Abstract
African ancestry alleles may contribute to CKD among Hispanics/Latinos, but whether associations differ by Hispanic/Latino background remains unknown. We examined the association of CKD measures with African ancestry–specific APOL1 alleles that were directly genotyped and sickle cell trait (hemoglobin subunit β gene [HBB] variant) on the basis of imputation in 12,226 adult Hispanics/Latinos grouped according to Caribbean or Mainland background. We also performed an unbiased genome-wide association scan of urine albumin-to-creatinine ratios. Overall, 41.4% of participants were male, 44.6% of participants had a Caribbean background, and the mean age of all participants was 46.1 years. The Caribbean background group, compared with the Mainland background group, had a higher frequency of two APOL1 alleles (1.0% versus 0.1%) and the HBB variant (2.0% versus 0.7%). In the Caribbean background group, presence of APOL1 alleles (2 versus 0/1 copies) or the HBB variant (1 versus 0 copies) were significantly associated with albuminuria (odds ratio [OR], 3.2; 95% confidence interval [95% CI], 1.7 to 6.1; and OR, 2.6; 95% CI, 1.8 to 3.8, respectively) and albuminuria and/or eGFR<60 ml/min per 1.73 m2 (OR, 2.9; 95% CI, 1.5 to 5.4; and OR, 2.4; 95% CI, 1.7 to 3.5, respectively). The urine albumin-to-creatinine ratio genome-wide association scan identified associations with the HBB variant among all participants, with the strongest association in the Caribbean background group (P=3.1×10−10 versus P=9.3×10−3 for the Mainland background group). In conclusion, African-specific alleles associate with CKD in Hispanics/Latinos, but allele frequency varies by Hispanic/Latino background/ancestry.
Keywords: Hispanic, Latino, albuminuria, chronic kidney disease, genetic variants, African ancestry
In the United States population, Hispanics/Latinos make up 15.1% of all adults ≥20 years of age and remain one of the fastest growing minority groups in the United States.1 Hispanic/Latino ethnicity is associated with higher risk for several chronic medical conditions including CKD.2–6 However, the Hispanic/Latino population within the United States is genetically heterogeneous, and CKD risk and its risk factors (hypertension, diabetes) vary by country of origin,7–9 and likely with Hispanic/Latino continental ancestry background.
The three predominant sources of ancestry for the majority of United States Hispanics/Latinos correspond to European, Amerindian, and African founder populations with two major arms broadly representing Mainland (Mexican, Central, or South American) versus Caribbean (Puerto Rican, Dominican, or Cuban) origin.9 Whereas genetic ancestry is mainly European and Amerindian in the Mainland background group, genetic ancestry among the Caribbean group (Puerto Rican, Dominican, or Cuban) is mainly European and African.9,10 The proportion of African genetic ancestry among Hispanics/Latinos has been associated with CKD risk.7,8,11 Recent studies have demonstrated that homozygosity or compound heterozygosity for the APOL1 risk variants (G1 and G2) alleles and the heterozygous HBB rs334 missense mutation (p.Glu6Val) related to sickle cell trait (SCT) (one copy) confer increased risk of nondiabetic kidney disease among black populations.11–21 These variants are found in chromosomal regions of African ancestry. The APOL1 G1 is a two-allele haplotype consisting of two nonsynonymous coding variants (rs73885319 [S342G] and rs60910145 [I384M]) in complete linkage disequilibrium (LD), and G2 is a 6 bp deletion (rs7178513) with G2 being mutually exclusive of G1.14 A few studies have reported strong associations between nondiabetic CKD and APOL1 variants among Hispanic/Latino adults receiving dialysis22,23 or in biobank data from a clinical institution11 but to our knowledge no study has examined the association between CKD measures and the sickle cell variant in a Hispanic/Latino population. This is the first report on the association between APOL1 alleles and albuminuria and decreased eGFR in Hispanics/Latinos not selected for prior evidence of CKD, and the first report on the association between the HBB rs334 missense mutation related to SCT and CKD measures in a Hispanic/Latino study population. We hypothesized that the African ancestry–specific variants APOL1 G1 and G2 and HBB rs334 are associated with CKD and urine albumin-to-creatinine ratio (UACR) variation in Hispanics/Latinos. To detect additional genetic variants associated with UACR variation in Hispanic/Latinos, a genome-wide association study (GWAS) of UACR was performed. Because the sickle cell variant rs334 was identified as being significantly associated with UACR among the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) participants, we also report the GWAS results of UACR in this manuscript.
Results
Frequency of eGFR<60 ml/min per 1.73 m2 Is Higher among Caribbean Versus Mainland Group
Among the 12,226 participants included in the analysis, 41.4% were men, mean age was 46.1 years, and 44.6% had a Caribbean background (Table 1). The mean percentage of African ancestry was 14.7% overall, 24.8% in the Caribbean Hispanic/Latino background group, and 6.0% in the Mainland background group. Detailed information on the genetic diversity of the HCHS/SOL participants has been previously described.24 Descriptive characteristics of the HCHS/SOL study population along with CKD prevalence in the HCHS/SOL study have been previously reported.25 Overall, the frequency of albuminuria was similar between Caribbean and Mainland Hispanic/Latino background groups but the frequency of eGFR<60 ml/min per 1.73 m2 was approximately two-fold higher among the Caribbean versus the Mainland group (4.1% versus 2.1%, respectively; P=5.2×10−9).
Table 1.
Descriptive characteristics of the genotyped HCHS/SOL participants 2008–2011, n=12,226
| Characteristics | Total (n=12,226) | Caribbeana (n=5458) | Mainlanda (n=6734) |
|---|---|---|---|
| Age, yr | 46.1 (13.9) | 47.7 (14.0) | 44.8 (13.8) |
| Male, % | 41.4 | 42.8 | 40.3 |
| African ancestry,b % | 14.7 | 24.8 | 6.0 |
| Diabetes, % | 19.9 | 21.0 | 19.1 |
| Hypertension, % | 28.1 | 35.8 | 21.9 |
| Obese (BMI≥30 kg/m2), % | 42.9 | 43.4 | 42.5 |
| Albuminuria,c % | 14.1 | 15.0 | 13.4 |
| Decreased eGFR,d % | 3.0 | 4.1 | 2.0 |
| Decreased eGFR and/or albuminuria, % | 15.7 | 17.4 | 14.4 |
| Decreased eGFR and albuminuria, % | 1.6 | 2.1 | 1.3 |
Age shown as mean (SD) BMI, body mass index.
Hispanic/Latino group defined using self-reported background group and PCs and was missing in 34 participants.
Percent African ancestry calculated on an unrelated subset of HCHS/SOL participants.
Albuminuria defined as UACR≥17 mg/g in men and ≥25 mg/g in women.
Decreased eGFR defined as <60 ml/min per 1.73 m2 using the combined cystatin C and creatinine equation.
Presence of Two Copies of the APOL1 Risk Alleles and the HBB-rs334 Risk Allele Is Associated with CKD
Sixty individuals had two copies of the APOL1 high risk alleles (G1/G1 n=21; G1/G2 n=22; and G2/G2 n=17). Caribbean Hispanics/Latinos had a ten-fold higher frequency of two APOL1 risk alleles versus zero/one copy (1% versus 0.1%) and nearly three-fold higher frequency of the imputed SCT rs334 risk allele versus zero copies (2% versus 0.7%) compared with Mainland Hispanics/Latinos (Central American, Mexican, South American, n=6539). On the basis of imputation, there were four individuals whose genotype probabilities indicated that they are likely to have two copies of the rs334 risk allele (sickle cell anemia), with probabilities ranging from 0.50 to 0.70. Three of these individuals were of Caribbean background, and the other was a genetic outlier in the Mainland group due to high amounts of African ancestry. Although no HCHS/SOL participant reported having sickle cell anemia, one of these individuals had a hemoglobin concentration <8 g/dl.
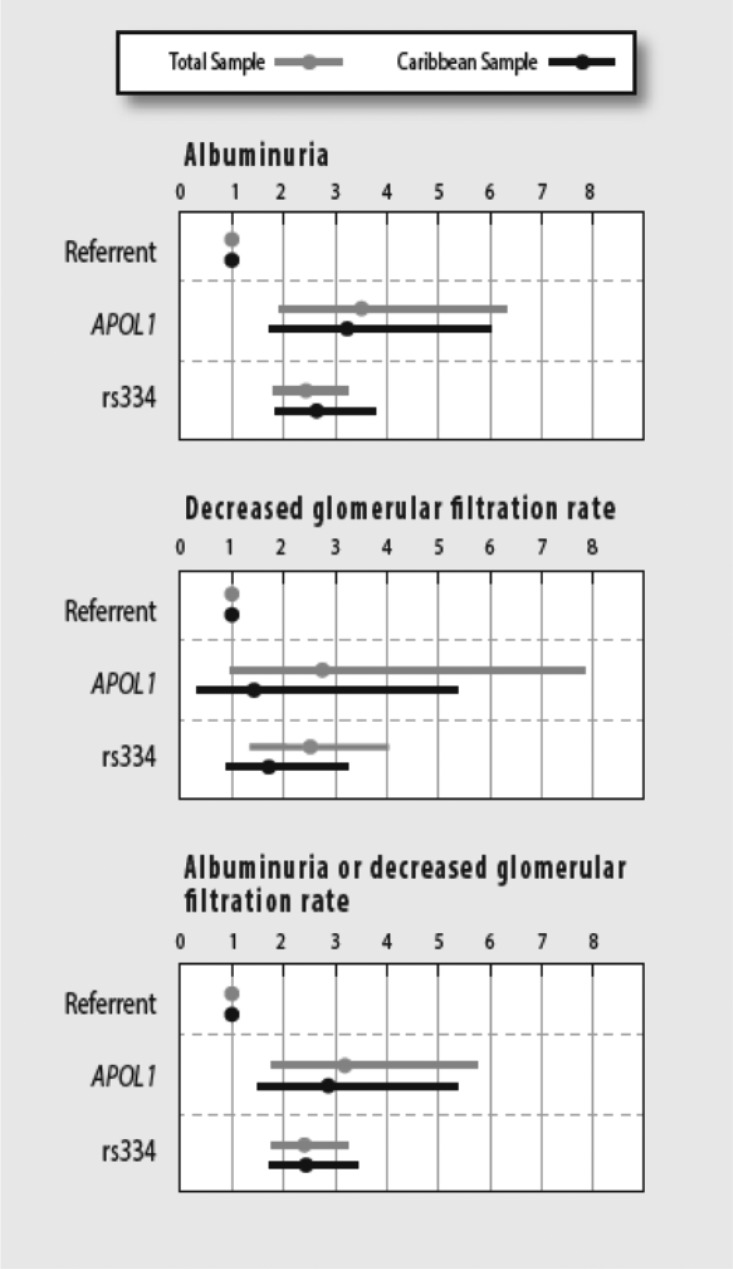
Overall, presence of two APOL1 variants versus zero/one copy was associated with 3.5-fold higher odds of albuminuria (95% CI, 1.9 to 6.4; P=3.3×10−5), 2.8-fold higher odds of low eGFR (95% CI, 1.0 to 7.9; P=0.06), and 3.2-fold higher odds of having decreased eGFR and/or albuminuria (95% CI, 1.8 to 5.7; P=0.001) among the overall sample (Figure 1, Supplemental Table 1). Among the Caribbean Hispanic/Latino background group, presence of two versus zero/one APOL1 variants was associated with 3.2-fold higher odds of albuminuria (95% CI, 1.7 to 6.1; P=2.7×10−4), a nonsignificant 1.4-fold higher odds of low eGFR (95% CI, 0.4 to 5.4; P=0.61), and 2.9-fold higher odds of having decreased eGFR and/or albuminuria (95% CI, 1.5 to 5.4; P=0.001) (Figure 1, Supplemental Table 1).
Figure 1.
Adjusted ORs (circles) and 95% CIs (horizontal bars) for CKD measures in the total HCHS/SOL sample (n=12,226) and in the Caribbean sample (n=5458) by presence of two APOL1 risk variants (referent =0 or 1 APOL1 risk variants) or the imputed sickle cell variant rs334. Models were adjusted for age, sex, diabetes, systolic BP, use of antihypertensive medications, and the first five PCs calculated using data from all HCHS/SOL participants. Hispanic/Latino group defined using self-reported background group and PCs; albuminuria defined as UACR≥17 mg/g in men and ≥25 mg/g in women; reduced GFR defined as eGFR<60 ml/min per 1.73 m2 using the combined cystatin C and creatinine equation.40
The imputed sickle cell variant rs334 was associated with CKD outcomes in the overall sample and among the Caribbean Hispanic/Latino background group for albuminuria (OR, 2.4 [95% CI, 1.8 to 3.3]; P=1.1×10−8; and OR, 2.6 [95% CI, 1.8 to 3.8]; P=2.1×10−7, respectively), and for the combined CKD outcome of albuminuria and/or decreased eGFR (OR, 2.4 [95% CI, 1.8 to 3.2]; P=6.6×10−9; and OR, 2.4 [95% CI, 1.7 to 3.5]; P=8.5×10−7, respectively). The sickle cell variant rs334 was significantly associated with lower eGFR only in the overall sample. Results did not change for the total sample or the Caribbean sample when analyses were repeated after excluding four individuals imputed to have two copies of the sickle cell variant (Supplemental Figure 1).
Genome-Wide Association Analysis of UACR Identifies an Association of HBB rs334 Risk Allele with UACR Variation
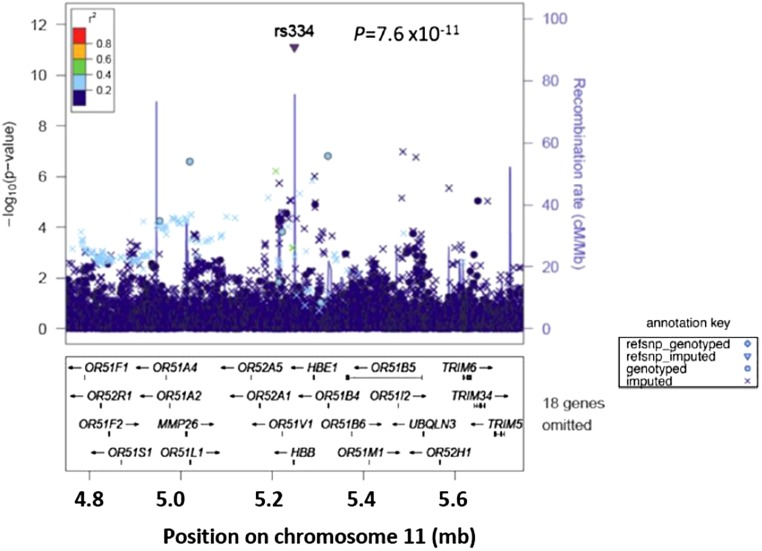
To detect genetic variants associated with interindividual variation in UACR, both overall and by Hispanic/Latino backgrounds, we performed a GWAS of UACR (log-transformed due to skewed distribution) in the overall sample and in samples stratified by Caribbean and Mainland background. There was little evidence for genomic inflation (λ=1.035) after filtering variants by minor allele frequency (MAF) ≥1% and imputation quality (oevar≥0.3) (Supplemental Figure 2). We identified significant associations with UACR for the sickle cell variant (rs334 on chromosome 11) and CUBN (chromosome 10) (Supplemental Figure 3, Table 2). The sickle cell variant rs334 was associated with UACR variation overall (P=7.6×10−12) (Figure 2) but the association was substantially stronger among the Caribbean background group (P=3.1×10−10) versus the Mainland background group (P=0.01). To further evaluate if the association of UACR was present in the Caribbean Hispanic/Latino background group with the sickle cell variant rs334 (one copy of the risk allele) as recently reported in black populations,13 we excluded individuals imputed to be homozygous for the rs334 risk allele. The evidence for association between rs334 and UACR was similar with (P=3.1×10−10) and without (P=5.5×10−10) inclusion of individuals with two copies of the risk alleles.
Table 2.
GWAS results for UACR in the HCHS/SOL population 2008–2011, n=12,226.
| Hispanic/Latino Background (Sample Size) | Genetic Variant (Chr:Position) | Coded/Other Allele | MAF | β | SEM | P Value |
|---|---|---|---|---|---|---|
| CUBN-rs144250387 | ||||||
| Overall (n=12,226) | (10:16966414) | GA/G | 0.14 | −0.11 | 0.02 | 2.1×10−8 |
| Caribbean (n=5458) | 0.14 | −0.12 | 0.03 | 2.5×10−5 | ||
| Mainland (n=6734) | 0.14 | −0.10 | 0.03 | 1.2×10−4 | ||
| HBB-rs334 | ||||||
| Overall (n=12,226) | (11:5248232) | T/A | 0.01 | −0.44 | 0.06 | 7.6×10−12 |
| Caribbean (n=5458) | 0.02 | −0.50 | 0.08 | 3.1×10−10 | ||
| Mainland (n=6734) | 0.01 | −0.31 | 0.12 | 9.3×10−3 | ||
All alleles are relative to the plus strand. Chr, chromosome.
Figure 2.
Regional plot of the association of the imputed sickle cell related HBB variant with UACR in genome-wide association using the overall sample of Hispanics/Latinos (n=12,226). The y axis shows the −log10 P values of single nucleotide polymorphisms, and the x axis shows their chromosomal position on chromosome 11 and genes are listed below. The significance threshold is a P value <5.0×10−8 based on multiple testing (see Concise Methods).
At the chromosome 10 locus, the associated variant is an imputed intronic insertion/deletion variant of CUBN (rs144250387; MAF=0.14; P=2.1×10−8), a gene previously associated with UACR in European ancestry populations.26 In contrast to the sickle cell variant findings, this CUBN indel had similar allele frequency and association estimates with UACR in Mainland and Caribbean Hispanics/Latinos (Table 2). The LD of rs144250387 with the previously described CUBN variant rs1801239 is 0.38 in the HCHS/SOL study (Supplemental Figure 4). After conditioning on the known variant (rs1801239) in the CUBN locus, the potentially new CUBN variant association with UACR was attenuated but still present (P=1.3 × 10−4), thus suggesting that it may be an independent signal at this locus in the Hispanic/Latino population or perhaps in stronger LD with the causal variant(s). The P value for rs1801239 after conditioning on the new CUBN variant was no longer significant in the HCHS/SOL study (P>0.05).
Discussion
This study demonstrates that African ancestral genetic variants associated with CKD risk are associated with CKD in Hispanic/Latino adults. The frequencies of these CKD-related genetic variants vary by Hispanic background, with risk allele frequency up to 14-fold higher among Hispanics/Latinos with a Caribbean background versus those with a Mainland background. According to the 2010 United States census, a Caribbean background is present in approximately 15% of the United States Hispanic/Latino population.1 However, even within the Mainland Hispanic/Latino background group, some individuals may have substantial amounts of African ancestry, although this is uncommon.10,27 Findings from this study suggest that the link between African ancestry and CKD risk may be attributed in part to known CKD risk alleles that reside in chromosomal regions of African ancestry including the APOL1 G1 and G2 alleles and the rs334 in HBB.
The frequency of APOL1 alleles associated with CKD has been examined in Hispanic/Latino adults receiving dialysis.22,23 A genetic analysis of 229 Hispanic adults, mostly of Caribbean background (Puerto Rico and Dominican Republic), receiving dialysis in facilities in New York City for management of ESRD found two APOL1 risk alleles present in 20% and 6% of patients with nondiabetic and diabetic ESRD, respectively.22 Among black patients with ESRD, the prevalence of two APOL1 risk alleles was somewhat higher at 39% and 22% with nondiabetic and diabetic ESRD, respectively.22 The association of APOL1 alleles with prevalent albuminuria was reported in the population-based Atherosclerosis Risk in Communities study (OR, 2.14; 95% CI, 1.55 to 2.97)15 and in the Dallas Heart Study (OR, 2.8; 95% CI, 1.8 to 4.1).28 Our study showed somewhat higher odds of albuminuria in Hispanics/Latinos with two APOL1 alleles (OR, 3.2; 95% CI, 1.7 to 6.3, among Caribbeans), but overlap existed in the 95% CIs with these previously reported cross-sectional associations in black populations. More recently, the Coronary Artery Development in Young Adults study showed an association of two APOL1 risk alleles versus zero/one copy with albuminuria among young black individuals (OR, 2.32; 95% CI, 1.73 to 3.13) which preceded the decline in eGFR over a period of 9 years.29 Future studies should examine whether APOL1 alleles are associated with incident albuminuria and progression of CKD in the Hispanic/Latino population.
An additional African ancestry–specific and CKD risk genetic variant is the HBB rs334 missense mutation (p.Glu6Val) in the β-globin chain of hemoglobin related to SCT (one copy) and sickle cell anemia (two copies). To our knowledge, no previous study has examined the association between the sickle cell variant rs334 and CKD among Hispanic/Latino adults. In an analysis that included >15,000 black individuals from five different cohort studies, SCT (carriers of one rs334 risk allele) was present in 7.8% and was associated with higher odds of albuminuria (OR, 1.86; 95% CI, 1.49 to 2.31) and a higher annual rate of eGFR decline (0.218 ml/min per 1.73 m2; 95% CI, 0.06 to 0.37).13
In the United States, all newborns are genotyped for rs334 to screen for sickle cell disease, which affects one in 16,300 Hispanic/Latino Americans. Although it remains unclear whether individuals with SCT are at higher risk for ESRD, screening for CKD and CKD risk factors (hypertension and diabetes) among individuals with SCT could inform the care of individuals if it leads to earlier interventions to mitigate kidney disease progression. Cost and benefits of this approach should be evaluated in future studies. The most common manifestation of CKD is albuminuria, which is also highly prevalent in Hispanic/Latino adults5,30 including HCHS/SOL participants.25 Although the prevalence of APOL1/SCT variants associated with albuminuria varied by Hispanic background group, HCHS/SOL participants had similar frequency of albuminuria between Caribbean and Mainland groups. Our GWAS of UACR identified the CUBN locus, previously associated with UACR variation in adults with European or African ancestry26 and with diabetes-associated ESRD risk in populations with recent African ancestry.31 This CUBN indel variant associated with UACR had similar allele frequency and association estimates across Hispanic/Latino backgrounds, thus representing a shared genetic risk across Hispanic/Latino admixed populations. Although this CUBN variant is in moderate LD with the CUBN SNP previously identified in European ancestry, our conditional analysis provides evidence that this variant is an independent signal at the locus. CUBN encodes cubilin, a high-affinity apo A-1 receptor present in the intestines and kidney that facilitates endocytosis of high-density lipoprotein and micronutrients including albumin.32 It remains unclear how variants in CUBN lead to increased urine albumin excretion and whether these variants confer risk for ESRD.
The strengths of this study include the large number of HCHS/SOL participants and the inclusion of Hispanic/Latino adults from six different Hispanic/Latino background groups who have different African admixtures. Limitations include use of imputation analysis to determine the predicted probabilities for rs334 alleles, and lack of information on other hemoglobin mutations that may coexist with sickle cell disease and trait. We cannot determine the prevalence of APOL1 and SCT in the United States Hispanic/Latino population as our sample is limited to specific subgroups targeted for recruitment in the HCHS/SOL. However, this may be the first study to examine both the APOL1 and sickle cell rs334 variants in a large number of Hispanic/Latino adults with Mainland or Caribbean background.
In summary, genetic variants that confer CKD risk and are associated with African ancestry are also associated with increased CKD risk and UACR variation among Hispanic/Latino adults.
Concise Methods
For a complete description of the methods see Supplemental Material.
Study Population
The details of the design and implementation of the HCHS/SOL cohort have been previously described.33,34 Briefly, HCHS/SOL is a community-based cohort study of 16,415 self-identified Hispanic/Latino adults aged 18–74 years from randomly selected households in four United States field centers (Chicago, Bronx, Miami, and San Diego) with baseline examination (2008–2011) and yearly telephone follow-up assessment for at least 3 years. The HCHS/SOL cohort includes participants self-identified as having Hispanic/Latino background, with the largest groups being Central American (n=1730), Cuban (n=2348), Dominican (n=1460), Mexican (n=6471), Puerto-Rican (n=2728), and South American (n=1068). The study was approved by the Institutional Review Board at each participating institution. All participants provided written informed consent. Approximately 13,000 participants consented to have DNA extracted for genetic studies, and this study is based on the sample with GWAS genotyping (12,803). Participants with missing information on UACR or serum creatinine and cystatin C for estimation of GFR, and 19 individuals with 39%–100% East Asian ancestry, were excluded.24 Participants were assigned genetic analysis groups on the basis of self-reported background and principal components (PCs).10 A total of 12,226 individuals with data on UACR, estimated GFR, and genetic analysis group were available for analyses.
Genotyping and Imputation
HCHS/SOL participants were genotyped using a Custom Illumina Omni2.5M array (HumanOmni2.5–8v1–1), which was called using GenomeStudio v2011.1, Genotyping Module v 1.9.4, and GenTrain version 2. The array contains a total of 2,536,661 SNPs of which 2,427,090 are from a standard Illumina Omni2.5M array (HumanOmni2.5–8v1–1) and the remaining 109,571 are custom SNPS including the APOL1 alleles G1 (rs73885319 and rs60910145) and G2 (rs71785313). Quality control was completed using methods described by Laurie et al.35 Untyped variants were imputed using the 1000 Genomes Project phase 1 cosmopolitan reference panel. SHAPEIT2 (v2.r644) was used for prephasing, followed by imputation with IMPUTE2 (v2.3.0) software.24 PC-AiR36 was used to estimate PCs in an unrelated subset of SOL subjects, excluding 19 subjects with substantial Asian ancestry, and project PC values for the remaining related subjects.24 Kinship coefficients were estimated using PC-Relate36 by conditioning on the top five PCs.24 High risk APOL1 genotypes were defined by the presence of two risk alleles (G1/G1, G2/G2, or G1/G2) on the basis of variants rs73885319 (G1) and rs71785313 (G2). Rs334 was imputed (imputation quality rsq=0.83), and genotype probabilities were used to calculate risk allele dosage.
Continental ancestry proportions were calculated using the ADMIXTURE software in supervised mode with three ancestral populations (West African, European, and Amerindian) for a subset of 10,642 mutually unrelated HCHS/SOL participants (kinship coefficient <0.044).37 The referent samples were from the Human Genome Diversity Project, 1000 Genomes, and controls genotyped with the HCHS/SOL subjects.38 Because of HCHS/SOL established protocols for genetic analyses, associations between the overall percentage of ancestry and CKD traits were not examined.
CKD Phenotypes
The baseline examination included collection of fasting blood samples and a midstream urine sample. CKD phenotypes included albuminuria defined as UACR≥17 mg/g in men and ≥25 mg/g in women,39 decreased eGFR<60 ml/min per 1.73 m2 on the basis of the serum creatinine and cystatin C CKD-EPI equation,40 and a composite outcome of decreased eGFR and/or albuminuria. Serum creatinine measurements were isotope dilution mass spectrometry traceable. Serum cystatin C was measured using a turbidimetric method on the Roche Modular P Chemistry Analyzer (Gentian AS, Moss, Norway). Albumin (mg/dl) and creatinine (g/dl) were measured in urine specimens using an immunoturbidimetric method and a creatinase enzymatic method, respectively.
Statistical Analyses
We used generalized linear mixed models to test the association of these variants with CKD case status while accounting for the correlation structure due to genetic relatedness and sampling design.24,41 All models were adjusted for age, sex, diabetes, systolic BP, use of antihypertensive medications, and the first five PCs calculated using data from all HCHS/SOL participants. Analyses were completed in the total sample set and after stratifying into Caribbean versus Mainland Hispanic/Latino background groups. Because of the low frequency of APOL1 risk variants in the Mainland group, we report association results for the Caribbean Hispanic/Latino background group only. For APOL1 analyses, we used a recessive model as previously described.18 For the GWAS of UACR, we examined UACR as a log-transformed variable and used additive genetic models, filtering by variants with an MAF≥1% and imputation quality rsq ≥0.3. These models were adjusted for age, sex, the first five PCs, and the HCHS/SOL study site. The significance threshold to account for multiple testing in the UACR GWAS was set at P<5×10−8. In sensitivity analyses for the association between the sickle cell variant rs334 and UACR, we repeated analyses in individuals of Caribbean background after the exclusion of individuals imputed to have two copies of rs334.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors thank the staff and participants of Hispanic Community Health Study/Study of Latinos (HCHS/SOL) for their important contributions. Investigators’ website: http://www.cscc.unc.edu/hchs/.
The baseline examination of HCHS/SOL was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Neurological Disorders and Stroke, National Institutes of Health Institution–Office of Dietary Supplements. The Genetic Analysis Center at Washington University was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Additional analysis support was provided by the NHLBI grant HL123677-01 (N.F.). Genotyping efforts were supported by NHLBI HSN 26220/20054C, National Center for Advancing Translational Sciences Clinical and Translational Science Institute grant UL1TR000124, and NIDDK Diabetes Research Center grant DK063491.
This manuscript has been reviewed by the HCHS/SOL Publications Committee for scientific content and consistency of data interpretation with previous HCHS/SOL publications. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the NHLBI, the National Institutes of Health, or the US Department of Health and Human Services.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016030357/-/DCSupplemental.
References
- 1.United States Census Bureau: 2010 U.S. census. Available at: www.census.gov. Accessed October 1, 2015
- 2.Peralta CA, Shlipak MG, Fan D, Ordoñez J, Lash JP, Chertow GM, Go AS: Risks for end-stage renal disease, cardiovascular events, and death in Hispanic versus non-Hispanic white adults with chronic kidney disease. J Am Soc Nephrol 17: 2892–2899, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Levey AS, Coresh J, Balk E, Kausz AT, Levin A, Steffes MW, Hogg RJ, Perrone RD, Lau J, Eknoyan G; National Kidney Foundation : National Kidney Foundation practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Ann Intern Med 139: 137–147, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Jones CA, Francis ME, Eberhardt MS, Chavers B, Coresh J, Engelgau M, Kusek JW, Byrd-Holt D, Narayan KM, Herman WH, Jones CP, Salive M, Agodoa LY: Microalbuminuria in the US population: third National Health and Nutrition Examination Survey. Am J Kidney Dis 39: 445–459, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Bryson CL, Ross HJ, Boyko EJ, Young BA: Racial and ethnic variations in albuminuria in the US Third National Health and Nutrition Examination Survey (NHANES III) population: associations with diabetes and level of CKD. Am J Kidney Dis 48: 720–726, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Kramer H, Jacobs DR Jr, Bild D, Post W, Saad MF, Detrano R, Tracy R, Cooper R, Liu K; The Multi-Ethnic Study of Atherosclerosis : Urine albumin excretion and subclinical cardiovascular disease. Hypertension 46: 38–43, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Pabon-Nau LP, Cohen A, Meigs JB, Grant RW: Hypertension and diabetes prevalence among U.S. Hispanics by country of origin: the National Health Interview Survey 2000-2005. J Gen Intern Med 25: 847–852, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peralta CA, Li Y, Wassel C, Choudhry S, Palmas W, Seldin MF, Risch N, Siscovick D, Arnett D, Psaty B, Shlipak MG: Differences in albuminuria between Hispanics and whites: an evaluation by genetic ancestry and country of origin: the multi-ethnic study of atherosclerosis. Circ Cardiovasc Genet 3: 240–247, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González Burchard E, Borrell LN, Choudhry S, Naqvi M, Tsai HJ, Rodriguez-Santana JR, Chapela R, Rogers SD, Mei R, Rodriguez-Cintron W, Arena JF, Kittles R, Perez-Stable EJ, Ziv E, Risch N: Latino populations: a unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am J Public Health 95: 2161–2168, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manichaikul A, Palmas W, Rodriguez CJ, Peralta CA, Divers J, Guo X, Chen WM, Wong Q, Williams K, Kerr KF, Taylor KD, Tsai MY, Goodarzi MO, Sale MM, Diez-Roux AV, Rich SS, Rotter JI, Mychaleckyj JC: Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet 8: e1002640, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udler MS, Nadkarni GN, Belbin G, Lotay V, Wyatt C, Gottesman O, Bottinger EP, Kenny EE, Peter I: Effect of genetic African ancestry on eGFR and kidney disease. J Am Soc Nephrol 26: 1682–1692, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freedman BI, Divers J, Palmer ND: Population ancestry and genetic risk for diabetes and kidney, cardiovascular, and bone disease: modifiable environmental factors may produce the cures. Am J Kidney Dis 62: 1165–1175, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naik RP, Derebail VK, Grams ME, Franceschini N, Auer PL, Peloso GM, Young BA, Lettre G, Peralta CA, Katz R, Hyacinth HI, Quarells RC, Grove ML, Bick AG, Fontanillas P, Rich SS, Smith JD, Boerwinkle E, Rosamond WD, Ito K, Lanzkron S, Coresh J, Correa A, Sarto GE, Key NS, Jacobs DR, Kathiresan S, Bibbins-Domingo K, Kshirsagar AV, Wilson JG, Reiner AP: Association of sickle cell trait with chronic kidney disease and albuminuria in African Americans. JAMA 312: 2115–2125, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster MC, Coresh J, Fornage M, Astor BC, Grams M, Franceschini N, Boerwinkle E, Parekh RS, Kao WH: APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol 24: 1484–1491, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, Feldman HI, Parekh RS, Kusek JW, Greene TH, Fink JC, Anderson AH, Choi MJ, Wright JT Jr, Lash JP, Freedman BI, Ojo A, Winkler CA, Raj DS, Kopp JB, He J, Jensvold NG, Tao K, Lipkowitz MS, Appel LJ; AASK Study Investigators; CRIC Study Investigators : APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 369: 2183–2196, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, Mokrzycki MH, Kimmel PL, Limou S, Ahuja TS, Berns JS, Fryc J, Simon EE, Smith MC, Trachtman H, Michel DM, Schelling JR, Vlahov D, Pollak M, Winkler CA: APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 22: 2129–2137, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K: Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guasch A, Navarrete J, Nass K, Zayas CF: Glomerular involvement in adults with sickle cell hemoglobinopathies: Prevalence and clinical correlates of progressive renal failure. J Am Soc Nephrol 17: 2228–2235, 2006 [DOI] [PubMed] [Google Scholar]
- 20.McPherson Yee M, Jabbar SF, Osunkwo I, Clement L, Lane PA, Eckman JR, Guasch A: Chronic kidney disease and albuminuria in children with sickle cell disease. Clin J Am Soc Nephrol 6: 2628–2633, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosmanova EO, Zaidi S, Wan JY, Adams-Graves PE: Prevalence and progression of chronic kidney disease in adult patients with sickle cell disease. J Investig Med 62: 804–807, 2014 [DOI] [PubMed] [Google Scholar]
- 22.Tzur S, Rosset S, Skorecki K, Wasser WG: APOL1 allelic variants are associated with lower age of dialysis initiation and thereby increased dialysis vintage in African and Hispanic Americans with non-diabetic end-stage kidney disease. Nephrol Dial Transplant 27: 1498–1505, 2012 [DOI] [PubMed] [Google Scholar]
- 23.Behar DM, Rosset S, Tzur S, Selig S, Yudkovsky G, Bercovici S, Kopp JB, Winkler CA, Nelson GW, Wasser WG, Skorecki K: African ancestry allelic variation at the MYH9 gene contributes to increased susceptibility to non-diabetic end-stage kidney disease in Hispanic Americans. Hum Mol Genet 19: 1816–1827, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, Sofer T, Fernández-Rhodes L, Justice AE, Graff M, Young KL, Seyerle AA, Avery CL, Taylor KD, Rotter JI, Talavera GA, Daviglus ML, Wassertheil-Smoller S, Schneiderman N, Heiss G, Kaplan RC, Franceschini N, Reiner AP, Shaffer JR, Barr RG, Kerr KF, Browning SR, Browning BL, Weir BS, Avilés-Santa ML, Papanicolaou GJ, Lumley T, Szpiro AA, North KE, Rice K, Thornton TA, Laurie CC: Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet 98: 165–184, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricardo AC, Flessner MF, Eckfeldt JH, Eggers PW, Franceschini N, Go AS, Gotman NM, Kramer HJ, Kusek JW, Loehr LR, Melamed ML, Peralta CA, Raij L, Rosas SE, Talavera GA, Lash JP: Prevalence and Correlates of CKD in Hispanics/Latinos in the United States. Clin J Am Soc Nephrol 10: 1757–1766, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Böger CA, Chen MH, Tin A, Olden M, Köttgen A, de Boer IH, Fuchsberger C, O’Seaghdha CM, Pattaro C, Teumer A, Liu CT, Glazer NL, Li M, O’Connell JR, Tanaka T, Peralta CA, Kutalik Z, Luan J, Zhao JH, Hwang SJ, Akylbekova E, Kramer H, van der Harst P, Smith AV, Lohman K, de Andrade M, Hayward C, Kollerits B, Tönjes A, Aspelund T, Ingelsson E, Eiriksdottir G, Launer LJ, Harris TB, Shuldiner AR, Mitchell BD, Arking DE, Franceschini N, Boerwinkle E, Egan J, Hernandez D, Reilly M, Townsend RR, Lumley T, Siscovick DS, Psaty BM, Kestenbaum B, Haritunians T, Bergmann S, Vollenweider P, Waeber G, Mooser V, Waterworth D, Johnson AD, Florez JC, Meigs JB, Lu X, Turner ST, Atkinson EJ, Leak TS, Aasarød K, Skorpen F, Syvänen AC, Illig T, Baumert J, Koenig W, Krämer BK, Devuyst O, Mychaleckyj JC, Minelli C, Bakker SJ, Kedenko L, Paulweber B, Coassin S, Endlich K, Kroemer HK, Biffar R, Stracke S, Völzke H, Stumvoll M, Mägi R, Campbell H, Vitart V, Hastie ND, Gudnason V, Kardia SL, Liu Y, Polasek O, Curhan G, Kronenberg F, Prokopenko I, Rudan I, Arnlöv J, Hallan S, Navis G, Parsa A, Ferrucci L, Coresh J, Shlipak MG, Bull SB, Paterson NJ, Wichmann HE, Wareham NJ, Loos RJ, Rotter JI, Pramstaller PP, Cupples LA, Beckmann JS, Yang Q, Heid IM, Rettig R, Dreisbach AW, Bochud M, Fox CS, Kao WH; CKDGen Consortium : CUBN is a gene locus for albuminuria. J Am Soc Nephrol 22: 555–570, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL: The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet 96: 37–53, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR: Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol 22: 2098–2105, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peralta CA, Bibbins-Domingo K, Vittinghoff E, Lin F, Fornage M, Kopp JB, Winkler CA: APOL1 genotype and race differences in incident albuminuria and renal function decline. J Am Soc Nephrol 27: 887–893, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolly SE, Burrows NR, Chen SC, Li S, Jurkovitz CT, Narva AS, Norris KC, Shlipak MG: Racial and ethnic differences in albuminuria in individuals with estimated GFR greater than 60 mL/min/1.73 m(2): results from the Kidney Early Evaluation Program (KEEP). Am J Kidney Dis 55[Suppl 2]: S15–S22, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J, Guan M, Bowden DW, Ng MC, Hicks PJ, Lea JP, Ma L, Gao C, Palmer ND, Freedman BI: Association analysis of the cubilin (CUBN) and megalin (LRP2) genes with ESRD in African Americans. Clin J Am Soc Nephrol 11: 1034–1043, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozyraki R, Fyfe J, Kristiansen M, Gerdes C, Jacobsen C, Cui S, Christensen EI, Aminoff M, de la Chapelle A, Krahe R, Verroust PJ, Moestrup SK: The intrinsic factor-vitamin B12 receptor, cubilin, is a high-affinity apolipoprotein A-I receptor facilitating endocytosis of high-density lipoprotein. Nat Med 5: 656–661, 1999 [DOI] [PubMed] [Google Scholar]
- 33.Sorlie PD, Avilés-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, Giachello AL, Schneiderman N, Raij L, Talavera G, Allison M, Lavange L, Chambless LE, Heiss G: Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol 20: 629–641, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anonymous: Hispanic Community Health Study/Study of Latinos study manuals: Study protocol, general description and study management. Available at: https://www2.cscc.unc.edu/hchs/manuals-forms. Accessed
- 35.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, Gabriel SB, Harris EL, Hu FB, Jacobs KB, Kraft P, Landi MT, Lumley T, Manolio TA, McHugh C, Painter I, Paschall J, Rice JP, Rice KM, Zheng X, Weir BS; GENEVA Investigators : Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol 34: 591–602, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conomos MP, Miller MB, Thornton TA: Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol 39: 276–293, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexander DH, Novembre J, Lange K: Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavalli-Sforza LL: The Human Genome Diversity Project: past, present and future. Nat Rev Genet 6: 333–340, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Mattix HJ, Hsu CY, Shaykevich S, Curhan G: Use of the albumin/creatinine ratio to detect microalbuminuria: implications of sex and race. J Am Soc Nephrol 13: 1034–1039, 2002 [DOI] [PubMed] [Google Scholar]
- 40.Inker LA, Schmid CH, Tighiouart H, Eckfeldt JH, Feldman HI, Greene T, Kusek JW, Manzi J, Van Lente F, Zhang YL, Coresh J, Levey AS; CKD-EPI Investigators : Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 367: 20–29, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, Wang C, Conomos MP, Stilp AM, Li Z, Sofer T, Szpiro AA, Chen W, Brehm JM, Celedón JC, Redline S, Papanicolaou GJ, Thornton TA, Laurie CC, Rice K, Lin X: Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet 98: 653–666, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.