ABSTRACT
An association between oral bacteria and atherosclerosis has been postulated. A limited number of studies have used 16S RNA gene sequencing-based metagenomics approaches to identify bacteria at the species level from atherosclerotic plaques in arterial walls. The objective of this study was to establish detailed oral microbiome profiles, at both genus and species level, of clinically healthy coronary and femoral artery tissues from patients with atherosclerosis. Tissue specimens were taken from clinically non-atherosclerotic areas of coronary or femoral arteries used for attachment of bypass grafts in 42 patients with atherosclerotic cardiovascular disease. Bacterial DNA was sequenced using the MiSeq platform, and sequence reads were screened in silico for nearly 600 oral species using the HOMINGS ProbeSeq species identification program. The number of sequence reads matched to species or genera were used for statistical analyses. A total of 230 and 118 species were detected in coronary and femoral arteries, respectively. Unidentified species detected by genus-specific probes consisted of 45 and 30 genera in coronary and in femoral artery tissues, respectively. Overall, 245 species belonging to 95 genera were detected in coronary and femoral arteries combined. The most abundant species were Porphyromonas gingivalis, Enterococcus faecalis, and Finegoldia magna based on species probes. Porphyromonas, Escherichia, Staphylococcus, Pseudomonas, and Streptococcus genera represented 88.5% mean relative abundance based on combined species and genus probe detections. Porphyromonas was significantly more abundant than Escherichia (i.e. 46.8% vs. 19.3%; p = 0.0005). This study provides insight into the presence and types of oral microbiome bacterial species found in clinically non-atherosclerotic arteries.
KEYWORDS: Oral microbiome, metagenomics, HOMINGS, atherosclerosis, P. gingivalis
An association between oral bacteria and atherosclerosis has been postulated [1–4]. Epidemiological studies have shown an association between periodontal pathogens, namely Aggregatibacter actinomycetemcomitans (previously Actinobacillus actinomycetemcomitans) and Porphyromonas gingivalis, and coronary artery disease and stroke. Proposed mechanisms include: (1) a distant site colonization by oral microbiota, resulting in heightened immune reactions from various origins (e.g. increase in proinflammatory cytokines), and (2) generation of cross-reactive antibodies against periodontal pathogens [5–8]. Additionally, although many studies have shown the relationship between various gingival tissue manipulations and bacteremia of oral species [9], the precise mechanisms by which oral periodontal pathogens colonize and proliferate in human artery walls has not yet been elucidated [5–8,10,11].
Metagenomics studies have largely characterized the nature of the oral microbiome in dental plaque associated with periodontal disease [12,13], so that comprehensive correspondence with bacterial profiles of arteries from atherosclerotic patients may be established in downstream investigations.
Scientific evidence supporting a possible role of oral bacterial species in atherosclerosis relies to a large extent on the detection and identification of bacterial DNA in human arterial wall tissues or atherosclerotic plaque in cross-sectional study designs [14]. Most importantly, Kozarov et al. demonstrated that viable A. actinomycetemcomitans and P. gingivalis could be isolated from atherosclerotic plaque [10].
Several metagenomics studies, using different technologies, have determined bacterial profiles in atherosclerotic plaque. These studies, however, have provided limited information at the species level or have shown little overlap at the species level with regard to the listings of taxa described [15–19]. Only one of these studies describes the presence of A. actinomycetemcomitans in atherosclerotic plaque [18], but none of them reported finding P. gingivalis. Another shortfall is the lack of confirmatory studies to determine the absolute and relative abundance for some of the more predominant species that had previously been detected by other techniques (e.g. culture, microscopy, targeted polymerase chain reaction) in arterial tissues from atherosclerotic patients and controls [14].
Confirmatory studies are thus needed to determine the number and abundance of species present in atherosclerotic plaque and clinically non-atherosclerotic artery walls of atherosclerotic patients, in particular those species associated with periodontal disease. The purpose of this study was to establish detailed oral microbiome profiles for genera and species found within clinically non-atherosclerotic coronary and femoral artery tissues using Human Oral Microbe Identification applying Next Generation Sequencing (HOMINGS) [20].
Methods
Patient population
Full-thickness tissue specimens were taken from clinically non-atherosclerotic healthy areas of two different arteries used for attachment of bypass grafts. The designation of ‘clinically non-atherosclerotic healthy’ areas is based on the clinician surgeon’s judgment made during macroscopic selection of an area to attach the graft, such area having no obvious atherosclerotic pathology. Coronary artery tissue from patients with atherosclerotic cardiovascular disease (CVD; n = 32) and femoral artery tissue from patients with atherosclerotic vascular disease-related blockages distant to the site of tissue acquisition vascular diseases (n = 10) were collected between 2002 and 2003 at the authors’ hospital through a previous Institutional Review Board–approved study. A waiver of informed consent was obtained in 2014 for this non-interventional retrospective study on de-identified specimens, involving minimal risk and appropriate PHI protection. A total of 23 coronary and eight femoral artery specimens were collected from the male cohort (n = 31), and nine coronary and two femoral artery specimens were collected from the female cohort (n = 11). Demographic information (age, sex, and ethnicity) were collected for all patients. Systolic and diastolic blood pressure information was collected for all patients except for four males. Body mass index, diabetes status, lipids panel, and C-reactive protein levels data were collected for 33, 33, 17, and 17 patients, respectively (see Table 1). Periodontal disease measurements and tissue histology data related to inflammation were not collected.
Table 1.
Demographic and clinical characteristics.
| Characteristics | Data | Subjects included |
| Age (years), M (SD) | 64 (9.86) | All (n = 42) |
| Males, n (%); age (years), M (SD) | 31 (73.8%); 62.3 (9.46) | |
| Females, n (%); age (years), M (SD) | 11 (26.2%); 68.7 (9.84) | |
| Ethnicity, n (%) | Subset (n = 41) | |
| White | 38 (90.3%) | |
| African American | 3 (7.3%) | |
| Hispanic | 1 (2.4%) | |
| Other | 0 (0%) | |
| SBP, M (SD), range | 133.29 (25.37), 100–190 | Subset (n = 38) |
| DBP, M (SD), range | 71.87 (13.40), 48–100 | Subset (n = 38) |
| MAP, M (SD), range | 92.34 (14.56), 67.33–120 | Subset (n = 38) |
| BMI, M (SD), range | 27.93 (4.75), 16.18–38.55 | Subset (n = 33) |
| Diabetes, n (%) | 12 (36.4%) | Subset (n = 33) |
| Lipids panel, M (SD), range | Subset (n = 17) | |
| Cholesterol | 184.76 (52.25), 119–337 | |
| Triglycerides | 184 (119.72), 64–478 | |
| HDL | 105.59 (42.16), 62–242 | |
| C-reactive protein, M (SD), range | 42.41 (15.65), 27–91 | Subset (n = 17) |
Clinical characteristics relevant to atherosclerosis were obtained for patients for whom clinically healthy coronary and femoral artery tissues were recovered from bypass surgery and used for microbiome profile determination by HOMINGS. Males and females were of a similar age (M [SD]). Available data indicated that based on mean values, the patient cohort was characterized by prehypertension systolic blood pressure (SBP; mmHg) between 120 and 140, normal diastolic blood pressure (DBP; mmHg) <80, high metric body mass index in ‘overweight’ category (BMI; kg/m2), cholesterol levels below borderline high threshold of 200 mg/dL, borderline high triglycerides (mg/dL) between 150 and 200, near optimal low density lipoproteins (LDL; mg/dL), and levels of C-reactive protein indicative of active inflammation (>10 mg/L).
HDL, high-density lipoprotein; MAP, mean arterial pressure; SD, standard deviation.
HOMINGS analysis
Arterial tissue specimens were snap-frozen and stored in liquid nitrogen. DNA was extracted using the Epicentre MasterPureTM DNA Purification Kit according to the manufacturer’s instructions (Epicentre, Madison, WI, USA). The HOMINGS technology was used to enable the identification of nearly 600 oral bacterial species, as previously described [21–23]. Briefly, 16S rRNA genes were amplified (V3–V4 hypervariable region) and processed using a modified MiSeq (Illumina, Inc., San Diego, CA, USA) platform, as described by Caporaso et al. [24]. Species taxa identification and frequency were determined using the ProbeSeq program [22,23]. Sequence reads that were uniquely electronically hybridized to one species- or one genus-specific probe were counted as a ‘hit’ and accumulated. Genus probes can provide an estimate of (1) unidentified oral and non-oral species, or (2) the presence of genera of known species for which a specific species probe within ProbeSeq against the V3–V4 16S rRNA gene sequence has not been designed. More than 20% of unmatched reads is reflective of significant bacterial DNA degradation if proportional loss of bacterial identification can be determined (i.e. increased underrepresentation of species or genera hits as the amount of unaccounted reads increases). Total hits by species or genus probe by patient were used for statistical analyses.
Statistical analyses
Shannon and Simpson alpha diversity indexes were determined using an online open source program (http://www.comparingpartitions.info/index.php?link=Tool). Differences between groups (i.e. coronary vs. femoral, male vs. female, male/female patients with younger than 60 years of age vs. older than 60 years of age) were analyzed using the Mann–Whitney U-test with adjustment for differences in sample sizes, using an online tool (http://www.Vassarstats. net/utest.html). PERMANOVA (unrestricted permutation of raw data, 999 permutations, type III partial sum of squares) in the PRIMER 7 program (PRIMER-E Ltd., Ivybridge, UK) was used to compare beta diversity between groups based on Bray–Curtis similarity matrixes derived from bacterial species and genera abundance data (p < 0.05). Spearman correlations of bacterial species or genus hits with unmatched reads were determined using an online open source program (http://www.socscistatistics.com/tests/spearman/Default2.aspx; p < 0.05).
Results
Demographic data and clinical characteristics for the 42-patient cohort (n = 31 males, n = 11 females) are presented in Table 1.
HOMINGS initial aggregate raw data analysis showed that the percentage of sequence reads that e-hybridized to species probes ranged from 5.7% to 84.1% and e-hybridized to genus probes from 1.0% to 65.7% of the total reads by patient (Table 2). The frequency of unmatched reads ranged from 5.9% to 83.1% (M [SD] = 29,091.7 [20,309.9]; Table 2), reflective of the presence of (1) non-oral bacterial species, (2) novel oral species for which ProbeSeq probes have not been developed, or (3) low-quality reads, possibly resulting from bacterial DNA degradation. HOMINGS data obtained for coronary (n = 32) and femoral (n = 10) artery tissues showed the presence of 230 and 118 oral bacterial species in these tissues, respectively, with 103 species in common (Table 3). In addition, unknown species detected by genus-specific probes represented 45 and 30 genera for the coronary and femoral artery tissues, respectively (Table 3). Overall, 95 genera (i.e. the sum of 86 genera detected by species probes and an additional nine detected by genus probes) and 245 species were uniquely present in coronary and femoral arteries combined (Table 3).
Table 2.
HOMINGS aggregate raw data obtained from clinically non-atherosclerotic coronary and femoral artery tissue samples (n = 42).
| Reads category | Total | Range | Median | Mean | Standard deviation | Mean relative abundancea(%) | Range relative abundanceb (%) |
|---|---|---|---|---|---|---|---|
| Species Probes hits | 1,490,502 | 4,782–111,251 | 28,234 | 35,488.1 | 27,572.4 | 39.79 | 5.7–84.1 |
| Genus Probes hits | 1,033,630 | 967–86,927 | 21,326 | 24,610.2 | 17,385.1 | 27.59 | 1.0–65.7 |
| Unmatched reads | 1,221,851 | 4,754–78,298 | 21,700 | 29,091.7 | 20,309.9 | 32.62 | 5.9–83.1 |
The HOMINGS species identification process is based on an iterative in silico hybridization process. Within ProbeSeq program, each 16S rRNA bacterial gene sequence read is matched in silico against each unique species probe. A perfect species probe match is recorded as one ‘hit’. Following the species probe matching process, as yet unmatched reads are matched to genus probes. Sequence reads matching neither a species nor genus probe are, finally, accumulated as ‘Unmatched Reads’.
Calculations were based on all patient samples (n=42), i.e., coronary (n=32) or femoral (n=10); a Mean Relative Abundance is shown as percentage based on the ratio mean/total hits or reads; b Relative Abundance Range is shown as percentage based on the ratio minimum or maximum hits or reads/total hits or reads, respectively.
Table 3.
Species and genera detection data.
| Coronary tissue |
Femoral tissue |
Common coronary and femoral |
Unique coronary and femoral |
|
|---|---|---|---|---|
| Species and/or genus probe-based detection | (n = 32) | (n = 10) | (n = 42) | (n = 42) |
| Species count per species probes | 230 | 118 | (103)# | 245 |
| Genera count per genus probes | 45 | 30 | (28)# | 47 |
| Genera count per species probes | 84 | 58 | (56)# | 86 |
| Genera count unique to genus probes | 8 | 7 | (6)# | 9 |
| Total genera count | 92 | 65 | (62)# | 95 |
A species or genus is considered detected if at least one hit is accounted for that species or genus. Species probe hits may be combined with corresponding genus probe hits to determine total number of genera detected. # Number of common species or genera is represented by (number).
The top three unique species among 245 detected in coronary and femoral arteries combined were (in decreasing order of abundance) P. gingivalis, Enterococcus faecalis, and Finegoldia magna (Table 4), while the top three genera among the 95 detected were Porphyromonas, Escherichia, and Staphylococcus (Table 5). In addition, the top five most abundant genera (i.e. Porphyromonas, Escherichia, Staphylococcus, Pseudomonas, and Streptococcus) were consistently detected across all patients (n = 42), altogether representing 88.5% of the total hits (Table 5). The taxa P. gingivalis, Rothia mucilaginosa, Escherichia, Staphylococcus, Pseudomonas, Streptococcus, and Granulicatella were detected in all patient samples (n = 42; Tables 4 and 5). The rest of the taxa exhibited relatively scarce data with occasional high outlier values. The number of hits across patients for these five most abundant genera correlated negatively with their respective number of unmatched reads (data not shown). All r correlation values determined by Spearman’s rank correlation were negative (mostly p < 0.05), indicating that a significant amount of unmatched reads most likely corresponds to low-quality sequences, leading to stochastic proportional underrepresentation of taxa hits. Although there was no abundance data correlation found between the five most abundant genera in pairwise comparisons, no correlation analyses were performed with any of the clinical variables. Indeed, such correlations were deemed unwarranted or coincidental due to the significant impact by unaccounted low quality reads on absolute total hits.
Table 4.
Ten most prominent species and genera identified by HOMINGS.
| Species Name | No. positives | Total hits | Hits range | Median hits | Mean hits | Standard deviation | % Relative abundancea |
|---|---|---|---|---|---|---|---|
| 1. Porphyromonas gingivalis | 42 | 1,180,493 | 103–101,244 | 18,523.5 | 28,107.0 | 27,625.7 | 79.20% |
| 2. Enterococcus faecalis | 32 | 46,100 | 0–18,537 | 1 | 1,097.6 | 3,529.0 | 3.09% |
| 3. Finegoldia magna | 22 | 32,795 | 0–13,180 | 1 | 780.8 | 2,831.9 | 2.20% |
| 4. Pseudomonas aeruginosa | 25 | 30,857 | 0–7,848 | 17 | 734.7 | 1,445.7 | 2.07% |
| 5. Haemophilus parainfluenzae | 19 | 7,685 | 0–4,773 | 0 | 183.0 | 765.3 | 0.52% |
| 6. Rothia mucilaginosa | 42 | 6,675 | 17–2,088 | 45 | 158.9 | 374.2 | 0.45% |
| 7. Gemella haemolysans | 33 | 6,609 | 0–5,932 | 1.5 | 157.4 | 916.2 | 0.44% |
| 8. Brevundimonas diminuta | 8 | 6,560 | 0–3,324 | 0 | 156.2 | 562.1 | 0.44% |
| 9. Stenotrophomonasmaltophilia | 11 | 4,800 | 0–2,845 | 0 | 114.3 | 500.7 | 0.32% |
| 10. Gemella morbillorum | 16 | 4,369 | 0–4,094 | 0 | 104.0 | 631.4 | 0.29% |
Calculations were based on all patient samples (n=42), i.e., coronary (n=32) or femoral (n=10); a Mean Relative Abundance is shown as percentage based on the ratio mean/total hits per total 245 species detected (1,490,502 hits).
Table 5.
Most prominent genera.
| Genus name | No. positives | Total hits | Hits range | Median hits | Mean hits | Standard deviation | % Relative abundancea |
|---|---|---|---|---|---|---|---|
| 1. Porphyromonas | 42 | 1,180,564 | 103–101,245 | 18,523.5 | 28,108.7 | 27,625.3 | 46.77% |
| 2. Escherichia | 42 | 488,103 | 112–83,578 | 4,351.5 | 11,621.5 | 18,261.4 | 19.34% |
| 3. Staphylococcus | 42 | 264,248 | 11–43,098 | 3,688.5 | 6,291.6 | 8,475.1 | 10.47% |
| 4. Pseudomonas | 42 | 194,684 | 8–26,997 | 2,280.5 | 4,635.3 | 6,010.3 | 7.71% |
| 5. Streptococcus | 42 | 105,195 | 52–19,802 | 577 | 2,504.6 | 4,226.8 | 4.17% |
| 6. Enterococcus | 32 | 46,765 | 0–18,537 | 1 | 1,113.5 | 3,525.4 | 1.85% |
| 7. Acinetobacter | 39 | 46,382 | 0–32,132 | 4 | 1,104.3 | 4,982.6 | 1.84% |
| 8. Finegoldia | 22 | 32,795 | 0–13,180 | 1 | 780.8 | 2,831.9 | 1.30% |
| 9. Anaerococcus | 20 | 25,337 | 0–12,884 | 0 | 603.3 | 2,096.5 | 1.00% |
| 10. Granulicatella | 42 | 20,038 | 7–13,066 | 18 | 477.1 | 2,157.4 | 0.79% |
Calculations were based on all patient samples (n=42), i.e., coronary (n=32) or femoral (n=10); a Mean Relative Abundance is shown as percentage based on the ratio mean/total hits per total 95 genera detected (2,524,132 hits).
Also, no significant differences were found in bacterial alpha or beta diversity measurement between coronary and femoral artery tissues (p ≥ 0.05). Therefore, the data were pooled for further analysis. In addition, no significant differences were found in alpha or beta diversity between males (n = 31) and females (n = 11), or between patients age 60 or younger (n = 15) and those older than 60 years of age (n = 27; p ≥ 0.05; data not shown). However, despite the fact that there were no differences in alpha and beta diversity between the above-mentioned subgroups, P. gingivalis and the genus Escherichia were the most abundant taxa. These two taxa had similar hits per patient distributions in combined coronary and femoral artery tissues data sets (Figure 1(a) and (b)). However, P. gingivalis was significantly more abundant (p = 0.0005; Figure 1(a) and (b)). P. gingivalis alone accounted for 79.2% of the total species probe hits (Table 4), and genus Porphyromonas accounted for 46.8% of the total combined species and genus probe hits (Table 5). The second most abundant genus, Escherichia, for which no E. coli probe had been designed, accounted for 19.3% of total combined species and genus probe hits. The z-score distributions showed that patient outliers with regard to the abundance data for these two taxa were different (Figure 1(a) and (b)). The potential implications of these results are discussed below.
Figure 1.
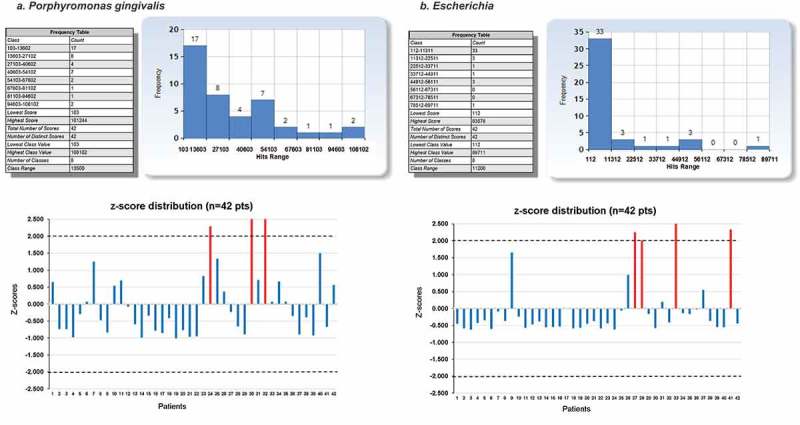
Abundance frequency plots and z-score distribution for species Porphyromonas gingivalis and genus Escherichia in clinically non-atherosclerotic coronary and femoral artery tissues (a) P. gingivalis (b) Genus Escherichia. Frequency distributions for most abundant taxa, P. gingivalis (a) and genus Escherichia (b), are shown. Both taxa were detected in significant amounts in virtually all coronary and femoral artery tissue patient samples (n = 42). The distributions of both taxa were similar. However, the z-score distributions showed no outliers overlap (red bars), i.e. patients 24, 30, and 32 for P. gingivalis (z-score plot in (a)) and patients 27, 28, 33, and 41 for Escherichia (z-score plot in (b)). Thus, P. gingivalis abundance data do not correlate with those of Escherichia (r = 0.02552; p = 0.87252).
Discussion
This is the first study to report using HOMINGS targeted metagenomics to screen for nearly 600 known oral bacterial species in artery tissues clinically unaffected with atherosclerosis that were obtained from patients with atherosclerotic CVD who underwent coronary or femoral artery bypass surgery. P. gingivalis was by far the most abundant species, representing nearly 80% of all bacterial species counts.
Three other species (i.e. E. faecalis, F. magna, and P. aeruginosa) were detected with a relative abundance of 2% to just over 3% compared with all remaining species detected at <1%. All three species have been identified as pathogens involved in cardiovascular complications. For instance, E. faecalis is associated with infective endocarditis [25,26]. F. magna has been shown to invade skin cells [21], and is one of the most frequent pathogens in the etiology of postoperative and prosthetic implant-related infection [22]. It is also associated with mediastinitis following coronary artery bypass [23]. F. magna has been detected in rare cases in atherosclerotic plaques from carotid arteries [27]. Finally, chronic P. aeruginosa infection has been shown to increase aorta and coronary artery thickness and wall rigidity in a rat model of atherosclerosis [28].
In this study, the genus Escherichia was the second most abundant behind Porphyromonas, while relative abundances of both taxa did not correlate across patients. No species probes were available within the HOMINGS platform to ascertain species identification within the Escherichia genus. Nevertheless, such findings would indicate detection of taxa of non-oral origin. Staphylococcus was the third most abundant genus detected, with the dual species probe for S. aureus and S. gallinarum accounting for nearly 27% of the total hits for this genus in 41/42 patients, respectively. Staphylococcus spp. have been detected in atherosclerotic plaques of patients with coronary heart disease [16]. Although HOMINGS focuses mainly on oral taxa, approximately 70–90% of the taxa were accounted for, with significant detection at the genus level by species and genus probes. Unmatched reads could correspond to species that are exclusively of non-oral origin, strain variation, or sequencing error.
The present results differ from previous metagenomics studies that focused on the analysis of atherosclerotic plaque and did not find P. gingivalis as a predominant species [15–19]. These studies did not use clinically non-atherosclerotic arterial walls from patients diagnosed with atherosclerosis as control tissues. For instance, Ott et al. [16] used control coronary artery tissues from patients who died from malignancies and used tissues from heart-beating tissue donors, both without presence of atherosclerosis of any coronary artery per macroscopic examination [16]. The authors did not, however, detect bacterial DNA in the control tissues as opposed to atherosclerotic plaque samples, and are hence in contrast with the current findings.
In addition, since the present study used clinically healthy tissues from patients with atherosclerosis, the results differ significantly from those obtained from atherosclerotic plaque material through metagenomic approaches [15–19]. These studies showed little overlap at the species level regarding the identified predominant taxa. Also, as results are inherently dependent on the technological platform used, reproducibility needs to be assessed using complementary approaches in larger cohorts. While the use of HOMINGS has enabled a unique bacterial profile to be identified of clinically non-atherosclerotic artery tissues for oral species, future investigations will benefit from alternative approaches such as the Pathochip array technology or sequencing algorithms that allow for comprehensive analysis of all bacterial taxa [29,30].
Although the periodontal disease status of our patient population is unknown, C-reactive protein (CRP) levels recorded for 17/42 patients ranged from 27 to 91 mg/L (Table 1), which could possibly correlate with periodontal disease in patients with cardiovascular disease [8]. Overall, the significant detection of P. gingivalis in the arterial tissues of all patients (n = 42) in this study is in agreement with other studies, implicating P. gingivalis in the development of atherosclerosis. However, A. actinomycetemcomitans was not detected, except in one sample with 11 hits, which might reflect fundamental colonization differences between plaque material and clinically non-atherosclerotic arterial tissues. Similarly, Chlamydia pneumonia was detected in four patients at very low levels.
A large body of evidence has implicated the periodontal pathogen P. gingivalis in atherosclerosis [11]. However, the role of P. gingivalis as an etiological agent of atherosclerosis is yet to be elucidated. P. gingivalis was shown to reside in diseased atherosclerotic tissues and the aneurysmal wall of blood vessels and to induce atherosclerosis in pigs following bacteremia regardless of cholesterol level [16,31]. It has been suggested that periodontal disease predisposes for atherosclerosis due to an association between the presence of antibodies against P. gingivalis and atherosclerosis in large cohort studies [3,4,7,8,32]. Based on epidemiological and experimental studies, several mechanisms have been proposed [7,8,10]: (1) intracellular invasion of arteries at the sites of atherosclerotic disease by periodontal pathogens [10]; (2) increased responsiveness of the immune response to the presence of P. gingivalis, with possible involvement of proinflammatory bacterial products such as lipopolysaccharides or phosphorylated dihydroceramides [7,33,34]; and (3) cross-reactivity of antibacterial heat-shock protein antibodies with human heat-shock proteins [7,8].
Overall, the vastly predominant presence of P. gingivalis in clinically healthy arteries observed in this study is in line with the concept that P. gingivalis possesses unique properties to invade the arterial walls to survive intracellularly and escape the immune system [28]. It is unknown, however, whether the bacterial colonization of healthy tissues occurs prior to atherosclerosis disease onset or whether diseased tissues form before the colonization, thereby enabling subsequent colonization. It is also unknown for how long bacteria remain alive within tissue, and which species act as bystanders or contribute to the development of atherosclerosis in the initial stages of the disease. Longitudinal studies in animal models for atherosclerosis will be required to identify biomarkers useful in human studies for the determination of whether inflammation precedes or follows bacterial colonization of damaged or non-damaged tissues and how colonization evolves during the course of the disease. Understanding of the different stages of colonization could provide some explanation as to why certain species would be present in clinically ‘healthy’ tissues and absent in atherosclerotic plaque. In particular, investigation of mechanisms of periodontal disease that would influence such distant site colonization will be necessary to develop improved prevention and therapies for atherosclerosis [35,36].
In conclusion, this study provides insight into the presence and types of bacteria found in clinically healthy artery tissue. These species may be associated with the initiation and/or exacerbation of atherosclerosis, with or without any role in causation. The role of P. gingivalis, the most predominant species identified in this study, and other oral bacteria potentially involved in the disease process may be further elucidated in future studies.
Acknowledgments
This study was funded by Carolinas HealthCare System’s Research fund. We thank Dr. Joseph Cook, Dr. Jeremiah Holleman, Jenene Noll, Louise Kent, and other clinical staff for their help with patient samples and data collection, and Dr. Craig Murdoch laboratory, Shirley Coleman, Darla Morton, and Alexis Kokaras for DNA isolation/processing. We also thank Drs. Gillian Smith, Marcello Riggio, and Martin Thornhill for helpful discussions.
Biographies
Jean-Luc C. Mougeot received his Master’s degree in Molecular Biology/Virology in 1991 and his doctoral degree in Molecular and Cellular Biology/Virology in 1995 from the University of Louis Pasteur, Strasbourg, France. He trained as a postdoctoral fellow in Friedrich Miescher Institute/ Novartis, Switzerland for two years followed by a three-year European Molecular Biology Organization fellowship at the Medical Research Council, Cambridge, UK. He then worked as senior research scientist leading the protein expression core laboratory at deCODE Genetics, Inc., Reykjavik, Iceland from 2000 to 2002. Since 2002, Dr. Mougeot directs translational genomics and computational biology research at Carolinas HealthCare System in Charlotte, NC. He is also an adjunct professor in the departments of Biology and Bioinformatics and Genomics at the University of North Carolina at Charlotte. Dr. Mougeot has extensive experience in drug target characterization, stem cell research, computational biology and data mining for the study of etiological mechanisms of disease, genetic regulation and biomarker discovery in neuroscience, gene expression studies in cancer and oral complications of cancer therapy, and role of oral microbiome in the onset and development of local and systemic diseases.
Bruce J. Paster is presently Senior Member of the Staff and Chair of the Department of Microbiology, director of the HOMINGS Oral Bacterial Identification Core, and director of the Sequencing Core Facility at The Forsyth Institute in Cambridge, Massachusetts. He is also Professor in the Department of Oral Medicine, Infection & Immunity, Harvard School of Dental Medicine and Professor II in the Dental Faculty in the University of Oslo. In 1975, he received a BS in Microbiology at the University of Rhode Island, in 1981, a PhD in Microbiology at the University of Massachusetts, Amherst and, in 2014, an honorary PhD at the University of Oslo. He was a post-doc for Dr. Carl Woese at the University of Illinois from 1981–1983 and later in 1983–1986 for Dr. Ronald Gibbons at The Forsyth Institute.
Craig B. Stevens graduated from the University of Connecticut in 1969 with a degree in accounting and went on to become a CPA working in the paper industry until retirement at the corporate vice-president level in 2007. He then earned a degree in chemistry and biology from Winthrop University, S. Carlina. Following graduation with honors recognition, he began working at Carolinas HealthCare System in the Department of Oral Medicine Microbiome laboratory as a research analyst.
Michael T. Brennan, DDS, MHS, attended the University of Iowa for undergraduate and dental training. He completed a General Practice Residency at Loyola University Hospital outside of Chicago. Following his residency, he received four years of additional training at the National Institutes of Health (NIH) in Bethesda, Maryland. He received a Certificate in Oral Medicine from NIH and a Masters in Clinical Research from Duke University in Durham, North Carolina. He joined the Department of Oral Medicine at the Carolinas Medical Center in June 2000, and is currently Professor and Chairman with the Department of Oral Medicine. He also serves as the Director of the Sjögren’s Syndrome and Salivary Disorders Center at Carolinas Medical Center. He has extensive research experience with over 120 peer reviewed articles and book chapters and currently is the co-PI on an NIH U01 grant examining the oral sequela in head and neck cancer patients treated with radiation therapy.
Peter B. Lockhart, DDS, a native of Massachusetts, graduated from University of North Carolina School of Dentistry. After completing a two-year General Practice Residency (GPR) at Medical Center Hospital of Vermont, he joined the faculty at Brigham and Women’s Hospital in Boston where he directed the GPR program, practiced general dentistry and oral medicine, and conducted clinical research for 10 years. He was Chairman of the Department of Oral Medicine at Carolinas Medical Center, in Charlotte, NC for 26 years, where he is now a Research Professor focusing on clinical research studies. He received a World Health Organization Fellowship in Oral Medicine in the United Kingdom, and later a TC White Visiting Professorship at University of Glasgow, Scotland. He holds Fellowship in Dental Surgery diplomas from the Royal College of Physicians and Surgeons of Glasgow, and the Royal College of Surgeons of Edinburgh. He has a variety of research interests in oral medicine, particularly the dental management of medically complex patients. He has had funding for bacteremia research for over 25 years, most recently a 5-year NIDCR\NIH R01 study concerning associations between oral disease and infective endocarditis. Since 2004, he has been the American Dental Association liaison to the American Heart Association’s Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease. He also served on three recent multidisciplinary Writing Committees on prophylaxis guidelines for patients with prosthetic joints. He has over 130 textbook chapters and peer reviewed journal articles and he is the editor of the text book, Oral Medicine and Medically Complex Patients.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Mattila KJ, Nieminen MS, Valtonen VV. Association between dental health and acute myocardial infarction. BMJ. 1989;298(6676):779–9. doi: 10.1136/bmj.298.6676.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JD, Slade G, Offenbacher S. Oral disease, cardiovascular disease and systemic inflammation. Periodontol 2000. 2000;23:110–120. doi: 10.1034/j.1600-0757.2000.2230111.x. [DOI] [PubMed] [Google Scholar]
- Pussinen PJ, Jousilahti P, Alfthan G. Antibodies to periodontal pathogens are associated with coronary heart disease. Arterioscler Thromb Vasc Biol. 2003;23(7):1250–1254. doi: 10.1161/01.ATV.0000072969.71452.87. [DOI] [PubMed] [Google Scholar]
- Pussinen PJ, Alfthan G, Rissanen H. Antibodies to periodontal pathogens and stroke risk. Stroke. 2004;35(9):2020–2023. doi: 10.1161/01.STR.0000136148.29490.fe. [DOI] [PubMed] [Google Scholar]
- Rosenfeld ME, Campbell LA. Pathogens and atherosclerosis: update on the potential contribution of multiple infectious organisms to the pathogenesis of atherosclerosis. Thromb Haemost. 2011;106(5):858–867. doi: 10.1160/TH11-06-0392. [DOI] [PubMed] [Google Scholar]
- Chistiakov DA, Bobryshev YV, Kozarov E. Role of gut microbiota in the modulation of atherosclerosis-associated immune response. Front Microbiol. 2015;6:671. doi: 10.3389/fmicb.2015.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford PJ, Gemmell E, Hamlet SM. Cross-reactivity of GroEL antibodies with human heat shock protein 60 and quantification of pathogens in atherosclerosis. Oral Microbiol Immunol. 2005;20(5):296–302. doi: 10.1111/j.1399-302X.2005.00230.x. [DOI] [PubMed] [Google Scholar]
- Seymour GJ, Ford PJ, Cullinan MP. Relationship between periodontal infections and systemic disease. Clin Microbiol Infect. 2007;13(Suppl 4):3–10. doi: 10.1111/j.1469-0691.2007.01798.x. [DOI] [PubMed] [Google Scholar]
- Lockhart PB, Brennan MT, Sasser HC. Bacteremia associated with toothbrushing and dental extraction. Circulation. 2008;117(24):3118–3125. doi: 10.1161/CIRCULATIONAHA.107.758524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarov EV, Dorn BR, Shelburne CE. Human atherosclerotic plaque contains viable invasive Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis . Arterioscler Thromb Vasc Biol. 2005;25(3):e17–8. doi: 10.1161/01.ATV.0000155018.67835.1a. [DOI] [PubMed] [Google Scholar]
- Olsen I, Progulske-Fox A. Invasion of Porphyromonas gingivalis strains into vascular cells and tissue. J Oral Microbiol. 2015;7:28788. doi: 10.3402/jom.v7.28788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes BP, Berber VB, Kokaras AS. Microbiomes of endodontic-periodontal lesions before and after chemomechanical preparation. J Endod. 2015;41(12):1975–1984. doi: 10.1016/j.joen.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford A, Hoffman GS. Evidence for a vascular microbiome and its role in vessel health and disease. Curr Opin Rheumatol. 2015;27(4):397–405. doi: 10.1097/BOR.0000000000000184. [DOI] [PubMed] [Google Scholar]
- Lehtiniemi J, Karhunen PJ, Goebeler S. Identification of different bacterial DNAs in human coronary arteries. Eur J Clin Invest. 2005;35(1):13–16. doi: 10.1111/j.1365-2362.2005.01440.x. [DOI] [PubMed] [Google Scholar]
- Ott SJ, El Mokhtari NE, Musfeldt M. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation. 2006;113(7):929–937. doi: 10.1161/CIRCULATIONAHA.105.579979. [DOI] [PubMed] [Google Scholar]
- Koren O, Spor A, Felin J. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandrini CA, Ribeiro AC, Gonnelli AC. Microbial composition of atherosclerotic plaques. Oral Dis. 2014;20(3):e128–34. doi: 10.1111/odi.12205. [DOI] [PubMed] [Google Scholar]
- Mitra S, Drautz-Moses DI, Alhede M. In silico analyses of metagenomes from human atherosclerotic plaque samples. Microbiome. 2015;3:38. doi: 10.1186/s40168-015-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougeot JL, Stevens CB, Cotton SL. Concordance of HOMIM and HOMINGS technologies in the microbiome analysis of clinical samples. J Oral Microbiol. 2016;8:30379. doi: 10.3402/jom.v8.30379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EC, Mörgelin M, Reinhardt DP. Identification of molecular mechanisms used by Finegoldia magna to penetrate and colonize human skin. Mol Microbiol. 2014;94(2):403–417. doi: 10.1111/mmi.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy PY, Fenollar F, Stein A. Finegoldia magna: a forgotten pathogen in prosthetic joint infection rediscovered by molecular biology. Clin Infect Dis. 2009;49(8):1244–1247. doi: 10.1086/605672. [DOI] [PubMed] [Google Scholar]
- Kernéis S, Matta M, Hoï AB. Postoperative mediastinitis due to Finegoldia magna with negative blood cultures. J Clin Microbiol. 2009;47(12):4180–4182. doi: 10.1128/JCM.01192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg R, Schmied C, Falk V. Molecular evidence of Enterococcus faecalis in an occluding coronary thrombus from a patient with late prosthetic valve endocarditis. Clin Res Cardiol. 2012;101(12):1021–1023. doi: 10.1007/s00392-012-0483-8. [DOI] [PubMed] [Google Scholar]
- Pericás JM, Zboromyrska Y, Cervera C. Enterococcal endocarditis revisited. Future Microbiol. 2015;10(7):1215–1240. doi: 10.2217/fmb.15.46. [DOI] [PubMed] [Google Scholar]
- Kedzia A, Ciecierski M, Kufel A. Isolation of anaerobic bacteria from atherosclerotic plaques from carotid arteries. Acta Angiologica. 2012;18(2):59–67. [Google Scholar]
- Turkay C, Saba R, Sahin N. Effect of chronic Pseudomonas aeruginosa infection on the development of atherosclerosis in a rat model. Clin Microbiol Infect. 2004;10(8):705–708. doi: 10.1111/j.1469-0691.2004.00920.x. [DOI] [PubMed] [Google Scholar]
- Baldwin DA, Feldman M, Alwine JC. Metagenomic assay for identification of microbial pathogens in tumor tissues. MBio. 2014;5(5):e01714–14. doi: 10.1128/mBio.01714-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hebshi NN, Nasher AT, Idris AM. Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: application to oral carcinoma samples. J Oral Microbiol. 2015;7:28934. doi: 10.3402/jom.v7.28934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodala N, Merricks EP, Bellinger DA. Porphyromonas gingivalis bacteremia induces coronary and aortic atherosclerosis in normocholesterolemic and hypercholesterolemic pigs. Arterioscler Thromb Vasc Biol. 2005;25(7):1446–1451. doi: 10.1161/01.ATV.0000167525.69400.9c. [DOI] [PubMed] [Google Scholar]
- Gibson FC, III, Genco CA. Porphyromonas gingivalis mediated periodontal disease and atherosclerosis: disparate diseases with commonalities in pathogenesis through TLRs. Curr Pharm Des. 2007;13(36):3665–3675. doi: 10.2174/138161207783018554. [DOI] [PubMed] [Google Scholar]
- Nichols FC, Riep B, Mun J. Structures and biological activity of phosphorylated dihydroceramides of Porphyromonas gingivalis . J Lipid Res. 2004;45(12):2317–2330. doi: 10.1194/jlr.M400278-JLR200. [DOI] [PubMed] [Google Scholar]
- Zahlten J, Riep B, Nichols FC. Porphyromonas gingivalis dihydroceramides induce apoptosis in endothelial cells. J Dent Res. 2007;86(7):635–640. doi: 10.1177/154405910708600710. [DOI] [PubMed] [Google Scholar]
- Hussain M, Stover CM, Dupont A. P. gingivalis in periodontal disease and atherosclerosis - Scenes of action for antimicrobial peptides and complement. Front Immunol. 2015;6:45. doi: 10.3389/fimmu.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W, Taubert KA, Gewitz M. Prevention of infective endocarditis. guidelines from the American Heart Association. A guideline from the American Heart Association Rheumatic Fever, Endocarditis, and Kawasaki Disease Committee, Council on Cardiovascular Disease in the Young, and the Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and the Quality of Care and Outcomes Research Interdisciplinary Working Group. Circulation. 2007;116(15):1736–1754. doi: 10.1161/CIRCULATIONAHA.106.183095. [DOI] [PubMed] [Google Scholar]


