Abstract
Long-term kidney transplant outcomes remain suboptimal, delineating an unmet medical need. Although current immunosuppressive therapy in kidney transplant recipients is effective, dosing is conventionally adjusted empirically on the basis of time after transplant or altered in response to detection of kidney dysfunction, histologic evidence of allograft damage, or infection. Such strategies tend to detect allograft rejection after significant injury has already occurred, fail to detect chronic subclinical inflammation that can negatively affect graft survival, and ignore specific risks and immune mechanisms that differentially contribute to allograft damage among transplant recipients. Assays and biomarkers that reliably quantify and/or predict the risk of allograft injury have the potential to overcome these deficits and thereby, aid clinicians in optimizing immunosuppressive regimens. Herein, we review the data on candidate biomarkers that we contend have the highest potential to become clinically useful surrogates in kidney transplant recipients, including functional T cell assays, urinary gene and protein assays, peripheral blood cell gene expression profiles, and allograft gene expression profiles. We identify barriers to clinical biomarker adoption in the transplant field and suggest strategies for moving biomarker-based individualization of transplant care from a research hypothesis to clinical implementation.
Keywords: transplant outcomes, transplantation, biomarker
Despite advancements in understanding immune responses induced in transplanted organs and despite diminution in acute rejection (AR) rates,1 lifelong immunosuppression is required after kidney transplantation, and long-term allograft survival rates remain suboptimal.2 The causes of late allograft loss are multiple and include late rejection as well as recipient death with a functioning graft.3 The prevailing immunosuppression strategies are center-based protocols, with potential for overimmunosuppression (predisposing to infection or drug toxicity) or underimmunosuppression (predisposing to immunologic graft injury) of individual transplant recipients. Currently used tactics used to guide immunosuppression choices and dosing are relatively rudimentary and include epidemiologic parameters (e.g., age and self-declared race), HLA mismatching, and donor-specific, anti-HLA antibody (DSA) screening. Identification and validation of biomarkers that correlate with and/or predict allograft injury and that could improve therapeutic decision making are priorities for the transplantation community.4 Herein, we review available data on immune monitoring assays that are moving toward clinical use in kidney transplantation, highlighting assays approved by the US Food and Drug Administration (FDA) and those that are validated in external cohorts but are not FDA approved (Table 1). We also discuss selected promising assays that require additional work before clinical implementation. Finally, we offer a framework monitoring strategy for consideration, and we outline likely barriers and potential solutions to clinical adoption of biomarker testing in transplantation.
Table 1.
Selected biomarkers for outcomes in kidney transplantation: summary of current status
| Authors | Assay Name | Assay Type | Timing Post-Transplant | Outcome | Discovery Set: Se/sp/ppv/npv | Validation Set: Se/sp/ppv/npv | Training/Test Sample | Biomarker Lifecycle |
|---|---|---|---|---|---|---|---|---|
| FDA-approved assays | ||||||||
| Patel and Terasaki19 | CDC crossmatch | Microcytotoxicity assay | Pretransplant | Hyper-AR/early graft loss | 0.75/0.97/0.80/0.97 | Not applicable, no validation set | 225 | FDAA: Y; Comm: Y |
| Mahoney et al.149 | Flow crossmatch | Flow cytometry | Pretransplant | Early graft loss (<2 mo) | 0.71/0.74/0.33/0.93 | Not applicable, no validation set | 90 | FDAA: Y; Comm: Y |
| Pei et al.150 | Lumine | HLA beads; flow cytometry | Variable pre-/post-transplant | Anti-HLA Ab | Details not available in manuscript | Not applicable, no validation set | 10 | FDAA: Y; Comm: Ya |
| Ashokkumar et al.61 | Pleximmune | T cytotoxic memory cell assay | Rejection episodes | Biopsy-proven AR | 0.88/0.94/0.93/0.88 | 1.0/0.86/0.80/1.0 | 32/11 | FDAA: Y; Comm: Y |
| He et al.63 | Cylex-Immuknow | Lymphocyte ATP generation assay | Serial <30 mo | CD4-T cell function | Details not available in manuscript | Not applicable, no validation set | 42 | FDAA: Y; Comm: Y |
| Loupy et ald.32 | C1q binding assay | Flow cytometric C1q bindingb | Baseline, 1 yr, rejection episode | TCMR/ABMR/graft loss | Details not available in manuscript | Not applicable, no validation set | 1016 | FDAA: Y; Comm: Yb |
| Selected externally validated assays in kidney transplantation (pending FDA approval) | ||||||||
| Hricik et al.47 | IFN-γ ELISPOT | Donor-reactive memory T cell | Preatransplant | De novo DSA and/or rejection | 1.0/0.67/0.67/1.0 | Not applicable, no validation set | 21 | FDAA: N; Comm: N |
| Hricik et al.70 | Urine CXCL9 | Urine ELISA | Serial <6 mo, rejection episode | TCMR | 0.85/0.81/0.68/0.92 | Not applicable, no validation set | 258 | FDAA: N; Comm: N |
| Suthanthiran et al.86 | Urine three-gene signature | Urinary RNA by qPCR | Serial <12 mo, rejection episode | AR | Details not available in manuscript | Details not available in manuscript | 485 (4300 urine samples) | FDAA: N; Comm: N |
| Roedder et al.119 | KSORT | Peripheral blood RNA by qPCR | Serial <24 mo, rejection episode | Biopsy-proven AR | 0.83/0.91/0.81/0.91 | 0.91/0.99/0.95/0.98 | 143/124 | FDAA: N; Comm: N |
| Halloran et al.112 | ENDAT | Graft biopsy RNA by microarray | Variable (1 wk to 31 yr) | ABMR | 0.69/0.85/0.50/0.94 | 0.67/0.90/0.64/0.91 | 300/403 | FDAA: N; Comm: N |
| O’Connell et al.18 | GoCAR score | Graft biopsy RNA by microarray (3-mo protocol biopsies) | 3 mo biopsy | Prediction of fibrosis/histologic progression | 0.93/0.95/0.93/0.95 | 0.67/0.92/0.86/0.81 | 159/45 | FDAA: N; Comm: N |
Se, sensitivity; sp, specificity; ppv, positive predictive value; npv, negative predictive value; CDC, complement dependent cytotoxicity; FDAA, US FDA approved; Y, yes; Comm, commercialized assay available; TCMR, T cell–mediated rejection (Banff ≥1A); N, no; KSORT, kidney solid organ response test; ENDAT, endothelial-associated transcripts; GoCAR, Genomics of Chronic Allograft Rejection.
Assay approved for detection of DSAs (not for quantification).
C1q binding Bindazyme assay is FDA approved, but flow cytometric C1qScreen (One-Lambda) used in this study is not FDA approved.
Biomarker Overview
Clinically useful biomarkers may be prognostic, be predictive, and/or serve as surrogate end points,5–7 and they may be within or outside the causal pathway of the disease process for which they are being used. In kidney transplantation, biomarkers can be used to predict/diagnose AR, immunologic/operational tolerance,8,9 overimmunosuppression, nonadherence, opportunistic infections,10,11 progressive, chronic graft injury,12 and graft loss. We focus on biomarkers for AR, subclinical alloimmune injury, and graft loss.
An ideal immune biomarker should rapidly, accurately, inexpensively, and noninvasively identify subjects with or at risk for incipient allograft injury (e.g., rejection), discern the type of injury (e.g., antibody mediated versus cellular rejection), and differentiate rejection from other causes of graft damage (e.g., infection). Clinically useful assays have high sensitivity, specificity, and negative/positive predictive values and a diagnostic area under the receiver operator characteristic curve nearing 1.0.7,13 Serial biomarker measurements should correlate with remission of the pathologic process, especially in response to therapeutic interventions. For most transplant trials, statistical analyses of biomarkers have been on the basis of comparison with the gold standard of histologic detection of rejection on an allograft biopsy. Because pathologic definitions of rejection are subject to intra-/interobserver and/or sampling error–related variability in biopsy readouts,14,15 such variability must be considered when interpreting biomarker study results. Although graft loss is an undisputable outcome for any biomarker comparison, it is challenging to design prospective studies of sufficient length and power to adequately study graft loss.16–18
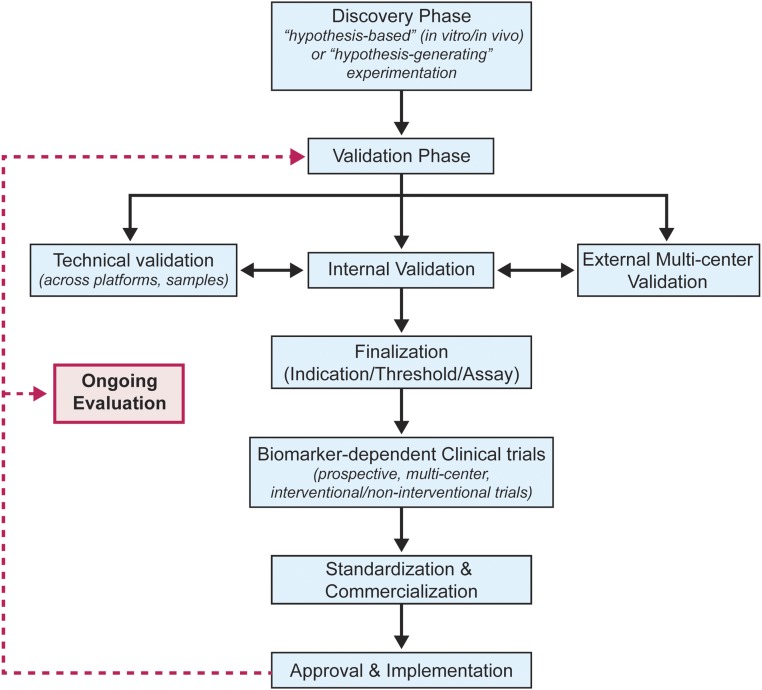
Biomarker development occurs via a lifecycle (Figure 1) that includes discovery, internal single-center, and external multicenter validation, standardization, commercialization, and ultimately, adoption into clinical care. After entrance into the clinical arena, widespread use tends to generate new questions regarding assay utility, potentially spawning second-order, controlled trials.
Figure 1.
Biomarker development should proceed through a lifecyle that includes external validation. A proposed template depicting the various steps involved from biomarker discovery and validation to clinical application in transplantation.
Anti-HLA Antibody Testing by Solid-Phase Assays
As initially published by Patel and Terasaki,19 preexisting recipient serum antidonor HLA antibodies are associated with early rejection/graft loss (hyper-AR) after kidney transplantation. Accurate detection of these antibodies is essential; crossmatch testing by FDA-approved assays, including solid-phase assays (e.g., luminex based), is now routinely used clinically for pretransplant risk assessments.19–24 The strengths and weaknesses of each approach have been reviewed elsewhere in detail,25–34 but conclusions suggest that solid-phase assays (e.g., luminex based) have the highest sensitivity for predicting post-transplant antibody-mediated rejection (ABMR) and/or graft loss. Standardization of solid-phase assay techniques among laboratories is achievable,35 although the assay is not designed (or FDA approved) to be quantitative. Although transplant centers routinely avoid transplantation with DSAs as detected by solid-phase assays with mean fluorescence intensity (MFI) of >10,000 (high risk of hyper-AR), the thresholds for defining positivity and the clinical implications of pretransplant DSAs with lower MFI remain controversial.36
Accumulating evidence from multiple studies associates development of de novo post-transplant DSAs with an elevated risk of late graft loss,34,37 particularly in the context of medication nonadherence.38 To improve the prognostic utility of de novo DSAs for incipient graft injury, investigators have examined whether various DSA characteristics, including time of development post-transplant, specificity (class 1 versus 2 HLA), isotype (IgG subtypes), strength (MFI or titer), and function (e.g., complement fixing),34,37 confer increased clinical risk. Reports suggest a higher risk for kidney allograft loss in subjects with serum DSAs that bind/activate complement as measured by a standard solid-phase testing assay that additionally detects C1q binding.32–34 C1q-positive de novo DSA was associated with a shorter time to graft loss than C1q-negative de novo DSA or the absence of any DSA.33 Although it was postulated that C1q positivity indicates antibodies preferentially capable of initiating complement-dependent allograft rejection, additional work suggests that C1q positivity is a consequence of higher serum DSA titers33 rather than complement-activating activity per se.39 Although solid-phase DSA testing is widely used, the clinical utility of C1q binding DSAs (among other innovations)39,40 as a risk assessment tool and the timing/frequency of DSA testing for detection of de novo DSAs remain unclear. One barrier to implementing routine post-transplant DSA testing is the absence of evidence that available therapies can prevent/reverse incipient allograft injury/loss in DSA-positive transplant recipients.
Assessing Pretransplant Risk for Development of Post-Transplant DSAs
Building on the above-noted observations, research teams have attempted to identify pretransplant biomarkers that predict high likelihood of developing post-transplant DSAs.
Epitope mismatch analysis of donor and recipient HLA polymorphisms builds on current HLA typing to identify donor-recipient mismatches for both class 1 (triplets) and 2 (eplets) HLA at the molecular epitope level. The HLAMatchmaker software is an epitope analysis tool that integrates knowledge of HLA molecule three-dimensional structures41 with known correlations among sero- or genotyping results at HLA loci to identify polymorphic amino acid differences, which when located on exposed regions, are potential immunogens that stimulate antibody production.42,43 Studies showed that high numbers of epitope mismatches between donor and recipient44,45 are associated with an elevated risk of developing de novo DSAs, particularly in kidney transplant recipients nonadherent to immune suppressants46 or recipients undergoing immunosuppression withdrawal.47 One implication is that individuals with high epitope mismatches may require more immunosuppression to prevent de novo DSAs. Although epitope mismatch analysis requires high-resolution HLA genotyping, which incurs an additional expense, the software is freely available, making this a readily implementable risk assessment strategy that could be used by any transplant center today. Remaining issues requiring attention are multicenter validation of optimal thresholds for positivity and testing the hypothesis that differential treatment strategies on the basis of epitope mismatching will prevent DSA and graft loss in those at highest risk.
Anti-HLA alloantibodies are produced by antibody-producing plasma cells and long-lived memory B cells (Bmems), the latter of which differentiate into plasma cells on re-encountering antigen.48 Donor-specific Bmems are detectable in humans independent of whether serum anti-HLA antibodies are demonstrable.49–51 The B cell ELISPOT assay52 detects IgG-producing B cells, including Bmems.53 Frequencies of circulating donor-HLA–reactive Bmems correlate with degree of sensitization and ABMR episodes.50 Large European observational studies are ongoing to assess the value of quantifying HLA-specific Bmems in kidney transplantation (O. Bestard, personal communication). Commercialization efforts are underway and may become available to United States transplant centers in the near future.
Assays of T Cell Alloreactivity
Alloreactive T cells are essential mediators of allograft rejection,54–57 spawning efforts to quantify alloreactive T cell immune responses as potential biomarkers of transplant outcome.
In vitro T cell responses to alloantigens are detectable by proliferative mixed lymphocyte reactions, in which recipient T cells are tested for reactivity to donor cells using various readouts.57 Although weak proliferative responses (3H-thymidine incorporation) suggest overimmunosuppression and strong responses suggest elevated risk of allograft rejection, the proliferative mixed lymphocyte reaction has limited predictive value in clinical transplantation.58,59 As an alternative, Ashokkumar et al.60 showed that pretransplant mitogen stimulation of alloreactive T cells induces upregulated surface expression of T cell CD154. Higher expression levels associate with higher risk of liver transplantation rejection in children. Limited data from adult kidney transplant recipients suggest diagnostic potential for ongoing cellular rejection.61 Although T cell expression of CD154 is a commercially available and FDA-approved assay and can be performed by any clinical laboratory with a flow cytometer, multicenter validation of its diagnostic/prognostic biomarker utility in kidney transplantation remains to be determined.
Measuring ATP generation by mitogen-stimulated CD4 lymphocytes (Immuknow assay)62 is an FDA-approved biomarker that is potentially informative in transplant recipients. Results are reported as “within normal limits,” high (suggesting underimmunosuppression), or low (suggesting overimmunosuppression). Observational studies performed in small kidney transplant cohorts provide inconsistent conclusions63–65 but implied that the assay may be best at detecting overimmunosuppression and an elevated risk of infection. The most informative study was a prospective trial of 202 liver transplant recipients66 randomized to receive standard of care immunosuppression versus immunosuppression guided by ATP release assay results. One-year patient survival rates were higher and infection rates were lower in the group receiving ATP release biomarker-guided immunosuppression. Randomized, controlled trials in kidney transplantation have not been performed.
A portion of the alloreactive T cell repertoire derives from the memory pool, and memory donor–reactive T cells can negatively affect transplant outcomes.67,68 Donor-reactive memory T cells are detectable by flow cytometry67 and cytokine enzyme–linked immunosorbent spot (ELISPOT).69 IFN-γ ELISPOT positivity before transplantation was shown to correlate with elevated risk of developing post-transplant acute cellular rejection (ACR) and/or poor graft function in multiple cohorts of renal recipients, particularly those who were not given induction therapy with T cell–depleting agents.70–76 The ELISPOT procedure requires donor cells and takes 24–36 hours, which limits application in deceased donor recipients. An alternative T cell reactivity index (panel of reactive T cells [PRT]) is analogously performed using a pool of donor cells reflective of the organ donor pool; pretransplant PRT results correlate with an elevated risk of post-transplant allograft injury.77–79 The complexities of performing ELISPOT assays require careful standardization80 for reproducibility. Commercialization for donor-specific assays requires obtaining donor cells, but commercialization efforts for PRT assays are underway. Prospective ELISPOT-guided interventional trials are needed to determine whether targeting specific therapies to subjects exhibiting ELISPOT positivity will improve short- and long-term outcomes in kidney transplant recipients.
Urine Biomarkers
Suthanthiran and colleagues81–86 first recognized the potential of studying urine as a window into kidney allograft inflammation. Multiple single-center and small observational studies showed that preselected immune transcripts in urinary cell RNA measured by quantitative PCR (qPCR) can noninvasively differentiate ACR from non-ACR in kidney transplant recipients.81–86 Detailed summaries of the published findings are summarized elsewhere.5,13 The most informative findings derive from a large, multicenter, prospective, National Institutes of Health (NIH)-funded, observational study that showed that an 18S-ribosomal RNA normalized 3-gene signature (CD3ε, IP-10, and 18S-rRNA) distinguished ACR from non-ACR with high accuracy. The biomarker was detectable before the clinical recognition of the rejection episode.86,87 Standardization of urinary gene expression has been accomplished,88 and optimal control genes have been identified,89 making it feasible to adopt urinary gene expression profiling by qPCR in the clinic. Importantly, RNA must be produced rapidly from the collected urine (within 4 hours) to be informative.88 Urine cell qPCR testing is not yet widely commercially available. Whether clinical outcomes are positively affected by urine qPCR-driven interventions in the absence of biopsies remains to be determined via prospective, randomized, multicenter studies.5
Among other testing strategies close to commercialization is urinary protein measurement of the chemoattractant chemokines CXCL9 and/or CXCL10.47,90–92 Early cross-sectional and relatively small single-center studies showed strong correlations between urinary CXCL9 and T cell–mediated rejection,90,91,93 with some indicating strong correlations between urinary CXCL10 and ABMR.92,94,95 In an NIH-funded, prospective, multicenter trial, urinary CXCL9 (ELISA) outperformed CXCL9-mRNA levels (qPCR) for the diagnosis of ACR.90 In a separate, prospective, randomized, tacrolimus-withdrawal study, elevations in urinary CXCL9 predicted incipient acute cellular rejection, providing the first evidence that serial monitoring of a urinary biomarker could be used to accurately guide clinical decision making.47 Results reported at the American Transplant Congress 201696 suggest that serial urinary CXCL9 measurements can be informative with regard to efficacy of antirejection therapy. ELISAs can be performed by most clinical laboratories, making this approach readily adoptable in clinical care, although commercialized assays are yet to be routinely offered in the United States. Whether treatment decisions on the basis of urinary CXCL9 affect clinical outcomes in kidney transplantation remains to be tested.
Molecular Approaches
High-throughput technologies for broadly analyzing gene transcripts in blood or tissue, including microarray and RNA-sequencing (RNA-seq), can simultaneously map differential expression patterns of thousands of transcripts and associate them with particular outcomes or disease states. Microarrays5,97 are reported as fold changes in reference to a standard. Falling costs make it possible that microarray-based approaches could be adapted to clinical care.
RNA-seq provides single-base resolution, absolute transcript quantification from total or fractionated RNA, detection of splice variants, a large dynamic range for detection of expression levels (8000-fold changes), and the ability to discover novel transcripts.98 The enormous amount of data obtained from each RNA-seq experiment, the time involved in analysis, and the relative inability to analyze data regarding one or a few candidate transcripts alone could limit the use of this technology as a clinical biomarker assay.
Biopsy Transcript Assessment as Biomarkers
The biopsy transcriptome as interrogated by qPCR99–101 or high-throughput techniques102–107 confirms allograft upregulation of T cell–related/immune transcripts (e.g., chemokines, CD3, FOXP3, IFN-γ, Fas/FasL, perforin, and Granzyme B) compared with those without rejection. Beyond these observations, allograft transcriptome studies suggest that molecular phenotyping adds to the histologic diagnosis of ACR,103,104,108,109 in part by identifying histologically undifferentiated lesions,105 improving prognostication,105 and providing insights into the clinical importance of borderline ACR106 and isolated “v-lesions.”110 Allograft transcriptomic data may be relevant for diagnosing and prognosticating ABMR in the absence of classic histologic criteria and/or C4d positivity.107,111–113
As one example of how “molecular microscope” analyses of graft tissue improve prognostication,114 Halloran and colleagues16 developed a biopsy-derived molecular classifier that independently associated with an elevated risk of graft loss within 1 year. The molecular risk score had a higher hazard ratio for graft loss than histologic or clinical factors. These intriguing results have the potential to overcome variability in interpretation of kidney allograft pathology115 but will require prospective validation, standardization, and commercialization before they will be adopted into clinical care.
Data from the prospective, NIH-funded Genomics of Chronic Allograft Rejection Study provide additional insight.18 Microarray analysis was performed on 3-month surveillance biopsies to identify genes and pathways associated with 3- and 12-month Chronic Allograft Dysfunction Index scores. A biomarker comprised of a refined 13-gene set accurately predicted impending fibrosis at 12 months, predicted graft loss, independent of simultaneous ACR was validated in external cohorts, which included for-cause biopsies,12,16 and was independently associated with histologic progression and graft loss.116
Together, these genomic analyses suggest that, if transcriptome assessments were performed on surveillance or for-cause biopsies, prognostic gene sets could risk stratify allografts for histologic progression or graft loss and thereby, guide treatment decisions. Ultimately, prospective studies will be required to test whether therapeutic interventions on the basis of biopsy-derived molecular signatures will improve outcomes compared with therapeutic interventions driven by results of standard histology.
Blood Transcriptome Assessments
Assessment of peripheral blood gene expression profiles circumvents the need for a biopsy and could serve as an attractive immune monitoring tool.117 High-throughput transcriptional profiling of total RNA from peripheral blood validated across three microarray platforms identified a specific five-gene set that correlated well with AR/no AR status on simultaneously procured allograft biopsies.118 The team subsequently developed and validated a 17-gene signature that accurately diagnosed AR in a multicenter adult and pediatric cohort.119 Other studies by Murphy et al.120 identified a peripheral blood gene signature that showed diagnostic accuracy for subclinical rejection, including borderline patients, early post-transplant. Although these approaches could potentially be used to guide therapeutic decisions in the absence of biopsy tissue, multicenter validation and commercialization are required. Adoption of such assays into clinical care will require clinical experience and additional studies to support the hypothesis that interventions on the basis of peripheral blood signatures can positively influence patient outcomes.
Cutting Edge Biomarkers
Although not yet commercially available, several novel approaches reported since 2010 have potential to become clinically useful transplant biomarkers.
Quantification of donor-origin cellfree DNA (cfDNA) in recipient blood or urine (by filtration/secretion or injury) has potential diagnostic utility for detecting rejection.121–124 Donor-origin cfDNA is likely released into recipient plasma during allograft injury and quantifiable as a fraction of total cfDNA from blood or urine.125 The donor-recipient cfDNA distinction is made by identifying genetic differences between donor and recipient.125–128 Small studies have shown associations between AR and the quantity of plasma and urinary donor cfDNA in cardiac129 or kidney transplant recipients.126,127 Available evidence suggests that elevations in cfDNA cannot distinguish AR from other causes of graft injury, and additional markers may need to be layered over this assay to improve specificity or accurately relay need for allograft biopsy.125,126,130 Although cfDNA commercialization is underway (Allosure; CareDx, Brisbane, CA), multicenter, prospective, observational studies (ongoing as part of the NIH-funded Clinical Trials in Organ Transplantation 19 study; www.CTOT.org) and ultimately, interventional studies will be required to delineate the utility of this promising approach as a transplantation biomarker.
Urine proteomic profiling studies performed in the early 2000s identified several protein signals that differentiated subjects with AR from those with no AR without additional characterization of the proteins associated with these signals.90,91,131–135 Studies published since 2010 using for-cause biopsies have identified numerous candidate proteins differentially identified in urine of subjects with AR compared with those with other pathologies.136–138 Urine proteomes of transplant recipients with AR contain upregulation of immune response–related proteins and acute-phase reactants, with downregulation of solute/ion transport proteins.138 The overlap among proteins in various studies has been minimal.132 Standardization, validation, and commercialization remain barriers to moving proteomic approaches to the clinic.
Metabolomics involves the simultaneous profiling of dozens of small molecule metabolites (<1500 D) in tissue and biologic fluids,139,140 and because the transcriptome or proteome can undergo additional regulatory alterations, the metabolome can reflect the downstream products of cellular processes. A composite urine metabolite-mRNA signature accurately differentiated cellular AR from non-AR in an adult cohort.140 The metabolite signature alone was less accurate, suggesting that combinations of markers applied sequentially or simultaneously may ultimately be more effective for noninvasive diagnosis and surveillance. Multicenter validation and commercialization remain to be done.
Heritable genomic changes in either donor or recipient DNA single-nucleotide polymorphisms (SNPs) are stable and measurable. Multiple SNPs in various genes have been shown to correlate strongly with certain transplant outcomes,141–144 and they have the potential to serve as risk stratifiers pretransplantation. Although these observations offer promise, small sample sizes, low odds ratios observed for outcomes between exposed and nonexposed, limited validation, and lack of evidence that immunosuppression choices on the basis of SNP risk stratification influence outcomes have thus far restricted the SNP analysis to research settings.
Sequencing of DNA sequences at the T cell receptor β-chain CDR3 region can identify a fingerprint of specifically donor-reactive T cell clones within recipients. In a small series, allograft tolerance was associated with the disappearance of these donor-reactive clones, whereas the lack of tolerance was associated with their persistence.145 If the appearance of these T cells in the blood or urine precedes the development of AR and if reproducible in larger cohorts of patients on usual immunosuppression, then this assay could help monitor donor-reactive T cell immunity specifically and serve as a surveillance tool.
Potential Framework for Kidney Transplant Monitoring
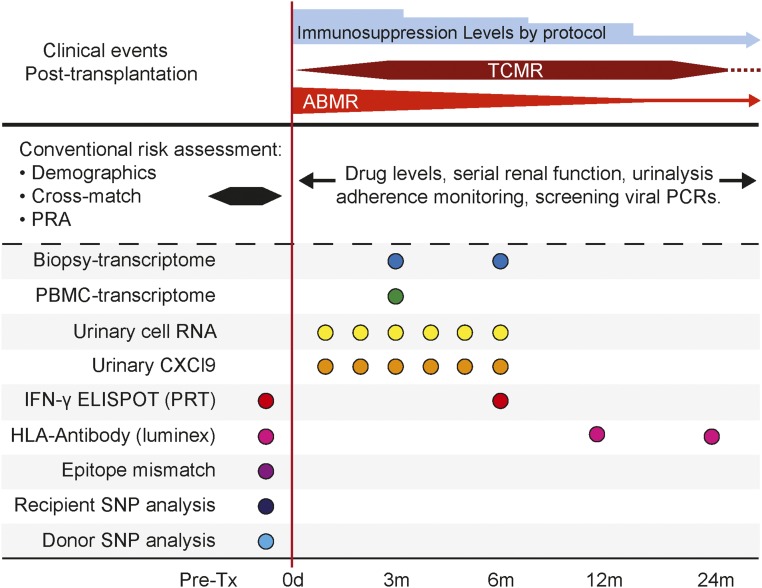
A clinically useful immune monitoring strategy must build on the known mechanisms of allograft injury, the data indicating time dependence of alloimmune injury,1,18,108,109,146–148 and the recognition that subclinical inflammation contributes to chronic allograft damage.109,147,148 We contend that multiple assays will need to be deployed pretransplant and recurrently post-transplant to optimally detect incipient injury and individualize therapy. As a framework (Figure 2), we envision comprehensive risk assessment via cellular assays, genomic technologies, and protein and metabolite profiling. SNP and epitope mismatch analysis, IFN-γ ELISPOT or PRT assay, and anti-HLA antibody assessment are likely to be informative pretransplantation and guide initial choices regarding immunosuppression. Noninvasive urinary gene expression, urine chemokine testing, and blood gene expression patterns are likely to replace or minimize biopsies to diagnose ongoing injury. Optimal post-transplant time points for other biomarkers need additional study. We envision that serial urinary CXCL9 or urinary RNA measurements will be required during the first few months post-transplantation (highest risk for AR), particularly during changes in immunosuppressive drug dosing. Peripheral blood expression profiling performed early (approximately 3 months) or during allograft dysfunction119 could indicate ongoing subclinical or clinical rejection and trigger a biopsy120; a biopsy transcriptome assessment could then help prognosticate allograft injury with greater granularity than histology,18 guiding maintenance therapy and/or follow-up. Although DSA measurements could be performed in all subjects, intense monitoring schedules might be better restricted to those individuals deemed at highest risk (e.g., with high epitope mismatches45,47 with positive donor-specific IFN-γ ELISPOT assays47) and/or at risk for nonadherence45 or who experienced prior AR episodes.147
Figure 2.
Biomarker-based individualization of immunosuppression of kidney transplant recipients will require use of multiple testing strategies. The x axis depicts the time points relative to the transplantation (Tx) when these biomarkers could be applied. The dashed horizontal line separates the conventional clinical and laboratory tests that are used currently. The continuous horizontal line describes common clinical immunologic events encountered in Tx with respect to times after Tx; the widths of the T cell–mediated rejection (TCMR) and ABMR lines refer to frequencies of these events. PRA, panel of reactive antibody.
Perspective and Future Directions
Although significant effort by numerous research groups has been focused on developing biomarkers to detect kidney allograft rejection and/or predict graft loss, few assays have moved from the research arena to a clinically implementable testing strategy that could guide therapeutic decision making.
The road to clinical biomarker implementation in transplantation is fraught with barriers that include high costs of commercializing assays for relatively small market sizes and a field that is being driven by physician-scientists rather than commercial entities. Improved partnering approaches among scientists, academic institutions, and various pharmaceutical and/or biotechnology companies will be required to commercialize biomarkers and make them available to transplant clinicians, particularly for assays with limited intellectual property rights, because methods are in the public domain (e.g., ELISAs for chemokines).
In addition, the relative lack of information on how to clinically use biomarkers remains problematic. The transplant field needs controlled trials, in which subjects are randomized to receive standard of care treatment or individualized care on the basis of prespecified biomarker-directed strategies. End points should include graft function/survival, rates of infectious complications, and biopsy-related complications. The hypothesis to be tested is that biomarker-driven care will decrease the need for biopsies and limit infectious complications while improving graft function/survival. Such controlled trials would provide the best evidence for how to use a given marker, but commercialized, FDA-approved biomarkers can also be broadly tested by the transplant community without hard evidence. Such real world experience can be hypothesis generating and drive improved trial design to address how an assay could be optimally used in a given setting.
Overall, although work remains to be done, the past decade witnessed significant progress in biomarker development, testing, and clinical implementation in transplantation. We look forward to the next decade as we anticipate that the kidney transplant community will move beyond protocol-based immunosuppression and begin to individualize treatment of transplant patients on the basis of results of objective and reliable biomarkers.
Disclosures
None.
Acknowledgments
The authors thank Jill K. Gregory (Icahn School of Medicine at Mount Sinai) for assistance with illustrations. The authors specifically acknowledge the late Dan Salomon for his leadership, dedication, and exceptional contributions to the field of genomic biomarkers in transplantation.
M.C.M. is supported by American Heart Association Scientist Development Award 15SDG25870018. This work is supported by National Institutes of Health grants R01DK102420 (to B.M.) and U01AI63594 (to P.S.H.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Hart A, Smith JM, Skeans MA, Gustafson SK, Stewart DE, Cherikh WS, Wainright JL, Boyle G, Snyder JJ, Kasiske BL, Israni AK: Kidney. Am J Transplant 16[Suppl 2]: 11–46, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meier-Kriesche HU, Schold JD, Kaplan B: Long-term renal allograft survival: Have we made significant progress or is it time to rethink our analytic and therapeutic strategies? Am J Transplant 4: 1289–1295, 2004 [DOI] [PubMed] [Google Scholar]
- 3.El-Zoghby ZM, Stegall MD, Lager DJ, Kremers WK, Amer H, Gloor JM, Cosio FG: Identifying specific causes of kidney allograft loss. Am J Transplant 9: 527–535, 2009 [DOI] [PubMed] [Google Scholar]
- 4.Cravedi P, Heeger PS: Immunologic monitoring in transplantation revisited. Curr Opin Organ Transplant 17: 26–32, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menon MC, Keung KL, Murphy B, O’Connell PJ: The use of genomics and pathway analysis in our understanding and prediction of clinical renal transplant injury. Transplantation 100: 1405–1414, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willis JC, Lord GM: Immune biomarkers: The promises and pitfalls of personalized medicine. Nat Rev Immunol 15: 323–329, 2015 [DOI] [PubMed] [Google Scholar]
- 7.Lo DJ, Kaplan B, Kirk AD: Biomarkers for kidney transplant rejection. Nat Rev Nephrol 10: 215–225, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Sagoo P, Perucha E, Sawitzki B, Tomiuk S, Stephens DA, Miqueu P, Chapman S, Craciun L, Sergeant R, Brouard S, Rovis F, Jimenez E, Ballow A, Giral M, Rebollo-Mesa I, Le Moine A, Braudeau C, Hilton R, Gerstmayer B, Bourcier K, Sharif A, Krajewska M, Lord GM, Roberts I, Goldman M, Wood KJ, Newell K, Seyfert-Margolis V, Warrens AN, Janssen U, Volk HD, Soulillou JP, Hernandez-Fuentes MP, Lechler RI: Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J Clin Invest 120: 1848–1861, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, Burlingham WJ, Marks WH, Sanz I, Lechler RI, Hernandez-Fuentes MP, Turka LA, Seyfert-Margolis VL; Immune Tolerance Network ST507 Study Group : Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest 120: 1836–1847, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saundh BK, Baker R, Harris M, Welberry Smith MP, Cherukuri A, Hale A: Early BK polyomavirus (BKV) reactivation in donor kidney is a risk factor for development of BKV-associated nephropathy. J Infect Dis 207: 137–141, 2013 [DOI] [PubMed] [Google Scholar]
- 11.Humar A, Lebranchu Y, Vincenti F, Blumberg EA, Punch JD, Limaye AP, Abramowicz D, Jardine AG, Voulgari AT, Ives J, Hauser IA, Peeters P: The efficacy and safety of 200 days valganciclovir cytomegalovirus prophylaxis in high-risk kidney transplant recipients. Am J Transplant 10: 1228–1237, 2010 [DOI] [PubMed] [Google Scholar]
- 12.Naesens M, Khatri P, Li L, Sigdel TK, Vitalone MJ, Chen R, Butte AJ, Salvatierra O, Sarwal MM: Progressive histological damage in renal allografts is associated with expression of innate and adaptive immunity genes. Kidney Int 80: 1364–1376, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anglicheau D, Naesens M, Essig M, Gwinner W, Marquet P: Establishing biomarkers in transplant medicine: A critical review of current approaches. Transplantation 100: 2024–2038, 2016 [DOI] [PubMed] [Google Scholar]
- 14.Furness PN, Philpott CM, Chorbadjian MT, Nicholson ML, Bosmans JL, Corthouts BL, Bogers JJ, Schwarz A, Gwinner W, Haller H, Mengel M, Seron D, Moreso F, Cañas C: Protocol biopsy of the stable renal transplant: A multicenter study of methods and complication rates. Transplantation 76: 969–973, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Elshafie M, Furness PN: Identification of lesions indicating rejection in kidney transplant biopsies: Tubulitis is severely under-detected by conventional microscopy. Nephrol Dial Transplant 27: 1252–1255, 2012 [DOI] [PubMed] [Google Scholar]
- 16.Einecke G, Reeve J, Sis B, Mengel M, Hidalgo L, Famulski KS, Matas A, Kasiske B, Kaplan B, Halloran PF: A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J Clin Invest 120: 1862–1872, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naesens M, Lerut E, Emonds MP, Herelixka A, Evenepoel P, Claes K, Bammens B, Sprangers B, Meijers B, Jochmans I, Monbaliu D, Pirenne J, Kuypers DR: Proteinuria as a noninvasive marker for renal allograft histology and failure: An observational cohort study. J Am Soc Nephrol 27: 281–292, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connell PJ, Zhang W, Menon MC, Yi Z, Schröppel B, Gallon L, Luan Y, Rosales IA, Ge Y, Losic B, Xi C, Woytovich C, Keung KL, Wei C, Greene I, Overbey J, Bagiella E, Najafian N, Samaniego M, Djamali A, Alexander SI, Nankivell BJ, Chapman JR, Smith RN, Colvin R, Murphy B: Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: A multicentre, prospective study. Lancet 388: 983–993, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel R, Terasaki PI: Significance of the positive crossmatch test in kidney transplantation. N Engl J Med 280: 735–739, 1969 [DOI] [PubMed] [Google Scholar]
- 20.Williams GM, Hume DM, Hudson RP Jr., Morris PJ, Kano K, Milgrom F: “Hyperacute” renal-homograft rejection in man. N Engl J Med 279: 611–618, 1968 [DOI] [PubMed] [Google Scholar]
- 21.Terasaki PI, McClelland JD: Microdroplet assay of human serum cytotoxins. Nature 204: 998–1000, 1964 [DOI] [PubMed] [Google Scholar]
- 22.Ettinger RB, Terasaki PI, Opelz G, Malekzadeh M, Uittenbogaart C, Pennisi AJ, Fine R: Successful renal allografts across a positive cross-match for donor B-lymphocyte alloantigens. Lancet 2: 56–58, 1976 [DOI] [PubMed] [Google Scholar]
- 23.Montgomery RA, Zachary AA: Transplanting patients with a positive donor-specific crossmatch: A single center’s perspective. Pediatr Transplant 8: 535–542, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Orandi BJ, Alachkar N, Kraus ES, Naqvi F, Lonze BE, Lees L, Van Arendonk KJ, Wickliffe C, Bagnasco SM, Zachary AA, Segev DL, Montgomery RA: Presentation and outcomes of C4d-negative antibody-mediated rejection after kidney transplantation. Am J Transplant 16: 213–220, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClelland J, Terasaki PI, Berne T, Brennan LP, Bretan P, Cohen A, Dainko E, Danovitch G, Ettenger R, Lieberman E, et al. : Evaluation of the UNOS point system for kidney recipient selection. Clin Transpl 1989: 471–476, 1989 [PubMed] [Google Scholar]
- 26.Israni AK, Salkowski N, Gustafson S, Snyder JJ, Friedewald JJ, Formica RN, Wang X, Shteyn E, Cherikh W, Stewart D, Samana CJ, Chung A, Hart A, Kasiske BL: New national allocation policy for deceased donor kidneys in the United States and possible effect on patient outcomes. J Am Soc Nephrol 25: 1842–1848, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizutani K, Terasaki P, Rosen A, Esquenazi V, Miller J, Shih RN, Pei R, Ozawa M, Lee J: Serial ten-year follow-up of HLA and MICA antibody production prior to kidney graft failure. Am J Transplant 5: 2265–2272, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Mao Q, Terasaki PI, Cai J, Briley K, Catrou P, Haisch C, Rebellato L: Extremely high association between appearance of HLA antibodies and failure of kidney grafts in a five-year longitudinal study. Am J Transplant 7: 864–871, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Cardarelli F, Pascual M, Tolkoff-Rubin N, Delmonico FL, Wong W, Schoenfeld DA, Zhang H, Cosimi AB, Saidman SL: Prevalence and significance of anti-HLA and donor-specific antibodies long-term after renal transplantation. Transpl Int 18: 532–540, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Zou Y, Stastny P, Süsal C, Döhler B, Opelz G: Antibodies against MICA antigens and kidney-transplant rejection. N Engl J Med 357: 1293–1300, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Sánchez-Zapardiel E, Castro-Panete MJ, Mancebo E, Morales P, Laguna-Goya R, Morales JM, Apaza J, de Andrés A, Talayero P, Paz-Artal E: Early renal graft function deterioration in recipients with preformed anti-MICA antibodies: Partial contribution of complement-dependent cytotoxicity. Nephrol Dial Transplant 31: 150–160, 2016 [DOI] [PubMed] [Google Scholar]
- 32.Loupy A, Lefaucheur C, Vernerey D, Prugger C, Duong van Huyen JP, Mooney N, Suberbielle C, Frémeaux-Bacchi V, Méjean A, Desgrandchamps F, Anglicheau D, Nochy D, Charron D, Empana JP, Delahousse M, Legendre C, Glotz D, Hill GS, Zeevi A, Jouven X: Complement-binding anti-HLA antibodies and kidney-allograft survival. N Engl J Med 369: 1215–1226, 2013 [DOI] [PubMed] [Google Scholar]
- 33.Guidicelli G, Guerville F, Lepreux S, Wiebe C, Thaunat O, Dubois V, Visentin J, Bachelet T, Morelon E, Nickerson P, Merville P, Taupin JL, Couzi L: Non-complement-binding de novo donor-specific anti-HLA antibodies and kidney allograft survival. J Am Soc Nephrol 27: 615–625, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Süsal C, Wettstein D, Döhler B, Morath C, Ruhenstroth A, Scherer S, Tran TH, Gombos P, Schemmer P, Wagner E, Fehr T, Živčić-Ćosić S, Balen S, Weimer R, Slavcev A, Bösmüller C, Norman DJ, Zeier M, Opelz G; Collaborative Transplant Study Report : Association of kidney graft loss with de novo produced donor-specific and non-donor-specific HLA antibodies detected by single antigen testing. Transplantation 99: 1976–1980, 2015 [DOI] [PubMed] [Google Scholar]
- 35.Reed EF, Rao P, Zhang Z, Gebel H, Bray RA, Guleria I, Lunz J, Mohanakumar T, Nickerson P, Tambur AR, Zeevi A, Heeger PS, Gjertson D: Comprehensive assessment and standardization of solid phase multiplex-bead arrays for the detection of antibodies to HLA-drilling down on key sources of variation. Am J Transplant 13: 3050–3051, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tait BD, Süsal C, Gebel HM, Nickerson PW, Zachary AA, Claas FH, Reed EF, Bray RA, Campbell P, Chapman JR, Coates PT, Colvin RB, Cozzi E, Doxiadis II, Fuggle SV, Gill J, Glotz D, Lachmann N, Mohanakumar T, Suciu-Foca N, Sumitran-Holgersson S, Tanabe K, Taylor CJ, Tyan DB, Webster A, Zeevi A, Opelz G: Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation 95: 19–47, 2013 [DOI] [PubMed] [Google Scholar]
- 37.Bamoulid J, Roodenburg A, Staeck O, Wu K, Rudolph B, Brakemeier S, Halleck F, Lehner L, Schönemann C, Lachmann N, Budde K: Clinical outcome of patients with de novo C1q-binding donor-specific HLA antibodies after renal transplantation [published online ahead of print September 21, 2016]. Transplantation doi:10.1097/TP.0000000000001487 [DOI] [PubMed] [Google Scholar]
- 38.Wiebe C, Gareau AJ, Pochinco D, Gibson IW, Ho J, Birk PE, Blydt-Hasen T, Karpinski M, Goldberg A, Storsley L, Rush DN, Nickerson PW: Evaluation of C1q status and titer of de novo donor-specific antibodies as predictors of allograft survival [published online ahead of print August 19, 2016]. Am J Transplant doi:10.1111/ajt.14015 [DOI] [PubMed] [Google Scholar]
- 39.Tambur AR, Herrera ND, Haarberg KM, Cusick MF, Gordon RA, Leventhal JR, Friedewald JJ, Glotz D: Assessing antibody strength: Comparison of MFI, C1q, and titer information. Am J Transplant 15: 2421–2430, 2015 [DOI] [PubMed] [Google Scholar]
- 40.Lefaucheur C, Viglietti D, Bentlejewski C, Duong van Huyen JP, Vernerey D, Aubert O, Verine J, Jouven X, Legendre C, Glotz D, Loupy A, Zeevi A: IgG donor-specific anti-human HLA antibody subclasses and kidney allograft antibody-mediated injury. J Am Soc Nephrol 27: 293–304, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC: Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329: 506–512, 1987 [DOI] [PubMed] [Google Scholar]
- 42.Duquesnoy RJ: HLAMatchmaker: A molecularly based algorithm for histocompatibility determination. I. Description of the algorithm. Hum Immunol 63: 339–352, 2002 [DOI] [PubMed] [Google Scholar]
- 43.Duquesnoy RJ, Askar M: HLAMatchmaker: A molecularly based algorithm for histocompatibility determination. V. Eplet matching for HLA-DR, HLA-DQ, and HLA-DP. Hum Immunol 68: 12–25, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duquesnoy RJ, Takemoto S, de Lange P, Doxiadis II, Schreuder GM, Persijn GG, Claas FH: HLAmatchmaker: A molecularly based algorithm for histocompatibility determination. III. Effect of matching at the HLA-A,B amino acid triplet level on kidney transplant survival. Transplantation 75: 884–889, 2003 [DOI] [PubMed] [Google Scholar]
- 45.Wiebe C, Pochinco D, Blydt-Hansen TD, Ho J, Birk PE, Karpinski M, Goldberg A, Storsley LJ, Gibson IW, Rush DN, Nickerson PW: Class II HLA epitope matching-A strategy to minimize de novo donor-specific antibody development and improve outcomes. Am J Transplant 13: 3114–3122, 2013 [DOI] [PubMed] [Google Scholar]
- 46.Wiebe C, Nevins TE, Robiner WN, Thomas W, Matas AJ, Nickerson PW: The synergistic effect of class II HLA epitope-mismatch and nonadherence on acute rejection and graft survival. Am J Transplant 15: 2197–2202, 2015 [DOI] [PubMed] [Google Scholar]
- 47.Hricik DE, Augustine J, Nickerson P, Formica RN, Poggio ED, Rush D, Newell KA, Goebel J, Gibson IW, Fairchild RL, Spain K, Iklé D, Bridges ND, Heeger PS; CTOT-01 consortium : Interferon gamma ELISPOT testing as a risk-stratifying biomarker for kidney transplant injury: Results from the CTOT-01 ,multicenter study. Am J Transplant 15: 3166–3173, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarlinton D, Good-Jacobson K: Diversity among memory B cells: Origin, consequences, and utility. Science 341: 1205–1211, 2013 [DOI] [PubMed] [Google Scholar]
- 49.Bauer T, Jilg W: Hepatitis B surface antigen-specific T and B cell memory in individuals who had lost protective antibodies after hepatitis B vaccination. Vaccine 24: 572–577, 2006 [DOI] [PubMed] [Google Scholar]
- 50.Lúcia M, Luque S, Crespo E, Melilli E, Cruzado JM, Martorell J, Jarque M, Gil-Vernet S, Manonelles A, Grinyó JM, Bestard O: Preformed circulating HLA-specific memory B cells predict high risk of humoral rejection in kidney transplantation. Kidney Int 88: 874–887, 2015 [DOI] [PubMed] [Google Scholar]
- 51.Colvin RB: Antibody-mediated renal allograft rejection: Diagnosis and pathogenesis. J Am Soc Nephrol 18: 1046–1056, 2007 [DOI] [PubMed] [Google Scholar]
- 52.Czerkinsky CC, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A: A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J Immunol Methods 65: 109–121, 1983 [DOI] [PubMed] [Google Scholar]
- 53.Bernasconi NL, Traggiai E, Lanzavecchia A: Maintenance of serological memory by polyclonal activation of human memory B cells. Science 298: 2199–2202, 2002 [DOI] [PubMed] [Google Scholar]
- 54.Lechler R, Lombardi G: The structural basis of alloreactivity. Immunol Res 9: 135–146, 1990 [DOI] [PubMed] [Google Scholar]
- 55.Baker RJ, Hernandez-Fuentes MP, Brookes PA, Chaudhry AN, Cook HT, Lechler RI: Loss of direct and maintenance of indirect alloresponses in renal allograft recipients: Implications for the pathogenesis of chronic allograft nephropathy. J Immunol 167: 7199–7206, 2001 [DOI] [PubMed] [Google Scholar]
- 56.Vella JP, Spadafora-Ferreira M, Murphy B, Alexander SI, Harmon W, Carpenter CB, Sayegh MH: Indirect allorecognition of major histocompatibility complex allopeptides in human renal transplant recipients with chronic graft dysfunction. Transplantation 64: 795–800, 1997 [DOI] [PubMed] [Google Scholar]
- 57.Bestard O, Nickel P, Cruzado JM, Schoenemann C, Boenisch O, Sefrin A, Grinyó JM, Volk HD, Reinke P: Circulating alloreactive T cells correlate with graft function in longstanding renal transplant recipients. J Am Soc Nephrol 19: 1419–1429, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hernandez-Fuentes MP, Warrens AN, Lechler RI: Immunologic monitoring. Immunol Rev 196: 247–264, 2003 [DOI] [PubMed] [Google Scholar]
- 59.Tanaka Y, Tashiro H, Onoe T, Ide K, Ishiyama K, Ohdan H: Optimization of immunosuppressive therapy based on a multiparametric mixed lymphocyte reaction assay reduces infectious complications and mortality in living donor liver transplant recipients. Transplant Proc 44: 555–559, 2012 [DOI] [PubMed] [Google Scholar]
- 60.Ashokkumar C, Talukdar A, Sun Q, Higgs BW, Janosky J, Wilson P, Mazariegos G, Jaffe R, Demetris A, Dobberstein J, Soltys K, Bond G, Thomson AW, Zeevi A, Sindhi R: Allospecific CD154+ T cells associate with rejection risk after pediatric liver transplantation. Am J Transplant 9: 179–191, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashokkumar C, Shapiro R, Tan H, Ningappa M, Elinoff B, Fedorek S, Sun Q, Higgs BW, Randhawa P, Humar A, Sindhi R: Allospecific CD154+ T-cytotoxic memory cells identify recipients experiencing acute cellular rejection after renal transplantation. Transplantation 92: 433–438, 2011 [DOI] [PubMed] [Google Scholar]
- 62.Sottong PR, Rosebrock JA, Britz JA, Kramer TR: Measurement of T-lymphocyte responses in whole-blood cultures using newly synthesized DNA and ATP. Clin Diagn Lab Immunol 7: 307–311, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He J, Li Y, Zhang H, Wei X, Zheng H, Xu C, Bao X, Yuan X, Hou J: Immune function assay (ImmuKnow) as a predictor of allograft rejection and infection in kidney transplantation. Clin Transplant 27: E351–E358, 2013 [DOI] [PubMed] [Google Scholar]
- 64.Quaglia M, Cena T, Fenoglio R, Musetti C, Cagna D, Radin E, Roggero S, Amoroso A, Magnani C, Stratta P: Immune function assay (immunknow) drop over first 6 months after renal transplant: A predictor of opportunistic viral infections? Transplant Proc 46: 2220–2223, 2014 [DOI] [PubMed] [Google Scholar]
- 65.Libri I, Gnappi E, Zanelli P, Reina M, Giuliodori S, Vaglio A, Palmisano A, Buzio C, Riva G, Barozzi P, Luppi M, Cravedi P, Maggiore U: Trends in immune cell function assay and donor-specific HLA antibodies in kidney transplantation: A 3-year prospective study. Am J Transplant 13: 3215–3222, 2013 [DOI] [PubMed] [Google Scholar]
- 66.Ravaioli M, Neri F, Lazzarotto T, Bertuzzo VR, Di Gioia P, Stacchini G, Morelli MC, Ercolani G, Cescon M, Chiereghin A, Del Gaudio M, Cucchetti A, Pinna AD: Immunosuppression modifications based on an immune response assay: Results of a randomized, controlled trial. Transplantation 99: 1625–1632, 2015 [DOI] [PubMed] [Google Scholar]
- 67.Pearl JP, Parris J, Hale DA, Hoffmann SC, Bernstein WB, McCoy KL, Swanson SJ, Mannon RB, Roederer M, Kirk AD: Immunocompetent T-cells with a memory-like phenotype are the dominant cell type following antibody-mediated T-cell depletion. Am J Transplant 5: 465–474, 2005 [DOI] [PubMed] [Google Scholar]
- 68.Dinavahi R, Heeger PS: T-cell immune monitoring in organ transplantation. Curr Opin Organ Transplant 13: 419–424, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mashishi T, Gray CM: The ELISPOT assay: An easily transferable method for measuring cellular responses and identifying T cell epitopes. Clin Chem Lab Med 40: 903–910, 2002 [DOI] [PubMed] [Google Scholar]
- 70.Hricik DE, Rodriguez V, Riley J, Bryan K, Tary-Lehmann M, Greenspan N, Dejelo C, Schulak JA, Heeger PS: Enzyme linked immunosorbent spot (ELISPOT) assay for interferon-gamma independently predicts renal function in kidney transplant recipients. Am J Transplant 3: 878–884, 2003 [DOI] [PubMed] [Google Scholar]
- 71.Näther BJ, Nickel P, Bold G, Presber F, Schönemann C, Pratschke J, Volk HD, Reinke P: Modified ELISPOT technique--highly significant inverse correlation of post-Tx donor-reactive IFNgamma-producing cell frequencies with 6 and 12 months graft function in kidney transplant recipients. Transpl Immunol 16: 232–237, 2006 [DOI] [PubMed] [Google Scholar]
- 72.Nickel P, Presber F, Bold G, Biti D, Schönemann C, Tullius SG, Volk HD, Reinke P: Enzyme-linked immunosorbent spot assay for donor-reactive interferon-gamma-producing cells identifies T-cell presensitization and correlates with graft function at 6 and 12 months in renal-transplant recipients. Transplantation 78: 1640–1646, 2004 [DOI] [PubMed] [Google Scholar]
- 73.Kim SH, Oh EJ, Kim MJ, Park YJ, Han K, Yang HJ, Kim JY, Choi BS, Yang CW, Kim YS, Bang BK: Pretransplant donor-specific interferon-gamma ELISPOT assay predicts acute rejection episodes in renal transplant recipients. Transplant Proc 39: 3057–3060, 2007 [DOI] [PubMed] [Google Scholar]
- 74.Augustine JJ, Poggio ED, Heeger PS, Hricik DE: Preferential benefit of antibody induction therapy in kidney recipients with high pretransplant frequencies of donor-reactive interferon-gamma enzyme-linked immunosorbent spots. Transplantation 86: 529–534, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cherkassky L, Lanning M, Lalli PN, Czerr J, Siegel H, Danziger-Isakov L, Srinivas T, Valujskikh A, Shoskes DA, Baldwin W, Fairchild RL, Poggio ED: Evaluation of alloreactivity in kidney transplant recipients treated with antithymocyte globulin versus IL-2 receptor blocker. Am J Transplant 11: 1388–1396, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bestard O, Cruzado JM, Lucia M, Crespo E, Casis L, Sawitzki B, Vogt K, Cantarell C, Torras J, Melilli E, Mast R, Martinez-Castelao A, Gomà M, Reinke P, Volk HD, Grinyó JM: Prospective assessment of antidonor cellular alloreactivity is a tool for guidance of immunosuppression in kidney transplantation. Kidney Int 84: 1226–1236, 2013 [DOI] [PubMed] [Google Scholar]
- 77.Andree H, Nickel P, Nasiadko C, Hammer MH, Schönemann C, Pruss A, Volk HD, Reinke P: Identification of dialysis patients with panel-reactive memory T cells before kidney transplantation using an allogeneic cell bank. J Am Soc Nephrol 17: 573–580, 2006 [DOI] [PubMed] [Google Scholar]
- 78.Augustine JJ, Poggio ED, Clemente M, Aeder MI, Bodziak KA, Schulak JA, Heeger PS, Hricik DE: Hemodialysis vintage, black ethnicity, and pretransplantation antidonor cellular immunity in kidney transplant recipients. J Am Soc Nephrol 18: 1602–1606, 2007 [DOI] [PubMed] [Google Scholar]
- 79.Poggio ED, Augustine JJ, Clemente M, Danzig JM, Volokh N, Zand MS, Hricik DE, Heeger PS: Pretransplant cellular alloimmunity as assessed by a panel of reactive T cells assay correlates with acute renal graft rejection. Transplantation 83: 847–852, 2007 [DOI] [PubMed] [Google Scholar]
- 80.Ashoor I, Najafian N, Korin Y, Reed EF, Mohanakumar T, Ikle D, Heeger PS, Lin M: Standardization and cross validation of alloreactive IFNγ ELISPOT assays within the clinical trials in organ transplantation consortium. Am J Transplant 13: 1871–1879, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li B, Hartono C, Ding R, Sharma VK, Ramaswamy R, Qian B, Serur D, Mouradian J, Schwartz JE, Suthanthiran M: Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med 344: 947–954, 2001 [DOI] [PubMed] [Google Scholar]
- 82.Ding R, Li B, Muthukumar T, Dadhania D, Medeiros M, Hartono C, Serur D, Seshan SV, Sharma VK, Kapur S, Suthanthiran M: CD103 mRNA levels in urinary cells predict acute rejection of renal allografts. Transplantation 75: 1307–1312, 2003 [DOI] [PubMed] [Google Scholar]
- 83.Tatapudi RR, Muthukumar T, Dadhania D, Ding R, Li B, Sharma VK, Lozada-Pastorio E, Seetharamu N, Hartono C, Serur D, Seshan SV, Kapur S, Hancock WW, Suthanthiran M: Noninvasive detection of renal allograft inflammation by measurements of mRNA for IP-10 and CXCR3 in urine. Kidney Int 65: 2390–2397, 2004 [DOI] [PubMed] [Google Scholar]
- 84.Muthukumar T, Dadhania D, Ding R, Snopkowski C, Naqvi R, Lee JB, Hartono C, Li B, Sharma VK, Seshan SV, Kapur S, Hancock WW, Schwartz JE, Suthanthiran M: Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med 353: 2342–2351, 2005 [DOI] [PubMed] [Google Scholar]
- 85.Afaneh C, Muthukumar T, Lubetzky M, Ding R, Snopkowski C, Sharma VK, Seshan S, Dadhania D, Schwartz JE, Suthanthiran M: Urinary cell levels of mRNA for OX40, OX40L, PD-1, PD-L1, or PD-L2 and acute rejection of human renal allografts. Transplantation 90: 1381–1387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suthanthiran M, Schwartz JE, Ding R, Abecassis M, Dadhania D, Samstein B, Knechtle SJ, Friedewald J, Becker YT, Sharma VK, Williams NM, Chang CS, Hoang C, Muthukumar T, August P, Keslar KS, Fairchild RL, Hricik DE, Heeger PS, Han L, Liu J, Riggs M, Ikle DN, Bridges ND, Shaked A; Clinical Trials in Organ Transplantation 04 (CTOT-04) Study Investigators : Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med 369: 20–31, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matignon M, Ding R, Dadhania DM, Mueller FB, Hartono C, Snopkowski C, Li C, Lee JR, Sjoberg D, Seshan SV, Sharma VK, Yang H, Nour B, Vickers AJ, Suthanthiran M, Muthukumar T: Urinary cell mRNA profiles and differential diagnosis of acute kidney graft dysfunction. J Am Soc Nephrol 25: 1586–1597, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Keslar KS, Lin M, Zmijewska AA, Sigdel TK, Tran TQ, Ma L, Bhasin M, Rao P, Ding R, Iklé DN, Mannon RB, Sarwal MM, Strom TB, Reed EF, Heeger PS, Suthanthiran M, Fairchild RL: Multicenter evaluation of a standardized protocol for noninvasive gene expression profiling. Am J Transplant 13: 1891–1897, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Galichon P, Amrouche L, Hertig A, Brocheriou I, Rabant M, Xu-Dubois YC, Ouali N, Dahan K, Morin L, Terzi F, Rondeau E, Anglicheau D: Urinary mRNA for the diagnosis of renal allograft rejection: The issue of normalization [published online ahead of print May 27, 2016]. Am J Transplant doi:10.1111/ajt.13891 [DOI] [PubMed] [Google Scholar]
- 90.Hricik DE, Nickerson P, Formica RN, Poggio ED, Rush D, Newell KA, Goebel J, Gibson IW, Fairchild RL, Riggs M, Spain K, Ikle D, Bridges ND, Heeger PS; CTOT-01 Consortium : Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant 13: 2634–2644, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jackson JA, Kim EJ, Begley B, Cheeseman J, Harden T, Perez SD, Thomas S, Warshaw B, Kirk AD: Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am J Transplant 11: 2228–2234, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ho J, Rush DN, Karpinski M, Storsley L, Gibson IW, Bestland J, Gao A, Stefura W, HayGlass KT, Nickerson PW: Validation of urinary CXCL10 as a marker of borderline, subclinical, and clinical tubulitis. Transplantation 92: 878–882, 2011 [DOI] [PubMed] [Google Scholar]
- 93.Ho J, Rush DN, Nickerson PW: Urinary biomarkers of renal transplant outcome. Curr Opin Organ Transplant 20: 476–481, 2015 [DOI] [PubMed] [Google Scholar]
- 94.Schaub S, Nickerson P, Rush D, Mayr M, Hess C, Golian M, Stefura W, Hayglass K: Urinary CXCL9 and CXCL10 levels correlate with the extent of subclinical tubulitis. Am J Transplant 9: 1347–1353, 2009 [DOI] [PubMed] [Google Scholar]
- 95.Hirt-Minkowski P, Amico P, Ho J, Gao A, Bestland J, Hopfer H, Steiger J, Dickenmann M, Burkhalter F, Rush D, Nickerson P, Schaub S: Detection of clinical and subclinical tubulo-interstitial inflammation by the urinary CXCL10 chemokine in a real-life setting. Am J Transplant 12: 1811–1823, 2012 [DOI] [PubMed] [Google Scholar]
- 96.Gandolfini I, Harris C, Purroy C, Nair V, Reid-Adam J, Bestard O, Heeger P: Rapid detection of urinary CXCL9 as a diagnostic and prognostic tool for managing acute cellular rejection (ACR) in kidney transplantation [Abstract]. Am J Transplant 16: 252, 2016 [Google Scholar]
- 97.Ying L, Sarwal M: In praise of arrays. Pediatr Nephrol 24: 1643–1659, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Z, Gerstein M, Snyder M: RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strehlau J, Pavlakis M, Lipman M, Shapiro M, Vasconcellos L, Harmon W, Strom TB: Quantitative detection of immune activation transcripts as a diagnostic tool in kidney transplantation. Proc Natl Acad Sci USA 94: 695–700, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma VK, Bologa RM, Li B, Xu GP, Lagman M, Hiscock W, Mouradian J, Wang J, Serur D, Rao VK, Suthanthiran M: Molecular executors of cell death--differential intrarenal expression of Fas ligand, Fas, granzyme B, and perforin during acute and/or chronic rejection of human renal allografts. Transplantation 62: 1860–1866, 1996 [DOI] [PubMed] [Google Scholar]
- 101.Zheng XX, Schachter AD, Vasconcellos L, Strehlau J, Tian Y, Shapiro M, Harmon W, Strom TB: Increased CD40 ligand gene expression during human renal and murine islet allograft rejection. Transplantation 65: 1512–1515, 1998 [DOI] [PubMed] [Google Scholar]
- 102.Akalin E, Hendrix RC, Polavarapu RG, Pearson TC, Neylan JF, Larsen CP, Lakkis FG: Gene expression analysis in human renal allograft biopsy samples using high-density oligoarray technology. Transplantation 72: 948–953, 2001 [DOI] [PubMed] [Google Scholar]
- 103.Reeve J, Einecke G, Mengel M, Sis B, Kayser N, Kaplan B, Halloran PF: Diagnosing rejection in renal transplants: A comparison of molecular- and histopathology-based approaches. Am J Transplant 9: 1802–1810, 2009 [DOI] [PubMed] [Google Scholar]
- 104.Reeve J, Sellarés J, Mengel M, Sis B, Skene A, Hidalgo L, de Freitas DG, Famulski KS, Halloran PF: Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transplant 13: 645–655, 2013 [DOI] [PubMed] [Google Scholar]
- 105.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, Salvatierra O Jr.: Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med 349: 125–138, 2003 [DOI] [PubMed] [Google Scholar]
- 106.de Freitas DG, Sellarés J, Mengel M, Chang J, Hidalgo LG, Famulski KS, Sis B, Einecke G, Halloran PF: The nature of biopsies with “borderline rejection” and prospects for eliminating this category. Am J Transplant 12: 191–201, 2012 [DOI] [PubMed] [Google Scholar]
- 107.Sellarés J, Reeve J, Loupy A, Mengel M, Sis B, Skene A, de Freitas DG, Kreepala C, Hidalgo LG, Famulski KS, Halloran PF: Molecular diagnosis of antibody-mediated rejection in human kidney transplants. Am J Transplant 13: 971–983, 2013 [DOI] [PubMed] [Google Scholar]
- 108.Halloran PF, Chang J, Famulski K, Hidalgo LG, Salazar ID, Merino Lopez M, Matas A, Picton M, de Freitas D, Bromberg J, Serón D, Sellarés J, Einecke G, Reeve J: Disappearance of T cell-mediated rejection despite continued antibody-mediated rejection in late kidney transplant recipients. J Am Soc Nephrol 26: 1711–1720, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Modena BD, Kurian SM, Gaber LW, Waalen J, Su AI, Gelbart T, Mondala TS, Head SR, Papp S, Heilman R, Friedewald JJ, Flechner SM, Marsh CL, Sung RS, Shidban H, Chan L, Abecassis MM, Salomon DR: Gene expression in biopsies of acute rejection and interstitial fibrosis/tubular atrophy reveals highly shared mechanisms that correlate with worse long-term outcomes. Am J Transplant 16: 1982–1998, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Salazar ID, Merino López M, Chang J, Halloran PF: Reassessing the significance of intimal arteritis in kidney transplant biopsy specimens. J Am Soc Nephrol 26: 3190–3198, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cohen D, Colvin RB, Daha MR, Drachenberg CB, Haas M, Nickeleit V, Salmon JE, Sis B, Zhao MH, Bruijn JA, Bajema IM: Pros and cons for C4d as a biomarker. Kidney Int 81: 628–639, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Halloran PF, Pereira AB, Chang J, Matas A, Picton M, De Freitas D, Bromberg J, Serón D, Sellarés J, Einecke G, Reeve J: Microarray diagnosis of antibody-mediated rejection in kidney transplant biopsies: An international prospective study (INTERCOM). Am J Transplant 13: 2865–2874, 2013 [DOI] [PubMed] [Google Scholar]
- 113.Haas M, Sis B, Racusen LC, Solez K, Glotz D, Colvin RB, Castro MC, David DS, David-Neto E, Bagnasco SM, Cendales LC, Cornell LD, Demetris AJ, Drachenberg CB, Farver CF, Farris AB 3rd, Gibson IW, Kraus E, Liapis H, Loupy A, Nickeleit V, Randhawa P, Rodriguez ER, Rush D, Smith RN, Tan CD, Wallace WD, Mengel M; Banff Meeting Report Writing Committee : Banff 2013 meeting report: Inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant 14: 272–283, 2014 [DOI] [PubMed] [Google Scholar]
- 114.Loupy A, Lefaucheur C, Vernerey D, Chang J, Hidalgo LG, Beuscart T, Verine J, Aubert O, Dubleumortier S, Duong van Huyen JP, Jouven X, Glotz D, Legendre C, Halloran PF: Molecular microscope strategy to improve risk stratification in early antibody-mediated kidney allograft rejection. J Am Soc Nephrol 25: 2267–2277, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Furness PN, Taub N; Convergence of European Renal Transplant Pathology Assessment Procedures (CERTPAP) Project : International variation in the interpretation of renal transplant biopsies: Report of the CERTPAP project. Kidney Int 60: 1998–2012, 2001 [DOI] [PubMed] [Google Scholar]
- 116.Bromberg JS: What’s hot, what’s new at WTC--basic science. Am J Transplant 15: 320–326, 2015 [DOI] [PubMed] [Google Scholar]
- 117.Flechner SM, Kurian SM, Head SR, Sharp SM, Whisenant TC, Zhang J, Chismar JD, Horvath S, Mondala T, Gilmartin T, Cook DJ, Kay SA, Walker JR, Salomon DR: Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant 4: 1475–1489, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li L, Khatri P, Sigdel TK, Tran T, Ying L, Vitalone MJ, Chen A, Hsieh S, Dai H, Zhang M, Naesens M, Zarkhin V, Sansanwal P, Chen R, Mindrinos M, Xiao W, Benfield M, Ettenger RB, Dharnidharka V, Mathias R, Portale A, McDonald R, Harmon W, Kershaw D, Vehaskari VM, Kamil E, Baluarte HJ, Warady B, Davis R, Butte AJ, Salvatierra O, Sarwal MM: A peripheral blood diagnostic test for acute rejection in renal transplantation. Am J Transplant 12: 2710–2718, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roedder S, Sigdel T, Salomonis N, Hsieh S, Dai H, Bestard O, Metes D, Zeevi A, Gritsch A, Cheeseman J, Macedo C, Peddy R, Medeiros M, Vincenti F, Asher N, Salvatierra O, Shapiro R, Kirk A, Reed EF, Sarwal MM: The kSORT assay to detect renal transplant patients at high risk for acute rejection: Results of the multicenter AART study. PLoS Med 11: e1001759, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Menon M, Zhang W, Yi Z, Wei C, Woytovich C, Rosales I, Schroppel B, Chapman J, Nankivell B, O’Connell P, Smith RN, Colvin RB, Murphy B: A peripheral blood gene expression signature for subclinical acute rejection and associated long-term graft injury. Am J Transplant 14: 220–232, 2014 [Google Scholar]
- 121.Gielis EM, Ledeganck KJ, De Winter BY, Del Favero J, Bosmans JL, Claas FH, Abramowicz D, Eikmans M: Cell-free DNA: An upcoming biomarker in transplantation. Am J Transplant 15: 2541–2551, 2015 [DOI] [PubMed] [Google Scholar]
- 122.Beck J, Urnovitz HB, Riggert J, Clerici M, Schütz E: Profile of the circulating DNA in apparently healthy individuals. Clin Chem 55: 730–738, 2009 [DOI] [PubMed] [Google Scholar]
- 123.Suzuki N, Kamataki A, Yamaki J, Homma Y: Characterization of circulating DNA in healthy human plasma. Clin Chim Acta 387: 55–58, 2008 [DOI] [PubMed] [Google Scholar]
- 124.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R: DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 61: 1659–1665, 2001 [PubMed] [Google Scholar]
- 125.Beck J, Bierau S, Balzer S, Andag R, Kanzow P, Schmitz J, Gaedcke J, Moerer O, Slotta JE, Walson P, Kollmar O, Oellerich M, Schütz E: Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem 59: 1732–1741, 2013 [DOI] [PubMed] [Google Scholar]
- 126.Sigdel TK, Vitalone MJ, Tran TQ, Dai H, Hsieh SC, Salvatierra O, Sarwal MM: A rapid noninvasive assay for the detection of renal transplant injury. Transplantation 96: 97–101, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang J, Tong KL, Li PK, Chan AY, Yeung CK, Pang CC, Wong TY, Lee KC, Lo YM: Presence of donor- and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: Urinary DNA chimerism. Clin Chem 45: 1741–1746, 1999 [PubMed] [Google Scholar]
- 128.Bruno DL, Ganesamoorthy D, Thorne NP, Ling L, Bahlo M, Forrest S, Veenendaal M, Katerelos M, Skene A, Ierino FL, Power DA, Slater HR: Use of copy number deletion polymorphisms to assess DNA chimerism. Clin Chem 60: 1105–1114, 2014 [DOI] [PubMed] [Google Scholar]
- 129.Snyder TM, Khush KK, Valantine HA, Quake SR: Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci USA 108: 6229–6234, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.García Moreira V, Prieto García B, Baltar Martín JM, Ortega Suárez F, Alvarez FV: Cell-free DNA as a noninvasive acute rejection marker in renal transplantation. Clin Chem 55: 1958–1966, 2009 [DOI] [PubMed] [Google Scholar]
- 131.O’Riordan E, Orlova TN, Mei J J, Butt K, Chander PM, Rahman S, Mya M, Hu R, Momin J, Eng EW, Hampel DJ, Hartman B, Kretzler M, Delaney V, Goligorsky MS: Bioinformatic analysis of the urine proteome of acute allograft rejection. J Am Soc Nephrol 15: 3240–3248, 2004 [DOI] [PubMed] [Google Scholar]
- 132.Schaub S, Rush D, Wilkins J, Gibson IW, Weiler T, Sangster K, Nicolle L, Karpinski M, Jeffery J, Nickerson P: Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J Am Soc Nephrol 15: 219–227, 2004 [DOI] [PubMed] [Google Scholar]
- 133.Pelzl S, Opelz G, Daniel V, Wiesel M, Süsal C: Evaluation of posttransplantation soluble CD30 for diagnosis of acute renal allograft rejection. Transplantation 75: 421–423, 2003 [DOI] [PubMed] [Google Scholar]
- 134.Pelzl S, Opelz G, Wiesel M, Schnülle P, Schönemann C, Döhler B, Süsal C: Soluble CD30 as a predictor of kidney graft outcome. Transplantation 73: 3–6, 2002 [DOI] [PubMed] [Google Scholar]
- 135.Chen Y, Tai Q, Hong S, Kong Y, Shang Y, Liang W, Guo Z, He X: Pretransplantation soluble CD30 level as a predictor of acute rejection in kidney transplantation: A meta-analysis. Transplantation 94: 911–918, 2012 [DOI] [PubMed] [Google Scholar]
- 136.Sigdel TK, Kaushal A, Gritsenko M, Norbeck AD, Qian WJ, Xiao W, Camp DG 2nd, Smith RD, Sarwal MM: Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteomics Clin Appl 4: 32–47, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sigdel TK, Salomonis N, Nicora CD, Ryu S, He J, Dinh V, Orton DJ, Moore RJ, Hsieh SC, Dai H, Thien-Vu M, Xiao W, Smith RD, Qian WJ, Camp DG 2nd, Sarwal MM: The identification of novel potential injury mechanisms and candidate biomarkers in renal allograft rejection by quantitative proteomics. Mol Cell Proteomics 13: 621–631, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sigdel TK, Gao Y, He J, Wang A, Nicora CD, Fillmore TL, Shi T, Webb-Robertson BJ, Smith RD, Qian WJ, Salvatierra O, Camp DG 2nd, Sarwal MM: Mining the human urine proteome for monitoring renal transplant injury. Kidney Int 89: 1244–1252, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Blydt-Hansen TD, Sharma A, Gibson IW, Mandal R, Wishart DS: Urinary metabolomics for noninvasive detection of borderline and acute T cell-mediated rejection in children after kidney transplantation. Am J Transplant 14: 2339–2349, 2014 [DOI] [PubMed] [Google Scholar]
- 140.Suhre K, Schwartz JE, Sharma VK, Chen Q, Lee JR, Muthukumar T, Dadhania DM, Ding R, Ikle DN, Bridges ND, Williams NM, Kastenmüller G, Karoly ED, Mohney RP, Abecassis M, Friedewald J, Knechtle SJ, Becker YT, Samstein B, Shaked A, Gross SS, Suthanthiran M: Urine metabolite profiles predictive of human kidney allograft status. J Am Soc Nephrol 27: 626–636, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Freedman BI, Julian BA, Pastan SO, Israni AK, Schladt D, Gautreaux MD, Hauptfeld V, Bray RA, Gebel HM, Kirk AD, Gaston RS, Rogers J, Farney AC, Orlando G, Stratta RJ, Mohan S, Ma L, Langefeld CD, Hicks PJ, Palmer ND, Adams PL, Palanisamy A, Reeves-Daniel AM, Divers J: Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transplant 15: 1615–1622, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Menon MC, Chuang PY, Li Z, Wei C, Zhang W, Luan Y, Yi Z, Xiong H, Woytovich C, Greene I, Overbey J, Rosales I, Bagiella E, Chen R, Ma M, Li L, Ding W, Djamali A, Saminego M, O’Connell PJ, Gallon L, Colvin R, Schroppel B, He JC, Murphy B: Intronic locus determines SHROOM3 expression and potentiates renal allograft fibrosis. J Clin Invest 125: 208–221, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sankaran D, Asderakis A, Ashraf S, Roberts IS, Short CD, Dyer PA, Sinnott PJ, Hutchinson IV: Cytokine gene polymorphisms predict acute graft rejection following renal transplantation. Kidney Int 56: 281–288, 1999 [DOI] [PubMed] [Google Scholar]
- 144.Pelletier R, Pravica V, Perrey C, Xia D, Ferguson RM, Hutchinson I, Orosz C: Evidence for a genetic predisposition towards acute rejection after kidney and simultaneous kidney-pancreas transplantation. Transplantation 70: 674–680, 2000 [DOI] [PubMed] [Google Scholar]
- 145.Morris H, DeWolf S, Robins H, Sprangers B, LoCascio SA, Shonts BA, Kawai T, Wong W, Yang S, Zuber J, Shen Y, Sykes M: Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci Transl Med 7: 272ra10, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Park W, Griffin M, Grande JP, Cosio F, Stegall MD: Molecular evidence of injury and inflammation in normal and fibrotic renal allografts one year posttransplant. Transplantation 83: 1466–1476, 2007 [DOI] [PubMed] [Google Scholar]
- 147.Moreso F, Carrera M, Goma M, Hueso M, Sellares J, Martorell J, Grinyó JM, Serón D: Early subclinical rejection as a risk factor for late chronic humoral rejection. Transplantation 93: 41–46, 2012 [DOI] [PubMed] [Google Scholar]
- 148.Stegall MD, Gaston RS, Cosio FG, Matas A: Through a glass darkly: Seeking clarity in preventing late kidney transplant failure. J Am Soc Nephrol 26: 20–29, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mahoney RJ, Ault KA, Given SR, Adams RJ, Breggia AC, Paris PA, Palomaki GE, Hitchcox SA, White BW, Himmelfarb J, Leeber DA: The flow cytometric crossmatch and early renal transplant loss. Transplantation 49: 527–535, 1990 [DOI] [PubMed] [Google Scholar]
- 150.Pei R, Lee JH, Shih NJ, Chen M, Terasaki PI: Single human leukocyte antigen flow cytometry beads for accurate identification of human leukocyte antigen antibody specificities. Transplantation 75: 43–49, 2003 [DOI] [PubMed] [Google Scholar]