Abstract
Plasmodium parasites, the causative agents of malaria, have evolved a unique cell division cycle in the clinically relevant asexual blood-stage of infection1. DNA replication commences approximately halfway through the intracellular development following invasion and parasite growth. The schizont stage is associated with multiple rounds of DNA replication and nuclear division without cytokinesis resulting in a multinucleated cell. Nuclei divide asynchronously through schizogony, with only the final round of DNA replication and segregation being synchronous and coordinated with daughter cell assembly2,3. However, the control mechanisms for this divergent mode of replication are unknown. Here we show that the Plasmodium-specific kinase PfCRK4 is a key cell cycle regulator that orchestrates the multiple rounds of DNA replication throughout schizogony in P. falciparum. PfCRK4 depletion led to a complete block in nuclear division and profoundly inhibited DNA replication. Quantitative phosphoproteomic profiling identified a set of PfCRK4-regulated phosphoproteins with greatest functional similarity to CDK2 substrates, particularly proteins involved in origin of replication firing. PfCRK4 was required for the initial and subsequent rounds of DNA replication during schizogony, and in addition was essential for development in the mosquito vector. Our results identified an essential S phase promoting factor of the unconventional P. falciparum cell cycle. PfCRK4 is required for both a prolonged period of the intraerythrocytic blood-stage of malaria infection, as well as for transmission, revealing a broad window for PfCRK4-targeted chemotherapeutics.
Malaria parasites proliferate through schizogony in the blood-stage of infection: a series of rapid rounds of DNA replication and nuclear division produces a syncytial cell with approximately 20 nuclei. In contrast to the synchronous nuclear division observed in other multinucleated cells, such as the early drosophila embryo4, P. falciparum nuclei divide asynchronously during the blood-stage despite sharing the same cytoplasm2,3, suggesting that cell cycle progression is not governed by diffusible cytoplasmic factors. Knockout screens have identified non-essential blood-stage Plasmodium kinases, and suggested those that could be essential for the regulation of schizogony5,6. The molecular mechanisms regulating this diverged mode of replication are largely unknown yet the recent development of conditional gene expression technologies now allows to unequivocally demonstrate gene essentiality in P. falciparum7–9.
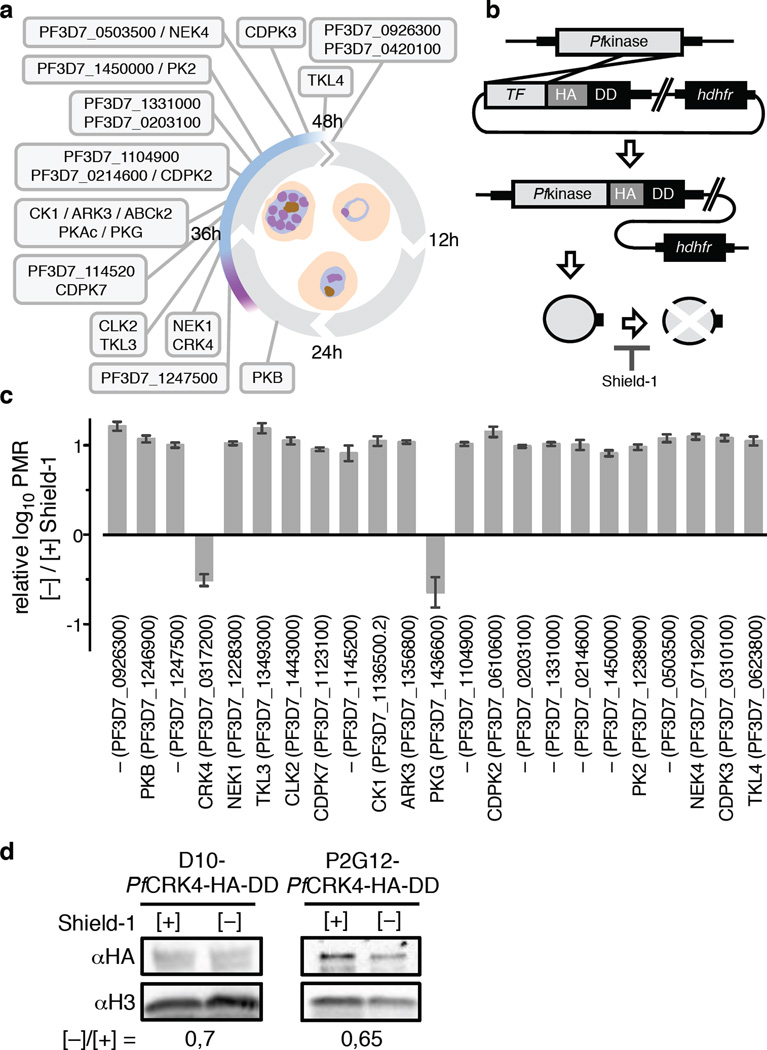
To directly identify critical regulators of schizogony and simultaneously determine protein function, we adopted the destabilization domain (DD) conditional knockdown approach7,10,11. We generated endogenous DD fusions of 23 schizont-stage kinases in the P. falciparum D10 strain and screened for vulnerability to destabilization by DD (Fig. 1a, b, Supplementary Information Fig. 1a). Using this approach, we found that two kinases, the cGMP-dependent protein kinase (PfPKG) and the cdc2-related protein kinase 4 (PfCRK4), had profound proliferation defects in absence of Shield-1 and showed dose-dependency (Fig. 1c, Supplementary Information Fig. 1b, c). Chemical inhibition of PfPKG has previously established its essential role in parasite egress from erythrocytes at the end of schizogony12,13, now confirmed by conditional destabilization.
Fig. 1. Conditional destabilization identified P. falciparum CRK4 as essential for asexual blood-stage development.
a, Schematic of the P. falciparum blood-stage cycle indicating peak transcript times of kinases targeted with a destabilization domain; purple, approximate onset of DNA replication; blue, approximate duration of schizogony. b, Single crossover 3’ replacement strategy to fuse candidate kinases with a hemagglutinin (HA) and a destabilization domain (DD) tag, which destabilizes kinases in absence of Shield-1. Note, PKAc, ABCk2, and PF3D7_0420100 were refractory to HA-DD fusion. c, Effect of conditional kinase destabilization on parasite growth over two cycles (± SD); PMR, parasite multiplication rate. d, Western blots demonstrate conditional destabilization of PfCRK4 in two genetic backgrounds; the level of knockdown is indicated below; H3, histone H3.
As previously observed14,15, we found little correlation between the level of protein knockdown and the ability to reveal essentiality of other kinases previously thought to be essential6 using the DD approach (Fig. 1d, Supplementary Information Fig. 1d, 9c, d). This could be due to insufficient destabilization of these proteins or because they are required at inherently different levels for asexual proliferation.
The biological function of PfCRK4 is unknown and we confirmed its essentiality in a different parental line, P. falciparum P2G1216, which also produces gametocytes (Supplementary Information Fig. 2). PfCRK4 is a member of an Apicomplexa-specific kinase subfamily related to cyclin-dependent kinases (CDK) (Supplementary Information Fig. 3, Supplementary Data table 1)17,18. In many organisms CDKs in complex with cyclins regulate key steps of the cell cycle and other cellular functions19,20. Compared to human CDK2, the kinase domain of PfCRK4 possesses multiple sequence inserts of unknown function (Fig. 2a, Supplementary Information Fig. 4).
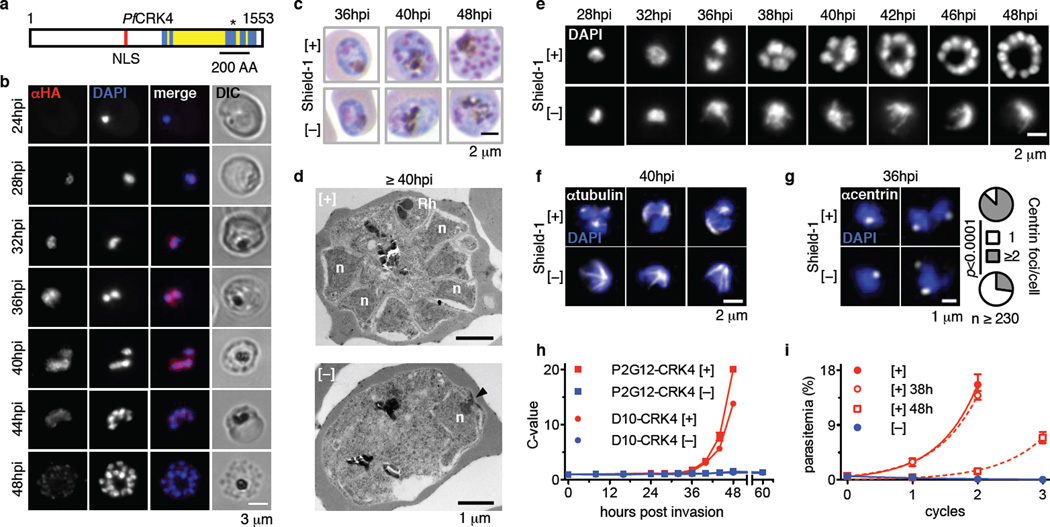
Fig. 2. Nuclear localized PfCRK4 is essential for trophozoite-to-schizont transition and DNA replication.
a, Domain structure of PfCRK4; blue, kinase domain; yellow, kinase domain insertions relative to human CDK2; red, predicted nuclear localization signal (NLS); asterisk, active site residues; numbers, first and last amino acid (AA). b, Immunofluorescence detection of PfCRK4-HA-DD in blood stage parasites (representative of three biological replicates). c, Light microscopy of May-Grünwald-Giemsa-stained PfCRK4-depleted parasites (D10 parent, representative of two biological replicates and results with P2G12-PfCRK4-HA-DD). d, Ultrastructure of PfCRK4-depleted parasites (D10 parent, representative of n ≥ 25 cells per condition); arrowhead, spindle pole body; n, nucleus; Rh, rhoptry. e, Nuclear development in PfCRK4-depleted parasites (D10 parent, representative of three biological replicates and results with P2G12-PfCRK4-HA-DD). f, Immunofluorescence detection of spindle structures in PfCRK4-depleted parasites (P2G12 parent, representative of two biological replicates). g, Quantification of centriolar plaques by immunofluorescence in PfCRK4-depleted parasites (D10 parent; p-value, Chi-Square test; representative of two biological replicates and results with P2G12-PfCRK4-HA-DD). h, DNA content of PfCRK4-depleted parasites from both parental lines; mean ± SD of triplicates (representative of three biological replicates); ring stage DNA content defined as 1 C. i, Proliferation of parasites when PfCRK4 was re-stabilized from 38 hpi or 48 hpi of the first cycle onwards, mean ± SD (representative of two biological replicates and results with P2G12-PfCRK4-HA-DD).
PfCRK4 is localized to the nucleus of late trophozoites and schizonts, with the signal greatly diminished in segmented schizonts that had undergone cytokinesis (Fig. 2b). Following conditional depletion of PfCRK4 (Fig. 1d) parasites arrested at the trophozoite-to-schizont transition (Fig. 2c). At ≥40 hours post invasion (hpi), parasites [+] Shield-1 (i.e., wild type PfCRK4 levels) segmented into daughter cells, containing nuclei and rhoptries (apical organelles required for merozoite invasion), which are characteristic of mature schizonts (Fig. 2d, top). In contrast, parasites cultured [−] Shield-1 (i.e., PfCRK4 is depleted) showed no nuclear division or apical organelle biogenesis (Fig. 2d, bottom, Supplementary Information Fig. 5a).
Analysis of nuclei stained with fluorescent DNA-specific dyes confirmed that the nuclei did not divide and revealed a substantially distorted nuclear morphology (Fig. 2e, Supplementary Information Fig. 5b). Hemispindle structures were evident in PfCRK4-depleted cells (Fig. 2f; Supplementary Information Fig. 5c, d); however, they were greatly enlarged relative to spindles in wild type parasites and might account for the nuclear distortion. Concordantly, division of the centriolar plaque3,21, the parasite’s microtubule organizing centre, was diminished (Fig. 2g). To ascertain whether PfCRK4 affects DNA replication, we quantified the parasite’s DNA content by flow cytometry22,23. While in wild type parasites DNA replication commences at 29–32 hpi, i.e., parasites with C-values >1 appear, we found DNA replication to be profoundly inhibited in PfCRK4-depleted parasites (Fig. 2h). In contrast, the development of the mitochondria and apicoplast organelles was unaffected following depletion of PfCRK4 (Supplementary Information Fig. 5e, f). The PfCRK4-dependent block was completely reversible up to 38 hpi by addition of Shield-1. However, reversion at 48 hpi led to very poor recovery (Fig. 2i), identifying a window within which PfCRK4 depletion is cytostatic before becoming cytotoxic.
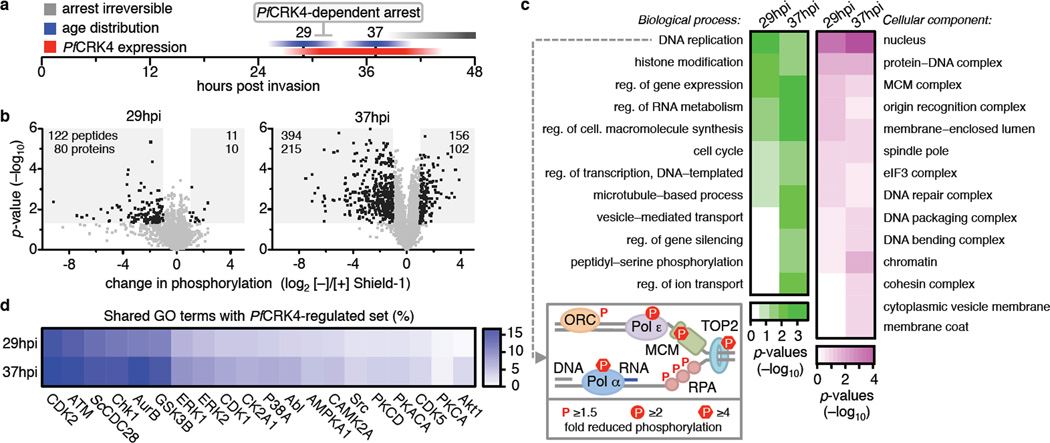
To elucidate processes regulated by PfCRK4, we assessed changes in the phosphoproteome of PfCRK4-depleted parasites at two time points (Fig. 3a): at 29 hpi, when PfCRK4 is already expressed at the onset of DNA replication (Fig. 2b, h), and also at 37 hpi, where in wild type parasites DNA replication is evident and when the PfCRK4-dependent arrest was still reversible (Fig. 2h, i) (Supplementary Information Fig. 6; Supplementary Data table 2–5). Large changes occur in the phosphoproteome at 29 hpi indicating that PfCRK4 is active when the first round of DNA replication commences (Fig. 2h). We used k-means clustering to identify the phosphosites most affected by the PfCRK4 depletion, which showed a reduction of about 2-fold (Supplementary Information Fig. 6b). Applying a threshold based on a ≥2-fold decrease in phosphorylation and p-values <0.05, we identified 220 proteins (80 from 29 hpi, 215 from 37 hpi with 75 proteins shared), suggesting that these belong to a PfCRK4-regulated set of phosphoproteins (Fig. 3b, Supplementary Data table 4). Western blot analysis confirmed differential phosphorylation of histone H3 (serine 28) in a different genetic background (Supplementary Information Fig. 6c). The most enriched phosphorylation motif within these proteins was a previously undescribed SΦxK motif (Φ, hydrophobic; x, any amino acid), with S being phosphorylated. The canonical CDK motif S/TP24,25, with S or T phosphorylated, was also enriched (Supplementary Information Fig. 6d, e).
Fig. 3. PfCRK4 regulates S phase.
a, Schematic illustrating the sampling time points for phosphoproteomic profiling. b, Differential phosphorylation in D10-PfCRK4-HA-DD parasites [−] Shield-1 relative to [+] Shield-1; 29 hpi in duplicate, 37 hpi in triplicate; p-values by Student`s t-test; grey-shaded regions indicate >2-fold changes in phosphorylation and p-values of <0.05, number of peptides and respective proteins are indicated c, GO term enrichment analysis of proteins with ≥2-fold reduced phosphorylation (p-values <0.05); p-values, Modified Fisher’s Exact Test. Box: P. falciparum homologues of S. cerevisiae factors required for origin of replication activation in vitro with reduced phosphorylation at 37 hpi in PfCRK4-depleted parasites; Pol, DNA polymerase; MCM, mini chromosome maintenance complex; TOP2, topoisomerase 2; RPA, replication protein A; ORC, origin recognition complex. d, Percentage of shared GO terms of proteins with ≥2-fold reduced phosphorylation (p-values <0.05) upon PfCRK4-depletion and of substrate sets of human and S. cerevisiae (Sc) kinases.
To examine the functional significance of the PfCRK4-regulated set of proteins, we used gene ontology (GO) term enrichment analysis (Fig. 3c). The most enriched biological process GO term was ‘DNA replication’, and ‘nucleus’ was the most enriched cellular component GO term. Strikingly, six of the seven identifiable P. falciparum homologues (or subunits thereof) of the 16 Saccharomyces cerevisiae factors required for origin licensing and firing in vitro26 showed reduced phosphorylation in PfCRK4 depleted parasites (Fig. 3c, box; Supplementary Information Fig. 6f, Supplementary Data table 3, 4). GO terms of proteins with a ≥2-fold increase in phosphorylation (p-value <0.05) were enriched for biosynthetic and intracellular transport processes at 37 hpi, perhaps indicating secondary effects; however, no statistically significant enrichment was observed at 29 hpi (Supplementary Information Fig. 6g).
We next compared the GO terms of the PfCRK4-regulated set with GO terms of other available kinase substrate sets–one S. cerevisiae kinase27 and 19 human kinases28 (www.phosphosite.org)–each with ≥30 confirmed substrates (Fig. 3d). The substrate set of human CDK2, a major S phase promoting factor, showed the highest percentage of shared annotations, suggesting that PfCRK4 acts in a related fashion to promote S phase in P. falciparum.
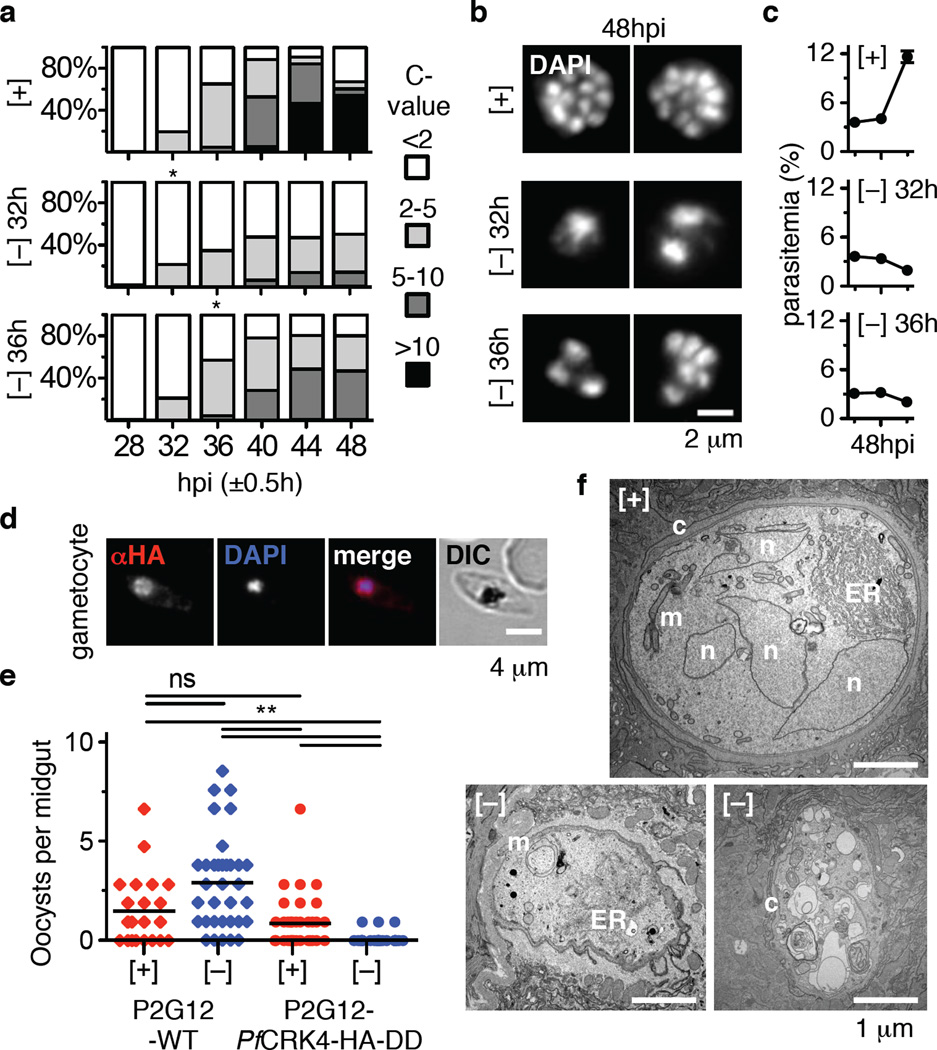
PfCRK4 expression increases through schizogony (Fig. 2b) and its transcripts peak the latest of all putative Plasmodium CDK-like kinases29. We therefore hypothesized that PfCRK4 is crucial for all rounds of DNA replication seen in schizogony. When we depleted PfCRK4 at points later in schizogony, we detected no further increase in DNA content (Fig. 4a), premature termination of nuclear division (Fig. 4b), and parasites failed to proliferate (Fig. 4c). Thus demonstrating that PfCRK4 function is critical throughout schizogony.
Fig. 4. PfCRK4 is essential throughout schizogony and critical for transmission.
a, DNA content analysis of D10-PfCRK4-HA-DD parasites when PfCRK4 was present or depleted from 32 hpi, or 36 hpi onwards; 1 C, ring stage DNA content; asterisks, time of Shield-1 removal (representative of two biological replicates and results with P2G12-PfCRK4-HA-DD). Note, second cycle ring-stage parasites re-appear at 48 hpi when PfCRK4 is present. b, DAPI-stained nuclei of parasites from Fig. 4a (representative of n ≥ 70 cells per condition). c, Proliferation of cultures from Fig. 4a, mean ± SD of triplicates. d, Immunofluorescence detects PfCRK4-HA-DD in gametocytes (P2G12 parent, representative of two biological replicates). e, Depletion of PfCRK4 during gametocytogenesis diminished oocyst numbers on An. gambiae female midguts; bars, mean oocyst numbers; ns, not significant; **, p≤ 0.01 (Kruskal-Wallis test). f, Ultrastructure of mosquito-stage parasites from infections with PfCRK4-depleted gametocytes (P2G12 parent, representative of ≥ ten cells from independent infection experiments); n, nucleus; ER, endoplasmic reticulum; m, mitochondria; c, capsule.
To determine a role for PfCRK4 in other life cycle stages, we used the P2G12-PfCRK4-HA-DD line that produces high levels of gametocytes to analyse PfCRK4 function during parasite transmission to the Anopheles gambiae mosquito vector. While we detected nuclear expression of PfCRK4 in gametocytes (Fig. 4d, Supplementary Information Fig. 7a), depletion during the latter portion of gametocyte development (i.e. from day 6 to 16 post-induction) had no effect on gametocytemia nor on the male-to-female gametocyte ratio (Supplementary Information Table 1). PfCRK4 depletion also did not impair exflagellation (Supplementary Information Fig. 7b, c, d). We observed a reduced PfCRK4 signal in ookinetes (Supplementary Information Fig. 7e), and the numbers of oocysts on An. gambiae female midguts was greatly diminished upon PfCRK4 depletion (Fig. 4e, Supplementary Information Fig. 8). Oocysts derived from gametocyte cultures in the presence of PfCRK4 showed multiple nuclei, well-expanded organelles, and a well-developed capsule (Fig. 4f, top). In contrast, infections with PfCRK4-depleted gametocytes resulted in necrotic parasites reminiscent of dead ookinetes, the motile zygote of Plasmodium parasites (Fig. 4f, bottom; Supplementary Information Fig. 8). This indicates that PfCRK4 is required at some point during ookinete formation and early oocyst development, and likely plays a role during DNA replication in zygotes.
Our data provide evidence that PfCRK4 is an essential protein kinase promoting S phase through the initiation of multiple rounds of DNA replication during schizogony. While phosphorylation is likely important for the regulation of PfCRK4 (Supplementary Information Fig. 3b, 4a), a potential cyclin-dependence is in question as the cyclin binding domain is poorly conserved (Supplementary Information Fig. 4a)17. Interestingly, homologues of Group I cyclins are not found in the Plasmodium genome, including cyclin E, which is critical for G1 to S transition in other systems30. Interventions targeting PfCRK4 function would be efficacious throughout the extended period of schizogony in the clinically relevant blood-stage, as well as at the transmission stages, which taken together are attractive features for future drug development.
Methods
Reagents and oligonucleotide primers
We purchased chemicals from Sigma-Aldrich (unless otherwise noted), primers Integrated DNA Technologies (sequences are available upon request), and restriction enzymes from New England Biolabs.
Construction of plasmids
The terminal 1–2 kb of PKB (PF3D7_1246900), TLK4 (PF3D7_0623800), and PKG (PF3D7_1436600) were sub-cloned into the 3’ replacement vector pJDD417, generating HA-DD single crossover tagging plasmids. The terminal ~1 kb of PF3D7_0420100 was amplified using an extended reverse primer, introducing two additional HA-tags. Cloning into pJDD41 resulted in a 3×HA-DD single crossover tagging plasmid. All other targeting fragments were sub-cloned into this 3×HA-DD plasmid (sequences available upon request).
Parasite culture, transfection and synchronization
P. falciparum D10 was obtained from the Walter and Eliza Hall Institute (Melbourne, Australia). P. falciparum P2G12 (clone of 3D7) was obtained from Harvard T.H. Chan School of Public Health (Boston, USA). Parasites were cultured as previously described in RPMI-1640 media supplemented with 0.5% Albumax-II (Invitrogen), 50 mg / l hypoxanthine, 0.21% Sodium Bicarbonate, and 25 mM HEPES (EMD Biosciences)31. Human O+ erythrocytes (Research Blood Components) were diluted to 2–4% haematocrit. Transgenic parasites were generated by electroporation of synchronized ring-stage parasites as previously described32. Single homologous recombination events were selected in presence of 500 nM Shield-1 by cycling on and off WR99210 (Jacobus Pharmaceutical Company) and cloned by limiting dilution. Parasites were synchronized by a combination of heparin and sorbitol treatments as previously described33. Unless otherwise noted, DD-tagged parasites were cultured in 250 nM Shield-1. When desired, Shield-1 was removed by triple washes with excess of RPMI.
Southern blot analysis
Genomic DNA was harvested with QIAamp Blood Mini Kit (Qiagen) and digested with the enzymes indicated in Supplementary Fig. 1a, resolved on 0.8% agarose gels, transferred to GeneScreen Plus (Perkin Elmer), and hybridized with specific radiolabeled probes.
May-Grünwald-Giemsa staining and imaging of blood-stage parasites
Air-dried thin-smear blood films were fixed and stained according to manufacturers instructions and imaged on a Zeiss AxioCam microscope equipped with a 100X oil-immersion objective. Raw images were analysed using ImageJ34.
Dose response experiments
Dose response curves were generated as previously described using SYBR® Green I35. In brief, triplicate 2-fold Shield-1 dilution series were set up in 100 µl parasite cultures (0.075% parasitemia in 2% haematocrit). After two cycles, parasites were lysed with 20 µl of 6× SYBR® green I lysis buffer (0.16% saponin; 20 mM Tris-HCl, 5 mM EDTA, 1.6% Triton X 100, pH 7.4), supplemented with 1:1.000 SYBR® green I (from 10.000× stock, Thermo Fisher). Fluorescence intensity was measured on a SpectraMax M5 plate reader (Molecular Devices).
Conditional destabilization screen of schizont-stage kinases
We used the relative log10 parasite multiplication rate (PMR) [−] over [+] Shield-1 as a measure of the effect of conditional destabilization of schizont-stage kinases. Synchronized ring-stage parasites of all P. falciparum kinase-DD fusion lines were seeded in triplicate at 0.05% parasitemia, 0.5% haematocrit [+] or [−] Shield-1. Parasitemia was monitored by flow cytometry (see below) and expressed as log10. Linear regression analysis (Prism, GraphPad) allowed calculating the PMR. The effect of protein knockdown ([−] Shield-1) relative to [+] Shield-1 was plotted as log10 PMR [−] Shield-1 over log10 PMR [+] Shield-1. The error (standard deviation) for
was ropagated as follows:
Where A is the log10 PMR [−] Shield-1, B is the log10 PMR [+] Shield-1 and sA and sB are the sample standard deviations in A and B, respectively36.
Parasitemia quantification by flow cytometry
Parasitized erythrocytes were fixed as previously described37, stained with 1:2.000 SYBR® green I (Life Technologies) in PBS, and analysed on a Miltenyi MACSQuant® flow cytometer. Data were analysed using FlowJo (Tree Star) and Prism (GraphPad) software.
Immunoblot analysis
Synchronized parasites were harvested and uninfected erythrocytes were removed using 0.05% saponin in PBS plus 1× cOmplete™ protease inhibitor cocktail (Roche). Upon washes, parasites were lysed in Laemmli sample buffer and proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with the following primary antibodies: anti-HA (1:1.000; Roche; clone 3F10), anti-Pflactate dehydrogenase (LDH) (1:2.000; gift of Michael T. Makler), anti-histone H3 (1:1.000; Abcam; ab1791), anti-pS28 histone H3 (1:1.000; Abcam; ab5169), anti-Pfaldolase (1:40.000, Abcam; ab38905). Signal intensities were quantitated using Imag J.
Gametocyte assays and An. gambiae infections
Gametocyte cultures were seeded at 0.5% parasitemia in 5 ml RPMI with 10% serum, 4% haematocrit, and 1 µM Shield-1 and kept in a candle jar at 37 °C. From day six onwards parasites were cultured [+] and [−] Shield-1. Estimates for the various stages and sexes were determined from a Giemsa-stained thin-smear blood film. On days 15 to 18 of culture, parasites were harvested and male gametocyte exflagellation was assessed by microscopy. For the infection of An. gambiae KEELE strain females, cultures were washed and gametocytemia was adjusted to 0.3% in normal human serum with 45% haematocrit. Subsequently, females were infected in a Standard Membrane Feeding Assay as previously described38.
Oocyst quantification
Midguts from infected An. gambiae females were dissected on days 7 or 8 post blood-feeding and subsequently the number of oocysts were enumerated following 0.1% mercurochrome staining and analysis by light microscopy (20X objective).
DNA content analysis
The DNA-content of parasites was determined by flow cytometry. Parasitized erythrocytes were fixed in 4% paraformaldehyde and 0.0075% glutaraldehyde for 45 min at room temperature37. To reduce RNA-derived background signal cells were permeabilized with 0.1% Triton X-100, treated with 0.3 mg / ml RNase A for 20’ at 37 °C, and subsequently washed. Cells were stained with 1:2.000 SYBR® green I (Life Technologies) in PBS, washed, and analysed on a Miltenyi MACSQuant® flow cytometer. Data were analysed using FlowJo (Tree Star), Excel (Microsoft), and Prism (GraphPad) software. Second cycle ring-stage parasites were omitted for DNA content analysis of 48 hpi schizont-stage parasites.
Immunofluorescence assays
Immunofluorescence assays of blood-stage parasites was done as previously described37,39. In brief, parasitized erythrocytes were air-dried, methanol-fixed39, and analysed using rat anti-HA (1:100, Roche, clone 3F10), mouse anti-centrin (1:500, Millipore, clone 20H5), mouse anti-α-tubulin (1:2.000, Sigma-Aldrich, clone B-5-1-2), rabbit anti-GAP45 (1:2.000, gift of Julian C. Rayner), or rabbit anti-PfEBA175 (1:500, Malaria Research and Reference Reagent Resource Center, R3347) antibodies. Alternatively, parasites were fixed in solution37 for 20 min at room temperature, immobilized on poly-L-lysine coated cover slips (Corning®), and analysed using mouse anti-α-tubulin (1:2.000, Sigma-Aldrich, clone B-5-1-2) or rabbit anti-P. falciparum acyl carrier protein40 (1:500, gift of Sean T. Prigge) antibodies. Primary antibodies were detected using Alexa Fluor® conjugated secondary antibodies (1:2.000, Thermo Fisher Scientific). Cells were mounted with DAPI Fluoromount-G® (SouthernBiotech) or cells were stained with Hoechst 33342 (Thermo Fisher Scientific) and mounted Fluoromount-G® (SouthernBiotech), imaged on a Nikon Eclipse TE300 microscope equipped with a Hamamatsu C10600 Orca R2 digital camera, and analysed employing ImageJ software.
For immunofluorescence analysis of exflagellation, mature gametocytes were resuspended in RPMI-1640 media supplemented with 20% human serum (AB+), 50 mg / l hypoxanthine, 0.21% Sodium Bicarbonate, 25 mM HEPES (EMD Biosciences), and 50 µM xanthurenic acid at room temperature and pH 8 to induce exflagellation41. Following 20 min incubation at room temperature, cells were washed, spotted on a glass slide, air-dried, methanol-fixed, and further processed as described above.
For immunofluorescence analysis of ookinetes, female An. gambiae mosquitoes were fed with late stage PfCRK4-HA-DD gametocyte cultures, which were maintained [+] and [−] Shield-1 from day 6 post-induction. Approximately 24 hours post-feeding, the blood bolus from the mosquito midgut lumen was isolated. Bolus material was spotted on glass slides and air-dried preceding immunofluorescence analysis.
Immunofluorescence analysis of P. falciparum late midgut oocysts was done as previously described42. In brief, dissected midguts were fixed in 4% paraformaldehyde for 1h at room temperature, washed, and permeabilized, and blocked using 0.2% Triton X-100 in 1% bovine serum albumin in PBS. Midgut oocysts were analysed using a mouse anti-circumsporozoite (CS) protein antibody (15 µg/ml, gift of Photini Sinnis), an Alexa Flour® 594 secondary antibody (1:1.000), and DAPI nuclear stain, mounted on a glass slide in 10% glycerol in PBS, and imaged on a Nikon Eclipse 90i microscope. Raw images were analysed using Volocity software (Perkin-Elmer).
JC-1 staining of P. falciparum mitochondria
Parasite mitochondria were stained with the cationic mitochondrial membrane potential sensor JC-1 (Life Technologies) as previously described43. In brief, 2 µM JC-1 in 37 °C warm RPMI was filtered using an Acrodisc® Syringe Filter (0.2 µm HT Tuffryn® Membrane, Pall) to removed preformed JC-1 aggregates. Parasites were incubated with JC-1 containing media for 20 min at 37 °C, washed, and imaged immediately on a Nikon Eclipse TE300 microscope with equipped a Hamamatsu C10600 Orca R2 digital camera and analysed employing ImageJ software.
Electron microscopy
Synchronized blood-stage parasites were isolated by magnetic affinity purification using a MACS LS column (Miltenyi Biotec), adjusted to ~50% parasitemia with uninfected erythrocytes, and resuspended in 200 µl 3% BSA in PBS. Cells were fixed for 1h at room temperature with an equal volume of 2.5% paraformaldehyde, 5.0% glutaraldehyde, and 0.06% picric acid in 0.2 M cacodylate buffer. Cells were washed in cacodylate buffer, post-fixed for 1 h with 1% osmium tetroxide (OsO4), 1.5% potassium ferrocyanide (KFeCN6), and washed in H2O. Following 1h incubation in 1% aqueous uranyl acetate solution, samples were washed in H2O and subsequently dehydrated in grades of alcohol. The samples were placed in propyleneoxide for 1 h and infiltrated in a 1:1 mixture of propyleneoxide and TAAB Epon (Marivac) over night. Samples were embedded in TAAB Epon and polymerized at 60 °C for 48 h. Ultrathin sections from a Reichert Ultracut-S microtome were mounted on copper grids and stained with lead citrate. Samples were imaged on a TecnaiG2 Spirit BioTWIN with an AMT 2k CCD camera under 80 kV.
For P. falciparum oocyst thin-section transmission electron microscopy, infected midguts were fixed at day 7 post infection in 2.5% glutaraldehyde (Electron Microscopy Sciences) and processed as described44. Stained sections were examined with a Philips CM120 EM under 80 kV.
Phosphoproteomic profiling
Cell culture and harvest
Synchronized D10-PfCRK4-HA-DD parasites were cultured for 29 or 37 hours post invasion (hpi) [+] or [−] Shield-1. At 29 hpi duplicate samples were harvested, at 37 hpi triplicate samples were harvested for each [+] and [−] Shield-1. For each of the ten samples ~1.25 × 109 parasites were isolated and uninfected erythrocytes were removed using saponin buffer (0.05% saponin in PBS plus 1× PhosSTOP (Roche), 1× cOmplete™ protease inhibitor cocktail (Roche), and 1 mM phenylmethanesulfonyl fluoride (PMSF)). Subsequently, cells were washed in saponin buffer to further remove erythrocytes and erythrocyte debris, followed by washes with PBS plus 1× PhosSTOP, 1× cOmplete™ protease inhibitor cocktail, and 1 mM PMSF.
Cell lysis and protein digestion
Lysates were prepared as previously described45,46. In brief, cells were lysed in 8 M urea, 100 mM sodium chloride, 25 mM TRIS pH 8 plus 1× PhosSTOP, 1× cOmplete™ protease inhibitor cocktail, and 1 mM PMSF in PBS. The protein concentration was estimated in the Bio-Rad Protein Assay (Bio-Rad). Disulfide bonds were reduced with 5 mM tris(2-chloroethyl) phosphate (TCEP) for 30 min at 37 °C. Cysteines were alkylated with 15 mM iodoacetamide for 30 min at room temperature in the dark, followed by incubation with 5 mM DTT for 15 min at room temperature in the dark to quench excess of iodoacetamide.
Chloroform-methanol precipitated proteins47 were resuspended in 8 M urea, 50 mM HEPES pH 8.5 and subsequently diluted to 1 M urea 50 mM HEPES pH 8.5 for digestion with LysC protease (1:100 protease-to-protein ratio, 3 h at 37 °C) before addition of trypsin (1:100 protease-to-protein ratio) and continued digest overnight at 37 °C. The reaction was quenched with 1% formic acid, subjected to C18 solid phase extraction (Sep-Pak, Waters), followed by vacuum-centrifugation.
Isobaric labelling with tandem mass tags (TMT)
Peptides (200 µg) from each sample were dissolved in 100 mM HEPES pH 8.5. Labelling with TMT reagents (Thermo Fisher Scientific) was done according the manufactures instructions and as previously described48. The TMT-labelled samples were combined at a 1:1 ratio across all samples, vacuum centrifuged to near dryness, and subjected to C18 solid phase extraction (Sep-Pak, Waters).
Phosphopeptide enrichment
Peptides were resuspended in 100 mM HEPES pH 8.5 and phosphopeptides were enriched using Titanosphere TiO2 5 µm particles (GL Biosciences) as previously described46,48,49. The flow-through was kept for proteome analysis. Enriched TMT-labelled phosphopeptides and peptides were dried via vacuum centrifugation.
Off-line basic pH reversed-phase (BPRP) fractionation
For protein level analysis, we fractionated the pooled TMT-labelled peptide sample via BPRP HPLC as previously described48,50. Fractions were desalted, dried, and reconstituted in 5% acetonitrile, 5% formic acid for LC-MS/MS processing.
For phosphopeptide analysis, we used the Pierce Off-line basic pH reversed-phase (BPRP) fractionation kit (Thermo Scientific). Fractions collected included organic buffer bumps with 5, 7.5, 10, 12.5, 15, 17.5, 20, 50, and 70% acetonitrile, in addition to the wash and flow through fractions. These fractions were vacuum-centrifuged to near dryness, desalted via StageTip, dried, and reconstituted in 5% acetonitrile, 5% formic acid for LC-MS/MS processing.
Liquid chromatography and tandem mass spectrometry
Data were collected using an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) coupled to a Proxeon EASY-nLC II liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were fractionated on a 75 µm inner diameter micro capillary column packed with ~0.5 cm of Magic C4 resin (5 µm, 100 Å, Michrom Bioresources) followed by ~35 cm of GP-18 resin (1.8 µm, 200 Å, Sepax). For each analysis, we loaded ~1 µg onto the column.
For total proteome analysis, peptides were separated using a 180 min gradient of 6–23% acetonitrile in 0.125% formic acid (flow rate: ~600 nl / min). Each analysis used the multi-notch MS3-based TMT method51. The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120.000; mass range 400–1,400 m/z; automatic gain control (AGC) target 2.0 × 105; maximum injection time 100 ms). Precursors for MS2/MS3 analysis were selected using a TopSpeed 2 seconds method. MS2 analysis consisted of collision-induced dissociation (quadrupole ion trap analysis; AGC 4 × 103; normalized collision energy (NCE) 35; maximum injection time 150 ms). Following acquisition of each MS2 spectrum, we collected an MS3 spectrum using a recently described method in which multiple MS2 fragment ions were captured in the MS3 precursor population using isolation waveforms with multiple frequency notches51. MS3 precursors were fragmented by high-energy collision-induced dissociation (HCD) and analysed (NCE 55; AGC 5 × 104; maximum injection time 150 ms, resolution was 60.000 at 200 Th).
For phosphoproteome analysis, peptides were separated using a 120 min gradient of 3–23% acetonitrile in 0.125% formic acid (flow rate: ~575 nl / min). Each analysis used the multi-notch MS3-based TMT method51. The scan sequence was identical to the proteome scan, except the maximum injection time was 200 ms for MS2 analysis and 250 ms for MS3 analysis.
Initial mass spectrometry data analysis
Mass spectra were processed using a SEQUEST-based in-house software pipeline52. Spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included all entries from a combined Uniprot human and Plasmodium database (February, 2015). This database was concatenated with one composed of all protein sequences in the reversed order. Total protein level analysis searches were performed using a 50 ppm precursor ion tolerance, product ion tolerance was set to 0.9 Da. These wide mass tolerance windows were chosen to maximize sensitivity in conjunction with SEQUEST searches and linear discriminant analysis52,53. TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification for both total proteome and phosphoproteome datasets. For phosphoprotein analysis, +79.966 Da was set as a variable modification on serine, threonine, and tyrosine residues.
Peptide-spectral matches (PSMs) were adjusted to a 1% false discovery rate (FDR)54,55. PSM filtering used a linear discriminant analysis as previously described52, while considering the following parameters: XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy. For the phosphorylation dataset, site localization was evaluated via AScore53. For TMT-based reporter ion quantitation, we extracted the summed signal-to-noise (S/N) ratio for each TMT channel and found the closest matching centroid to the expected mass of the TMT reporter ion.
The search space for each reporter ion was limited to a range of 0.003 Th to prevent overlap between the isobaric reporter ions. For protein-level comparisons, PSMs were identified, quantified, collapsed to a 1% peptide false discovery rate (FDR), and further collapsed to a final protein-level FDR of 1%. Protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for observed peptides.
Proteins were quantified by summing reporter ion counts across all matching PSMs using in-house software, as previously described52. Poor quality PSMs, MS3 spectra with >8 TMT reporter ion channels missing, MS3 spectra with TMT reporter summed signal-to-noise ratio <100, or no MS3 spectra were excluded from quantitation51. Protein quantitation values were exported for further analysis in Microsoft Excel or SAS JMP. Each reporter ion channel was summed across all quantified proteins and normalized assuming equal protein loading of all 10 samples.
Evolutionary relationship of PfCRK4
Protein kinase amino acid sequences from the cyclin-dependent kinase (CDK) families of apicomplexans and model organism and non-CDK out-group kinases were chosen for comparison with PfCRK4 (Supplementary Data table 1). Kinase domain sequences were trimmed, aligned using MUSCLE56 and analysed using RAxML57.
Domain drawings of PfCRK4 and its orthologues in Plasmodium spp
Domain boundaries were identified by searching sequences against Pfam58 using the HMMER package59. Domains were illustrated using Protdraw (http://sourceforge.net/projects/protdraw/).
Homology modelling of PfCRK4
The protein kinase domain of PfCRK4 was modelled using the co-crystal structure of the human CDK2-Cyclin A-substrate peptide complex (1QMZ.pdb)25. We used this structure as a template in order to model PfCRK4 in a potentially active form. The kinase domains of the target and query sequences were aligned using MUSCLE56 followed by manual review and adjustment. Homology models were made using MODELLER 9.1560. Structural alignments were reviewed and adjusted using Chimera61.
Identification of conditionally related clusters in proteomic and phosphoproteomic data
The total sum signal-to-noise values of peptides collapsed into proteins are given in Supplementary Data table 3. For phosphopeptides values are given in Supplementary Data table 4. Contaminants and false positives were removed, and values for each channel were normalized by the column sum. Values were placed on a scale of 0 – 1 by dividing by the maximum value for the protein or phosphopeptide. Scaling was done for all channels at once and separately for experiments at 29 hpi and 37 hpi. Phosphopeptides for which the total sum signal-to-noise sum ≤100 were not included in clustering analysis, and the total sum signal-to-noise values were adjusted to reflect protein levels from the proteome analysis. K-means clustering62 was done using R (http://cran.r-project.org/) with the Hartigan-Wong method using 1,000 starting configurations and 10,000 iterations. Clustering results, log2-fold changes, and –log10 p-values (Student’s t-test) are included in Supplementary Data tables 3 and 4.
Gene orthology relationships
Relationships between P. falciparum proteins and Saccharomyces cerevisiae orthologues for Figure 3F Box (Supplementary Data table 3) were investigated using a downloaded version of the OrthoMCL database, version 563. A comprehensive orthology analysis is provided in Supplementary Data table 6.
Functional annotation and enrichment analysis
The functional enrichment of sets of proteins with phosphopeptides most reduced in knockdown experiments at 29 hpi and 37 hpi was assessed using Ontologizer64. Gene association files containing gene ontology (GO) terms for Plasmodium falciparum, Saccharomyces cerevisiae, and Homo sapiens were from http://www.geneontology.org/gene-associations65 downloaded on May 21, 2015. To generate comparative heat maps of GO terms for both time points we selected the most specific terms, i.e. those without children, which we refer to as “tips” for convenience. Enrichment p-values were included in the heat map if they were present at either time point; p-values for non-tip terms were included if the term was a tip at the other time point. GO term annotations of the proteins detected in this study are given in Supplementary Data table 3 and defined in Supplementary Data table 7.
Functional comparison of the PfCRK4-regulated set of phosphoproteins
The functional similarity between PfCRK4 and other protein kinases was assessed by comparing GO terms of the PfCRK4-regulated set with GO terms of reference kinase substrate sets. Reference human kinase substrate sets were obtained from PhosphoSitePlus (http://www.phosphosite.org/staticDownloads.do)28 on May 4, 2015, the substrate set from Saccharomyces cerevisiae (Sc) cdc28 came from27. Reference kinases with less than 30 identified substrates were omitted in the analysis. Functional enrichment was assessed as described above. To calculate the relatedness of the substrate sets of kinases with PfCRK4, the number of terms in the intersect of the set of significantly enriched functions from the PfCRK4-regulated set and the substrate set from the reference kinases was divided by the number of significant functional terms in the substrate set from the reference kinase.
Identification and enrichment analysis of phosphorylation motifs
Alignments of phospho-peptides with reduced abundance in the absence of Shield-1 were manually inspected. The two motifs discernable in this set may be retrieved from tab-delimited tables of triskaidekapeptides by the regular expressions66: \t[A-Z]{6}[ST][IVLFMWA].[RKH] (the SϕxK motif; ϕ, hydrophobic amino acid; x, any amino acid) and \t[A-Z]{6}[ST]P (the S/TP motif). The set of phosphopeptides detected in this study (Supplementary Data table 4) was searched with these regular expressions and the percentage peptides comprised of these motifs was calculated. Phosphopeptides containing SϕxK and S/TP motifs are identified in Supplementary Data table 4. Sequence logos were made from alignments of phosphopeptides containing SϕxK and S/TP motifs from sets of phosphopeptides reduced in the absence of Shield-1 using Weblogo (http://weblogo.berkeley.edu)67.
Data availability
The data supporting the results of this study are available within the paper and its Supplementary Information and Supplementary Data. Sequences of oligonucleotides and plasmids generated for this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We gratefully thank the members of the Duraisingh Laboratory for helpful discussions and critical reading of the manuscript, and D.F. Wirth for continuous guidance and support. We thank S.T. Prigge, P. Sinnis, M.T. Makler, and J. C. Rayner for sharing reagents. We thank the Microscopy Facility at the Johns Hopkins School of Medicine. We thank H. Hurd and P. Eggleston for the An. gambiae KEELE strain. This work was supported by a NIH R21 1R21AIO88314-01A1 (M.T.D.), a Wellcome Trust Project grant #094752/Z/10/Z (D.A.B and M.T.D); a Deutsche Forschungsgemeinschaft research fellowship GA 1668/2-1 (M.G.); a Pediatric Scientist Development Program Fellowship award K12-HD000850 (J.D.D.); a NIH/NIDDK grant K01 DK098285 (J.A.P.); a HFSP award RGY0073/2012 (J.G.K. and R.R.D); the Bloomberg Family Foundation through the Johns Hopkins Malaria Research Institute, the NIH National Center for Research Resources UL1 RR 025005, the Malaria Research Institute of The Johns Hopkins Bloomberg School of Public Health (R.R.D).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nmicrobiol/
Author contributions
M.G. designed, performed, and interpreted much of the experimental work; J.M.G. analysed the phosphoproteomic data and provided bioinformatics support; J.D.D., J.A.P., J.G.K., A.K.T., A.S.P., and I.C. designed and performed specific experimental work; J.Y., constructed plasmids; R.H.Y.J. provided bioinformatics support; B.E. performed Western blots; D.A.B., R.R.D., and S.P.G. provided reagents and intellectual input into study design; M.G., J.D.D., and M.T.D. conceived the study; M.G., J.M.G, and M.T.D wrote the manuscript. All authors commented on the manuscript.
The authors declare that they have no competing financial interests.
References
- 1.Francia ME, Striepen B. Cell division in apicomplexan parasites. Nat Rev Microbiol. 2014 doi: 10.1038/nrmicro3184. [DOI] [PubMed] [Google Scholar]
- 2.Read M, Sherwin T, Holloway SP, Gull K, Hyde JE. Microtubular organization visualized by immunofluorescence microscopy during erythrocytic schizogony in Plasmodium falciparum and investigation of post-translational modifications of parasite tubulin. Parasitology. 1993;106(Pt 3):223–232. doi: 10.1017/s0031182000075041. [DOI] [PubMed] [Google Scholar]
- 3.Arnot DE, Ronander E, Bengtsson DC. The progression of the intra-erythrocytic cell cycle of Plasmodium falciparum and the role of the centriolar plaques in asynchronous mitotic division during schizogony. Int J Parasitol. 2011;41:71–80. doi: 10.1016/j.ijpara.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Farrell JA, O’Farrell PH. From egg to gastrula: how the cell cycle is remodeled during the Drosophila mid-blastula transition. Annu. Rev. Genet. 2014;48:269–294. doi: 10.1146/annurev-genet-111212-133531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tewari R, et al. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8:377–387. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solyakov L, et al. Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Nat Commun. 2011;2:565. doi: 10.1038/ncomms1558. [DOI] [PubMed] [Google Scholar]
- 7.Dvorin JD, et al. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010;328:910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrell A, et al. A DOC2 protein identified by mutational profiling is essential for apicomplexan parasite exocytosis. Science. 2012;335:218–221. doi: 10.1126/science.1210829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul AS, et al. Parasite Calcineurin Regulates Host Cell Recognition and Attachment by Apicomplexans. Cell Host Microbe. 2015;18:49–60. doi: 10.1016/j.chom.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat Methods. 2007;4:1007–1009. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- 11.Chu BW, Banaszynski LA, Chen L-C, Wandless TJ. Recent progress with FKBP-derived destabilizing domains. Bioorg. Med. Chem. Lett. 2008;18:5941–5944. doi: 10.1016/j.bmcl.2008.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor HM, et al. The malaria parasite cyclic GMP-dependent protein kinase plays a central role in blood-stage schizogony. Eukaryotic Cell. 2010;9:37–45. doi: 10.1128/EC.00186-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins CR, et al. Malaria Parasite cGMP-dependent Protein Kinase Regulates Blood Stage Merozoite Secretory Organelle Discharge and Egress. PLoS Pathog. 2013;9:e1003344. doi: 10.1371/journal.ppat.1003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muralidharan V, Oksman A, Pal P, Lindquist S, Goldberg DE. Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat Commun. 2012;3:1310. doi: 10.1038/ncomms2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck JR, Muralidharan V, Oksman A, Goldberg DE. PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature. 2014;511:592–595. doi: 10.1038/nature13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchholz K, et al. A high-throughput screen targeting malaria transmission stages opens new avenues for drug development. J Infect Dis. 2011;203:1445–1453. doi: 10.1093/infdis/jir037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doerig C, Endicott J, Chakrabarti D. Cyclin-dependent kinase homologues of Plasmodium falciparum. Int J Parasitol. 2002;32:1575–1585. doi: 10.1016/s0020-7519(02)00186-8. [DOI] [PubMed] [Google Scholar]
- 18.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coudreuse D, Nurse P. Driving the cell cycle with a minimal CDK control network. Nature. 2010;468:1074–1079. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- 20.Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aikawa M, Huff CG, Sprinz H. Fine structure of the asexual stages of Plasmodium elongatum. J Cell Biol. 1967;34:229–249. doi: 10.1083/jcb.34.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russo I, Oksman A, Vaupel B, Goldberg DE. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc Natl Acad Sci USA. 2009;106:1554–1559. doi: 10.1073/pnas.0806926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theron M, Hesketh RL, Subramanian S, Rayner JC. An adaptable two-color flow cytometric assay to quantitate the invasion of erythrocytes by Plasmodium falciparum parasites. Cytometry A. 2010;77:1067–1074. doi: 10.1002/cyto.a.20972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Songyang Z, et al. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 25.Lowe ED, et al. Specificity determinants of recruitment peptides bound to phospho-CDK2/cyclin A. Biochemistry. 2002;41:15625–15634. doi: 10.1021/bi0268910. [DOI] [PubMed] [Google Scholar]
- 26.Yeeles JTP, Deegan TD, Janska A, Early A, Diffley JFX. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ubersax JA, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 28.Hornbeck PV, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bozdech Z, et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roques M, et al. Plasmodium P-Type Cyclin CYC3 Modulates Endomitotic Growth during Oocyst Development in Mosquitoes. PLoS Pathog. 2015;11:e1005273. doi: 10.1371/journal.ppat.1005273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 32.Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci USA. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle MJM, et al. Isolation of viable Plasmodium falciparum merozoites to define erythrocyte invasion events and advance vaccine and drug development. Proc Natl Acad Sci USA. 2010;107:14378–14383. doi: 10.1073/pnas.1009198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JD, et al. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrobial agents and chemotherapy. 2007;51:1926–1933. doi: 10.1128/AAC.01607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuhauser C. Calculus For Biology and Medicine: Pearson New International Edition. Pearson Higher Ed; 2013. [Google Scholar]
- 37.Tonkin CJ, et al. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol. 2004;137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Dinglasan RR, et al. Plasmodium falciparum ookinetes require mosquito midgut chondroitin sulfate proteoglycans for cell invasion. PNAS. 2007;104:15882–15887. doi: 10.1073/pnas.0706340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flueck C, et al. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 2010;6:e1000784. doi: 10.1371/journal.ppat.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallagher JR, Matthews KA, Prigge ST. Plasmodium falciparum Apicoplast Transit Peptides are Unstructured in vitro and During Apicoplast Import. Traffic. 2011;12:1124–1138. doi: 10.1111/j.1600-0854.2011.01232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Billker O, et al. Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito. Nature. 1998;392:289–292. doi: 10.1038/32667. [DOI] [PubMed] [Google Scholar]
- 42.Thathy V, et al. Levels of circumsporozoite protein in the Plasmodium oocyst determine sporozoite morphology. EMBO J. 2002;21:1586–1596. doi: 10.1093/emboj/21.7.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasini EM, van den Ierssel D, Vial HJ, Kocken CHM. A novel live-dead staining methodology to study malaria parasite viability. Malar J. 2013;12:190. doi: 10.1186/1475-2875-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coppens I, Joiner KA. Host but not parasite cholesterol controls Toxoplasma cell entry by modulating organelle discharge. Mol Biol Cell. 2003;14:3804–3820. doi: 10.1091/mbc.E02-12-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villén J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulo JA, et al. Effects of MEK inhibitors GSK1120212 and PD0325901 in vivo using 10-plex quantitative proteomics and phosphoproteomics. Proteomics. 2014;15:462–473. doi: 10.1002/pmic.201400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 48.Paulo JA, Gygi SP. A comprehensive proteomic and phosphoproteomic analysis of yeast deletion mutants of 14-3-3 orthologs and associated effects of rapamycin. Proteomics. 2015;15:474–486. doi: 10.1002/pmic.201400155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kettenbach AN, Gerber SA. Rapid reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general phosphotyrosine-specific phosphoproteomics experiments. Anal. Chem. 2011;83:7635–7644. doi: 10.1021/ac201894j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paulo JA, Gaun A, Gygi SP. Global Analysis of Protein Expression and Phosphorylation Levels in Nicotine-Treated Pancreatic Stellate Cells. J. Proteome Res. 2015;14:4246–4256. doi: 10.1021/acs.jproteome.5b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McAlister GC, et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 54.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 55.Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn RD, et al. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics. 2014;47:5–32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 61.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Hartigan JA. Clustering. Annu. Rev. Biophys. Bioeng. 1973;2:81–101. doi: 10.1146/annurev.bb.02.060173.000501. [DOI] [PubMed] [Google Scholar]
- 63.Fischer S, et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011 doi: 10.1002/0471250953.bi0612s35. Chapter 6, Unit–Un19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bauer S, Grossmann S, Vingron M, Robinson P. N. Ontologizer 2.0--a multifunctional tool for GO term enrichment analysis and data exploration. 2008;24:1650–1651. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- 65.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Friedl JEF. Mastering Regular Expressions. O’Reilly Media, Inc.; 2006. [Google Scholar]
- 67.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the results of this study are available within the paper and its Supplementary Information and Supplementary Data. Sequences of oligonucleotides and plasmids generated for this study are available from the corresponding author upon request.