Abstract
The human immune system is highly variable between individuals but relatively stable over time within a given person. Recent conceptual and technological advances have enabled systems immunology analyses, which reveal the composition of immune cells and proteins in populations of healthy individuals. The range of variation and some specific influences that shape an individual’s immune system is now becoming clearer. Human immune systems vary as a consequence of heritable and non-heritable influences, but symbiotic and pathogenic microbes and other non-heritable influences explain most of this variation. Understanding when and how such influences shape the human immune system is key for defining metrics of immunological health and understanding the risk of immune-mediated and infectious diseases.
The immune system is intrinsic to health, but translating what we have learned about basic immunology from animal models to humans has been a major challenge, with many more failures than successes1–3. To improve our knowledge of the human immune system, immunologists are now looking at different ways to directly investigate the immune status of humans3–5. There has been a pressing need for new research strategies that could work within the constraints of humans, as many of the manipulations that are standard in mouse immunology cannot be directly translated to humans. One of the most promising strategies is adapted from systems biology and is referred to as systems vaccinology6 or systems immunology3. In general, systems biology approaches seek to identify the major components of a given system and measure how these components change in response to perturbations of the system. In the immune system, the main components are the different types of immune cells and the cytokines that they communicate with. Fortunately, the majority of these components can be measured with available technologies and a representation of these components is present in a blood sample — which is widely available in human studies.
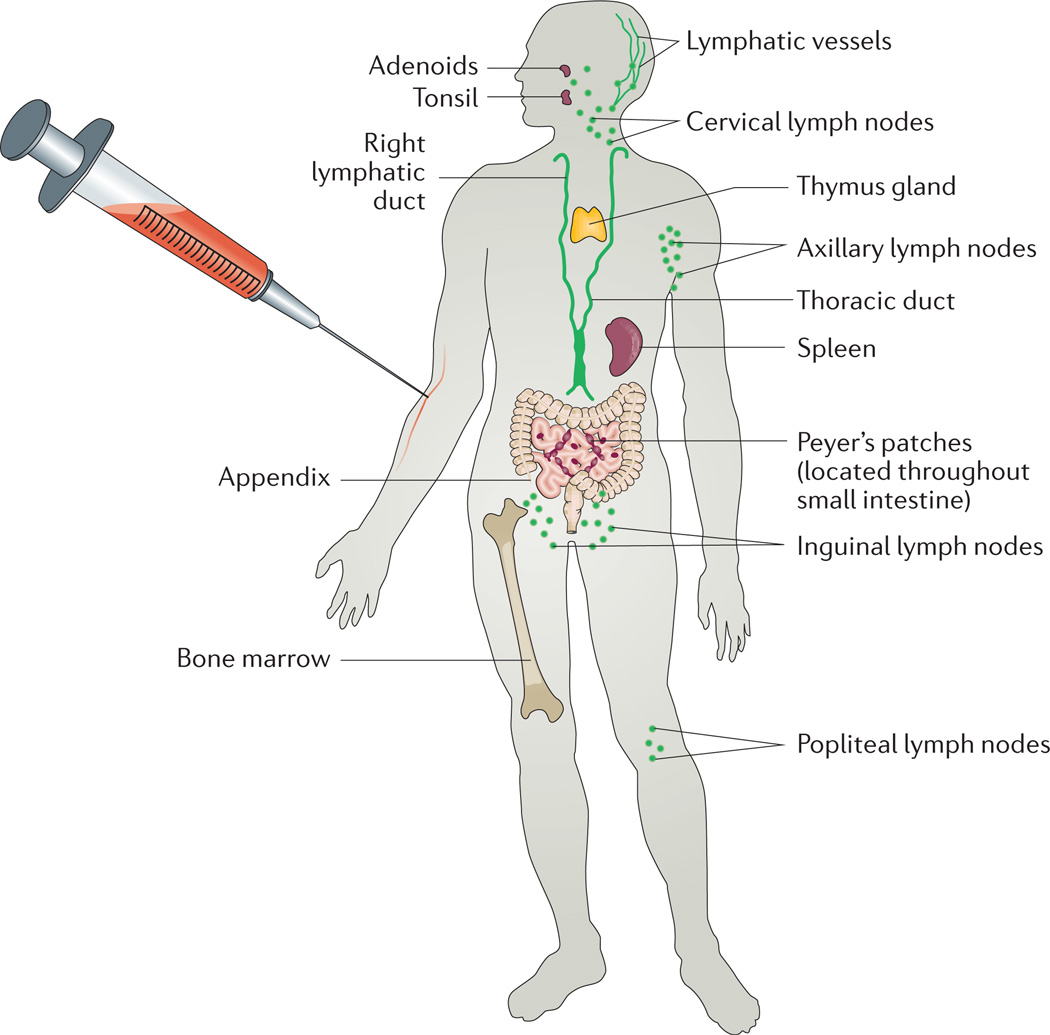
A wide range of factors can perturb the human immune system, but the most convenient to investigate for systems immunology is the response to standard vaccinations such as influenza virus vaccines and, in particular, the very effective and robust yellow fever vaccine. Systems vaccinology can reveal which components of the immune system change and how they change in response to perturbations, and this in turn yields information about the sensitivities of a given person’s immune system and the variation of immune responses between individuals. This information might predict responsiveness or non-responsiveness to vaccines, which is an important problem for less robust vaccines, such as the influenza vaccines, and especially when administered to very young or elderly individuals. By focusing mainly on blood, a systems immunology approach can be informative about both healthy and ill individuals, as well as young and old. In addition, systems approaches make use of the fact that specialized cells in the immune system are both the detectors and effectors of the immune system, that these cells communicate with each other through cytokines and direct interactions and that a global representation of what is happening in the immune system of a particular person at a given time can be estimated by analysing such interactions. Although blood is not an immunological organ per se, it is the conduit for most immune cells circulating in the body, especially after an immunological stimulus such as vaccination (FIG. 1). As an illustration of this, Wilson and colleagues found that 50–80% of circulating plasmablasts were specific for antigens in the vaccine seven days after an influenza virus vaccination7. A similar time course has been shown for gluten-specific CD4+ T cells following gluten challenge in patients with coeliac disease8,9.
Figure 1. The blood as a window for global immune system analysis in humans.
Although the blood is not an immunological organ per se, it is the conduit for most immune cells circulating in the body, especially after an immunological stimulus such as vaccination, allowing even distal processes to be reflected in a blood sample that is readily accessible even in humans.
The recent development of many new high-throughput technologies enables simultaneous measurements of many cell types, cytokines and other biomarkers of immune function in the same blood sample. Such advances provide an opportunity for studying human immune system variation at a global scale, taking co-variation of specific cell populations and proteins into account. Recent population studies have also showed that human immune system variation can now be studied globally, and the influences of age, sex and specific environmental factors can be addressed. These studies are timely and complementary to the many studies investigating genetic influences on immune system function and immunological diseases. A combined understanding of both the heritable and the non-heritable influences on immunity is necessary to fully understand inter-individual variation and its consequences on immunological health and disease. The immune system varies between different tissues within an organism, but in this Review we focus on peripheral blood since it is the most well characterized tissue in these early days of systems immunology. We focus on our current understanding of human immune system variation within individuals over time and between individuals in different age groups and of different sex, and we discuss the specific environmental exposures that shape human immune systems.
Technological advances
There have been a number of important advances in technologies that enable high-dimensional immune system analyses (BOX 1). The possibility to analyse many, if not all, immune system components in the blood allows novel questions to be answered, specifically relating to the interactions between the many components of human immune systems10. Such approaches are providing novel understanding of immune system regulation in health and disease11. Also, by globally profiling, for example all mRNA transcripts, unexpected pathways activated under a specific condition, such as after vaccination can be revealed12–14.
Box 1. Details of high-dimensional immune system analyses technologies.
Immune cell analyses
Flow cytometry
This technique enables single cell analysis by using fluorescently labelled antibodies measuring up to 30 simultaneous parameters in millions of individual cells in the most technically specialized laboratories98; however, most flow cytometric protocols detect 15 or fewer simultaneous parameters. The speed and versatility of the technology and the ability to sort viable cells makes flow cytometry a cornerstone technology in immunology research.
Mass cytometry
Single-cell analysis using antibodies tagged with unique mass-reporters are detectable at single-cell resolution using an inductively-coupled plasma mass spectrometer (ICP-MS) system99. Cytometry-by-time-of-flight (CyTOF; Fluidigm Inc) readily allows the simultaneous measurement of approximately 45 parameters, including proteins and nucleic acids100 in millions of individual immune cells, which enables a unique combination of width and depth of analysis into the cellular immune system37. Both phenotypic and functional measurements such as cytokines101 and intracellular signalling102,103 can be addressed simultaneously, which enables assessment of both phenotypes and functions.
Single-cell gene expression analyses
Sequencing analysis methods in single cells have developed rapidly in recent years. Currently, global transcriptome analyses in several thousands of individual immune cells are possible using the latest protocols, allowing analyses of gene-regulatory patterns in such cell populations and refined atlases of cell populations104. Gene expression analyses also provide the attractive possibility of analysing variable receptor genes, such as those encoding T cell and B cell receptors to determine the clonality of immune cells and their specificity, and combining such information with simultaneous analyses of functional properties. Both PCR-based105 and sequencing-based methodologies106 have been developed for such analyses.
Analyses of serum proteins
Bead array methods using fluorescent bead readouts are popular and commonly used for analysis of serum proteins. Another approach with sensitive detection due to dual recognition of proteins is offered by proximity-extension assays (ProSeek; Olink AB)38, in which affinity reagents are detected by associated nucleic acid probes. Also, mass spectrometry-based plasma proteomics have re-emerged in recent years owing to the developments in fractionation methods, instrumentation and analytical approaches, which enable the broadest analysis of the ~3,000 plasma proteins that are present at variable concentration in humans107.
Time-dependent immune system variation
Variation over time within individuals
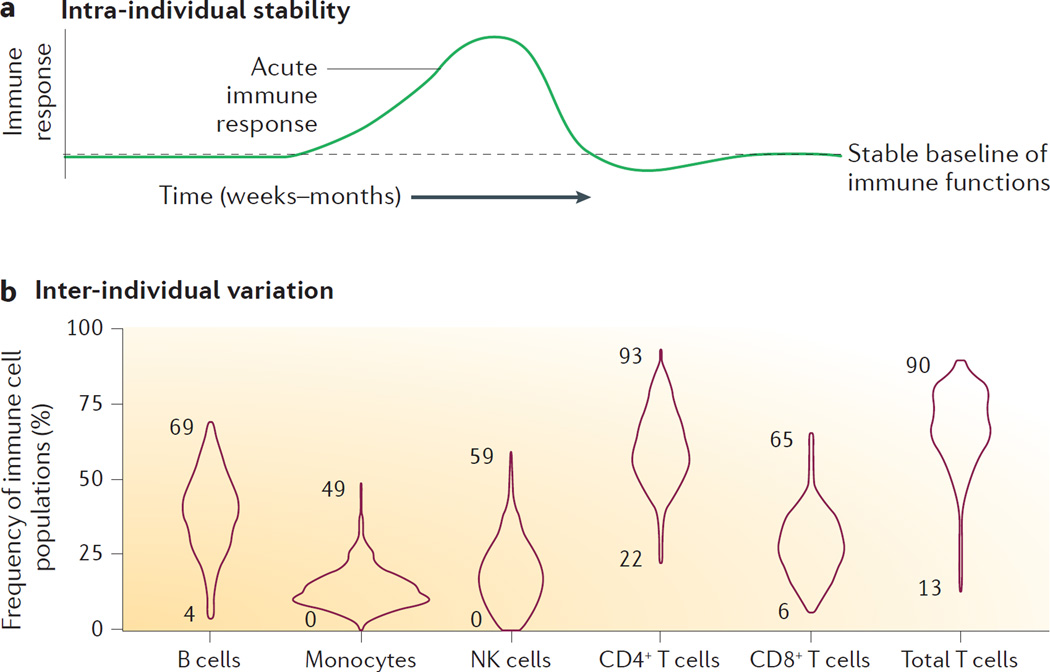
When studying the immune response in an individual during the course of an acute infection or some other perturbation, such as during a vaccine-induced immune response, the immune system would seem to be a continuously moving target. However, this is not the case outside of such episodes, at least not in healthy adults. Blood samples taken weeks to months apart from healthy adults show very stable immune cell frequencies and serum protein levels15,16. Our own analyses over the course of yearly samplings suggest that immune profiles remain stable even at longer intervals (up to 6 years) in healthy individuals17. This suggests that each individual has a baseline state of immune system composition in which cells and proteins are well regulated, and the balances between these are optimal for the current conditions (FIG. 2a). Immune responses to an acute challenge leads to drastic changes that involve expanding cell populations and stark increases in serum protein concentrations, which will quickly return to the same baseline state as before the challenge (FIG. 2a). The mechanisms that regulate such systems-level coordination and regulation are poorly understood but have become amenable for study in recent years thanks to the technological developments that enable simultaneous measurements of all system components in the same sample, and these analysis will help to improve our mechanistic understanding.
Figure 2. Variation in immune cells and proteins.
a | An illustration of the observed stability of most immune cell and protein measurements over the course of weeks to months. During acute immune responses drastic changes occur, but thereafter measurements seem to return to a stable baseline. b | Distributions of six principal immune cell populations from a Stanford cohort (n = 398) of healthy adults11,17. Numbers indicate minimum and maximal values observed.
Immune system variation with age
Young children and elderly individuals are more susceptible to infections than other age groups18,19. The infant has an immune system biased towards tolerance as a consequence of life in utero, and it consists of cells with mostly naive phenotypes that mature when exposed to the environment. The neonatal immune system depends on different protective cell populations compared with adults20, and qualitative differences in immune responses by shared cell populations between young children and adults have also been reported18,21. More work is needed to fully appreciate all the differences between the immune systems of children and adults, with the potential for improved vaccination strategies in the future22. The immune systems of very old individuals are characterized by loss of immune cells, lymphopenia and reduced diversity of variable receptor genes on B cells23 and T cells24, although this reduction in T cell diversity seems less pronounced than previously thought25. Still, it is possible that changes in relative frequencies of specific adaptive lymphocyte clones that differ in phenotype, could contribute to some of the changes in the immune system composition with advancing age. It is important to note that even if specific parameters are found to correlate positively or negatively with age, this cannot be taken as proof of their involvement in the process of ageing. Environmental factors can often influence individuals differently during different stages of life and adaptive changes in the immune system to such factors could explain age-correlated immune system parameters.
Increased concentrations of pro-inflammatory cytokines such as tumour necrosis factor (TNF) have been found in the circulation of some elderly individuals, which suggests there is a low-grade inflammatory state in these individuals26. A recent population analysis of over 1,000 individuals found that 24 out of 92 protein biomarkers in the serum of adults were strongly influenced by age27. Vaccine responses are poor in some older individuals, and baseline biomarkers that can predict poor responsiveness are beginning to emerge15,28,29. Although much work is needed to better understand the overall changes in immune system composition and function over the course of life, it is clear that age is an important factor to consider when assessing human immune variation.
Seasonal and circadian immune system variation
The incidence of autoimmune type 1 diabetes in children varies over the seasons, with lowest incidence in the summer months and highest incidence in the autumn and winter in the northern hemisphere30. Many patients with rheumatoid arthritis subjectively experience seasonal variation in joint symptoms, and one study performed in Japan found evidence of such seasonal variation in disease severity scores31. An analysis of blood gene expression profiles has revealed clear seasonal patterns, but the differences primarily involved genes expressed in platelets and red blood cells, the frequencies of which are known to vary seasonally32. Another study of gene expression patterns across multiple cohorts identified seasonal changes in the expression of genes thought to be unique to specific immune cell subpopulations, which suggests changes in the immune cell composition over the course of the year33. This finding needs to be confirmed by more direct measurements of immune cell frequencies over time.
Circadian variation in inflammatory manifestations, such as stiffness and pain being worst in the morning hours, is a defining symptom of several autoimmune conditions including rheumatoid arthritis34. This variation has been attributed to concomitant circadian regulation of endogenous hormones such as cortisol. In patients with rheumatoid arthritis, worsening of the symptoms has been shown to coincide with a spike in serum levels of interleukin-6 (IL-6) early in the morning35. In mice, antimicrobial responses have been shown to differ during certain times of the day; hence a pathogen encounters different immune responses in the daytime and at night36.
Inter-individual variation
It is widely accepted that human immune systems are variable between individuals, but the extent of variation is only starting to become clear. Recent advances in cytometry37 and multiplex serum protein measurements38,39 enable simultaneous analyses of the cells and proteins that constitute human immune systems. These analyses enable estimates of inter-individual variation, not only at the level of individual measurements but also at the systems-level across hundreds of individuals within a population. By considering inter-dependencies between these immune system components, we can also learn how these measurements co-vary in health and disease.
Quantifying inter-individual variation
Several cohort studies have analysed immune cell frequencies and serum protein concentrations in healthy adults in recent years11,15,16,40. Here we use data from two separate cohorts of healthy individuals recruited and sampled at the Clinical and Translational Research Unit at Stanford University Hospital, USA, and immune cell frequency measurements made using mass cytometry at the Human Immune Monitoring Center also at Stanford University11,17. Using this data, we illustrate the range of variation observed in the relative frequencies of six principal immune cell populations: B cells, monocytes, natural killer (NK) cells, CD4+ T cells, CD8+ T cells and total T cells (FIG. 2b). The range of variation among these healthy individuals is many orders of magnitude and individuals completely devoid of specific cell populations such as monocytes (CD33+ cells) and NK cells (CD3−CD56+ cells) were identified. The frequency of CD4+ T cells as a fraction of the total T cell population ranges between 22–90% and the fraction of CD8+ T cells ranges between 6–65% (FIG. 2b). The B cell fraction ranges between 4–69% of the total number of lymphocytes (FIG. 2b). The fact that seemingly healthy individuals display such a large degree of variation in specific immune system components suggests novel avenues for future studies into the mechanisms ensuring robustness and redundancy in the immune system. A complex system, such as the immune system, probably uses adaptive strategies, compensatory pathways and functional redundancy to maintain its vital functions even in ‘outlier’ individuals.
The structure of immune system variation in human populations
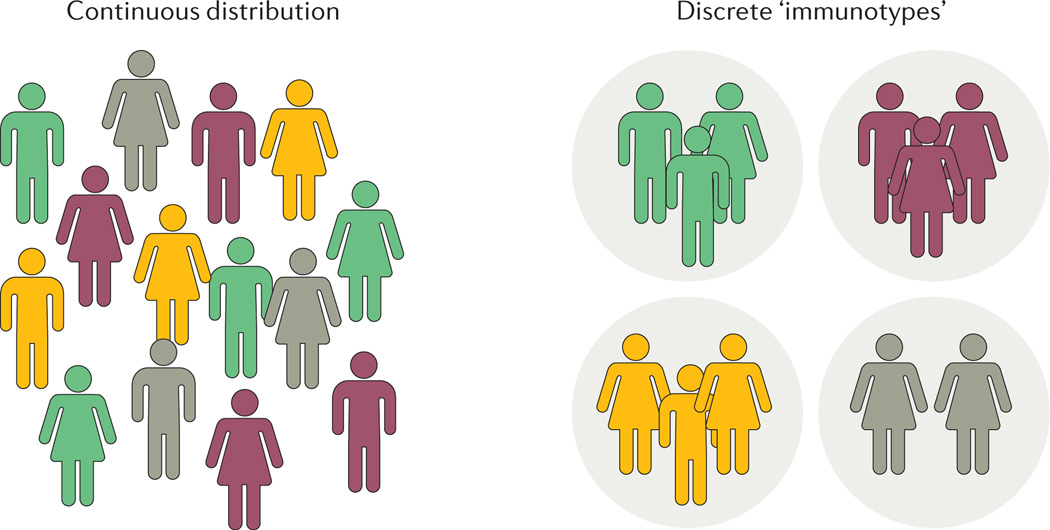
When analysing an increasing number of individuals with respect to the composition of immune cells and proteins in their immune systems, it is important not to catalogue only the range of variation for individual measurements, but also to investigate novel associations between immune system components and the structure of variation between individuals. If the composition of cells and proteins that make up an individual’s immune system is referred to as the individual’s ‘immunotype’, it is interesting to study whether such immunotypes are distributed as discrete groups or as a continuum (FIG. 3). Such a global understanding of human immune system variation could help identify individuals with outlier immunotypes and immunotypes associated with increased risk of severe infections or immune-mediated disease. Many studies have used global gene expression profiles to define variation between patients, for example patients with systemic lupus erythematosus (SLE) can be grouped into seven discrete groups of patients based on global gene expression profiles and disease severity41. SLE is a disease notorious for its heterogeneous clinical presentation, and it is unknown whether such discrete groups can be defined also in healthy individuals.
Figure 3. Distribution of immune system variation in human populations.
There are two possibilities for human immune system variation, either individuals are distributed continuously with respect to their immune system composition or in discrete groups, so called ‘immunotypes’.
Immune system variation by sex
Many immune-mediated disorders show different incidence rates between men and women — for example, 80% or more of the patients with autoimmune diseases, such as Sjogren syndrome, SLE and autoimmune thyroid disease, are women42, whereas the incidence and severity of ankylosing spondylitis is higher in men43. Differences in baseline states of immune cell frequencies, serum protein concentrations and functional properties between men and women are not well defined16. In a study of 1,005 Swedish individuals, a few serum proteins such as E-selectin, growth hormone, fatty acid-binding protein 4 and tartrate-resistant acid phosphatase type 5, differed significantly between females and men at the baseline state, but the consequences of these differences on immunity are unclear27. Gene expression analysis of whole blood samples has indicated clear differences between men and women, both for autosomal genes and for genes expressed on X and Y chromosomes44. Our current understanding of differences in baseline immune profiles between men and women is incomplete, but taking sex into account when analysing immune system variation is important, and if sex is reported in publications together with immunological data, the situation can become clearer in the future. The influence of sex on functional immune responses, such as the response to vaccination, is also unclear but is under investigation. It is often thought that women typically mount stronger immune responses than men owing to the immunomodulatory effects of oestrogen as an enhancer45, and testosterone as a suppressor, of humoral immunity46. Alternatively, such sex differences could be explained by differences in the kinetics of the immune responses, with men showing a peak in immune responses at day 1 after challenge, whereas women elicit their peak responses at day 3 post challenge as suggested by one recent analysis of gene expression responses to influenza virus vaccination13. As data from multiple studies are being reported the, often subtle, effects of age and sex on immune cell and protein profiles will become clearer and we should have sufficient statistical power to disentangle their effects on human immune system variation. Public repositories for immunological data, such as the ImmPort database47, will be useful in this respect by allowing for meta-analyses across studies and cohorts.
Heritable influences
The manifestation of infection in individual patients is known to be influenced by host genetics48,49, with severe infections occurring in childhood often representing a monogenic immunodeficiency and severe manifestation during secondary infection being a result of more complex genetic predisposition50. The contribution of heritable and non-heritable factors to the composition and function of specific immune system components is less clear. Many studies have been performed exploring possible heritable traits associated with specific immune system measurements. Typically, genome-wide association studies (GWAS) are designed to associate genetic loci with individual immune system measurements, such as specific immune cell frequencies or the concentration of a specific cytokine. A separate line of investigation focuses on associating genetic loci with the occurrence of immune-mediated diseases and provides genetic leads for further aetiological studies. In particular the ~80 autoimmune conditions affecting humans have been extensively studied, and many genetic risk variants have been found51,52. Here we focus our discussion on the heritable influences explaining the inter-individual variation of immune system components in the blood, such as frequencies of immune cell populations and serum protein concentrations. We also discuss some analyses performed to discern heritable influences on immune cell functions.
The genetics of immune cell frequencies
White blood cell (WBC) counts are key diagnostic measurements because of their sharp increase during acute infections. Several population studies have found a moderate heritability of WBCs of about 0.38 (REF. 53), and specific loci that could partially explain the variation between individuals have been identified54. Also, the total frequencies of lymphocytes, monocytes, neutrophils, eosinophils and basophils are moderately heritable from 0.14 for basophils to 0.4 for monocytes53. Some of the specific loci that regulate immune cell frequencies have also been associated with immune-mediated disorders, such as a locus on chromosome 2 containing ITGA4, which is associated with monocyte counts54 and coeliac disease55. Using a slightly different approach, two studies analysed smaller populations but measured many more concurrent immune cell population frequencies by high-dimensional flow cytometry, which revealed an additional 24 loci associated with >20 different immune cell populations40,56. Together these studies clearly show that a fraction of the total inter-individual variation in immune cell frequencies can be explained by specific genetic variants.
The genetics of serum protein concentrations
Many immune-mediated disorders are characterized by dysregulated cytokine profiles, for example SLE57,58, which has a gene expression signature dominated by interferon-inducible genes in the blood59. The key pathogenic cytokine interferon-α (IFNα) in SLE is increased in the serum of patients with SLE but also in their healthy first-degree relatives, which suggests genetic influences on the IFNα serum levels60. Furthermore, the concentration of other cytokines can be influenced by genetic variants, such as the levels of IL-18 in older individuals61. Several additional genetic associations have been made between genetic variants within cytokine genes and immune mediated diseases, but whether such genetic variants actually contribute to the variation in serum concentration of the cytokine itself is often difficult to determine62. In our own analyses of twins, we found that serum cytokines and chemokines ranged from 0 to 1 in estimated heritability, but with an average heritability slightly higher than what was found for immune cell frequencies11. This finding could be explained by the fact that cell frequencies might be regulated by more complex and polygenic influences, whereas serum proteins are the direct products of individual genes.
Heritable influences on functional immune responses
The ImmVar project is a cohort study involving individuals of African-American, East Asian and European ancestry in the Boston metropolitan area, and within this project detailed analyses were performed to investigate variability in functional responses between individuals, specifically responses by T cells and dendritic cells (DCs). Gene expression profiling of these cells revealed that 22% of the overall variation in gene expression between individuals could be attributed to heritable factors63. This is in line with previous analyses showing a minor to moderate contribution of heritable factors on the inter-individual variation of blood transcriptomes64,65. Additional reports from the ImmVar cohort revealed substantial inter-individual variation of gene expression profiles, with minor to moderate heritable influence on gene expression patterns in DCs upon pathogen sensing66 and in CD4+ T cells after activation of T cell receptor signalling in vitro67. Interestingly, several of the identified variants have been previously associated with immune-mediated diseases66,67. In a twin study, analysis of cytokine-induced signalling responses across multiple immune cell populations was shown to be highly variable between individuals11. Most of these responses, which were induced by cytokines like IL-6, IL-10, IFNα and IFNγ, showed very limited heritability. By contrast, the phosphorylation of signal transducer and activator of transcription 5 (STAT5) in both CD4+ and CD8+ T cells after stimulation with homeostatic cytokines IL-2 and IL-7 was almost completely explained by heritable factors11. Together these results show important differences in the heritable and non-heritable influences that regulate different functional properties of human immune systems.
Non-heritable influences
The immune system is a sensory system for internal and external stimuli. Similar to other sensory systems, the cells of the immune system must adapt to inputs received in order to maintain its responsiveness to relative, rather than absolute, changes in stimuli over time68,69. It is therefore conceivable that adaptive changes induced by environmental influences would be important in shaping the composition and function of an individual’s immune system. Non-heritable influences are typically interpreted as environmental influences, such as infections and vaccines, but should rather be considered a common denominator for all relevant influences that do not have germline inheritance, including de novo mutations and stochastic epigenetic changes, in addition to the influences exerted by pathogenic and symbiotic microorganisms. Such stochastic epigenetic changes are interesting and poorly understood, but they occur in immune cells with every cell division, owing to the imperfect fidelity of the replication machinery, and have the potential for real influence on immune cell phenotypes. Stochastic changes can give rise to globally different epigenetic patterns between monozygotic twins during a lifetime70. However, given that epigenetic changes can also be induced by environmental stimuli, distinguishing cause and effects for epigenetic changes observed in immune cells is very difficult. To improve such studies, better study design focusing on interventions and longitudinal profiling is needed71.
Influences of the microbiota
In mice the contribution of the microbiota, similar to the influence of other environmental factors, can be investigated in isolation owing to the controlled environments in animal facilities. The use of germ-free mice has indicated several important effects of the microbiota on the mouse immune system, even at the level of specific strains of bacteria. For example, normal development of lymphoid tissues in the gut depend on interactions with gut bacteria, and several immune cell populations show numerical and functional deficiencies in germ-free mice72. The effect of the microbiota seems to be species specific, which suggests co-evolution between specific bacterial strains and their hosts73. The controlled environment in animal facilities represents both an opportunity and a curse — specifically when trying to understand normal microbiota–immune system interactions. Recent studies have found that wild mice or pet-shop mice, which have a more natural pathogenic exposure history, exhibited immune system profiles that were more comparable to human immune systems than normal laboratory mice74. Furthermore, when mice are infected with common pathogens before vaccination it induces altered gene expression in response to the vaccine and this is comparable to human vaccine responses75. These findings illustrate the difficulty in translating findings made in germ-free and specific-pathogen-free mice housed in clean facilities to humans3.
The microbiota has an important role in shaping the human immune system. In 1989, the hygiene hypothesis was proposed by Strachan to explain epidemiological data showing an increased incidence of in immune-mediated conditions such as hay-fever, asthma and eczema coinciding with increased hygiene in the post-industrial society76. Also, autoimmune conditions such as type 1 diabetes, multiple sclerosis and Crohn disease are thought to share some of these mechanisms of immune perturbation as a consequence of improved hygiene and the ensuing reduction in pathogens encountered early in life77. Strong evidence supports a protective effect of early-life exposure to farm environments and, in particular, support a role of endotoxin in inducing tolerance to common environmental antigens78. In fact, exposures to different strains of bacteria carrying different versions of endotoxin can exert different influences on developing immune systems, which could possibly explain some of the striking differences in incidence of immune-mediated diseases observed between different populations79.
Bacterial dysbiosis and its effect on human immune systems
Apart from the effect of endotoxin, microbial dysbiosis — which is defined as an imbalance between specific species in the colonizing microbiome — has been linked to an increased risk of asthma, suggesting an immune system-perturbation during the first 100 days of life in humans80. More locally in the gut, the intestinal microbiota has been linked to initiating and maintaining inflammation as well as determining the presentation of inflammatory bowel diseases such as Crohn disease and ulcerative colitis81. This crosstalk between immune cells and microorganisms in the gut is also illustrated in patients undergoing allogeneic stem cell transplantation and suffering from intestinal graft–versus–host disease (GVHD). The inflammation induced by alloreactive T cells during GVHD seems to give rise to a dysbiosis among gut microorganisms, which can influence the duration and severity of the inflammatory response82. Although the effects of the microbiota on intestinal immune responses can seem obvious, perhaps a more surprising finding is the link between the microbiota and the humoral immune response to non-adjuvanted vaccines. For example, Toll-like receptor 5 (TLR5) — which mediates sensing of flagellin on bacteria — is necessary for optimal plasma cell activation and antibody production in response to vaccination83. Thus, inter-individual variation in vaccine responses could be influenced by differences in the gut micro biota83. Furthermore, the gut microbiota can influence the microenvironment surrounding tumours, which has implications for the responsiveness to chemotherapy84 and immune-modulatory agents85,86.
The influence of viruses on the human immune system
Humans have co-evolved with viruses for millennia, during which some viruses have integrated into our genomes whereas others have established life-long chronic infections. Broad serological profiling has revealed that at any given time, an individual carries antibodies to about 10 different viral species87. Several viruses such as the cytomegalovirus (CMV) have been extensively studied as modulators of host immune systems. CMV is thought to reactivate regularly after the primary infection and each time it reactivates it induces changes in the host immune system such that about 10% of the T cell repertoire is CMV specific88, and other cell types, such as NK cells89, also adapt their phenotypes to the presence of CMV. An analysis of monozygotic twins discordant for CMV showed that 119 out of 204 immune cell frequencies and serum proteins had a reduced twin–twin correlation compared with CMV-negative monozygotic twins, which suggests that this virus broadly influences the composition of an individual’s immune system11. In another analysis, the presence of CMV in younger adults (20–30 years of age) was associated with stronger immune responses to the flu-vaccine in healthy individuals, suggesting beneficial effects of CMV infection for immunocompetent individuals90. Humans are constantly reinfected by low-virulence viruses that can induce immune responses and probably also adaptive changes in cell frequencies and functions, which can shape an individual’s immune system and influence the risk of immune mediated disease91. Furthermore, the interactions between viruses and host immune cells have been supported by an analysis of dynamical changes in the blood virome (by sequencing cell-free DNA) of patients who had undergone organ transplantation and were treated with immunosuppressive drugs. Many viruses, for example members of the Anelloviridae family, varied with the extent of immunosuppression and clinical outcome, which illustrates a relationship between these viruses and host immune competence92. Although it is important to consider the presence of common viruses such as CMV when assessing human immune variation, an individual’s immune system is shaped by the complete viral history and this should be taken into account.
Non-microbial environmental factors
Humans live in a complex environment, and although the influences of microorganisms in shaping human immune systems are the most well-described factors, many other environmental factors can influence our immune systems. Cigarette smoke and its ~4,500 components exert broad and damaging effects both on local immune parameters in the lung and systemically93. For example, current smokers have increased total leukocyte counts, a phenomenon that is reversible upon smoking cessation94. Smokers also have reduced overall levels of serum immunoglobulins and reduced NK-cell functional activity93,95. The specificities of antibodies are also altered in smokers with a higher general abundance of autoantibodies96 and antibodies specific to post-translational modified peptides, such as citrullinated peptides, that are of clinical importance in autoimmune diseases such as rheumatoid arthritis97.
Conclusion
With the advent of systems analyses of human immunity, we can broadly assess human immune system variation in increasing numbers of individuals and consider inter-dependencies between immune system components and analyse their variation between individuals in health and disease. So far most studies have been performed in blood, but as we expand the use of systems immunology analyses we can assess the global structure of variation in human populations and this will have implications for the understanding of immunological health and prediction of disease risk. In cancer research, the success of the new immunological therapies has inspired a surge in treatments aiming to modulate immune systems for the treatment of cancers and such developments will benefit greatly from a better general understanding of human immune system variation and the mechanisms underlying this variation. The idea of personalized therapy or precision medicine stems from the realization that individual patients vary with respect to their disease mechanisms and requirements for successful treatment and by determining what these requirements are for the individual patient, better outcomes can be achieved. Here, the issue of human immune variation, both during health and disease will be essential to take into account.
We also believe that better understanding of the mechanisms by which individuals’ immune systems vary might help to develop therapies that target such mechanisms to modulate the immune response, either to alleviate an immune mediated disorder, such as chronic inflammatory disease or allergy, or to potentiate a desired immune response against vaccines, pathogens or tumours. In the more long-term perspective, understanding when and how an individual’s stable immune system state is established might help us promote the long-term immunological health for all populations through the optimization of modifiable environmental conditions. More generally, systems immunology will also help us understand the immune system as a whole, not just in the fragments that are typical of modern biology. This is likely to reveal novel interactions and lead to more effective modelling of immune function and dysfunction than is currently possible.
Acknowledgments
P.B. is supported by a starting grant from the European Research Council, the Swedish Research Council, the Swedish Society for Medical Research, and Karolinska Institutet. M.M.D. is supported by NIH grants U19 AI090019, U19 AI057229 and the Howard Hughes Medical Institute.
Glossary
- Rheumatoid arthritis
An immunological disorder that is characterized by symmetrical polyarthritis, often progressing to crippling deformation after years of synovitis. It is associated with systemic immune activation, with acute-phase reactants being present in the peripheral blood, as well as rheumatoid factor (immunoglobulins specific for IgG), which forms immune complexes that are deposited in many tissues.
- Cortisol
A steroid hormone produced by the adrenal glands and released in response to stress and has a generally suppressive function on the immune system.
- Systemic lupus erythematosus
(SLE). An autoimmune disease in which autoantibodies that are specific for DNA, RNA or proteins associated with nucleic acids form immune complexes that damage small blood vessels, especially in the kidney. Patients with SLE generally have abnormal B cell and T cell functions.
- Sjogren syndrome
A long-term autoimmune disease affecting mucous membranes and moisture-secreting glands of the eyes and mouth, resulting in decreased production of tears and saliva, but there are also systemic manifestations such as muscle and joint pain and fatigue.
- Ankylosing spondylitis
A long-term inflammatory disease, more common in men than women, affecting the joints of the spine causing vertebrae to fuse together.
- Hygiene hypothesis
A hypothesis stating that the lack of early childhood exposure to infectious and symbiotic microorganisms increases the susceptibility to allergic diseases later in life, by altering the normal development of the immune system.
- Graft–versus–host disease
(GVHD). An immune response mediated by donor T cells contained in a transplanted allograft and directed against the recipient. GVHD is not associated with solid-organ transplantation but can occur with bone marrow or haematopoietic stem cell transplants.
Footnotes
Competing interests statement
The authors declare no competing interests.
DATABASES
ImmPort: https://immport.niaid.nih.gov/
FURTHER INFORMATION
Interactive twin data visualization: http://brodinlab.com/twins.html
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Herrath, von MG, Nepom GT. Lost in translation: barriers to implementing clinical immunotherapeutics for autoimmunity. J. Exp. Med. 2005;202:1159–1162. doi: 10.1084/jem.20051224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinman RM, Mellman I. Immunotherapy: bewitched, bothered, and bewildered no more. Science. 2004;305:197–200. doi: 10.1126/science.1099688. [DOI] [PubMed] [Google Scholar]
- 3.Davis MMA. Prescription for human immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Germain RN, Schwartzberg PL. The human condition: an immunological perspective. Nat. Immunol. 2011;12:369–372. doi: 10.1038/ni0511-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayday AC, Peakman M. The habitual, diverse and surmountable obstacles to human immunology research. Nat. Immunol. 2008;9:575–580. doi: 10.1038/ni0608-575. [DOI] [PubMed] [Google Scholar]
- 6.Hagan T, Nakaya HI, Subramaniam S, Pulendran B. Systems vaccinology: enabling rational vaccine design with systems biological approaches. Vaccine. 2015;33:5294–5301. doi: 10.1016/j.vaccine.2015.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han A, et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease. Proc. Natl Acad. Sci. USA. 2013;110:13073–13078. doi: 10.1073/pnas.1311861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ráki M, et al. Tetramer visualization of gut-homing gluten-specific T cells in the peripheral blood of celiac disease patients. Proc. Natl Acad. Sci. USA. 2007;104:2831–2836. doi: 10.1073/pnas.0608610104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodin P. Powerful populations respond to viruses and vaccines. Immunity. 2015;43:1035–1037. doi: 10.1016/j.immuni.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 11. Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. This systems immunology analysis of healthy human twins revealed that non-heritable influences mainly shape human immune systems and that the influence of heritable factors is limited in most cases. This influence is cumulative over the course of life, which leads to the divergence of the immune systems of monozygotic twin with time.
- 12. Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2008;10:116–125. doi: 10.1038/ni.1688. A pioneering systems vaccinology study that revealed novel gene expression signatures, which indicated previously unappreciated pathways activated by vaccination.
- 13.Andres-Terre M, et al. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity. 2015;43:1199–1211. doi: 10.1016/j.immuni.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobolev O, et al. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat. Immunol. 2016;17:204–213. doi: 10.1038/ni.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang JS, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carr EJ, et al. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 2016;17:461–468. doi: 10.1038/ni.3371. This study revealed a strong effect of co-habitation on immune system variation, emphasizing the importance of shared environmental factors.
- 17.Shen-Orr SS, et al. Defective signaling in the JAK-STAT pathway tracks with chronic inflammation and cardiovascular risk in aging humans. Cell Syst. 2016;3:374.e4–384.e4. doi: 10.1016/j.cels.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowling DJ, Levy O. Ontogeny of early life immunity. Trends Immunol. 2014;35:299–310. doi: 10.1016/j.it.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proc. Biol. Sci. 2015;282:20143085. doi: 10.1098/rspb.2014.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbons DL, et al. Neonates harbour highly active gammadelta T cells with selective impairments in preterm infants. Eur. J. Immunol. 2009;39:1794–1806. doi: 10.1002/eji.200939222. [DOI] [PubMed] [Google Scholar]
- 21.Gibbons D, et al. Interleukin-8 (CXCL8) production is a signatory T cell effector function of human newborn infants. Nat. Med. 2014;20:1206–1210. doi: 10.1038/nm.3670. [DOI] [PubMed] [Google Scholar]
- 22.Amenyogbe N, Levy O, Kollmann TR. Systems vaccinology: a promise for the young and the poor. Phil. Trans. R. Soc. B, Biol. Sci. 2015;370:20140340. doi: 10.1098/rstb.2014.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang N, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med. 2013;5:171ra19. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goronzy JJ, Weyand CM. Aging, autoimmunity and arthritis: T-cell senescence and contraction of T-cell repertoire diversity - catalysts of autoimmunity and chronic inflammation. Arthritis Res. Ther. 2003;5:225–234. doi: 10.1186/ar974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl Acad. Sci. USA. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaud M, et al. Proinflammatory cytokines, aging, and age-related diseases. J. Am. Med. Dir. Assoc. 2013;14:877–882. doi: 10.1016/j.jamda.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Enroth S, Johansson Å, Enroth SB, Gyllensten U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat. Commun. 2014;5:4684. doi: 10.1038/ncomms5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furman D, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol. Syst. Biol. 2013;9:659–659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakaya HI, et al. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity. 2015;43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moltchanova EV, Schreier N, Lammi N, Karvonen M. Seasonal variation of diagnosis of Type 1 diabetes mellitus in children worldwide. Diabet. Med. 2009;26:673–678. doi: 10.1111/j.1464-5491.2009.02743.x. [DOI] [PubMed] [Google Scholar]
- 31.Iikuni N, et al. What’s in season for rheumatoid arthritis patients? Seasonal fluctuations in disease activity. Rheumatology. 2007;46:846–848. doi: 10.1093/rheumatology/kel414. [DOI] [PubMed] [Google Scholar]
- 32.De Jong S, et al. Seasonal changes in gene expression represent cell-type composition in whole blood. Hum. Mol. Genet. 2014;23:2721–2728. doi: 10.1093/hmg/ddt665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dopico XC, et al. Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat. Commun. 2015;6:7000. doi: 10.1038/ncomms8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Straub RH, Cutolo M. Circadian rhythms in rheumatoid arthritis: implications for pathophysiology and therapeutic management. Arthritis Rheum. 2007;56:399–408. doi: 10.1002/art.22368. [DOI] [PubMed] [Google Scholar]
- 35.Cutolo M, Straub RH. Circadian rhythms in arthritis: hormonal effects on the immune/inflammatory reaction. Autoimmun. Rev. 2008;7:223–228. doi: 10.1016/j.autrev.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 36.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LAJ. Circadian clock proteins and immunity. Immunity. 2014;40:178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christiansson L, et al. The use of multiplex platforms for absolute and relative protein quantification of clinical material. EuPA Open Proteom. 2014;3:37–47. [Google Scholar]
- 40.Roederer M, et al. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161:387–403. doi: 10.1016/j.cell.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banchereau R, et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165:551–565. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitacre CC. Sex differences in autoimmune disease. Nat. Immunol. 2001;2:777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 43.Lee W, et al. Are there gender differences in severity of ankylosing spondylitis? Results from the PSOAS cohort. Ann. Rheum. Dis. 2007;66:633–638. doi: 10.1136/ard.2006.060293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitney AR, et al. Individuality and variation in gene expression patterns in human blood. Proc. Natl Acad. Sci. USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cutolo M, et al. Sex hormones influence on the immune system: basic and clinical aspects in autoimmunity. Lupus. 2004;13:635–638. doi: 10.1191/0961203304lu1094oa. [DOI] [PubMed] [Google Scholar]
- 46.Furman D, et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc. Natl Acad. Sci. USA. 2014;111:869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharya S, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol. Res. 2014;58:234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 48.Sørensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N. Engl. J. Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 49.Casanova J-L, Abel L. The human model: a genetic dissection of immunity to infection in natural conditions. Nat. Rev. Immunol. 2004;4:55–66. doi: 10.1038/nri1264. [DOI] [PubMed] [Google Scholar]
- 50.Alcaïs A, et al. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann. NY Acad. Sci. 2010;1214:18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
- 51.Kochi Y. Genetics of autoimmune diseases: perspectives from genome-wide association studies. Int. Immunol. 2016;28:155–161. doi: 10.1093/intimm/dxw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu. Rev. Immunol. 2009;27:363–391. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pilia G, et al. Heritability of Cardiovascular and Personality Traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nalls MA, et al. Multiple loci are associated with white blood cell phenotypes. PLoS Genet. 2011;7:e1002113. doi: 10.1371/journal.pgen.1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garner CP, et al. Replication of celiac disease UK genome-wide association study results in a US population. Hum. Mol. Genet. 2009;18:4219–4225. doi: 10.1093/hmg/ddp364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orrù V, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pascual V, Farkas L, Banchereau J. Systemic lupus erythematosus: all roads lead to type I interferons. Curr. Opin. Immunol. 2006;18:676–682. doi: 10.1016/j.coi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 58.Kariuki SN, Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Translat. Res. 2010;155:109–117. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baechler EC, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl Acad. Sci. USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niewold TB, Hua J, Lehman TJA, Harley JB, Crow MK. High serum IFN-α activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matteini AM, et al. Novel gene variants predict serum levels of the cytokines IL-18 and IL-1ra in older adults. Cytokine. 2014;65:10–16. doi: 10.1016/j.cyto.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ollier WER. Cytokine genes and disease susceptibility. Cytokine. 2004;28:174–178. doi: 10.1016/j.cyto.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 63.De Jager PL, et al. ImmVar project: insights and design considerations for future studies of ‘healthy’ immune variation. Semin. Immunol. 2015;27:51–57. doi: 10.1016/j.smim.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 64.Price AL, et al. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 2011;7:e1001317. doi: 10.1371/journal.pgen.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao T, Brodin P, Davis MM, Jojic V. Drug-induced mRNA signatures are enriched for the minority of genes that are highly heritable; Pac. Symp. Biocomput; 2015. pp. 395–406. [PubMed] [Google Scholar]
- 66.Lee MN, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye CJ, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345:1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grossman Z, Paul WE. Adaptive cellular interactions in the immune system: the tunable activation threshold and the significance of subthreshold responses. Proc. Natl Acad. Sci. USA. 1992;89:10365–10369. doi: 10.1073/pnas.89.21.10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brodin P, Kärre K, Höglund P. NK cell education: not an on-off switch but a tunable rheostat. Trends Immunol. 2009;30:143–149. doi: 10.1016/j.it.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 70.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Birney E, Smith GD, Greally JM. Epigenome-wide association studies and the interpretation of disease-omics. PLoS Genet. 2016;12:e1006105. doi: 10.1371/journal.pgen.1006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beura LK, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532:512–516. doi: 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reese TA, et al. Sequential infection with common pathogens promotes human-like immune gene expression and altered vaccine response. Cell Host Microbe. 2016;19:713–719. doi: 10.1016/j.chom.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. The original paper proposing the hygiene hypothesis.
- 77.Bach J-F. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 78.Braun-Fahrländer C, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N. Engl. J. Med. 2002;347:869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 79.Vatanen T, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arrieta M-C, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 81.Dalal SR, Chang EB. The microbial basis of inflammatory bowel diseases. J. Clin. Invest. 2014;124:4190–4196. doi: 10.1172/JCI72330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jenq RR, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J. Exp. Med. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oh JZ, et al. TLR5-mediated sensing of gut microbiota Is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vétizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu GJ, et al. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sylwester AW, et al. Broadly targeted human cytomegalovirus-specific CD4 + and CD8 + T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rölle A, Brodin P. Immune adaptation to environmental influence: the case of NK cells and HCMV. Trends Immunol. 2016;37:233–243. doi: 10.1016/j.it.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 90.Furman D, et al. Cytomegalovirus infection enhances the immune response to influenza. Sci Transl Med. 2015;7:281ra43. doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Vlaminck I, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sopori M. Effects of cigarette smoke on the immune system. Nat. Rev. Immunol. 2002;2:372–377. doi: 10.1038/nri803. [DOI] [PubMed] [Google Scholar]
- 94.Higuchi T, et al. Current cigarette smoking is a reversible cause of elevated white blood cell count: cross-sectional and longitudinal studies. Prev. Med. Rep. 2016;4:417–422. doi: 10.1016/j.pmedr.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferson M, Edwards A, Lind A, Milton GW, Hersey P. Low natural killer-cell activity and immunoglobulin levels associated with smoking in human subjects. Int. J. Cancer. 1979;23:603–609. doi: 10.1002/ijc.2910230504. [DOI] [PubMed] [Google Scholar]
- 96.Mathews JD, Whittingham S, Hooper BM, Mackay IR, Stenhouse NS. Association of autoantibodies with smoking, cardiovascular morbidity, and death in the Busselton population. Lancet. 1973;2:754–758. doi: 10.1016/s0140-6736(73)91037-4. [DOI] [PubMed] [Google Scholar]
- 97.Padyukov L, et al. A gene-environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum. 2004;50:3085–3092. doi: 10.1002/art.20553. [DOI] [PubMed] [Google Scholar]
- 98.Chattopadhyay PK, Roederer M. A mine is a terrible thing to waste: high content, single cell technologies for comprehensive immune analysis. Am. J. Transplant. 2015;15:1155–1161. doi: 10.1111/ajt.13193. [DOI] [PubMed] [Google Scholar]
- 99.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 100.Frei AP, et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat. Methods. 2016;13:269–275. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bodenmiller B, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stubbington MJT, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pernemalm M, Lehtiö J. Mass spectrometry-based plasma proteomics: state of the art and future outlook. Expert. Rev. Proteom. 2014;11:431–448. doi: 10.1586/14789450.2014.901157. [DOI] [PubMed] [Google Scholar]