenetic relationships between established phyla, and the discovery and characterization of new phyla will enable further evaluation of these hypotheses67,68. In fact, a substantially expanded tree of life was recently published69. Although it highlights the rich diversity of life that remains to be discovered and characterized, the phylogenetic analysis was not optimized to discern ancient relationships of well-characterized phyla as was the analysis by Raymann et al.46; therefore, it remains more appropriate for us to use the tree by Raymann et al. for the construction of rooted phylogenetic trees that represent relationships between bacterial phyla (FIG. 4). However, both trees show that neither the monoderm nor diderm species are monophyletic. Thus, if the diderm cell plan evolved only once, and was not passed by horizontal transfer across phyla, the last common ancestor of all branches with diderm members must have been a diderm, and the monoderm members of those same branches are best explained by the loss of the ancestral outer membrane. The future discovery of even more new phyla will not change that — it will only add new branches.
Conclusions
In summary, we have presented the hypothesis that the diderm cell plan arose from a sporulation-like event. We have reviewed the images of sporulating monoderm and diderm species and Gram-negative and Gram-positive cell walls that support the hypothesis. We have considered the implications of this hypothesis in light of the increasingly well-characterized phylogenetic tree of life. Although the root of the tree is still unknown, there are two conclusions that can be made regardless of which root is chosen: the first being that most, if not all, monoderms have not always been monoderm, but rather are the result of a diderm losing its outer membrane; and the second being that sporulation and the diderm cell plan are extremely ancient, preceding all or at most one branch point in the tree of life.
The history of the early Earth remains unclear, but we argue here that one of the earliest common ancestors of all cellular life formed endospores. Interestingly, spores have long been recognized as exquisitely robust life forms that are known to be able to withstand all kinds of environmental insults, such as extremes of temperature and pH and dehydration70. One possible explanation for our model is that during the cataclysmic early conditions on Earth, cellular evolution went through a bottleneck in which only robust spores survived; therefore, the last universal common ancestor was a spore. An alternative explanation is panspermia71: life arrived on Earth as a spore, for example, in one of the frozen comets that some theorize created the oceans. As the Earth became more clement, for efficiency, major branches of the bacterial tree of life lost the ability to sporulate, which resulted in numerous diderm non-sporulating phyla today. Other explanations are also obviously possible.
In any case, our analysis at least challenges the notion that complexity gradually increases through evolutionary time: instead it is a case of high complexity (a diderm endospore-forming cell) existing very early, followed by billions of years of losses with comparatively minor modifications such as the exchange of mycolic acids for LPS in the outer membrane of mycobacteria or the evolution of LPS from diderms that lack LPS. This is in agreement with other recent evidence that complexity increased rapidly in early life on Earth followed by a long period of simplification72. However, we acknowledge that it will be difficult, if not impossible, to test these speculations, and that some investigators believe that genes have been swapped so frequently and in such numbers across so many species that trees of life do not make sense73. Nevertheless, we believe our idea that the outer membrane arose through a sporulation event is intriguing, and that if it happened in this way, parsimony argues it must have been a very ancient, perhaps even initiatory, event.
Acknowledgments
Cryotomography in the Jensen laboratory is supported, in part, by the US National Institutes of Health (including grant RO1 GM101425), the Howard Hughes Medical institute (HHMI) and the Beckman Institute at California Institute of Technology (Caltech), Pasadena, USA. Further support for this publication came from the John Templeton Foundation. The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation. The authors thank C. Cleland, T. Nordheim, L. Brengman, K. Willford and J. Eisen for helpful discussions about the early Earth and possible roots of the tree of life.
Footnotes
Competing interests statement
The authors declare no competing interests.
Contributor Information
Elitza I. Tocheva, Department of Stomatology and the Department of Biochemistry and Molecular Medicine, Université de Montréal, P. O. Box 6128 Station Centre-Ville, Montreal, Québec H3C 3J7, Canada
Davi R. Ortega, Department of Biology and Biological Engineering, California Institute of Technology, 1200 East California Boulevard, Pasadena, California 91125, USA
Grant J. Jensen, Howard Hughes Medical Institute, Department of Biology and Biological Engineering, California Institute of Technology, 1200 East California Boulevard, Pasadena, California 91125, USA
References
- 1.Gram C. Ueber die isolirte firbung der schizomyceten iu schnitt-und trockenpriparate. Fortschitte Med. 1884;2:185–189. (in German) [Google Scholar]
- 2.Sutcliffe IC. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 2010;18:464–470. doi: 10.1016/j.tim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Gupta RS. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin W, Baross J, Kelley D, Russell MJ. Hydrothermal vents and the origin of life. Nat Rev Microbiol. 2008;6:805–814. doi: 10.1038/nrmicro1991. [DOI] [PubMed] [Google Scholar]
- 6.Gupta RS, Golding GB. Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- 7.Koch AL. Were Gram-positive rods the first bacteria? Trends Microbiol. 2003;11:166–170. doi: 10.1016/s0966-842x(03)00063-5. [DOI] [PubMed] [Google Scholar]
- 8.Lake JA, Herbold CW, Rivera MC, Servin JA, Skophammer RG. Rooting the tree of life using nonubiquitous genes. Mol Biol Evol. 2007;24:130–136. doi: 10.1093/molbev/msl140. [DOI] [PubMed] [Google Scholar]
- 9.Skophammer RG, Servin JA, Herbold CW, Lake JA. Evidence for a Gram-positive, eubacterial root of the tree of life. Mol Biol Evol. 2007;24:1761–1768. doi: 10.1093/molbev/msm096. [DOI] [PubMed] [Google Scholar]
- 10.Tocheva EI, et al. Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell. 2011;146:799–812. doi: 10.1016/j.cell.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swithers KS, Fournier GP, Green AG, Gogarten JP, Lapierre P. Reassessment of the lineage fusion hypothesis for the origin of double membrane bacteria. PLoS ONE. 2011;6:e23774. doi: 10.1371/journal.pone.0023774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta RS. Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie Van Leeuwenhoek. 2011;100:171–182. doi: 10.1007/s10482-011-9616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lake JA. Evidence for an early prokaryotic endosymbiosis. Nature. 2009;460:967–971. doi: 10.1038/nature08183. [DOI] [PubMed] [Google Scholar]
- 14.Kay D, Warren SC. Sporulation in Bacillus subtilis. Morphological changes. Biochem J. 1968;109:819–824. doi: 10.1042/bj1090819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchandin H, et al. Negativicoccus succinicivorans gen. nov., sp nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. nov. and Acidaminococcaceae fam. nov. in the bacterial phylum Firmicutes. Int J Syst Evol Microbiol. 2010;60:1271–1279. doi: 10.1099/ijs.0.013102-0. [DOI] [PubMed] [Google Scholar]
- 17.Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kane MD, Breznak J. A Acetonema longum gen. nov. sp. nov., an H2/CO2 acetogenic bacterium from the termite, Pterotermes occidentis. Arch Microbiol. 1991;156:91–98. doi: 10.1007/BF00290979. [DOI] [PubMed] [Google Scholar]
- 19.Campbell C, Sutcliffe IC, Gupta RS. Comparative proteome analysis of Acidaminococcus intestini supports a relationship between outer membrane biogenesis in Negativicutes and Proteobacteria. Arch Microbiol. 2014;196:307–310. doi: 10.1007/s00203-014-0964-4. [DOI] [PubMed] [Google Scholar]
- 20.Mergenhagen SE. Polysaccharide–lipid complexes from Veillonella parvula. J Bacteriol. 1965;90:1730–1734. doi: 10.1128/jb.90.6.1730-1734.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tocheva EI, et al. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013;88:673–686. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan L, Jensen GJ. Electron tomography of cells. Q Rev Biophys. 2012;45:27–56. doi: 10.1017/S0033583511000102. [DOI] [PubMed] [Google Scholar]
- 23.Errington J. L-form bacteria, cell walls and the origins of life. Open Biol. 2013;3:120143. doi: 10.1098/rsob.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vollmer W. Bacterial outer membrane evolution via sporulation? Nat Chem Biol. 2011;8:14–18. doi: 10.1038/nchembio.748. [DOI] [PubMed] [Google Scholar]
- 25.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Verwer RW, Nanninga N, Keck W, Schwarz U. Arrangement of glycan chains in the sacculus of Escherichia coli. J Bacteriol. 1978;136:723–729. doi: 10.1128/jb.136.2.723-729.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dmitriev BA, et al. Tertiary structure of bacterial murein: the scaffold model. J Bacteriol. 2003;185:3458–3468. doi: 10.1128/JB.185.11.3458-3468.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollmer W, Holtje JV. The architecture of the murein (peptidoglycan) in Gram-negative bacteria: vertical scaffold or horizontal layer(s)? J Bacteriol. 2004;186:5978–5987. doi: 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci USA. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 32.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPherson DC, Driks A, Popham DL. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J Bacteriol. 2001;183:6046–6053. doi: 10.1128/JB.183.20.6046-6053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 35.Foster S, Popham D. In: Bacillus subtilis And Its Close Relatives: From Genes to Cells. Sonenshein AL, Hoch JA, Losick R, editors. American Society for Microbiology; 2002. pp. 21–41. [Google Scholar]
- 36.Verwer RW, Nanninga N. Electron microscopy of isolated cell walls of Bacillus subtilis var. niger. Arch Microbiol. 1976;109:195–197. doi: 10.1007/BF00425135. [DOI] [PubMed] [Google Scholar]
- 37.Dominguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 38.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc Natl Acad Sci USA. 2008;105:14603–14608. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beeby M, Gumbart JC, Roux B, Jensen GJ. Architecture and assembly of the Gram-positive cell wall. Mol Microbiol. 2013;88:664–672. doi: 10.1111/mmi.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer P, Gutierrez J, Pogliano K, Dworkin J. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol Microbiol. 2010;76:956–970. doi: 10.1111/j.1365-2958.2010.07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Pedro MA, Cava F. Structural constraints and dynamics of bacterial cell wall architecture. Front Microbiol. 2015;6:449. doi: 10.3389/fmicb.2015.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoiczyk E, Baumeister W. Envelope structure of four gliding filamentous cyanobacteria. J Bacteriol. 1995;177:2387–2395. doi: 10.1128/jb.177.9.2387-2395.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desmarais SM, De Pedro MA, Cava F, Huang KC. Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly. Mol Microbiol. 2013;89:1–13. doi: 10.1111/mmi.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 46.Raymann K, Brochier-Armanet C, Gribaldo S. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci USA. 2015;112:6670–6675. doi: 10.1073/pnas.1420858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puigbo P, Wolf YI, Koonin EV. Seeing the Tree of Life behind the phylogenetic forest. BMC Biol. 2013;11:46. doi: 10.1186/1741-7007-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lake JA, Skophammer RG, Herbold CW, Servin JA. Genome beginnings: rooting the tree of life. Philos Trans R Soc Lond B Biol Sci. 2009;364:2177–2185. doi: 10.1098/rstb.2009.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gogarten JP, et al. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gouy R, Baurain D, Philippe H. Rooting the tree of life: the phylogenetic jury is still out. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140329. doi: 10.1098/rstb.2014.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cavalier-Smith T. Rooting the tree of life by transition analyses. Biol Direct. 2006;1:19. doi: 10.1186/1745-6150-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valas RE, Bourne PE. Structural analysis of polarizing indels: an emerging consensus on the root of the tree of life. Biol Direct. 2009;4:30. doi: 10.1186/1745-6150-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lake JA, Sinsheimer JS. The deep roots of the rings of life. Genome Biol Evol. 2013;5:2440–2448. doi: 10.1093/gbe/evt194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nigou J, Gilleron M, Brando T, Puzo G. Structural analysis of mycobacterial lipoglycans. Appl Biochem Biotechnol. 2004;118:253–267. doi: 10.1385/abab:118:1-3:253. [DOI] [PubMed] [Google Scholar]
- 57.Uenishi Y, Fujita Y, Kusunose N, Yano I, Sunagawa M. Comprehensive analysis of mycolic acid subclass and molecular species composition of Mycobacterium bovis BCG Tokyo 172 cell wall skeleton (SMP-105) J Microbiol Methods. 2008;72:149–156. doi: 10.1016/j.mimet.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 58.Senaratne RH, et al. Expression of a gene for a porin-like protein of the OmpA family from Mycobacterium tuberculosis H37Rv. J Bacteriol. 1998;180:3541–3547. doi: 10.1128/jb.180.14.3541-3547.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H. Mycobacterial outer membranes: in search of proteins. Trends Microbiol. 2010;18:109–116. doi: 10.1016/j.tim.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hillmann D, Eschenbacher I, Thiel A, Niederweis M. Expression of the major porin gene mspA is regulated in Mycobacterium smegmatis. J Bacteriol. 2007;189:958–967. doi: 10.1128/JB.01474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis. 2008;88:526–544. doi: 10.1016/j.tube.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Remmert M, Biegert A, Linke D, Lupas AN, Soding J. Evolution of outer membrane β-barrels from an ancestral ββ hairpin. Mol Biol Evol. 2010;27:1348–1358. doi: 10.1093/molbev/msq017. [DOI] [PubMed] [Google Scholar]
- 63.Cavalier-Smith T. The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life. Cold Spring Harb Perspect Biol. 2014;6:a016006. doi: 10.1101/cshperspect.a016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sutcliffe IC. Cell envelope architecture in the Chloroflexi: a shifting frontline in a phylogenetic turf war. Environ Microbiol. 2011;13:279–282. doi: 10.1111/j.1462-2920.2010.02339.x. [DOI] [PubMed] [Google Scholar]
- 65.Houben EN, Korotkov KV, Bitter W. Take five — Type VII secretion systems of Mycobacteria. Biochim Biophys Acta. 2014;1843:1707–1716. doi: 10.1016/j.bbamcr.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 66.Lombard J, Lopez-Garcia P, Moreira D. The early evolution of lipid membranes and the three domains of life. Nat Rev Microbiol. 2012;10:507–515. doi: 10.1038/nrmicro2815. [DOI] [PubMed] [Google Scholar]
- 67.Brown CT, et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 68.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 69.Hug LA, et al. Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages. Environ Microbiol. 2016;18:159–173. doi: 10.1111/1462-2920.12930. [DOI] [PubMed] [Google Scholar]
- 70.Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 71.Nicholson WL. Ancient micronauts: interplanetary transport of microbes by cosmic impacts. Trends Microbiol. 2009;17:243–250. doi: 10.1016/j.tim.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 72.Wolf YI, Koonin EV. Genome reduction as the dominant mode of evolution. Bioessays. 2013;35:829–837. doi: 10.1002/bies.201300037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]


