Abstract
Genome-wide association studies have identified >50 common variants associated with kidney function, but these variants do not fully explain the variation in eGFR. We performed a two-stage meta-analysis of associations between genotypes from the Illumina exome array and eGFR on the basis of serum creatinine (eGFRcrea) among participants of European ancestry from the CKDGen Consortium (nStage1: 111,666; nStage2: 48,343). In single-variant analyses, we identified single nucleotide polymorphisms at seven new loci associated with eGFRcrea (PPM1J, EDEM3, ACP1, SPEG, EYA4, CYP1A1, and ATXN2L; PStage1<3.7×10−7), of which most were common and annotated as nonsynonymous variants. Gene-based analysis identified associations of functional rare variants in three genes with eGFRcrea, including a novel association with the SOS Ras/Rho guanine nucleotide exchange factor 2 gene, SOS2 (P=5.4×10−8 by sequence kernel association test). Experimental follow-up in zebrafish embryos revealed changes in glomerular gene expression and renal tubule morphology in the embryonic kidney of acp1- and sos2-knockdowns. These developmental abnormalities associated with altered blood clearance rate and heightened prevalence of edema. This study expands the number of loci associated with kidney function and identifies novel genes with potential roles in kidney formation.
Keywords: renal function, human genetics, kidney development
CKD is considered a complex phenotype with a genetic predisposition.1 Previous genome-wide association studies (GWAS) have successfully identified multiple common genetic risk variants associated with the CKD-defining measures of eGFR and urinary albumin-to-creatinine ratio (UACR).2–5 Together, these variants explain only a small proportion of the variation in eGFR and UACR.6 To comprehensively interrogate protein-coding regions and assess the effects of rare variants (minor allele frequency [MAF]<1%), we carried out a two-stage meta-analysis of the association between eGFR on the basis of serum creatinine (eGFRcrea) and variants genotyped on the Illumina HumanExome chip (http://genome.sph.umich.edu/wiki/Exome_Chip_Design) among 111,666 European ancestry (EA) participants from the CKDGen Consortium and assessed the role of genes significantly associated with eGFRcrea in kidney development using embryonic zebrafish models. In secondary analyses, we examined associations with eGFRcrea stratified by diabetes status, and in a smaller subset of EA participants, we also tested eGFR on the basis of cystatin C (eGFRcys) and UACR. An additional 9624 participants of African ancestry (AA) were also used in an independent exome-chip discovery meta-analysis.
Results
Up to 120,357 participants from 27 studies of EA and up to 11,386 participants from seven studies of AA were included in stage 1 analyses for eGFRcrea, eGFRcys, or UACR. An additional 48,343 participants from 12 studies of EA were included in stage 2 analysis of eGFRcrea. All participants provided informed consent and each of the studies was approved by its governing ethics committee or Institutional Review Board. Sample characteristics and genotyping information for each study are summarized in Supplemental Tables 1 and 2.
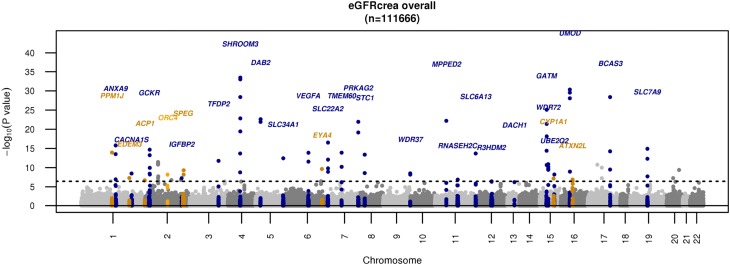
In stage 1 single-variant EA analyses, we identified 33 loci associated with eGFRcrea (Supplemental Table 3, Table 1) that met our a priori chip-wide significance threshold of P<3.7×10−7(0.05/134299 variants). Of these, eight had not been identified in association with eGFRcrea from previous GWAS analyses; six were missense variants: rs34611728 (PPM1J), rs78444298 (EDEM3), rs11553746 (ACP1), rs2307394 (ORC4), rs55760516 (SPEG), and rs9493627 (EYA4); and two were GWAS tag single nucleotide polymorphisms (SNPs) included on the exome chip due to prior associations in the National Human Genome Research Institute (NHGRI) GWAS Catalog for caffeine/coffee intake:7,8 rs2472297 (intergenic, near CYP1A1) and inflammatory bowel disease9 rs8049439 (intronic, ATXN2L); all were common variants (MAF>1%). A Manhattan plot displaying all 33 chip-wide significant loci is shown in Figure 1. Quantile-quantile plots indicated no inflation of the overall P value distribution (Supplemental Figure 1). Regional association plots show the associations of other variants within the 500 Mb region of the index variant of the eight newly identified loci in Supplemental Figure 2.
Table 1.
Novel variants associated with eGFRcrea in EA participants from single-variant analysis in stage 1 meeting chip-wide significance (P<3.7×10−7) and associations in stage 2 and combined analysis
| Locusa | dbSNPID | Chr | Positionb | Variation (Substitution) | Stage 1c | Stage 2d | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1/A2 (A1 AF) | β (SEM) | P Value | I2 | β (SEM) | 1-sided P | β (SEM) | P Value | Prop Var Exp (%) | |||||
| PPM1J | rs34611728 | 1 | 113255456 | c.639G>T (L213F) | A/C (0.13) | −0.0103 (0.0013) | 1.2E-14 | 13.2 | −0.0059 (0.0023) | 4.7E-03 | −0.0092 (0.0011) | 3.3E-16 | 0.05 |
| EDEM3 | rs78444298 | 1 | 184672098 | c.2236C>T (P746S) | A/G (0.02) | −0.0183 (0.0034) | 5.2E-08 | 15.3 | −0.0225 (0.0055) | 1.8E-05 | −0.0195 (0.0029) | 1.5E-11 | 0.03 |
| ACP1 | rs11553746 | 2 | 272203 | c.129C>T (T95l) | T/C (0.35) | −0.0049 (0.0009) | 2.0E-07 | 20.7 | −0.0032 (0.0016) | 2.2E-02 | −0.0045 (0.0008) | 1.0E-08 | 0.02 |
| ORC4e | rs2307394 | 2 | 148716428 | c.233A>G (N78S) | C/T (0.32) | −0.0058 (0.0010) | 6.8E-09 | 14.3 | −0.0025 (0.0016) | 6.0E-02 | −0.0049 (0.0009) | 8.4E-09 | 0.03 |
| SPEG | rs55760516 | 2 | 220354108 | c.8191A>G (R2731G) | G/A (0.33) | 0.0059 (0.0009) | 4.8E-10 | 0.5 | 0.0054 (0.0016) | 3.7E-04 | 0.0058 (0.0008) | 1.7E-13 | 0.04 |
| EYA4 | rs9493627 | 6 | 133789728 | c.829G>A (G223S) | A/G (0.31) | 0.0061 (0.0010) | 2.3E-10 | 0.0 | 0.0049 (0.0016) | 1.4E-03 | 0.0058 (0.0009) | 1.4E-11 | 0.04 |
| CYP1A1 | rs2472297 | 15 | 75027880 | intergenic | T/C (0.24) | 0.0057 (0.0010) | 7.0E-08 | 0.0 | 0.0059 (0.0017) | 3.2E-04 | 0.0058 (0.0009) | 3.0E-11 | 0.03 |
| ATXN2L | rs8049439 | 16 | 28837515 | intronic | C/T (0.40) | 0.0048 (0.0009) | 1.3E-07 | 7.1 | 0.0045 (0.0016) | 1.8E-03 | 0.0047 (0.0008) | 1.2E-09 | 0.03 |
A1, effect allele; A2, non-effect allele; A1 AF, effect allele frequency; Chr, chromosome; Prop Var Exp, proportion of variance in ln(eGFRcrea) explained.
Loci are named according to the closest gene on the basis of the position of the lead SNP for new loci.
Position is reported in UCSC Genome Browser build hg19.
Sample size for stage 1 analysis: n=111,666.
Sample size for stage 2 analysis: n=48,343.
This variant reached chip-wide significance (P<3.7×10−7) in the stage 1 samples but did not meet validation criteria in stage 2.
Figure 1.
Manhattan plot for single-variant analysis in eGFRcrea among 111,666 EA participants. Newly identified variants are in dark orange. The gene, ORC4, not successfully replicated, is in orange. Known loci are in blue.
In stage 2 analyses, we followed up the eight newly identified eGFRcrea loci meeting our significance threshold among an additional 48,343 EA participants from 12 studies (sample characteristics and genotyping information are summarized in Supplemental Tables 2 and 4). All loci but ORC4 met criteria for replication (a direction of effect consistent with stage 1 analysis; a P1-sided<0.05 from stage 2 analysis; and P<3.7×10−7 from a combined stage 1 and stage 2 analysis; Table 1). Because diabetes mellitus is a major risk factor for CKD, we assessed the genetic associations at these loci in the presence or absence of diabetes to obtain additional insights into potential mechanistic pathways. In total, 11,040 and 94,677 participants were included in the diabetes- and nondiabetes-specific analyses, respectively. In analyses stratified by diabetes status, the β-coefficients were directionally consistent and of similar magnitude between the strata for six out of seven newly identified loci (Supplemental Table 5).
To identify additional novel loci associated with alternative measures of kidney function, we tested the association of single variants with eGFRcys (n=32,861 EA participants) and UACR (n=31,164 EA participants). All observed associations achieving the significance threshold (P<3.7×10−7; Supplemental Table 6) have been previously reported in association with eGFRcrea, eGFRcys, or UACR.3,4,6
To further investigate the role of rare variants in kidney function, we performed gene-based tests for 9990 autosomal genes that contained at least two nonsynonymous or splice-site variants with MAF<1%. No evidence of inflation was observed in quantile-quantile plots for gene-based tests (Supplemental Figure 3). Three genes, SOS2, SLC47A1, and LRP2, met the experiment-wide threshold for gene-based significance (P<2.5×10−6; 0.05/19,922 tests [9961 genes × 2 tests]; Table 2; Supplemental Figure 4). The association for SLC47A1, a renal solute transporter, was driven by the presence of a single variant, rs111653425, with MAF approximately 1% (Supplemental Table 7). Common variants at SLC47A1 and LRP2 have been previously reported in association with eGFRcrea,4,6 thus implicating both rare and common variants at both loci.4,6 Conversely, the association with SOS2 was novel (PSKAT=5.38×10−8; PT1=3.25×10−6). No genes reached the threshold for chip-wide significance in gene-based associations for eGFRcrea stratified by diabetes status, eGFRcys, or UACR.
Table 2.
Genes associated with eGFRcrea in EA participants from gene-based analyses meeting chip-wide significance thresholds (P<2.5×10−6)
| Genea | Chr | cMAF | N Variantsb | T1c | SKATd | ||
|---|---|---|---|---|---|---|---|
| β | SEM | P Value | P Value | ||||
| LRP2 | 2 | 0.070 | 38 | 0.003 | 0.002 | 6.7E-02 | 3.5E-7e |
| SLC47A1f | 17 | 0.033 | 4 | −0.033 | 0.004 | 7.8E-15e | 3.4E-12e |
| SOS2g | 14 | 0.040 | 8 | 0.020 | 0.004 | 3.3E-06 | 5.4E-08e |
Chr, chromosome; cMAF, cumulative MAF used in analysis; SKAT, sequence kernel association test.
Gene name.
Number of variants used in analysis.
The standard burden test collapses the variants with MAF<1% into a single variable and tests the association between this variable with a phenotype.30
The SKAT aggregates individual variant score test statistics.32
Meets chip-wide significant threshold, P<2.5×10−6.
Gene-based association results driven by one variant.
Novel gene.
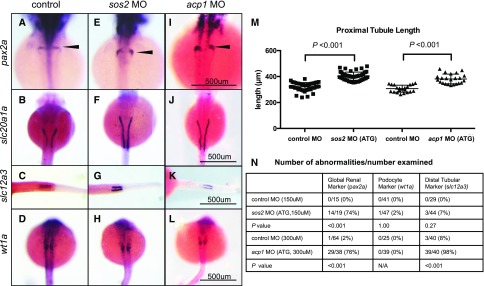
To identify genes that may play a role during kidney development, we used knockdown zebrafish embryos generated by injecting morpholino oligonucleotides (MOs) into single-cell stage embryos. Morpholinos are a commonly used tool in zebrafish screens because they enable an efficient identification of genes that may have a role in developmental processes. The morpholinos targeted genes with nonsynonymous variants (ppm1j, acp1, eya4, speg, and edem3) and the novel gene-based finding, sos2 (Supplemental Figure 5A and Supplemental Table 8). General defects in the pronephros or embryonic kidney structure (marked by expanded pax2a expression) were observed in acp1 ATG MO- and sos2 ATG MO-injected embryos compared with controls (Figure 2, A, E, and I; P<0.001 for both). Both acp1 ATG- and sos2 ATG-knockdowns showed elongated proximal tubules (increased slc20a1a expression) compared with the control group (Figure 2, B, F, and J; mean difference in proximal tubule length: sos2=81.7 μm; P<0.001; and acp1=74.7 μm; P<0.001; Figure 2M). The increase in proximal tubule length was not a consequence of increased embryo length, as both sos2 ATG and acp1 ATG-morphants had significantly reduced body length relative to controls (Supplemental Figure 5B). Additionally, acp1 ATG-knockdowns showed shorter distal tubule length (slc12a3 expression), which may be a consequence of reduced body length (Figure 2, C, G, and K; P<0.001). No abnormalities were observed for podocytes (wt1a expression) for sos2 or acp1 compared with controls (Figure 2, D, H, and L; P>0.05).
Figure 2.
sos2 and acp1 knockdowns result in defective kidney development. (A–D) Whole mount in situ hybridization in control embryos demonstrates normal expression of kidney markers, including pax2a (global kidney, A), slc20a1a (proximal tubules, B), and slc12a3 (distal tubules, C) at 48 hpf, and wt1a (podocytes, D) at 24 hpf. (E–L) sos2 and acp1 ATG morpholino (MO) knockdown embryos develop glomerular gene expression defects (E, I, arrowheads) and display elongated proximal tubules (F, J). Knockdown of acp1 shortened the distal tubules, whereas sos2 knockdown left distal tubule slc12a3 expression unaffected (G, K). No abnormalities in podocyte marker wt1a were observed for sos2 ATG- and acp1 ATG-MOs (H, L). (M) Quantitative assessment of proximal tubule length (slc20a1a expression) shows that proximal tubules are elongated in sos2 ATG- and acp1 ATG-MO injected embryos. t test used to calculate P values. (N) Table of observed abnormal embryos and total number examined by kidney markers pax2a, wt1a, and slc12a3, and MO-injected or control status. Fisher exact test used to calculate P values.
Because of the potential off-target effects of morpholinos, these developmental findings were validated with secondary splice-site morpholinos designed to target the sos2 and acp1 pre-mRNA (Supplemental Figure 5A). The sos2 and acp1 splice morpholino-injected embryos had significantly increased proximal tubule length relative to controls (Supplemental Figure 5, C–E). Furthermore, the acp1 splice-site morpholino-injected embryos had shortened distal tubules (slc12a3 expression) consistent with the phenotype induced by the ATG morpholino (Suppemental Figure 5F). The reproducibility of the proximal tubule developmental defects with ATG and splice-site morpholinos adds confidence to the specificity of the morpholino-induced tubule phenotype.
Follow-up in situ hybridization experiments to determine expression patterns of sos2 and acp1 during zebrafish development did not reveal kidney-specific expression of sos2 or acp1; however, sos2 and acp1 were broadly expressed throughout embryogenesis at key stages of kidney development and may be acting to control kidney development (Supplemental Figure 6). In humans, both sos2 and acp1 protein are detected in adult renal tubules.10 No significant developmental abnormalities were observed among MO knockdowns for the remaining genes (Supplemental Figure 6C). These findings suggest that both SOS2 and ACP1 may influence embryonic renal development, and that genetic influences on kidney development may contribute to variation in kidney function.
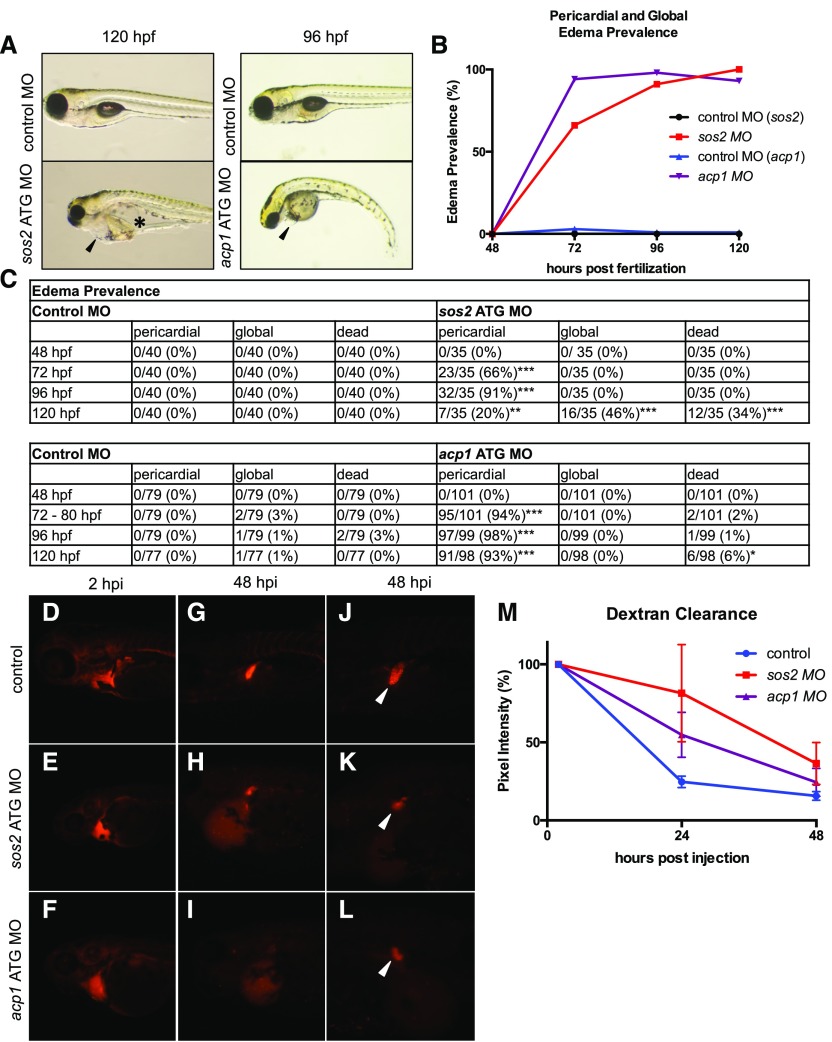
We next sought to determine whether sos2 and acp1-mediated developmental alterations led to abnormalities in kidney function. In zebrafish, edema is a common sign of kidney failure.11 We first performed an edema prevalence study and identified incidence rates of pericardial and global edema in sos2 and acp1 morphant larva. Both sos2 and acp1 ATG morpholino-injected embryos had a heightened incidence of pericardial edema beginning at 72 hours post fertilization (hpf) (Figure 3, A–C). The sos2 morphants developed severe global edema by 120 hpf, whereas the acp1 morphants presented with only pericardial edema (Figure 3, A–C). Both sos2 and acp1 morphants showed indications of embryonic lethality by 120–144 hpf (Figure 3, A–C).
Figure 3.
sos2 and acp1 knockdowns result in altered kidney function. (A) sos2 and acp1 morphants develop edema, which is a sign of kidney failure in zebrafish. sos2 morphants display severe global edema at 120 hpf, with fluid accumulation in the pericardium (black arrow) and intestinal tract (black star). At 96 hpf, acp1 morphants have severe pericardial edema (black arrow). (B–C) Incidence of edema and embryonic lethality in acp1 and sos2 morphants with Fisher exact test. *Indicates P value <0.05; **indicates P value <0.01; ***indicates P value <0.001. (D–I) Embryos were injected with control, sos2, or acp1 morpholino at the single cell stage and subsequently injected with 70 kDa molecular mass fluorescent rhodamine dextran at 72 hpf. Dextran fluorescence intensity was measured over 48 hpi. Dextran-injected embryos show equal loading at 2 hpi. Compared with control embryos, sos2 and acp1 MO injected embryos have reduced dextran clearance in the cardiovascular region over time. (J–L) Morphant embryos have altered convoluted tubule morphology at 120 hpf (48 hpi) (white arrows). (M) Dextran fluorescence intensity over time as normalized to starting fluorescence intensity.
An additional metric for kidney function is the assessment of glomerular filtration and fluid flow by fluorescent dextran clearance.12 Control, sos2, or acp1 morphant embryos were injected with equal volumes of rhodamine-labeled 70 kDa molecular mass dextran in the cardiac sinus venosus at 72 hpf. Dextran clearance rate was assessed by the quantification of rhodamine fluorescence intensity in a standardized area of the cardiac region at 2, 24, and 48 hours post injection (hpi). sos2 and acp1 morphant embryos exhibited decreased dextran clearance at both 24 and 48 hpi relative to controls (Figure 3, D–I, and M). Furthermore, renal tubules marked by fluorescent dextran in sos2 and acp1 morphant larva displayed an abnormal morphology (Figure 3, J–L). Specifically, the proximal convoluted tubules were reduced in size and lacked coiling depth. Failed clearance of 70 kDa molecular mass dextran and abnormal tubular structure suggests that morphants may have defects in tubular fluid flow and glomerular filtration.
We also evaluated heart rate in the sos2 and acp1 morphants relative to controls because compromised cardiovascular function could contribute to defects in dextran clearance. At 96 hpf (24 hours after dextran injection), the sos2 (145.5±3.854 bpm; P=0.003) and acp1 (151.2±2.653 bpm; P<0.001) morphants had an elevated mean heart rate relative to controls (127.8±3.353 bpm) (Supplemental Figure 2G). Although altered kidney development, abnormal tubular structure, edema, and decreased dextran clearance all support the conclusion that both sos2 and acp1 are regulators of kidney development and function, it remains possible that altered hemodynamics contribute to the edema and dextran clearance phenotypes observed.
In separate single variant analyses among 9624 AA participants, we identified three loci in association with eGFRcrea at chip-wide significance (Supplemental Table 9; P<3.7×10−7). These variants were rare and the limited availability of AA cohorts prevented replication of these findings. The APOL1 G1 variant, rs73885319, that was a known risk factor for kidney function decline and ESRD in AA populations, was included on the exome array. However, it was not associated with eGFRcrea using an additive genetics model in the AA participants here (P=0.70).
To investigate if the newly identified eGFRcrea loci were also associated with diabetes mellitus and arterial hypertension, major risk factors for CKD, we tested for associations between the seven validated eGFRcrea loci with BP and type 2 diabetes (T2D) among EA participants in collaborations with the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE) Blood Pressure13 and CHARGE Glycemia-T2D14 Working Groups. Consistent with prior observations, the majority of variants were not associated with BP traits or T2D (Supplemental Table 10). The exception was rs2472297 at the CYP1A1 locus, a GWAS tag SNP previously associated with coffee/caffeine intake.7,8 This SNP was also associated with systolic and diastolic BP (P=7.4×10−7, and 4.6×10−11, respectively).13
Discussion
Our main findings are four-fold. First, we identified and validated seven loci associated with eGFRcrea through genotyping on the exome-chip among 160,009 participants of EA in a two-stage study design. Second, the majority of the newly uncovered associations were for common variants with modest effect sizes, which argues against the presence of rare protein-coding variants with large effect sizes represented on the exome chip. Third, we identified one novel association for SOS2 through gene-based testing. Fourth, we demonstrated altered kidney development and function in zebrafish sos2 and acp1 knockdowns.
Our study emphasizes the continued success of efforts combining population-based genetics and model organisms to identify genes underlying kidney function.4,15 The zebrafish knockdowns provide a systematic model to examine the consequences of gene perturbations in the embryonic renal system of the fish. Our zebrafish morpholino experiments revealed a potential role for sos2 and acp1 in kidney development and function. Mutations in the SOS gene family (SOS1 and SOS2) lead to Noonan syndrome, a congenital RASopathy (syndromes caused by germline mutations controlling signal transduction pathways) that can feature mild kidney dysfunction16 and renal anomalies.17,18 The SOS2 protein is expressed in the glomeruli and tubules of kidneys from adult humans.10 Together, these findings provide further evidence for a potential role for SOS proteins in kidney development and function. There are no prior reports of an association between kidney function and ACP1, a gene that encodes for an acid phosphatase involved in the immune response and found in erythrocytes.19 Our observations of kidney abnormalities in sos2- and acp1-knockdowns provide genes and target tissues for prioritization in future studies of more extensive functional follow-up, diagnostic screening, and potentially drug development.
Although morpholinos are an efficient tool for the rapid evaluation of GWAS hits, they have the potential for off-target effects, and morphant phenotypes are not always recapitulated in genetic mutant models. In this study, we evaluated sos2 and acp1 kidney development phenotypes using two independent morpholinos, which we believe adds confidence in the specificity of our phenotypes. Furthermore, we provided evidence that kidney developmental changes correlate with edema and reduced dextran clearance from the blood. We believe that our morpholino screen has allowed us to clarify promising candidates for further study. Simultaneously, we acknowledge that future studies in genetic mutants will enhance and clarify these findings. Because genetic knockout techniques do not necessarily recapitulate exact features of the identified human variants, future experiments are also needed to evaluate the effect of specific variants on kidney development and function.
The majority of identified novel variants were common. The strongest single-variant association with eGFRcrea to date is the common variant rs13329952 at the UMOD locus with an effect size of 0.016 ln(ml/min per 1.73 m2) (MAF=0.19).15 Given our large discovery sample, our study was adequately powered (>80%) to detect effect sizes of 0.11–0.008 ln(ml/min per 1.73 m2) for very rare variants to more common variants (0.0005>MAF>0.10) (details of the power calculation can be found in the Supplemental Material). Although the selection of nonsynonymous content was expected to enrich for functional variants with large effect sizes, a possible reason for the lack of these findings might be the design-based, limited coverage of rare variants on the exome chip. The exome chip was primarily designed to assess nonsynonymous rare variants that had been observed among approximately 12,000 sequenced participants that were not selected on the basis of kidney disease (http://genome.sph.umich.edu/wiki/Exome_Chip_Design), and thus, we would not expect kidney disease–causing mutations to be well represented on the exome chip. Although the convenience and low cost of chip-based genotyping arrays focused on exonic variants were influential in facilitating a large number of cohorts to participate, as is necessary for a well powered study, the limited coverage of the exome affected the ability of both single-variant and gene-based tests to assess all exonic variation in association with kidney function. Thus, we cannot rule out bona fide rare variant associations with eGFRcrea. Large-scale whole exome or whole genome sequencing will be able to adequately address the unresolved issue of the contribution of rare variants to the variation of kidney function in the general population. Because of limited sample size in the diabetes stratum compared with the nondiabetes stratum, we did not have adequate power to assess unique genetic associations in the diabetes stratified analysis. We did not implement a random effects meta-analysis as an alternative screening procedures for novel loci because the between-study heterogeneity was small. The analysis module that we used cannot correctly account for the different allele dosage between women and men and therefore we were unable to implement association analysis for chromosome X variants. Although our study supports the observation that gene-based analyses aggregating rare nonsynonymous variants are able to identify new loci in association with common complex phenotypes14,20–22 and can additionally uncover new rare missense variants within known loci, the number of loci identified through gene-based methods remains a small minority of the findings compared with single-variant results. Finally, the zebrafish knockdowns helped us to screen novel loci that appear to have a role in kidney development or function. However, extrapolation of the relationship between the novel loci among patients with more advanced CKD remains to be determined; follow up studies are needed to assess the association of these loci with ESRD and incident CKD.
In summary, we identified eight novel loci (seven common single variants and one gene with multiple rare coding variants) associated with kidney function. Functional experiments in zebrafish highlighted potential roles for SOS2 and ACP1 in embryonic kidney development. Future whole exome and whole genome sequencing studies will be needed to assess the full spectrum of rare genetic variants on kidney function.
Concise Methods
Study Participants
Across all traits analyzed, a total of 120,357 participants from 27 studies of EA were included in stage 1 of this study. An additional 48,343 participants from 12 studies of EA were included in stage 2. A total of 11,386 participants from seven studies of AA were also included in separate analyses. Study-specific characteristics are summarized in Supplemental Tables 1, 2, and 4. All participants provided informed consent and each study was approved by its governing ethics committee or Institutional Review Board.
Phenotype Definitions
Serum creatinine was measured in each study as described in the Study-Specific Methods section in the Supplemental Material, and calibrated to the National Health and Nutrition Examination Study data to account for between-laboratory variation.23,24 eGFRcrea was estimated using the four-variable Modification of Diet in Renal Disease (MDRD) Study Equation.25 Cystatin C, an alternative biomarker of kidney function, was measured in a sub-set of participating studies. eGFRcys was estimated as 76.7×(serum cystatin C).25 All eGFRcrea and eGFRcys values <15 ml/min per 1.73m2 were set to 15, and those >200 ml/min per 1.73m2 were set to 200 to avoid undue influence from outliers. UACR was defined as urinary albumin (mg/L)/ urinary creatinine (mg/dl)*100. All analyzed traits (eGFRcrea, eGFRcys, and UACR) were natural log (ln)-transformed. Diabetes was defined as fasting glucose ≥126 mg/dl, pharmacologic treatment for diabetes, or by self-report. Hypertension was defined as systolic BP ≥140 mmHg, diastolic BP ≥90 mmHg, or pharmacologic treatment for hypertension.15
Genotypes
Genotyping was conducted in each study using the Illumina Human Exome BeadChip (http://genome.sph.umich.edu/wiki/Exome_Chip_Design). This genotyping array containing 247,870 markers focuses on exonic variants discovered through exome sequencing of approximately 12,000 individuals. Illumina’s GenTrain version 2.0 clustering algorithm in GenomeStudio, zCall,26 or a combination of both procedures were used to call genotypes. To improve genotype calling of low frequency and rare variants, genotypes from eight of the contributing cohorts (Atherosclerosis Risk in Communities [ARIC], Age, Gene/Environment Susceptibility-Reykjavik Study [AGES], Cardiovascular Health Study [CHS], Framingham Heart Study [FHS], Rotterdam Study [RS], Health, Aging and Body Composition Study [Health ABC], Family Heart Study [FamHS], and Jackson Heat Study [JHS]) were jointly clustered and called via the CHARGE Consortium algorithm.27 Other participating cohorts were called individually. Among them, CROATIA-Korcula and Generation Scotland (GS) applied the cluster file from the CHARGE Consortium for genotype calling. Details regarding genotyping and quality control within each study are summarized in Supplemental Table 2.
Statistical Methods for Stage 1
By following a centralized analysis plan, each study performed two sets of analyses: single-variant analysis and gene-based analysis. The primary meta-analyses were focused on the EA population and a secondary set of meta-analyses were focused on the AA population due to a substantially smaller sample size. Where not specified otherwise, R software was used for data management, statistical analyses, and graphing.28
Single-Variant Analysis
Each study performed association analyses of the following phenotypes and models: (1) ln-transformed eGFRcrea, (2) ln-transformed eGFRcys, (3) ln-transformed UACR, and (4) ln-transformed eGFRcrea stratified by diabetes status. These association analyses were based on linear regression models adjusting for age, sex, study site (if applicable), family structure (if applicable), and the first ten principal components to control for population stratification. All analyses were performed assuming an additive genetic effect and all analyses were stratified by ancestry.
For single-variant meta-analysis, study-specific results were combined for each trait in a fixed-effects model using METAL.29 We restricted single-variant meta-analyses to (1) autosomal variants, (2) polymorphic variants, (3) variants existing in the joint calling effort within the CHARGE consortium27, and (4) with minor allele count ≥20 across all cohorts in each ancestry group. Bonferroni correction for the number of variants tested within each ancestry was used to set the significance threshold for each analysis (chip-wide significance), corresponding to P<3.7×10−7 (0.05/134299 variants) for EA analysis and P<5.5×10−7 (0.05/91187 variants) for AA analysis.
Gene-Based Analysis
Single-variant analysis methods have limited power to detect association for rare variants. Recently developed gene-based methods, where defined variants contained in a gene region are collapsed into one unit for analysis, provide additional statistical power30 to investigate the role of rare variants on kidney function traits.
For gene-based meta-analysis, study-specific results were combined using the seqMeta package for R.31 The same phenotypes and covariates were used as for single-variant testing and analyses were again stratified by ancestry. We used two gene-based tests for aggregated analysis of rare single nucleotide variants (SNVs): (1) T1,30 which is more powerful when all variants within the gene region affect the phenotype in the same direction; and (2) the sequence kernel association test,32 which allows for bidirectional effects and is more powerful when there are both protective and deleterious variants within the same gene. Both gene-based tests were restricted to variants with MAF<1% and variants likely to exert a major effect on the gene product (stop gain/loss, nonsynonymous, or splice-site variants on the basis of annotation with dbNSFP [v.2.0]).33 Genes containing at least two variants with a cumulative minor allele count ≥2020 were included in the analysis. In total, we tested 9990 autosomal genes meeting our established thresholds and filters. Bonferroni correction for the number of genes and tests performed was used to set the significance threshold for the gene-based analysis corresponding to P<2.5×10−6 (0.05/19,922 tests [9961 genes × 2 tests]) in EA analysis and P<2×10−6 (25,378 tests [12,689 genes × 2 tests]) in AA analysis.
Statistical Methods for Stage 2
Chip-wide significant results from single-variant meta-analyses were brought forward for testing in stage 2 where each new study followed the same methodology as stage 1. Details regarding the genotyping and population characteristics of each cohort can be found in Supplemental Tables 2 and 4.
Study-specific results from stage 2 cohorts were combined using the same meta-analysis approach and software as in stage 1 (fixed-effects model in METAL29). Criteria for validation were: (1) a direction of effect consistent with stage 1 analysis, (2) a one-sided P value <0.05 from stage 2 analysis, and (3) P<3.7×10−7 from combined stage 1 and stage 2 analysis.
The percentage of phenotypic variance explained by each novel locus was estimated as R2=β2var(SNP)/var(ln[eGFRcrea]), where β is the estimated effect of the SNP on ln(eGFRcrea), and var(SNP)=2*MAFSNP*(1-MAFSNP). Var(ln[eGFRcrea]) was estimated in the ARIC study. All loci were assumed to have independent effects on the phenotype.
Associations with Other Traits
To examine potential associations of the novel eGFR-associated variants with other correlated traits and conditions, we performed external look ups for systolic and diastolic BP in collaboration with the CHARGE Blood Pressure Working Group (n=145,872)13 and for T2D in collaboration with the CHARGE Glycemia-T2D Working Group (n=10,240 T2D cases and 63,105 controls)20 among EA participants.
NHGRI GWAS Catalog and PubMed Queries
For all newly identified and validated variants, we interrogated the NHGRI GWAS Catalog (https://www.ebi.ac.uk/gwas/;10/12/15) for the lead SNP at each locus and for SNPs in linkage disequilibrium with the lead SNP (within 1 Mbp and r2>0.5 from 1000 Genomes Pilot in CEU; http://www.broadinstitute.org/mpg/snap/) to assess association with other traits (see Supplemental Table 11 for full listing of GWAS Catalog associations). Additionally, for each locus we searched PubMed (http://www.ncbi.nlm.nih.gov/pubmed;10/12/15) for publications on kidney/renal function or CKD.
Functional Studies in Zebrafish
To investigate a potential role of the newly identified genes during kidney development, we assessed the functional consequences of gene knockdown in zebrafish embryos. We used antisense MO technology to knock down genes identified on the basis of validated associations for nonsynonymous variants and for novel gene-based loci. Two independent MO probes were used for each gene. Zebrafish were maintained in accordance with established Institutional Animal Care and Use Committee protocols.
Morpholinos (Gene Tools) were designed against zebrafish target genes. Morpholino sequences are as follows: sos2 (5′ GCACCGGGAACAACCACACAACTTT 3′), sos2 (exon 2) (5′ CCTGCACCTATAAACACAGAATAGA 3′), ppm1j (5′ AATTTGTGACATCAGCGGCACGGTA 3′), acp1 ATG (5′ TCCGCTGGAAGCCGCCATATTGGTC 3′), acp1 (exon 1) (5′ TATAGCATTTCTTACCCAAGCACAC 3′), speg ATG (5′ TCTTCTCTTCAGTAACTTTTCTCAT 3′), edem3 (exon 1) (5′ AGTCCTCACACAGACACATACCTCA 3′), and eya4 ATG (5′ CAGATCCTGTGTATTCTCCATCAGT 3′). Zebrafish embryos were injected with various concentrations of MO (sos2 ATG–150 μM, sos2 (exon 2)– 400 μM, ppm1j ATG, acp1 ATG, and acp1 (exon1)– 400 μM, edem3 ATG– 300 μM, eya4 ATG– 400 μM, speg ATG– 100 μM) at the one-cell stage. We fixed embryos in 4% paraformaldehyde at relevant developmental stages for analysis by in situ hybridization (http://zfin.org/ZFIN/Methods/ThisseProtocol.html). Distinct pronephros (embryonic kidney) structures were visualized using a series of established markers: pax2a (global), wt1a (podocyte), slc20a1a (proximal tubule), and slc12a3 (distal tubule). acp1 probe was generated from zebrafish cDNA using the following primers: 5′ TGGAGAATAGACAGTGCCGC 3′ (forward 1) and 5′ TTTTCACGCTGCTTGCCTTC 3′ (reverse 1), and 5′ GTGGAGAATAGACAGTGCCG 3′ (forward 2) and 5′ CAGGAAGGCTTTGCATC 3′ (reverse 2). sos2 probe was generated from zebrafish cDNA using the following primers: 5′ GTGTTCGAGGAAGGAGCACA 3′ (forward) and 5′ TGATGTTCCACCCACTGACG 3′ (reverse).
Abnormal gene expression patterns were identified by direct comparison to control embryos that were injected with a standard control MO designed by GeneTools (SynGene, Cambridge, UK). Developmental phenotypes were scored by two independent researchers. Fisher exact tests were used to test for normal and abnormal embryonic phenotypes for the pax2a, wt1a, and slc12a3 markers and t test was used to test for differences in proximal tubule length for the slc20a1a marker; P<0.05 was set as a threshold for statistical significance. To evaluate proximal tubule length, the distance between the most anterior and the most posterior tip of the right proximal tubule (from standardized dorsal-view images) was measured using the imageJ measurement tool. Fisher exact test was used to test for differences in edema prevalence; P<0.05 was set as a threshold for statistical significance.
Dextran clearance experiments were performed following a previously described protocol.12 Seventy-two hours after morpholino injection, embryos were anesthetized in a 1:20 dilution of 4 mg/ml Tricane in embryo water and placed on a 2% agarose injection mold. An equal volume of tetramethylrhodamine dextran (70 kDa molecular mass; Invitrogen, Carlsbad, CA) was injected into the cardiac sinus venosus of each embryo, and individual embryos were sorted into designated wells for timelapse imaging. Fluorescent microscopy images were taken at 2 hpi (74 hpf), 24 hpi (96 hpf), and 48 hpi (120 hpf) for each sorted embryo to assess loading fluorescence and the dextran clearance over time. Fluorescence intensity in the cardiac region was measured as the mean grayscale value using Image J as previously described.34 Remaining fluorescence intensity at each time point was normalized to the starting intensity and plotted as a percentage of the initial fluorescence intensity. To evaluate edema, morpholinos were injected into the single-cell stage embryo and embryos were examined every 24 hours for evidence of edema. The number of affected embryos was recorded as a fraction of total number of injected embryos.
Power Calculation
Power for association was evaluated for eGFRcrea assuming a mean of 4.5 ln(ml/min per 1.73 m2) with standard deviation of 0.2 ln(ml/min per 1.73 m2), estimates from the EA samples in the ARIC study, using QUANTO power calculator, version 1.2.4 (http://biostats.usc.edu/Quanto.html) at the significance level of 3.7×10−7 for a variant with MAF of 0.1, 0.05, 0.03, 0.01, 0.005, 0.001, or 0.0005. In stage 1 with sample size n=111,666, there was at least 80% power to detect effect sizes of 0.008, 0.011, 0.015, 0.026, 0.036, 0.08, or 0.11 ln(ml/min/1.73m2), respectively. For stage 2, using one-sided tests, there was 92% power to detect an effect size of 0.018 for a variant with MAF of 0.02 (minimum MAF among all eight SNPs tested) on the basis of 48,343 samples in stage 2 and correcting for the eight SNPs tested.
Disclosures
Caroline S. Fox and Audrey Y. Chu are employed by Merck Research Laboratories as of December 14, 2015 and July 18, 2016, respectively.
Supplementary Material
Acknowledgments
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute, the National Institutes of Health, or the US Department of Health and Human Services. Acknowledgments and funding information for all contributing studies are presented in the Supplemental Material. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The full list of consortium members is provided in the supplemental material.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016020131/-/DCSupplemental.
Contributor Information
Collaborators: Jennifer Wessel, Audrey Y. Chu, Sara M. Willems, Shuai Wang, Hanieh Yaghootkar, Jennifer A. Brody, Marco Dauriz, Marie-France Hivert, Sridharan Raghavan, Leonard Lipovich, Bertha Hidalgo, Keolu Fox, Jennifer E. Huffman, Ping An, Yingchang Lu, Laura J. Rasmussen-Torvik, Niels Grarup, Margaret G. Ehm, Li, Abigail S. Baldridge, Alena Stančáková, Ravinder Abrol, Ce´line Besse, Anne Boland, Jette Bork-Jensen, Myriam Fornage, Daniel F. Freitag, Melissa E. Garcia, Xiuqing Guo, Kazuo Hara, Aaron Isaacs, Johanna Jakobsdottir, Leslie A. Lange, Jill C. Layton, Man Li, Jing Hua Zhao, Karina Meidtner, Alanna C. Morrison, Mike A. Nalls, Marjolein J. Peters, Maria Sabater-Lleal, Claudia Schurmann, Angela Silveira, Albert V. Smith, Lorraine Southam, Marcus H. Stoiber, Rona J. Strawbridge, Kent D. Taylor, Tibor V. Varga, Kristine H. Allin, Najaf Amin, Jennifer L. Aponte, Tin Aung, Caterina Barbieri, Nathan A. Bihlmeyer, Michael Boehnke, Cristina Bombieri, Donald W. Bowden, Sean M. Burns, Yuning Chen, Yii-DerI Chen, Ching-Yu Cheng, Adolfo Correa, Jacek Czajkowski, Abbas Dehghan, Georg B. Ehret, Gudny Eiriksdottir, Stefan A. Escher, Aliki-Eleni Farmaki, Mattias Frånberg, Giovanni Gambaro, Franco Giulianini, William A. Goddard, III, Anuj Goel, Omri Gottesman, Megan L. Grove, Stefan Gustafsson, Yang Hai, Go¨ran Hallmans, Jiyoung Heo, Per Hoffmann, Mohammad K. Ikram, Richard A. Jensen, Marit E. Jørgensen, Torben Jørgensen, Maria Karaleftheri, Chiea C. Khor, Andrea Kirkpatrick, Aldi T. Kraja, Johanna Kuusisto, Ethan M. Lange, I, T. Lee, Wen-Jane Lee, Aaron Leong, Jiemin Liao, Chunyu Liu, Yongmei Liu, Cecilia M. Lindgren, Allan Linneberg, Giovanni Malerba, Vasiliki Mamakou, Eirini Marouli, Nisa M. Maruthur, Angela Matchan, Roberta McKean-Cowdin, Olga McLeod, Ginger A. Metcalf, Karen L. Mohlke, Donna M. Muzny, Ioanna Ntalla, Nicholette D. Palmer, Dorota Pasko, Andreas Peter, Nigel W. Rayner, Frida Renstro¨m, Ken Rice, Cinzia F. Sala, Bengt Sennblad, Ioannis Serafetinidis, Jennifer A. Smith, Nicole Soranzo, Elizabeth K. Speliotes, Eli A. Stahl, Kathleen Stirrups, Nikos Tentolouris, Anastasia Thanopoulou, Mina Torres, Michela Traglia, Emmanouil Tsafantakis, Sundas Javad, Lisa R. Yanek, Eleni Zengini, Diane M. Becker, Joshua C. Bis, James B. Brown, L. Adrienne Cupples, Torben Hansen, Erik Ingelsson, Andrew J. Karter, Carlos Lorenzo, Rasika A. Mathias, Jill M. Norris, Gina M. Peloso, Wayne H.-H. Sheu, Daniela Toniolo, Dhananjay Vaidya, Rohit Varma, Lynne E. Wagenknecht, Heiner Boeing, Erwin P. Bottinger, George Dedoussis, Panos Deloukas, Ele Ferrannini, Oscar H. Franco, Paul W. Franks, Richard A. Gibbs, Vilmundur Gudnason, Anders Hamsten, Tamara B. Harris, Andrew T. Hattersley, Caroline Hayward, Albert Hofman, Jan-Håkan Jansson, Claudia Langenberg, Lenore J. Launer, Daniel Levy, Ben A. Oostra, Christopher J. O’Donnell, Stephen O’Rahilly, Sandosh Padmanabhan, James S. Pankow, Ozren Polasek, Michael A. Province, Stephen S. Rich, Paul M. Ridker, Igor Rudan, Matthias B. Schulze, Blair H. Smith, Andre´ G. Uitterlinden, Mark Walker, Hugh Watkins, Tien Y. Wong, Eleftheria Zeggini, The EPIC-InterAct Consortiumy, Markku Laakso, Ingrid B. Borecki, Daniel I. Chasman, Oluf Pedersen, Bruce M. Psaty, E. Shyong Tai, Cornelia M. van Duijn, Nicholas J. Wareham, Dawn M. Waterworth, Eric Boerwinkle, W.H. Linda Kao, Jose C. Florez, Ruth J.F. Loos, James G. Wilson, Timothy M. Frayling, David S. Siscovick, Jose´e Dupuis, Jerome I. Rotter, James B. Meigs, Robert A. Scott, Mark O. Goodarzi, Chunyu Liu, Aldi T. Kraja, Jennifer A. Smith, Jennifer A. Brody, Nora Franceschini, Joshua C. Bis, Kenneth Rice, Alanna C. Morrison, Yingchang Lu, Stefan Weiss, Xiuqing Guo, Walter Palmas, Lisa W. Martin, Yii-Der Ida Chen, Praveen Surendran, Fotios Drenos, James P. Cook, Paul L. Auer, Audrey Y. Chu, Ayush Giri, Wei Zhao, Johanna Jakobsdottir, Li-An Lin, Jeanette M. Stafford, Najaf Amin, Hao Mei, Jie Yao, Arend Voorman, CHD Exome Plus Consortium, ExomeBP Consortium, GoT, D Consortium, GoT, DGenes consortium, Martin G. Larson, Megan L. Grove, Albert V. Smith, Shih-Jen Hwang, Han Chen, Tianxiao Huan, Gulum Kosova, Nathan O. Stitziel, Sekar Kathiresan, Nilesh Samani, Heribert Schunkert, Panos Deloukas, Myocardial Infarction Genetics, CARDIoGRAM Exome Consortia Investigators, Man Li, Christian Fuchsberger, Cristian Pattaro, Mathias Gorski, CKDGen Consortium, Charles Kooperberg, George J. Papanicolaou, Jacques E. Rossouw, Jessica D. Faul, Sharon L.R. Kardia, Claude Bouchard, Leslie J. Raffel, André G. Uitterlinden, Oscar H. Franco, Ramachandran S. Vasan, Christopher J. O'Donnell, Kent D. Taylor, Kiang Liu, Erwin P. Bottinger, Omri Gottesman, E. Warwick Daw, Franco Giulianini, Santhi Ganesh, Elias Salfati, Tamara B. Harris, Lenore J. Launer, Marcus Dörr, Stephan B. Felix, Rainer Rettig, Henry Völzke, Eric Kim, Wen-Jane Lee, I, Te Lee, Wayne H-H Sheu, Krystal S. Tsosie, Digna R. Velez Edwards, Yongmei Liu, Adolfo Correa, David R. Weir, Uwe Völker, Paul M Ridker, Eric Boerwinkle, Vilmundur Gudnason, Alexander P. Reiner, Cornelia M. van Duijn, Ingrid B. Borecki, Todd L. Edwards, Aravinda Chakravarti, Jerome I. Rotter, Bruce M. Psaty, Ruth J.F. Loos, Myriam Fornage, Georg Ehret, Christopher Newton-Cheh, Daniel Levy, and Daniel I. Chasman
References
- 1.Satko SG, Sedor JR, Iyengar SK, Freedman BI: Familial clustering of chronic kidney disease. Semin Dial 20: 229–236, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Böger CA, Chen MH, Tin A, Olden M, Köttgen A, de Boer IH, Fuchsberger C, O’Seaghdha CM, Pattaro C, Teumer A, Liu CT, Glazer NL, Li M, O’Connell JR, Tanaka T, Peralta CA, Kutalik Z, Luan J, Zhao JH, Hwang SJ, Akylbekova E, Kramer H, van der Harst P, Smith AV, Lohman K, de Andrade M, Hayward C, Kollerits B, Tönjes A, Aspelund T, Ingelsson E, Eiriksdottir G, Launer LJ, Harris TB, Shuldiner AR, Mitchell BD, Arking DE, Franceschini N, Boerwinkle E, Egan J, Hernandez D, Reilly M, Townsend RR, Lumley T, Siscovick DS, Psaty BM, Kestenbaum B, Haritunians T, Bergmann S, Vollenweider P, Waeber G, Mooser V, Waterworth D, Johnson AD, Florez JC, Meigs JB, Lu X, Turner ST, Atkinson EJ, Leak TS, Aasarød K, Skorpen F, Syvänen AC, Illig T, Baumert J, Koenig W, Krämer BK, Devuyst O, Mychaleckyj JC, Minelli C, Bakker SJ, Kedenko L, Paulweber B, Coassin S, Endlich K, Kroemer HK, Biffar R, Stracke S, Völzke H, Stumvoll M, Mägi R, Campbell H, Vitart V, Hastie ND, Gudnason V, Kardia SL, Liu Y, Polasek O, Curhan G, Kronenberg F, Prokopenko I, Rudan I, Arnlöv J, Hallan S, Navis G, Parsa A, Ferrucci L, Coresh J, Shlipak MG, Bull SB, Paterson NJ, Wichmann HE, Wareham NJ, Loos RJ, Rotter JI, Pramstaller PP, Cupples LA, Beckmann JS, Yang Q, Heid IM, Rettig R, Dreisbach AW, Bochud M, Fox CS, Kao WH, Kao WH; CKDGen Consortium : CUBN is a gene locus for albuminuria. J Am Soc Nephrol 22: 555–570, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Paré G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tönjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstätter A, Kollerits B, Kedenko L, Mägi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Völzke H, Kroemer HK, Nauck M, Völker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Krämer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS: New loci associated with kidney function and chronic kidney disease. Nat Genet 42: 376–384, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pattaro C, Köttgen A, Teumer A, Garnaas M, Böger CA, Fuchsberger C, Olden M, Chen MH, Tin A, Taliun D, Li M, Gao X, Gorski M, Yang Q, Hundertmark C, Foster MC, O’Seaghdha CM, Glazer N, Isaacs A, Liu CT, Smith AV, O’Connell JR, Struchalin M, Tanaka T, Li G, Johnson AD, Gierman HJ, Feitosa M, Hwang SJ, Atkinson EJ, Lohman K, Cornelis MC, Johansson Å, Tönjes A, Dehghan A, Chouraki V, Holliday EG, Sorice R, Kutalik Z, Lehtimäki T, Esko T, Deshmukh H, Ulivi S, Chu AY, Murgia F, Trompet S, Imboden M, Kollerits B, Pistis G, Harris TB, Launer LJ, Aspelund T, Eiriksdottir G, Mitchell BD, Boerwinkle E, Schmidt H, Cavalieri M, Rao M, Hu FB, Demirkan A, Oostra BA, de Andrade M, Turner ST, Ding J, Andrews JS, Freedman BI, Koenig W, Illig T, Döring A, Wichmann HE, Kolcic I, Zemunik T, Boban M, Minelli C, Wheeler HE, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Nöthlings U, Jacobs G, Biffar R, Endlich K, Ernst F, Homuth G, Kroemer HK, Nauck M, Stracke S, Völker U, Völzke H, Kovacs P, Stumvoll M, Mägi R, Hofman A, Uitterlinden AG, Rivadeneira F, Aulchenko YS, Polasek O, Hastie N, Vitart V, Helmer C, Wang JJ, Ruggiero D, Bergmann S, Kähönen M, Viikari J, Nikopensius T, Province M, Ketkar S, Colhoun H, Doney A, Robino A, Giulianini F, Krämer BK, Portas L, Ford I, Buckley BM, Adam M, Thun GA, Paulweber B, Haun M, Sala C, Metzger M, Mitchell P, Ciullo M, Kim SK, Vollenweider P, Raitakari O, Metspalu A, Palmer C, Gasparini P, Pirastu M, Jukema JW, Probst-Hensch NM, Kronenberg F, Toniolo D, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Siscovick DS, van Duijn CM, Borecki I, Kardia SL, Liu Y, Curhan GC, Rudan I, Gyllensten U, Wilson JF, Franke A, Pramstaller PP, Rettig R, Prokopenko I, Witteman JC, Hayward C, Ridker P, Parsa A, Bochud M, Heid IM, Goessling W, Chasman DI, Kao WH, Fox CS, Goessling W, Chasman DI, Kao WH, Fox CS; CARDIoGRAM Consortium; ICBP Consortium; CARe Consortium; Wellcome Trust Case Control Consortium 2 (WTCCC2) : Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet 8: e1002584, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okada Y, Sim X, Go MJ, Wu JY, Gu D, Takeuchi F, Takahashi A, Maeda S, Tsunoda T, Chen P, Lim SC, Wong TY, Liu J, Young TL, Aung T, Seielstad M, Teo YY, Kim YJ, Lee JY, Han BG, Kang D, Chen CH, Tsai FJ, Chang LC, Fann SJ, Mei H, Rao DC, Hixson JE, Chen S, Katsuya T, Isono M, Ogihara T, Chambers JC, Zhang W, Kooner JS, Albrecht E, Yamamoto K, Kubo M, Nakamura Y, Kamatani N, Kato N, He J, Chen YT, Cho YS, Tai ES, Tanaka T; KidneyGen Consortium; CKDGen Consortium; GUGC consortium : Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet 44: 904–909, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, Garnaas M, Tin A, Sorice R, Li Y, Taliun D, Olden M, Foster M, Yang Q, Chen M-H, Pers TH, Johnson AD, Ko Y-A, Fuchsberger C, Tayo B, Nalls M, Feitosa MF, Isaacs A, Dehghan A: d/'Adamo, P, Adeyemo, A, Dieffenbach, AK, Zonderman, AB, Nolte, IM, van der Most, PJ, Wright, AF, Shuldiner, AR, Morrison, AC, Hofman, A, Smith, AV, Dreisbach, AW, Franke, A, Uitterlinden, AG, Metspalu, A, Tonjes, A, Lupo, A, Robino, A, Johansson, A, Demirkan, A, Kollerits, B, Freedman, BI, Ponte, B, Oostra, BA, Paulweber, B, Kramer, BK, Mitchell, BD, Buckley, BM, Peralta, CA, Hayward, C, Helmer, C, Rotimi, CN, Shaffer, CM, Muller, C, Sala, C, van Duijn, CM, Saint-Pierre, A, Ackermann, D, Shriner, D, Ruggiero, D, Toniolo, D, Lu, Y, Cusi, D, Czamara, D, Ellinghaus, D, Siscovick, DS, Ruderfer, D, Gieger, C, Grallert, H, Rochtchina, E, Atkinson, EJ, Holliday, EG, Boerwinkle, E, Salvi, E, Bottinger, EP, Murgia, F, Rivadeneira, F, Ernst, F, Kronenberg, F, Hu, FB, Navis, GJ, Curhan, GC, Ehret, GB, Homuth, G, Coassin, S, Thun, G-A, Pistis, G, Gambaro, G, Malerba, G, Montgomery, GW, Eiriksdottir, G, Jacobs, G, Li, G, Wichmann, HE, Campbell, H, Schmidt, H, Wallaschofski, H, Volzke, H, Brenner, H, Kroemer, HK, Kramer, H, Lin, H, Leach, IM, Ford, I, Guessous, I, Rudan, I, Prokopenko, I, Borecki, I, Heid, IM, Kolcic, I, Persico, I, Jukema, JW, Wilson, JF, Felix, JF, Divers, J, Lambert, J-C, Stafford, JM, Gaspoz, J-M, Smith, JA, Faul, JD, Wang, JJ, Ding, J, Hirschhorn, JN, Attia, J, Whitfield, JB, Chalmers, J, Viikari, J, Coresh, J, Denny, JC, Karjalainen, J, Fernandes, JK, Endlich, K, Butterbach, K, Keene, KL, Lohman, K, Portas, L, Launer, LJ, Lyytikainen, L-P, Yengo, L, Franke, L, Ferrucci, L, Rose, LM, Kedenko, L, Rao, M, Struchalin, M, Kleber, ME, Cavalieri, M, Haun, M, Cornelis, MC, Ciullo, M, Pirastu, M, de Andrade, M, McEvoy, MA, Woodward, M, Adam, M, Cocca, M, Nauck, M, Imboden, M, Waldenberger, M, Pruijm, M, Metzger, M, Stumvoll, M, Evans, MK, Sale, MM, Kahonen, M, Boban, M, Bochud, M, Rheinberger, M, Verweij, N, Bouatia-Naji, N, Martin, NG, Hastie, N, Probst-Hensch, N, Soranzo, N, Devuyst, O, Raitakari, O, Gottesman, O, Franco, OH, Polasek, O, Gasparini, P, Munroe, PB, Ridker, PM, Mitchell, P, Muntner, P, Meisinger, C, Smit, JH, Consortium, I, Consortium, A, Cardiogram, Group, CH-HF, Consortium, EC, Kovacs, P, Wild, PS, Froguel, P, Rettig, R, Magi, R, Biffar, R, Schmidt, R, Middelberg, RPS, Carroll, RJ, Penninx, BW, Scott, RJ, Katz, R, Sedaghat, S, Wild, SH, Kardia, SLR, Ulivi, S, Hwang, S-J, Enroth, S, Kloiber, S, Trompet, S, Stengel, B, Hancock, SJ, Turner, ST, Rosas, SE, Stracke, S, Harris, TB, Zeller, T, Zemunik, T, Lehtimaki, T, Illig, T, Aspelund, T, Nikopensius, T, Esko, T, Tanaka, T, Gyllensten, U, Volker, U, Emilsson, V, Vitart, V, Aalto, V, Gudnason, V, Chouraki, V, Chen, W-M, Igl, W, Marz, W, Koenig, W, Lieb, W, Loos, RJF, Liu, Y, Snieder, H, Pramstaller, PP, Parsa, A, O/'Connell, JR, Susztak, K, Hamet, P, Tremblay, J, de Boer, IH, Boger, CA, Goessling, W, Chasman, DI, Kottgen, A, Kao, WHL, Fox, CS: Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 7: 10023, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis MC, Monda KL, Yu K, Paynter N, Azzato EM, Bennett SN, Berndt SI, Boerwinkle E, Chanock S, Chatterjee N, Couper D, Curhan G, Heiss G, Hu FB, Hunter DJ, Jacobs K, Jensen MK, Kraft P, Landi MT, Nettleton JA, Purdue MP, Rajaraman P, Rimm EB, Rose LM, Rothman N, Silverman D, Stolzenberg-Solomon R, Subar A, Yeager M, Chasman DI, van Dam RM, Caporaso NE: Genome-Wide Meta-Analysis Identifies Regions on 7p21 AHR and 15q24 CYP1A2 As Determinants of Habitual Caffeine Consumption. PLoS Genet 7: e1002033, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulem P, Gudbjartsson DF, Geller F, Prokopenko I, Feenstra B, Aben KKH, Franke B, den Heijer M, Kovacs P, Stumvoll M, Mägi R, Yanek LR, Becker LC, Boyd HA, Stacey SN, Walters GB, Jonasdottir A, Thorleifsson G, Holm H, Gudjonsson SA, Rafnar T, Björnsdottir G, Becker DM, Melbye M, Kong A, Tönjes A, Thorgeirsson T, Thorsteinsdottir U, Kiemeney LA, Stefansson K: Sequence variants at CYP1A1-CYP1A2 and AHR associate with coffee consumption. Hum Mol Genet 20: 2071–2077, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, Kugathasan S, Bradfield JP, Walters TD, Sleiman P, Kim CE, Muise A, Wang K, Glessner JT, Saeed S, Zhang H, Frackelton EC, Hou C, Flory JH, Otieno G, Chiavacci RM, Grundmeier R, Castro M, Latiano A, Dallapiccola B, Stempak J, Abrams DJ, Taylor K, McGovern D, Silber G, Wrobel I, Quiros A, Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmuda MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwillam R, Tremelling M, Delukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ, Heyman MB, Ferry GD, Kirschner B, Lee J, Essers J, Grand R, Stephens M, Levine A, Piccoli D, Van Limbergen J, Cucchiara S, Monos DS, Guthery SL, Denson L, Wilson DC, Grant SF, Daly M, Silverberg MS, Satsangi J, Hakonarson H; Western Regional Alliance for Pediatric IBD; International IBD Genetics Consortium; NIDDK IBD Genetics Consortium; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium : Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet 41: 1335–1340, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F: Proteomics. Tissue-based map of the human proteome. Science 347: 1260419, 2015 [DOI] [PubMed] [Google Scholar]
- 11.Hanke N, Staggs L, Schroder P, Litteral J, Fleig S, Kaufeld J, Pauli C, Haller H, Schiffer M: “Zebrafishing” for novel genes relevant to the glomerular filtration barrier. BioMed Res Int 2013: 658270, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hentschel DM, Mengel M, Boehme L, Liebsch F, Albertin C, Bonventre JV, Haller H, Schiffer M: Rapid screening of glomerular slit diaphragm integrity in larval zebrafish. Am J Physiol Renal Physiol 293: F1746–F1750, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Liu C, Kraja AT, Smith JA, Brody JA, Franceschini N, Bis JC, Rice K, Morrison AC, Lu Y, Weiss S, Guo X, Palmas W, Martin LW, Chen YD, Surendran P, Drenos F, Cook JP, Auer PL, Chu AY, Giri A, Zhao W, Jakobsdottir J, Lin LA, Stafford JM, Amin N, Mei H, Yao J, Voorman A, Larson MG, Grove ML, Smith AV, Hwang SJ, Chen H, Huan T, Kosova G, Stitziel NO, Kathiresan S, Samani N, Schunkert H, Deloukas P, Li M, Fuchsberger C, Pattaro C, Gorski M, Kooperberg C, Papanicolaou GJ, Rossouw JE, Faul JD, Kardia SL, Bouchard C, Raffel LJ, Uitterlinden AG, Franco OH, Vasan RS, O'Donnell CJ, Taylor KD, Liu K, Bottinger EP, Gottesman O, Daw EW, Giulianini F, Ganesh S, Salfati E, Harris TB, Launer LJ, Dorr M, Felix SB, Rettig R, Volzke H, Kim E, Lee WJ, Lee IT, Sheu WH, Tsosie KS, Edwards DR, Liu Y, Correa A, Weir DR, Volker U, Ridker PM, Boerwinkle E, Gudnason V, Reiner AP, van Duijn CM, Borecki IB, Edwards TL, Chakravarti A, Rotter JI, Psaty BM, Loos RJ, Fornage M, Ehret GB, Newton-Cheh C, Levy D, Chasman DI: Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet 48: 1162–1170, 2016 [DOI] [PMC free article] [PubMed]
- 14.Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA, Dauriz M, Hivert M-F, Raghavan S, Lipovich L, Hidalgo B, Fox K, Huffman JE, An P, Lu Y, Rasmussen-Torvik LJ, Grarup N, Ehm MG, Li L, Baldridge AS, Stancakova A, Abrol R, Besse C, Boland A, Bork-Jensen J, Fornage M, Freitag DF, Garcia ME, Guo X, Hara K, Isaacs A, Jakobsdottir J, Lange LA, Layton JC, Li M, Zhao H: J, Meidtner, K, Morrison, AC, Nalls, MA, Peters, MJ, Sabater-Lleal, M, Schurmann, C, Silveira, A, Smith, AV, Southam, L, Stoiber, MH, Strawbridge, RJ, Taylor, KD, Varga, TV, Allin, KH, Amin, N, Aponte, JL, Aung, T, Barbieri, C, Bihlmeyer, NA, Boehnke, M, Bombieri, C, Bowden, DW, Burns, SM, Chen, Y, Chen, Y-D, Cheng, C-Y, Correa, A, Czajkowski, J, Dehghan, A, Ehret, GB, Eiriksdottir, G, Escher, SA, Farmaki, A-E, Franberg, M, Gambaro, G, Giulianini, F, Goddard, WA, Goel, A, Gottesman, O, Grove, ML, Gustafsson, S, Hai, Y, Hallmans, G, Heo, J, Hoffmann, P, Ikram, MK, Jensen, RA, Jorgensen, ME, Jorgensen, T, Karaleftheri, M, Khor, CC, Kirkpatrick, A, Kraja, AT, Kuusisto, J, Lange, EM, Lee, IT, Lee, W-J, Leong, A, Liao, J, Liu, C, Liu, Y, Lindgren, CM, Linneberg, A, Malerba, G, Mamakou, V, Marouli, E, Maruthur, NM, Matchan, A, McKean-Cowdin, R, McLeod, O, Metcalf, GA, Mohlke, KL, Muzny, DM, Ntalla, I, Palmer, ND, Pasko, D, Peter, A, Rayner, NW, Renstrom, F, Rice, K, Sala, CF, Sennblad, B, Serafetinidis, I, Smith, JA, Soranzo, N, Speliotes, EK, Stahl, EA, Stirrups, K, Tentolouris, N, Thanopoulou, A, Torres, M, Traglia, M, Tsafantakis, E, Javad, S, Yanek, LR, Zengini, E, Becker, DM, Bis, JC, Brown, JB, Adrienne Cupples, L, Hansen, T, Ingelsson, E, Karter, AJ, Lorenzo, C, Mathias, RA, Norris, JM, Peloso, GM, Sheu, WHH, Toniolo, D, Vaidya, D, Varma, R, Wagenknecht, LE, Boeing, H, Bottinger, EP, Dedoussis, G, Deloukas, P, Ferrannini, E, Franco, OH, Franks, PW, Gibbs, RA, Gudnason, V, Hamsten, A, Harris, TB, Hattersley, AT, Hayward, C, Hofman, A, Jansson, J-H, Langenberg, C, Launer, LJ, Levy, D, Oostra, BA, O/'Donnell, CJ, O/'Rahilly, S, Padmanabhan, S, Pankow, JS, Polasek, O, Province, MA, Rich, SS, Ridker, PM, Rudan, I, Schulze, MB, Smith, BH, Uitterlinden, AG, Walker, M, Watkins, H, Wong, TY, Zeggini, E, The, E-IC, Laakso, M, Borecki, IB, Chasman, DI, Pedersen, O, Psaty, BM, Shyong Tai, E, van Duijn, CM, Wareham, NJ, Waterworth, DM, Boerwinkle, E, Linda Kao, WH, Florez, JC, Loos, RJF, Wilson, JG, Frayling, TM, Siscovick, DS, Dupuis, J, Rotter, JI, Meigs, JB, Scott, RA, Goodarzi, MO: Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun 6: 5897, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, Garnaas M, Tin A, Sorice R, Li Y, Taliun D, Olden M, Foster M, Yang Q, Chen MH, Pers TH, Johnson AD, Ko YA, Fuchsberger C, Tayo B, Nalls M, Feitosa MF, Isaacs A, Dehghan A, d’Adamo P, Adeyemo A, Dieffenbach AK, Zonderman AB, Nolte IM, van der Most PJ, Wright AF, Shuldiner AR, Morrison AC, Hofman A, Smith AV, Dreisbach AW, Franke A, Uitterlinden AG, Metspalu A, Tonjes A, Lupo A, Robino A, Johansson Å, Demirkan A, Kollerits B, Freedman BI, Ponte B, Oostra BA, Paulweber B, Krämer BK, Mitchell BD, Buckley BM, Peralta CA, Hayward C, Helmer C, Rotimi CN, Shaffer CM, Müller C, Sala C, van Duijn CM, Saint-Pierre A, Ackermann D, Shriner D, Ruggiero D, Toniolo D, Lu Y, Cusi D, Czamara D, Ellinghaus D, Siscovick DS, Ruderfer D, Gieger C, Grallert H, Rochtchina E, Atkinson EJ, Holliday EG, Boerwinkle E, Salvi E, Bottinger EP, Murgia F, Rivadeneira F, Ernst F, Kronenberg F, Hu FB, Navis GJ, Curhan GC, Ehret GB, Homuth G, Coassin S, Thun GA, Pistis G, Gambaro G, Malerba G, Montgomery GW, Eiriksdottir G, Jacobs G, Li G, Wichmann HE, Campbell H, Schmidt H, Wallaschofski H, Völzke H, Brenner H, Kroemer HK, Kramer H, Lin H, Leach IM, Ford I, Guessous I, Rudan I, Prokopenko I, Borecki I, Heid IM, Kolcic I, Persico I, Jukema JW, Wilson JF, Felix JF, Divers J, Lambert JC, Stafford JM, Gaspoz JM, Smith JA, Faul JD, Wang JJ, Ding J, Hirschhorn JN, Attia J, Whitfield JB, Chalmers J, Viikari J, Coresh J, Denny JC, Karjalainen J, Fernandes JK, Endlich K, Butterbach K, Keene KL, Lohman K, Portas L, Launer LJ, Lyytikäinen LP, Yengo L, Franke L, Ferrucci L, Rose LM, Kedenko L, Rao M, Struchalin M, Kleber ME, Cavalieri M, Haun M, Cornelis MC, Ciullo M, Pirastu M, de Andrade M, McEvoy MA, Woodward M, Adam M, Cocca M, Nauck M, Imboden M, Waldenberger M, Pruijm M, Metzger M, Stumvoll M, Evans MK, Sale MM, Kähönen M, Boban M, Bochud M, Rheinberger M, Verweij N, Bouatia-Naji N, Martin NG, Hastie N, Probst-Hensch N, Soranzo N, Devuyst O, Raitakari O, Gottesman O, Franco OH, Polasek O, Gasparini P, Munroe PB, Ridker PM, Mitchell P, Muntner P, Meisinger C, Smit JH, Kovacs P, Wild PS, Froguel P, Rettig R, Mägi R, Biffar R, Schmidt R, Middelberg RP, Carroll RJ, Penninx BW, Scott RJ, Katz R, Sedaghat S, Wild SH, Kardia SL, Ulivi S, Hwang SJ, Enroth S, Kloiber S, Trompet S, Stengel B, Hancock SJ, Turner ST, Rosas SE, Stracke S, Harris TB, Zeller T, Zemunik T, Lehtimäki T, Illig T, Aspelund T, Nikopensius T, Esko T, Tanaka T, Gyllensten U, Völker U, Emilsson V, Vitart V, Aalto V, Gudnason V, Chouraki V, Chen WM, Igl W, März W, Koenig W, Lieb W, Loos RJ, Liu Y, Snieder H, Pramstaller PP, Parsa A, O’Connell JR, Susztak K, Hamet P, Tremblay J, de Boer IH, Böger CA, Goessling W, Chasman DI, Köttgen A, Kao WH, Fox CS; ICBP Consortium; AGEN Consortium; CARDIOGRAM; CHARGe-Heart Failure Group; ECHOGen Consortium : Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 7: 10023, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto GL, Aguena M, Gos M, Hung C, Pilch J, Fahiminiya S, Abramowicz A, Cristian I, Buscarilli M, Naslavsky MS, Malaquias AC, Zatz M, Bodamer O, Majewski J, Jorge AAL, Pereira AC, Kim CA, Passos-Bueno MR, Bertola DR: Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J Med Genet 52: 413–421, 2015 [DOI] [PubMed] [Google Scholar]
- 17.Romano AA, Allanson JE, Dahlgren J, Gelb BD, Hall B, Pierpont ME, Roberts AE, Robinson W, Takemoto CM, Noonan JA: Noonan syndrome: clinical features, diagnosis, and management guidelines. Pediatrics 126: 746–759, 2010 [DOI] [PubMed] [Google Scholar]
- 18.George CD, Patton MA, el Sawi M, Sharland M, Adam EJ: Abdominal ultrasound in Noonan syndrome: a study of 44 patients. Pediatr Radiol 23: 316–318, 1993 [DOI] [PubMed] [Google Scholar]
- 19.Mancini F, Rigacci S, Berti A, Balduini C, Torti M: The low-molecular-weight phosphotyrosine phosphatase is a negative regulator of FcgammaRIIA-mediated cell activation. Blood 110: 1871–1878, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, Brody JA, Khetarpal SA, Crosby JR, Fornage M, Isaacs A, Jakobsdottir J, Feitosa MF, Davies G, Huffman JE, Manichaikul A, Davis B, Lohman K, Joon AY, Smith AV, Grove ML, Zanoni P, Redon V, Demissie S, Lawson K, Peters U, Carlson C, Jackson RD, Ryckman KK, Mackey RH, Robinson JG, Siscovick DS, Schreiner PJ, Mychaleckyj JC, Pankow JS, Hofman A, Uitterlinden AG, Harris TB, Taylor KD, Stafford JM, Reynolds LM, Marioni RE, Dehghan A, Franco OH, Patel AP, Lu Y, Hindy G, Gottesman O, Bottinger EP, Melander O, Orho-Melander M, Loos RJ, Duga S, Merlini PA, Farrall M, Goel A, Asselta R, Girelli D, Martinelli N, Shah SH, Kraus WE, Li M, Rader DJ, Reilly MP, McPherson R, Watkins H, Ardissino D, Zhang Q, Wang J, Tsai MY, Taylor HA, Correa A, Griswold ME, Lange LA, Starr JM, Rudan I, Eiriksdottir G, Launer LJ, Ordovas JM, Levy D, Chen YD, Reiner AP, Hayward C, Polasek O, Deary IJ, Borecki IB, Liu Y, Gudnason V, Wilson JG, van Duijn CM, Kooperberg C, Rich SS, Psaty BM, Rotter JI, O’Donnell CJ, Rice K, Boerwinkle E, Kathiresan S, Cupples LA; NHLBI GO Exome Sequencing Project : Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet 94: 223–232, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auer PL, Teumer A, Schick U, O’Shaughnessy A, Lo KS, Chami N, Carlson C, de Denus S, Dubé MP, Haessler J, Jackson RD, Kooperberg C, Perreault LP, Nauck M, Peters U, Rioux JD, Schmidt F, Turcot V, Völker U, Völzke H, Greinacher A, Hsu L, Tardif JC, Diaz GA, Reiner AP, Lettre G: Rare and low-frequency coding variants in CXCR2 and other genes are associated with hematological traits. Nat Genet 46: 629–634, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huffman JE, de Vries PS, Morrison AC, Sabater-Lleal M, Kacprowski T, Auer PL, Brody JA, Chasman DI, Chen MH, Guo X, Lin LA, Marioni RE, Müller-Nurasyid M, Yanek LR, Pankratz N, Grove ML, de Maat MP, Cushman M, Wiggins KL, Qi L, Sennblad B, Harris SE, Polasek O, Riess H, Rivadeneira F, Rose LM, Goel A, Taylor KD, Teumer A, Uitterlinden AG, Vaidya D, Yao J, Tang W, Levy D, Waldenberger M, Becker DM, Folsom AR, Giulianini F, Greinacher A, Hofman A, Huang CC, Kooperberg C, Silveira A, Starr JM, Strauch K, Strawbridge RJ, Wright AF, McKnight B, Franco OH, Zakai N, Mathias RA, Psaty BM, Ridker PM, Tofler GH, Völker U, Watkins H, Fornage M, Hamsten A, Deary IJ, Boerwinkle E, Koenig W, Rotter JI, Hayward C, Dehghan A, Reiner AP, O’Donnell CJ, Smith NL: Rare and low-frequency variants and their association with plasma levels of fibrinogen, FVII, FVIII, and vWF. Blood 126: e19–e29, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox CS, Larson MG, Leip EP, Culleton B, Wilson PW, Levy D: Predictors of new-onset kidney disease in a community-based population. JAMA 291: 844–850, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Selvin E, Manzi J, Stevens LA, Van Lente F, Lacher DA, Levey AS, Coresh J: Calibration of serum creatinine in the National Health and Nutrition Examination Surveys (NHANES) 1988-1994, 1999-2004. Am J Kidney Dis 50: 918–926, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Levey AS, de Jong PE, Coresh J, El Nahas M, Astor BC, Matsushita K, Gansevoort RT, Kasiske BL, Eckardt KU: The definition, classification, and prognosis of chronic kidney disease: a KDIGO Controversies Conference report. Kidney Int 80: 17–28, 2011 [DOI] [PubMed] [Google Scholar]
- 26.Goldstein JI, Crenshaw A, Carey J, Grant GB, Maguire J, Fromer M, O’Dushlaine C, Moran JL, Chambert K, Stevens C, Sklar P, Hultman CM, Purcell S, McCarroll SA, Sullivan PF, Daly MJ, Neale BM; Swedish Schizophrenia Consortium; ARRA Autism Sequencing Consortium : zCall: a rare variant caller for array-based genotyping: genetics and population analysis. Bioinformatics 28: 2543–2545, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, Gudnason V, Harris TB, Kathiresan S, Kraaij R, Launer LJ, Levy D, Liu Y, Mosley T, Peloso GM, Psaty BM, Rich SS, Rivadeneira F, Siscovick DS, Smith AV, Uitterlinden A, van Duijn CM, Wilson JG, O’Donnell CJ, Rotter JI, Boerwinkle E: Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One 8: e68095, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R: A Language and Environment for Statistical Computing [computer program]. Version 3.2.1 (June 18, 2015). Vienna, Austria: R Foundation for Statistical Computing; 2008
- 29.Willer CJ, Li Y, Abecasis GR: METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B, Leal SM: Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet 83: 311–321, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voorman A, Brody J, Chen H, Lumley T: seqMeta: An R package for meta-analyzing region-based tests of rare DNA variants. R Package version 14, 2014
- 32.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X: Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 89: 82–93, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Jian X, Boerwinkle E: dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat 34: E2393–E2402, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christou-Savina S, Beales PL, Osborn DP: Evaluation of zebrafish kidney function using a fluorescent clearance assay. J Vis Exp 96: e52540, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.