ABSTRACT
This study compared salivary bacterial profiles in two groups having a 10-fold difference in levels of caries experience, as it was hypothesized that the composition of the salivary microbiota might associate with the levels of caries experience.
Bacterial profiles in stimulated saliva samples from 85 individuals with low levels of caries experience (healthy group) and 79 individuals with high levels of caries experience (caries group) were analyzed by means of the Human Oral Microbiome Identification Next Generation Sequencing (HOMINGS) technique. Subsequently, saliva samples from caries-free individuals in the healthy group (n = 57) and the caries group (n = 31) were compared.
A significantly higher α-diversity (p < 0.0001) and a twofold higher relative abundance of Neisseria, Haemophilus, and Fusobacterium were recorded in saliva samples from the healthy group compared with the caries group. Differences observed were more pronounced when limiting the analyses to caries-free individuals in each group.
Data from this cross-sectional analysis suggest that low levels of caries experience might associate with a characteristic salivary bacterial composition different from that in individuals with high caries experience. Consequently, longitudinal studies are required to determine if the composition of the salivary microbiota might be a predictive factor of caries risk at the individual level.
KEYWORDS: Caries susceptibility, microbiology, HOMINGS, oral diagnosis, clinical studies/trials
Introduction
Although dental caries has affected the human race for millennia [1], most comprehensive data on ancient frequencies are low, with only 4–6% of the teeth being affected [2]. Experiences half a century ago far exceeded these frequencies, with 80–90% of children and adolescents having treatment requiring caries [3]. Since up to 90% of the population can develop caries, caries seems to be a common condition among humans. While ancient frequencies show that poor oral hygiene alone cannot account for caries, nowadays a huge intake of sucrose combined with poor oral hygiene can result in catastrophic caries development requiring extensive dental restoration. Meticulous oral hygiene, refraining from sucrose, and daily exposure to fluoride have reduced caries effectively in most of the developed world during the past decades [4]. Today, the severe and rampant caries of times past is mainly seen in selected patient groups, and it is evident that subnormal saliva flow rates accelerate the progression of dental caries considerably [5]. However, an interesting phenomenon is the very few percent of a population who do not develop caries, most likely amounting to some 10% given the past frequencies on dental caries [3].
Whole saliva, being a mixture of secretions from all salivary glands combined with a broad spectrum of oral bacteria, is a biological fluid that can be easily collected in sufficient quantities for state-of-the-art sophisticated microbial analyses [6,7]. Recently, it has been reported that the composition of the microbiota from such whole saliva is influenced by smoking, life-style, and the salivary secretion rate [8–11]. Likewise, differences in salivary bacterial profiles have been shown in patients with oral diseases, such as caries and periodontitis, when compared with orally healthy individuals [12–14]. However, at present, it is not known if different levels of present and past caries experience associates with characteristic salivary bacterial profiles.
Thus, the purpose of the present study was to compare microbiotas in saliva samples from two groups that previously have been physicochemically characterized with a 10-fold difference in caries experience [15]. The null hypothesis was that salivary bacterial profiles would not associate with highly significant different levels of caries experience.
Materials and methods
Oral examination
Oral examinations were performed in 2008 and 2009 as part of the Danish Health Examination Survey (DANHES), which has been presented in detail elsewhere [16]. In brief, the oral examination was performed in fully equipped mobile dental settings or local municipal clinics by three dental hygienists who were calibrated on two occasions with inter- and intra-examiner kappa coefficients between 0.85 and 0.93 [16]. Caries was registered according to Moller and Poulsen [17] as manifest caries on tooth and surface level expressed as DMFT (decayed, missed, filled tooth) and DMFS (decayed, missed, filled surfaces) [16].
Study population
The study population, which is a subpopulation of the DANHES, has been described previously [16]. In brief, from a total of 4,402 individuals enrolled in the DANHES cohort, three groups of age-, sex-, and geographically matched individuals with normal salivary flow rate but different oral health status were identified for comparative analysis of salivary physicochemical characteristics [15]. Subsequently, two of these groups were enrolled in this study: a group with low levels of caries experience (healthy group: n = 85) and a group with high levels of caries experience (caries group: n = 85). A total of six samples failed the initial quality control steps of Human Oral Microbiome Identification Next Generation Sequencing (HOMINGS), which meant that a total of 164 saliva samples could be analyzed by HOMINGS: 85 samples from the healthy group and 79 from the caries group. Finally, subsequent analysis was performed on the active caries-free part of both groups (healthy: n = 57; caries: n = 31).
Ethical approval
The study was carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans, and was approved by the Ethical Committees for the Region of Copenhagen, Denmark (H-C-2007–0118). All participants signed informed consent. The establishment of a biobank was approved by the Danish Data Protection Authority (2007–41-1567), and the manuscript complies with the STROBE guidelines.
Collection of paraffin chewing stimulated whole saliva samples
Collection of stimulated saliva samples was performed according to a previously described protocol [15,16]. In brief, stimulated saliva sampling was started by flushing with tap water followed by chewing on 1 g of sterile paraffin wax for 1 min, after which the initially collected saliva was discarded and not used for analyses. Subsequently, the participant was instructed to chew and spit continuously for 3 min into a sterile plastic cup, after which time the collected saliva from the cup was stored at –80°C. The aliquot used for the present study had not been previously thawed in order to ensure minimal neuraminidase and protease activities.
HOMINGS
DNA isolation was done as previously described [11], and bacterial identification was performed using HOMINGS. HOMINGS is a next-generation sequencing (Illumina)–based molecular technique, which in combination with a customized BLAST program (ProbeSeq for HOMINGS, Forsyth Institute, Cambridge, MA, USA) enables simultaneous identification of around 600 oral taxa [18]. The Probeseq database contains sequences of custom-made 16S rDNA probes (17–40 bases long) based on the HOMD database [19]. Specifically, bacterial identification is based on 598 species-specific probes and 94 genus-specific probes (probes targeting two or more closely related taxa within the same genus) present in the Probeseq database. A complete list of probes present in the Probeseq database is presented in the Supplementary Material (S1). The laboratory procedures of HOMINGS was performed according to a protocol modified from Caporaso et al. [20], as 10–50 ng of DNA was initially polymerase chain reaction (PCR)-amplified using primers targeting the V3–V4 region (341 F and 806 R) of the 16S rRNA gene. Subsequently, AMPure beads were employed for purification of PCR amplicons, and 100 ng of each library was pooled and gel purified. Finally, qPCR was used for quantification, followed by MiSeq (Illumina, San Diego, CA, USA) sequencing of 12 pM of the library mixture and 20% Phix. In this study, an average of 91,531 (range 38,086–146,049) sequences (441 bp long) was obtained after bad reads sequences had been removed from analyses, out of which 78% (range 46–90%) and 36.0% (range 16–57%) were identified at species level and genus level, respectively. A total of 618 probe-targets were observed, with a mean number of 240 (range 137–341) per sample. A complete list of all identified probe-targets is presented in the Supplementary Material (S2).
Statistical analyses
Epidemiological data, clinical data and α-diversity were compared using an unpaired t-test and chi-square test. For these analyses, a p-value of <0.05 was considered significant.
Relative abundance of species identified were compared between groups using the Mann–Whitney U-test, and Benjamini–Hochberg correction was used to control for multiple comparisons [21]. For this analysis, an adjusted p-value of <0.0001 was considered statistically significant. Correspondence analysis was used for data reduction. GraphPad Prism v5 (San Diego, CA, USA) and MeV 4_8_1 were used as statistical software [22].
Results
Epidemiological data and clinical characteristics
Epidemiological data and clinical characteristics of the healthy group with low levels of caries experience (n = 85) and the caries group (n = 79) with high levels of caries experience are presented in Table 1. As can be seen, these groups expressed comparable compositions in terms of age and sex (p = 0.77 and p = 0.19, respectively) but major differences in number of surfaces with caries present (DS; p < 0.0001) and number of filled surfaces (FS; p < 0.0001).
Table 1.
Epidemiological and clinical data.
| Healthy (n = 85) | Caries (n = 79) | p-Value | |
|---|---|---|---|
| Age (years), mean (range) | 47 (18–75) | 48 (22–76) | 0.77 |
| Sex, % M/F | 41/59 | 30/70 | 0.19 |
| DS, M ° SD | 0.7 ± 1.3 (0–6) | 2.7 ± 4.1 (0–18) | <0.0001 |
| FS, M ° SD | 6.4 ± 6.4 (0–23) | 67.0 ± 24.4 (22–110) | <0.0001 |
| DFS, M ° SD | 5.5 ± 5.2 (0–19) | 57.1 ± 21.2 (20–97) | <0.0001 |
Statistically significant values shown in bold.
DS, decayed surfaces; SD, standard deviation; FS, filled surfaces.
Likewise, epidemiological data and clinical characteristics of caries-free individuals in each group are presented in Table 2, which demonstrates insignificantly different compositions in age (p = 0.14) and sex (p = 0.10) but a major difference in the number of filled surfaces (FS; p < 0.0001).
Table 2.
Epidemiological and clinical data on individuals without decayed surfaces.
| Healthy (n = 57) | Caries (n = 31) | p-Value | |
|---|---|---|---|
| Age (years), mean (range) | 45 (18–75) | 50 (22–76) | 0.14 |
| Sex, % M/F | 40/60 | 22/78 | 0.10 |
| DS, M ° SD | 0.0 ± 0.0 (0–0) | 0.0 ± 0.0 (0–0) | 1.0 |
| FS, M ° SD | 6.2 ± 6.3 (0–23) | 73.2 ± 22.5 (33–110) | <0.0001 |
| DFS, M ° SD | 4.8 ± 4.8 (0–18) | 61.2 ± 21.2 (24–97) | <0.0001 |
Statistically significant values shown in bold.
Salivary bacterial profiles differ between individuals with different levels of caries and caries experience
The top 10 most predominant genera are displayed in Figure 1(a). In both groups, the five most predominant genera were Streptococcus, Prevotella, Rothia, Veillonella, and Granulicatella, accounting for approximately 50% of all bacterial DNA present. Saliva samples from the healthy group were associated with a twofold higher relative abundance of Neisseria, Haemophilus, and Fusobacterium, whereas a slightly higher relative abundance of Streptococcus was noted in samples from the caries group.
Figure 1.
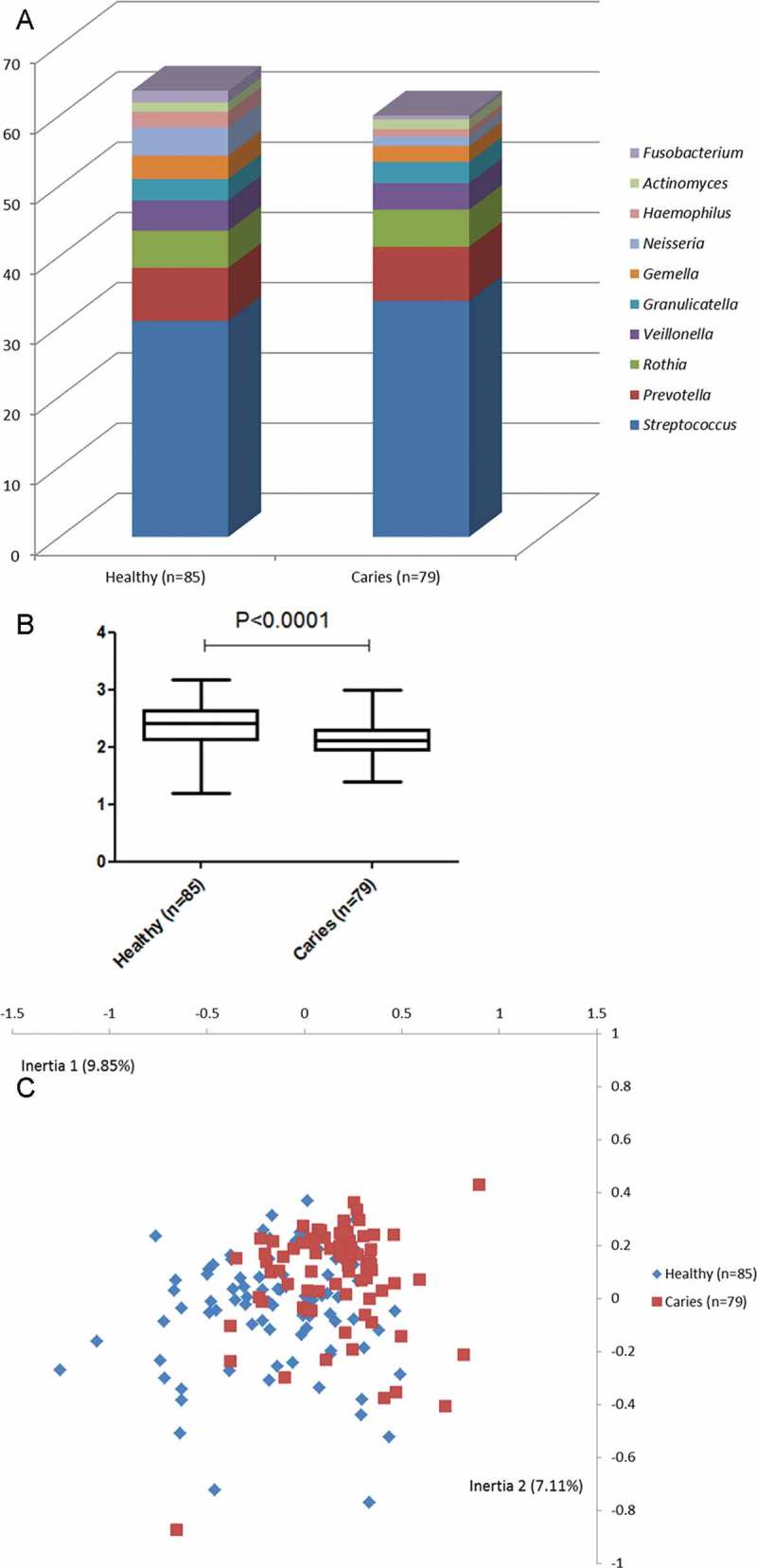
Salivary bacterial profiles in individuals with different levels of caries and caries experience. (a) Relative abundance of the 10 most predominant bacterial genera identified in saliva samples in each group. (b) Shannon index expressed as mean and range in each group. (c) Correspondence analysis visualized two-dimensionally with axes expressed as the two most crucial inertia values (cumulative inertia: 16.96%). Blue: healthy group; red: caries group.
In addition, when comparing microbial diversity by means of the Shannon diversity index, samples from the healthy group (M ± SD = 2.36 ± 0.35) displayed a significantly higher α-diversity than samples from the caries group did (M ± SD = 2.12 ± 0.32; Figure 1(b); p < 0.0001). Furthermore, when applying correspondence analysis (CoA) for data reduction, a tendency for separation between groups was evident (Figure 1(c)).
Finally, comparisons between groups at species level revealed that 26 bacterial species were present in significantly different relative abundance in the two groups (12 health associated and 14 caries associated; adjusted p-value <0.0001). In general, bacterial species found to associate with either group constituted <0.01% of salivary microbiota. However, traditional caries-associated bacteria such as Streptococcus mutans and Lactobacillus salivarius were identified with higher relative abundance in samples from the caries group. A complete list of bacterial species identified with significantly different relative abundance in the two groups is presented in Table 3.
Table 3.
List of species identified with significantly different relative abundance in samples from individuals with different levels of caries and caries experience.
| Relative abundance (%) |
||||
|---|---|---|---|---|
| Species | Healthy (n = 85) | Caries (n = 79) | Raw p-value | Adjusted p-value |
| Bifidobacterium dentium | 0.00185 | 0.00704 | 8.65 × 10–11 | 2.67*10–8 |
| Fusobacterium periodonticum | 1.65575 | 0.52774 | 2.02 × 10–10 | 4.16*10–8 |
| Parascardovia denticolens | 0.00013 | 0.00388 | 6.19 × 10–10 | 9.57*10–8 |
| Lactobacillus salivarius | 0.00001 | 0.09681 | 2.26 × 10–8 | 2.79 × 10–6 |
| Stomatobaculum sp ot097 | 0.39023 | 0.14618 | 3.10 × 10–8 | 3.19 × 10–6 |
| Mitsuokella sp ot131 | 0.00053 | 0.00297 | 9.46 × 10–8 | 6.50 × 10–6 |
| Porphyromonas sp ot279 | 1.15374 | 0.35327 | 1.75 × 10–7 | 1.08 × 10–5 |
| Streptococcus mutans | 0.01418 | 0.04534 | 2.07 × 10–7 | 1.16 × 10–5 |
| Bergeyella sp ot322 | 0.11329 | 0.05635 | 2.42 × 10–7 | 1.22 × 10–5 |
| Cryptobacterium curtum | 0.00442 | 0.01567 | 2.68 × 10–7 | 1.22 × 10–5 |
| Actinomyces sp ot181 | 0.04063 | 0.10343 | 6.02 × 10–7 | 2.32 × 10–5 |
| Haemophilus parainfluenzae | 2.26257 | 1.05409 | 7.25 × 10–7 | 2.64 × 10–5 |
| Veillonella rogosae | 1.25629 | 0.63628 | 8.29 × 10–7 | 2.85 × 10–5 |
| Veillonella sp ot917 | 0.06596 | 0.15617 | 1.46 × 10–6 | 4.75 × 10–5 |
| Neisseria flavescens | 2.16957 | 0.29464 | 1.79 × 10–6 | 5.43 × 10–5 |
| Lactobacillus vaginalis | 0.00001 | 0.01389 | 1.85 × 10–6 | 5.43 × 10–5 |
| Atopobium parvulum | 0.02330 | 0.04696 | 2.22 × 10–6 | 6.01 × 10–5 |
| Leptotrichia sp ot223 | 0.01805 | 0.00756 | 9.10 × 10–6 | 2.25 × 10–4 |
| Megasphaera sp ot123 | 0.00096 | 0.00655 | 9.76 × 10–6 | 2.32 × 10–4 |
| Lachnoanaerobaculum sp ot083 | 0.01101 | 0.00043 | 1.24 × 10–5 | 2.74 × 10–4 |
| Tannerella sp ot808 | 0.03441 | 0.00419 | 1.31 × 10–5 | 2.80 × 10–4 |
| Streptococcus sp ot068 | 0.00368 | 0.00627 | 1.65 × 10–5 | 3.40 × 10–4 |
| Prevotella shahii | 0.03324 | 0.00718 | 1.94 × 10–5 | 3.88 × 10–4 |
| Scardovia wiggsiae | 0.01458 | 0.03128 | 2.28 × 10–5 | 4.40 × 10–4 |
| Prevotella sp ot396 | 0.00892 | 0.00172 | 4.44 × 10–5 | 8.07 × 10–4 |
| Veillonella denticariosi | 0.09186 | 0.11177 | 5.17 × 10–5 | 9.13 × 10–4 |
Compositional differences in salivary microbiota in caries-free individuals with different levels of caries experience
In general, salivary microbial differences observed between the healthy group and the caries group were more pronounced when limiting the analysis to caries-free individuals in both groups. This was illustrated by a higher relative abundance of Neisseria (threefold), Fusobacterium (threefold), Gamella (twofold), and Haemophilus (twofold) in the healthy group (Figure 2(a)). In line with this, a more marked difference in α-diversity (2.39 ± 0.32 vs. 2.00 ± 0.29; p < 0.0001; Figure 2(b)) and almost complete separation using CoA between samples from caries-free individuals with different levels of caries experience were evident (Figure 2(c)).
Figure 2.
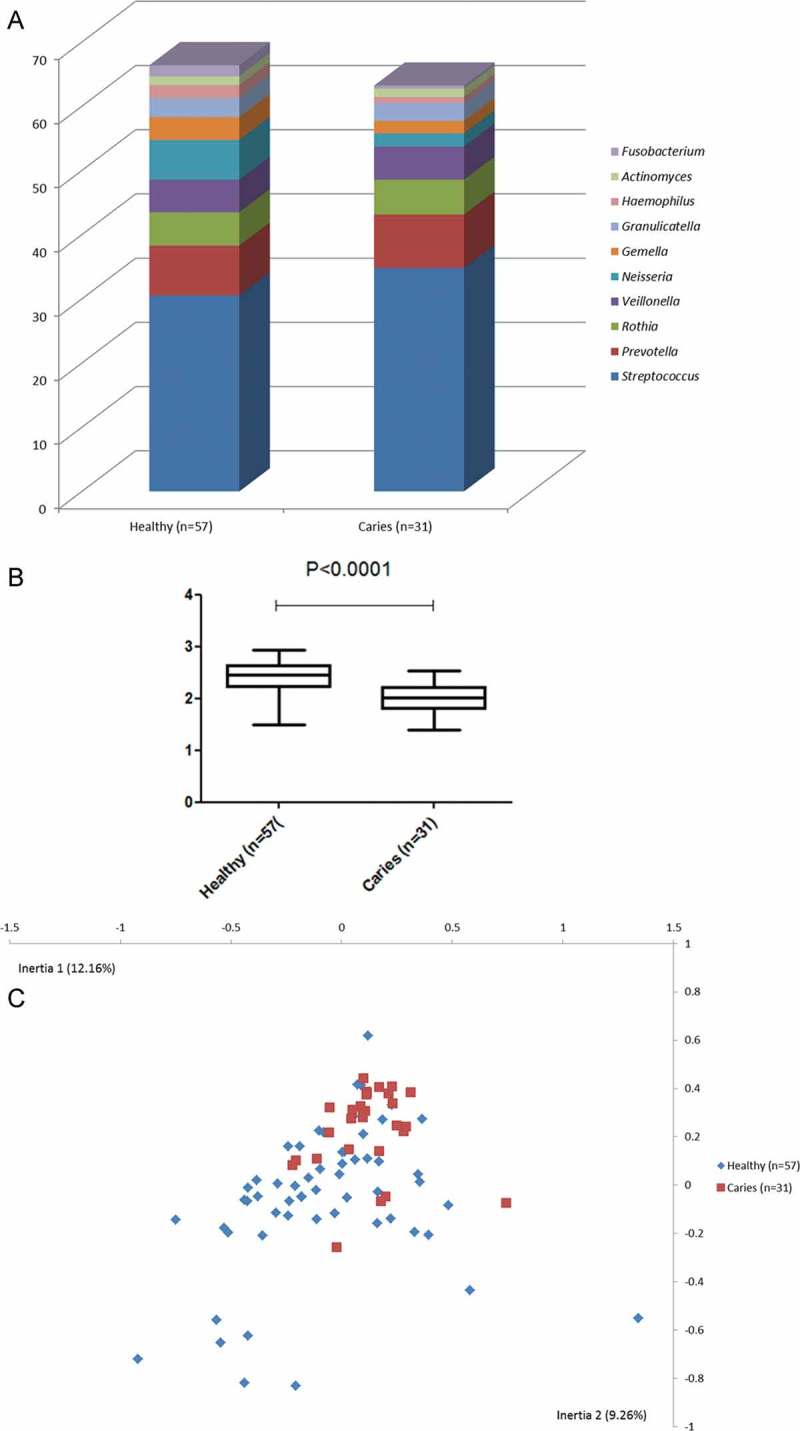
Salivary bacterial profiles in caries-free individuals with different levels of caries experience. (a) Relative abundance of the 10 most predominant bacterial genera identified in saliva samples in caries-free individuals in each group. (b) Shannon index expressed as mean and range in caries-free individuals in each group. (c) Correspondence analysis visualized two-dimensionally with axes expressed as the with most crucial inertia values (cumulative inertia: 21.42%). Blue: healthy group; red: caries group.
In contrast, only 11 bacterial species were identified with significantly different relative abundance (seven health associated and four caries-associated; adjusted p-value <0.0001). A complete list of bacterial species identified with significantly different relative abundance is presented in Table 4.
Table 4.
List of species identified with significantly different relative abundance in samples from caries-free individuals with different levels of caries experience.
| Relative abundance (%) |
||||
|---|---|---|---|---|
| Species | Healthy (n = 57) | Caries (n = 31) | Raw p-value | Adjusted p-value |
| Bifidobacterium dentium | 0.00351 | 0.00561 | 1.46 × 10–7 | 2.77 × 10–5 |
| Fusobacterium periodonticum | 1.26922 | 0.45017 | 1.49 × 10–7 | 2.77 × 10–5 |
| Porphyromonas sp ot279 | 0.86330 | 0.15509 | 2.27 × 10–7 | 2.77 × 10–5 |
| Bergeyella sp ot322 | 0.08561 | 0.04039 | 2.80 × 10–7 | 2.77 × 10–5 |
| Lactobacillus salivarius | 0.00347 | 0.00983 | 5.50 × 10–7 | 4.66 × 10–5 |
| Streptococcus sp ot068 | 0.00469 | 0.00738 | 7.79 × 10–6 | 5.13 × 10–4 |
| Parascardovia denticolens | 0.00083 | 0.00213 | 1.36 × 10–5 | 6.71 × 10–4 |
| Alloprevotella sp ot914 | 0.00941 | 0.00098 | 1.54 × 10–5 | 7.02 × 10–4 |
| Stomatobaculum sp ot097 | 0.32110 | 0.13459 | 1.75 × 10–5 | 7.34 × 10–4 |
| Leptotrichia sp ot223 | 0.01186 | 0.00370 | 1.94 × 10–5 | 7.34 × 10–4 |
| Lachnoanaerobaculum umeaense | 0.12965 | 0.07754 | 1.98 × 10–5 | 7.34 × 10–4 |
Discussion
The purpose of the present study was to compare salivary bacterial profiles in two groups with a major difference in caries experience. The main finding was that a significantly higher bacterial diversity (p < 0.0001) and considerably higher salivary proportions of the genera Neisseria, Fusobacterium, and Haemophilus was associated with samples from individuals with low levels of caries experience when compared with age-, sex-, and geographically matched individuals having a 10 times higher caries experience.
Interestingly, these results are in part comparable to the data on salivary physicochemical characteristics of the same individuals. These data showed that low levels of caries experience were associated with a more favorable physicochemical saliva composition, especially in terms of its remineralizing potential, due to higher calcium and phosphorous levels [15]. Thus, the findings of the present study support the concept that dental caries is a multifactorial disease where salivary physicochemical properties and the composition of the oral and salivary microbiota, among other factors, play a role in the presence or absence of disease [23]. Accordingly, samples from healthy individuals with low caries experience were associated with higher abundance of the genera of Neisseria, Haemophilus, and Fusobacterium, of which most species in these genera only slightly ferment sugar, whereas saliva from individuals with high caries experience was associated with higher salivary abundance of Streptococcus, which includes species that heavily degrades sugars and forms extracellular and impermeable polysaccharides as well as utilizes lactate (Figure 1(a)) [24].
One limitation of the present study was that some individuals in both groups had decayed tooth surfaces (Table 1), which might have influenced data on the salivary bacterial composition. In fact, it has previously been reported that salivary bacterial composition in patients with dental caries differentiates from healthy controls [12], and that higher salivary abundance of specific oral bacteria such as S. mutans and L. salivarius can be identified in saliva samples from patients with dental caries [13]. It is therefore noteworthy that the majority of bacterial species, which in this study was identified with significantly higher relative abundance in samples from the caries group, constituted <0.01% of the salivary microbiota identified (Table 3). Accordingly, these bacteria were most likely shed from caries lesions and then dispersed into whole saliva, which emphasizes the potential of using salivary presence of specific oral bacteria such as S. mutans as a biomarker of dental caries.
To exclude the impact of decayed tooth surfaces present, an additional analysis was performed limited to samples from caries-free individuals in each group (Table 2). Interestingly, when comparing the salivary microbiota in caries-free individuals with different levels of caries experience, only a limited number of bacterial species was identified with different relative abundances in the two groups (Table 4). On the other hand, even more pronounced differences in bacterial diversity (Figure 2(b); p < 0.0001) and microbial community profiling (CoA analysis; Figure 2(c)) were evident when limiting the analysis to caries-free individuals.
It is therefore speculated that pronounced salivary bacterial diversity, in combination with relatively high salivary abundance of the genera Neisseria, Fusobacterium, and Haemophilus, and favorable salivary psychochemical properties may potentially influence individual caries risk among healthy individuals with normal saliva flow rates. Along with these measures, diet and lifestyle will always be key parameters in any assessment of caries risk.
Obviously, the cross-sectional design of this investigation hampers the possibility of addressing solid causal aspects of the observed associations. Thus, future longitudinal studies are warranted to learn if the composition of the salivary microbiota might be a predictive factor of caries risk at the individual level.
Supplementary Material
Acknowledgements
The authors wish to thank professor Morten Grønbæk, National Institute of Public Health, University of Southern Denmark for access to the DANHES database. This study was supported by external financial support from the Danish Dental Association, the Danish Foundation of Mutual Efforts in Dental Care, Trygfonden, and the Simon Spies Foundation.
Biographies
Dr. Palle Holmstrup, DDS, PhD and dr. odont from the University of Copenhagen. Currently holds a position as Professor in Periodontology at Department of Odontology, University of Copenhagen.
Dr. Nils-Erik Fiehn, PhD and dr. odont from the University of Copenhagen. Currently holds a position as Associate Professor at the University of Copenhagen.
Dr. Nikolai Kirkby, MD and PhD from the University of Copenhagen. Currently employed at Department of Medical Microbiology, Copenhagen University Hospital, Copenhagen.
Ms. Alexis Kokaras, MSc. Lab Technician at HOMINGS core facility, Forsyth Institute, Cambridge, MA, USA.
Dr. Bruce J. Paster, PhD, Head of Department of Microbiology, Forsyth Institute. Currently holds a position as Pat Harvard School of Dental Medicine, Department of Oral Medicine, Infection and Immunity, Boston, MA, USA.
Dr. Allan Bardow, DDS and PhD from the University of Copenhagen. Currently holds a position as Associate Professor in Oral Biology. Primary research focuses on salivary biology in relation to dental caries.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary data
Supplementary data for this article can be accessed here.
References
- Fejerskov O, Guldager Bilde P, Bizzarro M. Dental caries in Rome, 50-100 AD. Caries Res. 2012;46:467–8. doi: 10.1159/000339664. [DOI] [PubMed] [Google Scholar]
- Manzi G, Salvadei L, Vienna A. Discontinuity of life conditions at the transition from the Roman Imperial age to the early middle ages: example from central Italy evaluated by pathological dento-alveolar lesions. Am J Hum Biol. 1999;11:327–341. doi: 10.1002/(SICI)1520-6300(1999)11:3<327::AID-AJHB5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Birkeland JM, Haugejorden O, von der Fehr FR. Some factors associated with the caries decline among Norwegian children and adolescents: age-specific and cohort analyses. Caries Res. 2000;34:109–116. doi: 10.1159/000016577. [DOI] [PubMed] [Google Scholar]
- Petersen PE. The world oral health report 2003: continuous improvement of oral health in the 21st century - the approach of the WHO global oral health programme. Community Dent Oral Epidemiol. 2003;31:3–24. doi: 10.1046/j..2003.com122.x. [DOI] [PubMed] [Google Scholar]
- Bardow A, Ten Cate JM, Nauntofte B. Effect of unstimulated saliva flow rate on experimental root caries. Caries Res. 2003;37:232–236. doi: 10.1159/000070450. [DOI] [PubMed] [Google Scholar]
- Giannobile WV, Beikler T, Kinney JS. Saliva as a diagnostic tool for periodontal disease: current state and future directions. Periodontol 2000. 2009;50:52–64. doi: 10.1111/j.1600-0757.2008.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannobile WV, McDevitt JT, Niedbala RS. Translational and clinical applications of salivary diagnostics. Adv Dent Res. 2011;23:375–380. doi: 10.1177/0022034511420434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almståhl A, Wikström M. Oral microflora in subjects with reduced salivary secretion. J Dent Res. 1999;78:1410–1416. doi: 10.1177/00220345990780080601. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Li J, Quinque D. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasidze I, Li J, Schroeder R. High diversity of the saliva microbiome in Batwa Pygmies. PLoSOne. 2011;6:e23352. doi: 10.1371/journal.pone.0023352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belstrom D, Holmstrup P, Nielsen CH. Bacterial profiles of saliva in relation to diet, lifestyle factors, and socioeconomic status. J Oral Microbiol. 2014;6:23609. doi: 10.3402/jom.v6.23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belstrom D, Fiehn NE, Nielsen CH. Altered bacterial profiles in saliva from adults with caries lesions: a case-cohort study. Caries Res. 2014;48:368–375. doi: 10.1159/000357502. [DOI] [PubMed] [Google Scholar]
- Belstrøm D, Paster BJ, Fiehn NE. Salivary bacterial fingerprints of established oral disease revealed by the Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS) technique. J Oral Microbiol. 2016 doi: 10.3402/jom.v8.30170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belstrøm D, Fiehn NE, Nielsen CH. Differences in bacterial saliva profile between periodontitis patients and a control cohort. J Clin Periodontol. 2014;41:104–112. doi: 10.1111/jcpe.12190. [DOI] [PubMed] [Google Scholar]
- Bardow A, Lykkeaa J, Qvist V. Saliva composition in three selected groups with normal stimulated salivary flow rates, but yet major differences in caries experience and dental erosion. Acta Odontol Scand. 2014;72:466–473. doi: 10.3109/00016357.2013.860621. [DOI] [PubMed] [Google Scholar]
- Kongstad J, Ekstrand K, Qvist V. Findings from the oral health study of the danish health examination survey 2007-2008. Acta Odontol Scand. 2013;71:1560–1569. doi: 10.3109/00016357.2013.776701. [DOI] [PubMed] [Google Scholar]
- Moller IJ, Poulsen S. A standardized system for diagnosing, recording and analyzing dental caries data. Scand J Dent Res. 1973;81:1–11. doi: 10.1111/j.1600-0722.1973.tb01488.x. [DOI] [PubMed] [Google Scholar]
- Gomes BP, Berber VB, Kokaras AS. Microbiomes of endodontic-periodontal lesions before and after chemomechanical preparation. J Endod. 2015;41:1975–1984. doi: 10.1016/j.joen.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Selwitz RH, Ismail A, Pitts NB. Dental caries. Lancet. 2007;369:51–59. doi: 10.1016/S0140-6736(07)60031-2. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


