Abstract
Central carbon metabolism is controlled by modulating the protein abundance profiles of enzymes that maintain the essential systems in living organisms. In this study, metabolic adaptation mechanisms in the model organism Saccharomyces cerevisiae were investigated by direct determination of enzyme abundance levels in 30 wild type and mutant strains. We performed a targeted proteome analysis using S. cerevisiae strains that lack genes encoding the enzymes responsible for central carbon metabolism. Our analysis revealed that at least 30% of the observed variations in enzyme abundance levels could be explained by global regulatory mechanisms. A enzyme-enzyme co-abundance analysis revealed that the abundances of enzyme proteins involved in the trehalose metabolism and glycolysis changed in a coordinated manner under the control of the transcription factors for global regulation. The remaining variations were derived from local mechanisms such as a mutant-specific increase in the abundances of remote enzymes. The proteome data also suggested that, although the functional compensation of the deficient enzyme was attained by using more resources for protein biosynthesis, available resources for the biosynthesis of the enzymes responsible for central metabolism were not abundant in S. cerevisiae cells. These results showed that global and local regulation of enzyme abundance levels shape central carbon metabolism in S. cerevisiae by using a limited resource for protein biosynthesis.
Introduction
Central carbon metabolism is an essential system of the living organisms. Active ethanol fermentation by the budding yeast, Saccharomyces cerevisiae, is a model of the eukaryotic central metabolism due to its importance in brewing industries, as well as its similarity to the Warburg effect in cancer cells [1]. It has been revealed that glycolytic flux is maintained via a complex control of metabolic reactions. For example, the allosteric regulation of phosphofructokinase (PFK) is important in glycolysis control [2]. A glycerol pathway has been considered as a drain for excess reducing power to avoid a redox imbalance inside the cells [3]. The post-translational regulation of enzymes also controls the central carbon metabolism [4–6].
Furthermore, it has been reported that the central carbon metabolism is controlled through a modulation of enzyme abundances in response to environmental conditions [7–11]. The regulation of enzyme abundance levels also works in mutant strains lacking genes of central carbon metabolism related enzymes. It is because metabolic imbalances caused by the gene deletions are compensated by increase and decrease in the abundance of other enzyme proteins to avoid a breakdown of metabolic systems [12]. The targeted proteome analysis showed that the expression of several glycolytic enzymes, including Pgi1, Tdh2, and Eno1 were increased in BY4742 pfk1Δ strain lacking PFK1 gene, suggesting that the loss of the α subunit of PFK was counteracted by an activation of glycolysis [13]. The distant parts of the metabolic network were modulated to compensate the loss of a single enzyme protein since the expression of several TCA cycle related enzymes, such as Cit1, Kgd1, and Fum1, were increased in the BY4742 zwf1Δ strain [13]. The metabolic flux analysis also revealed that knockout of glucose-6-phosphate dehydrogenase gene (ZWF1) caused an up-regulation of TCA cycle flux level [14].
The protein abundance profiles is likely regulated and constrained by several factors. First, the regulation of enzyme abundance is constrained by the cost of amino acid for protein biosynthesis [15, 16]. Given that glycolytic enzymes such as Tdh3 are one of the most abundant proteins in S. cerevisiae, the increase and decrease in these abundance should largely affect the composition and resource allocation of the S. cerevisiae proteome [17]. Second, transcriptome analyses indicated coordinated expressions of genes encoding enzymes [18–20]. For instance, a number of genes encoding glycolytic enzymes, such as TDH3 and TPI1, form a co-expression module through the regulation of transcriptional activators, such as Gcr1 and Gcr2 [18, 21]. Third, central carbon metabolism could be controlled by local regulation of a small number of enzyme genes. For instance, the promoter activity of a minor isoform of the pyruvate decarboxylase gene (PDC5) is specifically increased in a mutant strain lacking the major isoform gene (PDC1) to functionally compensate the reduced pyruvate decarboxylase activity [22]. Enzyme abundance levels, however, do not always correlate with their gene expression levels [19, 23, 24], and the relative contribution of these regulation to the maintenance of central carbon metabolism remains unclear.
In this study, we have addressed this issue by the direct determination of enzyme abundance in the single gene-deletion mutant strains of S. cerevisiae by targeted proteomics using liquid chromatography-tandem quadrupole mass spectrometry (LC-MS/MS). The selected reaction monitoring (SRM) assay for the central metabolism related enzymes of S. cerevisiae has been extensively developed by excellent efforts of the ETH group [25–27]. The analysis of the enzyme abundance dataset revealed that the central carbon metabolism in S. cerevisiae is controlled through specific and coordinated regulations of enzyme abundance levels by using a limited resource for protein biosynthesis.
Results and discussion
Targeted proteome analysis of single gene deletion mutants of S. cerevisiae
Protein abundance profiles of the central metabolism-related enzymes were obtained from the wild type (BY4742), 29 single-gene deletion strains lacking enzyme genes responsible for the central carbon metabolism and a gene encoding transcriptional activator (Fig 1 and S1 Table). The single-gene deletion strains were arbitrarily selected based on the growth phenotype in the Saccharomyces genome database (SGD, http://www.yeastgenome.org/). The single-gene deletion strains were used without checking these genotype in detail [28]. These strains were batch cultivated in shake flasks with triplicate, from which crude protein mixtures were extracted at an exponential growth phase. The trypsin-digested peptides were analyzed by the SRM assay methods (S2 Table). In order to compare protein levels accurately, fully 13C-labeled peptide samples were prepared from yeast strain S288C cells grown in SD medium containing [U-13C] glucose as the sole carbon source. The fully 13C-labeled peptide samples were used as the internal standards for all analyses in this study.
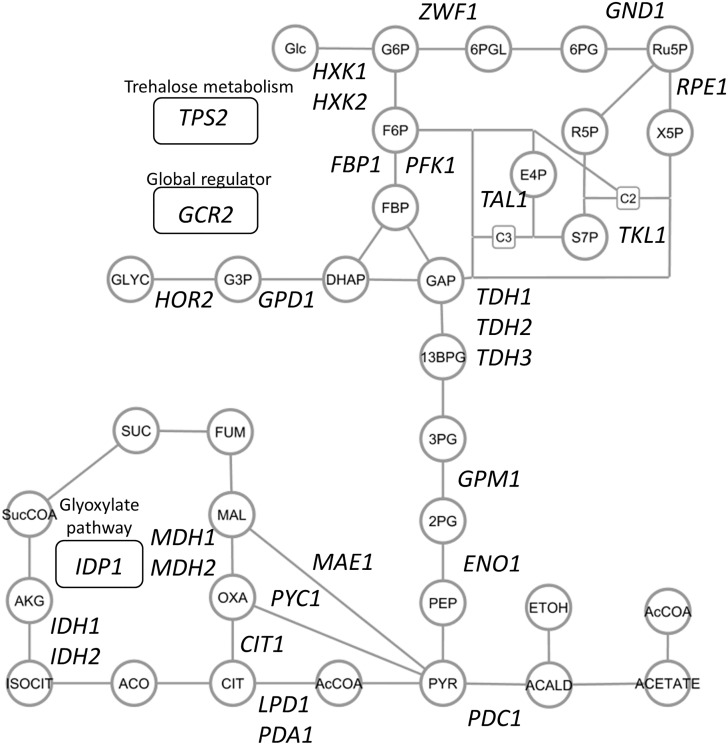
Fig 1. Central metabolism related genes whose single-gene deletion mutants were investigated in this study.
Deleted genes were shown in the simplified metabolite pathways. Circles and lines represent metabolites and reactions, respectively. Genes related to the trehalose metabolism, glyoxylate pathway, and the global regulation were shown in round rectangles. Abbreviations: Glc: glucose, G6P: glucose 6-phosphate, F6P: fructose 6-phosphate, FBP: fructose 1,6-bisphosphate,DHAP: dihydroxyacetone phosphate, GAP: glyceraldehyde 3-phosphate, 13BPD: 1,3-bisphosphoglycerate, 3PG: 3-phosphoglycerate, 2PG: 2-phosphoglycerate, PEP: phosphoenolpyruvate, Pyr: pyruvate, AcCoA: acetyl-coenzyme A, 6PGL: 6-phosphogluconolactone, 6PG: 6-phosphogluconate, Ru5P: ribulose 5-phosphate, R5P: ribose 5-phosphate, Xu5P: xylulose 5-phosphate, S7P: sedoheptulose 7-phosphate, E4P: erythrose 4-phosphate, ACALD: acetoaldehyde, ETOH: ethanol, CIT: citrate, ISOCIT: isocitrate, ACO: anonitate, AKG: alpha-ketoglutarate, SucCOA: succinyl coenzyme A, SUC: succinate, FUM: fumarate, MAL: malate, OXA: oxaloacetate, G3P: glycerol-3-phosphate, GLYC: glycerol.
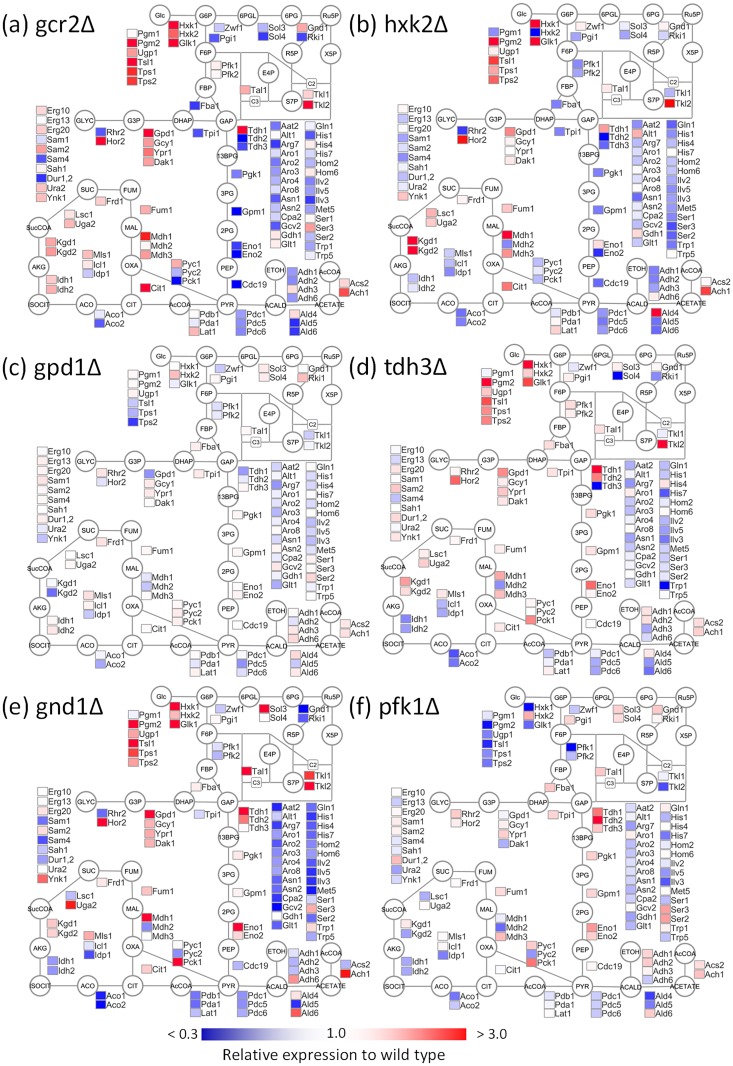
The SRM assays successfully determined the levels of peptides derived from the 110 enzymes (S3 Table and S1 Fig). For example, a comparison of the protein abundance profiles of the wild type (BY4742) with those of the mutant strains lacking a transcriptional activator of glycolysis, Gcr2 (denoted as gcr2Δ, Fig 2a) and a major isoform of hexokinase, Hxk2 (hxk2Δ, Fig 2b) showed significant changes in abundances of 37 and 27 enzymes, respectively (two-side Welch’s t-test, α = 0.01, S1 Table). The pattern of enzymes abundance in gcr2Δ strain was essentially similar to the literature reported transcriptome data of glycolytic genes [21]. The large perturbation of the enzyme abundance profile caused a decrease in the specific growth rate of gcr2Δ strain (73% of the wild type and S1 Table). Furthermore, the levels of trehalose metabolism-related enzymes such as Pgm2, Ugp1, Tps1, Tps2, and hexokinases, such as Hxk1 and Glk1, commonly increased in both strains [21]. On the other hand, the abundance levels of glycolytic enzymes, such as Fba1, Tpi1, and Cdc19, decreased. These results suggested that the single deletions of central metabolism-related enzyme genes such as Hxk2 caused an expressional modulation of numbers of enzymes.
Fig 2. Protein abundance profiles of the central metabolism related enzymes in Saccharomyces cerevisiae.
The data obtained from (a) gcr2Δ, (b) hxk2Δ, (c) gpd1Δ, (d) tdh3Δ, (e) gnd1Δ, and (f) pfk1Δ strains lacking GCR2, HXK2, GPD1, TDH3, GND1, and PFK1 genes were represented, respectively. Enzyme abundance levels relative to that of wild type (BY4742) were shown as a heat map of simplified metabolite pathways. All abbreviations were described in the legend for Fig 1.
Comparison with specific growth rates
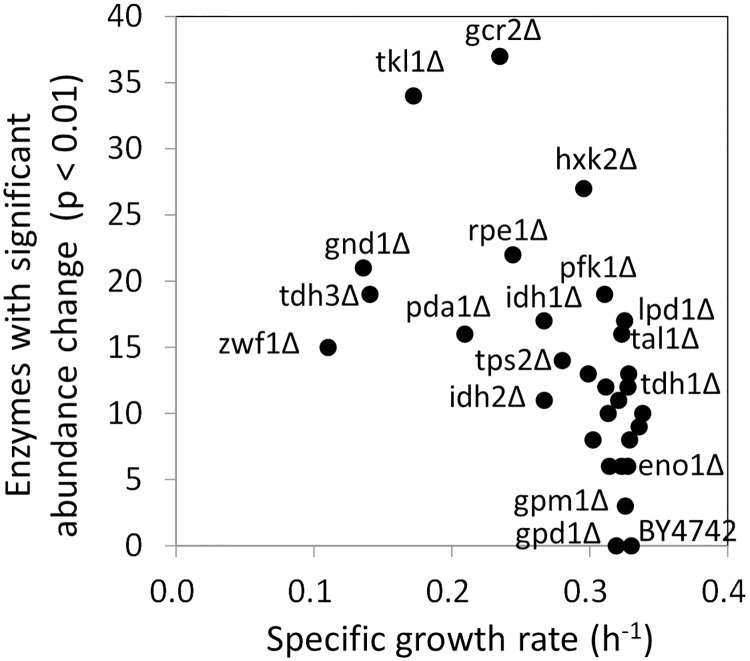
The specific growth rates of each strain were compared with the number of significantly modulated enzymes (S1 Table and Fig 3). The scatter plot (Fig 3) showed that there was a negative relationship between the specific growth rates and the number of significantly modulated enzymes. While the mutant strains with smaller number of significantly modulated enzymes showed firster specific growth rates such as in BY4742 and gpd1Δ, the mutant strains with slower growth rates tended to show larger modulation in the enzyme abundance.
Fig 3. Relationship between specific growth rates and numbers of enzymes whose expression levels were significantly changed.
The specific growth rates were determined from the flask-scale batch cultivation data.
For instance, the gpd1Δ strain lacking an isoform of glycerol-3-phosphate dehydrogenase, Gpd1, showed a similar specific growth rate to that of wild type, since level of no enzyme was significantly modulated (two-side Welch’s t-test, α = 0.01) (Figs 3 and 2c). The results also indicated that Gpd1 plays a minor role in the culture conditions since it has been reported that Gpd2 plays the major role under anaerobic conditions, and the deletion of GPD2 reduced growth under anaerobiosis [29–31]. In contrast, a slow growth phenotype as well as a large perturbation in the enzyme abundance profile was observed with deletion mutants, such as those lacking oxidative pentose phosphate pathway (gnd1Δ, tkl1Δ, zwf1Δ, and rpe1Δ) and glycolysis (tdh3Δ and pda1Δ) genes. For instance, tdh3Δ strain lacking a major isoform of glyceraldehyde 3-phosphate dehydrogenase, Tdh3, showed a 58% decrease in the specific growth rate (Fig 3) and a large perturbation in the enzyme abundance profile (Fig 2d). Moreover, the abundances of 21 enzymes significantly changed in gnd1Δ strain lacking a major isoform of 6-phosphoglucolate dehydrogenase, Gnd1 (Fig 2e).
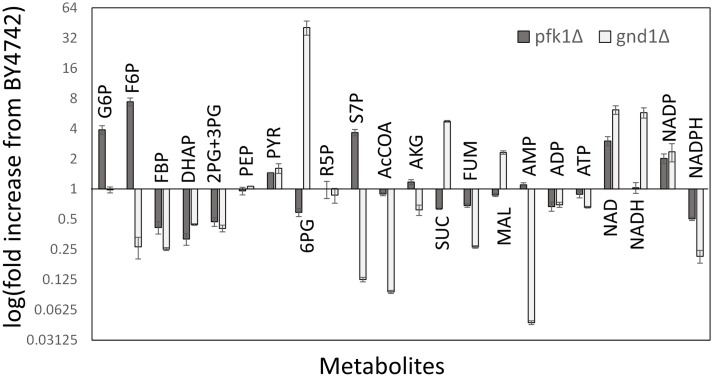
However, the negative relationship was not the case for lpd1Δ, hxk2Δ, and pfk1Δ because the mutants showed a large perturbation in the enzyme abundance profile and similar specific growth rates to that of wild type (Fig 3). The results suggested that the loss of deleted gene functions could be backed up or compensated by the modulation of abundance of a number of enzymes. For example, while the abundances of 27 enzymes significantly changed in hxk2Δ (Fig 2b), the specific growth rate was 90% of that of wild type (Fig 3). In the case of pfk1Δ strain lacking α subunit of phosphofruktokinase, Pfk1, significant variations were observed for 19 enzymes in the enzyme abundance profile data (Fig 2f). In order to investigate a relationship between the enzyme abundance profile and cell metabolism, a metabolic profiling analysis was conducted for BY4742 and pfk1Δ strains (Fig 4). The metabolic profile data showed that the loss of Pfk1 caused an accumulation of up-stream metabolites, including fructose-6-phosphate (F6P) and glucose-6-phosphate (G6P), as well as a decrease in down-stream metabolites such as fructose-1,6-bisphosphate (FBP) and dihydroxyacetone phosphate (DHAP). The level of sedoheptulose-7-phosphate (S7P) was also drastically increased in pfk1Δ strain by unknown reasons. However, since the metabolite levels in lower part of glycolysis were similar to those of the wild type, the effect of Pfk1 deletion on the metabolism was likely to be restricted in the upper part of the glycolysis by the modulation of expression of 19 enzymes. These results suggested that the single deletions of central metabolism-related genes caused a modulation of numbers of enzyme abundances to maintain the metabolic homeostasis in deleted gene dependent manners.
Fig 4. Metabolite profiles in pfk1Δ and gnd1Δ strains.
Fold increase with respect to the BY4742 strain is shown for each metabolite. Data are shown as the means of triplicate analyses with standard deviations. All abbreviations were described in the legend for Fig 1.
Resource allocation for enzyme biosynthesis
The cost of protein investment in cellular metabolic function can be estimated from the absolute protein abundance data [32]. The absolute protein abundance or copy numbers of expressed proteins per cell have been determined comprehensively for the wild type S. cerevisiae strains in previous studies [17, 33–35]. Here, the copy number means a molecular number of expressed protein in this study. For instance, the dataset by Kulak et al showed that the total copy number of the 110 enzymes analyzed in this study was 1.03 × 107 copies/cell [17]. The dataset also indicated that glycolysis related enzymes such as Tdh3 are among the most abundantly expressed of the 110 enzymes. An essentially similar trend was also observed in other datasets [33–35]. Here, using the dataset by Kulak et al as the copy numbers of the 110 enzymes in the wild type strain, the copy numbers in each mutant strain were calculated from the targeted proteome data obtained in this study (Fig 5 and S2 Fig).
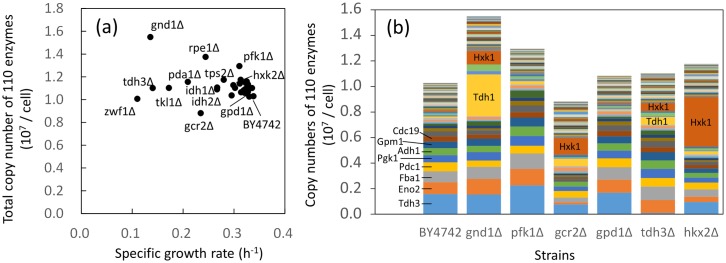
Fig 5. The composition of enzyme proteins.
(a) Relationship between specific growth rates and total copy numbers of the 110 enzymes investigated in this study. (b) Composition of the 110 enzymes in selected strains. In this study, the copy number indicates a molecular number of expressed protein. All data were calculated from the dataset by Kulak et al [17] and the targeted proteome data obtained in this study. Data of other strains are shown in S2 Fig. Data are shown as means of triplicate analyses. Strain and enzyme names were represented in the Figures.
The estimated total copy numbers of the 110 enzymes were compared with the specific growth rates (Fig 5a). The result showed that the total copy numbers increased exceptionally in several strains, such as gnd1Δ strain lacking the gene encoding glucose-6-phosphate dehydrogenase (1.55 × 107 copies/cell) and the rpe1Δ strain lacking the gene encoding ribulose-5-phosphate 3-epimerase (1.38 × 107 copies/cell) (Fig 5a). The composition of the protein abundance profile showed that the copy numbers of Tdh1 and Hxk1 drastically increased in gnd1Δ (Fig 5b). The copy numbers of Tdh1, Hxk1, and Tdh3 also increased in rpe1Δ (Fig 5b). The increase and decrease in the abundance of other enzymes, such as those involved in amino acid biosynthesis and the TCA cycle, had little effect on the copy number profiles, since the copy numbers of these enzymes in the wild type were significantly lower than those of glycolytic enzymes. In gnd1Δ and rpe1Δ, the specific growth rates were reduced partly because a large resource was additionally allocated for protein biosynthesis in response to serious metabolic perturbations caused by deletion of GND1 and RPE1. The comparison between the total copy numbers and the specific growth rate also indicated that gcr2Δ strain contained smaller copy numbers of the 110 enzymes than the wild type strain did (Fig 5a). Although the abundance of Hxk1 showed an increase, the abundances of other glycolytic enzymes decreased uniformly due to loss of the global regulator of glycolysis (Gcr2) as mentioned earlier (Fig 5b).
In the case of other mutant strains, the comparison between the total copy numbers and the specific growth rates (Fig 5a) showed that the total copy numbers of the 110 enzymes tended to increase but less than 115% of that of the wild type (Fig 5a). For example, the total copy numbers in gpd1Δ, tdh3Δ, and hxk2Δ were 105, 107, and 114% of that of the wild type, respectively (Fig 5a). Similar trends were observed for the numbers of amino acids required for the biosynthesis of the 110 enzymes (S2 Fig) as well as from the analyses using datasets produced by other researches (S3 Fig) [33, 35]. The results suggested that functional compensation of the deleted enzyme was attained by using more resources for protein biosynthesis under a constraint in the available resources for the synthesis of glycolytic enzymes.
The presence of a constraint was also evident from the enzyme compositions (Fig 5b). For example, Tdh3, a major isoform of glyceraldehyde-3-phosphate dehydrogenase, was one of the most abundant enzyme in BY4742 (Fig 5b). However, the total copy numbers of 110 enzymes in the tdh3Δ strain increased to 107% of that of the wild type, since the copy numbers of Tdh1, Hxk1, and other major glycolytic enzymes increased evenly in tdh3Δ (Fig 5b). Furthermore, in the hxk2Δ strain, the copy number of Hxk1 drastically increased for functional compensation of the isozyme (Fig 5b). The specific up-regulation of Hxk1 expression in hxk2Δ seemed reasonable as a backup for Hxk2 deletion, since the compensatory expression of HXK1 and HXK2 was observed under conditions having different carbon sources [36]. Genetic analysis also found that Hxk2 functions in the nucleus to repress HXK1 and GLK1 expression [37]. However, the total copy number was kept at 114% of the wild type, because the expressions of other enzymes were uniformly reduced in hxk2Δ (Fig 5b).
Enzyme-enzyme co-abundance network uncovered coordinated regulations of enzyme levels
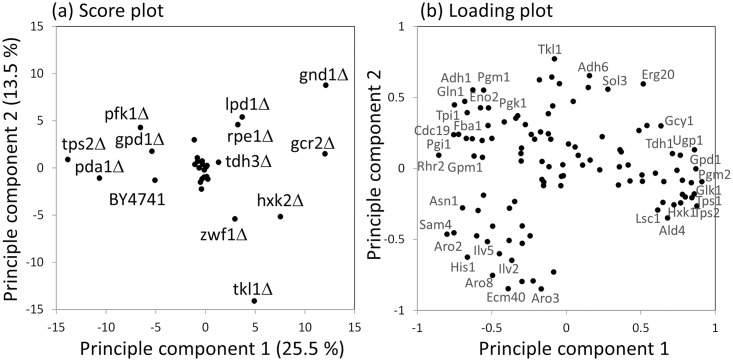
Effects of gene deletions on the enzyme abundance profiles were compared by the principal component analysis (PCA). The result showed that a cluster of mutant strains was observed in the center of the PCA score plot (Fig 6a). Since the wild type (BY4742) was outside the cluster, a common change in enzyme abundance was shared among the strains in the cluster. The enzyme expression profile data indicated that the abundances of enzymes, such as Hxk1, Pgm2, and Tps2 were commonly increased in these strains (Fig 6b and S1 Fig). The score plot also suggested that the enzyme abundance profiles were significantly changed in the deletion strains of genes related to pentose phosphate pathway (gnd1Δ, zwf1Δ, tkl1Δ, and rpe1Δ), entry point of glycolysis (hxk2Δ and pfk1Δ), pyruvate dehydrogenase complex (pda1Δ, lpd1Δ), trehalose biosynthesis (tps1Δ and tps2Δ), and gcr2Δ (Fig 5a). Although these strains failed to form clusters in the score plot of PCA, coordinated regulations somehow work in these mutant strains, since the first and second principal components explained the 30.5% of total variations in the enzyme expression profiles (Fig 6a).
Fig 6. Principle Component Analysis (PCA) of the enzyme abundance data.
(a) PCA score plot for mutants. The mean, log2-transformed and z-scored enzyme abundance data was used for the analysis. The score plots of principle component 1 and 2 are shown in the figure. (b) Loading plot of principle component 1 and 2. Strain and enzyme names were represented in the Figures.
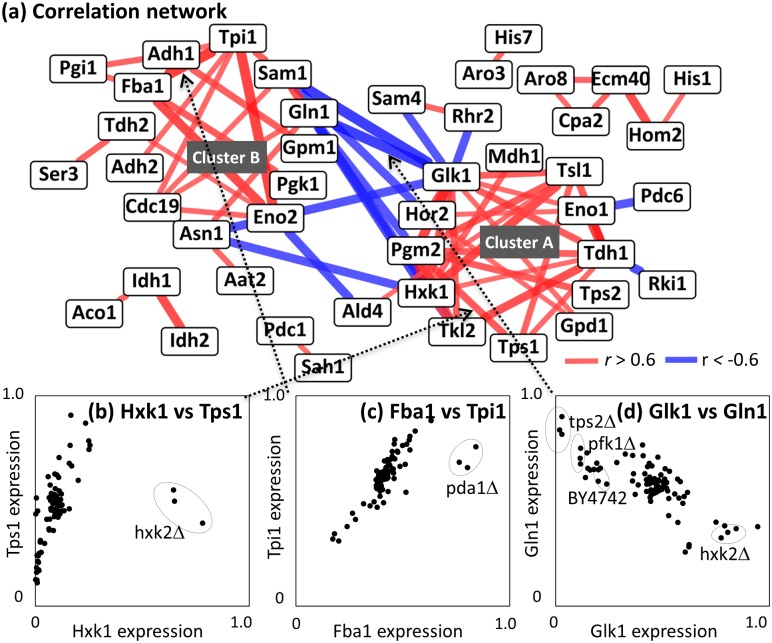
Coordination behind the regulation was investigated by construction of a co- abundance network of enzyme-enzyme pairs (Fig 7a). In this section, Spearman's rank-order correlation coefficients (r) were calculated among enzymes. In order to investigate effects derived from the deletion of other enzyme genes, the analysis was performed without using the data obtained from gcr2Δ and mutant strains of the protein of interest. For example, data obtained from gcr2Δ, hxk1Δ, and tps1Δ strains was omitted in the determination of correlation coefficients (r) between Hxk1 (a minor isoform of hexokinase) and Tps1 (a subunit of trehalose-6-phosphate synthase/phosphatase complex). It was because GCR2 did not encode enzyme and the abundances of Hxk1 in hxk1Δ and Tps1 in tps1Δ did not reflect effects derived from the deletion of other enzyme genes. The results showed that a linear correlation was observed between Hxk1 and Tps1 abundances, whose correlation coefficient was determined to be 0.61 (Fig 7b). The correlation coefficients of Fba1/Tpi1 and Glk1/Gln1 pairs were also calculated to be 0.79 and -0.67, respectively (Fig 7c and 7d).
Fig 7. Enzyme-enzyme co-abundance analysis.
(a) Enzyme-enzyme co-abundance network. Nodes and edges indicate enzymes and their correlations. Positive (r > 0.6) and negative (r < -0.6) correlations were indicated as red and blue, respectively. (b-d) Scatter plot between two enzyme expression data including (b) Hxk1 vs. Tps1, (c) Fba1 and Tpi1, and (d) Glk1 vs. Gln1. Dotted arrows indicate corresponding edges between two enzymes. Dotted circles represent data derived from specific mutants.
The positive and negative correlations were derived from coordinated regulations of enzyme abundance levels, since the distribution of correlation coefficients of all enzyme pairs is wider than null distribution (S4 Fig). The coordinated regulation of the abundance levels of enzyme proteins seems reasonable for controlling the glycolysis flux because an over-expression of the single enzyme failed to affect cell metabolism [38].
Coordinated regulations by global regulators
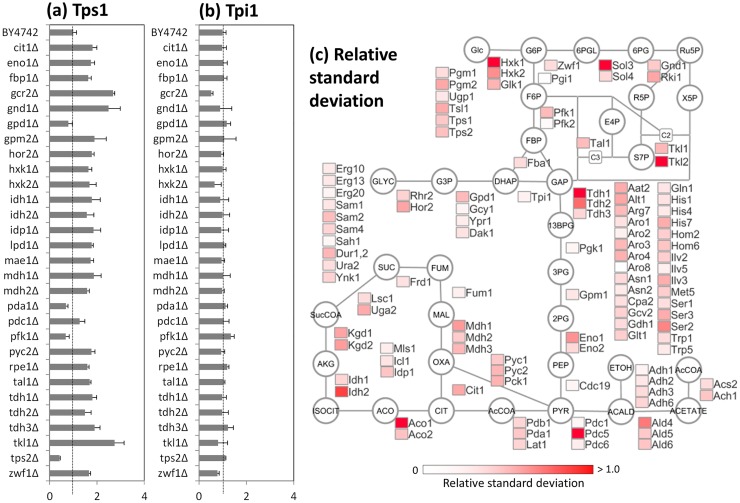
The co-abundance network contains two large and several small clusters of positively correlated enzymes. The largest cluster (Cluster A in Fig 7a) contains enzymes responsible for the trehalose and glycerol metabolism, such as Tps1, Pgm2, Tsl1, and Gpd1, in addition to the glycolytic enzymes, including Tdh1, Eno1, and Hxk1. The coordinated regulation of enzyme abundance levels seemed to play a role in the metabolic adaptation, since the expression levels of Tps1 (a subunit of trehalose-6-phosphate synthase/phosphatase complex) were significantly increased in 14 single deletion mutants, including gcr2Δ and hxk2Δ (Fig 2a and 2b) with large variations among 30 strains (0.48–2.68 fold of the wild type strain, Fig 8a). A relative standard deviation (RSD) of Tps1 abundance was 0.58. The large variations in the expression levels were also observed for other enzymes in the cluster A (Fig 8c).
Fig 8. Variation of enzyme abundances among S. cerevisiae mutants.
(a and b) Abundances of (a) Tps1 and (b) Tpi1 proteins in single gene deletion mutants. Lines indicate abundances of wild type (BY4742). Data are shown as the mean of triplicate analysis with standard deviations. (c) Relative standard deviations of enzyme expression levels among 30 wild type strain and mutants. The data were represented as a heat map in the simplified metabolic pathways.
A comparison with the other proteomics data suggested that the enzymes in Cluster A is involved in the cellular response to osmotic stress under regulation of transcriptional activators (Msn2/4) mediating a general response to multiple stresses [8, 39, 40]. The results suggested that the coordinated regulation of enzyme abundance levels observed in the single gene deletion mutants would be connected with the stress response mechanisms in S. cerevisiae. It has also been reported that Msn2/4 controls the expression of all enzyme-encoding genes in the pathways for trehalose biosynthesis from glucose-6-phosphate (G6P) and its degradation to glucose [18, 39, 41]. The trehalose biosynthesis and degradation pathways form a metabolic shunt or a futile cycle consuming ATP. While trehalose itself protects subcellular structures against osmotic and other stresses [42, 43], it has been recently reported that the futile cycling of the trehalose shunt has a role in the regulation of glycolysis stability [44, 45]. The reports showed that the trehalose shunt is a mechanism to reduce the probability of unbalanced state glycolysis, because the glycolysis in S. cerevisiae is intrinsically unstable. The larger variation in the abundance of the trehalose metabolism related enzymes observed in this study suggested that the regulation is an important mechanism for metabolic adaptation of S. cerevisiae (Fig 8c).
Second largest cluster (Cluster B) includes several glycolytic enzymes, such as Cdc19, Tpi1, Eno2, Fba1, and Pgi1 (Fig 7a). Amino acid biosynthesis related enzymes, such as Gln1 and Asn1, were also included in this module. In contrast to the cluster A, these enzymes showed small variations in the abundance levels. For instance, the variation of triose-phosphate isomerase (Tpi1) abundance was within 0.51–1.34 fold of the wild type strain (Fig 8b), whose RSD level (0.13) was one of the lowest levels among all enzymes (Fig 8c). The fine-tuning occurred in the single gene deletion strains with significantly perturbed enzyme abundance profiles, because Tpi1 abundances were decreased in hxk2Δ, tkl1Δ, and zwf1Δ strains, while increased in pfk1Δ and tdh3Δ strains (Figs 2b, 2d, 2f and 8b).
The enzymes in the cluster B was partly coincidence with the enzymes whose abundances were decreased in the gcr2Δ strains (Figs 7a and 2a). These results suggested a fine-tuning of these enzyme abundance levels are under the control of Gcr1/2 transcription factor [21, 46–48]. Furthermore, the tight co-expression of enzymes might reflect additional functions of enzymes, such as a glycolytic metabolon complex using filamentous actin for stabilization [49].
In addition to the two large clusters, small clusters containing enzymes related to TCA cycle (Aco1, Idh1, and Idh2) and amino acids biosynthesis (Cpa1, Ecm40, Aro8, Hom2, and His1) were also found in the co-abundance network (Fig 7a). The retrograde control by RTG genes [50] and the general amino acid control by GCN4 [51] seem to have some role behind the coordinated regulation.
The co-abundance network also showed that there were negative correlations between the enzymes in cluster A and B, such as the Glk1 (glucokinase)/Gln1 (glutamine synthetase) pair (r = -0.67, Fig 7d). These results suggested that there was an exclusive trend between the abundance of enzymes in the clusters. Although detailed mechanism remains unclear, the loading plot of PCA showed that the enzymes in cluster A (such as Tdh1, Tps1, and Glk1) and B (such as Tpi1, Cdc19, and Gln1) were plotted at the positive and negative ends of the principle component 1 (Fig 6b). Moreover, the enzyme abundance profile of hxk2Δ and gnd1Δ showed an increased abundance of Glk1 and the trehalose metabolism related enzymes as well as a decreased abundance of Gln1 and the glycolysis related enzymes (Figs 2b, 2e and 7d). Opposite profiles (increased and decreased abundances of Gln1 and Glk1, respectively) were observed for pfk1Δ and tps2Δ strains (Figs 2f and 7d).
Mutant-specific regulation of enzyme expression levels
The scatter plot of the Hxk1 and Tps1 abundances showed that the data obtained from the hxk2Δ were outliers (Fig 7b). The specific increase in Hxk1 abundance in hxk2Δ seemed reasonable, as mentioned previously. However, roles of expression modulation of other distant enzymes shown in hxk2Δ, such as the up-regulation of two subunits of α-ketoglutarate dehydrogenase (Kgd1 and Kgd2) in the TCA cycle, were unclear (Fig 2b). Since the abundances of Kgd1 and Kgd2 in the hxk2Δ were not only 3.3 and 3.0 times larger than that of BY4742 but also the most abundant among the strains examined in this study. It has been reported that the hxk2-null mutant strain showed a reduction of the glucose repression and fully oxidative growth at high glucose concentrations [52]. The increase in the abundance of Kgd1 and Kgd2 proteins should be responsible for the oxidative metabolism. The targeted proteome data indicated that the regulation mechanism is likely to be specifically working in the hxk2Δ strain.
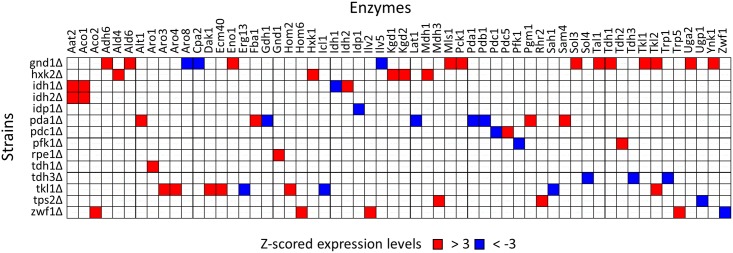
Similar mutant-specific and extreme modulations were investigated in the enzyme abundance profile data. Here, the variations in abundances of each enzyme among the 30 strains were normalized to z-scores. For example, the z-scored levels of Hxk1 and Kgd1 in hxk2Δ were 4.7 and 3.7, respectively, indicating that the abundance of Hxk1 and Kgd1 increased extraordinarily in hxk2Δ, among the 30 strains (Fig 9). The analysis of the whole dataset revealed that although extreme increase (z-score > 3.0) in enzyme expressions were observed for 42 cases, extreme decrease (z-score < -3.0) were observed for 19 cases in the 14 strains. Among them, the compensation by an extreme increase of an isozyme was observed for 4 cases, namely, Hxk1 in hxk2Δ, Idh2 in idh1Δ (two subunits of mitochondrial NAD(+)-dependent isocitrate dehydrogenase), Tkl2 in tkl1Δ (two isoforms of transketolase), and Pdc5 in pdc1Δ strains. The auto-regulation of isozymes was reported for the suppression of PDC5 expression by Pdc1 [22]. Other cases were found to be the modulation of distant enzymes, such as over-expression of Fba1 (Fructose 1,6-bisphosphate aldolase) in the pda1Δ strain, which was lacking the E1 α subunit of the pyruvate dehydrogenase (PDH) complex (Fig 7c). Distant modulations of larger numbers of enzymes were frequently observed for the mutant strains with reduced cell growth phenotypes such as gnd1Δ, rpe1Δ, tkl1Δ, zwf1Δ (lacking the pentose phosphate pathway related genes), and pda1Δ. For example, gnd1Δ showed a slow growth phenotype (Fig 2e). This was because the loss of Gnd1 is too critical to be backed up by modulation of other enzyme abundance profiles, i.e., an extreme increase and/or decrease of distant enzyme levels. The metabolome analysis of gnd1Δ also showed that the loss of Gnd1 caused an accumulation of its substrate, 6-phosphogluconate (6PG), and a drastic perturbation in other metabolite levels (Fig 4). These results suggest that extreme regulations of enzyme expressions work in a mutant-specific manner, although detailed roles and mechanisms in the control of central carbon metabolism are unknown.
Fig 9. Mutant-specific regulation of enzyme abundance levels.
Extreme increase and decrease in enzymes abundance among single gene deletion mutants. Colored cells indicate enzymes for which the z-scored abundance levels were over 3 (red) and below -3 (blue) in this strain. The blank strains and enzymes with no extreme increase and decrease are not shown in the Figure.
Conclusion
The targeted proteome analysis successfully produced an enzyme expression dataset from the 30 S. cerevisiae wild type and mutant strains. The data analysis uncovered that functional compensation of the deficient enzyme was attained by using more resources for protein biosynthesis (Fig 5a). However, it was also suggested that available resources for enzyme biosynthesis in central carbon metabolism were not abundant in S. cerevisiae cells. The enzyme-enzyme co-abundance network showed that at least 30% of the variations in the enzyme abundances were explained by coordinated and global regulation, one of the most frequently observed responses being the increase in trehalose metabolism related enzymes (Figs 6 and 7a). The remaining variations should be derived from mutant-specific and local mechanisms (Fig 9).
The comparison of the enzyme abundance profile also suggest that the mutant strains could be classified into four groups. First group includes gpd1Δ strains (Fig 2c), whose enzyme abundance profiles and specific growth rates were not changed by deletions of GPD1 genes (Fig 3). The second group forms a cluster in the center of PCA plot (Fig 6a), whose specific growth rates were similar to that of wild type (cit1Δ, eno1Δ, fbp1Δ, gpm2Δ, hor2Δ, hxk1Δ, idh1Δ, idh2Δ, idp1Δ, mae1Δ, mdh1Δ, mdh2Δ, pdc1Δ, pyc2Δ, tal1Δ, tdh1Δ, and tdh2Δ) (Fig 3). In the strains, the increase in trehalose metabolism related enzymes were commonly observed (S1 Fig). These suggested that metabolic homeostasis was maintained via the accumulation of trehalose and the regulation of glycolysis stability by the role of the trehalose shunt [44, 45]. Specific regulation of a small number of enzymes was also observed, such as the extreme increase in Pdc5 abundance in pdc1Δ (Fig 9).
A third group consists of hxk2Δ (Fig 2b), lpd1Δ, pfk1Δ (Fig 2f), and tps2Δ, in which the functions of deleted enzymes were recovered by additional modulation of enzyme abundance such as the Gcr1/2 dependent regulation of glycolytic enzymes. These strains also activated a strain-specific regulation for the extreme regulation of several enzymes (Fig 9). As demonstrated for pfk1Δ, these modulation of the enzyme abundance profile successfully compensated the functions of deleted gene and maintained metabolic homeostasis to keep cell growth (Figs 3 and 4).
A fourth group showed slower specific growth rate including gnd1Δ (Fig 2e), pda1Δ, rpe1Δ, tdh3Δ (Fig 2d), tkl1Δ, and zwf1Δ. In these mutants, the single gene deletions were too critical to be backed up by modulation of other enzyme expression profile, with an extreme increase and decrease of distant enzyme levels (Fig 9). For the case of gnd1Δ and rpe1Δ strains, an additional resource was allocated for protein biosynthesis to activate the metabolic function. Despite the employment of all possible means, however, the metabolic profile analysis showed that the metabolic homeostasis was hardly kept in the gnd1Δ strain (Fig 4). These results showed that global and local regulation of enzyme abundance levels shape central carbon metabolism in S. cerevisiae by using a limited resource for protein biosynthesis.
Although targeted proteomics is an essential tool for further dissection of the complex behavior of central carbon metabolism, the molecular mechanisms underlying the regulation of enzyme abundance, as well as their effects on metabolite accumulation and flux, need further investigation. For example, it was expected that a lack of either Zwf1 (glucose-6-phosphate dehydrogenase) or Gnd1 (a major isoform of 6-phosphogluconate dehydrogenase) would have a similar impact on metabolism, because these enzymes are both responsible for the oxidative pentose phosphate pathway (Fig 1). However, the score plot of PCA showed that the zwf1Δ and gnd1Δ strains each had different enzyme abundance profiles (Fig 6a). This could be explained by stating that the accumulation of 6-phosphogluconate in gnd1Δ (Fig 4) perturbed S. cerevisiae metabolism, for instance, via an allosteric inhibition of glucose-6-phosphate isomerase [53]. Furthermore, metabolite analysis showed that the metabolite levels, for instance, S7P accumulation in the pfk1Δ strain, were not simply explained by the enzyme abundance (Fig 4). These results show that an interaction between enzyme abundance and other layers of metabolism, such as metabolic flux, metabolite accumulation, and mRNA expression, has to be investigated considering the various post-translational regulation mechanisms of enzymes [5, 6, 54, 55].
Materials and methods
Yeast strains and growth conditions
S. cerevisiae strains, including S288C (MATα SUC2), BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0), and the single gene deletion mutants were purchased from Thermo Scientific (Pittsburgh, PA, USA) (S1 Table). All strains were cultured in yeast extract peptone dextrose (YPD) medium (1% bacto yeast extract, 2% bacto peptone, 2% glucose) and synthetic dextrose (SD) medium (6.7% yeast nitrogen base without amino acids and 5% non-labeled or [U-13C] glucose) containing required amino acids. [U-13C] glucose (99%) was purchased from Cambridge Isotope Laboratories (Andover, MA, USA).
S. cerevisiae cells from glycerol stocks were cultured in 5 mL of YPD medium at 30°C for 24 h with shaking at 120 rpm. The cells were inoculated into 100 mL of SD medium in 500-mL Sakaguchi flasks and cultured at 30°C for 24 h with shaking at 120 rpm. The precultures were transferred to the main culture (100 mL of SD medium in 500-mL Sakaguchi flasks at 30°C for 48 h with shaking at 120 rpm). The initial OD660 values for pre- and main cultures were set at 0.2. For the preparation of isotope-labeled standards from the S288C strain, SD medium containing [U-13C] glucose was used for the pre- and main cultures. The BY4742 and single gene deletion strains were cultured in SD medium containing unlabeled glucose. For 96 wells cultivation, the cells were inoculated into 5 mL of SD medium and cultured at 30°C with shaking at 120 rpm. The precultures were transferred to 200 μL of SD medium in 96-well plate (Costar 3595, Corning, NY, USA) containing 0.1 mg/mL of TTA. The initial OD600 was set at 0.1. The plates were incubated using the plate reader (Synergy HTX, Biotek instruments, VT, USA) at 30°C for 48 h with shake mode: orbital frequency: 237cpm (4mm), and orbital speed: slow.
Targeted proteome analysis using ultra fast mass spectrometry
Crude proteins were extracted from S. cerevisiae cells in the exponential growth phase (OD600 at 1.0), as previously described [9, 27], and protein concentrations were determined by the Bradford method. Trypsin digestion was performed as described previously [56, 57], and the resulting peptide solutions were desalted using GL-Tip GC micropipette tips (GL Science, Tokyo, Japan). In this manner, 60 μL of digested peptides were prepared from samples containing 50 μg of crude protein. Equal amounts of digested peptide derived from the target (cultured using nonlabeled glucose) and S288C (cultured in [U-13C] glucose medium as an internal standard) strains were mixed [13]. An identical 13C labeled peptide mixtures prepared from S288C strain was employed for all analyses in this study. The peptide samples (2 μL) were analyzed using a nano-LC-UFMS system (LC-20ADnano and LCMS-8040, Shimadzu, Kyoto, Japan), equipped with a valve system (FCV nano, Shimadzu), a nanospray interface (N8040, AMR, Tokyo, Japan), and a spray tip (Fortis tip 150–20, AMR). The analytical conditions were as follows. HPLC: column: L-column ODS (pore size: 5 μm, 0.1 × 150 mm, CERI, Tokyo Japan); trap column: L-column ODS (pore size: 5 μm, 0.3 × 5 mm, CERI); solvent system: 0.1% formic acid and 5% acetonitrile in water: 0.1% formic acid and 95% acetonitrile in water; gradient program: 0:100, v/v at 0 min; 0:100 at 7 min; 35:65 at 45 min; 50:50 at 50 min; and 100:0 at 65 min; flow rate: 400 nL/min. MS detection: interface temperature: 350°C; DL temperature: 150°C; heat block temperature: 200°C; drying gas flow: off; CID gas pressure: 310; interface voltage: +1.6 keV; detection mode: MRM positive. The data were recorded with the aid of LabSolutions LCMS version 5.6 (Shimadzu). The MRM assay method for the enzyme analysis of yeast central metabolism, described by Picotti et al. [9, 27], was used with modifications (S2 Table).
Data analysis
Chromatographic data was processed using Skyline version 3.1 [58, 59]. Relative peak areas were determined for all peptides using the 13C labeled peptides as the internal standards. In order to correct a systematic error derived from the protein quantification by the Bradford method, 55 proteins with smaller expression variations among 87 samples were selected considering relative standard deviations. Signal intensities of enzymes were normalized by dividing a mean relative peak area of 55 proteins of the samples. All data analysis was performed using the Multi experiment viewer (MeV) version 4.8 [60] and in-house scripts written in Perl. Statistical tests (Student’s t test) were performed for the raw abundances data. For principle component analysis, the mean abundances were converted to log 2 transformed and normalized data by using the functions of MeV. For the mutant-specific regulation of enzyme expression levels, the abundances were z-scored by subtracting the mean of 30 strains and then dividing by the standard deviation. The data obtained from gcr2Δ and mutant strains of the target protein (for example, Pdc1 expression data in pdc1Δ strains) were not used for the calculation of spearman correlation coefficients and relative standard deviations. The enzyme expression profiles and co-expression networks were visualized by Cytoscape3.0 [61, 62] and VANTED v2.2.0 [63].
LC-MS/MS analysis of intermediate metabolites
Culture broth was sampled rapidly and filtered through a 0.5-μm pore size filter (PTFE-type membrane, ADVANTEC, Japan). Cells on the filter were immediately immersed in 1.6 mL methanol (-80°C) and kept at -80°C until extraction. Following addition of 1.6 mL of chloroform (-30°C) and 640 μL of Milli-Q water (4°C) and vortexing for 1 min, the mixture was centrifuged at 15,000 g for 5 min at 4°C. Two hundred and fifty microliters of the aqueous layer was transferred to a 1.5-mL tube and dried using a SpeedVac SPD1010 (Thermo Scientific, Japan) at room temperature. The dried samples were suspended in 50 μL of Milli-Q water. LC-MS/MS analysis (LC: Agilent 1100 series; Agilent Technologies, MS/MS: API 2000; MA, AB SCIEX) was performed using the previously described method (17). The peak of each target metabolite was identified by comparing its chromatographic behavior with that of an authentic standard. Peak area was determined using the software Analyst (version 1.6.2, AB SCIEX).
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Dr. Katsuyuki Yugi, Dr. Atsushi Hatano (Universty of Tokyo), and Dr. Yoshihiro Toya (Osaka University) for helpful comment to this manuscript. This work was supported by the Industrial Technology Research Grant Program for 2011 of the New Energy and Industrial Technology Development Organization (NEDO) of Japan and by Grants in Aid for Scientific Research (C) No 15K06579. All raw data flies are deposited to jPOST (http://jpostdb.org/, ID: JPST000167 and PXD004714).
Data Availability
All raw data flies are deposited to jPOST (http://jpostdb.org/, ID: JPST000167 and PXD004714).
Funding Statement
This work was supported by the Industrial Technology Research Grant Program for 2011 of the New Energy and Industrial Technology Development Organization (NEDO, http://www.nedo.go.jp/) of Japan No 11B05004c and by JSPS (https://www.jsps.go.jp/) Grants in Aid for Scientific Research (C) No 15K06579.
References
- 1.Diaz-Ruiz R, Rigoulet M, Devin A. The Warburg and Crabtree effects: On the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim Biophys Acta. 2011;1807(6):568–76. 10.1016/j.bbabio.2010.08.010 [DOI] [PubMed] [Google Scholar]
- 2.Gustavsson AK, van Niekerk DD, Adiels CB, Kooi B, Goksor M, Snoep JL. Allosteric regulation of phosphofructokinase controls the emergence of glycolytic oscillations in isolated yeast cells. FEBS J. 2014;281(12):2784–93. 10.1111/febs.12820 [DOI] [PubMed] [Google Scholar]
- 3.Dijken JPv, Scheffers WA. Redox balances in the metabolism of sugars by yeasts FEMS Microbiol Rev. 1986;32:199–224. [Google Scholar]
- 4.Amoutzias GD, He Y, Lilley KS, Van de Peer Y, Oliver SG. Evaluation and properties of the budding yeast phosphoproteome. Mol Cell Proteomics. 2012;11(6):M111 009555 10.1074/mcp.M111.009555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliveira AP, Ludwig C, Picotti P, Kogadeeva M, Aebersold R, Sauer U. Regulation of yeast central metabolism by enzyme phosphorylation. Mol Syst Biol. 2012;8:623 10.1038/msb.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tripodi F, Nicastro R, Reghellin V, Coccetti P. Post-translational modifications on yeast carbon metabolism: Regulatory mechanisms beyond transcriptional control. Biochim Biophys Acta. 2015;1850(4):620–7. 10.1016/j.bbagen.2014.12.010 [DOI] [PubMed] [Google Scholar]
- 7.Bruckmann A, Hensbergen PJ, Balog CIA, Deelder AM, Brandt R, Snoek ISI, et al. Proteome analysis of aerobically and anaerobically grown Saccharomyces cerevisiae cells. J Proteomics. 2009;71(6):662–9. 10.1016/j.jprot.2008.11.012 [DOI] [PubMed] [Google Scholar]
- 8.Soufi B, Kelstrup CD, Stoehr G, Frohlich F, Walther TC, Olsen JV. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol Biosyst. 2009;5(11):1337–46. 10.1039/b902256b [DOI] [PubMed] [Google Scholar]
- 9.Costenoble R, Picotti P, Reiter L, Stallmach R, Heinemann M, Sauer U, et al. Comprehensive quantitative analysis of central carbon and amino-acid metabolism in Saccharomyces cerevisiae under multiple conditions by targeted proteomics. Mol Syst Biol. 2011;7:464 10.1038/msb.2010.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Hoek P, van Dijken JP, Pronk JT. Regulation of fermentative capacity and levels of glycolytic enzymes in chemostat cultures of Saccharomyces cerevisiae. Enzyme Microb Technol. 2000;26(9–10):724–36. [DOI] [PubMed] [Google Scholar]
- 11.van den Brink J, Canelas AB, van Gulik WM, Pronk JT, Heijnen JJ, de Winde JH, et al. Dynamics of glycolytic regulation during adaptation of Saccharomyces cerevisiae to fermentative metabolism. Appl Environ Microbiol. 2008;74(18):5710–23. 10.1128/AEM.01121-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bro C, Regenberg B, Nielsen J. Genome-wide transcriptional response of a Saccharomyces cerevisiae strain with an altered redox metabolism. Biotechnol Bioeng. 2004;85(3):269–76. 10.1002/bit.10899 [DOI] [PubMed] [Google Scholar]
- 13.Matsuda F, Ogura T, Tomita A, Hirano I, Shimizu H. Nano-scale liquid chromatography coupled to tandem mass spectrometry using the multiple reaction monitoring mode based quantitative platform for analyzing multiple enzymes associated with central metabolic pathways of Saccharomyces cerevisiae using ultra fast mass spectrometry. J Biosci Bioeng. 2015;119(1):117–20. 10.1016/j.jbiosc.2014.06.010 [DOI] [PubMed] [Google Scholar]
- 14.Blank LM, Kuepfer L, Sauer U. Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol. 2005;6(6):R49 10.1186/gb-2005-6-6-r49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kintaka R, Makanae K, Moriya H. Cellular growth defects triggered by an overload of protein localization processes. Sci Rep. 2016;6:31774 10.1038/srep31774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriya H, Makanae K, Watanabe K, Chino A, Shimizu-Yoshida Y. Robustness analysis of cellular systems using the genetic tug-of-war method. Mol Biosyst. 2012;8(10):2513–22. 10.1039/c2mb25100k [DOI] [PubMed] [Google Scholar]
- 17.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11(3):319–24. 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- 18.Ihmels J, Levy R, Barkai N. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat Biotechnol. 2004;22(1):86–92. 10.1038/nbt918 [DOI] [PubMed] [Google Scholar]
- 19.Fendt SM, Oliveira AP, Christen S, Picotti P, Dechant RC, Sauer U. Unraveling condition-dependent networks of transcription factors that control metabolic pathway activity in yeast. Mol Syst Biol. 2010;6:432 10.1038/msb.2010.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet. 2003;43(3):139–60. 10.1007/s00294-003-0381-8 [DOI] [PubMed] [Google Scholar]
- 21.Sasaki H, Uemura H. Influence of low glycolytic activities in gcr1 and gcr2 mutants on the expression of other metabolic pathway genes in Saccharomyces cerevisiae. Yeast. 2005;22(2):111–27. 10.1002/yea.1198 [DOI] [PubMed] [Google Scholar]
- 22.Eberhardt I, Cederberg H, Li H, Konig S, Jordan F, Hohmann S. Autoregulation of yeast pyruvate decarboxylase gene expression requires the enzyme but not its catalytic activity. Eur J Biochem. 1999;262(1):191–201. [DOI] [PubMed] [Google Scholar]
- 23.Arvas M, Pakula T, Smit B, Rautio J, Koivistoinen H, Jouhten P, et al. Correlation of gene expression and protein production rate—a system wide study. BMC Genomics. 2011;12:616 10.1186/1471-2164-12-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19(3):1720–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods. 2010;7(1):43–6. 10.1038/nmeth.1408 [DOI] [PubMed] [Google Scholar]
- 26.Picotti P, Lam H, Campbell D, Deutsch EW, Mirzaei H, Ranish J, et al. A database of mass spectrometric assays for the yeast proteome. Nat Methods. 2008;5(11):913–4. 10.1038/nmeth1108-913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138(4):795–806. 10.1016/j.cell.2009.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Shitrit T, Yosef N, Shemesh K, Sharan R, Ruppin E, Kupiec M. Systematic identification of gene annotation errors in the widely used yeast mutation collections. Nat Methods. 2012;9(4):373–8. 10.1038/nmeth.1890 [DOI] [PubMed] [Google Scholar]
- 29.Hubmann G, Guillouet S, Nevoigt E. Gpd1 and Gpd2 fine-tuning for sustainable reduction of glycerol formation in Saccharomyces cerevisiae. Appl Environ Microbiol. 2011;77(17):5857–67. 10.1128/AEM.05338-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjorkqvist S, Ansell R, Adler L, Liden G. Physiological response to anaerobicity of glycerol-3-phosphate dehydrogenase mutants of Saccharomyces cerevisiae. Appl Environ Microbiol. 1997;63(1):128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nissen TL, Hamann CW, Kielland-Brandt MC, Nielsen J, Villadsen J. Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synthesis. Yeast. 2000;16(5):463–74. [DOI] [PubMed] [Google Scholar]
- 32.Liebermeister W, Noor E, Flamholz A, Davidi D, Bernhardt J, Milo R. Visual account of protein investment in cellular functions. Proc Natl Acad Sci U S A. 2014;111(23):8488–93. 10.1073/pnas.1314810111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–41. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- 34.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25(1):117–24. 10.1038/nbt1270 [DOI] [PubMed] [Google Scholar]
- 35.de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455(7217):1251–4. 10.1038/nature07341 [DOI] [PubMed] [Google Scholar]
- 36.Herrero P, Galindez J, Ruiz N, Martinez-Campa C, Moreno F. Transcriptional regulation of the Saccharomyces cerevisiae HXK1, HXK2 and GLK1 genes. Yeast. 1995;11(2):137–44. 10.1002/yea.320110205 [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez A, De La Cera T, Herrero P, Moreno F. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem J. 2001;355(Pt 3):625–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenzweig RF. Regulation of fitness in yeast overexpressing glycolytic enzymes: parameters of growth and viability. Genet Res. 1992;59(1):35–48. [DOI] [PubMed] [Google Scholar]
- 39.Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J Bacteriol. 1998;180(5):1044–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, et al. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12(2):323–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zahringer H, Thevelein JM, Nwaka S. Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: variations of PKA effect during stress and growth. Mol Microbiol. 2000;35(2):397–406. [DOI] [PubMed] [Google Scholar]
- 42.Eleutherio E, Panek A, De Mesquita JF, Trevisol E, Magalhaes R. Revisiting yeast trehalose metabolism. Curr Genet. 2015;61(3):263–74. 10.1007/s00294-014-0450-1 [DOI] [PubMed] [Google Scholar]
- 43.Jain NK, Roy I. Effect of trehalose on protein structure. Protein Sci. 2009;18(1):24–36. 10.1002/pro.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Heerden JH, Wortel MT, Bruggeman FJ, Heijnen JJ, Bollen YJ, Planque R, et al. Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science. 2014;343(6174):1245114 10.1126/science.1245114 [DOI] [PubMed] [Google Scholar]
- 45.van Heerden JH, Bruggeman FJ, Teusink B. Multi-tasking of biosynthetic and energetic functions of glycolysis explained by supply and demand logic. Bioessays. 2015;37(1):34–45. 10.1002/bies.201400108 [DOI] [PubMed] [Google Scholar]
- 46.Carroll KM, Simpson DM, Eyers CE, Knight CG, Brownridge P, Dunn WB, et al. Absolute quantification of the glycolytic pathway in yeast: deployment of a complete QconCAT approach. Mol Cell Proteomics. 2011;10(12):M111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hauf J, Zimmermann FK, Muller S. Simultaneous genomic overexpression of seven glycolytic enzymes in the yeast Saccharomyces cerevisiae. Enzyme Microb Technol. 2000;26(9–10):688–98. [DOI] [PubMed] [Google Scholar]
- 48.Sun J, Shao Z, Zhao H, Nair N, Wen F, Xu JH, et al. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109(8):2082–92. 10.1002/bit.24481 [DOI] [PubMed] [Google Scholar]
- 49.Araiza-Olivera D, Chiquete-Felix N, Rosas-Lemus M, Sampedro JG, Pena A, Mujica A, et al. A glycolytic metabolon in Saccharomyces cerevisiae is stabilized by F-actin. FEBS J. 2013;280(16):3887–905. 10.1111/febs.12387 [DOI] [PubMed] [Google Scholar]
- 50.Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 1999;19(10):6720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–50. 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- 52.Diderich JA, Raamsdonk LM, Kruckeberg AL, Berden JA, Van Dam K. Physiological properties of Saccharomyces cerevisiae from which hexokinase II has been deleted. Appl Environ Microbiol. 2001;67(4):1587–93. 10.1128/AEM.67.4.1587-1593.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marchand M, Kooystra U, Wierenga RK, Lambeir AM, Van Beeumen J, Opperdoes FR, et al. Glucosephosphate isomerase from Trypanosoma brucei. Cloning and characterization of the gene and analysis of the enzyme. Eur J Biochem. 1989;184(2):455–64. [DOI] [PubMed] [Google Scholar]
- 54.Gruning NM, Rinnerthaler M, Bluemlein K, Mulleder M, Wamelink MM, Lehrach H, et al. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 2011;14(3):415–27. 10.1016/j.cmet.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hackett SR, Zanotelli VR, Xu W, Goya J, Park JO, Perlman DH, et al. Systems-level analysis of mechanisms regulating yeast metabolic flux. Science. 2016;354(6311). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uchida Y, Tachikawa M, Obuchi W, Hoshi Y, Tomioka Y, Ohtsuki S, et al. A study protocol for quantitative targeted absolute proteomics (QTAP) by LC-MS/MS: application for inter-strain differences in protein expression levels of transporters, receptors, claudin-5, and marker proteins at the blood-brain barrier in ddY, FVB, and C57BL/6J mice. Fluids Barriers CNS. 2013;10(1):21 10.1186/2045-8118-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamiie J, Ohtsuki S, Iwase R, Ohmine K, Katsukura Y, Yanai K, et al. Quantitative atlas of membrane transporter proteins: development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria. Pharm Res. 2008;25(6):1469–83. 10.1007/s11095-008-9532-4 [DOI] [PubMed] [Google Scholar]
- 58.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–8. 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bereman MS, MacLean B, Tomazela DM, Liebler DC, MacCoss MJ. The development of selected reaction monitoring methods for targeted proteomics via empirical refinement. Proteomics. 2012;12(8):1134–41. 10.1002/pmic.201200042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–93. 10.1016/S0076-6879(06)11009-5 [DOI] [PubMed] [Google Scholar]
- 61.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. 10.1007/978-1-60761-987-1_18 [DOI] [PubMed] [Google Scholar]
- 63.Rohn H, Junker A, Hartmann A, Grafahrend-Belau E, Treutler H, Klapperstuck M, et al. VANTED v2: a framework for systems biology applications. BMC Syst Biol. 2012;6:139 10.1186/1752-0509-6-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All raw data flies are deposited to jPOST (http://jpostdb.org/, ID: JPST000167 and PXD004714).