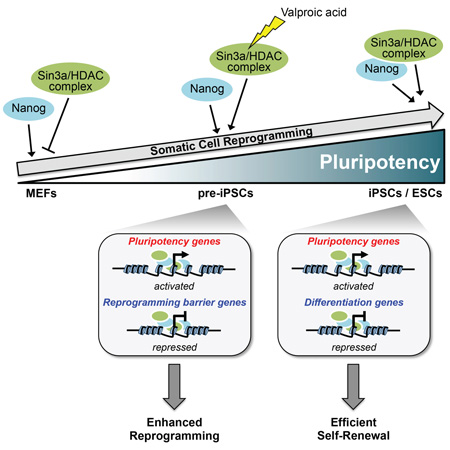
SUMMARY
Although Sin3a is required for survival of early embryos and embryonic stem cells (ESCs), the role of Sin3a in the maintenance and establishment of pluripotency remains unclear. Here we find that the Sin3a/HDAC corepressor complex maintains ESC pluripotency and promotes the generation of induced pluripotent stem cells (iPSCs). Members of the Sin3a/HDAC corepressor complex are enriched in an extended Nanog interactome and function in transcriptional coactivation in ESCs. We also identified a critical role for Sin3a and HDAC2 in efficient reprogramming of somatic cells. Mechanistically, Nanog and Sin3a co-occupy transcriptionally active pluripotency genes in ESCs and also co-localize extensively at their genome-wide targets in pre-iPSCs. Additionally, both factors are required to directly induce a synergistic transcriptional program wherein pluripotency genes are activated and reprogramming barrier genes are repressed. Our findings indicate a transcriptional regulatory role for a major HDAC-containing complex in promoting pluripotency.
Keywords: HDAC1, HDAC2, VPA, pre-iPSCs, iPSCs, EpiSCs, ESCs, Sin3b, Tet1/2
Graphical Abstract
INTRODUCTION
Sin3a is a large scaffold protein containing four paired amphipathic helix (PAH) domains and an internal HDAC interaction domain (HID). The PAH domains recognize and bind to sequence-specific transcriptional regulators such as Mad/Max, and the HID domain is responsible for tethering HDAC1 and HDAC2 (HDAC1/2) to Sin3a to mediate transcriptional repression of Sin3a target genes (Kadamb et al., 2013). Sin3a is required for early embryonic development, the viability of mouse embryonic fibroblasts (MEFs) and embryonic stem cells (ESCs), and the normal development and differentiation of male germ cells and muscle cells (rev. in Kadamb et al. 2013). Nanog is also required for embryonic and germline development as well as efficient ESC self-renewal, and importantly, Nanog is critical for executing the final stage of reprogramming to establish naïve pluripotency (rev. in Saunders et al., 2013).
Previous studies from our group (Costa et al., 2013; Wang et al., 2006) and others (Gagliardi et al., 2013; Liang et al., 2008) have established the physical interactions between Nanog and the Sin3a/HDAC complex in ESCs. These interactions are consistent with the requirement of Sin3a for early development and efficient ESC self-renewal, and also suggest a potential role of Sin3a in the final stage of reprogramming when Nanog is essential, although a detailed mechanistic understanding of this corepressor complex in transcriptional regulation of pluripotency and reprogramming is lacking. HDAC inhibitors have been routinely used to significantly enhance the efficiency of somatic cell nuclear transfer as well as the generation of mouse and human induced pluripotent stem cells (iPSCs) (Kretsovali et al., 2012), although the exact mechanism of action of these inhibitors is not fully understood. The three major HDAC-containing protein complexes, namely the CoREST, NuRD, and Sin3a complexes, are highly abundant in pluripotent stem cells. The CoREST complex has been shown to be critical for establishing and maintaining pluripotency (Yang et al., 2011), and the role of the NuRD complex in establishing pluripotency seems to be highly context-dependent (Rais et al., 2013; Santos et al., 2014). The function of the Sin3a/HDAC complex in somatic cell reprogramming, however, has not been investigated.
In this study, we address the significance of the Nanog/Sin3a functional partnership in stem cell maintenance and somatic cell reprogramming. We first report an increased transcriptional co-activator function of Sin3a, when partnered with Nanog, in ESCs underlying stem cell maintenance. We then document a requirement of Sin3a for reprogramming efficiency in a general context, followed by our findings that Sin3a can further co-localize and synergize with Nanog at the chromatin level to induce a transcriptional program that primes pre-iPSCs for efficient reprogramming.
RESULTS
Nanog and Sin3a co-occupy transcriptionally active promoters in ESCs
We recently expanded our Nanog interactome using enhanced affinity purification techniques, and identified an enrichment of members of the Sin3a/HDAC corepressor complex (Costa et al., 2013) (Figure S1A). We further confirmed that Nanog physically interacts with Sin3a in ESCs (Figure 1A). Although the Nanog-Sin3a partnership in ESCs was also reported in two previous studies (Gagliardi et al., 2013; Liang et al., 2008), the functional significance of this physical relationship in pluripotency has not been defined. To investigate how Sin3a might cooperate with Nanog in transcriptional regulation in ESCs, we analyzed the genome-wide occupancy of Sin3a, Nanog, HDAC1/2, and the chromatin marks H3K4me1, H3K4me3, H3K27ac, and H3K27me3 from the public domain (Table S1). Close inspection of Sin3a peak centers confirmed a substantial enrichment of the active promoter marks H3K4me3 and H3K27ac and a complete lack of the repressive promoter marks H3K4me1 and H3K27me3 (Figure S1B), indicating that Sin3a binds to open chromatin regions that are accessible to the transcriptional machinery in ESCs. In agreement with this, we found that 48% of Sin3a peaks lie directly at the transcription start sites (TSS) of its targets, with an additional 33% of peaks found within promoter regions (± 3 kb of TSS) (Figure S1C). The highest proportion of Nanog peaks was also centered at the TSS of its targets (Figure S1D), suggesting that Sin3a and Nanog may cooperatively regulate the expression of a subset of their common targets in ESCs.
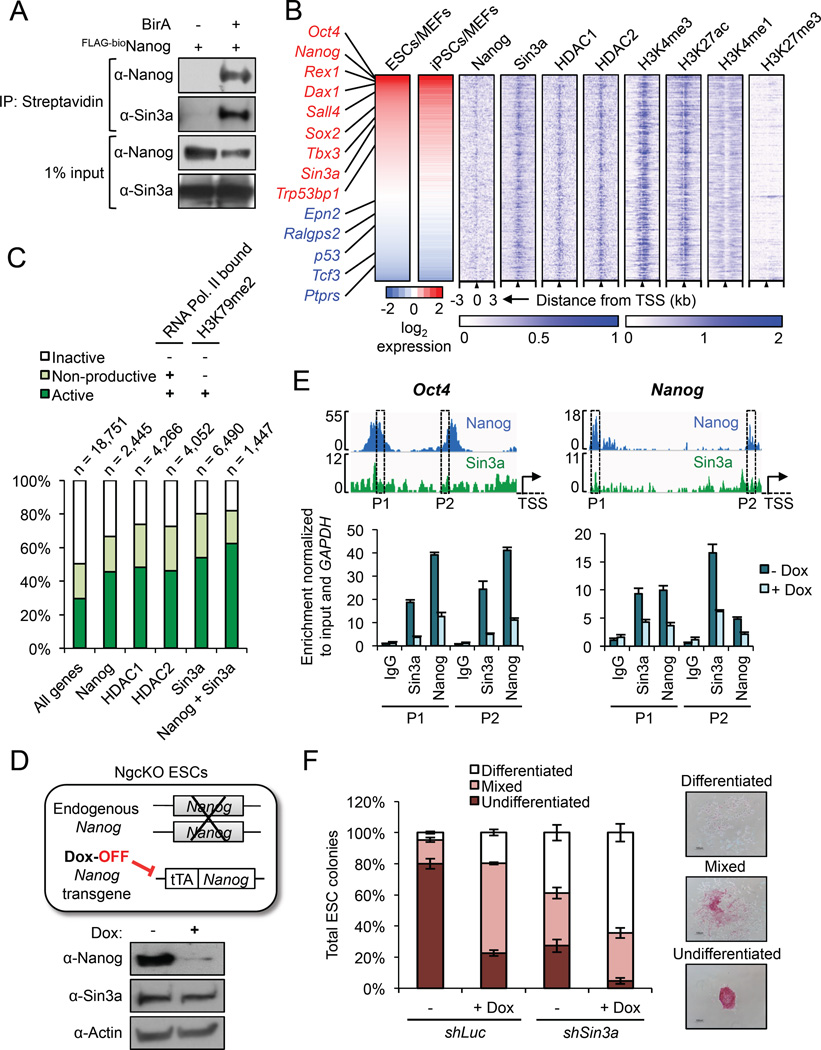
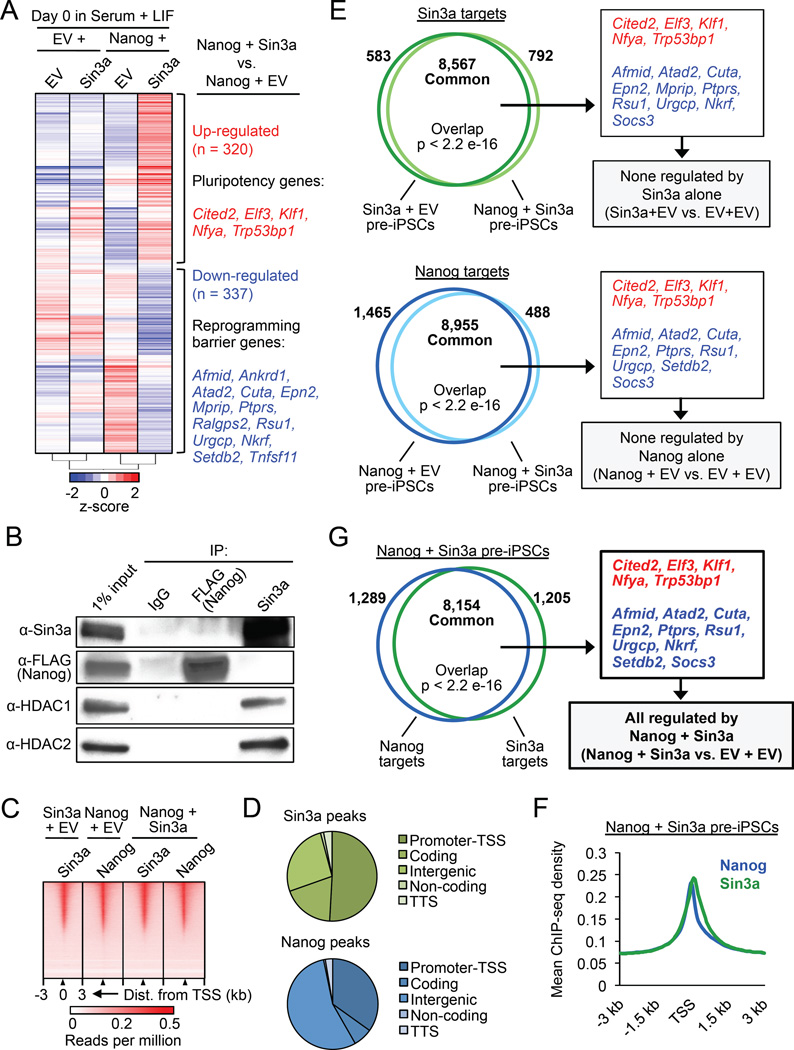
Figure 1. Nanog and Sin3a co-occupy active promoters in ESCs. See also Figure S1.
(A) IP/co-IP between FLAG-bioNanog and endogenous Sin3a in mouse ESCs. BirA = biotin ligase.
(B) Sin3a and Nanog co-bind highly expressed pluripotency genes in ESCs. Shown are all Nanog and Sin3a common target genes in ESCs (n = 1,447) based on ChIP-seq data, sorted by high (red) to low (blue) ESC/MEF expression ratios based on RNA-seq data. Pluripotency genes and reprogramming barrier genes are listed in red and blue text, respectively. Blue heat maps on the right show enrichment of Nanog, the Sin3a/HDAC complex, and four chromatin marks centered on the TSS.
(C) Percentages of active (RNA Pol. II bound + H3K79me2), non-productive (RNA Pol. II bound, no H3K79me2), and inactive (no RNA Pol. II bound, no H3K79me2) genes bound by each factor in ESCs, based on ChIP-seq data. “All genes” = genome-wide average of all genes, and numbers above chart indicate the number of target genes identified and analyzed for each individual factor or combination.
(D) Illustration of Nanog conditional knockout (NgcKO) ESCs, wherein endogenous Nanog is deleted and cells are maintained by a doxycycline (Dox)-suppressible Nanog transgene. Protein expression upon 8-hour Dox treatment (1 µg/mL) is shown.
(E) Nanog-dependent Sin3a binding to common target genes. ChIP-qPCR was performed for Sin3a and Nanog at two different peaks (P1, P2) in the Oct4 and Nanog promoters in NgcKO ESCs. Dox treatment (1 µg/mL) lasted for 8 hours.
(F) Colony formation assay after Sin3a knockdown, individually or combined with Nanog conditional knockout (+Dox), in NgcKO ESCs.
To determine the specific genes that are bound and potentially regulated by Nanog and Sin3a, we identified 1,447 Nanog/Sin3a common target genes from published ESC ChIP-seq data and compared their expression levels across ESCs, iPSCs, and MEFs from published RNA-seq data (Table S1). We found many key pluripotency genes (e.g., Nanog, Oct4, Sox2) among the group of genes with high ESC/MEF and iPSC/MEF expression ratios (labeled red) and several reprogramming barrier genes (e.g., p53, Tcf3, Ralgps2) among the group of genes with low ESC/MEF and iPSC/MEF expression ratios (labeled blue) (Figure 1B, left). Interestingly, we found that the active promoter marks H3K4me3 and H3K27ac were enriched at the TSS of all Nanog/Sin3a common targets, whereas the repressive promoter marks H3K4me1 and H3K27me3 were largely absent (Figure 1B, right).
To further understand the functional significance of Nanog and Sin3a co-occupancy in the ESC genome, we determined the proportions of genes bound by Nanog and/or Sin3a/HDAC complex members that are transcriptionally active (RNA Pol II bound + H3K79me2), nonproductive (RNA Pol II bound, no H3K79me2), or inactive (no RNA Pol II, no H3K79me2). We found that Nanog, Sin3a, and HDAC1/2 bound to higher proportions of active genes compared to the genome-wide average of all genes in ESCs (Figure 1C). More importantly, we found that Nanog and Sin3a co-occupied an even higher proportion of active genes than any of the individual factors examined (Figure 1C). This demonstrates that, in addition to its well-established corepressor functions, the Sin3a/HDAC complex can also act as a co-activator in ESCs. Using doxycycline (Dox)-inducible Nanog null (NgcKO) ESCs (Das et al., 2011) (Figure 1D), we further demonstrated the Nanog-dependent binding of Sin3a to two of their highest expressed common target genes, Oct4 and Nanog (Figure 1E), suggesting that Nanog and Sin3a cooperatively maintain the expression of these genes. This finding is in agreement with a previous report demonstrating a positive role of the Sin3a/HDAC complex on Nanog expression in ESCs (Baltus et al., 2009). Knockdown of Sin3a in these cells also confirmed that Sin3a is required for ESC self-renewal, as Sin3a knockdown individually, or in combination with conditional Nanog knockout (+Dox), resulted in a dramatic reduction in undifferentiated ESC colony formation (Figure 1F). Notably, this Sin3a loss-of-function phenotype may reflect a general defect in cell proliferation and/or viability (Figure S1E), consistent with previous reports describing the requirement of Sin3a for cell cycle progression as well as the derivation and survival of ESCs in culture (Kadamb et al., 2013). Importantly, this self-renewal defect caused by Sin3a knockdown was further exacerbated by Nanog depletion, as shown by the overall reduction of total colony numbers (Figure S1F, “shSin3a + Dox”) as well as the relative reduction and increase in undifferentiated and differentiated colonies, respectively (Figure 1F). Collectively, these results demonstrate that Nanog and the Sin3a/HDAC complex physically and functionally co-operate, and predominantly co-occupy actively transcribed genes in ESCs for their maintenance.
To identify other factors associated with Sin3a and Nanog that might promote the transcriptional co-activation function of this complex, we studied the Nanog (Costa et al., 2013) and Sin3a interactomes. We performed affinity purification of Sin3a protein complexes followed by mass spectrometry in ESCs, and identified 82 high-confidence Sin3a-interacting partners (Table S2), including 24 common interacting partners of Nanog, such as HDAC1/2, Sall4, Ogt, Tet1, and Tet2 (Tet1/2) (Figure S1G). The identification of these common partners is supported by the fact that our group as well as others have previously demonstrated the interactions between Tet1/2 and Nanog (Costa et al., 2013), and between Tet1/2 and Sin3a (McDonel et al., 2012; Williams et al., 2011). These results suggest that these Nanog/Sin3a common interacting partners might play important roles in promoting the functional cooperation between Nanog and Sin3a in activation, and to a lesser extent repression (Figure 1C), of their downstream targets for ESC maintenance.
Sin3a is required for efficient somatic cell reprogramming
To explore a potential role for Sin3a in somatic cell reprogramming, we first examined Sin3a and Nanog expression during MEF reprogramming (Sridharan et al., 2009). We found that Sin3a expression levels gradually increased during reprogramming, especially during the final pre-iPSC to iPSC transition stage, and as expected, Nanog expression was only detectable in iPSCs and ESCs (Figure S2A). This indicated that Sin3a might be limiting during the final stage of reprogramming, when Nanog function becomes critical (Silva et al., 2009). To assess the requirement of Sin3a in the larger context of somatic cell reprogramming, we knocked down Sin3a during OKSM-mediated MEF reprogramming (Vidal et al., 2014) (Figures 2A and S2B). We found that Sin3a knockdown dramatically reduced the efficiency of MEF reprogramming, with a significant reduction (~85%) in the number of alkaline phosphatase (AP)+ iPSC colonies after 10 days of reprogramming (Figure 2B). Similar to what we found in ESCs, this result is likely due to a proliferation defect, as others have reported that loss of Sin3a dramatically reduces MEF proliferation and survival (Cowley et al., 2005). Collectively, these data indicate that Sin3a is required for efficient somatic cell reprogramming by maintaining normal cell proliferation and/or survival during the reprogramming process.
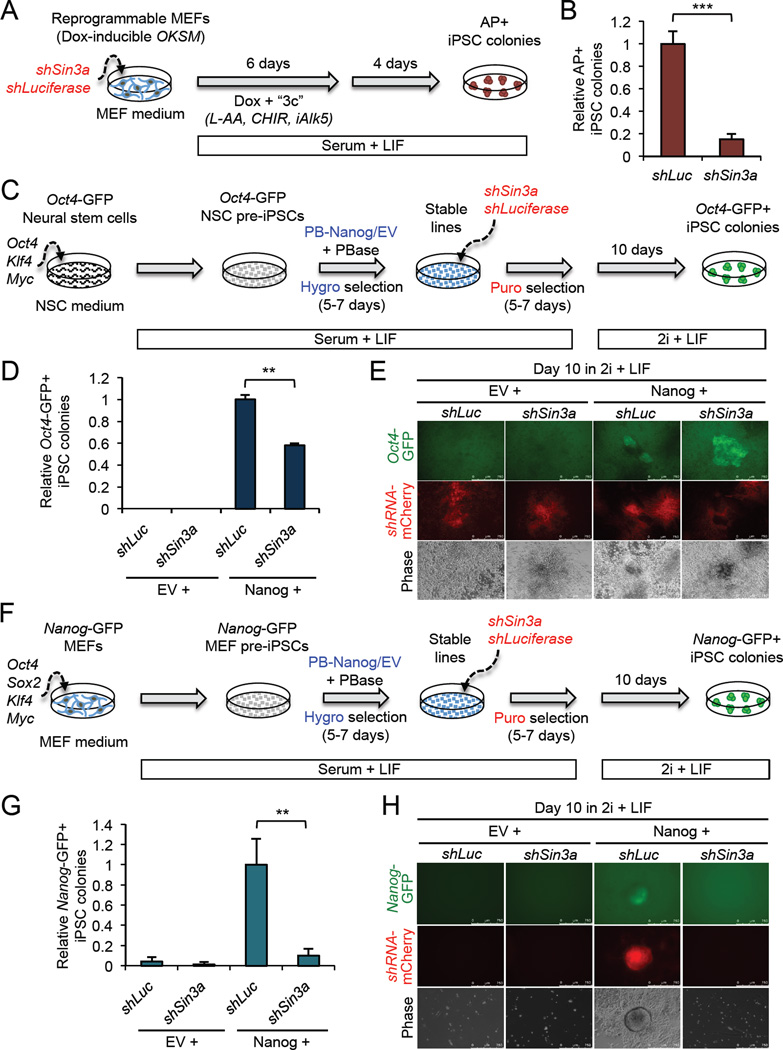
Figure 2. Sin3a is required for efficient somatic cell reprogramming. See also Figure S2.
(A) The procedure for assessing Sin3a knockdown in doxycycline (Dox)-inducible MEF reprogramming.
(B) Sin3a knockdown significantly decreases MEF reprogramming efficiency. Data are presented as average fold change of AP+ iPSC colonies ± SD (n = 3; *** p < 0.001).
(C) The procedure for assessing Sin3a knockdown in neural stem cell (NSC)-derived pre-iPSC reprogramming.
(D) Sin3a knockdown significantly decreases Nanog-mediated NSC pre-iPSC reprogramming efficiency. Data are presented as average fold change of Oct4-GFP+ iPSC colonies ± SD (n = 3; ** p < 0.01).
(E) Representative images of Oct4-GFP+ iPSC colonies.
(F) The procedure for assessing Sin3a knockdown in MEF-derived pre-iPSC reprogramming.
(G) Sin3a knockdown significantly decreases Nanog-mediated MEF pre-iPSC reprogramming efficiency. Data are presented as average fold change of Nanog-GFP+ iPSC colonies ± SD (n = 3; ** p < 0.01).
(H) Representative images of Nanog-GFP+ iPSC colonies.
To address the role of Sin3a in reprogramming in the context of its potential functional connection with Nanog, we first investigated the requirement of Sin3a in the Nanog-driven transition of partially reprogrammed neural stem cells (NSC-derived pre-iPSCs), containing a GFP reporter under the control of the Oct4 distal enhancer (Oct4-GFP), to fully reprogrammed iPSCs (Costa et al., 2013). We tested the effect of loss of Sin3a during reprogramming using a validated short hairpin (sh) RNA (Williams et al., 2011) (Figures 2C and S2C). As expected, cells expressing only empty vector (EV) control did not yield any Oct4-GFP+ iPSC colonies, regardless of Sin3a knockdown. However, in ectopic Nanog expressing cells, Sin3a knockdown resulted in a >40% reduction in Oct4-GFP+ iPSC colonies compared to Luciferase knockdown (shLuc) control (Figures 2D and E), an effect that is likely due to a proliferation defect in shSin3a cells (Figure S2D), and is consistent with the requirement of Sin3a for cell proliferation and survival (Kadamb et al., 2013). Inhibition of Nanog-driven reprogramming by Sin3a knockdown was also observed during the reprogramming of MEF-derived pre-iPSCs containing a Nanog-GFP reporter under the same reprogramming time line and culture conditions (Figures 2F–H). Moreover, we found that Sin3a knockdown also compromised Esrrb-mediated pre-iPSC reprogramming (Festuccia et al., 2012) (Figures S2E–G) due similarly to a proliferation defect of pre-iPSCs upon Sin3a loss (Figure S2H).
These results together demonstrate that Sin3a is required for efficient somatic cell reprogramming, most likely through its requisite functions in maintaining normal cell proliferation and/or viability, irrespective of transgene expression.
Sin3a over-expression promotes reprogramming in a context-dependent manner
To elucidate a potential function of Sin3a in reprogramming beyond its requirement for cell proliferation and/or survival, we tested the effect of over-expressing Sin3a during reprogramming. To do this, we established stable NSC-derived pre-iPSC lines ectopically expressing either Nanog or Sin3a alone, or both Nanog and Sin3a (Figures 3A and S3A). These pre-iPSCs are negative for Oct4-GFP expression in serum + LIF, and remain so until reprogrammed in 2i + LIF (Figure S3B). Over-expression of Sin3a alone had a minimal effect on reprogramming efficiency compared to EV control. However, when co-expressed with Nanog, Sin3a generated >3-fold more Oct4-GFP+ iPSC colonies compared to Nanog alone (Figures 3B and C), a profound reprogramming synergy that is not due to an increase in cell proliferation (Figure S3C). Upon transposase (PBase)-mediated PiggyBac transgene excision (see Experimental Procedures), we further confirmed the comparable pluripotency statuses of transgene-free Nanog + Sin3a iPSCs and Nanog + EV iPSCs by normal expression of pluripotency and differentiation markers at both RNA and protein levels (Figures S3D and E), silencing of retroviral Oct4, Klf4, and Myc transgenes (Figure S3F), reduced H3K27me3 foci in these female NSC pre-iPSCs upon full reprogramming in 2i + LIF (indicative of X chromosome reactivation) (Figure S3G), and multi-lineage differentiation propensities under three independent differentiation protocols (Figure S3H and Table S3). These results demonstrate that iPSCs generated by Nanog + Sin3a over-expression have indistinguishable characteristics to iPSCs generated by Nanog alone that have been validated with bona fide pluripotency (Silva et al., 2009).
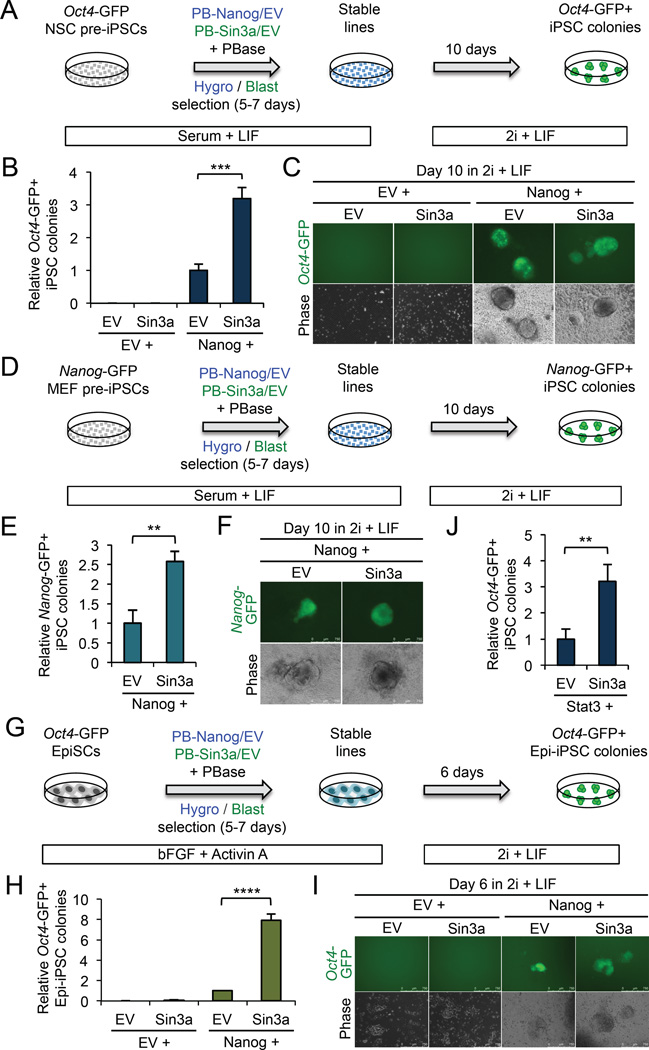
Figure 3. Sin3a over-expression promotes pre-iPSC and EpiSC reprogramming. See also Figure S3.
(A) The procedure for assessing Sin3a over-expression in NSC-derived pre-iPSC reprogramming.
(B) Sin3a can synergize with Nanog to significantly increase NSC-derived pre-iPSC reprogramming efficiency. Data are presented as average fold change of Oct4-GFP+ iPSC colonies ± SD (n = 3; *** p < 0.001).
(C) Representative images of Oct4-GFP+ iPSC colonies.
(D) The procedure for assessing Sin3a over-expression in MEF-derived pre-iPSC reprogramming.
(E) Sin3a can synergize with Nanog to significantly increase MEF pre-iPSC reprogramming efficiency. Data are presented as average fold change of Nanog-GFP+ iPSC colonies ± SD (n = 3; ** p < 0.01)
(F) Representative images of Nanog-GFP+ iPSC colonies.
(G) The procedure for assessing Sin3a over-expression in EpiSC reprogramming.
(H) Sin3a can synergize with Nanog to significantly increase EpiSC reprogramming efficiency. Data are presented as average fold change of Oct4-GFP+ Epi-iPSC colonies ± SD (n = 3; **** p < 0.0001).
(I) Representative images of Oct4-GFP+ Epi-iPSC colonies.
(J) Sin3a can synergize with Stat3 to significantly increase NSC-derived pre-iPSC reprogramming efficiency. Data are presented as average fold change of Oct4-GFP+ iPSC colonies ± SD (n = 3; ** p < 0.01)
The reprogramming synergy between Nanog and Sin3a was also observed in MEF-derived pre-iPSCs (Figures 3D–F) and in OEC-2 epiblast stem cells (EpiSCs) (Guo et al., 2009) (Figures 3G–I), yielding ~2.5-fold and 8-fold more iPSC colonies, respectively, by Nanog + Sin3a than by Nanog alone during reprogramming. Surprisingly, however, MEF reprogramming performed with combinations of OKSM plus retroviral Nanog, Sin3a, and/or HDAC2 revealed that ectopic Sin3a or HDAC2, with or without Nanog, significantly reduced MEF reprogramming efficiency (Figure S3I). While the exact mechanism underlying similar inhibitions of MEF reprogramming upon Sin3a knockdown (Figure 2B) and over-expression (Figure S3I) is unclear (see Discussion), our results nonetheless highlight a stage-specific effect of Sin3a over-expression in promoting reprogramming. In particular, Sin3a facilitates Nanog action in driving partially reprogrammed pre-iPSCs and primed pluripotent EpiSCs to full pluripotency.
Nanog and Sin3a induce parallel transcriptional programs that enhance reprogramming
To determine how Nanog and Sin3a may co-regulate gene expression in promoting pre-iPSC reprogramming, we performed microarray analysis on pre-iPSCs expressing Nanog and/or Sin3a. By examining significant global transcriptional changes (p < 0.05, fold change ≥ 1.5) immediately preceding the onset of reprogramming, we identified 320 up-regulated and 337 down-regulated genes in Nanog + Sin3a pre-iPSCs relative to Nanog + EV pre-iPSCs (Figure 4A). We identified up-regulated genes implicated in pluripotency and reprogramming including Cited2 (Kranc et al., 2015), Elf3 (Park et al., 2014), Klf1 (Nakagawa et al., 2008), Nfya (Dolfini et al., 2012), and Trp53bp1 (Marión et al., 2009) (Figure 4A, red text), and also identified several down-regulated genes known to act as barriers during the reprogramming process (Qin et al., 2014; Sakurai et al., 2014; Yang et al., 2014) (Figure 4A, blue text). We further validated the differential expression of a number of these reprogramming promoting and barrier genes by qRT-PCR (Figure S4A). In addition to these reprogramming barrier genes, we also found that Socs3, a negative regulator of the Jak/Stat3 pathway that was previously reported to be repressed by Nanog in ESCs and during reprogramming (Stuart et al., 2014), was further significantly down-regulated by Nanog + Sin3a (Figure S4B). Interestingly, we also observed a >3-fold reprogramming synergy by Stat3 + Sin3a (Figure 3J), similar to the synergy we observed for Nanog + Sin3a (Figure 3B). In contrast, we did not observe any reprogramming synergy by Esrrb + Sin3a or Klf2 + Sin3a (data not shown). These data suggest that the Jak/Stat3 signaling pathway, which is limiting during the final stage of reprogramming (van Oosten et al., 2012; Yang et al., 2010), may be involved in the Sin3a action in promoting the final stage of reprogramming to pluripotency, likely via its direct binding to Socs3 (see below) and transcriptional repression (Figure S4B). Collectively, these results demonstrate that co-expression of Nanog and Sin3a leads to the activation of pluripotency genes and the repression of reprogramming barrier genes, including Socs3, which primes pre-iPSCs for significantly enhanced reprogramming efficiency.
Figure 4. Nanog and Sin3a directly activate pluripotency genes and repress reprogramming barrier genes in pre-iPSCs. See also Figure S4.
(A) Microarray heat maps of significantly differentially expressed genes (p < 0.05, fold change ≥ 1.5) in NSC pre-iPSCs. Genes in red and blue are up- and down-regulated, respectively, in Nanog + Sin3a pre-iPSCs relative to Nanog + EV pre-iPSCs (right).
(B) IP/co-IP for 3×FLAGNanog, Sin3a, HDAC1, and HDAC2 in Nanog + Sin3a NSC pre-iPSCs.
(C) Heat maps for genome-wide binding of Nanog and/or Sin3a in indicated pre-iPSC populations.
(D) Genome-wide distribution of Nanog and Sin3a peaks in Nanog + Sin3a pre-iPSCs. Promoter-TSS = −1 kb to +100 bp from transcription start site (TSS), coding = exons, TTS = −100 bp to +1 kb from transcription termination site (TTS).
(E) Venn diagrams showing significantly overlapping Sin3a targets (top) and Nanog targets (bottom) in indicated pre-iPSC populations, as well as microarray genes of interest contained within common target sets (right).
(F) Average ChIP-seq read density for Nanog and Sin3a in Nanog + Sin3a pre-iPSCs.
(G) Venn diagram showing significant overlap of Nanog and Sin3a targets in Nanog + Sin3a pre-iPSCs (left), as well as microarray genes that are directly regulated by Nanog + Sin3a (right).
To explore whether Nanog and Sin3a may act together in a similar fashion as that in ESCs (Figure 1) to co-regulate the activity of these differentially expressed genes in promoting reprogramming, we first performed reciprocal immunoprecipitation/co-immunoprecipitation (IP/co-IP) experiments for Nanog and Sin3a in Nanog + Sin3a pre-iPSCs. Not unexpectedly, we confirmed the interaction of Sin3a with HDAC1/2 in pre-iPSCs, indicating the preservation of the Sin3a/HDAC complex in pre-iPSCs (Figure 4B). Surprisingly, however, we found that Nanog and Sin3a did not physically interact in pre-iPSCs (Figure 4B), suggesting that other co-factors such as pluripotency transcription factors and/or chromatin binding may be required to mediate their observed association in ESCs (Figure 1A) or reprogramming synergy in pre-iPSCs (Figure 3).
To explore whether Nanog and Sin3a, while lacking direct physical association (Figure 4B), cooperate to regulate the expression of genes of interest (Figure 4A) at the chromatin level, we performed ChIP-seq experiments to determine the genome-wide occupancy of Nanog and Sin3a in pre-iPSCs. We found that Nanog and Sin3a were heavily enriched at the TSS of their targets, with both factors also binding chromatin to a large extent within exons and intergenic regions (Figures 4C and D). We detected a significant overlap of Sin3a targets in Sin3a + EV and Nanog + Sin3a pre-iPSCs (Figure 4E, top), and a similar overlap of Nanog targets in Nanog + EV and Nanog + Sin3a pre-iPSCs (Figure 4E, bottom), indicating that no major reorganization of either Nanog or Sin3a binding occurs when the other factor is co-expressed. Interestingly, we found that Nanog and Sin3a co-bound a significant proportion (~86%) of target genes within 3 kb of their TSS in Nanog + Sin3a pre-iPSCs (Figures 4F and G), and that both factors target almost all microarray genes of interest, including Socs3 (Figures 4G and S4C). We detected a relatively high correlation among ChIP-seq targets for Nanog and Sin3a in Nanog + EV and Nanog + Sin3a pre-iPSCs (Figure S4D), and importantly, almost all microarray genes of interest as well as Socs3 were only significantly up- or down-regulated by the combined expression of Nanog and Sin3a (Figure 4G, right), but not by Sin3a or Nanog alone (Figure 4E, right).
Taken together, these data indicate that Nanog and Sin3a co-localize extensively at their targets to transcriptionally activate pluripotency genes and repress reprogramming barrier genes (Figures 4F–G and S4C), and suggest that Nanog and Sin3a may cooperate within distinct complexes at these loci in pre-iPSCs to elicit parallel transcriptional programs that promote reprogramming.
HDAC2 is critical for Nanog and Sin3a functional cooperation in pre-iPSC reprogramming
To further dissect the mechanistic action of Sin3a in promoting reprogramming, we explored the involvement of HDAC1/2 in the reprogramming-promoting function of Sin3a. By testing a Sin3a mutant lacking HDAC1/2 association (Sin3a ΔHID) in pre-iPSC reprogramming (Figures 5A and S5A), we found that the Sin3a HDAC interaction (HID) domain is required for the reprogramming synergy by Nanog + Sin3a, as deletion of HID completely abrogated the ability of Sin3a to synergize with Nanog during reprogramming (Figures 5B and S5B). Interestingly, the HID domain alone was able to modestly, but significantly, increase the reprogramming efficiency of Nanog, compared to EV; however, it was unable to recapitulate the full reprogramming activity of full-length, wild-type (WT) Sin3a (Figure 5B). These results clearly implicate HDAC1/2 in the functional cooperation between Nanog and Sin3a in enhancing pre-iPSC reprogramming, and suggest that additional factors associated with the Sin3a corepressor must also contribute to the reprogramming synergy of Nanog + Sin3a.
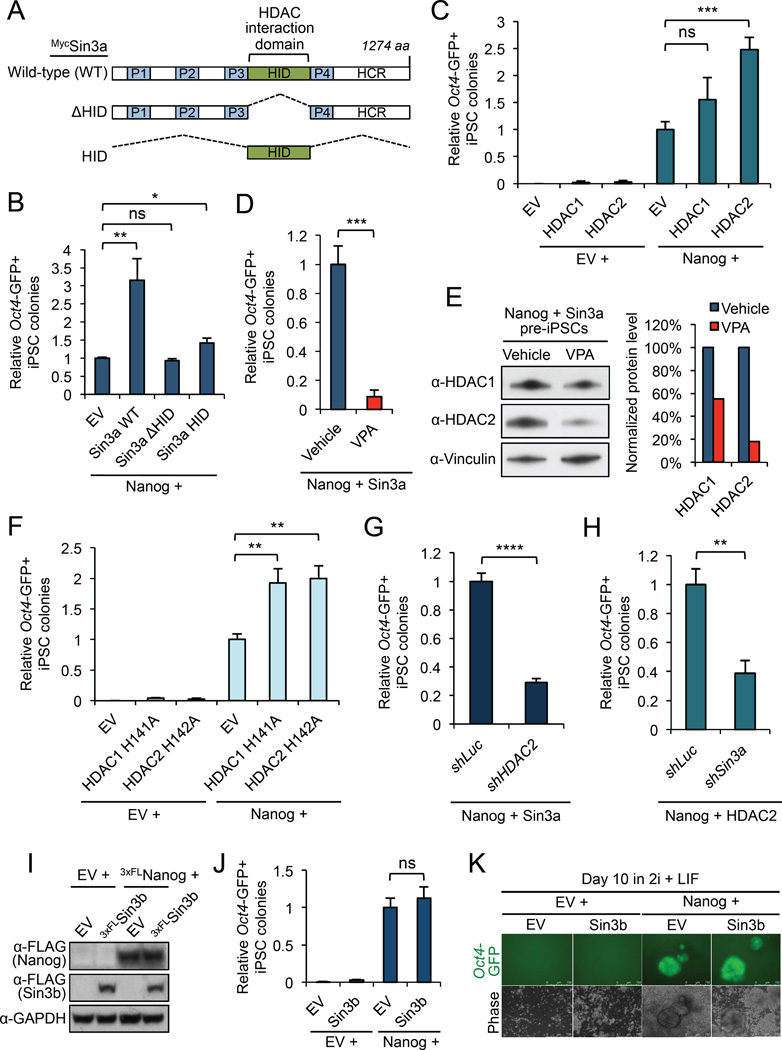
Figure 5. HDAC2 is critical for Nanog/Sin3a functional cooperation in NSC-derived pre-iPSC reprogramming. See also Figure S5.
(A) Myc-tagged Sin3a constructs used for assessing the requirement of HDAC1/2 association for Sin3a function in reprogramming.
(B) The Sin3a HID domain is critical for Sin3a function in synergizing with Nanog during pre-iPSC reprogramming. Data are presented as average fold change of Oct4-GFP+ iPSC colonies ± SD (n = 3; * p < 0.05, ** p < 0.01, ns = not significant).
(C) HDAC2, and to a minimal extent HDAC1, can synergize with Nanog during pre-iPSC reprogramming. Data are presented as average fold change of Oct4-GFP+ iPSC colonies ± SD (n = 3; *** p < 0.001, ns = not significant).
(D) VPA treatment (2 mM) completely abrogates the Nanog/Sin3a reprogramming synergy compared to vehicle treated control. Data are presented as average fold change of Oct4-GFP+ iPSC colonies ± SD (n = 3; *** p < 0.001).
(E) VPA treatment (2 mM; 24 hours) causes degradation of HDAC1/2 proteins. Western blot band intensities were quantified using ImageJ software and are relative to Vinculin loading control. HDAC1/2 protein levels were normalized to a vehicle treated sample.
(F) HDAC1/2 catalytic mutants can synergize with Nanog during pre-iPSC reprogramming. Data are presented as average fold change of Oct4-GFP+ iPSC colonies ± SD (n = 3; ** p < 0.01).
(G) HDAC2 is required for efficient pre-iPSC reprogramming by Nanog + Sin3a. Data are presented as average fold change of Oct4-GFP+ iPSC colonies ± SD (n = 3; **** p < 0.0001).
(H) Sin3a is required for efficient pre-iPSC reprogramming by Nanog + HDAC2. Data are presented as average fold change of Oct4-GFP+ iPSC colonies ± SD (n = 3; ** p < 0.01).
(I) Western blot for pre-iPSCs over-expressing 3×FLSin3b and 3×FLNanog in serum + LIF conditions.
(J) Co-expression of Sin3b and Nanog has no effect on pre-iPSC reprogramming efficiency compared to Nanog alone. Data are presented as average fold change of Oct4-GFP+iPSC colonies ± SD (n = 3; ns = not significant).
(K) Representative images of Oct4-GFP+ iPSC colonies.
We then directly tested HDAC1/2 function in the context of Nanog during the final stage of reprogramming. We found that, when acting together with Nanog, HDAC2, and to a minimal extent HDAC1, could significantly promote pre-iPSC reprogramming, whereas HDAC1 or HDAC2 alone had only minimal effects on reprogramming (Figures 5C and S5C–D). Our data thus far seem contradictory with the well-known function of small molecule HDAC inhibitors such as valproic acid (VPA) in promoting reprogramming of mouse and human fibroblasts (Huangfu et al., 2008a; 2008b). To resolve this issue and further explore a potentially novel function of HDAC1/2 in promoting reprogramming, we tested the effect of VPA on the final stage of reprogramming mediated by Nanog + Sin3a using the same concentration of VPA (2 mM) reported to significantly enhance the generation of mouse iPSCs (Huangfu et al., 2008a). Surprisingly, VPA treatment resulted in complete loss of the Nanog + Sin3a reprogramming synergy (Figure 5D), which was not due to a cell proliferation defect (Figure S5E). Interestingly, we noted that VPA treatment caused a 45% and 82% reduction in HDAC1 and HDAC2 proteins, respectively, in pre-iPSCs expressing Nanog + Sin3a (Figure 5E), suggesting that abrogation of the Nanog + Sin3a reprogramming synergy upon VPA treatment was likely due to the loss of HDAC1/2 expression and/or their intrinsic catalytic activity. To distinguish these two possibilities, we asked whether catalytic mutants of HDAC1 (H141A) (Mal et al., 2001) and HDAC2 (H142A) (Xu et al., 2010) could also synergize with Nanog. Interestingly, we found that both HDAC1 H141A and HDAC2 H142A catalytic mutants were able to synergize with Nanog in reprogramming (Figures 5F and S5F–G) to a similar extent as their wild-type counterparts (Figure 5C). Collectively, our results demonstrate the requirement of HDAC1/2 for the Nanog + Sin3a reprogramming synergy, and suggest that HDAC1/2 catalytic activity may be dispensable for this synergy.
To further address the role of HDAC2 in the Nanog + Sin3a reprogramming synergy, we knocked down HDAC2 during pre-iPSC reprogramming by Nanog + Sin3a (Figure S5H). Not surprisingly, we found that HDAC2 knockdown dramatically reduced the efficiency of reprogramming mediated by Nanog + Sin3a, indicating that HDAC2 is critical for the ability of Sin3a to functionally cooperate with Nanog in the final stage of reprogramming (Figures 5G and S5I). Similarly, Sin3a knockdown caused a significant reduction in the efficiency of reprogramming mediated by Nanog + HDAC2 (Figures 5H and S5J–K). These data demonstrate that a Sin3a/HDAC2 complex can promote pre-iPSC reprogramming in the context of Nanog, and that Sin3a and HDAC2 are mutually dependent for their functional cooperation with Nanog.
The mammalian Sin3 protein consists of two paralogs, Sin3a and Sin3b, which participate in distinct cellular functions despite the presence of a similar HID domain mediating their interactions with HDAC1/2 (Kadamb et al., 2013). Interestingly, we found that combined expression of Nanog + Sin3b had no effect on pre-iPSC reprogramming efficiency, compared to Nanog alone (Figures 5I–K), indicating that our observed reprogramming synergy with Nanog is specific to Sin3a.
DISCUSSION
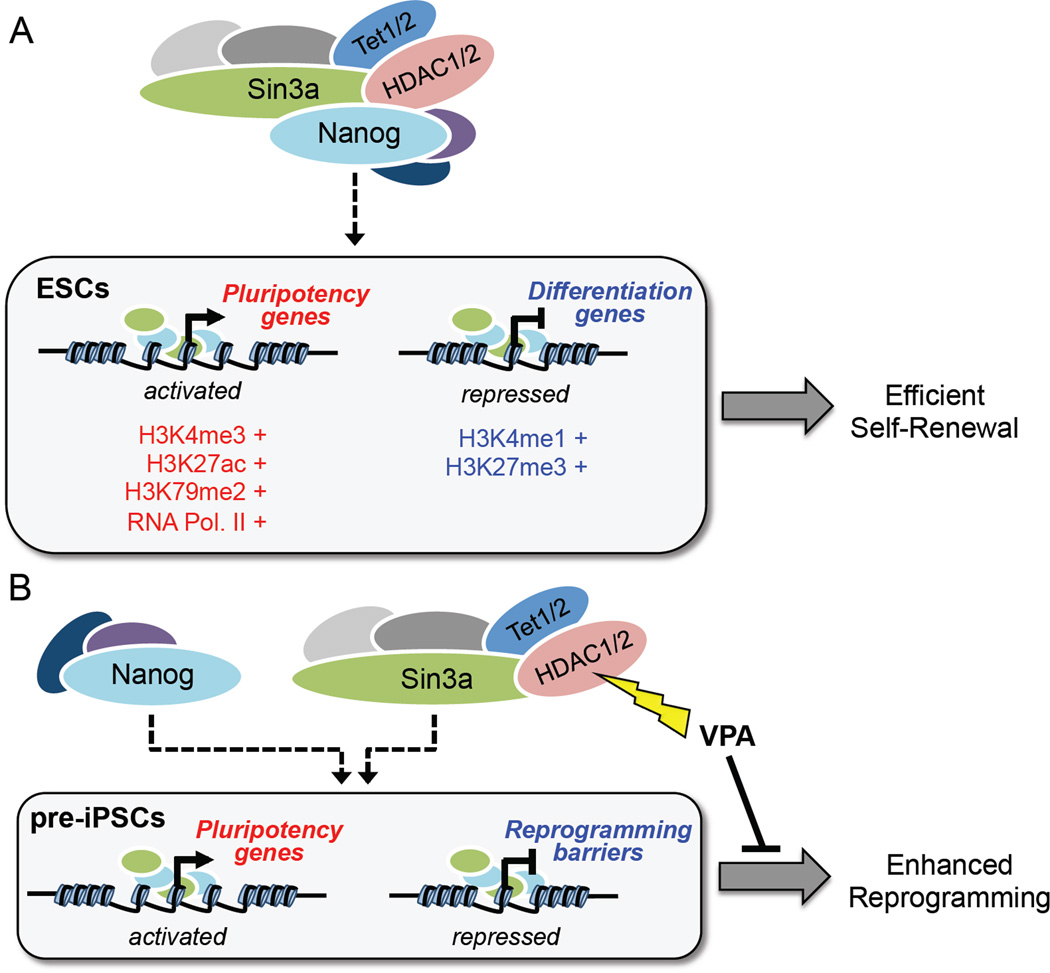
In this study, we report that despite its well-documented role as a corepressor complex, Sin3a/HDAC can also act as a transcriptional co-activator complex for regulation of ESC pluripotency (Figure 6A) and somatic cell reprogramming (Figure 6B). Through microarray and ChIP-seq analyses, we identified several reprogramming-promoting and barrier genes that are directly up-regulated and down-regulated, respectively, by Nanog and Sin3a in pre-iPSCs. We demonstrate that HDAC2 is critical for Sin3a function in promoting the final stage of Nanog-driven reprogramming, and that Sin3a and HDAC2 are mutually dependent for their reprogramming-promoting activities.
Figure 6. Proposed model of the Sin3a/HDAC complex in maintaining ESC self-renewal and promoting somatic cell reprogramming through functional cooperation with Nanog.
(A) Nanog and the Sin3a/HDAC complex co-occupy promoter regions of highly expressed pluripotency genes in ESCs and cooperate to promote ESC self-renewal.
(B) Nanog and the Sin3a/HDAC complex functionally cooperate in pre-iPSCs by directly activating pluripotency genes and repressing reprogramming barrier genes resulting in significantly enhanced reprogramming efficiency.
It is worth pointing out that our findings do not necessarily conflict with those reported by others regarding the beneficial effect of VPA on reprogramming, evident by a number of differences between this study and another report using VPA in promoting iPSC generation (Huangfu et al., 2008a). First, we used pre-iPSCs for our reprogramming experiments with VPA (Figure 5D), whereas Huangfu et al. used MEFs, resulting in distinctive chromatin states of starting cells used by each group. Second, our VPA treatment lasted for 10 days in 2i + LIF conditions, whereas that of Huangfu et al. lasted 7 days in serum + LIF conditions. Lastly, the final Nanog-dependent stage of reprogramming was employed in this study, whereas the 7-day VPA treatment in MEFs by Huangfu et al. corresponds to the early stage of reprogramming in which Nanog is dispensable. Our study thus reveals a stage-specific effect of the HDAC inhibitor VPA in reprogramming, that is, VPA treatment is beneficial during the early stage of reprogramming, but is detrimental for the final pre-iPSC to iPSC transition. It is also noteworthy that previous reports have also demonstrated that VPA treatment causes degradation of HDAC2 protein in MEF and human erythroleukemia cells (Anokye-Danso et al., 2011; Krämer et al., 2003).
Interestingly, our findings regarding the role of HDAC2 in pre-iPSC reprogramming are in contrast to a recent study (Wei et al., 2015), which identified HDAC2 as a barrier to the reprogramming process. Despite these seemingly contradictory results, however, there are several key differences between our study and theirs that should help explain these opposite findings. First, we used Oct4-GFP neural stem cell (NSC)-derived as well as Nanog-GFP MEF-derived pre-iPSCs, whereas Wei et al. used their own clonally derived Oct4-GFP MEF pre-iPSCs. Second, Wei et al. reprogrammed their MEF-derived pre-iPSCs in serum + LIF, a heterogeneous culture condition, whereas we reprogrammed our NSC/MEF-derived pre-iPSCs in 2i + LIF, a defined culture condition that promotes rapid transition to naïve pluripotency and that does not permit the survival of any non-reprogramming cells (Silva et al., 2008). The serum-containing medium used by Wei et al. therefore constitutes a heterogeneous culture condition that may permit some clonal pre-iPSC lines to spontaneously reprogram. Third, and most importantly, in reference to their pre-iPSC reprogramming experiments with HDAC2 KD, Wei et al. state in their paper that HDAC2 knockdown significantly blocked pre-iPSC reprogramming in 2i + LIF culture conditions, suggesting a positive role for HDAC2 in 2i + LIF induced reprogramming, which is consistent with our findings (Figures 5B–C and G). We did find, however, that HDAC2 over-expression had a detrimental effect on MEF reprogramming efficiency, consistent with Wei et al. Collectively, these findings demonstrate a stage-specific requirement for HDAC2 in reprogramming. That is, HDAC2 is detrimental for early-stage reprogramming, but is beneficial for the final stage of reprogramming, suggesting a switched function for the Sin3a/HDAC complex during the reprogramming process.
The similar reprogramming inhibition effects upon knockdown (Figure 2B) and over-expression (Figure S3I) of Sin3a during MEF reprogramming suggests that Sin3a levels may be dynamically regulated and properly balanced by OKSM during somatic cell reprogramming. Such an ‘ambivalent’ action of Sin3a in MEF reprogramming mimics the similar reprogramming inhibition by knockdown and over-expression of Mgarp, one of a few genes that are commonly up-regulated during OSKM-mediated reprogramming of multiple somatic cell types to pluripotency. In this case, counteracting activities of Oct4 and Klf4 on Mgarp expression during reprogramming provide a plausible explanation (Tiemann et al., 2014). Whether a similar expression control mechanism exists for Sin3a during MEF reprogramming is worthy of future investigation. Our study lays emphasis on Nanog-dependent functions of Sin3a in promoting reprogramming in a cell-state specific manner, demonstrating that Sin3a can synergize with Nanog to significantly enhance reprogramming of partially reprogrammed pre-iPSCs and primed pluripotent EpiSCs to full pluripotency.
Dissection of the protein complexes associated with Sin3a and Nanog at the interactome level has been instrumental in discovering many transcriptional co-regulators that might contribute to the Nanog-mediated transcriptional activation function of the Sin3a/HDAC complex (Figure S1G). As HDAC2 is necessary (Figure 5G) but not sufficient (Figures 5B and C) for the full reprogramming-promoting function of Sin3a, other shared partners of Nanog and Sin3a such as Tet1/2 (Figure S1G) likely contribute to the Nanog + Sin3a reprogramming synergy. This is supported by our previous findings demonstrating the Nanog-dependent function of Tet1/2 in transcriptional priming and reprogramming synergy (Costa et al., 2013) as well as our finding of the dramatic reduction in reprogramming efficiency of Nanog + Sin3a upon Tet1/2 knockdown (Figures S5L and M). We cannot rule out, however, that other Nanog and Sin3a common interacting partners such as Ogt or Sall4 may also contribute to the Nanog-mediated transcriptional function of Sin3a. Future studies are needed to elucidate the epigenetic mechanisms mediating the dual transcriptional activity of Sin3a, and to determine how non-catalytic functions of HDAC1/2 such as maintaining complex integrity (Dovey et al., 2013; Winter et al., 2013) and protein translation (Xu et al., 2010) may have contributed to pluripotency and reprogramming control.
EXPERIMENTAL PROCEDURES
Additional experimental procedures are provided in the Supplemental Experimental Procedures.
Reprogramming of pre-iPSCs, EpiSCs, and MEFs
Female neural stem cell (NSC)-derived pre-iPSCs were generated and used for reprogramming as described (Costa et al., 2013). For pre-iPSC reprogramming, 1.0 × 104 or 2.0 × 104 pre-iPSCs were seeded after selection onto gelatin-coated 12-well plates and grown in serum + LIF for 2 days before medium switch to 2i + LIF. MEF-derived Nanog-GFP (TNGA) pre-iPSCs were established and maintained as described (Costa et al., 2013), and were treated in the same manner as NSC pre-iPSCs described above.
For epiblast stem cell (EpiSC) reprogramming, 3.0 × 104 OEC-2 EpiSCs (Guo et al., 2009) were seeded onto fibronectin-coated 12-well plates and grown for 2 days in bFGF + Activin A. Medium was then switched from EpiSC medium to 2i + LIF, and Oct4-GFP+ iPSC colonies were scored 6 days after medium switch.
MEF reprogramming was performed as described (Vidal et al., 2014) with some modifications. Briefly, 3.0 × 104 reprogrammable MEFs containing a Dox-inducible OKSM cassette were infected with shSin3a or shLuciferase (shLuc) lentiviruses. The next day, 500 infected MEFs/well were seeded on top of a feeder layer of irradiated mouse embryonic fibroblast feeders on a 6-well plate coated with gelatin, in “Dox + 3c”-containing ESC medium (Vidal et al., 2014). On day 6, medium was switched to ESC medium without Dox or 3c, and plates were stained for alkaline phosphatase activity on day 10.
Statistical analysis
Statistical analyses for all reprogramming experiments were performed using an unpaired, two-tailed Student’s t-test, with significance values indicated in figure legends. Reprogramming experiments were performed with technical triplicates and were repeated at least 3 independent times. All error bars throughout figures represent standard deviation (SD).
Microarray analysis
Total RNA was extracted from pre-iPSCs maintained in serum + LIF and subjected to microarray gene expression analysis using Mouse WG-6 mouse v2.0 microarray chips (Illumina). Microarray was performed and raw expression data were generated at the Icahn Institute for Genomics and Multi Scale Biology Genomics Facility at the Icahn School of Medicine at Mount Sinai.
Affinity purification, mass spectrometry, and Sin3a interactome analysis
J1 mouse ESCs were expanded to 12 dishes (15-cm) in either SILAC “light” (L-arginine and L-lysine) or “heavy” (L-13C615N4-arginine and L-13C615N2-lysine) ESC medium for more than two weeks. The nuclear extracts (NE) were collected as previously described (Ding et al., 2015), and equal amounts of total proteins in NE were used for immunoprecipitation (IP). Heavy and light immunoprecipitates were then combined, separated by SDS-PAGE, and subjected to LC-MS/MS mass spectrometry analysis.
Supplementary Material
Acknowledgments
We thank Dr. Jose Silva (United Kingdom) for the pre-iPSCs, Dr. Andrew J. Paterson (UAB) for Sin3a mutant constructs, Dr. Asoke Mal (Roswell Park) and Dr. Jing Hu (Univ of Pittsburgh) for HDAC1 and HDAC2 catylatic activity mutants, respectively, and Dr. Matthias Stadtfeld (NYU) for the reprogrammable MEFs. This research was funded by grants from the National Institutes of Health (NIH) to J.W. (R01GM095942, R21HD087722), the Empire State Stem Cell Fund through New York State Department of Health (NYSTEM) to J.W. (C028103, C028121). J.W. is a recipient of Irma T. Hirschl and Weill-Caulier Trusts Career Scientist Award. A.S. is an awardee of the Traineeship of NIDCR-Interdisciplinary Training in Systems and Developmental Biology and Birth Defects (T32HD075735).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Experimental Procedures, 4 figures, and 3 tables that can be found with this article online.
AUTHOR CONTRIBUTIONS
A.S. designed and performed experiments, analyzed data, and wrote the manuscript; X.H. provided bioinformatics support; M.F., M.H.R., F.F., J.D., C.S-P., D.G., C.S., and D.L. provided technical assistance, reagents, and helpful discussions; J.W. conceived the project, designed the experiments, analyzed data, prepared and approved the manuscript.
REFERENCES
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltus GA, Kowalski MP, Tutter AV, Kadam S. A positive regulatory role for the mSin3A–HDAC complex in pluripotency through Nanog and Sox2. J. Biol. Chem. 2009;284:6998–7006. doi: 10.1074/jbc.M807670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, Levasseur DN, Li Z, Xu M, Reik W, Silva JCR, Wang J. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley SM, Iritani BM, Mendrysa SM, Xu T, Cheng PF, Yada J, Liggitt HD, Eisenman RN. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Molecular and Cellular Biology. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Jena S, Levasseur DN. Alternative splicing produces Nanog protein variants with different capacities for self-renewal and pluripotency in embryonic stem cells. Journal of Biological Chemistry. 2011;286:42690–42703. doi: 10.1074/jbc.M111.290189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Huang X, Shao N, Zhou H, Lee D-F, Faiola F, Fidalgo M, Guallar D, Saunders A, Shliaha PV, Wang H, Waghray A, Papatsenko D, Sánchez-Priego C, Li D, Yuan Y, Lemischka IR, Shen L, Kelley K, Deng H, Shen X, Wang J. Tex10 coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming. Cell Stem Cell. 2015;16:653–668. doi: 10.1016/j.stem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini D, Minuzzo M, Pavesi G, Mantovani R. The Short Isoform of NF-YA Belongs to the Embryonic Stem Cell Transcription Factor Circuitry. Stem Cells. 2012;30:2450–2459. doi: 10.1002/stem.1232. [DOI] [PubMed] [Google Scholar]
- Dovey OM, Foster CT, Conte N, Edwards SA, Edwards JM, Singh R, Vassiliou G, Bradley A, Cowley SM. Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood. 2013;121:1335–1344. doi: 10.1182/blood-2012-07-441949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb Is a Direct Nanog Target Gene that Can Substitute for Nanog Function in Pluripotent Cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi A, Mullin NP, Tan ZY, Colby D, Kousa AI, Halbritter F, Weiss JT, Felker A, Bezstarosti K, Favaro R, Demmers J, Nicolis SK, Tomlinson SR, Poot RA, Chambers I. A direct physical interaction between Nanog and Sox2 regulates embryonic stem cell self-renewal. The EMBO Journal. 2013:1–17. doi: 10.1038/emboj.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotech. 2008a;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotech. 2008b;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Kadamb R, Mittal S, Bansal N, Batra H, Saluja D. Sin3: insight into its transcription regulatory functions. Eur. J. Cell Biol. 2013;92:237–246. doi: 10.1016/j.ejcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Kranc KR, Oliveira DV, Armesilla-Diaz A, Pacheco-Leyva I, Catarina Matias A, Luisa Escapa A, Subramani C, Wheadon H, Trindade M, Nichols J, Kaji K, Enver T, Bragança J. Acute loss of Cited2 impairs Nanog expression and decreases self-renewal of mouse embryonic stem cells. Stem Cells. 2015;33:699–712. doi: 10.1002/stem.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, Brill B, Groner B, Bach I, Heinzel T, Göttlicher M. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. The EMBO Journal. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsovali A, Hadjimichael C, Charmpilas N. Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem Cells International. 2012;2012:184154. doi: 10.1155/2012/184154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. The EMBO Journal. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P, Demmers J, Tan DWM, Watt F, Hendrich BD. Sin3a is essential for the genome integrity and viability of pluripotent cells. Developmental Biology. 2012;363:62–73. doi: 10.1016/j.ydbio.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotech. 2008 doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Park S-W, Do H-J, Ha WT, Han M-H, Yang H-M, Lee S-H, Song H, Kim N-H, Kim J-H. Transcriptional regulation of OCT4 by the ETS transcription factor ESE-1 in NCCIT human embryonic carcinoma cells. Biochem Biophys Res Commun. 2014;450:984–990. doi: 10.1016/j.bbrc.2014.06.079. [DOI] [PubMed] [Google Scholar]
- Qin H, Diaz A, Blouin L, Lebbink RJ, Patena W, Tanbun P, LeProust EM, McManus MT, Song JS, Ramalho-Santos M. Systematic identification of barriers to human iPSC generation. Cell. 2014;158:449–461. doi: 10.1016/j.cell.2014.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, Maza I, Mor N, Baran D, Weinberger L, Jaitin DA, Lara-Astiaso D, Blecher-Gonen R, Shipony Z, Mukamel Z, Hagai T, Gilad S, Amann-Zalcenstein D, Tanay A, Amit I, Novershtern N, Hanna JH. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013:1–19. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Sakurai K, Talukdar I, Patil VS, Dang J, Li Z, Chang K-Y, Lu C-C, Delorme-Walker V, DerMardirossian C, Anderson K, Hanein D, Yang C-S, Wu D, Liu Y, Rana TM. Kinome-wide functional analysis highlights the role of cytoskeletal remodeling in somatic cell reprogramming. Cell Stem Cell. 2014;14:523–534. doi: 10.1016/j.stem.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Dos RL, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, Hendrich B, Silva JCR. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 2014;15:102–110. doi: 10.1016/j.stem.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Faiola F, Wang J. Concise review: pursuing self-renewal and pluripotency with the stem cell factor nanog. Stem Cells. 2013;31:1227–1236. doi: 10.1002/stem.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J. Promotion of reprogramming to ground state pluripotency by signal inhibition. Plos Biol. 2008 doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart HT, van Oosten AL, Radzisheuskaya A, Martello G, Miller A, Dietmann S, Nichols J, Silva JCR. NANOG amplifies STAT3 activation and they synergistically induce the naive pluripotent program. Curr. Biol. 2014;24:340–346. doi: 10.1016/j.cub.2013.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann U, Marthaler AG, Adachi K, Wu G, Fischedick GUL, Araúzo-Bravo MJ, Schöler HR, Tapia N. Counteracting activities of OCT4 and KLF4 during reprogramming to pluripotency. Stem Cell Reports. 2014;2:351–365. doi: 10.1016/j.stemcr.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten AL, Costa Y, Smith A, Silva JCR. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nature Communications. 2012;3:817. doi: 10.1038/ncomms1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal SE, Amlani B, Chen T, Tsirigos A, Stadtfeld M. Combinatorial modulation of signaling pathways reveals cell-type-specific requirements for highly efficient and synchronous iPSC reprogramming. Stem Cell Reports. 2014;3:574–584. doi: 10.1016/j.stemcr.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wei T, Chen W, Wang X, Zhang M, Chen J, Zhu S, Chen L, Yang D, Wang G, Jia W, Yu Y, Duan T, Wu M, Liu H, Gao S, Kang J. An HDAC2-TET1 switch at distinct chromatin regions significantly promotes the maturation of pre-iPS to iPS cells. Nucleic Acids Research. 2015;43:5409–5422. doi: 10.1093/nar/gkv430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M, Moser MA, Meunier D, Fischer C, Machat G, Mattes K, Lichtenberger BM, Brunmeir R, Weissmann S, Murko C, Humer C, Meischel T, Brosch G, Matthias P, Sibilia M, Seiser C. Divergent roles of HDAC1 and HDAC2 in the regulation of epidermal development and tumorigenesis. The EMBO Journal. 2013;32:3176–3191. doi: 10.1038/emboj.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Vatsyayan J, Gao C, Bakkenist CJ, Hu J. HDAC2 promotes eIF4E sumoylation and activates mRNA translation gene specifically. Journal of Biological Chemistry. 2010;285:18139–18143. doi: 10.1074/jbc.C110.131599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C-S, Chang K-Y, Rana TM. Genome-wide functional analysis reveals factors needed at the transition steps of induced reprogramming. Cell Rep. 2014;8:327–337. doi: 10.1016/j.celrep.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JCR, Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang Y, Chen J, Li H, Kang L, Zhang Y, Chen S, Zhu B, Gao S. RCOR2 is a subunit of the LSD1 complex that regulates ESC property and substitutes for SOX2 in reprogramming somatic cells to pluripotency. Stem Cells. 2011;29:791–801. doi: 10.1002/stem.634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.